The cellular prion protein (PrPC), a cell surface glycoprotein of 209 amino acids, has been considerably studied over the decades mainly due to its critical involvement in transmissible spongiform encephalopathies, or prion diseases. Indeed, it is the misfolding and aggregation of PrPC into pathological assemblies - named PrPSc – that constitute prions, the agents causing these unusual neurodegenerative diseases affecting humans and animals (Prusiner, 1982). Furthermore, increasing evidence support its relevance also in other neurodegenerative diseases (NDDs), such as Alzheimer’s and Parkinson’s diseases (Corbett et al., 2020).
Multiple physiological functions have been attributed to PrPC, ranging from neuronal differentiation and proliferation, myelin maintenance, neuroprotection, and homeostasis of divalent metals to receptor properties and cellular signaling participation; still, only a few have been confirmed (Kovac and Curin Serbec, 2022). PrPC undergoes several post-translational events - i.e. the addition of a glycosylphosphatidylinositol (GPI)-anchor, a disulfide bond, and N-linked sugar chains (a. a. 181 and 197, mouse sequence), as well as the endoproteolytic cleavages at four or more sites (α-, β-, γ-cleavages and shedding) - that widen the number of its biologically active forms and presumably contribute to the diversity of functions attributed to the mature form (Linsenmeier et al., 2017). The main proteolytic event of PrPC, whose responsible protease and exact cellular site are still uncertain, is α-cleavage, occurring between residues His109 and Lys110 (mouse sequence) and producing the membrane-bound C-terminal fragment C1, which encompasses the whole globular domain, and the secreted N-terminal unstructured fragment N1. An alternative endoproteolytic processing, but less active in physiological conditions, is β-cleavage, which occurs N-terminally to the α-cleavage site at multiple positions at the end of the octarepeat domain, around a. a. 90, and produces the membrane-bound C-terminal fragment C2 and the released N-terminal fragment N2. In physiological conditions, β-cleavage occurs at the cell membrane due to the direct activity of reactive oxygen species, suggesting an active role of PrPC in the cellular response against oxidative stress. Another relevant cleavage event is the shedding, occurring due to the action of the metalloprotease ADAM10 (a disintegrin and metalloproteinase) at the extreme C-terminus of PrPC and resulting in the release from the membrane of an almost full-length PrPC which lacks the GPI anchor and 3 to 4 C-terminal residues, named shed PrPC. Finally, a fourth newly discovered endoproteolytic event, γ-cleavage, occurs at the C-terminus of PrP, supposedly between a. a. 176 and 200. These post-translational events thus produce a plethora of variably glycosylated and/or GPI-anchored PrPC proteoforms that have not been systematically investigated. The multiple and specific endoproteolytic sites suggest that the endoproteolytic processing of PrPC might represent a key step for the production of functional proteins rather than a mere degradation step in PrPC turnover. Although increasing interest is now rising in these truncated proteoforms, not only for their physiological role but also because they appear – together with full-length PrPC (FL-PrPC) – to influence the course of prion and other NDDs (Linsenmeier et al., 2017), the physiological and pathological roles of PrPC-derived proteoforms are far from being understood.
