Abstract
Leaf growth initiates in the peripheral region of the meristem at the apex of the stem, eventually forming flat structures. Leaves are pivotal organs in plants, serving as the primary sites for photosynthesis, respiration, and transpiration. Their development is intricately governed by complex regulatory networks. Leaf development encompasses five processes: the leaf primordium initiation, the leaf polarity establishment, leaf size expansion, shaping of leaf, and leaf senescence. The leaf primordia starts from the side of the growth cone at the apex of the stem. Under the precise regulation of a series of genes, the leaf primordia establishes adaxial-abaxial axes, proximal-distal axes and medio-lateral axes polarity, guides the primordia cells to divide and differentiate in a specific direction, and finally develops into leaves of a certain shape and size. Leaf senescence is a kind of programmed cell death that occurs in plants, and as it is the last stage of leaf development. Each of these processes is meticulously coordinated through the intricate interplay among transcriptional regulatory factors, microRNAs, and plant hormones. This review is dedicated to examining the regulatory influences of major regulatory factors and plant hormones on these five developmental aspects of leaves.
Keywords: leaf primordium, leaf polarity, leaf size, leaf shape, leaf senescence
1. Introduction
Leaves serve as the primary photosynthetic organs in plants and are vital for capturing light energy and facilitating gas exchange. There are two types of simple and compound leaves, a leaf with only one leaflet on the petiole is referred to as a simple leaf, and its shape is determined by factors such as the tip, base, and margins of the leaf. Conversely, multiple single leaves (such as apical leaves, lateral leaves, and stipules) are collectively termed compound leaves. A typical leaf comprises three main parts: the blade, petiole, and stipules. Leaves are flat lateral structures that originate from the shoot apical meristem (SAM) (Zhang and Liu, 2022) and exhibit a high degree of morphological diversity (Tsukaya, 2014).
Leaf development is a complex, dynamic process that can be summarized into five processes: the first step is the leaf primordium initiation, the primordia begins in the peripheral zone of the apical meristem; the second step is the establishment of polarity, the initial leaf primordium develops in the direction of adaxial-abaxial growth axes, proximal-distal growth axes, medio-lateral growth axes; in the third step, the blade size is controlled, and the basic leaf shape is formed by the extension of the blade from the edge region of the adaxial plane and the abaxial plane; the fourth step is the regulation of leaf shape and leaf expansion to form the final shape (Yan et al., 2008; Xiong and Jiao, 2019; Wang et al., 2021a); the fifth step is leaf senescence which accompanied by programmed cell death (PCD) that occurs in plants (Tian et al., 2020)
Recently, the development of plant leaves has been explored from many angles such as anatomy, cytology and molecular biology, which makes the research more perfect. This review provides a review of the fundamental structure and developmental processes of leaves, along with an exploration of the molecular mechanisms involved in morphogenesis, leaf initiation, determination of leaf polarity, and the processes of leaf growth and senescence during development.
2. Initiation of leaf primordium
The leaf primordia of plants start from SAM, which is an embryonal cell population with a “tunica-corpus” structure. The structure can be divided into three layers. The tunica cells divide into L1 and L2 cambium peripherally, and the corpus cells divide into L3 cambium peripherally or vertically. The L1 layer is the promeristem layer, which develops into the epidermis. L1 layer cells control the division pattern of their inner layer cells. The L2 layer is divided into three parts: the promeristem (part) develops into the lower epidermis, the protocambium develops into a vascular bundle, and the basic meristem layer (part) develops into cortical and medullary rays. Layer 3 is the basic meristem layer (part), which develops into a pith ( Figure 1 ).
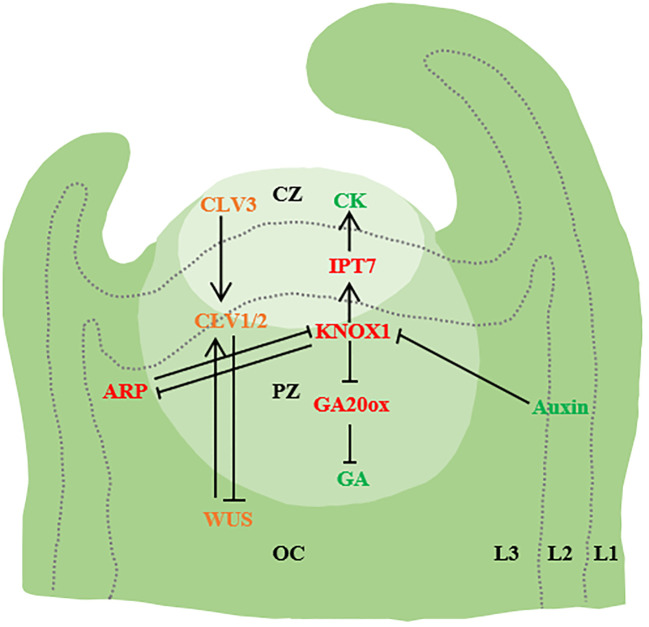
Figure 1.
Initiation of leaf primordium. SAM is divided into three functional regions [central region (CZ), peripheral region (PZ) and costal region (OC)] and Layer1 (L1), Layer2 (L2) and Layer3(L3). WUS activates CLV3, and CLV3 further binds to CLV1/2, thereby inhibiting WUS expression. Auxin accumulation in the flanks of SAM through PIN1/AUX1 mediated polar transport triggers primordium development. In addition, KNOX1 maintains the role of stem cells, positively regulates CK, negatively regulates GA signaling through IPT7 and GA20ox, and ARF regulates the emergence of young primordiae (→ represents positive regulation, and T-shaped arrows represent negative regulation. The same below.) (Barkoulas et al., 2007; Bar and Ori, 2014; Kalve et al., 2014; Wang et al., 2021a).
The shoot apical meristem (SAM) comprised by the central zone (CZ), the peripheral zone (PZ), and the organizing center (OC). At the apex of the SAM, there is a cluster of slowly dividing cells, constituting the central region of SAM. These cells are larger, possess stem cell functions, and play a pivotal role in maintaining the meristem’s integrity. The rate of cell proliferation and growth in this central region often differs significantly from that at the periphery. In the periphery of the central SAM region, cell division rates are notably accelerated. These rapidly dividing cells form the peripheral region of SAM, which serves as the origin for organ primordia such as leaf primordia. Below SAM lies the organizing center, also known as the Rib Meristem (RM), (Huang, 2003; Barton, 2010; Cao and Jiao, 2018; Xiong and Jiao, 2019). Within the SAM, the homeodomain transcription factor WUSCHEL (WUS) is expressed in the organizing center (OC) to uphold stem cells in the central zone (CZ). The migration of WUS to the central zone activates the accumulation level of the CLAVATA3 (CLV3). CLV3 acts as a negative regulator by encoding a secreted peptide. This peptide triggers the transmembrane receptor kinase CLV1 in the organizing center, resulting in the inhibition of WUS expression (Sassi and Vernoux, 2013).
During the formation of plant leaf primordium, the plant hormone auxin is the growth regulator of organ initiation. The highest local auxin concentration observed in the L1 cambium of the SAM. This localized increase in auxin concentration is facilitated by polarly localized PIN-FORMED1 (PIN1) efflux transporters. The rise in auxin levels coincides with the onset of leaf primordium formation, and the cellular response to auxin is mediated by AUXIN RESPONSE TRANSCRIPTION FACTORs (ARFs) (Ben‐Gera et al., 2012; Pinon et al., 2013; Bar and Ori, 2014; Dong and Huang, 2018; Wang et al., 2021). Auxin level has a negative effect on SAM size. Auxin transport of SAM in lateral organs can be inhibited by auxin transport switch, thereby maintaining SAM homeostasis and SAM size (Shi et al., 2018). AUX/PIN1 is involved in the regulation of simple leaf morphogenesis and compound leaf lobular initiation. Genes encoding AUXIN1 (AUX1) and LIKE-AUX1-2 (LAX2) are direct targets of auxin response transcription factor MONOPTEROS (MP) (Bhatia et al., 2016). Auxin also activates the expression of PIN1 through MP, in addition, auxin activates the expression of cytokinin signaling inhibitor ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN6 (AHP6) through MP, and AHP6 moves between cells to produce an inhibitory field that prevents premature growth of the primordium. AHP6 moves between cells to establish an inhibitory field that prevents premature primordium growth (Krogan et al., 2016; Du et al., 2018). In addition to auxin, cytokinin, another plant hormone, also plays a significant role in SAM maintenance (Werner et al., 2001; Kurakawa et al., 2007; Gordon et al., 2009; Shani et al., 2016).
Studies in Arabidopsis thaliana, Zea mays L., and Antirrhinum majus have unveiled two molecular mechanisms governing the initiation of leaf primordia (Barkoulas et al., 2007; Yan et al., 2008; Barton, 2010). The first mechanism involves the polar localization of the auxin transporter PIN1, ensuring the transport of auxin to the initial site of leaf primordium. Additionally, accumulated auxin inhibits the expression of the Class I KNOTTED1-like homeobox (KNOX1) gene (Reinhardt et al., 2003; Heisler et al., 2005; Jönsson et al., 2006; Wang et al., 2021a). The second mechanism is the mutual inhibition of the tip meristem maintenance gene KNOX1 and ARP[ASYMMETRIC LEAVES1(AS1)/ROUGH SHEATH2(RS2)/PHANTASTICA (ARP)] ( Figure 1 ). In single-leaf species, the expression of the KNOX1 in leaf primordium remains suppressed due to the inhibition of ARP, whereas in certain multiple-leaf species, reactivation of the KNOX1 expression occurs and contributes to lobule formation (Byrne et al., 2000; Lodha et al., 2013; Wang et al., 2021). Apart from ARP, the KNOX2 gene also promotes leaf development by counteracting KNOX1 (Furumizu et al., 2015). Analogous to ARPs, heterotopic expression of KNOX2 and its heterodimer partner BEL-LIKE HOMEODOMAIN (BELL) in the Arabidopsis relative Cardamine hirsuta inhibits SAM activity. During the development of compound leaves, KNOX1 expression is reestablished within the leaf primordium, thereby initiating the formation of distinct lobules (Hay and Tsiantis, 2006; Leiboff et al., 2021).
3. Establishment of leaf polarity
During leaf development, polarity establishment is an important process affecting leaf morphology. When leaf primordium emerges from the edge of the meristem, the three axes that determine the polar growth of leaf morphogenesis, namely, adaxial-abaxial axes (front-back of the leaf), proximal-distal axes (base-tip of the leaf) and medio-lateral axes (main vein-edge of the leaf), have been determined (Conklin et al., 2019; Muszynski et al., 2020). The development and morphogenesis of leaves along these three axes are regulated by plant genetic mechanisms and various environmental factors (Wang et al., 2021). The regulatory mechanism is shown in Figure 2 .
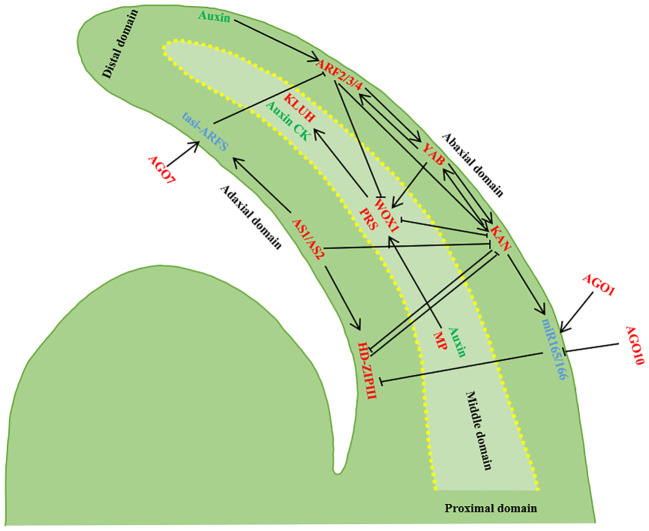
Figure 2.
Establishment of leaf polarity. The developing leaf primordium has three domains, and the transcription factors in the three domains inhibit each other’s expression and control each other. Transcripts of AS1/AS2, HD-ZIPIII and tasi-ARF accumulate in the adaxial domain of the leaf primordium, transcripts of ARF 3/ARF4, KAN and miR165/166 accumulate in the abaxia domain, and WOX1/PRS are expressed in the intermediate domain of the leaf primordium. AGO10 inhibits miR165/166, AGO1 regulates miR165/166 and miR165/166 inhibits HD-ZIPIII. AGO7 stabilizes ta-siR-ARF, and ta-siR-ARF degrades of ARF2/3/4. tasi-ARF and miR165/166 Rnas can move between cells and inhibit ARF2/3/4 and HD-ZIPIII after transcription, and ARF2/3/4 is controlled by auxin. KAN and HD-ZIPIII antagonized each other, and ARF2/3/4 and KAN were inhibited by AS1/AS2. KAN inhibited the expression of WOX1 and PRS, while WOX1 and PRS inhibited the expression of KAN. Adaxial expression of MP and off-axes enrichment of auxin together localized WOX1 and PRS expression in the intermediate domain. In addition, MP may be the direct target of positively expressed HD-ZIPIII, and YAB promotes the expression of WOX1/PRS with KAN and ARF2/3/4, while YAB promotes the expression of WOX1/PRS (Barkoulas et al., 2007; Kalve et al., 2014; Du et al., 2018; Wang et al., 2021a).
3.1. Adaxial-abaxial polarity
The near-distal axes, also referred to as the adaxial-abaxial axes, plays a pivotal role in establishing blade polarity and determining blade thickness. Palisade tissue develops on the adaxial plane of leaves, while spongy tissue forms on the abaxial plane. The maintenance of adaxial-abaxial polarity in the leaf primordium is determined by a complex gene regulatory network, while the characteristics of the adaxial plane and abaxial plane contribute to gene antagonism, thereby enhancing stability in maintaining two distinct cell fates. Adaxial-abaxial polarity is influenced by transcription factors, small RNAs, and auxin. The determination of adaxial-abaxial cell fate occurs before the basal rise of leaf primordium and is mediated by AS2 and KANADI1 (KAN1). The positional information established by the AS2-KAN1 prepattern transforms the non-polar distribution of auxin within the leaf primordium into a polar distribution, relying on ARF-dependent auxin signal transduction to establish adaxial-abaxial polarity (Burian et al., 2022). The adaxial-abaxial axes play a role in defining medio-lateral axes polarity by controlling the differential distribution of auxin and its downstream signaling molecules in the leaves, facilitating flat leaf growth (Qi et al., 2014). There is a partial overlap between auxin and the downstream response factor MP, resulting in a high auxin signal in the intermediate region between the adaxial-abaxial axes plane. Adaxial expression of MP directly activates the accumulation level of WUSCHEL-LIKE HOMEOBOX1 (WOX1) and PRESSED FLOWER (PRS), which are key determinants in the formation of the intermediate zone necessary for lateral blade expansion (Barkoulas et al., 2007; Kalve et al., 2014; Wang et al., 2021). Downstream auxin pathway response factors ARF2, 3, and 4 specifically expressed on the abaxial plane of leaves inhibit WOX1 and PRS expression in that domain. Regulatory genes expressed in the abaxial domain of the leaf exert a suppressive effect on the genes expressed in the adaxial domain. The combined action of MP and ARF2/3/4 enables specific WOX1 and PRS expression in the middle leaf domain, facilitating leaf unfolding (Shani et al., 2006; Barkoulas et al., 2007; Kalve et al., 2014; Guan et al., 2017; Wang et al., 2021a).
