Abstract
Background & aims
Although ferroptosis holds promise as a new strategy for treating hepatocellular carcinoma (HCC), there are several obstacles that need to be overcome. One major challenge is the lack of understanding about the mechanisms underlying ferroptosis. Additionally, while the m6A modification has been shown to regulate various forms of cell death, its role in regulating ferroptosis in HCC has been largely overlooked. Bridging this knowledge gap, our study aimed to elucidate the regulatory influence of m6A modification on HCC ferroptosis.
Materials
Dot blot and EpiQuik m6A RNA Methylation Quantitative kit detected changes in overall m6A modification level during ferroptosis in HCC. MeRIP-qPCR and RIP-qPCR identified that the m6A modification of ATG5 mRNA was significant changed. BALB/c nude mice were used to construct xenograft tumor models to verify the phenotypes upon YTHDC2 silencing. In addition, patient-derived organoid models were used to demonstrate that induction of ferroptosis was an effective strategy against HCC.
Results
Our study has shown that inducing ferroptosis is a promising strategy for combatting HCC. Specifically, we have found a significant correlation between ferroptosis and high levels of m6A modification in HCC. Notably, we discovered that the elevation of ATG5 mRNA m6A modification mediated by WTAP is dependent on the reading protein YTHDC2. Importantly, inhibition of either WTAP or YTHDC2 effectively prevented ferroptosis and suppressed HCC development in both in vitro and in vivo models.
Conclusion
Our study revealed that WTAP upregulates ATG5 expression post-transcriptionally in an m6A-YTHDC2-dependent manner, thereby promoting the translation of ATG5 mRNA during ferroptosis in HCC. These findings have significant implications for the development of innovative and effective therapeutic approaches for HCC treatment.
Keywords: Autophagy, Translation, Ferroptosis, Hepatocellular carcinoma, m6A
1. Introduction
Hepatocellular carcinoma (HCC) is a prevalent and life-threatening cancer worldwide. It is primarily attributed to factors such as excessive alcohol consumption, viral infections, and drug abuse [1]. Despite the availability of several therapeutic options, including surgical resection, immunotherapy, chemotherapy, and targeted therapy, the treatment outcomes for HCC remain unsatisfactory [2]. Therefore, in addition to addressing the causative factors, it is imperative to focus on the development of novel treatments to effectively reduce HCC-related mortality. Although research has shown that inducing apoptosis [3], pyroptosis [4], necrosis [5], and other forms of cell death can have a partial anti-carcinogenic effect on HCC, none of these methods have been found to be entirely effective in eradicating the disease. Our previous research revealed that dihydroartemisinin could induce ferroptosis in HCC, providing a promising avenue for treatment [6]. Furthermore, numerous studies have demonstrated the effectiveness of ferroptosis as an anti-cancer approach [[7], [8], [9]]. Despite substantial research efforts, the underlying epigenetic mechanisms driving the development of ferroptosis in HCC remain unclear. Therefore, the objective of the current study is to shed light on the epigenetic pathways implicated in HCC ferroptosis.
Unlike other types of programmed cell death, ferroptosis is a form of programmed cell death that is predominantly regulated by intracellular iron concentration [10]. This distinct form of programmed cell death is characterized by an imbalance in the intracellular redox system, leading to lipid peroxidation in the cell membrane and ultimately culminating in cell death [11]. The research found that multiple signaling pathways are involved in ferroptosis, including the RAS signaling pathway, p53 signaling pathway, Hippo signaling pathway, HIF1-α signaling pathway, etc. [[12], [13], [14]]. However, more and more studies have shown that the autophagy signaling pathway plays an important role in the process of ferroptosis [15]. Autophagy is a fundamental process for maintaining proper intracellular iron cycling. When cells accumulate excessive amounts of iron, an overactive autophagy process can trigger ferroptosis [16]. Our research had identified the potential of autophagy-dependent ferroptosis as a promising approach for treating HCC. While current studies have mainly focused on the transcriptional regulation of autophagic ferroptosis, there has been relatively little research on the post-transcriptional regulation of ferroptosis that is dependent on autophagy. Consequently, our investigation primarily focused on unraveling the post-transcriptional control of autophagy-dependent ferroptosis, aiming to enhance our understanding of its therapeutic potential for HCC in a more comprehensive manner.
The m6A RNA modification is the predominant post-transcriptional modification in eukaryotes. This intricate process involves the coordinated activity of methylation enzymes (referred to as writers), demethylases (referred to as erasers), and reader proteins, collectively working together to accomplish RNA modification [17]. This modification is crucial for various aspects of RNA function, including splicing, maturation, translation, and stability, making it an essential process for gene expression [18]. The writers of m6A modification including WT1 associated protein (WTAP), methyltransferase 3 (METTL3), METTL4, and METTL14, they form a complex to catalyze the m6A methylation modification [19]. Fat mass and obesity-associated protein (FTO) and alkB homolog 5 (ALKBH5) are currently known erasers that remove m6A modification from mRNA and thus keep the level of m6A modification in dynamic equilibrium [20]. The YTH domain is a known domain that can specifically recognize m6A site in mRNAs, mainly including the YTH N6-methyladenosine RNA binding protein (YTHDF) and YTH domain containing 1 (YTHDC) families [21]. They can recognize and bind m6A-modified RRACH sequences on mRNAs, and then influence the fate of mRNAs [22]. Conducting a comprehensive analysis of the alterations in m6A modification levels and critical enzymes during ferroptosis in hepatocellular carcinoma (HCC) can offer novel targets for the advancement of anti-HCC medication.
In our present study, we have identified ATG5 as a downstream target of WTAP-mediated m6A modification in ferroptotic HCC. Furthermore, we have provided evidence that YTHDC2 recognized and bound to the m6A site on ATG5 mRNA, leading to an enhancement in its translation efficiency. These findings underscore the importance of RNA epigenetic regulation as a distinct mechanism governing ATG5 expression during HCC ferroptosis. Consequently, our investigation has contributed to the advancement of our molecular comprehension of anti-HCC therapy.
2. Results
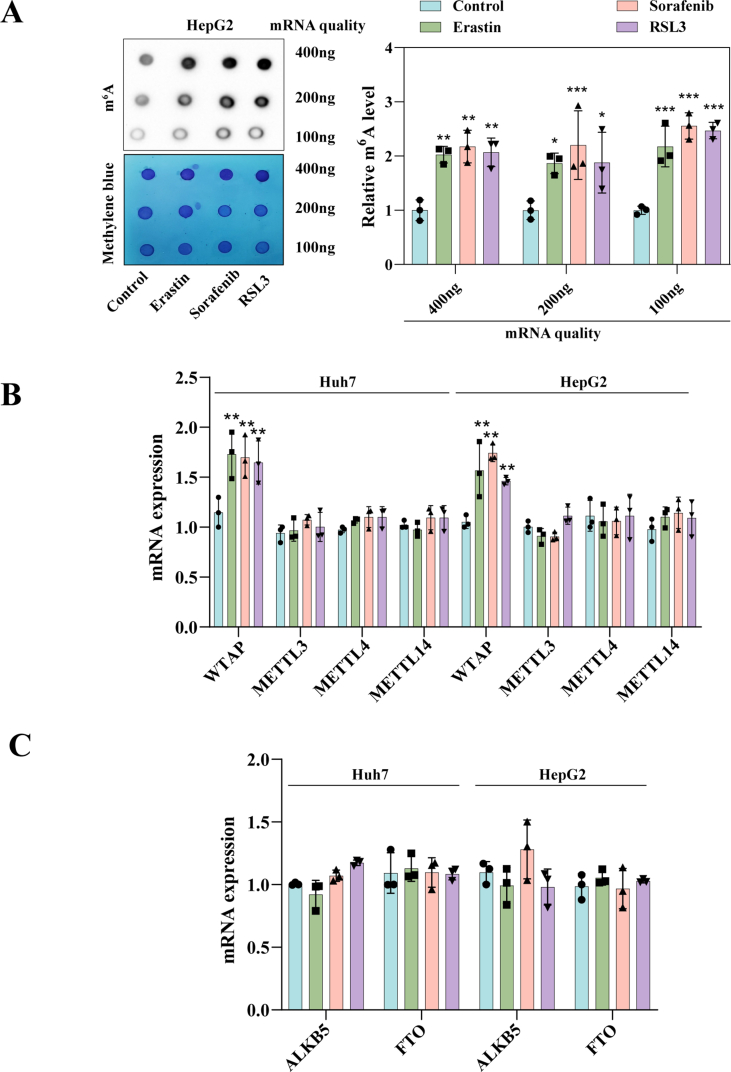
2.1. m6A modification is up regulated during ferroptosis in HCC
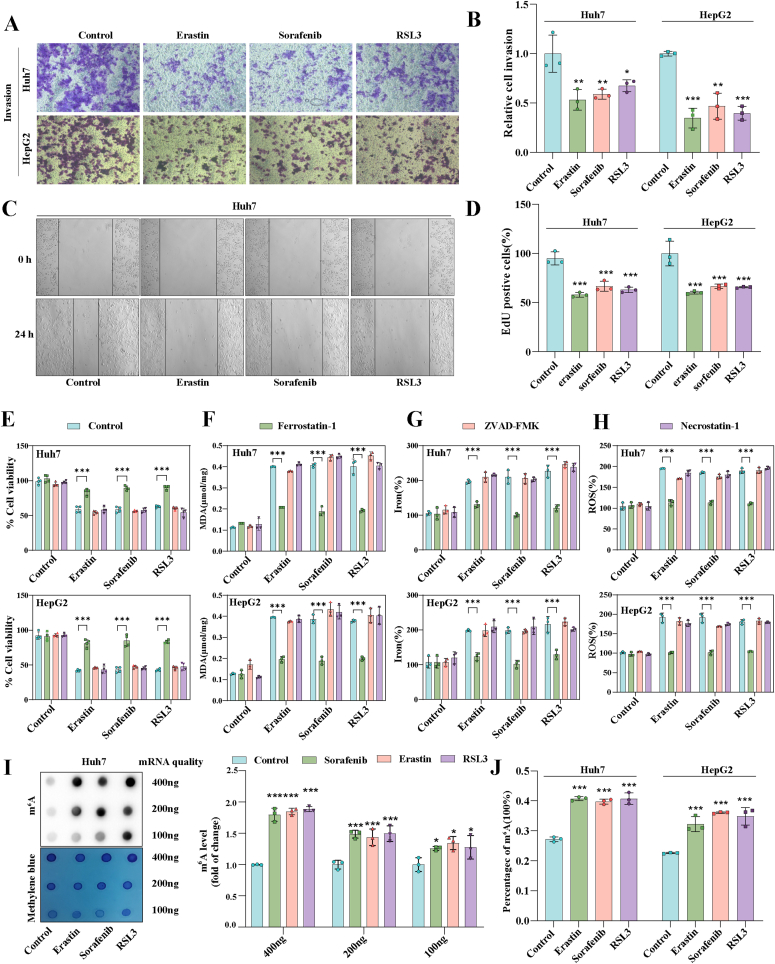
Our previous studies showed that ferroptosis inducers including erastin, sorafenib, and RSL3 can inhibit hepatic stellate cell (HSC) activation though ferroptosis pathway [23]. Similar results were obtained using HepG2 and Huh7. HCC cell lines were treated with ferroptosis inducers, and transwell assays were performed. Upon treatment with ferroptosis inducers, the ability of HepG2 and Huh7 to invade was markedly impaired (Fig. 1A and B). Wound healing experiments also showed that ferroptosis inducers could impair the migration ability of Huh7 cells (Fig. 1C). Additionally, all three ferroptosis inducers could inhibit the cells from multiplying (Fig. 1D). Next, we found that only ferroptosis inhibitor (ferrostatin-1) but not apoptosis inhibitor (ZVAD-FMK) and necrosis inhibitor (necrostatin-1) could restore the viability of HepG2 and Huh7 after treatment with erastin, sorafenib, and RSL3 (Fig. 1E). Consistent with our expectation, varying degrees elevation of ferroptosis markers MDA (Fig. 1F), iron (Fig. 1G), and ROS (Fig. 1H) were obtained in HCC cells after treatment with erastin, sorafenib, and RSL3. And only ferrostatin-1 could destroy the afore-mentioned classic ferroptosis event in HepG2 and Huh7 (Fig. 1F–H). Overall, these results confirmed that erastin, sorafenib, and RSL3 could trigger HCC ferroptosis in vitro. It suggested that ferroptosis induction was a feasible HCC prevention strategy.
