Our perspective on microbial diversity has improved enormously over the past few decades. In large part this has been due to molecular phylogenetic studies that objectively relate organisms. Phylogenetic trees based on gene sequences are maps with which to articulate the elusive concept of biodiversity. Thus, comparative analyses of small-subunit rRNA (16S or 18S rRNA) and other gene sequences show that life falls into three primary domains, Bacteria, Eucarya, and Archaea (51, 52). Based on rRNA trees, the main extent of Earth’s biodiversity is microbial. Our knowledge of the extent and character of microbial diversity has been limited, however, by reliance on the study of cultivated microorganisms. It is estimated that >99% of microorganisms observable in nature typically are not cultivated by using standard techniques (1).
Recombinant DNA and molecular phylogenetic methods have recently provided means for identifying the types of organisms that occur in microbial communities without the need for cultivation (see references 1, 20, and 35 for reviews). Results from application of these methods to a number of diverse environments confirm that our view of microbial diversity was limited and point to a wealth of novel and environmentally important diversity yet to be studied (34). It is the aim of this review to collate, compare, and incorporate the results of the environmental sequence-based studies into the context of known bacterial diversity. We discuss the sequence data at the taxonomic level of the phylogenetic division because divisions constitute first-order clades for describing the breadth of bacterial diversity. Although we have yet to determine even the outlines of the bacterial tree, common threads are beginning to emerge that revise our current views of bacterial diversity and distribution in the environment.
PHYLOGENETIC DIVERSITY IN THE BACTERIAL DOMAIN
In 1987, Woese described the bacterial domain as comprised of about 12 natural relatedness groups, based mainly on analyses of familiar cultivated organisms such as cyanobacteria, spirochetes, and gram-positive bacteria (all of which, based on rRNA sequence divergence, display greater evolutionary depth than plants, animals, and fungi) (51). These relatedness groups have variously been called “kingdoms,” “phyla,” and “divisions”; we use the latter term. For the purposes of this review we define a bacterial division purely on phylogenetic grounds as a lineage consisting of two or more 16S rRNA sequences that are reproducibly monophyletic and unaffiliated with all other division-level relatedness groups that constitute the bacterial domain. We judge reproducibility by the use of multiple tree-building algorithms, bootstrap analysis, and varying the composition and size of data sets used for phylogenetic analyses. The typical interdivisional rRNA sequence difference is 20 to 25%. For comparison, the 16S rRNAs of Escherichia coli and Pseudomonas aeruginosa, both representatives of the γ group of Proteobacteria, differ overall by about 15%; the 16S rRNAs of E. coli and Bacillus subtilis (“low-G+C gram-positive bacterial” division) differ by about 23%.
At the current stage in the phylogenetic classification of Bacteria, divisions are not consistently named or taxonomically ranked. rRNA-defined divisions are identified by classes (e.g., Proteobacteria [41] and Actinobacteria [42]), orders (e.g., Thermotogales and Aquificales), families (e.g., Chlorobiaceae), generic names such as the Nitrospira group (11), or common names such as the green nonsulfur (GNS) bacteria and low-G+C gram-positive bacteria (51). Division-level nomenclature has not even been consistent between studies, so some divisions are identified by more than one name. For instance, green sulfur bacteria is synonymous with Chlorobiaceae; high-G+C gram-positive bacteria is synonymous with Actinobacteria and Actinomycetales. Indeed, it probably is premature to standardize taxonomic rankings for bacterial divisions at this point when our picture of microbial diversity is likely still incomplete and the topology of the bacterial tree is still unresolved.
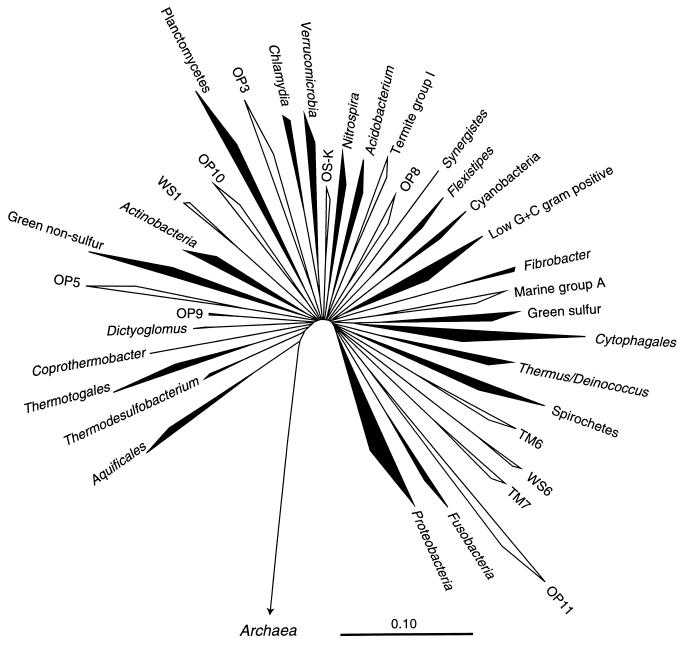
In the past decade the number of identifiable bacterial divisions has more than tripled to about 40 due in significant part to culture-independent phylogenetic surveys of environmental microbial communities (21, 34). These analyses rely on sequences of rRNA genes obtained by cloning directly from environmental DNA or, as in the majority of studies, after amplification by the PCR (1, 20, 35). Figure 1 represents the division-level diversity of the bacterial domain as inferred from representatives of the approximately 8,000 bacterial 16S rRNA gene sequences currently available. Although 36 divisions are shown in Fig. 1, several other division-level lineages are indicated by single environmental sequences (9, 21, 37), suggesting that the number of bacterial divisions may be well over 40. Several of the described divisions are well represented by cultivated strains and were the first to be characterized phylogenetically (51). The majority of the bacterial divisions, however, are poorly represented by cultured organisms. Indeed, 13 of the 36 divisions shown in Fig. 1 are characterized only by environmental sequences (shown outlined) and so are termed “candidate divisions” to indicate their unsubstantiated status as new bacterial divisions (21). One of these candidate divisions, OP11, is now sufficiently well represented by environmental sequences to conclude that it constitutes a major bacterial group (see below). Phylogenetic studies so far have not resolved branching orders of the divisions; bacterial diversity is seen as a fan-like radiation of division-level groups (Fig. 1). The exception to this, however, is the Aquificales division, which branches most deeply in the bacterial tree in most analyses.