New tools for a comprehensive analysis of PrPC endoproteolytic processing: Despite the suggested pathophysiological importance of these proteoforms, methods that allow a reliable analysis of all these fragments are still missing. Indeed, recently developed methods only allow a fine determination of the overall concentration of FL-PrPC and/or its main proteolytic C-terminal fragments C1 and C2 (Castle et al., 2019; Mortberg et al., 2022) and, most importantly, none of these methods can clearly discriminate between GPI-anchored FL-PrPC and shed PrPC, which is nowadays considered one of the most promising therapeutic targets and biomarkers. The recent generation of a cleavage site-specific antibody specifically detecting shed PrP (Linsenmeier et al., 2018) only partially solved the issue, as its use is limited to a few species and a quantitative analysis of this cleavage remains problematic. Given the urgency of a comprehensive representation of the PrPC proteoforms and of their possible alterations in NDDs, we developed an optimized western blot assay able to obtain the maximum information about PrPC endoproteolytic processing in a complex biological sample (Vanni et al., 2023). Combining PNGase F treatment of PrPC with discriminative electrophoresis conditions and extensive epitope mapping, we identified in brain homogenates all known PrPC proteoforms, including those shed by ADAM10, and refined previous knowledge about C-terminal γ-cleavage. Indeed, besides the known couples N- and C-terminal complementary proteoforms, such as C1/N1 or C2/N2, we identified two new couples that we named N3/C3 and N3’/C3’, with opposite glycosylation status, which depend on two alternative γ-cleavage-like sites, one N-terminal and one C-terminal to the two N-glycosylation sites of PrPC (Vanni et al., 2023). Overall, 11 PrPC proteoforms could be co-analyzed in brain homogenates, that is five GPI-anchored/C-terminal (FL, C1, C2, C3, and C3′), four released N-terminal (N1, N2, N3, and N3′), and two C-terminally shed (sFL and sC1) proteoforms. The shed and C-terminal proteoforms, except C3, are N-glycosylated, while N3 is the only glycosylated N-terminal proteoform (see Figure 1 for a representation of all PrPC proteoforms).
Figure 1.
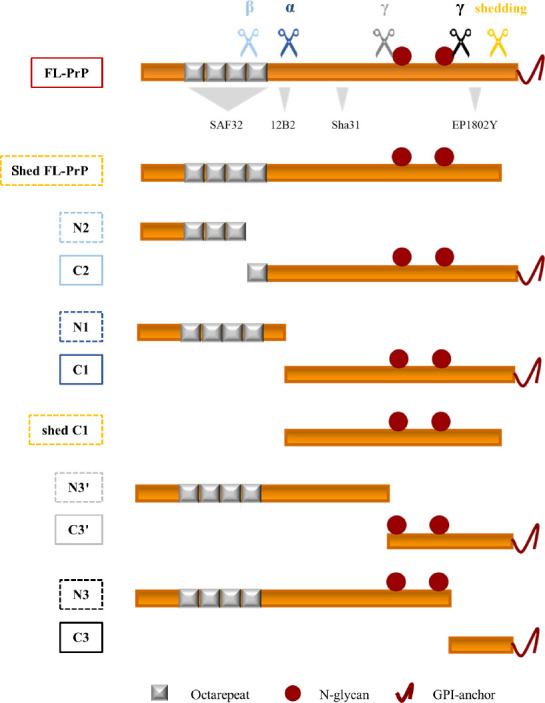
Schematic representation of the PrPC proteoforms detectable by western blot assay.
Linear representation of PrPC (23–230) showing (i) the octarepeat region (repeated grey boxes), (ii) the N-glycosylation sites (a. a. 181 and 197, red spheres), (iii) the GPI-anchor (red curved line, extreme C-terminus) and (iv) epitopes of pan-PrP antibodies (SAF32, 12B2, Sha31, and EP1802Y) which allow obtaining the maximum information about endoproteolytic processing with the optimized western blot method. Colored scissors indicate the main cleavage events of PrPC: light blue scissors represent β-cleavage processing, dark blue scissors represent α-cleavage, while orange scissors indicate shedding; grey and black scissors represent two γ-cleavage-like events. The respective cleavage products (the color of each rectangle indicates the generative cleavage event, while whole and dotted lines indicate if membrane-bound or soluble) are reported below. β-cleavage processing occurs at multiple sites at the end of the octarepeat domain of PrPC and produces the soluble N-terminal fragment N2 (~10 kDa) and the membrane-bound C-terminal fragment C2 (~18 kDa). α-cleavage, a proteolytic event that occurs between residues H109 and K110 producing the soluble N-terminal fragment N1 (~12 kDa) and the C-terminal membrane-anchored fragment C1 (~16 kDa). Shedding occurs at the extreme C-terminus of PrPC resulting in the release of almost full-length PrPC, i.e., FL-PrPC lacking 3-to-4 a. a. and the GPI-anchor (shed-PrP, ~25 kDa), or almost full-length C1 (shed-C1, ~14 kDa), from the plasma membrane. The two γ-cleavage-like events occur one before the first N-glycosylation site, producing the