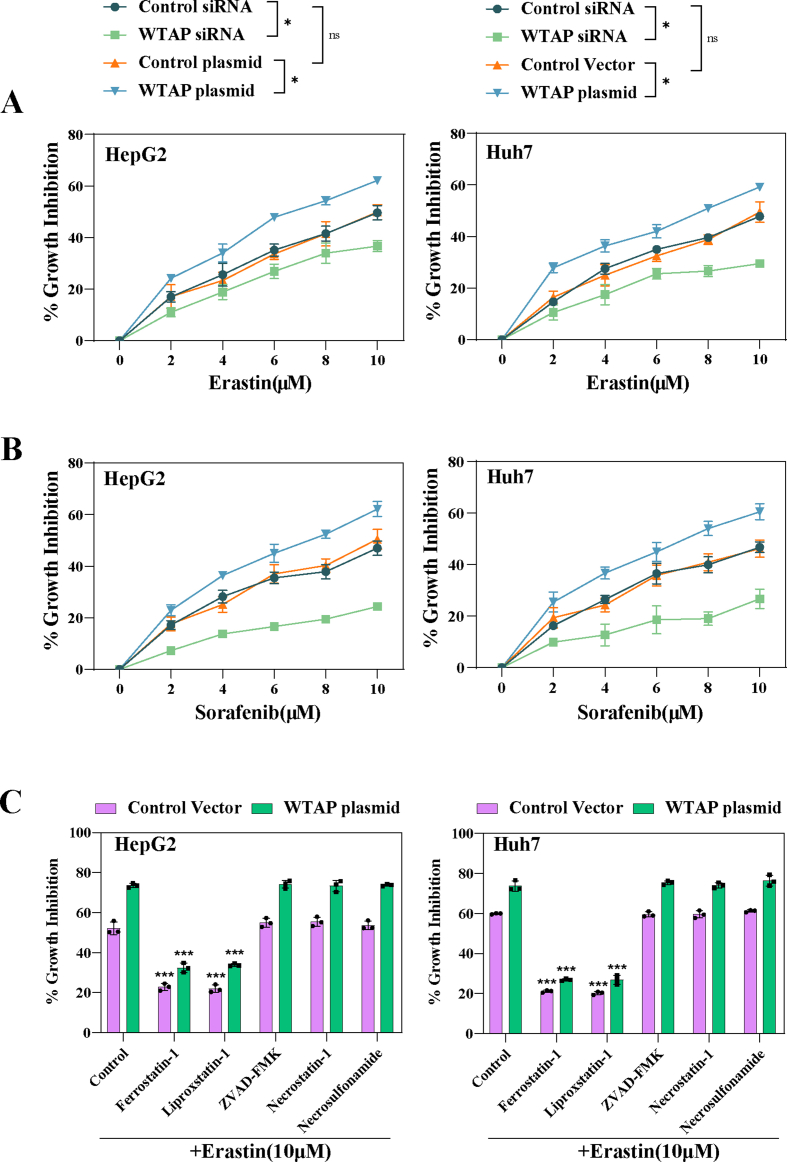
Fig. 1.
Levels of m6A modification is elevated in HCC ferroptosis. Ferroptosis inducers (10 μM Erastin, 10 μM Sorafenib, and 2.5 μM RSL3) were applied to Huh7 and HepG2 cells for 24 h with or without the addition of Ferrostatin-1 (1 μM), Necrostatin-1 (10 μM), and ZVAD-FMK (10 μM). A-B Transwell assays were used to assess the capacity for invasion. C Wound healing assay was performed to detect Huh7 cell migration. D Intracellular proliferation was detected using the Edu kit. E Overall cellular proliferation tested by MTT assay. F–H The level of MDA, iron, and GSH were tested detected by kits. I m6A modification level was detected by dot blot. J m6A modification level was detected by m6A RNA Methylation Quantitative kit. The mean ± SD was used as the value of three or four independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ns, no significance.
An increasing amount of evidence has revealed that m6A may regulate gene expression, thereby affecting various programmed cell death processes [24]. Interestingly, we found that m6A modification levels in Huh7 and HepG2 were significantly increased after ferroptosis inducer treatment compared to the untreated group through dotblot assay (Fig. 1I and Fig. S1A). At the same time, m6A kit detection also obtained the same result (Fig. 1J). Consequently, we hypothesize a potential link between HCC ferroptosis and alterations in m6A modification.
2.2. WTAP-mediated m6A modification contributes to HCC ferroptosis
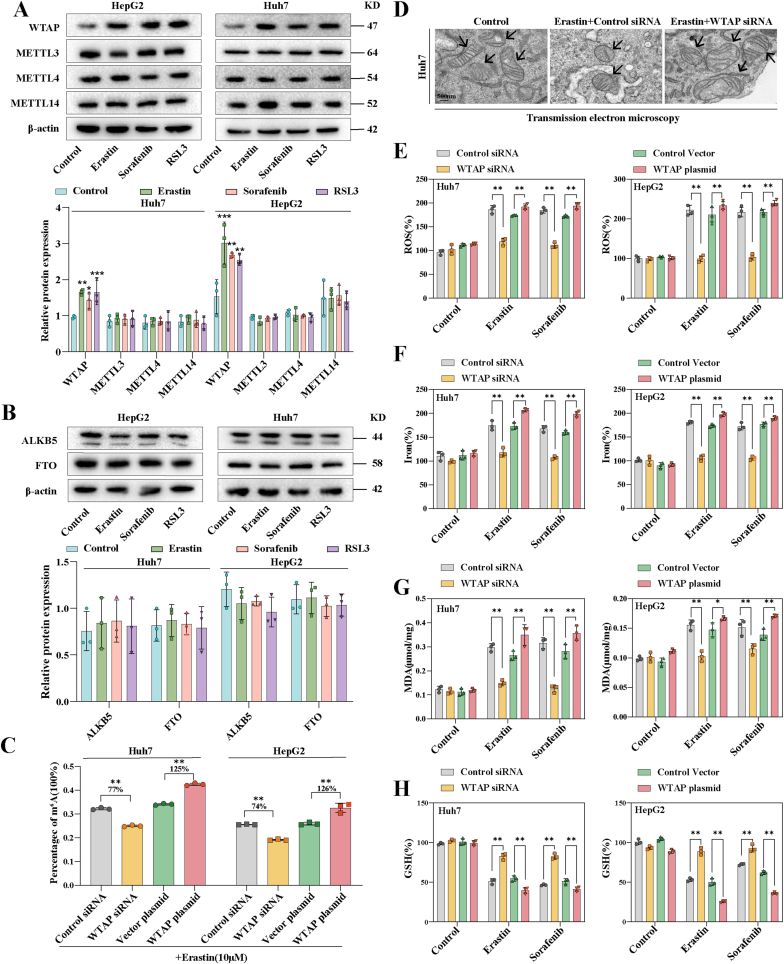
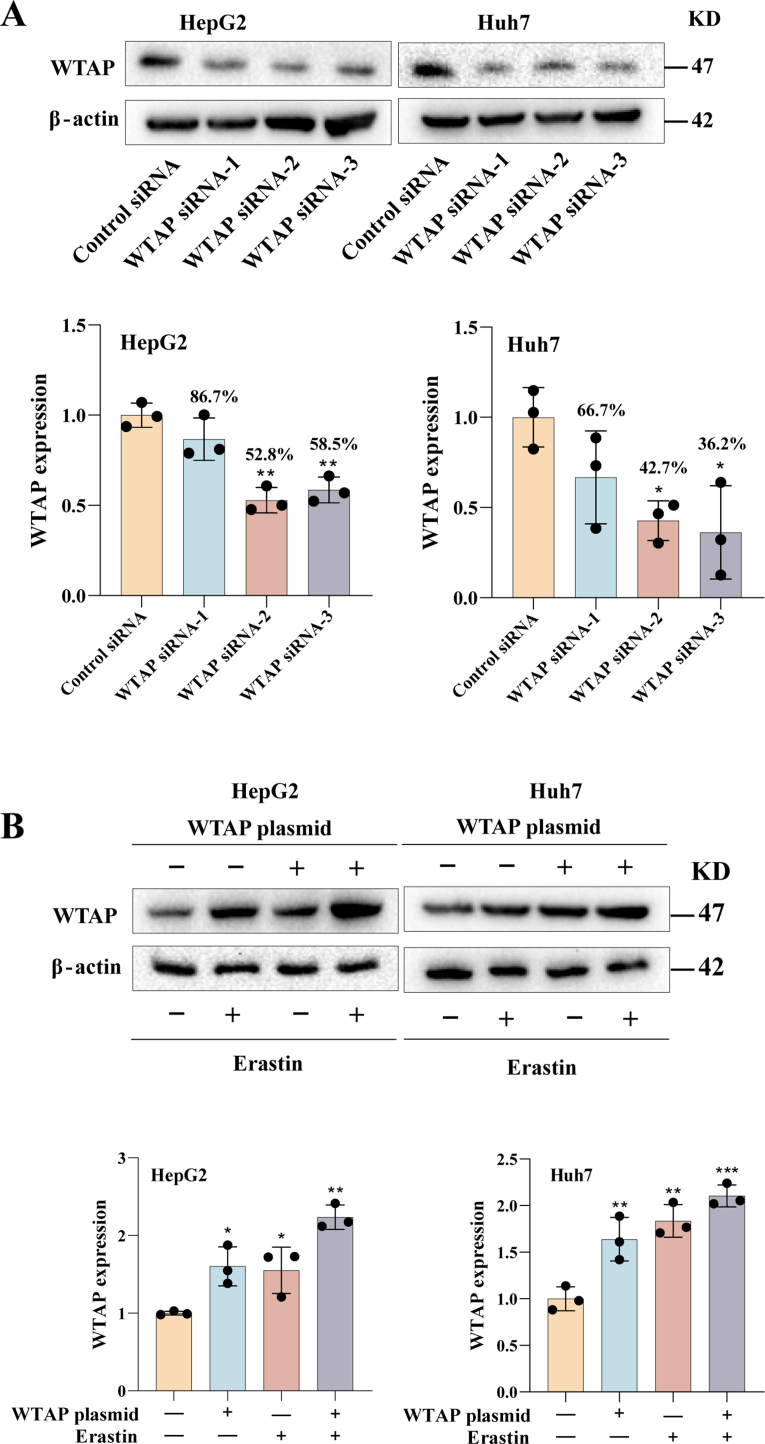
Next, we investigated the possible causes for the change of m6A modification in ferroptotic HCC. We found that both mRNA (Fig. S1B) and protein levels (Fig. 2A) of the writer WTAP were significantly increased, whereas the mRNA (Fig. S1C) and protein (Fig. 2B) levels of other writers and erasers did not show significant changes. We speculate that the heightened levels of the writer WTAP were the primary cause of increased m6A modification levels observed during HCC ferroptosis. Subsequently, we devised and synthesized WTAP siRNAs, among which we chose siRNA-2 to perform loss-of-function experiments (Fig. S2A). Concurrently, WTAP plasmid was used to performed gain-of-function experiments (Fig. S2B). As anticipated, the reduction of WTAP expression significantly decreased m6A modification levels in erastin-treated HCC cells (Huh7: 77 %; HepG2: 74 %), while overexpression of WTAP further increased m6A modification levels (Huh7: 125 %; HepG2: 126 %) (Fig. 2C). Furthermore, silencing WTAP mitigated the growth inhibition induced by erastin, whereas overexpressing WTAP further intensified the growth inhibition caused by erastin (Fig. S3A). Similar results were observed in Huh7 and HepG2 treated with sorafenib (Fig. S3B). Further research revealed that only ferrostatin-1 and liproxstatin-1, the two kinds of ferroptosis inhibitors, could attenuate the growth inhibition caused by over-expression of WTAP in erastin-treated Huh7 and HepG2, but not apoptosis, necrosis and necroptosis inhibitor (Fig. S3C).
Fig. 2.
WTAP-mediated m6A modification contributed to HCC ferroptosis. Ferroptosis inducers (10 μM Erastin, 10 μM Sorafenib, and 2.5 μM RSL3) were applied to Huh7 and HepG2 cells for 24 h. A-B Expression of m6A writers and erasers in ferroptoic HCC cells detected by Western blot. C WTAP siRNA and plasmid were utilized to transfect Huh7 and HepG2 cells respectively followed by 10 μM erastin. m6A RNA Methylation Quantitative kit was utilized to detect m6A modification level. D Transmission electron microscopic observation of mitochondrial morphology in Huh7. Scale bars: 500 nm. E-H The level of MDA, iron, and GSH were tested detected by kits. The mean ± SD was used as the value of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001; ns, no significance.