Adaxial polarity genes encompass AS1, AS2, and the Class III Homeodomain-leucine Zipper (HD-ZIPIII) family genes. The AS2 genes encode plant-specific LATERAL ORGAN BOUNDARIES (LOB) domain proteins. Research in Arabidopsis thaliana has revealed that HD-ZIP III is a pivotal gene for the development of the leaf adaxial plane. Genes within the HD-ZIPIII family, including PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV), work in concert to regulate the expression and differentiation of SAM cell division toward the leaf’s adaxial plane (Zhong and Ye, 1999; McConnell et al., 2001; Reinhart et al., 2002; Emery et al., 2003; Juarez et al., 2004). Mutations in PHB, PHV, and REV can lead to leaf abnormalities. The AS1-AS2 complex positively regulates HD-ZIPIII, inhibits the abundance of KNOXI and YABBY, and facilitates the establishment of adaxial plane polarity in leaves (Jönsson et al., 2006). The fate of leaf abaxial plane cells is governed by the KAN and YAB gene families, various ARFs, and small RNA molecules such as microRNAs and trans-acting short-interfering RNAs (ta-siRNA). The KANs encode proteins harbored with GARP domain, expressed in the distal domain, and they mutually inhibit the HD-ZIPIII expression, influencing the properties of the adaxial plane while regulating YAB gene expression (Kerstetter et al., 2001). The YABs playing a vital role in polarity maintenance and leaf development. The ARF family genes encode transcription factors responsive to plant auxin. All PHB, PHV, and REV genes have complementary sequences with miR165 and miR166 (Rhoades et al., 2002; Tang et al., 2003). Ectopic expression of microRNAs (miR165/166) inhibits the adaxial plane gene HD-ZIP III, resulting in leaf is apaxial (Otsuga et al., 2001; Emery et al., 2003; Juarez et al., 2004).
3.2. Proximal-distal polarity
The length of the leaves is determined by the proximal-distal axes, and BLADE-ON-PETIOLE1 (BOP1), BOP2, ROTUNDIFOLIA3 (ROT3), ROT4, LONGIFOLIA1 (LNG1), and LNG2 participate in proximal-distal axes formation (Kerstetter et al., 2001; Ha et al., 2003; Ha et al., 2004; Hepworth et al., 2005). The single mutant bop1 and double mutant bop1 and bop2 both exhibited leaf growth on the petiole with a reduced petiole region (Ha et al., 2003; Hepworth et al., 2005). The expression of BOP1/2s is localized to the basal and adaxial regions of the leaf primordium, exerting influence on leaf cell fate by inducing AS2 expression and inhibiting KNOXI (Ha et al., 2003; Jun et al., 2010). Moreover, the polarity of the proximal-distal axes is also under the regulation of auxin. Investigations conducted on Arabidopsis leaves have unveiled the roles of ARF6 and ARF8, two pivotal transcription factors within the auxin signal transduction pathway, in promoting proximal-distal growth of leaf reproductive organs, such as stem leaves and sepals. ARF6 and ARF8 instigate the synthesis and signal transduction of BRs by activating the expression of DWARF4 (DWF4), a pivotal enzyme gene in BR synthesis. BRs, in turn, facilitate the demethylation of cell wall pectin, resulting in isotropic in-plane cell wall loosening. By modulating BR biosynthesis, auxin influences cell wall mechanics and guides cell-oriented growth, ultimately giving rise to leaves with diverse shapes and overseeing the proximal-distal growth of leaf reproductive organs (Xiong et al., 2021).
3.3. Medio-lateral polarity
The medio-lateral axes determine the width of the leaf, and the WUSCHEL‐LIKE HOMEOBOX(WOX) family is involved in the establishment of the medio-lateral axes of the leaf, which promotes the growth of the medio-lateral axes while inhibiting the growth of the adaxial-abaxial axes. The WOX family can be divided into three branches: ancient branch, intermediate branch, and modern branch. WOX transcription factors belong to the homeodomain superfamily and have typical DNA-binding domains (Bürglin and Affolter, 2016). The conserved function of WOX1 is regulating the development of medio-lateral axes of leaves. Phylogenetic analysis of the entire WOX family in Solanum lycopersicum found that knocking out the SlLAM1, a modern branch of the WOX family, through CRISPR/Cas9-mediated genome editing resulted in a narrowing of the leaves and a reduction in the number of lobules (Wang et al., 2021c). Two different WOX transcription factors STF and WOX9 in Medicago truncatula and Nicotiana tabacum jointly regulate the expression of cytokinin oxidase gene CKX3 in plants through antagonistic effects. This regulation influences leaf development by affecting cytokinin content and cell division. (Wang et al., 2022a). M. truncatula STENOFOLIA(STF) and N. tabacum LAMINA1(LAM1) are homologous genes of AtWOX1, and stf/lam1 mutants show leaf narrowing phenotype. In addition, overexpression of STF in rice (Oryza sativa), Brachypodium distachyon, and the energy grass Panicum virgatum, respectively, showed the characteristics of wider leaves and thicker stems (Tadege et al., 2011; Wang et al., 2017). The leaf narrowing phenotype of stf/lam1 mutants can be restored to varying degrees by modern branch members of the WOX family (WUS, WOX1-WOX6), but the intermediate branch member WOX9 aggravates the leaf phenotype of stf/lam1 mutants, and studies have found that STF directly inhibits the expression of WOX9 (Lin et al., 2013; Wolabu et al., 2021). The recessive wide leaf mutant wl1(wide leaf 1), a novel DROUGHT AND SALT TOLERANCE(DST) allele, has also been found in rice. WIDE LEAF1(WL1) interacts with Tillering and Dwarf1(TAD1), a coactivator of anaphase-promoting complex/cyclosome (APC/C) multi-subunit E3 ligase and is degraded by the APC/CTAD1 complex via the ubiquitin/26S proteasome degradation pathway. In addition, WL1 further recruits Histone Deacetylase HDAC to inhibit the expression of narrow leaf gene NARROW LEAF1(NAL1) by binding to rice TPR-like transcriptional corepressors, thereby regulating leaf width (You et al., 2022).
In addition, leaf polarity establishment is under the regulation of ARGONAUTE (AGO) proteins and long non-coding RNAs (lncRNAs). AGO proteins belong to the RNA-binding protein class and play a pivotal role in small RNA-mediated gene silencing (Baumberger and Baulcombe, 2005). In Arabidopsis thaliana, AGO1 and AGO10 (PINHEAD/ZWILLE) are part of the same clade. AGO1 primarily governs the miRNA pathway and contributes to the post-transcriptional gene silencing of transgenes (Baumberger and Baulcombe, 2005), AGO10 maintains undifferentiated stem cells in stem meristem tissue (Liu et al., 2009b). AGO1, in conjunction with miR165/166 and tasiR-ARF, facilitates target cleavage (Mi et al., 2008), and the mutation of ago1 resulted in the lateral organs with adaxial and abaxial defects (Lynn et al., 1999). AGO10 inhibits miR165/166 expression in SAM and modulates the establishment of adaxial-abaxial polarity in leaves. The mutation of ago10/pnh/zll exhibit abnormal increases accumulation level of miR165/166 in leaves and SAM, leading to a repression of HD-ZIP III transcripts (Liu et al., 2009; Zhu et al., 2011). lncRNAs also play a significant role in leaf development. Studies in Liriodendron chinense have identified various lncRNA-transcription factor (TF) regulatory modules, including lch-lnc6026-BLH2, lch-lnc0809-ATHB4, lch-lnc4261/5500-GRF1, lch-lnc5465-bHLH30, and lch-lnc2601/3202/6972-TCPS, lch-lnc1857/4867/6438-AUX/IAAs. A variety of lncRNA-TF regulatory modules are involved in the establishment of leaf polarity and the regulation of leaf morphology (Tu et al., 2022). In rice, endogenous lncRNA TWISTED LEAF (TL) is transcribed by another strand of the R2R3-MYB coding locus OsMYB60. Down-regulating TL through RNA interference (RNAi) or overexpressing OsMYB60 can lead to leaf distortion in transgenic rice, underscoring the crucial role of lncRNAs in maintaining the flatness of rice leaves (Liu et al., 2018).
4. Regulation of leaf size
As the leaf primordium starts and its polarity is established, the leaf primordium begins to expand, forming its final size, under the regulation of plant hormones (auxin, gibberellin, cytokinin and brassinolide), microRNA [miR319-TEOSINTE BRANCHED, CYCLOIDEA and PCF1/2(TCP) and miR396-GROWTH‐REGULATING FACTOR(GRF) regulatory units] and APETALA2/ETHYLENE RESPONSIVE FACTOR(AP2/ERF) family gene. The regulatory pattern is shown in Figure 3 .
Figure 3.
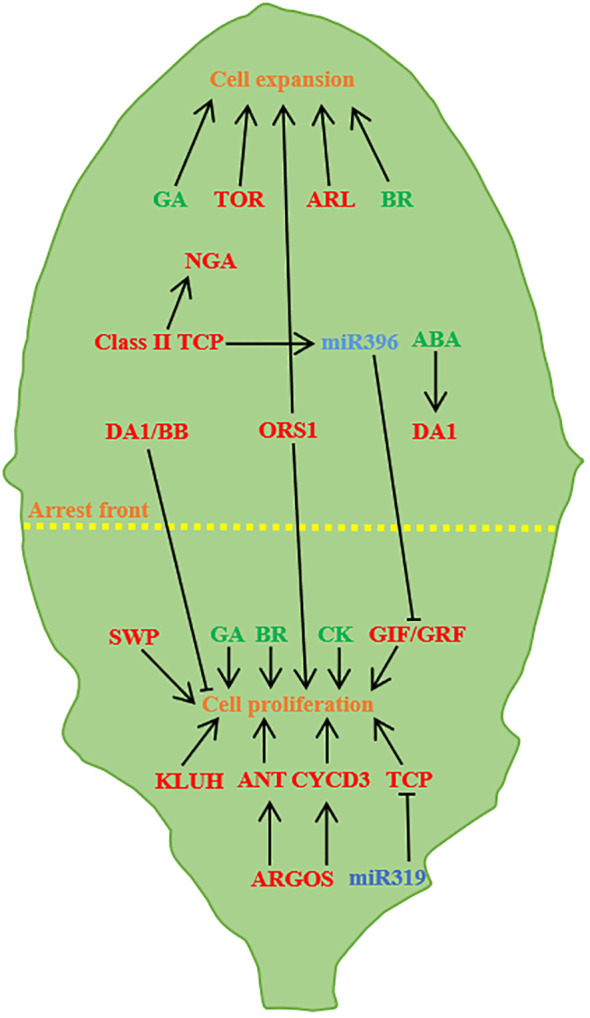
Regulation of leaf size (A case study of Arabidopsis leaves). Leaf size is controlled by cell proliferation and cell expansion. ARGOS promotes cell proliferation through ANT and CYCD3, and miR319 negatively regulates TCP and GRF/GIF transcription factors to promote proliferation. TOR and ARL promote cell expansion, TCP activates the NGA to promote cell expansion, BB and DA1 control the timing of proliferation, and abscisic acid partially facilitates the transition by regulating DA1. KLUH, SWP and CK promote cell proliferation, ORS1, GA and BR have positive effects on cell proliferation and cell expansion (Barkoulas et al., 2007; Kalve et al., 2014; Du et al., 2018; Wang et al., 2021a).
4.1. Regulation of leaf size by plant hormones
Auxin promotes cell proliferation and expansion (Leyser et al., 1996) and acts as a signal during cell proliferation to determine the shape and size of the final organ (Lincoln et al., 1990). The reverse regulatory balance of the two auxin-regulated genes AUXIN -GENE INVOLVED IN ORGANAIZE (ARGOS) and ARF2 is necessary for auxin to function properly in cell division, cell expansion and differentiation. Auxin can induce the expression of ARGOS genes, causing plants to exhibit larger leaves than normal via regulating or maintaining the transcription of AINTEGUMENTA(ANT) (Mizukami and Fischer, 2000; Hu et al., 2003; Spartz et al., 2012). ARF2 inhibits cell proliferation through the ANT pathway, and loss of ARF2 function promotes cell proliferation, resulted in leaf size increasing. Cytokinin (CK), gibberellin (GA) and brassinolide (BR) can also regulate leaf size. Both GA and BR promote leaf growth by promoting cell proliferation and expansion. Lack of GA and BR or insensitive mutants will lead to smaller leaves, and overexpression of GA and BR will make leaves larger (Li et al., 2001; Richards et al., 2001; Mitchum et al., 2006; Cheon et al., 2010; Oh et al., 2011; Zhiponova et al., 2013). CK manipulating the expression of IPT or HvCKX2 genes, thereby regulating cell proliferation mode and leaf size (Černý et al., 2013; Skalák et al., 2019). Studies on rice have found that CK is involved in the formation of rice plant type, CK accumulation mediated by CK OXIDASE/DEHYDROGENASE3 (OsCKX3) in rice controls the development of leaf pillow and negatively regulates the angle of leaves, and the loss of OsCKX3 can induce the asymmetric growth and development of leaf pillow (Huang et al., 2023).
4.2. Effects of genes on leaf size during leaf development
During leaf growth and development, TCPs play critical roles. Based on sequence differences in conserved domains, it can be divided into two subgroup including classI (promoting cell proliferation and plant growth) and class II (inhibiting cell proliferation) (Liu and Gao, 2016; Yang et al., 2019). Specifically, miR396 targets TCP genes, including TCP2, TCP3, TCP4, TCP10, and TCP24, which regulate cell proliferation (Palatnik et al., 2003). The tcp mutants increase leaf size and curvature by up-regulating cyclin-encoding genes (Schommer et al., 2008; Bresso et al., 2018). miR319 primarily regulates TCP transcriptional regulators and high miR319 expression causes severe leaf development defects, resulting in large and wrinkled leaves (Palatnik et al., 2003). Moreover, ERF4 interacts with TCP15, a key activator of the mitotic cell cycle, to promote intracellular replication by inhibiting CYCA2;3 expression. This interaction positively regulates cell enlargement and leaf expansion. The high abundance of TCP15 in young leaves promotes cell mitosis and proliferation, while ERF4 was highly expressed in mature leaves, and it essential for endokaryotic replication and cell expansion (Ding et al., 2022).