FIG. 1.
Evolutionary distance tree of the bacterial domain showing currently recognized divisions and putative (candidate) divisions. The tree was constructed using the ARB software package (with the Lane mask and Olsen rate-corrected neighbor-joining options) and a sequence database modified from the March 1997 ARB database release (43). Division-level groupings of two or more sequences are depicted as wedges. The depth of the wedge reflects the branching depth of the representatives selected for a particular division. Divisions which have cultivated representatives are shown in black; divisions represented only by environmental sequences are shown in outline. The scale bar indicates 0.1 change per nucleotide. The aligned, unmasked data sets used for this figure and Fig. 3 through 6 are available from http://crab2.berkeley.edu/pacelab/176.htm.
BACTERIAL DIVERSITY AND DISTRIBUTION IN THE ENVIRONMENT
Culture-dependent studies indicate that representatives of some bacterial divisions are cosmopolitan in the environment, whereas others appear restricted to certain habitats (39). Culture-independent studies so far conducted reflect and expand this view. Table 1 summarizes the environmental distribution of sequences by habitat type, compiled from most of the available 16S rRNA-based clonal analyses: 86 studies contributing nearly 3,000 sequences. An expanded version of this table that details division-level representation in the individual studies is available at http://crab2.berkeley.edu/pacelab/176.htm. Table 1 includes only divisions for which representatives have been detected in at least two independent studies and for which at least one near-complete 16S rRNA gene sequence is known. Table 1 is, therefore, not an exhaustive listing of potential division-level diversity for all studies.
TABLE 1.
Summary of 16S rRNA-based clonal analyses of diversity of uncultivated bacteriaa
| Habitat type | No. of studies | Bacterial divisionb
|
||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of sequencesc |
Proteobacteriad
|
|||||||||||||||||||||||||||||||||||||
| αc | β | γ | δd | ɛe | Cytophagales | Actinobacteria | Low-G+C gram positivese | Acidobacterium | Verrucomicrobia | Spirochetes | Nitrospira | GNS | OP11 | Planctomycetes | Green sulfur | TM7 | TM6 | Thermus/Deinococcus | Cyanobacteriac | Synergistes | OP8 | Termite group I | OS-K | Chlamydia | OP3 | OP10 | WS1 | OP5 | Marine group A | Fibrobacter | Flexistipes | Dictyoglomus | Thermotogales | Thermodesulfobacterium | Aquificales | |||
| Geothermal | 10 | 212 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||||||||||||||||||||
| Soil | 16 | 743 | • | ○ | ○ | ○ | ○ | • | ○ | • | • | ○ | ○ | |||||||||||||||||||||||||
| Marine | 23 | 687 | • | ○ | • | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ||||||||||||||||||||||||||
| Freshwater | 4 | 107 | • | ○ | ○ | ○ | ||||||||||||||||||||||||||||||||
| Wastewater | 5 | 430 | • | • | • | ○ | ○ | • | ○ | ○ | ○ | ○ | ||||||||||||||||||||||||||
| Pollutant associated | 7 | 202 | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | |||||||||||||||||||||||||||
| Acid metal leaching | 2 | 21 | • | ○ | • | ○ | ○ | ○ | ○ | • | ||||||||||||||||||||||||||||
| Subsurface | 6 | 229 | ○ | • | • | ○ | ○ | ○ | • | ○ | ○ | ○ | ○ | |||||||||||||||||||||||||
| Symbionts and commensals | 10 | 280 | • | ○ | ○ | ○ | ||||||||||||||||||||||||||||||||
| Disease associated | 3 | 7 | ○ | ○ | ○ | ○ | ○ | |||||||||||||||||||||||||||||||
| Totals | 86 | 2,918 | ||||||||||||||||||||||||||||||||||||
An expanded version of this table detailing individual studies is available at http://crab2.berkeley.edu/∼pacelab/176.htm.
Incidence of division-level representatives in studies of particular habitat types ranked from most represented to least represented divisions: >75% (•), 25 to 75% (○), or <25% (no symbol) of studies have representatives of division.
