Abstract
An Escherichia coli DNA fragment was identified that contained part of the β-glucoside (bgl) operon. This fragment was identified because it contained a promoter that was responsible for the expression of a reporter gene, the chloramphenicol acetyltransferase gene, in a mouse liver during bacterial infection but not when a bacterial clone was grown in vitro. This fragment contained a promoter and a rho-independent transcription terminator which were flanked by the 3′ end of bglG and the 5′ end of bglF. Reverse transcription-PCR confirmed that cat-specific mRNA was produced in infected mouse liver but not in vitro. mRNA encoding the positive regulator of the bgl operon, bglG, also was detected in mouse liver infected with an E. coli strain. These results demonstrated that expression of the bgl operon occurs in infected mouse liver and suggests a unique role for this operon in vivo.
The β-glucoside (bgl) operon of Escherichia coli encodes enzymes required for catabolism of aromatic β-glucosides. In E. coli K-12 the operon is cryptic, and wild-type cells cannot metabolize β-glucoside sugars, such as arbutin or salicin. The operon consists of three genes that map at 83.8 min on the E. coli chromosome (Fig. 1) (5). The three genes, bglG, bglF, and bglB, encode all the functions necessary for uptake and degradation of β-glucoside as well as regulation of the operon (23), while bglR is a cis-acting regulatory sequence. The bgl operon of E. coli K-12 has been extensively studied because, while the operon is cryptic, cis-active mutations have been identified that activate it. These activating mutations result from the insertion of IS1 or IS5 elements or various other point mutations within bglR (18–20). Mutations in another gene, bglJ, located at 99 min on the E. coli chromosome, also have been shown to activate the bgl operon (9). In addition, enhanced transcription from the bgl promoter P1 due to mutations in the genes encoding DNA gyrase (gyrA and gyrB) and histone-like nucleoprotein (hns) have been reported (7, 11).
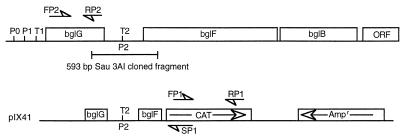
FIG. 1.
The bgl operon showing all three genes required for regulated uptake and utilization of β-glucosides. The identified promoters are designated P0, P1, and P2, and the rho-independent transcription terminators are designated T1 and T2. Two primers, FP2 and RP2, were used to identify bglG mRNA by RT-PCR. In plasmid pIX41 the bgl fragment that contains the 5′ end of bglG, promoter P2, terminator t2, and the 3′ end of bglF is cloned upstream of the promoterless cat gene. Primers designated FP1 and RP1 were used for RT-PCR identification of the cat gene. A reverse primer, SP1, was used for sequencing. ORF, open reading frame.
The functions of the various bgl genes have been determined. bglR is a cis-acting regulatory element located upstream of bglG. bglG encodes a positive regulator of the operon, BglG, which can be phosphorylated. In the dephosphorylated state BglG has antiterminator activity and prevents early transcription termination (1, 12, 21). Two rho-independent transcriptional terminators, t1 and t2, are located upstream of the bglG and bglF genes, respectively (14, 23). BglG can serve as an antiterminator for both terminator sequences. bglF encodes a β-glucoside-specific membrane-bound transport protein which is part of the phosphoenolpyruvate-sugar phosphotransferase system (22). BglF also senses the presence of β-glucosides and, in the absence of β-glucoside, is a negative regulator of the bgl operon (15). In the absence of β-glucosides, BglF phosphorylates BglG, thus inhibiting the antiterminator activity of BglG (1–3). The last gene of the operon, bglB, encodes a phospho-β-glucosidase that hydrolyzes phosphorylated β-glucosides.
The bgl operon is actively kept silent due to negative effects exerted by DNA structural elements near the promoter region (16, 24, 25). Expression of the bgl operon containing an activating mutation requires a β-glucoside inducer and is dependent on cyclic AMP and catabolite-activating protein (CAP) (17, 18). A CAP binding site is present upstream of bglG. In the presence of cyclic AMP and CAP, transcription initiates constitutively at the promoter proximal to bglG, but in the absence of β-glucosides it terminates at the rho-independent terminator t1. The cis-acting mutations in bglR increase transcription initiation, and in the presence of inducer β-glucosides, the negative effect of BglF is relieved, permitting expression of all three genes (1, 2, 21).