TCP family DE1CINCINNATA-LIKE TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTORS (CIN-TCP) redundancy of transcription factors inhibits growth, CIN-TCP is co-regulated by miR319 after transcription, and mutations in CIN-TCP or ectopic expression of miR319 result in larger leaves (Palatnik et al., 2003). TCP4 is the target of miR319 (Palatnik et al., 2003), in A. thaliana, point mutations in the miR319 target sites of TCP4 reduce interactions with miR319, resulting in higher levels of miR396, lower accumulation level of GRFs, and smaller leaf formation (Rodriguez et al., 2010). Overexpression of miR319 in Arabidopsis jaw-D mutants leads to decreased expression of TCP2, 3, 4, 10, and TCP24 and the formation of larger leaves (Palatnik et al., 2003).
GRF family regulates cell proliferation in a redundant manner, promotes leaf growth and development, and controls leaf size (Wang et al., 2020; Wu et al., 2021). Plants carrying GRF-mutated genes have small and narrow leaves, and GRF overexpressors tend to form excessively large leaves (Kim et al., 2003; Horiguchi et al., 2005; Kim and Lee, 2006). Arabidopsis GRF produces different leaf phenotypes, and overexpression of AtGRF1, AtGRF2, AtGRF3, or AtGRF5 results in larger leaves than normal leaves (Kim et al., 2003; Horiguchi et al., 2005; Debernardi et al., 2014). Mutants atgrf 1/2/3, atgrf 3, atgrf 4, or atgrf 5 lead to smaller and narrower leaves than normal leaves (Kim et al., 2003; Horiguchi et al., 2005; Kim and Lee, 2006; Lee et al., 2009; Debernardi et al., 2014). GRF9 restricts cell proliferation during leaf growth by controlling ORG3 expression (Omidbakhshfard et al., 2018). GRF transcription factors and GRF-INTERACTING FACTOR (GRF-GIF) transcriptional coactivators worked together to positively regulate leaf development. Arabidopsis GIF family consists of GIF1 (ANGUSTIFOLIA3, AN3), GIF2 and GIF3, which are positive regulators of cell proliferation. GIF family genes form functional complexes with GRF transcription factors and participate in cell proliferation activities during leaf development (Kim and Kende., 2004; Lee et al., 2009; Debernardi et al., 2014; Lee and Kim., 2014). The expression levels of CsGRF and CSGIF1 were high in young leaves of tea (Camellia sinensis), and the expression levels gradually decreasing with the increase of leaf permanent maturity, indicating that CsGRF and CSGIF1 genes may essential for early leaf tissue formation (Wu et al., 2017). In maize (Zea mays L.), ZmGRF10 regulates leaf size by restricting cell proliferation, and overexpression of ZmGRF10 can lead to impaired cell proliferation and reduced leaf length (Wu et al., 2014). In addition to the collaboration between GRFs and GIFs, the GRF is regulated by miR396 in the leaf development regulatory network (Debernardi et al., 2014). TCP can regulate miR396, while miR396 targets GRF (Huang et al., 2019), and overexpression of miR396 inhibits GRF, resulting in reduced cell number, smaller leaves, repression of miR396 or overexpression of GRF induces plants to grow larger leaves (Liu et al., 2009a; Rodriguez et al., 2010; Wang et al., 2011). GRFs are the upstream repressor of KNOX, and Hordeum vulgare BGRF1 can be used as the suppressor of the intron sequence of Hooded/Barley Knotted3 (Bkn3) of KNOX (Kuijt et al., 2014). However, AtGRF4, 5, and 6 can inhibit KNOTTED-LIKE FROM ARABIDOPSIS THALIANA2 (KNAT2) promoter activity (Kuijt et al., 2014).
Transcription levels of GRFs are affected by gibberellinic acid (GAs), GA3 in rice increased and decreased the expression levels of six (OsGRF1, OsGRF2, OsGRF3, OsGRF7, OsGRF8, OsGRF10, and OsGRF12) and one (OsGRF9) OsGRF gene, respectively (Choi et al., 2004). (Choi et al., 2004). In Chinese cabbage (Brassica rapa ssp. pekinensis), the transcript levels of most BrGRF are induced by exogenous GA3 treatment (Wang et al., 2014). In oilseed rape (Brassica napus), the expression level of BnGRFs under GA treatment may be negatively regulated (Ma et al., 2017). In peach (Prunus persica), the expression of six PpGRF genes (PpGRF1, PpGRF4, PpGRF5, PpGRF6, PpGRF7, and PpGRF10) was up-regulated and three PpGRF genes (PpGRF2, PpGRF6, and PpGRF7) were down-regulated after GA3 treatment (Liu et al., 2022). All CsGRF genes of sweet orange (Citrus sinensis) are involved in leaf development. After treatment with GA3, the transcription levels of CsGRF5 and CsGRF6 did not change, while the transcription levels of the remaining seven CsGRF genes showed significant improvement (Liu et al., 2016).
AP2/ERF family genes are unique transcription factors found in plants, playing pivotal roles in processes such as plant growth, hormone-induced development, ethylene response, and stress response. In L. chinense, three AP2 genes - LcERF94, LcERF96, and LcERF98 - are predominantly associated with early leaf development and morphogenesis, exhibiting high expression levels in both SAM and leaf primordia (Zong et al., 2021). Notably, overexpression of the AP2/ERF transcription factor BOLITA (BOL) in Arabidopsis and Tobacco results in reductions in both cell size and number, consequently yielding smaller leaves (Marsch-Martinez et al., 2006). Additionally, SsAP2/ERFs, exhibits widespread expression in mature sugarcane leaves, indicating its significant role in the growth and development of sugarcane (Li et al., 2020). Leaf size is intricately regulated by a multitude of genes, with microRNAs binding to AGO proteins to guide mRNA cleavage or suppress the translation of complementary RNAs. This targeted regulation of non-protein-coding transcripts by miRNAs can stimulate the generation of tasiRNA populations (Mallory et al., 2008). ARF3/ETTIN (ETT) and ARF4 are among the targets of TAS3 ta-siRNA, governing normal leaf development through the action of AGO7/ZIPPY, and, reciprocally, TAS3 ta-siRNA inhibits ARF3/4 expression (Adenot et al., 2006). ANT, a regulator in the AP2 family, emerges as a pivotal regulator of final leaf size. Overexpression of ANT leads to enlarged leaves, whereas ANT mutants yield smaller leaves (Mizukami and Fischer, 2000). In A. thaliana, the regulation of ANT on organ size is modulated by ARGOS; the loss of ARGOS function leads to smaller leaves. In plants overexpressing ARGOS, the loss of ANT function inhibits the macro-leaf phenotype (Hu et al., 2003). The DA1 regulatory factor encodes ubiquitin receptors that determine the organ size by limiting the cycle of cell proliferation. Point mutations in ubiquitin receptors result in the formation of larger leaves. Mutations in the phenotypic enhancer of da1-1 (EOD1)/BIG BROTHER (BB) in da1 mutants also caused in larger leaves (Li et al., 2008). Cytochrome P450 KLUH (KLU)/CYP78A5 acts as a stimulator of plant organ growth. Loss function of KLU lead to smaller organs due to premature cessation of cell proliferation. Conversely, KLU overexpression results in larger organs with an increased number of cells (Anastasiou et al., 2007).
In Arabidopsis, the growth is positively correlated with the expression level of TARGET OF RAPA-MYCIN (TOR) kinase. Decreased or increased gene expression in TOR leads to decreased or increased organ and cell size, respectively (Deprost et al., 2007). Overexpression of ARGOS-LIKE (ARL) leads to the enlargement of cotyledons, leaves, and other lateral organs in Arabidopsis (Hu et al., 2006).
5. Regulation of leaf shape
The shape of leaves varies significantly both within and between plant species, ranging from slender to oval. Leaf shape diversity is primarily driven by variations along the basic-apical axes, with further adjustments leading to a rich spectrum of leaf forms, which may include serrations, notches, and lobes predominantly situated along the leaf margins. Leaf shape is predominantly governed by the growth behaviour of the marginal blastozone, which possesses meristematic capabilities ( Figure 4 ).
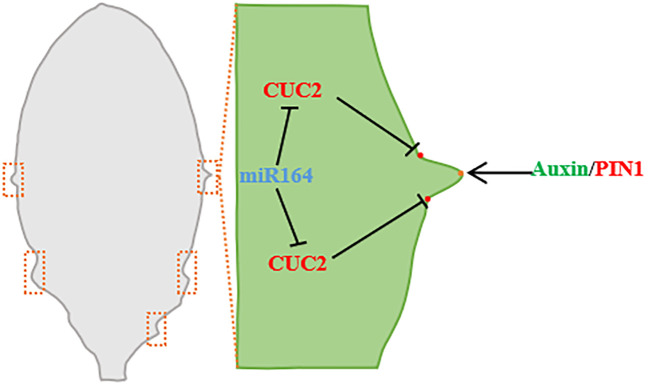
Figure 4.
Leaf edge development (taking Arabidopsis leaf as an example). During leaf margin development, miRNA164 and CUC2 are expressed in the overlapping regions of serrated sagging, CUC2 promotes the establishment of PIN1 convergence points, and PIN1 convergence points produce the maximum value of auxin along the serrated tip of leaf margin. Auxin maximously inhibits CUC2 at the tip of the tooth and promotes the growth of the tooth (Barkoulas et al., 2007; Wang et al., 2021a).
5.1. Regulation of plant hormones on leaf shape during leaf development
Many components of the auxin signaling pathway influence the formation of a compound leaf or leaf margin serration. Auxin regulates cell division by inducing the expression of AS1 and AS2, thereby regulating leaf shape, and mutants of as1 and as2 genes exhibit a distinct bilateral asymmetrical growth pattern, resulting in the formation of lobes and lobular structures from the petioles (Semiarti et al., 2001). As a member of the Aux/IAA family, the ENTIRE expression in the intermediate domain between the S. lycopersicum lobules inhibited the auxin response to maintain the leafless character on the axes. The ENTIRE interaction with ARF activators SIARF19A, SIARF19B, and SIMP, and ARF activation dosed to promote lobular growth to varying degrees (Xiong and Jiao, 2019). The NAC gene GOBLET(GOB) can interact with auxin, and its activity alters auxin signal distribution and, together with the ENTIRE auxin response, promotes compound leaf formation (Xiong and Jiao, 2019). The effect of GA on plant leaf shape development varies according to different species. Increasing the content of GA in single-leaf species can form leaf shapes with smooth leaf margins and no engraving, and GA can also promote the simplification of compound leaves (Achard et al., 2009; Bar and Ori, 2014). In leaf development, GA and CK play an antagonistic role. The activity of GA is related to KNOX1 and TCP. TCP can increase the GA level, while KNOX1 protein can reduce the GA level and increase the CK level. This interplay between the two hormones helps regulate the balance between them, ultimately controlling the development of leaf margin morphology. (Li et al., 2007).
5.2. Effects of genes on leaf shape during leaf development
Leaf shape traits are the outcome of complex gene networks, with key regulators like KNOX and REDUCED COMPLEXITY (RCO) homeobox genes playing pivotal roles in shaping leaf complexity and contributing to the evolutionary diversification of leaf morphology (Hay and Tsiantis, 2010; Vlad et al., 2014). The diversity in leaf geometry arises from two distinct processes. Firstly, the SHOOTMERISTEMLESS (STM) of the KNOX family enhances overall organ growth relative to leaf margin patterning, resulting in the formation of lobes. Secondly, RCO suppresses local growth in the vicinity of developing lobes, accentuating the differences in leaf shape stemming from the establishment of marginal patterns. Both processes are significant in Cardamine occulta but not in Arabidopsis leaves (Kierzkowski et al., 2019). KNOX1 are pivotal in maintaining undifferentiated cell fates and contributing to the development of complex leaf primordia in the shoot apical meristem (SAM). Their activity is suppressed by AS1-AS2 complexes, which are encoded by transcription factors that inhibit the expression of the KNOX1 in Arabidopsis leaves. Elevated levels of KNOX1 and RCO in as1 and as2 mutants collaboratively determine the formation of compound leaves. KNOX1 is expressed at the leaf midvein base, while RCO is symmetrically expressed at the base of lobular primordia. Both KNOX1 and RCO curtail local cell growth by extending the growth potential of leaf primordium cells, promoting anisotropic cell expansion, and giving rise to compound leaves (Wang et al., 2022c). KNOX1 expression is indispensable for the development of lobules in plants with compound leaves. The coordination between LATE MERISTEM IDENTITY1 (LMI1) and KNOX1 governs leaf development, resulting in a diverse range of leaf shapes, such as broad, shallow, and compound leaves, depending on the combinations of these genes (Chang et al., 2019). Leaf margin notches in plants are closely associated with KNOX1 expression, observed only in species featuring leaf margin notches (Rast-Somssich et al., 2015). Within the KNOX1 family, STM, BREVIPEDICELLUS (BP), and KNOTTED-1 (KN1) exert significant influence on the development of leaf margin shapes. Additionally, ARP genes and miRNAs are involved in regulating leaf shape development, with ARP down-regulation altering the number and shape of lobular lobes, while miR396 overexpression modulates leaf shape development by targeting the GRF genes (Jia et al., 2015).
