Abstract
Three open reading frames (ORFs) have been located downstream of cefE in the cephamycin C gene cluster of Streptomyces clavuligerus. ORF13 (pcd) encodes a 496-amino-acid protein (molecular weight [MW], 52,488) with an N-terminal amino acid sequence identical to that of pure piperideine-6-carboxylate dehydrogenase. ORF14 (cmcT) encodes a 523-amino-acid protein (MW, 54,232) analogous to Streptomyces proteins for efflux and resistance to antibiotics. ORF15 (pbp74) encodes a high molecular weight penicillin-binding protein (MW, 74,094).
Cephamycins are derived from the tripeptide δ-l-α-aminoadipyl-l-cysteinyl-d-valine. In bacteria, l-α-aminoadipic acid is formed by deamination of lysine by the lysine-6-aminotransferase that converts lysine into piperideine-6-carboxylic acid (P6C). The activity of a second enzyme, P6C dehydrogenase, has recently been shown to be required for conversion of P6C into l-α-aminoadipic acid (6). The genes for biosynthesis of antibiotics are frequently clustered with antibiotic resistance genes and sometimes with genes involved in precursor biosynthesis (12). The lat gene encoding lysine-6-aminotransferase is located in the cephamycin cluster of Streptomyces clavuligerus and Nocardia lactamdurans (2, 11). The purpose of this study was to investigate whether the pcd gene required for the second step in the formation of α-aminoadipic acid was clustered together with lat and other genes involved in cephamycin formation.
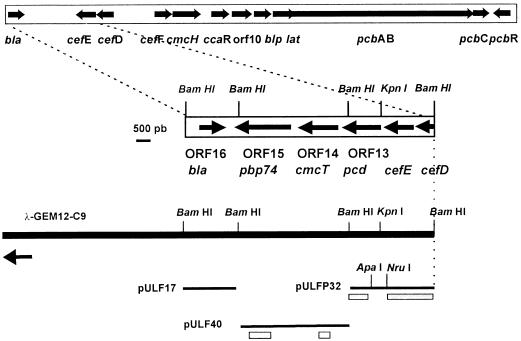
Cloning of the region downstream from cefE in the S. clavuligerus cephamycin cluster.
A genomic library of S. clavuligerus DNA in phage λGEM12 (15) was hybridized with plasmid pULF33 (inserting a 3.3-kb BglII-BamHI DNA fragment containing cefE and part of the cefD gene of S. clavuligerus). Nine phages gave positive hybridization, and one of them, λGEM-C9, contained the unknown region downstream of cefE (Fig. 1). Three BamHI DNA fragments from phage λGEM-C9 of 4.0, 3.2, and 1.7 kb were subcloned in pBSSK(+) to give plasmids pULF17, pULFP32, and pULF40. A total of 7.1 kb downstream from cefE was sequenced with the AutoRead Sequencing System (Pharmacia). Analysis of the nucleotide sequence of this 7.1-kb fragment (EMBL/GenBank/DDBJ accession no. AJ001743) with the Geneplot program (DNAstar) identified four open reading frames (ORFs). Three of them are in the same orientation as cefE (ORF13, ORF14, and ORF15), and ORF16, in the opposite orientation, corresponds to the bla gene (15).
FIG. 1.
Physical map of the DNA of S. clavuligerus encoding the cephamycin C gene cluster. A fragment of the phage λGEM-C9 DNA studied is indicated by a solid bar. The S. clavuligerus insert in the phage DNA follows in the direction of the arrows. DNA fragments in different plasmids used are indicated by thin lines, and the probes used for hybridization are shown as stippled bars.
ORF13 encodes the P6C-DH of S. clavuligerus.
The predicted start codon of 1,488-bp ORF13 is only 15 bp downstream from the stop codon of cefE, and ORF13 encodes a protein of 496 amino acids with a deduced Mr of 52,488. A search of the Swissprot data base showed a high similarity between the protein encoded by ORF13 and the aldehyde dehydrogenases (Fig. 2). The identity among amino acids was maximal in the region between amino acids 33 and 425. The highest similarity (48% identity in 389 amino acids) is found with an aldehyde dehydrogenase from Cenorhabditis elegans and with the betaine dehydrogenase responsible for turgor in Pisum sativum (45% identity in 392 amino acids). Because of its similarity with other dehydrogenases, the sequence TGSTRMGR (amino acids 239 to 246 in ORF13) corresponds to the NADH binding motif as calculated with the Prosite program. The protein has lower homology with 1-pyrroline-5-carboxylate dehydrogenase from Bacillus subtilis (29.3% identity) (8), an enzyme of proline catabolism that converts 1-pyrroline-5-carboxylic acid into glutamic acid.
