Summary
Background
The impact of previous vaccination on protective immunity, duration, and immune imprinting in the context of BA.5-XBB.1.9.1 reinfection remains unknown.
Methods
Based on a 2-year longitudinal cohort from vaccination, BA.5 infection and XBB reinfection, several immune effectors, including neutralizing antibodies (Nabs), antibody-dependent cellular cytotoxicity (ADCC), virus-specific T cell immunity were measured to investigate the impact of previous vaccination on host immunity induced by BA.5 breakthrough infection and BA.5-XBB.1.9.1 reinfection.
Findings
In absence of BA.5 Nabs, plasma collected 3 months after receiving three doses of inactivated vaccine (I-I-I) showed high ADCC that protected hACE2-K18 mice from fatality and significantly reduced viral load in the lungs and brain upon BA.5 challenge, compared to plasma collected 12 months after I-I-I. Nabs against XBB.1.9.1 induced by BA.5 breakthrough infection were low at day 14 and decreased to a GMT of 10 at 4 months and 28% (9/32) had GMT ≤4, among whom 67% (6/9) were reinfected with XBB.1.9.1 within 1 month. However, 63% (20/32) were not reinfected with XBB.1.9.1 at 5 months post BA.5 infection. Interestingly, XBB.1.9.1 reinfection increased Nabs against XBB.1.9.1 by 24.5-fold at 14 days post-reinfection, which was much higher than that against BA.5 (7.3-fold) and WT (4.5-fold), indicating an immune imprinting shifting from WT to XBB antigenic side.
Interpretation
Overall, I-I-I can provide protection against BA.5 infection and elicit rapid immune response upon BA.5 infection. Furthermore, BA.5 breakthrough infection effectively protects against XBB.1.9.1 lasting more than 5 months, and XBB.1.9.1 reinfection results in immune imprinting shifting from WT antigen induced by previous vaccination to the new XBB.1.9.1 antigen. These findings strongly suggest that future vaccines should target variant strain antigens, replacing prototype strain antigens.
Funding
This study was supported by R&D Program of Guangzhou National Laboratory (SRPG23-005), National Key Research and Development Program of China (2022YFC2604104, 2019YFC0810900), S&T Program of Guangzhou Laboratory (SRPG22-006), and National Natural Science Foundation of China (81971485, 82271801, 81970038), Emergency Key Program of Guangzhou Laboratory (EKPG21-30-3), Zhongnanshan Medical Foundation of Guangdong Province (ZNSA-2020013), and State Key Laboratory of Respiratory Disease (J19112006202304).
Keywords: SARS-CoV-2, Vaccinated-BA.5-XBB.1.9.1 reinfections, Immune imprinting, Protection-duration
Research in context.
Evidence before this study
Three doses of inactivated vaccines do not effectively induce Nabs against BA.5. However, it has been reported that individuals who received three doses of inactivated vaccine exhibited high efficacy against severe/critical COVID-19 caused by BA.2 infection after 4–6 months. Our previous study also demonstrated a gradual decline in vaccine-induced immunity over time. The effectiveness of inactivated vaccines against BA.5 infection after 1 year is currently unknown, which may help to explain why most of Chinese individuals experienced symptomatic infections during the BA.5 pandemic in December, 2022. Furthermore, the impact of inactivated vaccination on protective immunity, duration, and immune imprinting in the context of BA.5 infection and BA.5-XBB.1.9.1 reinfection requires further investigation.
Added value of this study
We conducted a 2-year longitudinal cohort study to investigate the impact of previous vaccination on host immunity induced by BA.5 breakthrough infection and BA.5-XBB.1.9.1 reinfection. We demonstrate the efficiency and duration of BA.5 breakthrough infection against XBB.1.9.1 infection. Furthermore, we indicate that XBB.1.9.1 reinfection results in immune imprinting shifting from WT antigen induced by previous vaccination to the new XBB.1.9.1 antigen.
Implications of all the available evidence
Our findings demonstrate the profound impact of vaccination-infection deeply on immune imprinting, and suggest future vaccines should target variant strain antigens, replacing prototype strain antigens.