The MYB gene family regulates leaf shape, and the MYB transcription factor has a highly conserved MYBDNA-BD (MYB domain). In plants, MYB proteins have multiple subfamilies, such as MYB1R, R2R3-typeMYB, MYB3R, and 4R-MYB factors, which are key factors in the regulatory network controlling growth, development, metabolism, and stress response (Huang et al., 2013). Nicotiana benthamiana PHANTASTICA (NbPHAN) is a novel R2R3-type MYB gene in Tobacco, NbPHAN silencing inhibits the expression of NTH20 gene, The leaves showed severe downward curling and abnormal leaf growth along the main vein (Huang et al., 2013). MYB transcription factor CLAUSA(CLAU) in S. lycopersicum promotes leaf morphogenesis by reducing cytokinin signal, CLAU facilitates differentiation by suppressing cytokinin (CK) signaling, while CK promotes morphogenesis by inhibiting CLAU expression and suppressing morphogenetic potential to regulate leaf development. Partly by weakening CK signaling (Bar et al., 2016). Trifoliate (Tf) gene in S. lycopersicum regulates leaf morphology, and Tf encodes the transcription factor R2R3MYB and the transcription factor LATERAL ORGAN FUSION1(LOF1) and LOF2 associated with Arabidopsis, Tf is expressed in leaf margins and leaf axils, Tf maintains leaf morphogenesis by inhibiting cell differentiation, Tf mutation leads to a single narrow leaf during the early stage of leaf development in tomato, and a pair of lateral lobules in the terminal and long petioles in the later stage (Naz et al., 2013). The OsMYB103L in rice encodes the R2R3-MYB transcription factor, and overexpression of OsMYB103L leads to leaf curl (Yang et al., 2014).
The interaction of auxin and NO APICAL MERISTEM/CUP-SHAPED COTYLEDON (NAM/CUC) transcription factors is involved in the regulation of leaf margin model (Žádníková and Simon, 2014). Reduced function of NAM/CUC inhibits the growth of leaf margins and leads to lobular reduction and fusion (Blein et al., 2008). NAM/CUC transcription factors promote the formation of leaf margin nicks in single leaves and regulate lobular differentiation and separation in compound leaves, and the silencing of NAM/CUC transcription factors leads to simplification of leaf shape (Bar and Ori, 2014). CUC inhibits growth between leaf notch and lobule (Hasson et al., 2011), and promotes the growth leaf teeth (Kawamura et al., 2010). The transcriptional levels of many leaves’ development regulatory genes, including CUC family genes, are regulated by NGALs, and the regulatory effects of NGALs on gene transcription are primarily negative and dependent on CUC2 (Shao et al., 2020). Arabidopsis NGAL1 can directly bind to the promoter of CUC2 and suppress the expression of CUC2 (Shao et al., 2020). In Arabidopsis, ectopic expression of CUC1 triggers lobular formation, CUC2 acts on the downstream of NGALs to regulate the lobed/serrated leaves, and the inactivation of CUC3 inhibits the serrated part (Kawamura et al., 2010; Hasson et al., 2011). In the B3 family NGATHA-LIKE (NGAL) subfamily of Arabidopsis, overexpression of the three transcription factors NGAL1-3, respectively, can lead to the formation of cup-shaped cotyledon and smoot-edged true leaves. ngaltri, a trimutant with NGAL1-3 function loss, showed an enhanced dentate leaf margin phenotype, indicating that NGALs were involved in leaf margin development (Shao et al., 2020). In addition, Zeng et al. (2022) studies on Citrus reticulata Blanco leaves also found that CiKN1 and CiKN6 have a significant impact on citrus leaf morphological development, CiKN1 and CiKN6 regulate the molecular mechanism of citrus leaf development, and they can combine with each other to form complexes. By binding to the miR164a promoter, it inhibits the expression of CimiR164a, thereby playing a role in the regulation of leaf development through the miR164a-CUC2 pathway (Zeng et al., 2022).
DNA methylation mediated by DNA methyltransferases (DMT) is an important epigenetic modification widely present in plant genomes, which regulates the shape of leaves during the development of leaves, and also controls cell division and expansion. In Populus simonii, the expression of DMF144 and DMF143 in the genomic regions of PtHT1 and PtHT2 was found to be higher in young leaves compared to mature leaves. This suggests that their expression may be suppressed by DNA methylation after cell division and expansion are completed, resulting in the differentiation of leaf shape in natural populations (Ci et al., 2016). DMF40, a methylation marker associated with leaf circumference in P. simonii, is located in the promoter region of PtPIN1, is situated within the promoter region of PtPIN1. It exerts a pronounced inhibitory effect on PtPIN1 and likely plays a crucial role in leaf development within natural populations (Ci et al., 2016). The transcriptional activity of plants is related to DNA methylation on the genome. There is a correlation between genomic methylation and histone H3.3 (H3 variant) enrichment in A.thaliana, knockdown of H3.3 will lead to various phenotypic defects and dysregulation of response genes. H3.3 knockdown results in serrated leaf margins (Wollmann et al., 2017).
In their recent study on rice, Zhou et al. (2023) discovered that OsSNF7.2, a subunit of the ESCRT-III complex in rice, interacts with the auxin biosynthesizer OsYUC8. This interaction plays a crucial role in regulating rice leaf curl by affecting the transport and endosomal degradation of OsYUC8. SERRATE (SE) serves as a core protein in both miRNA biogenesis and mRNA selective splicing. Another intriguing finding comes from Chen et al. (2023), who identified SEAIRa as an antisense intragenic lncRNA transcribed from the 3′end of SE. SEAIRa functions by inhibiting SE expression, resulting in serrated leaf margins. Research on maize leaves, as reported by Satterlee et al. (2023), revealed the role of the rim domain in leaf primordium edge plane growth. Genetically redundant WOX3 transcription factors regulate this rim domain, and high-order mutations of Wox3 genes in maize result in a significant reduction in leaf width and disruption of leaf growth and patterning. The gene expression signature linked to proximal-distal polarity in the initial lingual supports the concept that both lobes and lingua exhibit dorsoventral asymmetry and grow functionally through the juxtaposition of paraxial and distal domains. In Moso bamboo, the PheLBD29 (lateral organ boundaries domain 29) was highly expressed in leaves. Overexpression of PheLBD29 in Arabidopsis resulted in small and backward-curved leaves, with 35S:PheLBD29 Arabidopsis leaves exhibiting a transformation of cells from the positive side to the backside (Wu et al., 2023). The modification of N6-methyladenosine (m6A) in mRNA is a critical regulator of gene expression and plant growth and development. Huang et al. emphasized the significance of Vir-like m6A methyltransferase-associated (VIRMA) as a scaffold that connects the catalytic core components of the m6A methyltransferase complex. In their investigation of upland cotton (Gossypium hirsutum), they observed widespread expression of the genes GHVIR-A and GHVIR-D of VIRMA in various tissues. Disruption of the expression of GhVIR genes had a notable influence on the size, shape, and total cell number of leaf cells, thereby influencing the morphogenesis of cotton leaves (Huang et al., 2022).
6. Leaf senescence
Leaf senescence is a degenerative process that occurs during the final stage of leaf development, and it is regulated by a combination of internal factors (such as age, development, and nutrition) and external environmental factors (including light, temperature, and stress) (Tian et al., 2020). During leaf senescence, the expression profile of a large number of genes is changed, which forms a complex genetic regulatory network with other signal regulatory pathways in transcriptional regulation (Lim et al., 2007). Leaf senescence includes several processes such as chlorophyll decomposition and macromolecular degradation, at the appropriate age, plant leaves begin to senescence and decompose lipids, proteins, nucleic acids, and carbohydrates, thereby retransporting nutrients in the senescent leaves to seeds, storage organs or other growing tissues (Lim et al., 2007; Yu et al., 2021). The functional and regulatory interaction networks of many molecular components during leaf senescence vary with the degree of senescence (Lim et al., 2007) ( Figure 5 ).
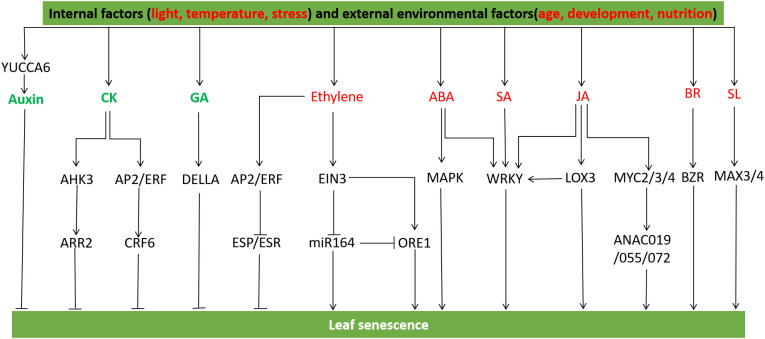
Figure 5.
Hormonal and gene regulation of leaf senescence. YCCA6 regulates auxin biosynthesis and inhibits leaf senescence. The AP2/ERF transcription factor CRF6 mediated by cytokinin inhibits senescence, and the cytokinin receptor AHK3 regulates leaf senescence through the regulatory factor ARR2. The abnormal accumulation of DELLA protein delayed leaf senescence by blocking GA biosynthesis. Ethylene activated AP2/ERF gene to regulate leaf senescence, ERF transcription factor inhibited the expression of ESP/ESR, a negative regulator of leaf senescence, ESR knockout promoted leaf senescence, while ESR overexpression did the opposite. EIN3 activates ORE1 and NAP to positively regulate leaf senescence. EIN3 inhibits miR164 transcription and up-regulates the transcription level of ORE1/NAC2, the target gene of miR164. ABA induces WRKY transcription factor and promotes early senescence of leaves under dark treatment. SA treatment induces SAGs expression, and WRKY influences plant aging and defense signaling pathways in SA-mediated signaling cascades. WRKY interacts with JA biosynthesis gene LOX3 to promote leaf senescence, MYC2/3/4 protein can activate JA-induced chlorophyll degradation, and the signaling pathway of MYC2/3/4 and NAC protein ANAC019/055/072 induces leaf senescence. BRs activates BZR family transcription factors to promote leaf senescence. SL genes MAX3 and MAX4 accelerate leaf senescence (Mayta et al., 2019; Guo et al., 2021a).
6.1. Regulation of plant hormones on leaf senescence
Plant hormones play a crucial role in the regulation of senescence. Among them, ethylene, brassinosteroids (BRs), jasmonic acid (JA), salicylic acid (SA), abscisic acid (ABA), and strigolactone (SLs) promote senescence, while auxin, gibberellins (GAs), and cytokinins delay senescence (Breeze et al., 2011; Guo and Gan, 2012; Jibran et al., 2013).
In the ethylene signaling pathway, key factors such as EIN3, miR164, and NAC (NAM, ATAF, and CUC) transcription factors like ORE1/NAC2, form regulatory networks that mediate leaf senescence (Qiu et al., 2015). EIN3, ORE1, and CCGs participate in ethylene-mediated chlorophyll degradation during leaf senescence in Arabidopsis. Specifically, EIN3 acts as a positive regulator of CCG expression in this process (Kim et al., 2009; Qiu et al., 2015). Ethylene can activate AP2/ERF and regulate the senescence of plant leaves, ERFs AtERF4 and AtERF8 which belongs to Class II increase with plant senescence, and the enhanced expression of NtERF3, AtERF4, or AtERF8 can lead to premature senescence of transgenic Arabidopsis leaves. terf4-terf 8 double mutants delay leaf senescence (Koyama et al., 2013). ERF transcription factors inhibiting leaf senescence occurs negative regulatory factors EPITHIOSPECIFIER PROTEIN/EPITHIOSPECIFYING SENESCENCE REGULATOR (ESP/ESR) gene expression (Miao and Zentgraf, 2007). ESR knockout can accelerate the senescence rate of leaves, and ESR overexpression leaves can slow down the senescence rate (Miao and Zentgraf, 2007). The mitogen-activated protein kinase (MAPK) cascade is a crucial signal transduction pathway in eukaryotic cells, consisting of three components: MAPK, MAPK kinase (MAPKK), and MAPKK kinase (MAPKKK). In ABA signaling, MAPKKK18 and MAPKKK are involved, with MAPKKK18 regulating plant senescence through ABA-dependent protein kinase activity, thereby controlling plant growth and the timing of senescence (Matsuoka et al., 2015). TaWRKY7 is induced by ABA, and TaWRKY7 overexpression can prevent water loss in leaves and enhance tolerance to drought, while TaWRKY7 expression is up-regulated during the senescence process of Arabidopsis leaves, promoting the early senescence of leaves under dark treatment, overexpression of TaWRKY7 leads to premature senescence of transgenic Arabidopsis leaves (Zhang et al., 2016b). The additional application of salicylic acid (SA) induces the expression of senescence-associated genes (SAGs), while the WRKY transcription factor AtWRKY70 plays a role in influencing plant senescence and defense signaling pathways within salicylic acid-mediated signaling cascades. The AtWRKY70 mutation results in the upregulation of developmental senescence-associated genes (SAGs) and defense genes in the salicylic acid (SA) (PR1 and PR2) and jasmonic acid/ethylene (JA/ethylene) (COR1 and PDF1.2) signaling pathways. This implies a connection between plant defense mechanisms and the developmental programs associated with leaf senescence (Ulker et al., 2007). WRKY53 serves as a positive regulator of senescence, while WRKY70 acts as a negative regulator of senescence. The interaction among WRKY53, WRKY54, WRKY70, and WRKY30 plays a role in regulating leaf senescence, facilitating the integration of the regulatory network involving internal and environmental signals during this process (Besseau et al., 2012). BRs activate BZs transcription through BRI1 receptor-like kinases and their well-defined signal transduction pathways, and WRKY6 in the WRKY family inhibits its own promoter activity as well as that of closely related WRKY family members, WRKY6 can regulate the expression of senescence-related genes in the senescence process (Robatzek and Somssich, 2002). WRKY transcription factor TaWRKY40-D is the promoter of leaf aging in transgenic Arabidopsis, and Arabidopsis plants overexpressed with TaWRKY40-D lead to premature leaf senescence after JA and ABA treatment (Zhao et al., 2020b). Senescence-related genes of Arabidopsis are up-regulated in wrky57 mutants, and auxin can antagonize the JA-induced leaf senescence process through WRKY57, in addition, JA down-regulates WRKY57 protein levels, while auxin up-regulates WRKY57 protein levels (Jiang et al., 2014). The WRKY transcription factor aWRKY42-B, a member of the WRKY family, actively participates in both developmental and dark-induced leaf senescence. TaWRKY42-B facilitates leaf senescence by interacting with the JA biosynthetic gene AtLOX3 and its homolog TaLOX3, thus promoting the accumulation of JA content (Zhao et al., 2020a). During dark treatment, the plant hormone ethylene significantly induced strigolactone biosynthesis genes MORE AXIALLY GROWTH3(MAX3) and MAX4, it showed that strigolactone was synthesized in leaves during senescence (Ueda and Kusaba, 2015).