FIG. 2.
Deduced amino acids sequence of the piperideine-6-carboxylate dehydrogenase encoded by the pcd gene. Comparison with aldehyde dehydrogenases from C. elegans and P. sativum and with the pyrroline-5-carboxylate dehydrogenase from B. subtilis is shown. Amino acids conserved in at least three of the sequences are indicated by gray shading. A bar indicates the putative NADH binding motif.
The piperideine-6-carboxylate dehydrogenase (P6C-DH), which converts 1-piperideine-6-carboxylic acid into α-aminoadipic acid, a precursor of cephamycin C, has recently been purified (6). This enzyme has an Mr of 56,200 as determined by gel filtration, which is similar to the deduced molecular weight (MW) of the protein encoded by ORF13. In order to test whether the protein encoded by ORF13 was the P6C-DH, we sequenced the N-terminal end of the pure protein with an Applied Biosystems protein sequencer (model 476A). The sequence found was VTAAISGTDEI(K/L)RARA, which matches perfectly the sequence of the protein encoded by ORF13 from amino acids 2 to 16, indicating that ORF13 encodes the P6C-DH in which the starting methionine residue is removed in vivo. Therefore, the gene was named pcd (for piperideine carboxylate dehydrogenase).
Hybridization of DNA from different Streptomyces species with a 1.3-kb BamHI-KpnI DNA fragment internal to pcd showed a positive hybridization signal with the cephamycin C producers Streptomyces jumonjinensis NRRL 5741, Streptomyces lipmanii NRRL 3584, and Streptomyces cattleya NRRL 8057 and with the Streptomyces griseus NRRL 3851 producer of cephamycins A and B, whereas no hybridizing band was found with the DNA of the nonproducer strain Streptomyces lividans JI1326, Streptomyces cacaoi subsp. asoensis ATCC 19094, or Streptomyces niveus NRRL 2466.
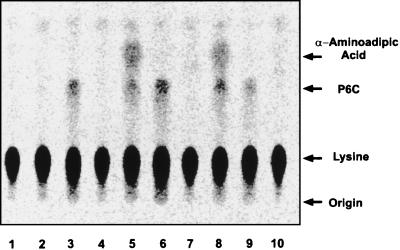
S. lividans lacks P6C-DH activity (6), and therefore, to confirm pcd function, the gene was expressed in S. lividans by fusing it to the saf promoter. The pcd gene was subcloned from plasmid pULF22, a pBSKS(+) derivative with a 2.2-kb insert containing pcd, as an Ecl36II-HindIII fragment extending from 266 bp upstream of the ORF to 490 bp downstream of the TGA termination codon. Plasmid pULS699, which contains a 300-bp insert with the saf promoter (4) in pBSKS(+), was digested with SmaI and SpeI, the cohesive ends were filled in, and both constructions were ligated to give pULSpcd, in which the pcd gene is expressed from the saf promoter. Cultures of S. lividans(pULSpcd) and control S. lividans(pULS699) (lacking the pcd insert) were grown in YEME medium (0.3% yeast extract, 0.3% malt extract), and cells were harvested at 36 h. S. clavuligerus NRRL 3585 was grown under the same conditions. P6C-DH activity was measured in the cell extracts by the coupled radioactive method (6). The assay mixture was then analyzed by thin-layer chromatography, and the labelled compounds formed were detected by autoradiography.
Reaction samples with S. clavuligerus NRRL 3585 extracts from 36-h-old cells showed the formation of a compound (Fig. 3, lane 5) which was not found in the absence of either lysine dehydrogenase from Agrobacterium tumefaciens (forming P6C) (lane 6) or cell extracts of S. clavuligerus (lane 7). This compound with an Rf of 0.44 is derived from lysine and corresponds to α-aminoadipic acid. When control S. lividans(pULS699) extracts were used, no α-aminoadipic acid was formed (lanes 3 and 4). Cell extracts from S. lividans(pULSpcd), in which the pcd gene is expressed, formed a labelled compound with an Rf of 0.44 that gave a positive reaction with ninhydrin. This compound cochromatographed with unlabelled α-aminoadipic acid when it was added to the assay after the reaction was stopped.