Excluding organelles.
Proteobacteria are presented at the subdivision level due to the extensive sequence representation of this division.
Cannot establish as a monophyletic group in all analyses.
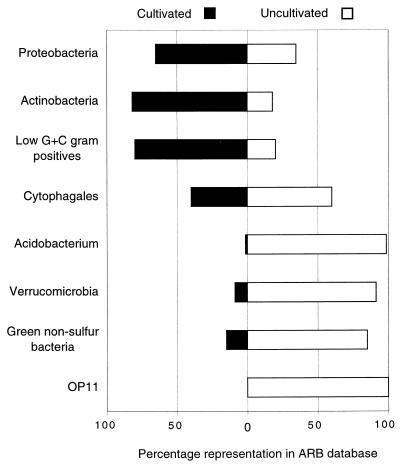
Sequence representatives of several bacterial divisions have been identified in a wide range of habitats, suggesting the cosmopolitan or ubiquitous distribution of the corresponding organisms in the environment and, potentially, their broad metabolic capabilities. Some of these cosmopolitan divisions are well-known from cultivation studies; however, others are little known or have not yet been detected by cultivation. Figure 2 summarizes the representation of selected cosmopolitan divisions by sequences of cultivated and uncultivated organisms. The Proteobacteria (purple photosynthetic bacteria and relatives), Cytophagales (Bacteroides-Cytophaga-Flexibacter group), and the two gram-positive divisions, Actinobacteria and low-G+C gram-positive bacteria, are well represented by cultivated organisms and therefore are familiar to us in principle. These four divisions account for 90% of all cultivated bacteria characterized by 16S rRNA sequences and approximately 70% of the environmental sequences collated in Table 1. By contrast, other cosmopolitan divisions revealed by clonal analyses, such as Acidobacterium, Verrucomicrobia, GNS bacteria, and OP11, are poorly represented by sequences from cultivated organisms (Fig. 2) and consequently are little known with regard to their general properties. Although many of the bacterial divisions occur widely, others seem to occupy a more limited range of habitats (Table 1). All cultivated representatives of Aquificales, for instance, are thermophilic hydrogen metabolizers, and all environmental sequences of Aquificales have been obtained only from high-temperature environments. This suggests a specialized habitat niche for this group. Alternatively, the apparently limited environmental distribution may simply reflect a sampling or methodological artifact and representatives of such divisions may be present in a wider range of habitats, but not yet detected.
FIG. 2.
Relative representation in selected cosmopolitan bacterial divisions of 16S rRNA sequences from cultivated and uncultivated organisms. Results were compiled from 5,224 and 2,918 sequences from cultivated and uncultivated organisms, respectively.
The database of environmental rRNA sequences is compromised in resolving some phylogenetic issues by a large number of relatively short sequences. More than half of the sequences collated in Table 1 are less than 500 nucleotides (nt) long, which represents only one-third of the total length of 16S rRNA. This is due to an unfortunate trend in many environmental studies of sequencing only a portion of the gene in the belief that a few hundred bases of sequence data is sufficient for phylogenetic purposes. Indeed, 500 nt is sufficient for placement if some longer sequence is closely related (>90% identity in homologous nucleotides) to the query sequence. In the case of novel sequences, <85% identical to known sequences, however, <500 nt is usually insufficient comparative information to place the sequence accurately in a phylogenetic tree and can even be misleading.
Since all but 4 (40, 46, 49, 50) of the 86 studies collated in Table 1 were conducted using PCR to amplify rDNA from extracted environmental DNA, the question arises as to whether molecular analyses accurately reflect the division-level diversity that occurs in the environment. It is well established that PCR-associated artifacts such as differential amplification of different rDNA templates (36, 44), sensitivity to rRNA gene copy number (12), PCR primer specificity (48), sensitivity to template concentration (6), amplification of contaminant rDNA (45), and formation of chimeric sequences (23) may skew our assessment of microbial diversity. Most of the studies collated in Table 1, however, analyzed tens to hundreds of clones, so it seems likely that these studies have sampled the main types of sequences in the communities examined. We believe, acknowledging the caveats of the methodology, that the clonal analyses collated in Table 1 probably include the most abundant (metabolically active) bacterial sequence types in the samples analyzed, likely representing the members of the communities that are involved in the principal metabolic activities, such as carbon cycling.
ABUNDANT BUT LITTLE-KNOWN BACTERIAL DIVISIONS
The rRNA sequence studies of environmental organisms probably identify the abundant organisms in the environments studied and, therefore, account for the organisms that participate significantly in the maintenance of the communities. Because of their abundance in the environment, representatives of some poorly studied phylogenetic divisions are predicted to play significant roles in environmental chemistry. Examples of such divisions, which because of their potential environmental significance merit study, are the Acidobacterium division, the Verrucomicrobia, the GNS bacteria, and candidate division OP11.
Acidobacterium division.