YUCCA encodes flavin-containing monooxygenases (FMO), facilitating the hydroxylation of the amino group of tryptamines (Zhao et al., 2001). With the increase of free IAA concentration, YUCCA6-activated mutants yuc6-1D and 35S:YUC6 Arabidopsis plants exhibit delayed senescence and reduced expression of senescence-associated genes (SAGs) in leaves (Kim et al., 2011). In the process of leaf senescence, the removal of DELLA protein inhibition results in premature leaf senescence, while the enhancement of DELLA protein delays leaf senescence. The removal or enhancement of DELLA protein leads to the upregulation or downregulation of SAG12 and SAG29. After DELLA inhibition was removed, the mutant ga1-3 gai-t6 rga-t2 rgl1-1 rgl2-1 (abbreviated as Q-DELLA/ga1-3) exhibited premature leaf senescence, while the mutant ga1-3, which blocks GA biosynthesis and accumulates abnormal DELLA protein, delayed leaf senescence (Chen et al., 2014). The cytokinin-mediated AP2/ERF transcription factor Cytokinin response factor6 (CRF6) has a negative regulatory effect on leaf developmental senescence (Zwack et al., 2013). There are three cytokinin receptors in Arabidopsis, namely AHK2, AHK3 and AHK4/CRE1WOL. AHK3 plays a significant role in regulating leaf lifespan through the regulatory factor ARR2. Missense mutations occurring in the extracellular domain of AHK3 have been observed to delay leaf senescence (Kim et al., 2006).
6.2. Regulation of genes during leaf senescence
Leaf senescence is regulated by a variety of genes, and age-dependent leaf senescence is an important research focus, mainly focusing on annual and perennial plant leaf senescence. Studies on age-dependent leaf senescence in annual plant Arabidopsis found that AtWDS1 (encoding WD repeat protein), as a novel REDOX homeostasis regulator, negatively regulates age-dependent and dark-induced leaf aging (Fu et al., 2019). The AtFer1 ferritin isoform is functionally implicated in processes leading to age-dependent senescence in Arabidopsis (Murgia et al., 2007). The ACBP3 (acyl-CoA binding protein) transgenic line exhibited age-dependent leaf senescence (Lee et al., 2011). The ACBP3 (acyl-CoA binding protein) transgentic line showed age-dependent leaf senescence (Xiao et al., 2010). The oresara9 (ore9) mutant leaves of Arabidopsis ORE9 prolong life during the age-dependent natural aging process by delaying the onset of various aging symptoms(Woo et al., 2001). The genome-wide H3K9 acetylation level of rice flag leaves increased with age-dependent aging, and the density and width of acetylated lysine residue 9 of histone H3 (H3K9ac) were positively correlated with gene expression and transcription elongation (Zhang et al., 2022b). Senescence-associated NAC (Sen-NAC) regulates the aging of autumn leaves of perennial poplar (Populus tomentosa). Age-dependent increases in intron retention (IR) splicing variants from Sen-NAC can fine-tune the molecular mechanisms of Populus leaf senescence (Wang et al., 2021b).
In addition, leaf senescence is also regulated by various transcription factors such as SQUAMOSA promoter binding protein (SBP), WRKY, C2H2, NAM/ATAF/CUC (NAC), bZIP, APETALA2 (AP2), MYB, etc. (Eulgem et al., 2000; Chen et al., 2002; Lin and Wu, 2004; Buchanan-Wollaston et al., 2005). NAC transcription factors (ANAC019, AtNAP, ANAC047, ANAC055, ORS1, and ORE1) are potential downstream elements of ETHYLENE INSENSITIVE2 (EIN2). EIN3, as a downstream signaling molecule of EIN2, binds to ORE1 and AtNAP promoters, stimulating their transcription. Consequently, EIN3 positively regulates leaf senescence by activating ORE1 and AtNAP (Li et al., 2013; Kim et al., 2014). EIN3 acted on ORESARA2 (ORE2)/ORE3/EIN2 downstream, inhibited miR164 transcription and up-regulated miR164 target gene ORE1/NAC2 transcription, miR164 overexpression or ORE1/NAC2 knockdown inhibits EIN3-induced early-senescence phenotype (Li et al., 2013). The expression of miR164 is negatively regulated by EIN2 and gradually decreases with age, resulting in upregulated expression of ORE1 (Kim et al., 2009). SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1) serves as a trans-regulator of Pheophytinase (PPH). SOC1 functions by inhibiting dark-induced leaf chlorophyll degradation and senescence, achieved through its negative regulation of Pheophytinase PPH expression in Arabidopsis (Chen et al., 2017). In the Basic helix-loop-helix (bHLH) subgroup IIIe, factors such as MYC2/3/4 proteins activate Chlorophyll catabolic genes (CCGs), facilitating JA-induced chlorophyll degradation. Furthermore, downstream of MYC2/3/4 protein, three NAC family proteins, ANAC019/055/072, directly promote the expression of CCGs (NYE1/SGR1, NYE2/SGR2, and NYC1) during chlorophyll degradation (Zhu et al., 2015). Overexpression of NAM transcription factor BnaNAM induces ROS production and leaf chlorosis in Rapeseed (Brassica napus) and positively regulates leaf senescence (Wang et al., 2022b). The WRKY transcription factor plays a crucial role in various aspects of plant physiology, including plant defense against pathogens, response to stress conditions, regulation of leaf senescence, and facilitation of plant growth and development. The overexpression of GhWRKY17 in cotton in Arabidopsis upregulates senescence related genes AtWRKY53, AtSAG12, and AtSAG13, and accelerates senescence in Arabidopsis leaves (Gu et al., 2018). WRKY gene CpWRKY71 in Chimonanthus praecox and CpWRKY71 transgenic plants showed premature leaf senescence (Huang et al., 2019). Leaf senescence1(LS1) gene encoding C2H2-type zinc finger protein, ls1 mutants will lead to premature senescence of leaves and decrease of chlorophyll content, and the expression of LS1 in young leaves is lower than that in mature and senescent leaves, destruction of LS1 function can promote ROS accumulation, accelerated leaf senescence, and cell death in rice (Zhang et al., 2022a). The C2H2-type zinc finger transcription factor MdZAT10 in Apple (Malus domestica) has been found to play a significant role in leaf senescence. It increases the expression of genes associated with senescence and promotes the acceleration of leaf aging. Additionally, MdZAT10 has the ability to enhance the transcriptional activity of MdABI5 on MdNYC1 and MdNYE1, leading to an accelerated leaf senescence process. This highlights the important regulatory role of MdZAT10 in controlling and modulating leaf senescence in Apple (Yang et al., 2021).
MYB-type transcription factors play a critical role in regulating plant growth and development, as well as in response to various abiotic stresses. Specifically, the expression of MYBH, a homologous gene of the MYB transcription factor MYBS3 in Arabidopsis, has been found to be upregulated in both aged leaves and leaves subjected to darkness treatment. Overexpression of MYBH has been shown to result in premature leaf senescence, while the MYBH mutant mybh-1 exhibited a delayed onset of plant senescence. Moreover, overexpression of MYBH has shown to enhance the expression of SAUR36, a key regulator of auxin that promotes leaf senescence. This phenomenon further accelerates leaf senescence induced by ABA and ethylene, highlighting the role of MYBH in mediating the senescence process (Huang et al., 2015). The findings have shed light on the intricate regulatory mechanisms by which MYB transcription factors, exemplified by MYBH, orchestrate plant responses to environmental cues and developmental processes, ultimately affecting leaf senescence (Zhang et al., 2011). The expression of the MYB-related transcription factor, Oryza sativa RADIALIS-LIKE3 (OsRL3), has been observed to be up-regulated in isolated leaves undergoing dark-induced senescence. This up-regulation of OsRL3 contributes to the promotion of leaf senescence process (Park et al., 2018). Under both dark and ABA-induced leaf senescence conditions, the knockout mutant of osmyb102 accelerated the senescence of rice, and overexpression of OsMYB102 controls the expression of SAGs (Piao et al., 2019). When MYBR1 is overexpressed under the control of OxMYBR1, leaf senescence is delayed, and plants with mybr1 gene function loss show faster chlorophyll loss and senescence (Jaradat et al., 2013).
7. Peroration
The regulatory network of leaf development has been gradually established and improved, and the development of leaves in more and more plants is continuously explored. Research on species is no longer limited to the single-leaf model plant Arabidopsis, and the leaf development process of compound-leaf model plants like tomatoes is also receiving increasing attention. The regulation of compound-leaf development is known to be incredibly intricate, with the involvement of various transcription factor families such as WOX, TCP, and MYB playing a pivotal role in the process in tomato plants. Leaf development itself is a complex and variable phenomenon, influenced by interactions between multiple transcription factors and hormones that ultimately govern leaf growth. Understanding these dynamics has been a central focus of research pertaining to leaf development models. One crucial aspect impacting leaf growth patterns is the establishment of an auxin concentration gradient facilitated by the auxin transport protein PIN1. Additionally, a class of KNOX genes acts as the primary genetic determinant in this process. Specifically, the KNOX1 protein regulates the balance between GA and CK during leaf development, and the inhibition of KNOX1 protein is necessary to drive forward the progression of leaf development.
In recent years, the single-cell transcriptome has allowed researchers to obtain a large amount of transcriptomic, epigenetic, and proteomic information from single cells, making spatial quantitative measurement of gene expression abundance possible. Spatial transcriptome can correlate the gene expression information of cells with their spatial location information. This makes it possible for us to study the developmental trajectories of various cells in the process of leaf development. The combination of single-cell transcriptome sequencing and spatial transcriptome sequencing is advantageous and complementary. It allows us to simultaneously obtain information on individual cell heterogeneity and the structural location of cells in tissue space. The combination of time and space multidimensional research techniques provides a method for in-depth understanding of the heterogeneity between cells in the process of leaf growth and development and for analyzing various growth phenomena during cell development.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author contributions
ZL: Investigation, Writing – original draft. WZ: Investigation, Writing – review & editing. SK: Investigation, Writing – review & editing. LL: Investigation, Writing – review & editing. SL: Resources, Supervision, Writing – review & editing.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research & Development Program of China (2021YFD2200503); the National Natural Science Foundation of China (32371977). We also appreciate all the authors for their valuable works.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Achard P., Gusti A., Cheminant S., Alioua M., Dhondt S., Coppens F., et al. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis . Curr. Biol. 19, 1188–1193. doi: 10.1016/j.cub.2009.05.059 [DOI] [PubMed] [Google Scholar]
- Adenot X., Elmayan T., Lauressergues D., Boutet S., Bouché N., Gasciolli V., et al. (2006). DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr. Biol. 16 (9), 927–932. doi: 10.1016/j.cub.2006.03.035 [DOI] [PubMed] [Google Scholar]
- Anastasiou E., Kenz S., Gerstung M., MacLean D., Timmer J., Fleck C., et al. (2007). Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 13 (6), 843–856. doi: 10.1016/j.devcel.2007.10.001 [DOI] [PubMed] [Google Scholar]
- Bar M., Israeli A., Levy M., Ben Gera H., Jiménez-Gómez J. M., Kouril S., et al. (2016). CLAUSA is a MYB transcription factor that promotes leaf differentiation by attenuating cytokinin signaling. Plant Cell 28 (7), 1602–1615. doi: 10.1105/tpc.16.00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M., Ori N. (2014). Leaf development and morphogenesis. Dev 141, 4219–4230. doi: 10.1242/dev.106195 [DOI] [PubMed] [Google Scholar]
- Barkoulas M., Galinha C., Grigg S. P., Tsiantis M. (2007). From genes to shape: regulatory interactions in leaf development. Curr. Opin. Plant Biol. 1, 660–666. doi: 10.1016/j.pbi.2007.07.012 [DOI] [PubMed] [Google Scholar]
- Barton M. K. (2010). Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 341, 95–113. doi: 10.1016/j.ydbio.2009.11.029 [DOI] [PubMed] [Google Scholar]
- Baumberger N., Baulcombe D. C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. U.S.A. 102 (33), 11928–11933. doi: 10.1073/pnas.0505461102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Gera H., Shwartz I., Shao M. R., Shani E., Estelle M., Ori N. (2012). ENTIRE and GOBLET promote leaflet development in tomato by modulating auxin response. Plant J. 70, 903–915. doi: 10.1111/j.1365-313X.2012.04939.x [DOI] [PubMed] [Google Scholar]
- Besseau S., Li J., Palva E. T. (2012). WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana . J. Exp. Bot. 63 (7), 2667–2679. doi: 10.1093/jxb/err450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia N., Bozorg B., Larsson A., Ohno C., Jönsson H., Heisler M. G. (2016). Auxin acts through MONOPTEROS to regulate plant cell polarity and pattern phyllotaxis. Curr. Biol. 26, 3202–3208. doi: 10.1016/j.cub.2016.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blein T., Pulido A., Vialette-Guiraud A., Nikovics K., Morin H., Hay A., et al. (2008). A conserved molecular framework for compound leaf development. Science 322 (5909), 1835–1839. doi: 10.1126/science.1166168 [DOI] [PubMed] [Google Scholar]
- Breeze E., Harrison E., McHattie S., Hughes L., Hickman R., Hill C., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell. 23 (3), 873–894. doi: 10.1105/tpc.111.083345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresso E. G., Chorostecki U., Rodriguez R. E., Palatnik J. F., Schommer C. (2018). Spatial control of gene expression by miR319-regulated TCP transcription factors in leaf development. Plant Physiol. 176, 1694–1708. doi: 10.1104/pp.17.00823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V., Page T., Harrison E., Breeze E., Lim P. O., Nam H. G., et al. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis . Plant J. 42 (4), 567–585. doi: 10.1111/j.1365-313X.2005.02399.x [DOI] [PubMed] [Google Scholar]