In this communication we provide evidence that the wild-type bgl operon is induced in mouse liver and is not linked to cis-acting mutations in the bglR locus. We have employed in vivo expression technology (IVET) to identify genes important in the pathogenesis of E. coli-induced septicemia. This technology allows the detection and identification of genes that are expressed in vivo during infection but not under standard laboratory conditions. The approach was employed to identify novel “silent” genes that may contribute to survival of the bacteria in the animal and to the development of disease. During the screening process, bglF was identified as a gene expressed in vivo. Our results suggest that the wild-type bgl operon is derepressed in the microenvironment of the liver.
E. coli i484, which is the focus of this study, was isolated from a patient with septicemia. Strain i484 has a serotype of O25:H autoagglutinating. A streptomycin-resistant mutant of i484 was selected by plating on Luria-Bertani (LB) agar containing streptomycin (30 μg/ml). To identify in vivo-induced genes, a modified plasmid-based IVET strategy was devised which used the promoter selection vector pKK232-8 (Pharmacia Biotech) (6). Plasmid pKK232-8 carries a promoterless chloramphenicol acetyltransferase (cat) gene cassette that serves as a reporter. Upstream of the cat cassette are transcriptional terminators and translational stop codons in each reading frame, followed by a multiple cloning site. A genomic library of E. coli i484 DNA was prepared by insertion of genomic fragments into the multiple cloning site of pKK232-8. This allowed the selection of active promoters based on the expression of cat. Total DNA was isolated from strain i484 by using cetyltrimethylammonium bromide (4) and was partially digested with the restriction endonuclease Sau3AI. Fragments (0.5 to 2.0 kb) were collected from a sucrose density gradient. The pooled DNA fragments were precipitated with ethanol, collected, and dried. The fragments were ligated into the BamHI site within the multiple cloning site upstream of the cat cassette. Recombinant plasmids were introduced into E. coli i484 by electroporation and plated on LB agar containing ampicillin (50 μg/ml).
A chloramphenicol-treated mouse infection model was developed to select the bacterial clones that expressed the cat gene in vivo. Mice (CD-1) (Harlan Sprague Dawley, Indianapolis, Ind.) were inoculated intraperitoneally with 108 cells from the library and immediately after challenge were treated with chloramphenicol (500 μg). Chloramphenicol was administered intraperitoneally six times at 4-h intervals. Thirty minutes after treatment, the concentration of chloramphenicol in the livers of the mice was 80 μg/ml, as measured by an agar diffusion assay. The concentration dropped to 25 μg/ml after 1 h. Two days postchallenge the mice were euthanized by CO2, and bacterial cells were recovered from infected mouse livers by plating on LB containing streptomycin (30 μg/ml) and ampicillin (100 μg/ml); 1.3 × 106 bacterial cells were recovered per gram of liver. Bacterial clones were replicated on LB agar containing chloramphenicol (20 μg/ml), and those clones sensitive to chloramphenicol in vitro were pooled. Eighty-two percent of the clones were sensitive to chloramphenicol in vitro. Subsequently, 108 in vitro chloramphenicol-sensitive cells were used to challenge a second group of chloramphenicol-treated mice. Once again bacterial cells sensitive to chloramphenicol in vitro were recovered from the mouse livers. After the second challenge the percentage of in vitro chloramphenicol-sensitive clones was 99.9%. These clones were presumed to be resistant to chloramphenicol in vivo. Random clones were selected, and limited sequencing of purified plasmids isolated from these clones was performed. The oligonucleotide primer SP1 (Fig. 1) (5′-GAAAATCTCGTCGAAGCTC-3′), which is specific for the 5′ end of cat, was used to sequence DNA flanking cat. Two clones, i673 and i678, were found to contain similar DNA fragments, and sequencing revealed a sequence located in the intracistronic region between bglG and bglF. This 593-bp Sau3AI fragment, nucleotide positions 1319 to 1911 (23), contained the 3′ end of bglG, promoter P2, the rho-independent transcription termination sequence t2, and the 5′ end of bglF (Fig. 1). Both clones were confirmed to be sensitive to chloramphenicol in vitro by replating on LB agar containing streptomycin and LB agar containing chloramphenicol, reaffirming that the cloned promoter was not active in vitro.