The Acidobacterium group is a newly recognized bacterial division with only three cultivated representatives: Acidobacterium capsulatum (18), Holophaga foetida (26), and Geothrix fermentans (28). Figure 3 is a phylogenetic dendrogram of this group, including selected environmental representatives. The limited physiological information known about these organisms provides few clues to properties that might be general throughout the division. Acidobacterium is a moderately acidophilic aerobic heterotroph; Holophaga and Geothrix are strict anaerobes that ferment aromatic compounds and acetate, respectively. The majority of sequences that make up this division, however, are from environmental clones. At least eight monophyletic subdivisions in the Acidobacterium group are identified by phylogenetic analyses (Fig. 3 [24, 29]). We define a subdivision as a lineage comprised of two or more 16S rRNA sequences within a division that are reproducibly monophyletic and unaffiliated with all other representatives of that division. Acidobacterium subdivisions 1, 3, 4, and 6 are well represented by environmental clone sequences from independent studies, yet no cultivated strains are known with the exception of subdivision 1, represented by A. capsulatum. The widespread occurrence of environmental sequences belonging to the Acidobacterium division (Table 1) suggests that members of this group are ecologically significant constituents of many ecosystems, particularly soil communities. They have been detected in every clonal analysis of soils (with a wide range of chemical properties), as well as in other habitats, including a peat bog, acid mine drainage, a contaminated aquifer, a hot spring, a freshwater lake, and a sample of the Atlantic ocean from a depth of 1,000 m (Fig. 3). In situ single-cell analyses with fluorescent hybridization probes specific for Acidobacterium subdivision 6 small-subunit rRNA indicate that this subdivision is morphologically diverse (29), as expected for a broad phylogenetic group. Members likely are metabolically diverse as well: the depth of phylogenetic diversity (depth of branching) in the Acidobacterium division is nearly as great as in the Proteobacteria.
FIG. 3.
Phylogenetic dendrogram of the Acidobacterium division. Names of cultivated organisms are shown in bold. The habitat source of each environmental sequence is indicated before the clone name. GenBank accession numbers are listed parenthetically. Subdivisions (see the text) are indicated by brackets at the right of the tree. Construction of the tree was as described for Fig. 1. The robustness of the topology presented was estimated by bootstrap resampling of independent distance, parsimony, and rate-corrected maximum-likelihood analyses as previously described (2). Distance and parsimony analyses were conducted using test version 4.0d61 of PAUP*, written by David L. Swofford. Branch points supported (bootstrap values of >75%) by most or all phylogenetic analyses are indicated by filled circles; open circles indicate branch points marginally supported (bootstrap values of 50 to 74%) by most or all analyses. Branch points without circles are not resolved (bootstrap values of <50%) as specific groups in different analyses. The scale bar indicates 0.1 change per nucleotide.
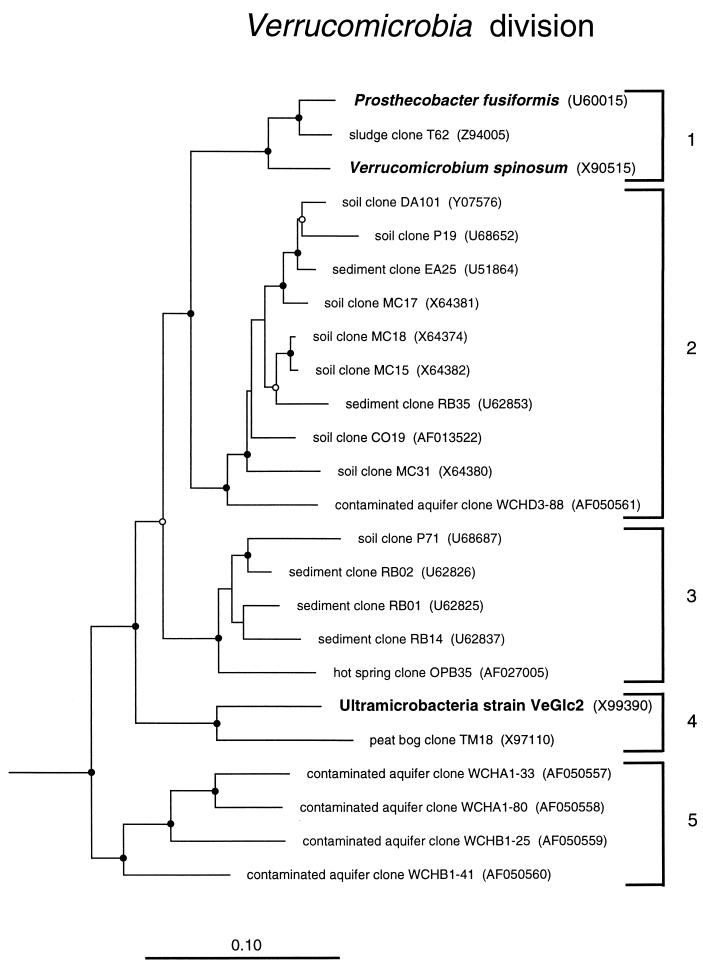
Verrucomicrobia.