Next, we sought to find out the effects of WTAP on classical ferroptosis-related events in Huh7 and HepG2. First, we discovered that silencing WTAP repaired mitochondrial damage brought on by erastin (Fig. 2D). In addition, Silencing WTAP disrupted ROS accumulation (Fig. 2E), intracellular iron overload (Fig. 2F), MDA content (Fig. 2G), and GSH depletion (Fig. 2H) induced by erastin and sorafenib in Huh7 and HepG2. In contrast, over-expression of WTAP could further enhance the occurrence of ferroptosis-related events in Huh7 and HepG2 treated with eastin (Fig. 2E–H). Taken together, these results suggest a potential association between m6A modification and HCC ferroptosis. Elevated WTAP levels may potentially contribute to ferroptosis in HCC induced by ferroptosis inducers.
2.3. m6A modification mediated ferroptosis is associated with autophagy activation
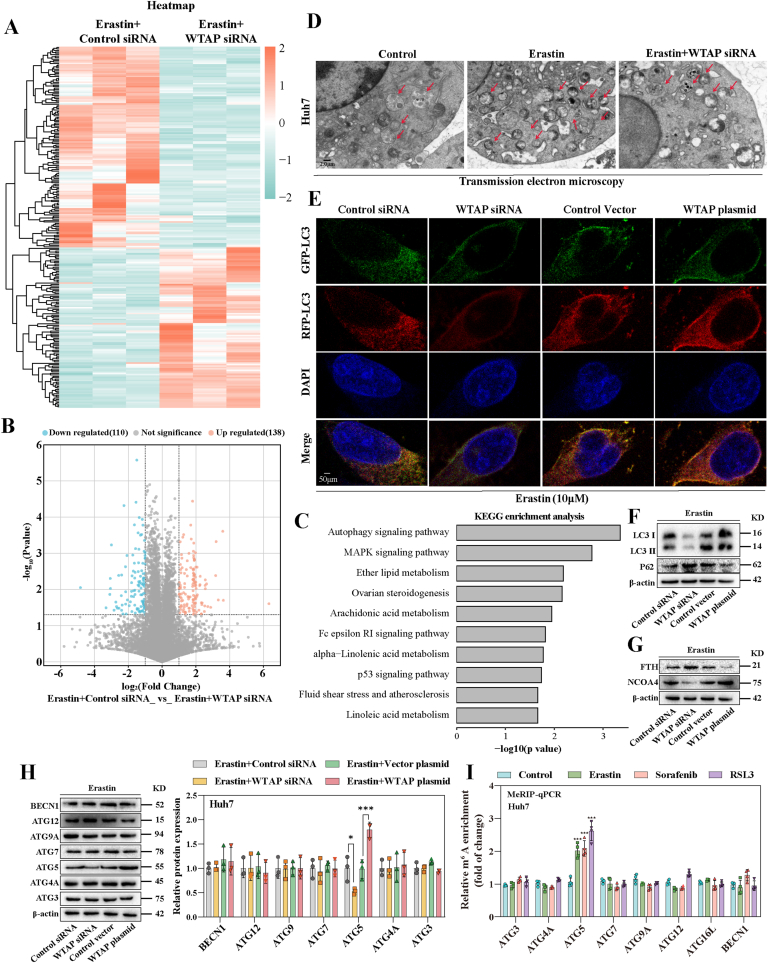
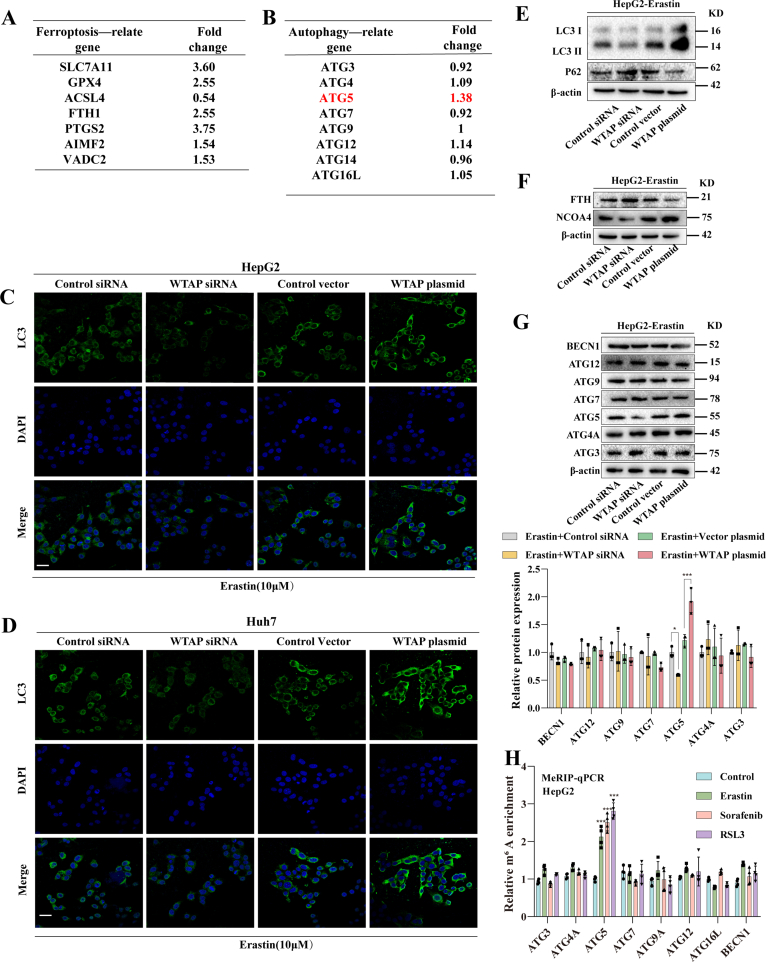
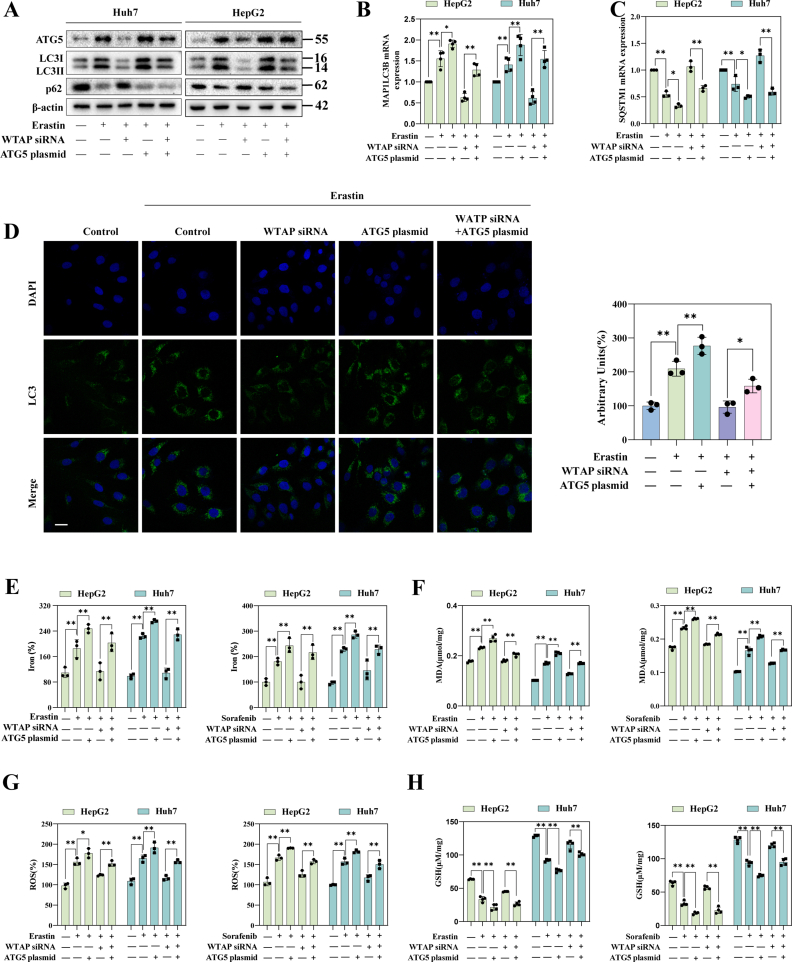
To gain a deeper understanding of the molecular mechanism by which m6A modification promotes HCC ferroptosis, we conducted RNA sequencing (RNA-seq) analysis. The heat map (Fig. 3A) and volcano plot (Fig. 3B) indicated that when WTAP was lowered, 138 mRNAs exhibited a significant rise (fold change >1.4), whereas 110 mRNAs showed a significant drop (fold change <0.65) in Huh7 treated with erastin. Based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) statistics, the data strongly support the potential regulatory role of autophagy signaling during HCC ferroptosis (Fig. 3C). As expected, among these genes, some ferroptosis-related genes, such as SLC7A11, GPX4, and ACSL4, were identified (Fig. S4A). Wet lab experiments were then carried out to investigate the involvement of autophagy in m6A modified-mediated ferroptosis in HCC. The number of autophagosomes (Fig. 3D) and the level of autophagic flow (Fig. 3E) decreased after silencing WTAP in ferroptotic HCC. In erastin-treated Huh7 and HepG2, the expression of LC3, a key marker of autophagy, was reduced after weakening m6A modification and further increased after elevating m6A modification (Figs. S4C and S4D), suggesting that autophagic activity was regulated by WTAP-mediated m6A modifications. The conversion of LC3-I to LC3-II were significantly reduced, while the expression of p62 was increased in ferroptoic HCC under the broken of WTAP expression, while the above autophagy events were significantly enhanced under the enhanced of WTAP expression (Fig. 3F and Fig. S4E). Notably, NCOA4-mediated ferritin autophagy is a key event in autophagy-induced ferroptosis [25]. We also observed a decrease in NCOA4 and an increase in FTH during Huh7 and HepG2 ferroptosis after WTAP silenced. This phenomenon was reversed by over-expression of WTAP (Fig. 3G and Fig. S4F).
Fig. 3.
m6A modification-mediated ferroptosis in HCC is autophagy-dependent. A Control siRNA and WTAP siRNA was transfected into Huh7 for12 h and then treated with erastin (10 μM) for 24 h. Total RNA was isolated for RNA-Seq. Clustering of Huh7 cells were demonstrated by microarray heat map. The significantly differentially expressed mRNAs were analysied by hierarchical cluster: white, no change; bright green, underexpression; bright orange, overexpression. B Enhanced Volcano was plotted by https://www.bioinformatics.com.cn, an online platform for data analysis and visualization. C Differentially expressed mRNAs were enriched by KEGG enrichment analysis in WTAP siRNA group (Erastin + Control siRNA, n = 3; Erastin + WTAP siRNA, n = 3). D The number of autophagosomes in Huh7 cells were observed by Transmission electron microscopic. Scale bars: 2.0 μm. E Huh7 cells transfected with WTAP siRNA and plasmid along with mRFP-EGFP-hLC3B-pKGX-Puro plasmid were treated with erastin (10 μM). Laser scanning confocal microscope was used to observe fluorescence intensity. Scale bars: 50 μm. F–H WTAP siRNA and plasmid were utilized to transfect Huh7 and HepG2 cells respectively followed by 10 μM erastin. Expression of autophagy-related genes was detected by Western blot. I Ferroptosis inducers (10 μM Erastin, 10 μM Sorafenib, and 2.5 μM RSL3) were applied to Huh7 for 24 h. MeRIP qPCR was used to detected the levels of m6A modification on autophagy-related genes. The mean ± SD was used as the value of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001.