non-glycosylated soluble N-terminal fragment N3’ (~17 kDa) and the glycosylated membrane-bound C3’ (~10 kDa), and one after the second N-glycosylation site, producing the glycosylated soluble N-terminal fragment N3 (~20 kDa) and the C-terminal non-glycosylated fragment C3 (~7 kDa). Independently of the rodent model analyzed, in the healthy brain, the most abundant PrPC proteoforms, i.e., FL-PrPC and the cleavage products of shedding and α- and β-cleavage, showed a specific and conserved rank order of abundance: C1 (43–52%) > FL-PrPC (21–29%) > C2 (11–18%) > sFL-PrPC (6–10%) > N1 (1–3.7%) > N2 (0.4–1.5%) (% of abundance intended as a “range” of abundance determined considering the whole set of rodent models analyzed (see Vanni et al., 2023 for single quantifications)). Created with Microsoft PowerPoint. α: α-Cleavage; β: β-cleavage; γ: γ-cleavage; FL-PrP: full-length prion protein; GPI: glycosylphosphatidylinositol.
PrPC endoproteolysis in physiology: As a first step, we compared PrPC endoproteolytic processing in different species (bank vole vs. mouse) as well as in transgenic mouse lines expressing PrPC from different species (bank vole, human, bovine and ovine). Among the tg lines, some differ at a single PrP amino acid (i.e., humanized tg mice expressing either M129 or V129 human PrP and ovinised tg mice expressing either A136 or V136 sheep PrP). This allowed disentangling the impact, if any, of PrP amino acid sequence, PrP heterologous expression, and PrP overexpression on its endoproteolytic processing. We observed that, in the healthy brain, the main PrPC proteoforms were in a specific and conserved rank order of abundance, i.e., C1 > FL > C2 > sFL > N1 > N2, independently of the rodent model analyzed. Thus, variation in PrP sequence or genetic background, level of expression, or heterogeneity between exogenous PrP and the mouse endoproteolytic machinery do not have a major impact on the constitutive processing of PrPC. The overall conservation – and fine regulation – of PrPC endoproteolytic processing support not only relevant physiological roles of the resulting fragments, but also of the proteolytic processing itself, fundamental to ensure a homeostatic balance of PrPC proteoforms. In this regard, the surprisingly low abundance of the FL membrane-anchored PrPC proteoform (20–25% of total PrPC) might represent a protective cellular strategy to produce controlled amounts of proteolytic proteoforms with physiological and neuroprotective roles (Linsenmeier et al., 2017). Importantly, this keeps under a physiological “safe” threshold the ratio between FL-PrPC and C1, where C1 is not considered a substrate for prion conversion and could act as a dominant-negative inhibitor of this process (Westergard et al., 2011).
Of note, almost 80–85% of total PrPC in the healthy brain was represented by GPI-anchored proteoforms, i.e. FL, C1, and C2; C1, rather than FL-PrPC, was by far the most abundant proteoform, as it accounted for up to the 50% of total PrPC, approximately 2 times more than the unprocessed FL proteoform. The soluble N-terminal fragments were much less represented than their GPI-anchored counterparts, with shed PrPC accounting for most of the residual 15–20% of total PrPC. Diffusion in the extracellular space and drainage via body fluids, inherent to the nature of these secreted proteoforms, as well as their low in vivo biostability (Mohammadi et al., 2020), might explain the huge difference in abundance between the GPI-anchored C-terminal fragments and their soluble N-terminal counterparts. Still being a released fragment, shed PrPC seems to be less affected by these phenomena, accounting for 7-to-10% of total PrPC and being only two-to-fourfold less than membrane-anchored FL-PrPC. This finding demonstrates a constitutively high PrPC shedding in the brain and suggests a high biostability of this proteoform that still harbors the globular domain, which is instead absent in N1 and N2 fragments.