FIG. 3.
Autoradiography of the products (separated by thin-layer chromatography) formed from 14C-lysine by the coupled lysine-ɛ-dehydrogenase-piperideine-6-carboxylate dehydrogenase assay. Lanes: 1, 14C-lysine (0.5 μCi); 2, 14C-lysine (0.5 μCi) and assay cofactors lacking cell extracts; 3, complete reaction with S. lividans(pIJ699) extracts; 4, as in lane 3 but without lysine-ɛ-dehydrogenase; 5, complete reaction with S. clavuligerus NRRL 3585 cell extracts; 6, as in lane 5 but without cell extract; 7, as in lane 5 but without lysine-ɛ-dehydrogenase; 8, complete reaction with S. lividans(pULSpcd) cell extract; 9, as in lane 8 but without cell extract; 10, as in lane 8 but without lysine-ɛ-dehydrogenase.
ORF14 encodes a protein with 14 transmembrane domains.
ORF14 is 1,569 bp long and is predicted to start with a GTG codon 69 bp downstream from the TGA codon of pcd. The protein encoded by ORF14 has 523 amino acids (Mr, 54,232). Hydrophobicity analysis of the protein encoded by ORF14 with the SOAP program (PCGene) indicated that this protein contains 14 transmembrane segments and shows a high similarity with membrane proteins involved in proton-dependent drug efflux (14). Some of these proteins, as well as transmembrane proteins of the ABC transporter superfamily, confer resistance to antibiotics (5) and are encoded by genes present in antibiotic biosynthesis clusters. The highest identity was found with the CmcT protein of N. lactamdurans (3) (60% identity in 486 amino acids) followed by proteins belonging to cluster e of the family of 14 transmembrane segments, such as PurT, for puromycin exclusion in Streptomyces alboniger (27% identity in 503 amino acids) and proteins involved in the exportation of tetracenomycin, lincomycin, or methylenomycin.
ORF15 encodes a high-MW PBP with a proline-rich amino-terminal region.
ORF15 contains 2,088 bp and is predicted to start with an ATG codon. Downstream from ORF15 there is an inverted sequence (nucleotides 5,856 to 5,870 and 5,884 to 5,899; ΔG25 of −38 kcal/mol) that may act as a transcriptional terminator.
ORF15 encodes a 696-amino-acid protein with a deduced MW of 74,094. This protein showed two different regions. Amino acids 1 to 300 showed homology to specific regions of proteins rich in proline such as glycoproteins for cell wall synthesis of Clostridium thermocellum (29.7% identical amino acids in 276 residues) (7) or with a protein secreted by Xenopus laevis skin (27.7% identity in 277 amino acids) (9). The C-terminal region (amino acids 301 to 696) showed a high similarity with penicillin-binding proteins (PBP) such as the PBP encoded by dacF of B. subtilis (27% identity) (16), Escherichia coli Pbp6 encoded by dacC (26% identity), and Pbp5 (25% identity) encoded by dacA (1). These proteins are d-alanyl-carboxypeptidases, produced during sporulation of B. subtilis or needed for cell wall synthesis in E. coli. The conserved motifs present in β-lactamases and the PBPs STAK, SGN, and KTG were found in the second half of the ORF15-encoded protein (amino acids 310 to 696). Hydropathy analysis of the protein with the RAOARGOS program (PCGene) detected the presence of an α-helix transmembrane motif (amino acids 286 to 309; LAMIAIPLAALLLVIAFVAVQLL). This region separates the proline-rich half of the protein and the PBP-like C-terminal half. This gene has been named pbp74.
A different gene (pbpR) encoding a PBP with a MW of 57,346, which confers partial resistance to benzylpenicillin, has been already located in the S. clavuligerus cephamycin C-clavulanic acid gene cluster (13). However, there is no substantial homology between both PBPs outside of the three well-conserved motifs found in all PBPs. The molecular weight of Pbp74 agrees quite well with the PBP with an MW of 79,000 detected by Horikawa et al. (10) in S. clavuligerus membranes.
Transcriptional analysis of the pcd-cmcT-pbp74 cluster.