Even though these clones did not express cat in vitro, we wanted to confirm that the bgl operon had not been permanently activated by growth in vivo due to alteration of important cis-active elements. Therefore, the wild-type E. coli strain i484 and the two in vivo-induced clones, i673 and i678, were grown on Bgl indicator plates. Indicator plates were prepared by adding 0.5% filter-sterilized salicin to a liter of sterile minimal medium followed by a 10-ml solution of 2% bromothymol blue, 50% ethanol, and 0.2 N NaOH. Ampicillin (100 μg/ml) was added to the medium if needed. All the strains tested exhibited a bgl-negative phenotype. This indicated that the chromosomal copy of the bgl operon was not activated by cis mutations after growth in vivo and that the regulation of the cat gene was under the control of a promoter in the cloned fragments (presumably P2). In addition, data obtained by employing reverse transcription (RT)-PCR also support this conclusion (see below).
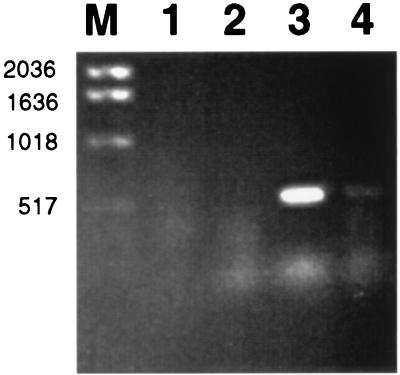
Because we were concerned that the clones selected in vivo might arise for reasons not related to production of Cat that allowed the organism to overcome the inhibitory effects of systemic chloramphenicol, an RT-PCR-based assay was used to determine the presence of cat-specific mRNA in vivo. Total RNA was isolated from the liver of a mouse infected with i673 and from i673 cells grown in LB medium. As a control, total RNA was also isolated from the liver of a mouse challenged with E. coli i484 containing pKK232-8 but with nothing inserted in the multiple cloning site. Two primers were designed for PCR with cat gene sequences: a reverse primer, 5′-TTCTGCCGACATGGAAGCCATCAC-3′ (RP1), and a forward primer, 5′-CCTATAACCAGACCGTTCAGCTGG-3′ (FP1), that could be used to transcribe and amplify a 516-bp fragment of cat mRNA (Fig. 1). Using primer RP1, we performed reverse transcription at 66°C for 20 min with Tth DNA polymerase (Boehringer Mannheim, Indianapolis, Ind.), followed by PCR amplification. The conditions of the first cycle included a denaturation step at 95°C for 2 min, annealing at 60°C for 1 min, and polymerization at 72°C for 1.5 min; this was followed by another 35 cycles of denaturation at 95°C for 1 min, annealing at 60°C for 1 min, and polymerization at 72°C for 1.5 min. At the end of the 36 cycles, the reaction mixtures were left at 72°C for 5.5 min. Ten microliters of each sample was subjected to agarose gel electrophoresis (Fig. 2). A product of 516 bp was detected when RNA was prepared from the liver of a mouse infected with i673 (Fig. 2, lane 3), whereas no RT-PCR products were detected in RNA isolated from mouse liver infected with i484 carrying pKK232-8 (Fig. 2, lane 1) or from in vitro-grown E. coli i673 (Fig. 2, lane 2). To eliminate the possibility that the 516-bp fragment was amplified from contaminating plasmid DNA, a second RT-PCR experiment was performed. Prior to RT-PCR amplification the RNA was subjected to RNase digestion. The RNase was boiled for 20 min prior to use to eliminate any contamination by DNase. After RNase digestion, cat-specific amplification did not occur (Fig. 2, lane 4).
FIG. 2.