Introduction
Since 2022, BA.5.2 and BF.7 have remained the dominant circulating Omicron strains in China, rapidly spreading across the country and infecting >80% of the population within 1 month. In China, approximately 89% of the population received two doses of inactivated SARS-CoV-2 vaccines (CoronaVac or BBIBP-CorV), with 71% receiving a third dose. A retrospective cohort study demonstrated that the real-world efficacy of three doses of inactivated vaccine against BA.2 in adults aged >18 years was 74% against pneumonia or worse and 93% against severe/critical COVID-19.1 Another case-control cohort study showed that the adjusted vaccine effectiveness (VE) of three-dose inactivated vaccines was 48.0% (95% confidence interval [CI]: 8.0–70.6%) against BA.2.2 infection.2 These results indicate that three doses of inactivated vaccines with low neutralizing antibody (Nab) titers could provide effective protection against BA.2 variants in the real world.3,4 Notably, it has been about 4–6 months since most people received their third dose of inactivated vaccines, and the BA.5 pandemic in China emerged in December 2022, meaning that it has been >12 months since the last inactivated vaccine booster. Therefore, it is essential to investigate whether an inactivated vaccine booster provides the same level of protection against BA.5 infection after 12 months as it does after 3 months.
Moreover, the inactivated COVID-19 vaccine was designed for the prototype SARS-CoV-2 virus, and the emerging variant BA.5 has antigens significantly different from the prototype.5, 6, 7 It has been reported that both three doses of inactivated vaccines and three doses of mRNA cannot effectively induce Nabs against BA.5.8,9 Thus, other immune effectors except Nabs must be involved in combating the new variant BA.5. Virus-specific IgG antibodies which play a critical role in stimulating antibody dependent cellular cytotoxicity (ADCC) and T cell responses have been shown to be essential components of the host immune responses against SARS-CoV-2 infection.10,11 Therefore, it is crucial to study the role of virus-specific ADCC and T cell responses in providing protection against the new variant BA.5.
The duration of population immunity established after BA.5.2 and BF.7 infection and its protection against new emerging strains, XBB and BQ1.1, remain uncertain. Epidemiological studies have suggested that hybrid immunity, resulting from a combination of vaccine and infection-induced immunity, may confer stronger and more durable protection against infections compared to vaccine- or infection-elicited immunity alone.12,13 Ma et al. reported that hybrid immunity and BA.1 breakthrough infection elicited neutralization for up to 4 months.14 Similarly, Wheatley et al. found that neutralization against the ancestral VIC 01 virus through BA.2 breakthrough infection was maintained over a 3–4-month period.15 While, from the end of April to the middle of May, XBB.1.9.1 variant infections surged rapidly, initially among individuals who had not previously experienced BA.5 infection, accounting for approximately 75% of cases. The increase in XBB reinfections indicated that the protective immunity derived from vaccine-infection is weakening. Thus, investigating the underlying mechanisms responsible for the chronological decay of protective immune effectors will be interesting.
Throughout the COVID-19 pandemic, individuals have acquired various types of immunity to the SARS-CoV-2 virus via either vaccination or infection based on the strains they have been exposed to, influencing the subsequent pattern of immunity. Immune imprinting which describes how the first exposure to a virus shapes the immunological outcome of subsequent exposures to antigenically related strains has been demonstrated among COVID-19 infection and vaccination based on Wuhan strain.16, 17, 18 A long-term study among UK healthcare workers who were triple-vaccinated and infected with Wuhan strain revealed that Omicron infection boosted immune responses to all other variants, but failed to boost Nabs and T cell responses against Omicron,19 indicating a profound imprinting effect that could impact the emerging variant types and outcomes of subsequent infections. This may be the reason for the breakthrough infection and reinfection of emerging new variants.
In this 2-year dynamic longitudinal study, we described the immune landscape from two doses of vaccine to BA.5 breakthrough infection and then to XBB.1.9.1 reinfection, investigating the immune effectors involved in each phase against subsequent infections and their protective duration. Our study also highlighted the dynamic kinetics of immune imprinting with changes in the vaccination-infection-reinfection pattern, focusing on how XBB reinfection could shift immune imprinting. These findings provide clear guidance for further vaccine design and pandemic preparedness.