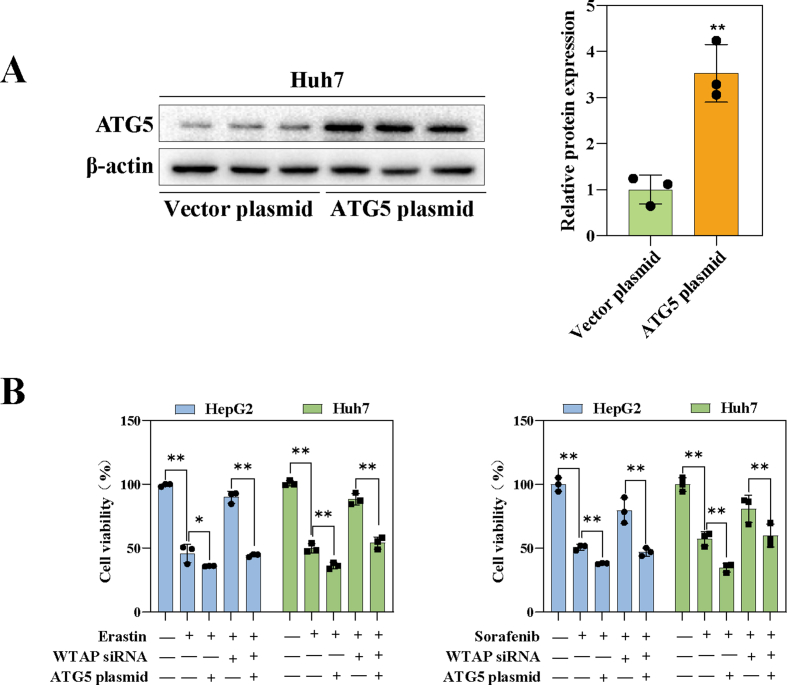
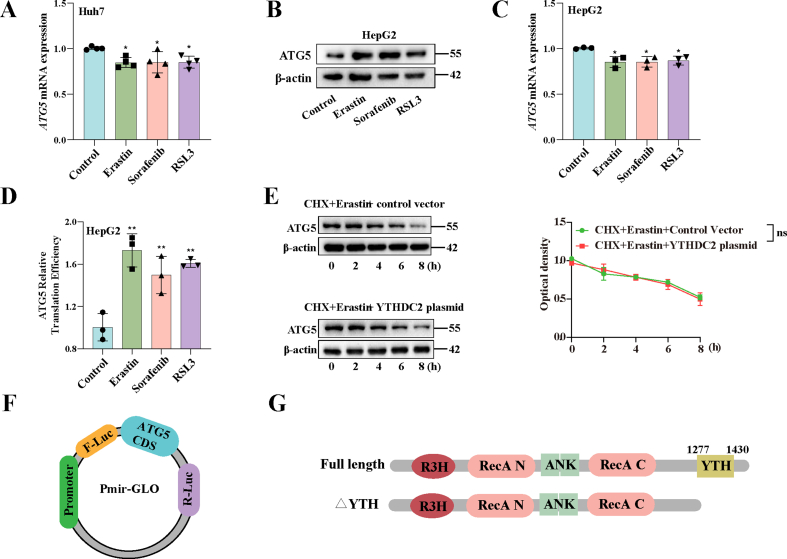
Remarkably, we found that the expression of ATG5, a crucial gene in autophagy signaling, decreased significantly upon WTAP decreased in Huh7 treated with erastin (Fold change = 0.18) (Fig. S4B). We further to estimate whether ATG5 was the critical target of m6A modification in ferroptoic HCC. Consistent with our sequencing results, after silencing WTAP, the expression of ATG5 was decreased, while the expression of ATG5 was increased after over-expression of WTAP in Huh7 and HepG2 after treated with erastin (Fig. 3H and Fig. S4G). We then performed MeRIP-qPCR to further validate this result. The results showed that only the m6A modification level of ATG5 mRNA was significantly increased after treatment with erastin, sorafenib, and RSL3 (Fig. 3I and Fig. S4H). Since ATG5 had been tentatively identified as a downstream target of WTAP-mediated m6A modification. An ATG5 plasmid was used to performed function-acquisition experiments to clarify this finding on this basis (Fig. S5A). Under the destruction of WTAP expression, ATG5 plasmid could restore the expression of ATG5, (Fig. S6A). In particular, we discovered that increased LC3 expression and decreased p62, which were autophagic hallmark events, that were recovered by overexpressing ATG5 (Figs. S6A and S6B). The same result was obtained by visualization of LC3 expression though immunofluorescence (Fig. S6C). Next, we tested cell viability to clarify the effect of autophagy on ferroptotic HCC. The MTT results showed that in ferroptotic HCC induced by erastin and sorafenib, activation of autophagy by overexpression of ATG5 could reversed the recovery of cell viability caused by silencing of WTAP (Fig. S5B). Finally, our study revealed that overexpression of ATG5, leading to the activation of autophagy, effectively reversed the attenuation of key ferroptosis events induced by WTAP silencing alone in Huh7 and HepG2 treated with erastin and sorafenib. Specifically, we observed the restoration of iron accumulation (Fig. S6E), MDA production (Fig. S6F), ROS generation (Fig. S6G), and GSH depletion (Fig. S6H). In summary, these results strongly suggested that elevated WTAP-mediated m6A modification promoted ATG5-initiated autophagy in ferroptotic HCC.
2.4. The m6A reader YTHDC2 initiated autophagic ferroptosis by promoting ATG5 translation
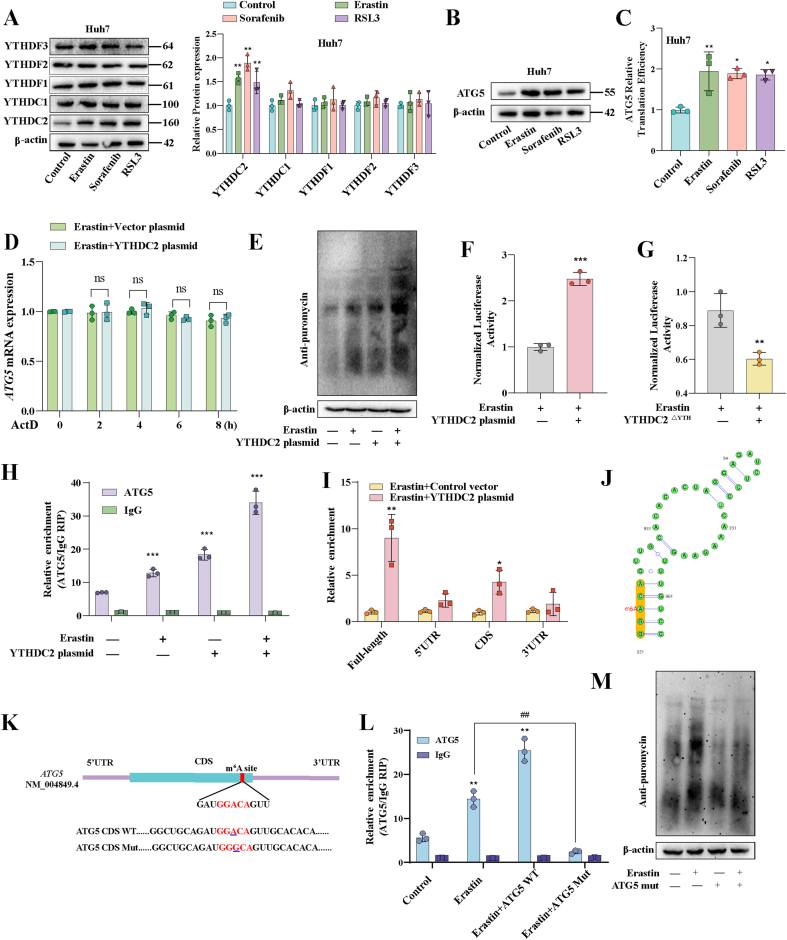
Notably, reader proteins played a crucial role in enabling m6A modification to exhibit a wide range of biological functions [18]. To seek out which reader was involved in m6A modification-dependent autophagy-related ferroptosis, we did an unbiased expression screen of five of the most common reader proteins. To our surprise, the expression of YTHDC2 was significantly increased after treatment of Erastin, Sorafenib and RSL3 (Fig. 4A and Fig. S7A). Next, we employed YTHDC2 siRNA-2 to conduct the loss-of-function experiments (Fig. S7B), and YTHDC2 plasmid was utilized for the gain-of-function experiment (Fig. S7C). As anticipated, the expression of ATG5 protein was reduced after the decreased of YTHDC2; on the contrary, ATG5 expression was restored after YTHDC2 re-expression (Fig. S7d). Furthermore, the results showed that the key events in the onset of ferroptosis such as GSH reduction (Fig. S7E), iron elevation (Fig. S7F), MDA increase (Fig. S7G), and ROS elevation (Fig. S7H) were all disrupted to varying degrees after the decreased of YTHDC2. Conversely, the introduction of exogenous ATG5 administration significantly restored the occurrence of ferroptosis (Figs. S7E–H). These results suggested that YTHDC2 was involved in m6A modification-mediated autophagy-related ferroptosis.
Fig. 4.
YTHDC2 interacts with ATG5 mRNA to promote its translation. A Expression of m6A readers in Huh7 detected by Western blot. B Expression of ATG5 protein detected by Western blot. C Translation efficiency of ATG5 in Huh7, calculated as: translation efficiency = ATG5 protein level/ATG5 mRNA expression level. D ActD was used to inhibit the transcription and RT-PCR was used to detected the mRNA of ATG5. E Puromycin labeling assay showed the protein synthesis in Huh7. F-G Huh7 cells were transfected with pmiR-GLO plasmids containing ATG5 CDS region for 24 h, and the translation efficiency of ATG5 was illustrated as the relative ratios between F-luc and R-luc. H RIP experiments to analyze the binding of ATG5 and YTHDC2. I The levels of m6A modification in ATG5 mRNA 3′-UTR, CDS and 5′-UTR were determined by MeRIP qPCR. J Diagram of the m6A site on ATG5 mRNA. K Schematic representation of the mutation in CDS on ATG5 mRNA. L RIP experiments to analyze the binding of ATG5 and YTHDC2. M Puromycin labeling assay showed the protein synthesis in Huh7. The mean ± SD was used as the value of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001.
We have initially shown that ATG5 was a downstream target of YTHDC2, thus highlighting the need for further investigation into the YTHDC2-mediated regulation of ATG5 expression. We observed a notable enhancement in the translational efficiency of ATG5 in ferroptotic HCC, as evidenced by an elevation in protein levels and a slight decrease in mRNA level (Fig. 4B–C and Figs. S8A–D). Simultaneously, we observed that YTHDC2 did not exert any influence on the degradation of ATG5 mRNA (Fig. 4D). Preliminary evidence from puromycin labeling of nascent polypeptides revealed that the overexpression of YTHDC2 had a pronounced effect on the overall mRNA translation at a global scale (Fig. 4E). Simultaneously, we ruled out the influence of YTHDC2 on ATG5 protein degradation, as the overexpression of YTHDC2 did not impact the degradation rate of ATG5 protein following inhibition of protein synthesis (Fig. S8E). The CDS section of ATG5 mRNA was cloned downstream of the portion of the pmirGLO plasmid that encoded firefly luciferase in order to examine if YTHDC2 influenced ATG5 translation (Fig. S8F). The ratio between firefly luciferase and internal Renin luciferase was significantly higher following YTHDC2 overexpression, suggesting that translation of ATG5 was enhanced by YTHDC2 (Fig. 4F). To confirm that the role of YTHDC2 was dependent on its YTH domain, we constructed the YTHDC2△YTH plasmid (Fig. S8G). This effect was significantly attenuated by deleted of the YTH domain of YTHDC2 (Fig. 4G). Previous studies have shown that the m6A reader is an RNA-binding protein [26]. Therefore, we examined the interaction of YTHDC2 with ATG5 mRNA using RIP-PCR experiments. The results indicated that ATG5 mRNA bound to YTHDC2, with a more pronounced interaction observed in the presence of erastin treatment (Fig. 4H). These results suggest that the function of YTHDC2 is to promote translation of ATG5 transcript in ferroptotic HCC.
Next, we delved into the m6A modification site on ATG5 mRNA. Firstly, MeRIP qPCR analysis demonstrated a significant enrichment of m6A modification in the CDS of ATG5 (Fig. 4I). Interestingly, a meticulous primary sequence analysis unveiled the presence of a potential m6A recognition site (GGACA) within the CDS region of the ATG5 mRNA (Fig. 4J). To confirm the functional significance of the m6A binding site in the ATG5 CDS region, we generated an ATG5-mut variant, as depicted in Fig. 5K. As anticipated, ATG5-mut disrupted the binding between YTHDC2 and ATG5 mRNA (Fig. 4L), leading to a significant reduction in the translation of ATG5 (Fig. 4M). Taken together, these findings indicate that YTHDC2 promoted the translation of ATG5 mRNA by binding to the m6A binding site in the CDS region at A921.