- Bürglin T. R., Affolter M. (2016). Homeodomain proteins: an update. Chromosoma 125, 497–521. doi: 10.1007/s00412-015-0543-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burian A., Paszkiewicz G., Nguyen K. T., Meda S., Raczyńska-Szajgin M., Timmermans M. C. (2022). Specification of leaf dorsiventrality via a prepatterned binary readout of a uniform auxin input. Nat. Plants. 8, 269–280. doi: 10.1038/s41477-022-01111-3 [DOI] [PubMed] [Google Scholar]
- Byrne M. E., Barley R., Curtis M., Arroyo J. M., Dunham M., Hudson A., et al. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. doi: 10.1038/35050091 [DOI] [PubMed] [Google Scholar]
- Cao X., Jiao Y. (2018). Auxin and DORNRÖSCHEN joint force in the shoot apex. Sci. China Life Sci. 61, 867–868. doi: 10.1007/s11427-018-9317-2 [DOI] [PubMed] [Google Scholar]
- Černý M., Kuklová A., Hoehenwarter W., Fragner L., Novák O., Rotková G., et al. (2013). Proteome and metabolome profiling of cytokinin action in Arabidopsis identifying both distinct and similar responses to cytokinin down-and up-regulation. J. Exp. Bot. 64, 4193–4206. doi: 10.1093/jxb/ert227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Mei G., Hu Y., Deng J., Zhang T. (2019). LMI1-like and KNOX1 genes coordinately regulate plant leaf development in dicotyledons. Plant Mol. Biol. 99, 449–460. doi: 10.1007/s11103-019-00829-7 [DOI] [PubMed] [Google Scholar]
- Chen M., Maodzeka A., Zhou L., Ali E., Wang Z., Jiang L. (2014). Removal of DELLA repression promotes leaf senescence in Arabidopsis . Plant Sci. 219, 26–34. doi: 10.1016/j.plantsci.2013.11.016 [DOI] [PubMed] [Google Scholar]
- Chen W., Provart N. J., Glazebrook J., Katagiri F., Chang H. S., Eulgem T., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 14 (3), 559–574. doi: 10.1105/tpc.010410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zhu X., Ren J., Qiu K., Li Z., Xie Z., et al. (2017). Suppressor of overexpression of CO1 negatively regulates dark-induced leaf degreening and senescence by directly repressing pheophytinase and other senescence-associated genes in Arabidopsis. Plant Physiol. 173 (3), 1881–1891. doi: 10.1104/pp.16.01457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zhu T., Shi Y., Chen Y., Li W. J., Chan R. J., et al. (2023). An antisense intragenic lncRNA SEAIRa mediates transcriptional and epigenetic repression of SERRATE in Arabidopsis . Proc. Natl. Acad. Sci. U.S.A. 120 (10), e2216062120. doi: 10.1073/pnas.2216062120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon J., Park S. Y., Schulz B., Choe S. (2010). Arabidopsis brassinosteroid biosynthetic mutant dwarf7-1exhibits slower rates of cell division and shoot induction. BMC Plant Biol. 10, 1–8. doi: 10.1186/1471-2229-10-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D., Kim J. H., Kende H. (2004). Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol. 45 (7), 897–904. doi: 10.1093/pcp/pch098 [DOI] [PubMed] [Google Scholar]
- Ci D., Song Y., Du Q., Tian M., Han S., Zhang D. (2016). Variation in genomic methylation in natural populations of Populus simonii is associated with leaf shape and photosynthetic traits. J. Exp. Bot. 67 (3), 723–737. doi: 10.1093/jxb/erv485 [DOI] [PubMed] [Google Scholar]
- Conklin P. A., Strable J., Li S., Scanlon M. J. (2019). On the mechanisms of development in monocot and eudicot leaves. New Phytol. 221, 706–724. doi: 10.1111/nph.15371 [DOI] [PubMed] [Google Scholar]
- Debernardi J. M., Mecchia M. A., Vercruyssen L., Smaczniak C., Kaufmann K., Inze D., et al. (2014). Post-transcriptional control of GRF transcription factors by micro RNA miR396 and GIF co-activator affects leaf size and longevity. Plant J. 79 (3), 413–426. doi: 10.1111/tpj.12567 [DOI] [PubMed] [Google Scholar]
- Deprost D., Yao L., Sormani R., Moreau M., Leterreux G., Nicolaï M., et al. (2007). The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep. 8 (9), 864–870. doi: 10.1038/sj.embor.7401043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding A. M., Xu C. T., Xie Q., Zhang M. J., Yan N., Dai C. B., et al. (2022). ERF4 interacts with and antagonizes TCP15 in regulating endoreduplication and cell growth in Arabidopsis. J. Integr. Plant Biol. 64, 1673–1689. doi: 10.1111/jipb.13323 [DOI] [PubMed] [Google Scholar]
- Dong J., Huang H. (2018). Auxin polar transport flanking incipient primordium initiates leaf adaxial-abaxial polarity patterning. J. Integr. Plant Biol. 60, 455–464. doi: 10.1111/jipb.12640 [DOI] [PubMed] [Google Scholar]
- Du F., Guan C., Jiao Y. (2018). Molecular mechanisms of leaf morphogenesis. Mol. Plant 11, 1117–1134. doi: 10.1016/j.molp.2018.06.006 [DOI] [PubMed] [Google Scholar]
- Emery J. F., Floyd S. K., Alvarez J., Eshed Y., Hawker N. P., Izhaki A., et al. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13 (20), 1768–1774. doi: 10.1016/j.cub.2003.09.035 [DOI] [PubMed] [Google Scholar]
- Eulgem T., Rushton P. J., Robatzek S., Somssich I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5 (5), 199–206. doi: 10.1016/S1360-1385(00)01600-9 [DOI] [PubMed] [Google Scholar]
- Fu M., Yuan C., Song A., Lu J., Wang X., Sun S. (2019). AtWDS1 negatively regulates age-dependent and dark-induced leaf senescence in Arabidopsis . Plant Sci. 285, 44–54. doi: 10.1016/j.plantsci.2019.04.020 [DOI] [PubMed] [Google Scholar]
- Furumizu C., Alvarez J. P., Sakakibara K., Bowman J. L. (2015). Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PloS Genet. 11, e1004980. doi: 10.1371/journal.pgen.1004980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. P., Chickarmane V. S., Ohno C., Meyerowitz E. M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. 106, 16529–16534. doi: 10.1073/pnas.0908122106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Li L., Wei H., Wang H., Su J., Guo Y., et al. (2018). Identification of the group IIa WRKY subfamily and the functional analysis of GhWRKY17 in upland cotton (Gossypium hirsutum L.). PloS One 13 (1), e0191681. doi: 10.1371/journal.pone.0191681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C., Wu B., Yu T., Wang Q., Krogan N. T., Liu X., et al. (2017). Spatial auxin signaling controls leaf flattening in Arabidopsis . Curr. Biol. 27, 2940–2950. doi: 10.1016/j.cub.2017.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Gan S. S. (2012). Convergence and divergence in gene expression profiles induced by leaf senescence and 27 senescence-promoting hormonal, pathological and environmental stress treatments. Plant Cell Environ. 35 (3), 644–655. doi: 10.1111/j.1365-3040.2011.02442.x [DOI] [PubMed] [Google Scholar]
- Guo Y., Ren G., Zhang K., Li Z., Miao Y., Guo H. (2021). Leaf senescence: Progression, regulation, and application. Mol. Hortic. 1 (1), 1–25. doi: 10.1186/s43897-021-00006-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C. M., Jun J. H., Nam H. G., Fletcher J. C. (2004). BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana . Plant Cell Physiol. 45 (10), 1361–1370. doi: 10.1093/pcp/pch201 [DOI] [PubMed] [Google Scholar]
- Ha C. M., Kim G. T., Kim B. C., Jun J. H., Soh M. S., Ueno Y., et al. (2003). The BLADE-ON-PETIOLE1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis . Dev, 161–172. doi: 10.1242/dev.00196 [DOI] [PubMed] [Google Scholar]
- Hasson A., Plessis A., Blein T., Adroher B., Grigg S., Tsiantis M., et al. (2011). Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell. 23 (1), 54–68. doi: 10.1105/tpc.110.081448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2006). The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta . Nat. Genet. 38, 942–947. doi: 10.1038/ng1835 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2010). KNOX genes: versatile regulators of plant development and diversity. Dev 137, 3153–3165. doi: 10.1242/dev.030049 [DOI] [PubMed] [Google Scholar]
- Heisler M. G., Ohno C., Das P., Sieber P., Reddy G. V., Long J. A., et al. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899–1911. doi: 10.1016/j.cub.2005.09.052 [DOI] [PubMed] [Google Scholar]
- Hepworth S. R., Zhang Y., McKim S., Li X., Haughn G. W. (2005). BLADE-ON-PETIOLE–dependent signaling controls leaf and floral patterning in Arabidopsis . Plant Cell 17 (5), 1434–1448. doi: 10.1105/tpc.104.030536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G., Kim G. T., Tsukaya H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana . Plant J. 43, 68–78. doi: 10.1111/j.1365-313X.2005.02429.x [DOI] [PubMed] [Google Scholar]
- Hu Y., Poh H. M., Chua N. H. (2006). The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J. 47 (1), 1–9. doi: 10.1111/j.1365-313X.2006.02750.x [DOI] [PubMed] [Google Scholar]
- Hu Y., Xie Q., Chua N. H. (2003). The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell. 15, 1951–1961. doi: 10.1105/tpc.013557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. (2003). Recent progresses from studies of leaf development. Chin. Bull. Botany. 20, 416–422. [Google Scholar]
- Huang X., Abuduwaili N., Wang X., Tao M., Wang X., Huang G. (2022). Cotton (Gossypium hirsutum) VIRMA as an N6-methyladenosine RNA methylation regulator participates in controlling chloroplast-dependent and independent leaf development. Int. J. Mol. Sci. 23 (17), 9887. doi: 10.3390/ijms23179887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Hu G., Li F., Li Y., Wu J., Zhou X. (2013). NbPHAN, a MYB transcriptional factor, regulates leaf development and affects drought tolerance in Nicotiana benthamiana. Physiol. Plant 149 (3), 297–309. doi: 10.1111/ppl.12031 [DOI] [PubMed] [Google Scholar]
- Huang R., Liu D., Huang M., Ma J., Li Z., Li M., et al. (2019). CpWRKY71, a WRKY transcription factor gene of Wintersweet (Chimonanthus praecox), promotes flowering and leaf senescence in Arabidopsis . Int. J. Mol. Sci. 20 (21), 5325. doi: 10.3390/ijms20215325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. K., Lo P. C., Huang L. F., Wu S. J., Yeh C. H., Lu C. A. (2015). A single-repeat MYB transcription repressor, MYBH, participates in regulation of leaf senescence in Arabidopsis. Plant Mol. Biol. 88, 269–286. doi: 10.1007/s11103-015-0321-2 [DOI] [PubMed] [Google Scholar]
- Huang P., Zhao J., Hong J., Zhu B., Xia S., Zhu E., et al. (2023). Cytokinins regulate rice lamina joint development and leaf angle. Plant Physiol. 191 (1), 56–69. doi: 10.1093/plphys/kiac401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaradat M. R., Feurtado J. A., Huang D., Lu Y., Cutler A. J. (2013). Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 13, 1–19. doi: 10.1186/1471-2229-13-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X. L., Li M. Y., Jiang Q., Xu Z. S., Wang F., Xiong A. S. (2015). High-throughput sequencing of small RNAs and anatomical characteristics associated with leaf development in celery. Sci. Rep. 5, 11093. doi: 10.1038/srep11093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Liang G., Yang S., Yu D. (2014). Arabidopsis WRKY57 functions as a node of convergence for jasmonic acid-and auxin-mediated signaling in jasmonic acid–induced leaf senescence. Plant Cell. 26 (1), 230–245. doi: 10.1105/tpc.113.117838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jibran R., Hunter D. A., Dijkwel P. P. (2013). Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 82, 547–561. doi: 10.1007/s11103-013-0043-2 [DOI] [PubMed] [Google Scholar]
- Jönsson H., Heisler M. G., Shapiro B. E., Meyerowitz E. M., Mjolsness E. (2006). An auxin-driven polarized transport model for phyllotaxis. Proc. Natl. Acad. Sci. 103, 1633–1638. doi: 10.1073/pnas.0509839103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez M. T., Kui J. S., Thomas J., Heller B. A., Timmermans M. C. (2004). microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88. doi: 10.1038/nature02363 [DOI] [PubMed] [Google Scholar]
- Jun J. H., Ha C. M., Fletcher J. C. (2010). BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2 . Plant Cell. 22, 62–76. doi: 10.1105/tpc.109.070763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalve S., De Vos D., Beemster G. T. (2014). Leaf development: a cellular perspective. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura E., Horiguchi G., Tsukaya H. (2010). Mechanisms of leaf tooth formation in Arabidopsis . Plant J. 62 (3), 429–441. doi: 10.1111/j.1365-313X.2010.04156.x [DOI] [PubMed] [Google Scholar]
- Kerstetter R. A., Bollman K., Taylor R. A., Bomblies K., Poethig R. S. (2001). KANADI regulates organ polarity in Arabidopsis . Nature 411, 706–709. doi: 10.1038/35079629 [DOI] [PubMed] [Google Scholar]
- Kierzkowski D., Runions A., Vuolo F., Strauss S., Lymbouridou R., Routier-Kierzkowska A. L., et al. (2019). A growth-based framework for leaf shape development and diversity. Cell 177, 1405–1418. doi: 10.1016/j.cell.2019.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Choi D., Kende H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis . Plant J. 36 (1), 94–104. doi: 10.1046/j.1365-313X.2003.01862.x [DOI] [PubMed] [Google Scholar]
- Kim H. J., Hong S. H., Kim Y. W., Lee I. H., Jun J. H., Phee B. K., et al. (2014). Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis . J. Exp. Bot. 65 (14), 4023–4036. doi: 10.1093/jxb/eru112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Kende H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis . Proc. Natl. Acad. Sci. U.S.A. 101 (36), 13374–13379. doi: 10.1073/pnas.0405450101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Lee B. H. (2006). GROWTH-REGULATING FACTOR4 of Arabidopsis thaliana is required for development of leaves, cotyledons, and shoot apical meristem. J. Plant Biol. 49, 463–468. doi: 10.1007/BF03031127 [DOI] [Google Scholar]