Verrucomicrobia is a newly proposed division of Bacteria (17) represented by a handful of isolates: Verrucomicrobium spinosum (after which the division is named) (47), four Prosthecobacter species (17), and three strains of ultramicrobacteria (22). Verrucomicrobia and Prosthecobacter are prosthecate bacteria isolated from freshwater, and the ultramicrobacteria, “dwarf-cell” strains only about 0.1 μm3 in volume, were isolated from a soil habitat. All of these isolates preferentially use sugars as growth substrates. Culture-independent analyses indicate that the Verrucomicrobia, like members of the Acidobacterium division, are widespread in the environment and abundant, particularly in soils (Table 1). Figure 4 shows a dendrogram of representatives of the Verrucomicrobia. Several monophyletic subdivisions are seen, only two of which are represented by the cultivated strains. Clone sequences of this division from soil are predominantly from members of the phylogenetically broad subdivisions 2 and 3. The abundance of these two groups suggests their ecological importance. For instance, the abundance of one representative of Verrucomicrobia subdivision 2 (EA25) was estimated by PCR at 107 to 108 cells per g of a pasture soil sample, 1 to 10% of the total microbial content (25).
FIG. 4.
Phylogenetic dendrogram of the Verrucomicrobia division. Names of cultivated organisms are shown in bold. The habitat source of each environmental sequence is indicated before the clone name. GenBank accession numbers are listed parenthetically. Subdivisions (see the text) are indicated by brackets at the right of the tree. Tree construction and support for branch points was as described for Fig. 1 and 3, respectively. The scale bar indicates 0.1 change per nucleotide.
In our phylogenetic analyses we consistently find that the division Chlamydia is a specific sister group of the Verrucomicrobia. We find no support for the notion (17, 30, 47) of a specific relatedness of the planctomycetes with the Verrucomicrobia.
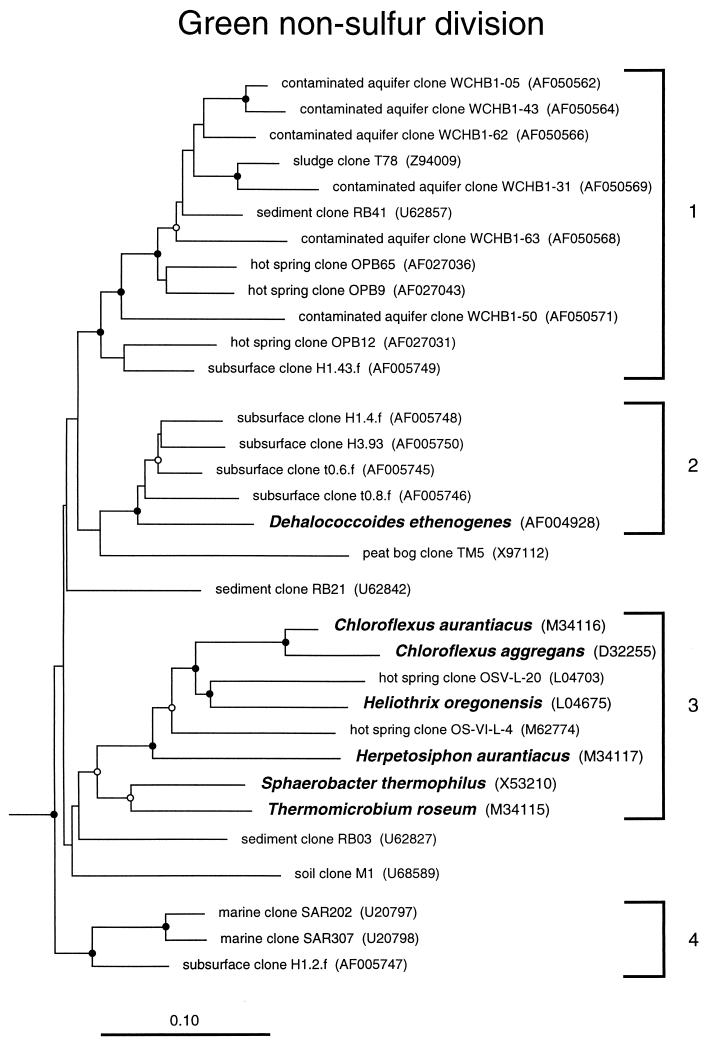
GNS bacteria.
The GNS bacteria have been recognized as a division-level bacterial group for over a decade (51). Even today, however, this division is still represented by only a few isolates. The cultured representatives have a wide range of phenotypes, from anoxygenic photosynthesis (Chloroflexus) to thermophilic organotrophy (Thermomicrobium). Figure 5 shows the relatedness groups of GNS bacteria detected in the environment. It is apparent from the dendrogram that all of the cultivated representatives except the chlorinated hydrocarbon-reducing Dehalococcoides ethenogenes (31) are related in subdivision 3, together with several clone sequences from a hot spring, a rice paddy, and activated sludge (data not shown). By contrast, most of the environmental sequences described to date fall into a different relatedness group, subdivision 1, with no cultivated representatives. Considering the wide variety of habitats that have contributed GNS sequences (Fig. 5; Table 1), particularly to GNS subdivision 1, members of this division likely play significant roles in the environment.
FIG. 5.
Phylogenetic dendrogram of the GNS division. Names of cultivated organisms are shown in bold. The habitat source of each environmental sequence is indicated before the clone name. GenBank accession numbers are listed parenthetically. Subdivisions (see the text) are indicated by brackets at the right of the tree. Tree construction and support for branch points was as described for Fig. 1 and 3, respectively. The scale bar indicates 0.1 change per nucleotide.