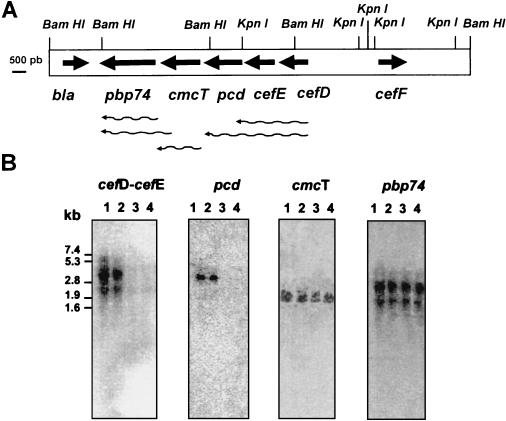
RNA from 24-, 48-, 72-, and 96-h cultures of S. clavuligerus was isolated from cultures in Trypticase soy broth medium as reported previously (15), separated by electrophoresis, and hybridized with (i) an ApaI-BamHI 0.56-kb DNA probe internal to pcd; (ii) a 0.4-kb SmaI-SacI DNA fragment internal to cmcT; (iii) a SacI-HindIII 1-kb DNA fragment internal to pbp74; and (iv) a 1.5-kb BamHI-NruI DNA fragment containing the 3′ end of cefD and the nearly complete cefE gene. As shown in Fig. 4A, when probes corresponding to the cefE-cefD region were used, transcripts of 2.6 and 4.1 kb were observed. The 2.6-kb mRNA corresponds to the transcription of cefE-cefD. The 4.1-kb transcript also contains information from the pcd gene, since a probe internal to pcd also hybridized with the 4.1-kb transcript. This larger transcript provides a mechanism to coordinate the expression of enzymes for both the early and middle steps of the pathway.
FIG. 4.
(A) Physical map of a 15.1-kb BamHI DNA fragment of S. clavuligerus. The putative transcription of the genes is indicated by wavy arrows. The DNA fragments used as probes are indicated by stippled bars in Fig. 1. (B) Hybridization of total RNA of S. clavuligerus NRRL 3585 grown in Trypticase soy broth medium for 24, 48, 72, and 96 h as shown in lanes 1, 2, 3, and 4, respectively. The markers correspond to the RNA type II of Boehringer-Mannheim.
cmcT was transcribed as a single mRNA of 1.9 kb, while the probe corresponding to pbp74 revealed the presence of two abundant transcripts of 1.9 and 3.1 kb. This result suggests that pbp74 is transcribed from two promoters, one of which is likely present inside the coding sequence of cmcT since the corresponding pbp74 ORF is 2.0 kb long. The lack of a 3.1-kb hybridizing band when the mRNA was probed with a fragment internal to cmcT may be due to the fact that this 0.4-kb probe corresponds to the 5′ region of the gene. The presence of a strong transcription termination signal 37 bp downstream from pbp74 and the presence of the bla gene in the opposite orientation make it less probable that the 3.1-kb transcript corresponds to regions downstream of pbp74.
The structural genes cefE, cefD, and pcd were transcribed early in the fermentation (at 24 and 48 h) (Fig. 4B), preceding the phase of intense cephamycin biosynthesis, which is maximal at 60 h of culture. A very small amount of transcript for these three genes (less than 20% as quantified by densitometry performed with an Instant Imager [Packard]) was observed thereafter. This rise and decline in the steady-state levels of transcripts for cefE, cefD, and pcd agrees well with the previously reported profile of cephamycin biosynthesis in S. clavuligerus and indicates that the decline in the rate of antibiotic synthesis is due to the decay of the mRNA for the biosynthetic genes. Interestingly, the cmcT and pbp74 genes are transcribed throughout the fermentation (until at least 96 h).
Acknowledgments
This research was supported by grants from the CICYT (Madrid, Spain) (Bio96-0827) and from Antibiotics S.A. (Madrid). F. J. Pérez-Llarena received a fellowship from the Education Council of the Basque Country Government (Spain).
We thank J. L. Fuente for providing a sample of pure piperideine-6-carboxylate dehydrogenase and J. García-Velasco for sequencing the N-terminal end of the protein.