Fig. 5.
YTHDC2 knockdown impairs HCC ferroptosis in vivo. A Images at the end points of Huh7 cell-derived subcutaneous xenograft tumors in BALB/c nude mice. B–C Changes in tumor volume and mouse body weight in each group during the whole experimental cycle. D-F Immunohistochemistry was used to detect the protein expression in tumor tissues (scale bar: 50 μM). G Weight of tumors in each group at the end points of xenograft tumors. H-J The level of GSH, MDA and iron in tumor tissues of each group. The mean ± SD was used as the value of five independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001.
2.5. Autophagy activation is required for m6A modification to regulate ferroptosis of xenograft tumor in vivo
To investigate the impact of m6A modification on the process of ferroptosis in HCC, we established stable YTHDC2-knockdown Huh7 cell lines (YTHDC2 KD). This is because YTHDC2, as a reader protein, serves as a crucial bridge between m6A modifications and downstream target genes. Additionally, to assess the regulatory effect of YTHDC2 on ferroptosis, Huh7 murine tumor model was constructed with nude mice through subcutaneous tumors xenografts. We found that both sorafenib and erastin could inhibit the growth of xenografts including tumor volume and weight (Fig. 5A, B and G) without the significant effect on the body weight of the mice (Fig. 5C), while YTHDC2 knockdown significantly weakened the anticancer effects of both (Fig. 5A–C and G). Then we found that sorafenib and erastin inhibited tumor proliferation, mainly manifested by decreased expression of ki67 and p16, while knockdown of YTHDC2 significantly increased the expression of ki67 and p16 (Fig. 5D). Consistent with previous experiments in vitro, the expression of ATG5 as well as YTHDC2 was significantly elevated in tumor tissues after induction of ferroptosis with erastin and sorafenib, while the levels of both were significantly reduced after YTHDC2 knockdown (Fig. 5E). PTGS2 is the main marker of ferroptosis in vivo [27]. Immunohistochemical staining showed that erastin and sorafenib could significantly increase its expression, while YTHDC2 knockdown significantly decreased its expression (Fig. 5F). Likewise, erastin and sorafenib significantly induced key ferroptotic events in tumor tissues, including GSH reduction (Fig. 5H), MDA accumulation (Fig. 5I), and iron elevation (Fig. 5J). Knockdown of YTHDC2 could significantly inhibit these processes. Based on the aforementioned findings, it has been observed that erastin and sorafenib have the ability to modulate autophagy-dependent ferroptosis through YTHDC2, thereby exerting anti-HCC effects.
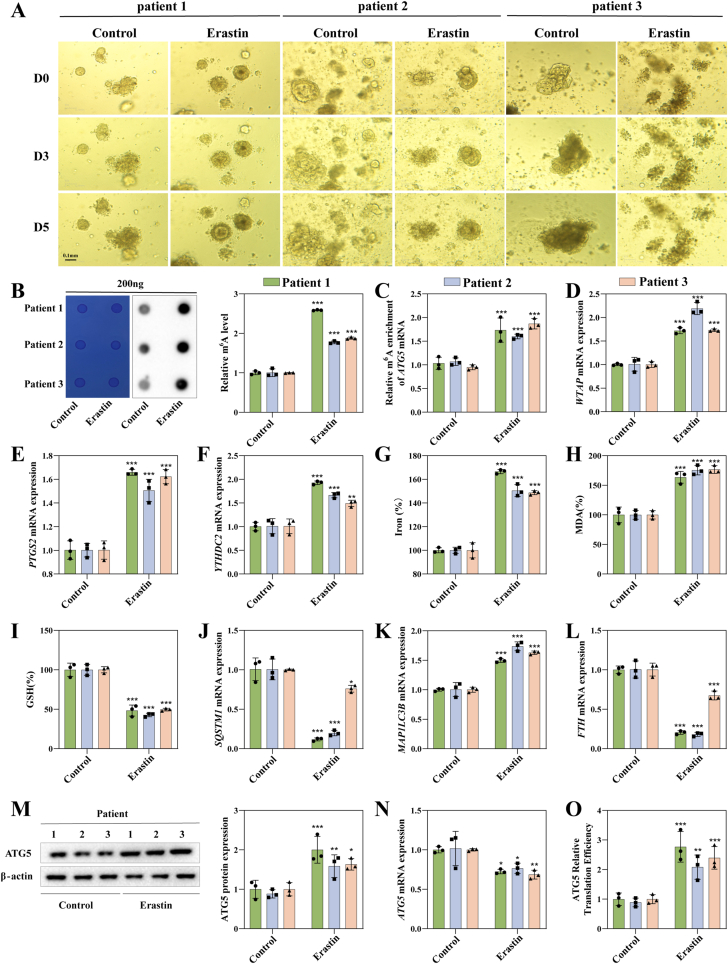
2.6. Induction of ferroptosis suppresses HCC PDO growth
PDOs have been recognized as a superior tool for simulating the three-dimensional structure and functionality of HCC in an in vivo-like environment [28]. Building upon this, we proceeded to investigate the susceptibility of HCC PDOs to ferroptosis using this model. Initially, we obtained freshly diagnosed HCC samples from individuals. Subsequently, we induced ferroptosis in HCC PDOs by treating with 10 μM erastin. Specifically, the HCC PDOs were exposed to erastin for a duration of 5 days. Following this, we monitored the growth of the PDOs over a period of five days within the same field of vision. Our observations revealed that PDOs in the Erastin group exhibited slow or no growth, while the control group demonstrated significant growth over the same time period (Fig. 6A). We extracted RNA and protein on the fifth day in order to detect related gene expression. First, we examined the overall m6A level in PDOs and found that it was significantly increased in the group treated with erastin (Fig. 6B), especially on ATG5 mRNA (Fig. 6C), which was consistent with our cell experiments. As anticipated, the erastin group exhibited significantly higher levels of WTAP and YTHDC2 expression compared to the control group (Fig. 6D and E). In line with what we expected, ferroptosis occurred in the HCC PDO as evidenced by the dramatically increased expression of PTGS2 (Fig. 6F), a crucial gene for the ferroptosis. Meanwhile, the changes in Iron, MDA, and GSH in PDOs after erastin treatment were also consistent with classical ferroptosis events (Fig. 6H and I). Furthermore, we conducted an examination of the expression of genes associated with autophagy. Consistent with the findings from cellular and animal studies, the expression of p62 was significant decreased and LC3 was significant increase in the group treated with erastin (Fig. 6 J and K). These results indicated that autophagy is involved in erastin induced PDOs ferroptosis. Additionally, a striking alteration in the expression of the ferritinophagy-related gene FTH, providing further evidence of the association between autophagy and ferroptosis in the erastin group (Fig. 6L). Finally. We found that the expression of ATG5 protein was elevated in PDOs (Fig. 6M), and its mRNA level was decreased (Fig. 6N), indicated that the translation level of ATG5 was significantly enhanced after erastin treatment (Fig. 6O), which was consistent with our in vitro results. According to the aforementioned findings, we preliminarily indicated that erastin could delay the growth of PDOs, which may be related to the increase of m6A modification level, activated autophagy and induced ferroptosis in PDOs.
Fig. 6.
Erastin promotes HCC PDO ferroptosis in an m6A-dependent manner. A Representative bright-field images of HCC PDOs from three individuals that were exposed to erastin at the indicated concentration at 0, 3, 5 days are shown. Scale bar, 0.1 mm. B m6A modification level in PDOs was detected by dot blot. C MeRIP-PCR detected m6A level on ATG5 mRNA. D-F The mRNA expression of WTAP, YTHDC2, PTGS2 was detected by RT-PCR. G-I The level of GSH, MDA and iron in PDOs was detected by kits. J-L The mRNA expression of FTH, MAP1LC3, SQSTM1 was detected by RT-PCR. M The protein expression of ATG5 in PDOs was detected by Western blot. N The mRNA expression of ATG5 was detected by RT-PCR. O Translation efficiency of ATG5 in PDOs. Calculated as: translation efficiency = ATG5 protein level/ATG5 mRNA expression level. The mean ± SD was used as the value of three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001.
3. Discussion
In light of the urgent need to gain a fresh perspective on the underlying mechanism of cell death in HCC, it is crucial to identify new avenues for treatment. One promising approach is exploring ferroptosis, an iron-dependent form of cell death characterized by lipid peroxidation [29]. It has been established by the current investigation that this type of cell death also depends on autophagy [30]. Previous research has indicated a correlation between elevated levels of iron ions and the development of HCC [31,32], potentially triggering the induction of ferroptosis. Our investigation has demonstrated that HCC exhibit susceptibility to ferroptosis when exposed to any of the three known ferroptosis inducer, such as erastin, sorafenib and RSL3. While existing research on ferroptosis has predominantly concentrated on the transcriptional level, the intricate precision, prolonged time course, and dynamic attributes of ferroptosis underscore the need for comprehensive investigation into its post-transcriptional modification level. Notably, the epigenetic modification termed m6A possesses the capacity to modulate gene expression at the post-transcriptional level, making it a compelling target for in-depth examination and scrutiny [33].
However, the role of m6A modification in HCC is controversial. Several studies have found that the level of m6A modification in HCC tissues was significantly higher than in normal liver tissues, owing to increased expression of the methylation enzyme METTL3 [34]. However, it had been discovered that the level of m6A modification was reduced during the development of HCC, owing primarily to a decrease in the methylation enzyme METTL14 [35]. The conflicting outcome could potentially be attributed to the influence of three reasons as follows: (1) Sample selection and heterogeneity: HCC exhibits a notable degree of heterogeneity, encompassing various subtypes and molecular subgroups. The utilization of diverse liver cancer tissue samples in research studies introduces the possibility of distinct molecular and clinical characteristics, which could contribute to the observed variations in m6A modification levels. (2) Cancer development and progression stages: Liver cancer undergoes a dynamic and progressive process, leading to alterations in cellular biological features and patterns of m6A modification throughout different stages of development and progression. Therefore, the inclusion of liver cancer patients at various stages in different studies may account for the inconsistencies observed. (3) Interacting networks and regulatory mechanisms: The m6A modification operates within a complex regulatory network, subject to the influence of other genes, proteins, and signaling pathways in liver cancer. Disparities in these interacting networks and regulatory mechanisms may underlie the observed inconsistencies in m6A modification levels across different studies. These factors can lead to complex alterations in the expression of m6A modification levels and regulatory factors during the development of HCC.
In this study, we concentrated on changes in m6A modification with the emergence of ferroptosis in HCC. Our investigation unveiled a remarkable increase in m6A modification levels within HCC following treatment with ferroptosis inducers. The increase of m6A modification level was also observed after erastin treatment in the PDO model. Subsequent analyses elucidated that this elevation was linked to the upregulation of WTAP, a methyltransferase enzyme. Intriguingly, contrary to previous reports implicating METTL14 in elevated m6A modification levels in HCC, our study identified WTAP as the primary driver of this observed increase. This disparity can be attributed to the specific focus of our research on the context of ferroptosis, in contrast to previous studies that directly assessed the expression of relevant enzymes in both tumor and adjacent tissues. Due to the specific cellular death mechanism of ferroptosis, its inducers may have a direct impact on m6A modification. Therefore, under the influence of ferroptosis inducers, the upregulation of WTAP expression is likely to result in a significant increase in m6A modification levels within HCC. Furthermore, this observation underscores the intricate nature of m6A modification, as the predominant methyltransferases involved can exhibit variations across diverse cellular contexts and pathological processes. In this study, we had provided compelling evidence supporting the crucial involvement of m6A modification in the occurrence of ferroptosis in HCC. Notably, we observed a significant increase in the level of m6A modification during the induction of ferroptosis in HCC. These results collectively suggest that there exists a dynamic balance of m6A modification levels in HCC, which, when disrupted, could adversely impact HCC development. By shedding light on the significance of m6A modification, our study offered a novel avenue for therapeutic interventions though m6A modification in the treatment of HCC.
The presence of m6A demethylases ensures the dynamic and reversible nature of m6A modifications. They can remove m6A modifications that have already occurred on mRNAs, thereby deregulating the m6A modifications on downstream target genes. There are only two well-characterized m6A demethylases, FTO [36] and ALKBH5 [37], and several studies have shown that they are involved in the regulation of HCC progression, but some of the conclusions are contradictory, and these conflicting functions may be related to the heterogeneity of the tumors or the small sample size used, which also suggests that m6A modification plays a complex role in HCC. Unfortunately, we did not find significant changes in m6A demethylase expression during HCC ferroptosis in current study, which ensured that m6A modification can be at a high-level during HCC ferroptosis. This leaded to the rapid recognition of m6A-modified mRNA by the reading protein YTHDC2, contributing to the regulation of the expression of downstream target genes by m6A modification.