Finally, the rank order of abundance of PrPC proteoforms and the surprisingly low abundance of FL-PrPC observed in our study highlight the complexity of setting up rapid methods able to reliably quantify PrPC and the need to account for the relative contribution of the different proteoforms to the quantitative output. As PrP is an important pharmacodynamic biomarker for PrP-lowering therapies, these considerations clearly apply to the methods that are being developed for the longitudinal quantification of PrP in body fluids.
PrPC endoproteolysis and its implications in disease: Given the importance of a finely regulated endoproteolytic processing of PrPC, alterations of the homeostatic balance of PrPC proteoforms might modulate the risk of the onset of pathological conditions. When we exploited quantitatively our method to assess the level of PrPC shedding, we unexpectedly found that it varies among the rodent models analyzed, mostly depending on the PrP amino acid sequence. Indeed, bank voles showed a constitutively lower PrPC shedding than mice, which was nicely mimicked by the heterologous expression of bank vole PrP in PrP-null mice. Furthermore, PrPC shedding was higher in humanized tg mice expressing M129 than in those expressing V129 human PrP, suggesting that a single amino acid variation outside the proteolytic cleavage site is sufficient to affect PrP shedding. Bank vole and wild-type mice are reported to have opposite prion strain preferences, and polymorphism at human PRNP codon 129 modulates disease susceptibility and phenotype in all categories of human prion diseases (Schmitz et al., 2017). Despite the limitation of a study conducted on a low number of animals and on transgenic mice rather than on the natural hosts, these results might represent a first insight into the implications of PrPC endoproteolysis in disease, suggesting a correlation between PrPC shedding and susceptibility to prions. Alteration in the steady-state levels of PrPC in pathological conditions would not be surprising, as a lowering of total PrPC in CSF, which is mainly soluble shed PrPC, has been reported in individuals with rare pathogenic PRNP variants (Mortberg et al., 2022) or with sporadic Creutzfeldt-Jakob disease (Villar-Pique et al., 2019), as well as in several other NDDs (Meyne et al., 2009). Together with our finding that single amino acid substitutions may affect PrPC shedding, these data raise the intriguing hypothesis that pathogenic PRNP mutations and PRNP polymorphisms known to modulate susceptibility to prions might increase PrPC propensity to misfold not only by directly influencing its conformational stability but also indirectly, by inducing an imbalance of PrPC proteoforms. Indeed, a decrease in PrPC shedding might induce an increase of the membrane-anchored proteoform – which is misfolding-prone and has been reported as a high-affinity binding partner for toxic oligomeric proteins abundantly produced in other NDDs – and potentially lead to pathological consequences. Such an imbalance in PrPC proteoforms could be reasonably implicated in the spontaneous conversion of wild-type PrPC to PrPSc that is supposed to occur in sporadic CJD, the most common human prion disease. In this context, it would be interesting to determine if aging, the main risk factor for sporadic NDDs, affects the PrPC proteoform composition in the brain.