Methods
Study design
The aim of this study was to evaluate the effect of three doses of inactivated vaccines (CoronaVac or BBIBP-CorV) against BA.5 infection. Individuals who received three doses of inactivated vaccines (I-I-I) between September and December 2021 at Guangzhou First People’s Hospital were recruited in this study. I-I-I individuals were excluded if they were previously infected with SARS-CoV-2. The I-I-I group consisted of 135 participants with a median age of 39 years (IQR: 21–60) and 177 plasma samples were collected from them at various time points after the third vaccination, and 107 participants (79%) in I-I-I group were females. Upon BA.5 pandemic in China at December 2022, 33 vaccinated-infected individuals who were diagnosed with SARS-CoV-2 infection through positive antigen testing with a median age of 29 (IQR: 24–45) were enrolled in this study (Table 1). The blood was collected at different time points (day 0, day 3, day 7, day 14, 1 month, 3 month and 4 month) post BA.5 breakthrough infection. As samples were collected at various time points post BA.5 infection, the timing of infection diagnosis may introduce bias when comparing the results across different time points. Therefore, we designated the day on which symptoms manifested and a positive antigen test was obtained as day 0. Meanwhile, to determine the impact of vaccination on the immunogenicity of BA.5 infection, a comparison was made between the neutralizing antibody titers of vaccinated and unvaccinated individuals who were infected with BA.5. Thus, 17 unvaccinated-infected individuals with a median age of 65 (IQR: 33–91) were also enrolled in this study. Thereafter, a longitudinal observation of the vaccinated-infected individuals was conducted upon XBB.1.9.1 reinfection until the end of June 2023. Among them, 16 individuals had been longitudinally followed up for two years since April 2021, starting with their first dose of vaccination.
Table 1.
Information of BA.5 breakthrough infected individuals.
| Patients | Gender/age | Symptomatic |
Re-infection | |||||
|---|---|---|---|---|---|---|---|---|
| Fever | Cough | Nasal congestion | Sore throat | Muscle soreness | Tiredness | |||
| P1 | F/27 | + | + | − | + | − | − | + |
| P2 | F/26 | + | − | − | − | − | − | − |
| P3 | F/27 | + | − | − | − | + | + | + |
| P4 | F/28 | + | + | − | − | + | − | − |
| P5 | F/27 | + | − | − | + | + | − | + |
| P6 | F/26 | + | + | − | − | + | − | − |
| P7 | F/27 | − | + | + | − | − | − | + |
| P8 | M/29 | + | − | + | − | − | − | + |
| P9 | M/33 | − | + | + | + | + | − | − |
| P10 | F/27 | + | + | + | + | − | − | − |
| P11 | F/29 | + | − | − | + | + | − | − |
| P12 | F/35 | − | + | + | + | − | − | − |
| P13 | M/31 | + | + | + | + | − | − | − |
| P14 | F/26 | + | + | + | + | − | − | − |
| P15 | F/26 | + | + | − | + | − | − | + |
| P16 | F/27 | + | + | + | + | + | − | − |
| P17 | F/30 | + | + | + | + | − | − | − |
| P18 | M/28 | + | − | + | + | − | − | − |
| P19 | F/25 | + | + | + | + | + | − | − |
| P20 | M/45 | − | + | + | + | − | − | − |
| P21 | M/30 | + | − | − | − | + | − | + |
| P22 | M/33 | + | + | − | + | + | − | − |
| P23 | F/27 | + | + | + | + | − | − | / |
| P24 | F/28 | + | + | + | + | − | − | + |
| P25 | M/26 | + | + | − | − | − | − | − |
| P26 | M/29 | + | + | − | + | + | − | − |
| P27 | M/24 | + | + | + | − | + | + | + |
| P28 | M/26 | + | + | − | + | + | − | − |
| P29 | M/28 | + | + | + | + | − | + | − |
| P30 | F/41 | + | − | − | + | + | + | − |
| P31 | M/33 | + | + | + | + | − | + | + |
| P32 | F/28 | + | + | − | + | − | + | + |
| P33 | F/34 | + | + | + | − | + | + | + |
+: positive; −: negative; /: not tested.