Identification of cat mRNA by RT-PCR. RT-PCR was performed with RNA isolated from mouse liver infected with i484 carrying pKK232-8 (lane 1), in vitro-grown i673 (lane 2), mouse liver infected with i673 (lane 3), and RNase-treated RNA from mouse liver infected with i673 (lane 4). Lane M, molecular weight marker.
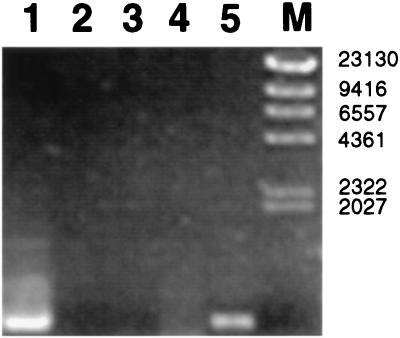
The DNA fragments cloned in pIX41 and pIX46 (strains i673 and i678, respectively) contain the promoter P2, which is required for transcription of bglF. However, for that to occur, the antiterminator activity of the dephosphorylated BglG is required. Moreover, the 5′ end of the bglG gene is absent in pIX41 and pIX46. Therefore, we hypothesized that bglG was supplied in these clones from the wild-type genomic copy and that growth in vivo promoted bglG expression. To determine whether bglG was derepressed in vivo, two PCR primers were designed that could be used to detect bglG mRNA. The reverse primer 5′-GCGGGTAATTTTGTTTTACTGC-3′ (RP2) and the forward primer 5′-CAACAGCGGGAAAAAGTCG-3′ (FP2), were used for reverse transcription and amplification of a 657-bp internal fragment of bglG. As shown in Fig. 3, a DNA fragment of 657 bp was detected when RNA prepared from infected mouse liver was used (Fig. 3, lane 5) while no RT-PCR amplification was observed when RNA prepared from in vitro-grown i673 and RNA from uninfected mouse liver were used (Fig. 3, lanes 2 and 3). As before, to eliminate the possibility that the amplification product was due to contaminating DNA in the RNA preparation from infected mouse liver, RNase-treated total RNA was used for PCR amplification. The subsequent PCR amplification did not yield any DNA fragments (Fig. 3, lane 4). To confirm that the RT-PCR-amplified product contained bglG, it was sequenced; as expected, the sequence of the PCR product contained the bglG sequence. These results indicate bglG-specific mRNA is produced in vivo, and they are consistent with our hypothesis that the bgl operon is derepressed when grown in infected mouse liver.
FIG. 3.
RT-PCR amplification of bglG mRNA. Lane 1, PCR-amplified bglG, using total DNA from strain i484; lane 2, RT-PCR with RNA isolated from uninfected mouse liver; lane 3, RT-PCR with RNA isolated from in vitro-grown strain i673; lane 4, PCR amplification of RNase-treated total RNA from infected mouse liver; lane 5, RT-PCR with total RNA from infected mouse liver; lane M, molecular weight marker.
Our results demonstrate that the induction of the bgl operon can occur in a specific in vivo habitat. This expression is not linked to activation of the cryptic bgl operon and does not result in constitutive expression. Expression occurs only in vivo, and the subsequent regrowth in vitro results in the repression of this operon. This result was confirmed by using RT-PCR, where cat-specific mRNA driven by the weak promoter P2 of bglF was detectable in the livers of mice infected with strain i673 (which contains pIX41) but was not produced when cells were grown in vitro.
Because the DNA fragment in pIX41 contained the 3′ end of bglG, a weak promoter, P2, located within or just downstream of the terminator t2 (21), and the 5′ end of bglF followed by cat, and because the expression of bglF, and therefore cat, requires antiterminator activities associated with BglG, it was assumed that BglG was supplied in trans from the chromosomal copy of the entire operon. Thus, BglG could act as an antiterminator for t2 located in the chromosome and the cloned DNA fragment. If this hypothesis was correct, bglG-specific mRNA should be produced by cells grown in mice but not by those grown in vitro. By using RT-PCR, this was found to be the case.