The role of N-terminal proteoforms in prion diseases: Despite representing only a minor fraction of total PrPC in the healthy brain, released N-terminal PrPC proteoforms might still play a role in prion diseases. As above mentioned, a protective role against prion diseases has been postulated for prion shedding. However, data obtained with transgenic mouse lines have been of considerable interest in showing the “double-face” of PrP shedding, since they showed that (i) GPI-less PrPC can misfold into PrPSc, (ii) the overexpression of GPI-less PrP induces the development of a spontaneous Gerstmann-Sträussler-Scheinker (GSS)-like disease (Stohr et al., 2011) and (iii) shedding can facilitate prion spreading and plaques formation (Linsenmeier et al., 2017). The data obtained with overexpressing GPI-less PrP transgenic mice resemble what was recently observed in patients carrying two rare mutations of GSS, i.e., Y226X and Q227X, which generate C-terminally truncated PrPs analogous to the physiological shed PrPC proteoform (Figure 2). Indeed, both patients showed PrP amyloid deposits, with the Y226X carrier associated with a prion protein cerebral amyloid angiopathy (PrP-CAA) and the Q227X one with an unusual GSS phenotype (Schmitz et al., 2017). The PrP-CAA and GSS-like diseases have been reported also in patients with three other rare nonsense mutations in PRNP, in which the nonsense substitution results in the production of truncated PrP. This is the case of mutations Y145X, Q160X, and Y163X (Schmitz et al., 2017). Interestingly, the truncated PrPs generated by these mutations resemble another physiological proteoform of PrPC, the ~15–17 kDa N-terminal fragment derived by the γ-cleavage-like processing that we named N3’ and that occurs N-terminally to the two N-glycosylation sites (Figure 2). Notably, N3 and N3’ include the 115–145 a. a. region that is reported to be necessary for amyloid formation (Vanni et al., 2020) and that is lacking in the other N-terminal proteoforms N1 and N2, meaning that they can undergo prion-like misfolding or mediate the toxic effect. They encompass the PrP region forming the protease-resistant core in GSS and thus, in principle, might represent novel misfolding-prone prion precursors, aside from FL-PrPC. Interestingly, a 16 kDa thermolysin-resistant fragment has been recently observed in GSS patients, whose electrophoretic features are similar to the physiological N3 fragment (Mercer et al., 2018).
Figure 2.
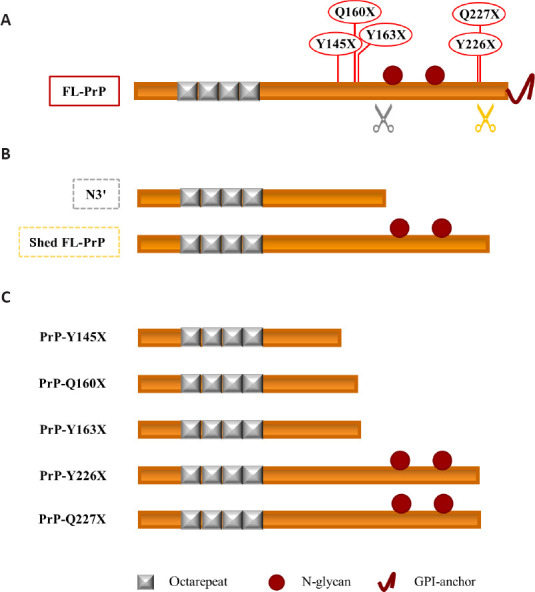
Soluble PrPC proteoforms and pathogenic truncated PrP: a schematic comparison.
(A) Linear representation of PrPC (23–230) showing (i) the octarepeat region (repeated grey boxes), (ii) the N-glycosylation sites (a. a. 181 and 197, red spheres), and (iii) the GPI-anchor (red curved line, extreme C-terminus). Grey and orange scissors indicate γ-cleavage and shedding of PrPC. The pathogenic stop-codon mutations (Y145X, Q160X, Y163X, Y226X, and Q227X) that generate truncated PrPs are shown in red above the linear representation of PrPC. (B) Linear representation of the physiological soluble PrP proteoforms N3’ (non-glycosylated PrP fragment generated by the γ-like-processing that occurs before the N-glycosylation site) and shed FL-PrPC (fully glycosylated and produced by shedding). (C) Linear representation of the pathological truncated PrPs. Truncated Y145X, Q160X, and Y163X PrPs resemble the physiological N-terminal fragment derived by the γ-cleavage-like processing, while truncated Y226X and Q227X PrPs look like the physiological shed PrPC proteoform. Created with Microsoft PowerPoint. FL-PrP: Full-length PrP; GPI: glycosylphosphatidylinositol; PrP: prion protein; PrP-Q160X: truncated Q160X PrP; PrP-Q227X: truncated Q227X PrP; PrP-Y145X: truncated Y145X PrP; PrP-Y163X: truncated Y163X PrP; PrP-Y226X: truncated Y226X PrP.