REFERENCES
- 1.Broome-Smith J K, Ionnidis I, Edelman A, Spratt B G. Nucleotide sequence of the penicillin-binding protein 5 and 6 genes of Escherichia coli. Nucleic Acids Res. 1988;16:1617. doi: 10.1093/nar/16.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coque J J R, Liras P, Láiz L, Martín J F. A gene encoding lysine 6-aminotransferase which forms α-aminoadipic acid, a precursor of β-lactam antibiotics, is located in the cluster of cephamycin biosynthetic genes in Nocardia lactamdurans. J Bacteriol. 1991;173:6258–6264. doi: 10.1128/jb.173.19.6258-6264.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coque J J R, Liras P, Martin J F. Genes for a β-lactamase, a penicillin-binding protein and a transmembrane protein are clustered with the cephamycin biosynthetic genes in Nocardia lactamdurans. EMBO J. 1993;2:631–639. doi: 10.1002/j.1460-2075.1993.tb05696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daza A, Martín J F, Vigal T, Gil J A. Analysis of the promoter region of saf, a Streptomyces griseus gene that increases production of extracellular enzymes. Gene. 1991;108:63–71. doi: 10.1016/0378-1119(91)90488-w. [DOI] [PubMed] [Google Scholar]
- 5.Fernández E, Lombó F, Méndez C, Salas J A. An ABC transporter is essential for resistance to the antitumor agent mithramycin in the producer Streptomyces argillaceus. Mol Gen Genet. 1996;251:692–698. doi: 10.1007/BF02174118. [DOI] [PubMed] [Google Scholar]
- 6.Fuente J L, Rumbero A, Martín J F, Liras P. δ-1-Piperideine-6-carboxylate dehydrogenase, a new enzyme that forms α-aminoadipate in Streptomyces clavuligerus and other cephamycin C producing actinomycetes. Biochem J. 1997;326:59–64. [PMC free article] [PubMed] [Google Scholar]
- 7.Fujino T, Béguin P, Aubert J-P. Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J Bacteriol. 1993;175:1891–1899. doi: 10.1128/jb.175.7.1891-1899.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glaser P, Kunst F, Arnaud M, Coudart M P, Gonzales W, Hullo M F, Ionescu M, Lubochinsky B, Marcelino L, Moszer I, Presecan E, Santana M, Schneider E, Schweizer J, Vertés A, Rapoport G, Danchin A. Bacillus subtilis genome project: cloning and sequencing of the 97 kb region from 325 to 333 degrees. Mol Microbiol. 1993;10:371–384. [PubMed] [Google Scholar]
- 9.Hauser F, Roeben C, Hoffmann W. XP2, a new member of the P-domain peptide family of potential growth factors, is synthesized in Xenopus laevis skin. J Biol Chem. 1992;267:14451. [PubMed] [Google Scholar]
- 10.Horikawa S, Nakazawa H, Ogawara H. Penicillin-binding proteins in Streptomyces cacaoi and Streptomyces clavuligerus. Kinetics of [14C]benzylpenicillin binding, temperature sensitivity and release of [14C]benzylpenicillin from the complex. J Antibiot. 1980;33:1363–1368. doi: 10.7164/antibiotics.33.1363. [DOI] [PubMed] [Google Scholar]
- 11.Madduri K, Stuttard C, Vining L C. Cloning and location of a gene governing lysine ɛ-aminotransferase, an enzyme initiating β-lactam biosynthesis in Streptomyces spp. J Bacteriol. 1991;173:985–988. doi: 10.1128/jb.173.3.985-988.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martín J F, Liras P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu Rev Microbiol. 1989;216:91–98. doi: 10.1146/annurev.mi.43.100189.001133. [DOI] [PubMed] [Google Scholar]
- 13.Paradkar A S, Aidoo K A, Jensen S E. Molecular analysis of a β-lactam resistance gene encoded within the cephamycin gene cluster of Streptomyces clavuligerus. J Bacteriol. 1996;178:6266–6274. doi: 10.1128/jb.178.21.6266-6274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pérez-Llarena F J, Martín J F, Galleni M, Coque J J R, Fuente J L, Frère J M, Liras P. The bla gene of the cephamycin cluster of Streptomyces clavuligerus encodes a class A β-lactamase of low enzymatic activity. J Bacteriol. 1997;179:6035–6040. doi: 10.1128/jb.179.19.6035-6040.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J-J, Schuch R, Piggot P J. Characterization of a Bacillus subtilis sporulation operon that includes genes for an RNA polymerase ς factor and for a putative dd-carboxypeptidase. J Bacteriol. 1992;174:4885–4892. doi: 10.1128/jb.174.15.4885-4892.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]