The purpose of this investigation was to identify E. coli genes that were exclusively expressed during the course of disease. Such an analysis will begin to describe the repertoire of genes expressed in the hostile in vivo environment and through this process define the adaptive ability of E. coli. The discovery that the cryptic bgl operon was exclusively induced in a murine infection model raises interesting questions about the regulation of this phenotypically silent operon and the importance of these genes in infection. A number of mutations that activate the bgl operon have been identified either in bglR or in other genes, like hns, gyrA, and gyrB. More recently the products of two genes, bglJ and leuO, have been identified as positive trans activators of the bgl operon (9, 26). However, an increased expression of these trans activators is required for activation of the bgl operon. We hypothesize that complex host-pathogen interactions in infected mouse liver may derepress certain bacterial genes that could directly relieve the silencing effect of the bgl operon or may in turn increase the expression of the trans activators, bglJ and leuO, leading to induction of the bgl operon. Growth condition-dependent induction of another cryptic operon, cel, has been reported (8). The growth of thermotolerant and mesothermophilic mutants of E. coli, Salmonella typhimurium, and Pseudomonas aeruginosa at 48 and 54°C results in the induction of the cel operon. While the bgl operon generally is not expressed except under specific activating conditions, a previous report demonstrated that it can be induced by growth under anaerobic conditions (13). In that study, expression was increased 22-fold but remained substantially below the levels achieved after activation. The results presented here confirm that other growth conditions (i.e., growth in mice) also induce this operon. We have not been able to quantify the degree of induction and therefore do not know whether expression in vivo achieves the levels obtained by growth under anaerobic conditions or activating conditions. The growth of bacteria in mammalian hosts also presents E. coli with unique stresses. These environmental changes could lead to the induction of a variety of genes during the disease process, including bgl. The importance of the bgl operon in the survival of the pathogen during infection in mouse liver is not clear. We have begun to ask whether bgl is required in vivo. Mice have been challenged with a mutant of i484 containing a deletion in bglF (P2) (unpublished results). No significant differences in mortality were observed compared to that of mice infected by the wild-type strain. However, in these experiments the challenge dose was quite high and more subtle effects on virulence might not have been detected. Since 90% of the natural isolates of E. coli carry the bgl operon (10), this cryptic metabolic system probably confers an advantage under specific growth conditions.
Acknowledgments
We thank Richard Wilson, Pennsylvania State University, for serotyping.
This work was supported by a grant from Johnson and Johnson, Corporate Office of Science and Technology.
REFERENCES
- 1.Amster-Choder O, Houman F, Wright A. Protein phosphorylation regulates transcription of the beta-glucoside utilization operon in E. coli. Cell. 1989;58:847–855. doi: 10.1016/0092-8674(89)90937-9. [DOI] [PubMed] [Google Scholar]
- 2.Amster-Choder O, Wright A. Regulation of activity of a transcriptional anti-terminator in E. coli by phosphorylation in vivo. Science. 1990;249:540–542. doi: 10.1126/science.2200123. [DOI] [PubMed] [Google Scholar]
- 3.Amster-Choder O, Wright A. Modulation of the dimerization of a transcriptional antiterminator protein by phosphorylation. Science. 1992;257:1395–1398. doi: 10.1126/science.1382312. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1997. pp. 2.4.1–2.4.2. [Google Scholar]