Interestingly, emerging research had revealed the regulatory role of autophagy in the occurrence of ferroptosis. Autophagy, a cellular degradation process, orchestrates the formation of autophagosomes to sequester damaged or surplus cellular components, subsequently facilitating their degradation within lysosomes to uphold cellular homeostasis [38]. Compelling evidence from various studies suggested that autophagy induction can heighten cellular susceptibility to ferroptosis, thereby amplifying the overall efficacy of ferroptosis. Notably, certain autophagy-related genes, including ATG5 and ATG7 [39], have been found to exert regulatory control over iron metabolism and the accumulation of lipid peroxides during the process of ferroptosis. In current study, RNA-seq found that the autophagy pathway changed most obviously after the change of m6A modification level. Further detection by MeRIP-PCR showed that ATG5 was the most important autophagy gene modified by m6A modification. Our study revealed that ATG5-mediated autophagy was a crucial step in HCC ferroptosis, as it facilitates the degradation of intracellular iron-storing proteins, leading to the release of a substantial amount of iron ions and initiating HCC ferroptosis. This crucial observation highlights the potential of promoting ferroptosis, particularly through autophagy-dependent pathways, as a promising therapeutic strategy for combating HCC.
Our investigation had unequivocally established the association between the upregulation of methyltransferase WTAP and the observed elevation in m6A modification levels during HCC ferroptosis. However, regarding the crucial involvement of reader proteins in the regulation of downstream target genes through m6A modification, one pertinent question arises: which specific reader protein assumes a pivotal role? By employing an unbiased screening approach, we have successfully identified YTHDC2 as the key mediator responsible for the m6A modification-mediated regulation of the downstream target gene ATG5 during HCC ferroptosis. Moreover, we identified a significant interaction between YTHDC2 and ATG5 mRNA, which was enhanced in the presence of ferroptosis inducers. Given that YTHDC2 has been shown to enhance the translation of target mRNAs [40], this finding suggests that it may promote the expression of target genes. Because of YTHDC2 structure contains DEAH/RNA helicase A family features, it can promote the translation of mRNA by binding to the m6A site in the CDS region and then resolving secondary structures [40]. Our research further demonstrated that YTHDC2 could bind to the CDS region of the ATG5 mRNA, facilitating its translation. Interestingly, we noticed a slight decrease in the expression of ATG5 mRNA, which may be related to its ability to interact with the 5′ → 3′ exonuclease [41]. Future research will delve into the molecular mechanism underlying this phenomenon and further illuminate the intricate role of YTHDC2, which we did not examine in detail here.
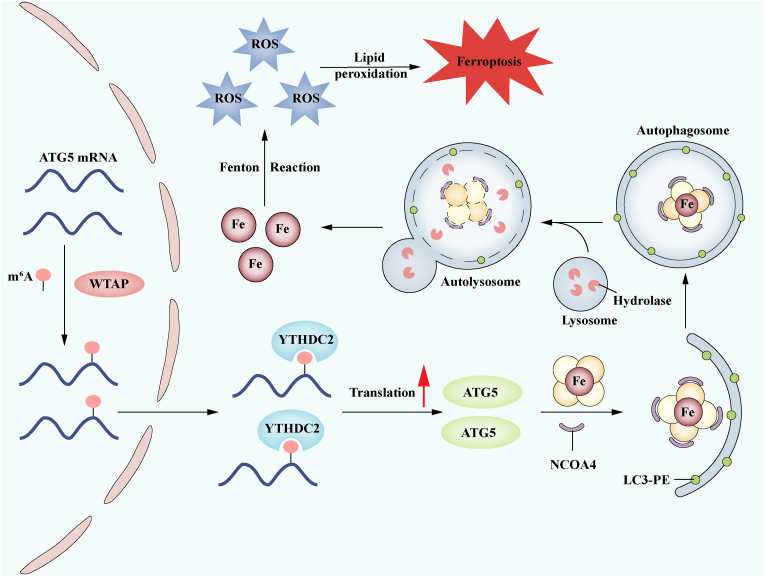
In summary, our research has demonstrated the crucial importance of WTAP-mediated m6A modification in the translation of ATG5 mRNA through a YTHDC2-associated mechanism during HCC ferroptosis induced by ferroptosis inducers (Fig. 7). These findings shed new light on the regulatory mechanisms governing ATG5 expression through epigenetic modifications, particularly emphasizing the significance of m6A modification in the context of HCC treatment.
Fig. 7.
WTAP-mediated m6A modification on ATG5 mRNA is dependent on YTHDC2 in autophagic iron death in HCC. Following the administration of HCC ferroptosis inducers, they triggered WTAP-mediated m6A modification on ATG5 mRNA, which was then recognized and bound by YTHDC2. This resulted in enhanced translation and elevated expression of ATG5, which then started ferritinophagy, an increase in the unstable iron pool, and eventually led to the onset of ferroptosis in HCC.
4. Materials and methods
4.1. Cell culture drug treatment
Human hepatoma cell lines Huh-7(Procell Cat#CL-0120) and HepG2 (Procell Cat#CL-0103) were cultured in Dulbecco's Modified Eagle Medium (DMEM, purchased from Servicebio, Wuhan, China) containing 10 % fetal bovine serum (FBS, purchased from biosharp, Chhina) and 1 % penicillin-streptomycin (purchased from Nanjing BioChannel Biotechnology Co., Ltd). By administering erastin (10 μM), sorafenib (10 μM), and RSL3 (2.5 μM) for 24 h, HCC ferroptosis was triggered. In the meantime, cells were given an equivalent amount of DMSO without any medicines as a control. Cells were authenticated by STR profiling and tested negative for mycoplasma contamination.
4.2. Construction of stable cell line
To construct a stable YTHDC2 knockdown cell line, lentiviral vectors harboring shRNA for knockdown of YTHDC2 (YTHDC2 KD), and negative control underwent syncretization and the cloned into pLKO.1 vector. Transfection was then started by adding the plasmid and Lipofectamine 3000 transfection reagent mixture (L3000015, Invitrogen) to the medium. In a nutshell, 10 μg/ml puromycin (Thermo Fisher Scientific Cat#A1113803) was used to select stably transfected cells for 3 weeks.
4.3. Plasmid construction
Lipofectamine 3000 (Invitrogen Cat#L3000015) was used for the siRNA and plasmid transfections in accordance with the manufacturer's recommendations. WTAP siRNA, pcDNA3.1-WTAP plasmid, ATG5 mutation plasmid (ATG5 mut), YTHDC2 plasmid, YTHDC2△YTH plasmid were constructed by Yormbio Tech. YTHDC2 siRNA, pcDNA3.1-ATG5 plasmid and mRFP-EGFP-hLC3B-pKGX-Puro was obtained from KeyGEN BioTECH.
4.4. Wound healing
For wound healing experiment, Huh7 and HepG2 cells were inoculated into 6-well plates, incubate overnight, and then scratch with a sterile 200 μl gun tip perpendicular to the cell plane. The dropped cells were washed off with PBS, and the width of the scratch was observed on the microscope and photographed. Then mitomycin C (MCE Cat#HY-13316) was added as well as the reagent, then continued inoculated in the incubator and observed and photographed again after 24 h under the same field of view.
4.5. Cell proliferation assay
Overall cell proliferation was measured by MTT-based cell viability assay. Put simply, cells were cultured in 96-well plates, and corresponding treatment was performed when the cell density reached about 50 %. After 24 h, 5 mg/ml of MTT (BioFroxx Cat #1334GR005) solution was added and the incubation was continued for 4 h at 37 °C. Finally, discard the supernatant fractions, add 200 μl DMSO to each well, incubated for 10 min at room temperature on a shaker, and detect the wavelength at 490 nm. For individual cell proliferation, Edu-488 cell proliferation kit was used to detect (Beyotime Biotechnology Cat# C0071S). HepG2 and Huh7 cells were inoculated into 96-well plates. Then labeled by 37 °C pre-warmed EdU 2 h prior to sample collection. After labeling, the cells were fixed with 4 % paraformaldehyde for 10 min and permeabilized with 0.3 % Triton X-100. Then assayed the fluorescence intensity according to the steps in the instructions.
4.6. Transwell assay
For transwell invasion assay, Huh7 and HepG2 cells were seeded into the upper chamber with Matrigel (Corning Cat#356234). Non-invaded cells were removed using a cotton swab after a 24-h incubation period, and cells on the chamber's bottom were fixed with 4 % paraformaldehyde for 10 min before being stained with Crystal Violet (KeyGEN Cat #KGA229). Next, 3 randomly chosen fields (200 magnification) were taken using an inverted microscope.
4.7. GSH, MDA, ROS, and iron content determination
The GSH was detected by a glutathione kit (BestBio Cat # BB-4711). The MDA was detected by a lipid oxidation detection kit (BestBio Cat # BB4709). ROS was detected by a ROS kit based on DCFH-DA probe (Beyotime Biotechnology Cat #S0033). Iron content was detected by an intracellular iron colorimetric assay kit (Applygen Cat #E1042). Bioassays were replicated three times.
4.8. m6A dot blot
The total RNA was isolated using phenol/chloroform extraction. Then the mRNA solution was mixed with the same volume of denaturant (20 × SSC buffer:37 % formaldehyde = 3:2) and incubated at 95 °C for 5 min. Then, 100 ng, 200 ng or 400 ng poly (A) + RNAs were added on nitrocellulose membrane (GE Healthcare). After drying at 37 °C for 30 min, the RNA was immobilized on the NC membrane by UV irradiation for 30 min. The NC membranes were soaked in 5 % milk for 1 h and then incubated with m6A antibody (ABclonal Cat#A19841) overnight. Then incubate with secondary antibody and expose by ECL. The same amount of RNA was spotted on NC membrane, stained with methylene blue and photographed.
4.9. Quantification of RNA m6A
The total RNA was isolated using phenol/chloroform extraction. Then used EpiQuik m6A RNA Methylation Quantitative kit (Epigentek, Farmingdale, NY, USA) to detect the total m6A level of RNA. All procedures were carried out according to manufacturer instructions. Briefly, 200 ng of RNA per sample was transferred to assay wells. Following the manufacturer's instructions, capture antibody solution and detection antibody solution were then added to assay wells separately in a suitable diluted concentration. The m6A levels were quantified colorimetrically by measuring the absorbance of each well at a wavelength of 450 nm, and calculations were performed based on the standard curve.
4.10. Western blot
Intracellular total protein extract by RIPA buffer (Beyotime, Shanghai, China) containing 1 % protease inhibitors and PMSF. After measuring the protein concentration, added 1/3 volume of 5 × protein loading buffer and heated at 95 °C for 15min. Then used the SDS-PAGE to separate protein, pageRuler prestained protein ladder was used for protein molecular weight estimation. After separation, transferred the protein to a 0.22 μm PVDF membrane. Block the membranes by incubating in 5 % skim milk for 2 h at room temperature. Membranes were then placed in primary antibody buffer and incubated overnight at 4 °C. The membranes were then blocked with the corresponding secondary antibody for 2 h at room temperature. Finally, the gel imaging system was used for analysis (Bio-Rad, Hercules, CA). Each antibody used in this study is listed in Table S1.
4.11. RNA extraction and real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen Cat #15596018) according to the manufacturer's instructions. Then used Nanodrop to measure the purity and concentration of RNA. Hifair® II 1st Strand CDNA Synthesis Super Mix (YEASEN Cat#11123ES60) was used to synthesize the template complementary DNA from total RNA. The QuantiTect SYBR Green PCR Kit Hieff qPCR SYBR Green Master Mix (YEASEN Cat#11202ES08) was used to analyze the cDNA by qRT-PCR on ABI 7500 system (Applied Biosystems). Gene expression was normalized using housekeeping gene GAPDH and relative quantification was calculated using 2−ΔΔCt.Used primers are presented in Table S2.