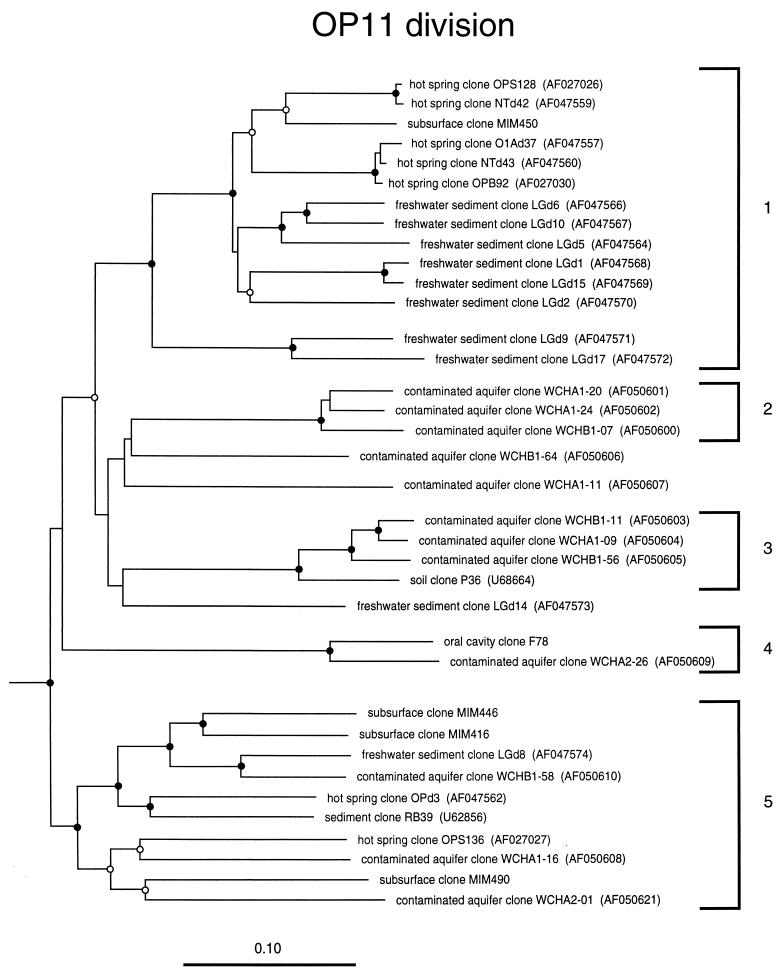
Candidate division OP11.
Candidate division OP11 is a recently proposed novel bacterial division for which there is no reported cultivated representative (19, 21). However, several independent clonal studies have reported environmental sequences that together form the OP11 clade. Figure 6 shows a dendrogram of the known environmental sequence representatives of the division, with five subdivisions currently identifiable. OP11 sequences all have highly atypical sequence signatures for the domain Bacteria (51), and they have low sequence identities, only about 80%, to sequences outside the OP11 division. This may be due to higher-than-average mutation rates in OP11 rRNAs, as has been suggested for other groups such as the planctomycetes (27). OP11 sequences have been obtained from a variety of habitats including several different types of soil, freshwater sediments, the deep subsurface, and hot springs (Table 1), suggesting that members of the division play significant ecological roles. Until cultivated representatives of the OP11 division are characterized, little beyond the general properties of Bacteria can be inferred about their physiology.
FIG. 6.
Phylogenetic dendrogram of the OP11 division. The habitat source of each environmental sequence is indicated before the clone name. GenBank accession numbers are listed parenthetically. Subdivisions (see the text) are indicated by brackets at the right of the tree. Tree construction and support for branch points was as described for Fig. 1 and 3, respectively. The four MIM clones and F78 clone are unreleased sequences generously made available to us by Pascale Durand (10) and Floyd Dewhirst (8). The scale bar indicates 0.1 change per nucleotide.
Additional candidate divisions.
Several additional candidate divisions have been identified based on environmental sequences alone, shown as outlined wedges in Fig. 1. These divisions comprise two or more sequences over 500 nt in length that were obtained mostly from independent studies, or at least from independent PCR events. An expanded view detailing representatives of each candidate division is available at http://crab2.berkeley.edu/pacelab/176.htm. The candidate divisions are identified according to the original source or clone names of the sequences that define the clade. Divisions designated OP were originally identified in an analysis of a Yellowstone hot spring, Obsidian Pool (21). Representatives of three of these divisions, OP5, -8, and -10, also have been encountered in a study of a hydrocarbon-contaminated aquifer at Wurtsmith Air Force Base in Michigan (9). The latter study also identified novel divisions WS1, now identified in a Siberian tundra soil (53), and WS6. Candidate division marine group A was originally identified and named based on partial sequences obtained from marine microbial communities in the Atlantic and Pacific oceans (13) and verified by full-length-sequence representatives of the group from similar marine samples (16). Abundance and depth profiles of marine group A sequences in the water column (16) suggest their global distribution in marine communities; no representatives of this candidate division outside of marine environments have yet been obtained (Table 1). Representatives of the termite group I candidate division originally were identified as a closely related clade of sequences from the termite gut (33) but now also have been identified in a contaminated aquifer (9). Candidate division OS-K was identified in a study of a Yellowstone hot spring, Octopus Spring (49) and bolstered by additional representative sequences from studies of a hydrothermal vent (32) and marine sediment (7). Candidate divisions TM6 and TM7 are named after sequences obtained in an environmental study of a peat bog (38), and other partial-length-sequence representatives of these candidate divisions were subsequently identified from activated sludges (4, 15) and soil (5).