- Kim J. I., Murphy A. S., Baek D., Lee S. W., Yun D. J., Bressan R. A., et al. (2011). YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana . J. Exp. Bot. 62 (11), 3981–3992. doi: 10.1093/jxb/err094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Ryu H., Hong S. H., Woo H. R., Lim P. O., Lee I. C., et al. (2006). Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis . Proc. Natl. Acad. Sci. U.S.A. 103 (3), 814–819. doi: 10.1073/pnas.0505150103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Woo H. R., Kim J., Lim P. O., Lee I. C., Choi S. H., et al. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis . Science 323 (5917), 1053–1057. doi: 10.1126/science.1166386 [DOI] [PubMed] [Google Scholar]
- Koyama T., Nii H., Mitsuda N., Ohta M., Kitajima S., Ohme-Takagi M., et al. (2013). A regulatory cascade involving class II ETHYLENE RESPONSE FACTOR transcriptional repressors operates in the progression of leaf senescence. Plant Physiol. 162 (2), 991–1005. doi: 10.1104/pp.113.218115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N. T., Marcos D., Weiner A. I., Berleth T. (2016). The auxin response factor MONOPTEROS controls meristem function and organogenesis in both the shoot and root through the direct regulation of PIN genes. New Phytol. 212, 42–50. doi: 10.1111/nph.14107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijt S. J., Greco R., Agalou A., Shao J., ‘t Hoen C. C., Övernäs E., et al. (2014). Interaction between the GROWTH-REGULATING FACTOR and KNOTTED1-LIKE HOMEOBOX families of transcription factors. Plant Physiol. 164 (4), 1952–1966. doi: 10.1104/pp.113.222836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655. doi: 10.1038/nature05504 [DOI] [PubMed] [Google Scholar]
- Lee I. C., Hong S. W., Whang S. S., Lim P. O., Nam H. G., Koo J. C. (2011). Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 52 (4), 651–662. doi: 10.1093/pcp/pcr026 [DOI] [PubMed] [Google Scholar]
- Lee B. H., Kim J. H. (2014). Spatio-temporal distribution patterns of GRF-INTERACTING FACTOR expression and leaf size control. Plant Signal. Behav. 9 (9), e29697. doi: 10.4161/psb.29697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. H., Ko J. H., Lee S., Lee Y., Pak J. H., Kim J. H. (2009). The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 151 (2), 655–668. doi: 10.1104/pp.109.141838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiboff S., Strable J., Johnston R., Federici S., Sylvester A. W., Scanlon M. J. (2021). Network analyses identify a transcriptomic proximodistal prepattern in the maize leaf primordium. New Phytol. 230 (1), 218–227. doi: 10.1111/nph.17132 [DOI] [PubMed] [Google Scholar]
- Leyser H. O., Pickett F. B., Dharmasiri S., Estelle M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10, 403–413. doi: 10.1046/j.1365-313x.1996.10030403.x [DOI] [PubMed] [Google Scholar]
- Li P., Chai Z., Lin P., Huang C., Huang G., Xu L., et al. (2020). Genome-wide identification and expression analysis of AP2/ERF transcription factors in sugarcane (Saccharum spontaneum L.). BMC Genomics 21 (1), 1–17. doi: 10.1186/s12864-020-07076-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. C., Kang D. M., Chen Z. L., Qu L. J. (2007). Hormonal regulation of leaf morphogenesis in Arabidopsis . J. Integr. Plant Biol. 49, 75–80. doi: 10.1111/j.1744-7909.2006.00410.x [DOI] [Google Scholar]
- Li J., Nam K. H., Vafeados D., Chory J. (2001). BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 127, 14–22. doi: 10.1104/pp.127.1.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Peng J., Wen X., Guo H. (2013). Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis . Plant Cell. 25 (9), 3311–3328. doi: 10.1105/tpc.113.113340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zheng L., Corke F., Smith C., Bevan M. W. (2008). Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana . Genes Dev. 22 (10), 1331–1336. doi: 10.1101/gad.463608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P. O., Kim H. J., Gil Nam H. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316 [DOI] [PubMed] [Google Scholar]
- Lin H., Niu L., McHale N. A., Ohme-Takagi M., Mysore K. S., Tadege M. (2013). Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants. Proc. Natl. Acad. Sci. 110, 366–371. doi: 10.1073/pnas.1215376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. F., Wu S. H. (2004). Molecular events in senescing Arabidopsis leaves. Plant J. 39 (4), 612–628. doi: 10.1111/j.1365-313X.2004.02160.x [DOI] [PubMed] [Google Scholar]
- Lincoln C., Britton J. H., Estelle M. (1990). Growth and development of the axr1 mutants of Arabidopsis . Plant Cell. 2, 1071–1080. doi: 10.1105/tpc.2.11.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. J., Gao H. (2016). Research progress on the family of TCP genes. Biol. Bull. 32, 14–22. doi: 10.13560/j.cnki.biotech.bull.1985.2016.09.003 [DOI] [Google Scholar]
- Liu X., Guo L. X., Jin L. F., Liu Y. Z., Liu T., Fan Y. H., et al. (2016). Identification and transcript profiles of citrus growth-regulating factor genes involved in the regulation of leaf and fruit development. Mol. Biol. Rep. 43, 1059–1067. doi: 10.1007/s11033-016-4048-1 [DOI] [PubMed] [Google Scholar]
- Liu L., Li X. J., Li B., Sun M. Y., Li S. X. (2022). Genome-wide analysis of the GRF gene family and their expression profiling in peach (Prunus persica). J. Plant Interact. 17 (1), 437–449. doi: 10.1080/17429145.2022.2045370 [DOI] [Google Scholar]
- Liu X., Li D., Zhang D., Yin D., Zhao Y., Ji C., et al. (2018). A novel antisense long noncoding RNA, TWISTED LEAF, maintains leaf blade flattening by regulating its associated sense R2R3-MYB gene in rice. New Phytol. 218 (2), 774–788. doi: 10.1111/nph.15023 [DOI] [PubMed] [Google Scholar]
- Liu D., Song Y., Chen Z., Yu D. (2009. a). Ectopic expression of miR396 suppresses GRF target gene expression and alters leaf growth in Arabidopsis . Physiol. Plant 136, 223–236. doi: 10.1111/j.1399-3054.2009.01229.x [DOI] [PubMed] [Google Scholar]
- Liu Q., Yao X., Pi L., Wang H., Cui X., Huang H. (2009. b). The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis . Plant J. 58 (1), 27–40. doi: 10.1111/j.1365-313X.2008.03757.x [DOI] [PubMed] [Google Scholar]
- Lodha M., Marco C. F., Timmermans M. C. (2013). The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes Dev. 27, 596–601. doi: 10.1101/gad.211425.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn K., Fernandez A., Aida M., Sedbrook J., Tasaka M., Masson P., et al. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Dev 126 (3), 469–481. doi: 10.1242/dev.126.3.469 [DOI] [PubMed] [Google Scholar]
- Ma J. Q., Jian H. J., Yang B., Lu K., Zhang A. X., Liu P., et al. (2017). Genome-wide analysis and expression profiling of the GRF gene family in oilseed rape (Brassica napus L.). Gene 620, 36–45. doi: 10.1016/j.gene.2017.03.030 [DOI] [PubMed] [Google Scholar]
- Mallory A. C., Elmayan T., Vaucheret H. (2008). MicroRNA maturation and action-the expanding roles of ARGONAUTEs. Curr. Opin. Plant Biol. 11 (5), 560–566. doi: 10.1016/j.pbi.2008.06.008 [DOI] [PubMed] [Google Scholar]
- Marsch-Martinez N., Greco R., Becker J. D., Dixit S., Bergervoet J. H., Karaba A., et al. (2006). BOLITA, an Arabidopsis AP2/ERF-like transcription factor that affects cell expansion and proliferation/differentiation pathways. Plant Mol. Biol. 62, 825–843. doi: 10.1007/s11103-006-9059-1 [DOI] [PubMed] [Google Scholar]
- Matsuoka D., Yasufuku T., Furuya T., Nanmori T. (2015). An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol. Biol. 87, 565–575. doi: 10.1007/s11103-015-0295-0 [DOI] [PubMed] [Google Scholar]
- Mayta M. L., Hajirezaei M. R., Carrillo N., Lodeyro A. F. (2019). Leaf senescence: The chloroplast connection comes of age. Plants 8 (11), 495. doi: 10.3390/plants8110495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell J. R., Emery J., Eshed Y., Bao N., Bowman J., Barton M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. doi: 10.1038/35079635 [DOI] [PubMed] [Google Scholar]
- Mi S., Cai T., Hu Y., Chen Y., Hodges E., Ni F., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133 (1), 116–127. doi: 10.1016/j.cell.2008.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Zentgraf U. (2007). The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell. 19 (3), 819–830. doi: 10.1105/tpc.106.042705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum M. G., Yamaguchi S., Hanada A., Kuwahara A., Yoshioka Y., Kato T., et al. (2006). Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 45, 804–818. doi: 10.1111/j.1365-313X.2005.02642.x [DOI] [PubMed] [Google Scholar]
- Mizukami Y., Fischer R. L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. 97, 942–947. doi: 10.1073/pnas.97.2.942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia I., Vazzola V., Tarantino D., Cellier F., Ravet K., Briat J. F., et al. (2007). Knock-out of ferritin AtFer1 causes earlier onset of age-dependent leaf senescence in arabidopsis. Plant Physiol. Biochem. 45 (12), 898–907. doi: 10.1016/j.plaphy.2007.09.007 [DOI] [PubMed] [Google Scholar]
- Muszynski M. G., Moss-Taylor L., Chudalayandi S., Cahill J., Del Valle-Echevarria A. R., Alvarez-Castro I., et al. (2020). The Maize Hairy Sheath Frayed1 (Hsf1) mutation alters leaf patterning through increased cytokinin signaling. Plant Cell. 32, 1501–1518. doi: 10.1105/tpc.19.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz A. A., Raman S., Martinez C. C., Sinha N. R., Schmitz G., Theres K. (2013). Trifoliate encodes an MYB transcription factor that modulates leaf and shoot architecture in tomato. Proc. Natl. Acad. Sci. U.S.A. 110 (6), 2401–2406. doi: 10.1073/pnas.1214300110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M. H., Sun J., Oh D. H., Zielinski R. E., Clouse S. D., Huber S. C. (2011). Enhancing Arabidopsis leaf growth by engineering the BRASSINOSTEROID INSENSITIVE1 receptor kinase. Plant Physiol. 157, 120–131. doi: 10.1104/pp.111.182741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omidbakhshfard M. A., Fujikura U., Olas J. J., Xue G. P., Balazadeh S., Mueller-Roeber B. (2018). GROWTH-REGULATING FACTOR 9 negatively regulates arabidopsis leaf growth by controlling ORG3 and restricting cell proliferation in leaf primordia. PloS Genet. 14, e1007484. doi: 10.1371/journal.pgen.1007484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga D., DeGuzman B., Prigge M. J., Drews G. N., Clark S. E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25, 223–236. doi: 10.1111/j.1365-313X.2001.00959.x [DOI] [PubMed] [Google Scholar]
- Palatnik J. F., Allen E., Wu X., Schommer C., Schwab R., Carrington J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958 [DOI] [PubMed] [Google Scholar]
- Park D. Y., Shim Y., Gi E., Lee B. D., An G., Kang K., et al. (2018). The MYB-related transcription factor RADIALIS-LIKE3 (OsRL3) functions in ABA-induced leaf senescence and salt sensitivity in rice. Environ. Exp. Bot. 156, 86–95. doi: 10.1016/j.envexpbot.2018.08.033 [DOI] [Google Scholar]
- Piao W., Kim S. H., Lee B. D., An G., Sakuraba Y., Paek N. C. (2019). Rice transcription factor OsMYB102 delays leaf senescence by down-regulating abscisic acid accumulation and signaling. J. Exp. Bot. 70 (10), 2699–2715. doi: 10.1093/jxb/erz095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V., Prasad K., Grigg S. P., Sanchez-Perez G. F., Scheres B. (2013). Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis . Proc. Natl. Acad. Sci. 110, 1107–1112. doi: 10.1073/pnas.1213497110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J., Wang Y., Yu T., Cunha A., Wu B., Vernoux T., et al. (2014). Auxin depletion from leaf primordia contributes to organ patterning. Proc. Natl. Acad. Sci. 111, 18769–18774. doi: 10.1073/pnas.1421878112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu K., Li Z., Yang Z., Chen J., Wu S., Zhu X., et al. (2015). EIN3 and ORE1 accelerate degreening during ethylene-mediated leaf senescence by directly activating chlorophyll catabolic genes in. Arabidopsis. PloS Genet. 11 (7), e1005399. doi: 10.1371/journal.pgen.1005399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast-Somssich M. I., Broholm S., Jenkins H., Canales C., Vlad D., Kwantes M., et al. (2015). Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta . Genes Dev. 29, 2391–2404. doi: 10.1101/gad.269050.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E. R., Stieger P., Mandel T., Baltensperger K., Bennett M., et al. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260. doi: 10.1038/nature02081 [DOI] [PubMed] [Google Scholar]
- Reinhart B. J., Weinstein E. G., Rhoades M. W., Bartel B., Bartel D. P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. doi: 10.1101/gad.1004402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M. W., Reinhart B. J., Lim L. P., Burge C. B., Bartel B., Bartel D. P. (2002). Prediction of plant microRNA targets. Cell 110 (4), 513–520. doi: 10.1016/S0092-8674(02)00863-2 [DOI] [PubMed] [Google Scholar]
- Richards D. E., King K. E., Ait-Ali T., Harberd N. P. (2001). How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu. Rev. Plant Biol. 52, 67–88. doi: 10.1146/annurev.arplant.52.1.67 [DOI] [PubMed] [Google Scholar]
- Robatzek S., Somssich I. E. (2002). Targets of AtWRKY6 regulation during plant senescence and pathogen defense. Genes Dev. 16 (9), 1139–1149. doi: 10.1101/gad.222702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez R. E., Mecchia M. A., Debernardi J. M., Schommer C., Weigel D., Palatnik J. F. (2010). Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Dev 137, 103–112. doi: 10.1242/dev.043067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassi M., Vernoux T. (2013). Auxin and self-organization at the shoot apical meristem. J. Exp. Bot. 64, 2579–2592. doi: 10.1093/jxb/ert101 [DOI] [PubMed] [Google Scholar]
- Satterlee J. W., Evans L. J., Conlon B. R., Conklin P., Martinez-Gomez J., Yen J. R., et al. (2023). A Wox3-patterning module organizes planar growth in grass leaves and ligules. Nat. Plants., 1–13. doi: 10.1038/s41477-023-01405-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C., Palatnik J. F., Aggarwal P., Chételat A., Cubas P., Farmer E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PloS Biol. 6, e230. doi: 10.1371/journal.pbio.0060230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semiarti E., Ueno Y., Tsukaya H., Iwakawa H., Machida C., Machida Y. (2001). The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Dev 128, 1771–1783. doi: 10.1242/dev.128.10.1771 [DOI] [PubMed] [Google Scholar]