4.12. RNA sequencing
Total RNA was isolated from Huh7 by TRIzol reagent (15596018, Invitrogen). For each sample, TruSeq Stranded Total RNA with 1 μg RNA was used for library preparation with Ribo-Zero Gold kit (MRZG12324, Illumina). TopHat v1.4.1 was used to align the reads of each sample with the human reference genome (GRCh38.p10). EdgeR version 3.08 was performed for differential gene expression. The Benjamini-Hochburg method was used to calculate the adjusted P values. We used fold change <0.65 for downregulation and fold change >1.4 for upregulation as cutoffs.
4.13. MeRIP qPCR
According to our previous reports [23], MeRIP qPCR was performed to quantify the enrichment of m6A in target gene. In briefly, 5 μg rabbit IgG and m6A antibody were conjugated to 50 μl Protein A/G Plus Agarose (ABclonal Cat#RM09008) overnight at 4 °C, respectively. The antibody-beads compound was then incubated with 100 μg RNA overnight at 4 °C. The precipitated products were incubated with elution buffer at 50 °C for 1.5 h. Finally, RNA was re-extracted by the phenol/chloroform method and analyzed by qPCR.
4.14. Transmission electron microscopy
According to our previous reports [42], using transmission electron microscopy to observe the morphology of mitochondrial and the number of autophagosomes. In brief, discard the medium in the large dish, directly added the electron microscope fixative solution, gently scrape the cells with a cell scraper and collected them into a centrifuge tube. Then centrifuged for 3 min at 1000 rpm, discarded the supernatant, added new electron microscope fixative, and fixed at room temperature for 2 h. Images were acquired by servicebio (Wuhan, China).
4.15. RIP-RT-PCR
RNA immunoprecipitation (RIP) was performed as previously described [23]. Put simply, added 0.75 % formaldehyde to the culture medium and rotate gently for 10 min at room temperature to cross-link in vivo protein-RNA complexes. Then isolated nuclei and lysed the nuclear pellets. The chromatin was mechanically sheared using a dounce homogenizer with 15–20 strokes. Add YTHDC2 antibody (Abcam Cat#ab220160) (5 μg) to supernatant (10 mg) and incubate for 2 h at 4 °C with gentle rotation. Add protein A/G beads (40 μL) and incubate for 1 h at 4 °C with gentle rotation. Washing off unbound material. Eventually, purification of RNA and analyzed by RT-PCR. Normalization of the relative enrichment was done to the input as: %Input = 1/10 × 2Ct [IP] – Ct [input].
4.16. Immunofluorescence
For immunofluorescence experiments [23], cells were seeded in 24-well plates and treated with compounds at indicated times. Then cells were fixed with paraformaldehyde and incubated with LC3 (Abcam Cat#ab48394) antibody overnight. Whereafter, cells incubated with FITC anti-rabbit IgG. Following this, the nuclei were stained with DAPI (KGA215-50, KeyGEN BioTECH). Finally, the images were taken by confocal laser.
4.17. Animal experiments
The animal care Committee of Nanjing University of Chinese Medicine approved all animal experiments. Four-week-old BALB/C-nu nude male mice were used for animal studies, and all animals were maintained in the specific pathogen-free (SPF) conditions at our institution. Huh-7 and stable YTHDC2 knockdown Huh-7 cells (approximately 1 × 107) resuspended with 50 μl of PBS and 50 μl of stromal gel were injected subcutaneously into the axilla of BALB/c nude mice to establish the subcutaneous xenograft model. When the volume of xenograft tumors up to 100 mm3, the mice were randomly divided into six groups, with five mice in each group. Erastin and sorafenib were dissolved in 10 % DMSO and 90 % corn oil and injected intraperitoneally into the mice every other day. After 28 days, mice were deeply anesthetized by intraperitoneal injection of sodium thiopental before decapitation, followed by tumors extraction, and stored in a −80 °C refrigerator for subsequent experiments. There were no occurrences of mouse mortality throughout the entire experimental period.
4.18. PDO culture
The culture of PDO was carried out according to the method in a previous report [43]. In brief, HCC samples were chopped, digested with trypsin, filtered, and centrifuged at 300 g for 5 min. The supernatant was removed, and the cells combined with Corning's Matrigel Matrix which contain Growth Factor Reduction (GFR) and seeded in 96-well plates (Corning). PDOs were cultured in DMEM containing 10 % FBS, and the corresponding growth factors were added to the supernatant to promote its growth.
4.19. Calculations and statistics
All data subjected to analysis had no less than three samples and are presented as means ± SD. Using GraphPad Prism 8 (GraphPad Software, La Jolla, CA, USA) to statistically analyze all data. Statistical analysis was performed using either one-way or Student's t-test (two-group comparison) analyses of variance followed by Student-Newman-Keuls test (more than two groups). A p-value of 0.05 or lower was regarded as statistically significant.
Authors’ contributions
Conceptualization and design: Yujia Li, Data acquisition (provided animals and PDO, provided access to facilities, etc.): Yang Wu, Zili Zhang, Shizhong Zheng; Data analysis and interpretation (e.g., statistical analysis, bioinformatics, and other computational analyses): Yujia Li, Mengran Li; Writing the manuscript: Yujia Li; Revision of the manuscript: Mei Guo, Yingqian Wang, Min Shen, Yangling Qiu; Feng Zhang, Jiangjuan Shao; Other (e.g. supervision of in vivo animal work): Xuefen Xu, Zili Zhang, Shizhong Zheng.
Funding
The work was supported by the National Natural Science Foundation of China (82374124, 82073914, 82000572, 82173874, 82274185, 82305046, 82304902), the Natural Science Foundation of Jiangsu Province (BK20200840, BK20220467), the Major Project of the Natural Science Research of Jiangsu Higher Education Institutions (20KJB310003, 22KJB310013), the Joint Project of Jiangsu Key Laboratory for Pharmacology and Safety Evaluation of Chinese Materia Medica and Yangtze River Pharmaceutical (JKLPSE202005), the Open Project of Chinese Materia Medica First-Class Discipline of Nanjing University of Chinese Medicine (2020YLKX023, 2020YLKX022), Jiangsu Provincial Double-Innovation Doctor Program (JSSCBS20220452, JSSCBS20220472), Young Elite Scientists Sponsorship Program by CACM (2022-QNRC2-B15), Outstanding Young Doctoral Training Program (2023QB0124), the Natural Science Foundation of Nanjing University of Chinese Medicine (NZY82000572, NZY82305046). The work was sponsored by Qing Lan Project.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional files.
Ethics approval and consent to participate
Hepatocellular carcinoma specimens were obtained from patients undergoing surgical resection at the First Affiliated Hospital of Nanjing Medical University. All human experiments were approved by the Ethical Review Committee of the First Affiliated Hospital of Nanjing Medical University. Written informed consent from the donors for research use of tissue in this study was obtained prior to acquisition of the specimen.
The animal study was approved by the Ethics Committee of the Nanjing University of Chinese Medicine. All animal experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animal by International Committees. Every effort was made to minimize the numbers and suffering of the included animals.
Consent for publication
The authors confirm that they have obtained written consent from each patient to publish the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102971.
Contributor Information
Xuefen Xu, Email: 460103@njucm.edu.cn.
Zili Zhang, Email: zilizhang@njucm.edu.cn.
Shizhong Zheng, Email: nytws@njucm.edu.cn.
Abbreviations
- HCC
hepatocellular carcinoma
- m6A
N6-methyladenosine
- METTL3
Methyltransferase like 3
- WTAP
Wilms tumor 1 associating protein
- FTO
fat mass and obesity associated protein
- ALKBH5
a-ketoglutarate-dependent dioxygenase alk B homolog 5
- YTH
YT521-B homology domain
- YTHDF
YTH N6-methyladenosine RNA binding protein
- YTHDC
YTH domain containing
- ATG5
autophagy related 5
- DMEM
Dulbecco's Modified Eagle Medium
- FBS
fetal bovine serum
- DMSO
dimethyl sulfoxide
- PBS
Phosphate Buffered Saline
- MTT
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- GSH
glutathione
- MDA
Malondialdehyde
- ROS
Reactive oxygen species
- PDO
Patient-derived organoid
- HSC
Hepatic stellate cell
- NCOA4
Nuclear receptor coactivator 4
- LC3
Microtubule associated protein 1 light chain 3 alpha
- p16
biomineralization protein SpP16
- ki67
antigen identified by monoclonal antibody Ki 67
- Ptgs2
prostaglandin-endoperoxide synthase 2
- XRN1
5′-3′ exoribonuclease 1
Appendix A. Supplementary data
The following are the Supplementary data to this article.
figs1.
The level of m6A modification in HepG2. A Ferroptosis inducers (10 μM Erastin, 10μM Sorafenib, and 2.5 μM RSL3) were applied to HepG2 cells for 24 h. m6A modification level was detected by dot blot. B-C Ferroptosis inducers (10 μM Erastin, 10 μM Sorafenib, and 2.5 μM RSL3) were applied to Huh7 and HepG2 cells for 24 h. mRNA expression of m6A writers and erasers in ferroptoic HCC cells detected by RT-PCR. The mean±SD was used as the value of three independent experiments. *p<0.05; **p<0.01; ***p<0.001.
figs2.
Transfection efficiency of WTAP siRNA and plasmid. A Intracellular WTAP expression levels after transfection with WTAP siRNA detected by Western blot. B The expression level of intracellular WTAP after transfection of WTAP plasmid detected by Western blot. The mean±SD was used as the value of three independent experiments. *p<0.05; **p<0.01; ***p<0.001.
figs3.
WTAP-mediated m6A modification contributed to HCC ferroptosis. WTAP siRNA and plasmid were utilized to transfect Huh7 and HepG2 cells respectively followed by 10 μM erastin or sorafenib. A-B Overall cellular proliferation tested by MTT assay. C Huh7 and HepG2 cells transfected with WTAP plasmid were treated with erastin with or without cell death inhibitors (Ferrostatin-1, 1 μM; Liproxstatin-1, 0.1 μM; Necrostatin-1, 10 μM; ZVAD-FMK, 10 μM; Necrosulfonamide, 0.5 μM) for 24 h. MTT assay was used to test overall cellular proliferation. The mean±SD was used as the value of three independent experiments. *p<0.05; **p<0.01; ***p<0.001.
figs4.
Autophagy activation mediated by m6A modification contributes to ferroptosis in HCC. A-B The fold change of ferrptosis- and autophagy-relate gene from RNA-Seq. C-D HepG2 and Huh7 were treated with erastin for 24 h after transfection with WATP siRNA and plasmid. Immunofluorescence detection of endogenous LC3 expression. E-G Expression of autophagy-related genes was detected by western blot. H MeRIP qPCR was used to detected the levels of m6A modification on autophagy-related genes. The mean±SD was used as the value of three independent experiments. *p<0.05; **p<0.01; ***p<0.001.
figs5.
ATG5 can restore the inhibitory effect of Erastin on HCC. A The ATG5 Plasmid was transfected into Huh7 cells for 12h, and the expression level of ATG5 was detected by Western blot. B Erastin was applied to HuH7 or HepG2 cells that had been transfected with WTAP siRNA and ATG5 plasmid alone or together. Overall cellular proliferation of HepG2 tested by MTT assay. The mean±SD was used as the value of three independent experiments. *p<0.05; **p<0.01; ***p<0.001.
figs6.
ATG5 is a downstream target gene of WTAP-mediated m6A modification in HCC ferroptosis. Erastin was applied to HuH7 or HepG2 cells that had been transfected with WTAP siRNA and ATG5 plasmid alone or together. A Expression of autophagy-related genes was detected by western blot. B-C The mRNA expression of MAP1LC3B and SQSTM1 was detected by RT-PCR. D Immunofluorescence detection of endogenous LC3 expression in Huh7. E-H The level of iron, MDA, ROS, and GSH in HCC were tested detected by kits. The mean±SD was used as the value of three or four independent experiments. *p<0.05; **p<0.01; ***p<0.001.
figs7.
YTHDC2 KD impaired HCC ferroptosis. A Expression of m6A readers in HepG2 cells detected by western blot. B The YTHDC2 siRNA was transfected into Huh7 cells for 12 h, and the expression level of YTHDC2 was detected by Western blot. C The YTHDC2 plasmid was transfected into Huh7 cells for 12 h, and the expression level of YTHDC2 was detected by Western blot. D Expression of ATG5 and YTHDC2 in Huh7 and HepG2 cells detected by western blot. E-F The level of GSH, iron, MDA, and ROS were tested detected by kits. The mean±SD was used as the value of three independent experiments. *p<0.05; **p<0.01; ***p<0.001.
figs8.