This study was approved and monitored by the GMUH Ethics Committee (No. 2021-78).
Mice
6–8-week-old K18-ACE2 mice (on C57BL/6 background) were used for challenge study and were purchased from GemPhamatech.
SARS-CoV-2 conventional virus neutralization test
The neutralizing activity of plasma was evaluated using a cytopathic effect (CPE)-based assay, as described previously.3 Two-fold serially diluted plasma samples were combined with a SARS-CoV-2 Wuhan-1 (WT) or Omicron BA.5 or XBB.1.9.1 viral solution, respectively, containing 100 50% tissue culture infective dose (TCID50) of the virus, in a 96-well plate, and incubated for 2 h at 37 °C and 5% CO2. Next, 1.2 × 104 Vero E6 cells were added, and the plates were incubated for 4 days at 37 °C with 5% CO2, and examined for CPE using the Celigo Imaging Cytometer (Nexcelom Bioscience, Lawrence, MA, USA).
Sera transfers and SARS-CoV-2 BA.5 infection
In order to determine if plasma from individuals who received three doses of inactivated vaccines at three-month (I-I-I 3 M) and twelve-month (I-I-I 12 M) post the third shot could provide protection against BA.5 infection, 200 μL plasma collected from I-I-I 3 M to I-I-I 12 M participants was intravenously injected into K18-hACE2 mice, respectively, while PBS was used as a control. Five hours after the transfer, the mice were anesthetized with isoflurane and challenged with 104 focus forming units (FFU) of BA.5 intranasally. The survival rate was monitored post challenge. Meanwhile, the mice in each group were sacrificed every two days and viral loads from lung and brain were measured by focus forming assay (FFA), and the cytokines from the lung were tested with cytokines & chemokines PCR array plate according to the manufacturer’s instruction (Wogene Biotech, Shanghai, China). The mice infections were conducted in Biosafety Level 3 (BSL-3) laboratories of Guangzhou Customs District Technology Center.
Focus forming assay (FFA)
The FFA was performed to determine the viral titer using a previously described protocol.16 Lung or brain homogenates were serially diluted and used to infect Vero E6 cells at 37 °C for 1 h. After removal of the inoculum, 125 μL of 1.6% carboxymethylcellulose warmed to 37 °C was added to each well. After 24 h, the cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. The cells were then incubated with a polyclonal antibody against the SARS-CoV-2 nucleocapsid protein (40143-T62, Sino Biological), followed by a secondary antibody labeled with HRP (109-035-088, Jackson ImmunoResearch Laboratories). The foci were visualized using TrueBlue Peroxidase Substrate (KPL, Gaithersburg, MD) and counted using an ELISPOT reader (Cellular Technology Ltd., Cleveland, OH). The viral titer was calculated as FFU per mL.
ADCC NK cell activation assay
A virus-specific assay for ADCC was conducted on I-I-I plasma samples.17,18 96-well plates were coated with either the SARS-CoV-2 N protein or S protein. Diluted plasma (1:10) collected from 75 individuals from different time points post the third inactivated vaccine were incubated with coated proteins for 2 h at 37 °C. After removing unbound antibodies through wash cycles, peripheral blood mononuclear cells (PBMC) from a healthy donor were added and incubated for 5 h at 37 °C with 5% CO2 in presence of anti-human CD107a APC (Table 2), brefeldin (BD bioscience, San Diego, CA), and monensin (BD bioscience, San Diego, CA). PBMC were then stained with surface antibodies (Table 2), fixed, permeabilized and further incubated with anti-human IFNγ PE antibody (Table 2). Data acquisition was performed using the Verse flow cytometer (BD bioscience, San Diego, CA) and analyzed with FlowJo software (Treestar).
Table 2.