CONCLUSION
Phylogenetic trees based on rRNA sequences show that bacterial diversity is represented by natural relatedness groups, the phylogenetic divisions (51). About 36 such divisions are currently identifiable. The final extent of division-level diversity in the bacterial domain is still unknown but clearly will be more than 40 divisions. Culture-independent studies have resulted in multiple hits on the majority of described divisions in different habitat types (Table 1), suggesting that the final number of divisions will be within the same order of magnitude as the present estimate.
The molecular analyses of environmental DNA have revealed substantial phylogenetic diversity with little or no representation among organisms previously studied. Because of their abundance and wide distribution, some of the organisms represented by the sequences likely contribute significantly to the global chemical cycles. Descriptions of newly identified, but apparently important, bacterial divisions such as the Acidobacterium and Verrucomicrobia, are presently confounded by too few cultivated representatives and only rudimentary descriptions of the strains. Cultivation efforts need to be directed at new representatives of the diverse groups for further study. Continued work to sequence the 16S rDNAs of all deposited type cultures (<50% sequenced to date [14]) may also result in detection of additional cultivated representatives of newly described divisions. It is a challenge to microbial biologists to determine the physiological diversity and environmental roles of these recently articulated divisions of Bacteria.
The phylogenetic differences between the bacterial divisions probably are reflected in substantial physiological differences. Some properties, the general properties of Bacteria, are expected to be distributed among all the divisions. Division-specific novelties are known as well, for instance, endospore formation by the low-G+C gram-positive bacteria or axial filaments (endoflagella) in the spirochetes. Some biochemical properties evidently have transferred laterally among the divisions. For example, the two types of photosynthetic complexes, photosystem I (PSI) and PSII, are each distributed sporadically among the divisions, consistent with lateral transfer (3). Lateral transfer may also have resulted in combinatorial novelty among the divisions; PSI and PSII, for instance, apparently came together in the cyanobacteria to create oxygenic photosynthesis, with profound consequences to the biosphere (3). Many more such division-specific qualities and cooperations should become evident at the molecular level as comparative genomics gives us a sharper phylogenetic picture of bacterial diversity.
ACKNOWLEDGMENTS
We thank John A. Fuerst for providing useful comments on the manuscript and Pascale Durand, Floyd Dewhirst, and Bruce Paster for supplying unreleased sequences. We also thank Michael Tanner and Scott Dawson for assistance in establishing the website of additional information.
The authors’ research is supported by grants to N.R.P. from the National Institutes of Health and Department of Energy.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blankenship R E. Origin and early evolution of photosynthesis. Photosynth Res. 1992;33:91–111. [PubMed] [Google Scholar]
- 4.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler D P, Fredrickson J K, Brockman F J. Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol Ecol. 1997;6:475–482. doi: 10.1046/j.1365-294x.1997.00205.x. [DOI] [PubMed] [Google Scholar]
- 7.Devereux R, Mundfrom G W. A phylogenetic tree of 16S rRNA sequences from sulfate-reducing bacteria in a sandy marine sediment. Appl Environ Microbiol. 1994;60:3437–3439. doi: 10.1128/aem.60.9.3437-3439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dewhirst, F. 1998. Personal communication.
- 9.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 10.Durand, P. 1998. Personal communication.