Expression of ATG5 in HCC ferroptosis. Huh7 and HepG2 cells treated with erastin, sorafenib and RSL3. A RT-PCR was used to detected ATG5 mRNA expression in Huh7. B Expression of ATG5 protein detected by western blot. C RT-PCR was used to detected ATG5 mRNA expression in HepG2. D Translation efficiency of ATG5 in HepG2, calculated as: translation efficiency = ATG5 protein level / ATG5 mRNA expression level. E Huh7 cells transfected with control or YTHDC2 plasmid were treated with CHX (100 μg/ml) and erastin (10 μM) for 24 h. The expression of ATG5 protein was detected at 0, 2, 4, 6 and 8 h respectively. F Schematic generation strategy for the Pmir-GLO luciferase reporters containing ATG5 CDS fragment. G Schematic view of the defined domains of YTHDC2 and YTH deletion truncations used. The mean±SD was used as the value of three independent experiments. *p<0.05; **p<0.01; ***p<0.001.
Data availability
The datasets generated or analyzed during this study areavailable from the corresponding author on reasonablerequest.
References
- 1.Luo J., Liu H., Luan S., He C., Li Z. Aberrant regulation of mRNA m⁶A modification in cancer development. Int. J. Mol. Sci. 2018;19(9) doi: 10.3390/ijms19092515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du D., Liu C., Qin M., Zhang X., Xi T., Yuan S., Hao H., Xiong J. Metabolic dysregulation and emerging therapeutical targets for hepatocellular carcinoma. Acta Pharm. Sin. B. 2022;12(2):558–580. doi: 10.1016/j.apsb.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y., Chen H.-N., Wang K., Zhang L., Huang Z., Liu J., Zhang Z., Luo M., Lei Y., Peng Y., Zhou Z.-G., Wei Y., Huang C. Ketoconazole exacerbates mitophagy to induce apoptosis by downregulating cyclooxygenase-2 in hepatocellular carcinoma. J. Hepatol. 2019;70(1):66–77. doi: 10.1016/j.jhep.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 4.Liang W.-F., Gong Y.-X., Li H.-F., Sun F.-L., Li W.-L., Chen D.-Q., Xie D.-P., Ren C.-X., Guo X.-Y., Wang Z.-Y., Kwon T., Sun H.-N. Curcumin activates ROS signaling to promote pyroptosis in hepatocellular carcinoma HepG2 cells. In Vivo. 2021;35(1):249–257. doi: 10.21873/invivo.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visalli M., Bartolotta M., Polito F., Oteri R., Barbera A., Arrigo R., Di Giorgio R.M., Navarra G., Aguennouz M.h. miRNA expression profiling regulates necroptotic cell death in hepatocellular carcinoma. Int. J. Oncol. 2018;53(2):771–780. doi: 10.3892/ijo.2018.4410. [DOI] [PubMed] [Google Scholar]
- 6.Su Y., Zhao D., Jin C., Li Z., Sun S., Xia S., Zhang Y., Zhang Z., Zhang F., Xu X., Shao J., Zhang B., Zheng S. Dihydroartemisinin induces ferroptosis in HCC by promoting the formation of PEBP1/15-LO. Oxid. Med. Cell. Longev. 2021;2021 doi: 10.1155/2021/3456725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang M., Wu X., Hu J., Wang Y., Wang Y., Zhang L., Huang W., Wang X., Li N., Liao L., Chen M., Xiao N., Dai Y., Liang H., Huang W., Yuan L., Pan H., Li L., Chen L., Liu L., Liang L., Guan J. COMMD10 inhibits HIF1α/CP loop to enhance ferroptosis and radiosensitivity by disrupting Cu-Fe balance in hepatocellular carcinoma. J. Hepatol. 2022;76(5):1138–1150. doi: 10.1016/j.jhep.2022.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y., Li L., Lan J., Cui Y., Rao X., Zhao J., Xing T., Ju G., Song G., Lou J., Liang J. CRISPR screens uncover protective effect of PSTK as a regulator of chemotherapy-induced ferroptosis in hepatocellular carcinoma. Mol. Cancer. 2022;21(1):11. doi: 10.1186/s12943-021-01466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bekric D., Ocker M., Mayr C., Stintzing S., Ritter M., Kiesslich T., Neureiter D. Ferroptosis in hepatocellular carcinoma: mechanisms, drug targets and approaches to clinical translation. Cancers. 2022;14(7) doi: 10.3390/cancers14071826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mou Y., Wang J., Wu J., He D., Zhang C., Duan C., Li B. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J. Hematol. Oncol. 2019;12(1):34. doi: 10.1186/s13045-019-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D., Li Y. The interaction between ferroptosis and lipid metabolism in cancer. Signal Transduct. Targeted Ther. 2020;5(1):108. doi: 10.1038/s41392-020-00216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y., Cao Y., Xiao J., Shang J., Tan Q., Ping F., Huang W., Wu F., Zhang H., Zhang X. Inhibitor of apoptosis-stimulating protein of p53 inhibits ferroptosis and alleviates intestinal ischemia/reperfusion-induced acute lung injury. Cell Death Differ. 2020;27(9):2635–2650. doi: 10.1038/s41418-020-0528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahoney-Sánchez L., Bouchaoui H., Ayton S., Devos D., Duce J.A., Devedjian J.-C. Ferroptosis and its potential role in the physiopathology of Parkinson's Disease. Prog. Neurobiol. 2021;196 doi: 10.1016/j.pneurobio.2020.101890. [DOI] [PubMed] [Google Scholar]
- 14.Macías-Rodríguez R.U., Inzaugarat M.E., Ruiz-Margáin A., Nelson L.J., Trautwein C., Cubero F.J. Reclassifying hepatic cell death during liver damage: ferroptosis-A novel form of non-apoptotic cell death? Int. J. Mol. Sci. 2020;21(5) doi: 10.3390/ijms21051651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou B., Liu J., Kang R., Klionsky D.J., Kroemer G., Tang D. Ferroptosis is a type of autophagy-dependent cell death. Semin. Cancer Biol. 2020:66. doi: 10.1016/j.semcancer.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Chen X., Yu C., Kang R., Kroemer G., Tang D. Cellular degradation systems in ferroptosis. Cell Death Differ. 2021;28(4):1135–1148. doi: 10.1038/s41418-020-00728-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y., You Y., Lu Z., Yang J., Li P., Liu L., Xu H., Niu Y., Cao X. -methyladenosine RNA modification-mediated cellular metabolism rewiring inhibits viral replication. Science. 2019;365(6458):1171–1176. doi: 10.1126/science.aax4468. [DOI] [PubMed] [Google Scholar]
- 18.He L., Li H., Wu A., Peng Y., Shu G., Yin G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer. 2019;18(1):176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei W., Sun J., Zhang H., Xiao X., Huang C., Wang L., Zhong H., Jiang Y., Zhang X., Jiang G. Interaction with WTAP promotes assembly and activity of the mA methyltransferase complex and promotes Cisplatin resistance in bladder cancer. Cancer Res. 2021;81(24):6142–6156. doi: 10.1158/0008-5472.CAN-21-1518. [DOI] [PubMed] [Google Scholar]
- 20.Jiang X., Liu B., Nie Z., Duan L., Xiong Q., Jin Z., Yang C., Chen Y. The role of m6A modification in the biological functions and diseases. Signal Transduct. Targeted Ther. 2021;6(1):74. doi: 10.1038/s41392-020-00450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y., Zhang W., Shen F., Yang X., Liu H., Dai S., Sun X., Huang J., Guo Q. YTH domain proteins: a family of mA readers in cancer progression. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.629560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X.-Y., Zhang J., Zhu J.-S. The role of mA RNA methylation in human cancer. Mol. Cancer. 2019;18(1):103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen M., Li Y., Wang Y., Shao J., Zhang F., Yin G., Chen A., Zhang Z., Zheng S. N-methyladenosine modification regulates ferroptosis through autophagy signaling pathway in hepatic stellate cells. Redox Biol. 2021;47 doi: 10.1016/j.redox.2021.102151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L., Li H., Hu D., Wang Y., Shao W., Zhong J., Yang S., Liu J., Zhang J. Insights into N6-methyladenosine and programmed cell death in cancer. Mol. Cancer. 2022;21(1):32. doi: 10.1186/s12943-022-01508-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin X., Zhang J., Wang B., Xu G., Yang X., Zou Z., Yu C. Ferritinophagy is involved in the zinc oxide nanoparticles-induced ferroptosis of vascular endothelial cells. Autophagy. 2021;17(12):4266–4285. doi: 10.1080/15548627.2021.1911016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T., Kong S., Tao M., Ju S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol. Cancer. 2020;19(1):88. doi: 10.1186/s12943-020-01204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., Cheah J.H., Clemons P.A., Shamji A.F., Clish C.B., Brown L.M., Girotti A.W., Cornish V.W., Schreiber S.L., Stockwell B.R. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clevers H. Modeling development and disease with organoids. Cell. 2016;165(7):1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 29.Lei G., Zhuang L., Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer. 2022;22(7):381–396. doi: 10.1038/s41568-022-00459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C., Zhang Y., Liu J., Kang R., Klionsky D.J., Tang D. Mitochondrial DNA stress triggers autophagy-dependent ferroptotic death. Autophagy. 2021;17(4):948–960. doi: 10.1080/15548627.2020.1739447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao H., Shi J., Wen K., Lin J., Liu Q., Shi B., Yan Y., Xiao Z. Molecular targets of ferroptosis in hepatocellular carcinoma. J. Hepatocell. Carcinoma. 2021;8:985–996. doi: 10.2147/JHC.S325593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Recalcati S., Correnti M., Gammella E., Raggi C., Invernizzi P., Cairo G. Iron metabolism in liver cancer stem cells. Front. Oncol. 2019;9:149. doi: 10.3389/fonc.2019.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H., Shi X., Huang T., Zhao X., Chen W., Gu N., Zhang R. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 2020;48(11):6251–6264. doi: 10.1093/nar/gkaa347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen M., Wei L., Law C.-T., Tsang F.H.-C., Shen J., Cheng C.L.-H., Tsang L.-H., Ho D.W.-H., Chiu D.K.-C., Lee J.M.-F., Wong C.C.-L., Ng I.O.-L., Wong C.-M. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67(6):2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 35.Ma J.-Z., Yang F., Zhou C.-C., Liu F., Yuan J.-H., Wang F., Wang T.-T., Xu Q.-G., Zhou W.-P., Sun S.-H. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65(2):529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 36.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.-G., He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.-M., Li C.J., Vågbø C.B., Shi Y., Wang W.-L., Song S.-H., Lu Z., Bosmans R.P.G., Dai Q., Hao Y.-J., Yang X., Zhao W.-M., Tong W.-M., Wang X.-J., Bogdan F., Furu K., Fu Y., Jia G., Zhao X., Liu J., Krokan H.E., Klungland A., Yang Y.-G., He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamoto H., Zhang S., Mizushima N. Autophagy genes in biology and disease. Nat. Rev. Genet. 2023;24(6):382–400. doi: 10.1038/s41576-022-00562-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou W., Xie Y., Song X., Sun X., Lotze M.T., Zeh H.J., Kang R., Tang D. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12(8):1425–1428. doi: 10.1080/15548627.2016.1187366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mao Y., Dong L., Liu X.-M., Guo J., Ma H., Shen B., Qian S.-B. mA in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 2019;10(1):5332. doi: 10.1038/s41467-019-13317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kretschmer J., Rao H., Hackert P., Sloan K.E., Höbartner C., Bohnsack M.T. The mA reader protein YTHDC2 interacts with the small ribosomal subunit and the 5'-3' exoribonuclease XRN1. RNA. 2018;24(10):1339–1350. doi: 10.1261/rna.064238.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z., Guo M., Li Y., Shen M., Kong D., Shao J., Ding H., Tan S., Chen A., Zhang F., Zheng S. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy. 2020;16(8):1482–1505. doi: 10.1080/15548627.2019.1687985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S., Wang Y., Xun X., Zhang C., Xiang X., Cheng Q., Hu S., Li Z., Zhu J. Hedgehog signaling promotes sorafenib resistance in hepatocellular carcinoma patient-derived organoids. J. Exp. Clin. Cancer Res. 2020;39(1):22. doi: 10.1186/s13046-020-1523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.
The datasets generated or analyzed during this study areavailable from the corresponding author on reasonablerequest.