Conclusion and future directions: In conclusion, new research tools are beginning to enlighten the endoproteolytic processing of PrPC and the generated proteoforms, opening a promising new area of research in the field of NDDs. Our first results highlight the complexity of the PrPC proteoforms and suggest a fine regulation of the cellular processes leading to multiple endoproteolytic processing. The study of PrPC constitutive processing in different species, tissues, cell lines, and transgenic lines expressing pathogenic PRNP mutations, as well as in rodent models used for aging and other neurodegenerative diseases studies, will lead to a better understanding of its physiological and pathological implications. Recent evidence suggests that therapeutic approaches able to impact PrP proteolytic processing can be developed, for example by increasing PrP shedding (Linsenmeier et al., 2021), further highlighting the need for methods able to differentiate PrP proteoforms. Finally, our approach can be useful for further development of PrP-lowering therapies, in the screening of such drugs as well as in allowing a comprehensive appraisal of the drug target itself - not limited to FL-PrPC but widened to the whole plethora of functional proteoforms.
The authors would like to apologize in advance for not being able to accommodate all studies related to PrPC endoproteolysis in health and diseases in the current manuscript, due to space and formatting regulations for a perspective.
This work was supported by the Ministero della Salute (grant No. RF-2016-02364498, to NR).
Additional file: Open peer review report 1 (93.9KB, pdf) .
Footnotes
Open peer reviewer: Mariana Juliani Do Amaral, Universidade Federal do Rio de Janeiro, Brazil.
P-Reviewer: do Amaral MJ; C-Editors: Zhao M, Liu WJ, Qiu Y; T-Editor: Jia Y
References
- 1.Castle AR, Daude N, Gilch S, Westaway D. Application of high-throughput capillary-based Western analysis to modulated cleavage of the cellular prion protein. J Biol Chem. (2019);294:2642–2650. doi: 10.1074/jbc.RA118.006367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corbett GT, Wang Z, Hong W, Colom-Cadena M, Rose J, Liao M, Asfaw A, Hall TC, Ding L, DeSousa A, Frosch MP, Collinge J, Harris DA, Perkinton MS, Spires-Jones TL, Young-Pearse TL, Billinton A, Walsh DM. PrP is a central player in toxicity mediated by soluble aggregates of neurodegeneration-causing proteins. Acta Neuropathol. (2020);139:503–526. doi: 10.1007/s00401-019-02114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kovac V, Curin Serbec V. Prion protein:the molecule of many forms and faces. Int J Mol Sci. (2022);(1232);23 doi: 10.3390/ijms23031232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Linsenmeier L, Altmeppen HC, Wetzel S, Mohammadi B, Saftig P, Glatzel M. Diverse functions of the prion protein - Does proteolytic processing hold the key? Biochim Biophys Acta Mol Cell Res. (2017);1864:2128–2137. doi: 10.1016/j.bbamcr.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 5.Linsenmeier L, Mohammadi B, Wetzel S, Puig B, Jackson WS, Hartmann A, Uchiyama K, Sakaguchi S, Endres K, Tatzelt J, Saftig P, Glatzel M, Altmeppen HC. Structural and mechanistic aspects influencing the ADAM10-mediated shedding of the prion protein. Mol Neurodegener. (2018);13:18. doi: 10.1186/s13024-018-0248-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linsenmeier L, Mohammadi B, Shafiq M, Frontzek K, Bär J, Shrivastava AN, Damme M, Song F, Schwarz A, Da Vela S, Massignan T, Jung S, Correia A, Schmitz M, Puig B, Hornemann S, Zerr I, Tatzelt J, Biasini E, Saftig P, et al. Ligands binding to the prion protein induce its proteolytic release with therapeutic potential in neurodegenerative proteinopathies. Sci Adv. (2021);7:eabj1826. doi: 