- Shani E., Yanai O., Ori N. (2006). The role of hormones in shoot apical meristem function. Curr. Opin. Plant Biol. 9, 484–489. doi: 10.1016/j.pbi.2006.07.008 [DOI] [PubMed] [Google Scholar]
- Shao J., Meng J., Wang F., Shou B., Chen Y., Xue H., et al. (2020). NGATHA-LIKEs control leaf margin development by repressing CUP-SHAPED COTYLEDON2 transcription. Plant Physiol. 184, 345–358. doi: 10.1104/pp.19.01598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B., Guo X., Wang Y., Xiong Y., Wang J., Hayashi K. I., et al. (2018). Feedback from lateral organs controls shoot apical meristem growth by modulating auxin transport. Dev. Cell. 44, 204–216. doi: 10.1016/j.devcel.2017.12.021 [DOI] [PubMed] [Google Scholar]
- Skalák J., Vercruyssen L., Claeys H., Hradilová J., Černý M., Novák O., et al. (2019). Multifaceted activity of cytokinin in leaf development shapes its size and structure in Arabidopsis. Plant J. 97, 805–824. doi: 10.1111/tpj.14285 [DOI] [PubMed] [Google Scholar]
- Spartz A. K., Lee S. H., Wenger J. P., Gonzalez N., Itoh H., Inzé D., et al. (2012). The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 70, 978–990. doi: 10.1111/j.1365-313X.2012.04946.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadege M., Lin H., Bedair M., Berbel A., Wen J., Rojas C. M., et al. (2011). STENOFOLIA regulates blade outgrowth and leaf vascular patterning in Medicago truncatula and Nicotiana sylvestris . Plant Cell. 23, 2125–2142. doi: 10.1105/tpc.111.085340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G., Reinhart B. J., Bartel D. P., Zamore P. D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17, 49–63. doi: 10.1101/gad.104810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Ma L., Liu Y., Xu D., Chen Q., Li G. (2020). Arabidopsis FAR-RED ELONGATED HYPOCOTYL3 integrates age and light signals to negatively regulate leaf senescence. Plant Cell. 32 (5), 1574–1588. doi: 10.1105/tpc.20.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. (2014). Comparative leaf development in angiosperms. Curr. Opin. Plant Biol. 17, 103–109. doi: 10.1016/j.pbi.2013.11.012 [DOI] [PubMed] [Google Scholar]
- Tu Z., Xia H., Yang L., Zhai X., Shen Y., Li H. (2022). The roles of microRNA-long non-coding RNA-mRNA networks in the regulation of leaf and flower development in Liriodendron chinense . Front. Plant Sci. 13. doi: 10.3389/fpls.2022.816875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Kusaba M. (2015). Strigolactone regulates leaf senescence in concert with ethylene in Arabidopsis . Plant Physiol. 169 (1), 138–147. doi: 10.1104/pp.15.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B., Shahid Mukhtar M., Somssich I. E. (2007). The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226, 125–137. doi: 10.1007/s00425-006-0474-y [DOI] [PubMed] [Google Scholar]
- Vlad D., Kierzkowski D., Rast M. I., Vuolo F., Dello Ioio R., Galinha C., et al. (2014). Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Sci 343, 780–783. doi: 10.1126/science.1248384 [DOI] [PubMed] [Google Scholar]
- Wang L., Gu X., Xu D., Wang W., Wang H., Zeng M., et al. (2011. a). miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis . J. Exp. Bot. 62, 761–773. doi: 10.1093/jxb/erq307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Kong F., Zhou C. (2021. a). From genes to networks: The genetic control of leaf development. J. Integr. Plant Biol. 63, 1181–1196. doi: 10.1111/jipb.13084 [DOI] [PubMed] [Google Scholar]
- Wang H., Li X., Wolabu T., Wang Z., Liu Y., Tadesse D., et al. (2022. a). WOX family transcriptional regulators modulate cytokinin homeostasis during leaf blade development in Medicago truncatula and Nicotiana sylvestris . Plant Cell. 34, 3737–3753. doi: 10.1093/plcell/koac188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Niu L., Fu C., Meng Y., Sang D., Yin P., et al. (2017). Overexpression of the WOX gene STENOFOLIA improves biomass yield and sugar release in transgenic grasses and display altered cytokinin homeostasis. PloS Genet. 13, e1006649. doi: 10.1371/journal.pgen.1006649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Qiu N., Ding Q., Li J., Zhang Y., Li H., et al. (2014). Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genom 15 (1), 1–12. doi: 10.1186/1471-2164-15-807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Rehmani M. S., Chen Q., Yan J., Zhao P., Li C., et al. (2022. b). Rapeseed NAM transcription factor positively regulates leaf senescence via controlling senescence-associated gene expression. Plant Sci. 323, 111373. doi: 10.1016/j.plantsci.2022.111373 [DOI] [PubMed] [Google Scholar]
- Wang Y., Strauss S., Liu S., Pieper B., Lymbouridou R., Runions A., et al. (2022. c). The cellular basis for synergy between RCO and KNOX1 homeobox genes in leaf shape diversity. Curr. Biol. 32, 3773–3784. doi: 10.1016/j.cub.2022.08.020 [DOI] [PubMed] [Google Scholar]
- Wang H. L., Zhang Y., Wang T., Yang Q., Yang Y., Li Z., et al. (2021. b). An alternative splicing variant of PtRD26 delays leaf senescence by regulating multiple NAC transcription factors in Populus. Plant Cell. 33 (5), 1594–1614. doi: 10.1093/plcell/koab046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Zhao B., He L., Zhou S., Liu Y., Zhao W., et al. (2021. c). The WOX family transcriptional regulator SlLAM1 controls compound leaf and floral organ development in Solanum lycopersicum . J. Exp. Bot. 72, 1822–1835. doi: 10.1093/jxb/eraa574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhou H., Zhao Y., Sun P., Tang F., Song X., et al. (2020). Characterization of poplar growth-regulating factors and analysis of their function in leaf size control. BMC Plant Biol. 20 (1), 1–11. doi: 10.1186/s12870-020-02699-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Motyka V., Strnad M., Schmülling T. (2001). Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. 98, 10487–10492. doi: 10.1073/pnas.171304098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolabu T. W., Wang H., Tadesse D., Zhang F., Behzadirad M., Tvorogova V. E., et al. (2021). WOX9 functions antagonistic to STF and LAM1 to regulate leaf blade expansion in Medicago truncatula and Nicotiana sylvestris . New Phytol. 229, 1582–1597. doi: 10.1111/nph.16934 [DOI] [PubMed] [Google Scholar]
- Wollmann H., Stroud H., Yelagandula R., Tarutani Y., Jiang D., Jing L., et al. (2017). The histone H3 variant H3. 3 regulates gene body DNA methylation in Arabidopsis thaliana. Genome Biol. 18, 1–10. doi: 10.1186/s13059-017-1221-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H. R., Chung K. M., Park J. H., Oh S. A., Ahn T., Hong S. H., et al. (2001). ORE9, an F-box protein that regulates leaf senescence in Arabidopsis . Plant Cell. 13 (8), 1779–1790. doi: 10.1105/TPC.010061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., He W., Wang L., Zhang X., Wang K., Xiang Y. (2023). Phelbd29, an lbd transcription factor from moso bamboo, causes leaf curvature and enhances tolerance to drought stress in transgenic arabidopsis. J. Plant Physiol. 280, 153865. doi: 10.1016/j.jplph.2022.153865 [DOI] [PubMed] [Google Scholar]
- Wu W., Li J., Wang Q., Lv K., Du K., Zhang W., et al. (2021). Growth-Regulating Factor 5 (GRF5)-mediated gene regulatory network promotes leaf growth and expansion in poplar. New Phytol. 30 (2), 612–628. doi: 10.1111/nph.17179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. J., Wang W. L., Zhuang J. (2017). Developmental processes and responses to hormonal stimuli in tea plant (Camellia sinensis) leaves are controlled by GRF and GIF gene families. Funct. Integr. Genomics 17, 503–512. doi: 10.1007/s10142-017-0553-0 [DOI] [PubMed] [Google Scholar]
- Wu L., Zhang D., Xue M., Qian J., He Y., Wang S. (2014). Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. J. Integr. Plant Biol. 56 (11), 1053–1063. doi: 10.1111/jipb.12220 [DOI] [PubMed] [Google Scholar]
- Xiao S., Gao W., Chen Q. F., Chan S. W., Zheng S. X., Ma J., et al. (2010). Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell. 22 (5), 1463–1482. doi: 10.1105/tpc.110.075333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Jiao Y. (2019). The diverse roles of auxin in regulating leaf development. Plants 8 (7), 243. doi: 10.3390/plants8070243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Wu B., Du F., Guo X., Tian C., Hu J., et al. (2021). A crosstalk between auxin and brassinosteroid regulates leaf shape by modulating growth anisotropy. Mol. Plant 14, 949–962. doi: 10.1016/j.molp.2021.03.011 [DOI] [PubMed] [Google Scholar]
- Yan S., Yan C. J., Gu M. H. (2008). Molecular mechanism of leaf development. Hereditas 30, 1127–1135. doi: 10.3724/SP.J.1005.2008.01127 [DOI] [PubMed] [Google Scholar]
- Yang K., An J. P., Li C. Y., Shen X. N., Liu Y. J., Wang D. R., et al. (2021). The apple C2H2-type zinc finger transcription factor MdZAT10 positively regulates JA-induced leaf senescence by interacting with MdBT2. Hortic. Res. 8. doi: 10.1038/s41438-021-00593-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Li D., Liu X., Ji C., Hao L., Zhao X., et al. (2014). OsMYB103L, an R2R3-MYB transcription factor, influences leaf rolling and mechanical strength in rice (Oryza sativaL.). BMC Plant Biol. 14 (1), 1–15. doi: 10.1186/1471-2229-14-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Zeng H. X., Zhang N., Ren J., Tang M., Liu Y. H., et al. (2019). Advances in molecular genetic mechanisms of plant leaf development. J.Changjiang Vegetables. 18, 44–48. doi: 10.3865/j.issn.1001-3547.2019.18.017 [DOI] [Google Scholar]
- You J., Xiao W., Zhou Y., Shen W., Ye L., Yu P., et al. (2022). The APC/CTAD1-WIDE LEAF 1-NARROW LEAF 1 pathway controls leaf width in rice. Plant Cell. 34, 4313–4328. doi: 10.1093/plcell/koac232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Qi Y., Xu J., Dai X., Chen J., Dong C. H., et al. (2021). Arabidopsis WRKY71 regulates ethylene-mediated leaf senescence by directly activating EIN2, ORE1 and ACS2 genes. Plant J. 107 (6), 1819–1836. doi: 10.1111/tpj.15433 [DOI] [PubMed] [Google Scholar]
- Žádníková P., Simon R. (2014). How boundaries control plant development. Curr. Opin. Plant Biol. 17, 116–125. doi: 10.1016/j.pbi.2013.11.013 [DOI] [PubMed] [Google Scholar]
- Zeng R. F., Fu L. M., Deng L., Liu M. F., Gan Z. M., Zhou H., et al. (2022). CiKN1 and CiKN6 are involved in leaf development in citrus by regulating CimiR164. Plant J. 110, 828–848. doi: 10.1111/tpj.15707 [DOI] [PubMed] [Google Scholar]
- Zhang X., Ju H. W., Chung M. S., Huang P., Ahn S. J., Kim C. S. (2011). The RR-type MYB-like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol. 52 (1), 138–148. doi: 10.1093/pcp/pcq180 [DOI] [PubMed] [Google Scholar]
- Zhang C., Li N., Hu Z., Liu H., Hu Y., Tan Y., et al. (2022. a). Mutation of leaf senescence 1 encoding a C2H2 zinc finger protein induces ROS accumulation and accelerates leaf senescence in rice. Int. J. Mol. Sci. 23 (22), 14464. doi: 10.3390/ijms232214464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li Y., Zhang Y., Zhang Z., Zhang D., Wang X., et al. (2022. b). Genome-wide H3K9 acetylation level increases with age-dependent senescence of flag leaf in rice. J. Exp. Bot. 73 (14), 4696–4715. doi: 10.1093/jxb/erac155 [DOI] [PubMed] [Google Scholar]
- Zhang X. Y., Liu X. (2022). Research progress of transcription factors and leaf development. Plant Physiol. J. 58 (1), 91–100. doi: 10.13592/j.cnki.ppj.2021.0253 [DOI] [Google Scholar]
- Zhang H., Zhao M., Song Q., Zhao L., Wang G., Zhou C. (2016). Identification and function analyses of senescence-associated WRKYs in wheat. Biochem. Biophys. Res. Commun. 474 (4), 761–767. doi: 10.1016/j.bbrc.2016.05.034 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Christensen S. K., Fankhauser C., Cashman J. R., Cohen J. D., Weigel D., et al. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291 (5502), 306–309. doi: 10.1126/science.291.5502.306 [DOI] [PubMed] [Google Scholar]
- Zhao M. M., Zhang X. W., Liu Y. W., Li K., Tan Q., Zhou S., et al. (2020. a). A WRKY transcription factor, TaWRKY42-B, facilitates initiation of leaf senescence by promoting jasmonic acid biosynthesis. BMC Plant Bio. 20 (1), 1–22. doi: 10.1186/s12870-020-02650-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Zhang W., Song Q., Xuan Y., Li K., Cheng L., et al. (2020. b). A WRKY transcription factor, TaWRKY40-D, promotes leaf senescence associated with jasmonic acid and abscisic acid pathways in wheat. Plant Biol. 22 (6), 1072–1085. doi: 10.1111/plb.13155 [DOI] [PubMed] [Google Scholar]
- Zhiponova M. K., Vanhoutte I., Boudolf V., Betti C., Dhondt S., Coppens F., et al. (2013). Brassinosteroid production and signaling differentially control cell division and expansion in the leaf. New Phytol. 197, 490–502. doi: 10.1111/nph.12036 [DOI] [PubMed] [Google Scholar]
- Zhong R., Ye Z. H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11, 2139–2152. doi: 10.1105/tpc.11.11.2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Chen S., Cai M., Cui S., Ren Y., Zhang X., et al. (2023). ESCRT-III component OsSNF7. 2 modulates leaf rolling by trafficking and endosomal degradation of auxin biosynthetic enzyme OsYUC8 in rice. J. Integr. Plant Biol. doi. doi: 10.1111/jipb.13460 [DOI] [PubMed] [Google Scholar]
- Zhu X., Chen J., Xie Z., Gao J., Ren G., Gao S., et al. (2015). Jasmonic acid promotes degreening via MYC 2/3/4-and ANAC 019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 84 (3), 597–610. doi: 10.1111/tpj.13030 [DOI] [PubMed] [Google Scholar]
- Zhu H., Hu F., Wang R., Zhou X., Sze S. H., Liou L. W., et al. (2011). Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145 (2), 242–256. doi: 10.1016/j.cell.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong Y. X., Hao Z. Y., Tu Z. H., Shen Y. F., Zhang C. G., Wen S. Y., et al. (2021). Genome-wide survey and identification of AP2/ERF genes involved in shoot and leaf development in Liriodendron chinense. BMC Genomics 22 (1), 1–17. doi: 10.1186/s12864-021-08119-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwack P. J., Robinson B. R., Risley M. G., Rashotte A. M. (2013). Cytokinin response factor 6 negatively regulates leaf senescence and is induced in response to cytokinin and numerous abiotic stresses. Plant Cell Physiol. 54 (6), 971–981. doi: 10.1093/pcp/pct049 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.