- 5.Bachmann B J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990;54:130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brosius J. Plasmid vector for the selection of promoters. Gene. 1984;27:151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- 7.DiNardo S, Voelkel K A, Sternglanz R, Reynolds A E, Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 8.Droffner M L, Yamamoto N. Demonstration of cel operon expression of Escherichia coli, Salmonella typhimurium, and Pseudomonas aeruginosa at elevated temperatures refractory to their growth. Appl Environ Microbiol. 1992;58:1784–1785. doi: 10.1128/aem.58.5.1784-1785.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giel M, Desnoyer M, Lopilato J. A mutation in a new gene, bglJ, activates the bgl operon in Escherichia coli K-12. Genetics. 1996;143:627–635. doi: 10.1093/genetics/143.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall B G, Betts P W. Cryptic genes for cellobiose utilization in natural isolates of Escherichia coli. Genetics. 1987;115:431–439. doi: 10.1093/genetics/115.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgins C F, Dorman C J, Stirling D A, Waddell L, Booth I R, May G, Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 12.Houman F, Diaz-Torres M R, Wright A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell. 1990;62:1153–1163. doi: 10.1016/0092-8674(90)90392-r. [DOI] [PubMed] [Google Scholar]
- 13.Lopilato J, Wright A. Mechanisms of activation of the cryptic bgl operon. In: Drlica K, Riley M, editors. The bacterial chromosome. Washington, D.C: American Society for Microbiology; 1990. pp. 439–444. [Google Scholar]
- 14.Mahadevan S, Wright A. A bacterial gene involved in transcription antitermination: regulation at a rho-independent terminator in the bgl operon of E. coli. Cell. 1987;50:485–494. doi: 10.1016/0092-8674(87)90502-2. [DOI] [PubMed] [Google Scholar]
- 15.Mahadevan S, Reynolds A E, Wright A. Positive and negative regulation of the bgl operon in Escherichia coli. J Bacteriol. 1987;169:2570–2578. doi: 10.1128/jb.169.6.2570-2578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukerji M, Mahadevan S. Characterization of the negative elements involved in silencing the bgl operon of Escherichia coli: possible roles for DNA gyrase, H-NS, and CRP-cAMP in regulation. Mol Microbiol. 1997;24:617–627. doi: 10.1046/j.1365-2958.1997.3621725.x. [DOI] [PubMed] [Google Scholar]
- 17.Prasad I, Schaefler S. Regulation of the beta-glucoside system in Escherichia coli K-12. J Bacteriol. 1974;120:638–650. doi: 10.1128/jb.120.2.638-650.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynolds A E, Mahadevan S, LeGrice S F, Wright A. Enhancement of bacterial gene expression by insertion elements or by mutation in a CAP-cAMP binding site. J Mol Biol. 1986;191:85–95. doi: 10.1016/0022-2836(86)90424-9. [DOI] [PubMed] [Google Scholar]
- 19.Reynolds A E, Felton J, Wright A. Insertion of DNA activates the cryptic bgl operon of E. coli. Nature. 1981;293:625–629. doi: 10.1038/293625a0. [DOI] [PubMed] [Google Scholar]
- 20.Schnetz K, Rak B. IS5: a mobile enhancer of transcription in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:1244–1248. doi: 10.1073/pnas.89.4.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schnetz K, Rak B. Regulation of the bgl operon of Escherichia coli by transcriptional antitermination. EMBO J. 1988;7:3271–3277. doi: 10.1002/j.1460-2075.1988.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnetz K, Sutrina S, Saier M H, Jr, Rak B. Identification of catalytic residues in the beta-glucoside permease of Escherichia coli by site-specific mutagenesis and demonstration of interdomain cross-reactivity between the beta-glucoside and glucose systems. J Biol Chem. 1990;265:13464–13471. [PubMed] [Google Scholar]
- 23.Schnetz K, Toloczyki C, Rak B. Beta-glucoside (bgl) operon of Escherichia coli K-12: nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J Bacteriol. 1987;169:2579–2590. doi: 10.1128/jb.169.6.2579-2590.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnetz K. Silencing of Escherichia coli bgl promoter by flanking sequence elements. EMBO J. 1995;14:2545–2550. doi: 10.1002/j.1460-2075.1995.tb07252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh J, Mukerji M, Mahadevan S. Transcriptional activation of the Escherichia coli bgl operon: negative regulation by DNA structural elements near the promoter. Mol Microbiol. 1995;17:1085–1092. doi: 10.1111/j.1365-2958.1995.mmi_17061085.x. [DOI] [PubMed] [Google Scholar]
- 26.Ueguchi C, Ohta T, Seto C, Suzuki T, Mizuno T. The leuO gene product has a latent ability to relieve bgl silencing in Escherichia coli. J Bacteriol. 1998;180:190–193. doi: 10.1128/jb.180.1.190-193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]