10.1126/sciadv.abj1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer RCC, Daude N, Dorosh L, Fu ZL, Mays CE, Gapeshina H, Wohlgemuth SL, Acevedo-Morantes CY, Yang J, Cashman NR, Coulthart MB, Pearson DM, Joseph JT, Wille H, Safar JG, Jansen GH, Stepanova M, Sykes BD, Westaway D. A novel Gerstmann-Straussler-Scheinker disease mutation defines a precursor for amyloidogenic 8 kDa PrP fragments and reveals N-terminal structural changes shared by other GSS alleles. PLoS Pathog. (2018);14:e1006826. doi: 10.1371/journal.ppat.1006826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyne F, Gloeckner SF, Ciesielczyk B, Heinemann U, Krasnianski A, Meissner B, Zerr I. Total prion protein levels in the cerebrospinal fluid are reduced in patients with various neurological disorders. J Alzheimers Dis. (2009);17:863–873. doi: 10.3233/JAD-2009-1110. [DOI] [PubMed] [Google Scholar]
- 9.Mohammadi B, Linsenmeier L, Shafiq M, Puig B, Galliciotti G, Giudici C, Willem M, Eden T, Koch-Nolte F, Lin YH, Tatzelt J, Glatzel M, Altmeppen HC. Transgenic overexpression of the disordered prion protein N1 fragment in mice does not protect against neurodegenerative diseases due to impaired ER translocation. Mol Neurobiol. (2020);57:2812–2829. doi: 10.1007/s12035-020-01917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mortberg MA, Zhao HT, Reidenbach AG, Gentile JE, Kuhn E, O'Moore J, Dooley PM, Connors TR, Mazur C, Allen SW, Trombetta BA, McManus A, Moore MR, Liu J, Cabin DE, Kordasiewicz HB, Mathews J, Arnold SE, Vallabh SM, Minikel EV. Regional variability and genotypic and pharmacodynamic effects on PrP concentration in the CNS. JCI Insight. (2022);7:e156532. doi: 10.1172/jci.insight.156532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. (1982);216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 12.Schmitz M, Dittmar K, Llorens F, Gelpi E, Ferrer I, Schulz-Schaeffer WJ, Zerr I. Hereditary human prion diseases:an update. Mol Neurobiol. (2017);54:4138–4149. doi: 10.1007/s12035-016-9918-y. [DOI] [PubMed] [Google Scholar]
- 13.Stohr J, Watts JC, Legname G, Oehler A, Lemus A, Nguyen HO, Sussman J, Wille H, DeArmond SJ, Prusiner SB, Giles K. Spontaneous generation of anchorless prions in transgenic mice. Proc Natl Acad Sci U S A. (2011);108:21223–21228. doi: 10.1073/pnas.1117827108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanni I, Pirisinu L, Acevedo-Morantes C, Kamali-Jamil R, Rathod V, Di Bari MA, D'Agostino C, Marcon S, Esposito E, Riccardi G, Hornemann S, Senatore A, Aguzzi A, Agrimi U, Wille H, Nonno R. Isolation of infectious non-fibrillar and oligomeric prions from a genetic prion disease. Brain. (2020);143:1512–1524. doi: 10.1093/brain/awaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanni I, Iacobone F, D'Agostino C, Giovannelli M, Pirisinu L, Altmeppen HC, Castilla J, Torres JM, Agrimi U, Nonno R. An optimized Western blot assay provides a comprehensive assessment of the physiological endoproteolytic processing of the prion protein. J Biol Chem. (2023);299:102823. doi: 10.1016/j.jbc.2022.102823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villar-Piqué A, Schmitz M, Lachmann I, Karch A, Calero O, Stehmann C, Sarros S, Ladogana A, Poleggi A, Santana I, Ferrer I, Mitrova E, Žáková D, Pocchiari M, Baldeiras I, Calero M, Collins SJ, Geschwind MD, Sánchez-Valle R, Zerr I, et al. Cerebrospinal fluid total prion protein in the spectrum of prion diseases. Mol Neurobiol. (2019);56:2811–2821. doi: 10.1007/s12035-018-1251-1. [DOI] [PubMed] [Google Scholar]
- 17.Westergard L, Turnbaugh JA, Harris DA. A naturally occurring c-terminal fragment of the prion protein (PrP) delays disease and acts as a dominant-negative inhibitor of PrPSc formation. J Biol Chem. (2011);286:44234–44242. doi: 10.1074/jbc.M111.286195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