- 11.Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E. A new obligately chemolithoautotrophic, nitrate-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship. Arch Microbiol. 1995;164:16–23. doi: 10.1007/BF02568729. [DOI] [PubMed] [Google Scholar]
- 12.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrity G M, Tiedje J, Searles D. Report on a workshop on the phylogeny of prokaryotes based upon sequence similarity of the small ribosomal subunit. 1998. Unpublished report. [Google Scholar]
- 15.Godon J-J, Zumstein E, Dabert P, Habouzit F, Moletta R. Molecular microbial diversity of an anaerobic digester as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol. 1997;63:2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon D A, Giovannoni S J. Detection of stratified microbial populations related to Chlorobium and Fibrobacter in the Atlantic and Pacific oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedlund B P, Gosink J J, Staley J T. Verrucomicrobia div. nov., a new division of the Bacteria containing three new species of Prosthecobacter. Antonie Leeuwenhoek. 1997;72:29–38. doi: 10.1023/a:1000348616863. [DOI] [PubMed] [Google Scholar]
- 18.Hiraishi A, Kishimoto N, Kosako Y, Wakao N, Tano T. Phylogenetic position of the menaquinone-containing acidophilic chemoorganotroph Acidobacterium capsulatum. FEMS Microbiol Lett. 1995;132:91–94. doi: 10.1111/j.1574-6968.1995.tb07816.x. [DOI] [PubMed] [Google Scholar]
- 19.Hugenholtz P, Hershberger K L, Flanagan J L, Kimmel B, Pace N R. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Widespread distribution of a novel phylum-depth bacterial lineage in nature, abstr. N-23; p. 385. [Google Scholar]
- 20.Hugenholtz P, Pace N R. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 1996;14:190–197. doi: 10.1016/0167-7799(96)10025-1. [DOI] [PubMed] [Google Scholar]
- 21.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janssen P H, Schuhmann A, Morschel E, Rainey F A. Novel anaerobic ultramicrobacteria belonging to the Verrucomicrobiales lineage of bacterial descent isolated by dilution culture from anoxic rice paddy soil. Appl Environ Microbiol. 1997;63:1382–1388. doi: 10.1128/aem.63.4.1382-1388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kopczynski E D, Bateson M M, Ward D M. Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl Environ Microbiol. 1994;60:746–748. doi: 10.1128/aem.60.2.746-748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuske C R, Barns S M, Busch J D. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl Environ Microbiol. 1997;63:3614–3621. doi: 10.1128/aem.63.9.3614-3621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S-Y, Bollinger J, Bezdicek D, Ogram A. Estimation of the abundance of an uncultured soil bacterial strain by a competitive quantitative PCR method. Appl Environ Microbiol. 1996;62:3787–3793. doi: 10.1128/aem.62.10.3787-3793.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liesack W, Bak F, Kreft J U, Stackebrandt E. Holophaga foetida gen. nov., sp. nov., a new homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch Microbiol. 1994;162:85–90. doi: 10.1007/BF00264378. [DOI] [PubMed] [Google Scholar]
- 27.Liesack W, Soller R, Stewart T, Haas H, Giovannoni S, Stackebrandt E. The influence of tachytelically (rapidly) evolving sequences on the topology of phylogenetic trees—intrafamily relationships and the phylogenetic position of Planctomycetaceae as revealed by comparative analysis of 16S ribosomal RNA sequences. Syst Appl Microbiol. 1992;15:357–362. [Google Scholar]
- 28.Lonergan D J, Jenter H L, Coates J D, Phillips E J P, Schmidt T, Lovely D R. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K-H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 30.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maymo-Gatell X, Chien Y-T, Gossett J, Zinder S. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- 32.Moyer C L, Dobbs F C, Karl D M. Phylogenetic diversity of the bacterial community from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1995;61:1555–1562. doi: 10.1128/aem.61.4.1555-1562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohkuma M, Kudo T. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:461–468. doi: 10.1128/aem.62.2.461-468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 35.Pace N R, Stahl D A, Lane D J, Olsen G J. The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol. 1986;9:1–55. [Google Scholar]
- 36.Reysenbach A-L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reysenbach A-L, Wickham G S, Pace N R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rheims H, Rainey F A, Stackebrandt E. A molecular approach to search for diversity among bacteria in the environment. J Ind Microbiol. 1996;17:159–169. [Google Scholar]
- 39.Schlegel H G, Jannasch H W. Prokaryotes and their habitats. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. I. New York, N.Y: Springer-Verlag; 1992. pp. 75–125. [Google Scholar]
- 40.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stackebrandt E, Murray R G E, Truper H G. Proteobacteria classis nov., a name for the phylogenetic taxon that includes the “purple bacteria and their relatives.”. Int J Syst Bacteriol. 1988;38:321–325. [Google Scholar]
- 42.Stackebrandt E, Rainey F A, Ward-Rainey N L. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–491. [Google Scholar]
- 43.Strunk O, Gross O, Reichel B, May M, Hermann S, Struckmann N, Nonhoff B, Lenke M, Vilbig A, Ludwig T, Bode A, Schleifer K H, Ludwig W. ARB: a software environment for sequence data. 1998. Submitted for publication. [Google Scholar]
- 44.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanner M, Goebel B M, Dojka M A, Pace N R. Specific rDNA sequences from diverse environmental settings correlate with experimental contaminants. 1998. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ward D M, Weller R, Bateson M M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 47.Ward-Rainey N, Rainey F A, Schlesner H, Stackebrandt E. Assignment of hitherto unidentified 16S rDNA species to a main line of descent within the domain Bacteria. Microbiology. 1995;141:3247–3250. [Google Scholar]
- 48.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weller R, Bateson M M, Heimbuch B K, Kopczynski E D, Ward D M. Uncultivated cyanobacteria, Chloroflexus-like inhabitants, and spirochete-like inhabitants of a hot spring microbial mat. Appl Environ Microbiol. 1992;58:3964–3969. doi: 10.1128/aem.58.12.3964-3969.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weller R, Weller J, Ward D M. 16S rRNA sequences of uncultivated hot spring cyanobacterial mat inhabitants retrieved as randomly primed cDNA. Appl Environ Microbiol. 1991;57:1146–1151. doi: 10.1128/aem.57.4.1146-1151.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woese C R, Winker S, Gutell R R. Architecture of ribosomal RNA: constraints on the sequence of tetra-loops. Proc Natl Acad Sci USA. 1990;87:8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou J, Davey M E, Figueras J B, Rivkina E, Gilichinsky D, Tiedje J M. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology. 1997;143:3913–3919. doi: 10.1099/00221287-143-12-3913. [DOI] [PubMed] [Google Scholar]