Antibodies used in ADCC assay and flow cytometry.
| Target | Fluorochrome | Clone | Dilution | Vendor | RRID |
|---|---|---|---|---|---|
| IFNγ | PE | B27 | 1:150 | BD Biosciences | AB_395518 |
| CD3 | FITC | UCHT1 | 1:200 | BioLegend | AB_314060 |
| CD3 | BUV395 | SK7 | 1:200 | BD Biosciences | AB_2744382 |
| CD4 | APC-H7 | RPA-T4 | 1:200 | BD Biosciences | AB_1645478 |
| CD4 | PerCP/Cyanine5.5 | RPA-T4 | 1:200 | Biolegend | AB_893328 |
| CD8 | PerCP/Cyanine5.5 | RPA-T8 | 1:200 | BD Biosciences | AB_1727513 |
| CD8 | FITC | HIT8a | 1:200 | BD Biosciences | AB_395996 |
| CD45RA | Alexa Fluor 700 | HI100 | 1:150 | BD Biosciences | AB_1727496 |
| CCR7 | APC | G043H7 | 1:150 | Biolegend | AB_10917387 |
| TNFα | PE-Cy7 | MAb11 | 1:150 | BD Biosciences | AB_396764 |
| IFNγ | APC | B27 | 1:150 | BD Biosciences | AB_398580 |
| IFNγ | BV786 | 4S.B3 | 1:150 | Biolegend | AB_11219192 |
| CD107a | APC | H4A3 | 1:150 | Biolegend | AB_1279057 |
PBMC isolation and ex vivo stimulation
PBMCs were isolated from heparinized whole blood using Ficoll–Paque (GE Healthcare, Singapore). Then, PBMCs from I-I-I were treated with the peptide pool containing 384 15-mer peptides spanning the antigen region of spike (S) protein (250 nM per peptide) in presence of 10 U/mL recombinant interleukin-2 (rIL-2) and 1 μM GolgiPlug (BD Biosciences, San Diego, CA) for 16 h at 37 °C, 5% CO2. PBMCs from BA.5 infection were treated with the peptide pool containing 487 15-mer peptides spanning the antigen region of spike (S), membrane (M), nucleocapsid (N), and envelope (E) proteins (250 nM per peptide) in presence of 10 U/mL rIL-2 and 1 μM GolgiPlug (BD Biosciences, San Diego, CA) for 16 h at 37 °C, 5% CO2. RPMI 1640 medium (Gibco, Waltham, MA) supplemented with 10% heat-inactivated FBS (Biological Industries, Israel Beit-Haemek), 100 U/mL penicillin (Gibco, Waltham, MA), 0.1 mg/mL streptomycin (Gibco, Waltham, MA), 10 U/mL rIL-2, and 0.01% DMSO (Sigma, Saint Louis, MO) was used as subtraction control.
In vitro PBMC expansion, cultivation and stimulation
PBMCs from I-I-I were treated with SARS-CoV-2 peptide pool (250 nM per peptide) and incubated for 10 days at 37 °C, 5% CO2. During cultivation, half of the medium was replaced every three days with fresh RPMI 1640 medium (Gibco, Waltham, MA) supplemented with 10% heat-inactivated FBS (Biological Industries, Israel Beit-Haemek), 100 U/mL penicillin (Gibco, Waltham, MA), 0.1 mg/mL streptomycin (Gibco, Waltham, MA) and 10 U/mL rIL-2. On day 10, cells were re-stimulated with peptides for 16 h at 37 °C, 5% CO2 and then stained for FACS analysis.
Flow cytometry
Cells harvested from ex vivo or in vitro stimulation were incubated with Live/dead aqua V510 or FVS440 for 15 min on ice. Then surface-staining was performed for 30 min on ice with antibodies (Table 2). Following fixation/permeabilization with Cytofix and Perm (BD Bioscience, Cat# 554714), cells were stained with intracellular antibodies (Table 2). Finally, cells were resuspended in FACS buffer, acquired using a FACSAria III instrument (BD Bioscience, San Diego, CA) and analyzed with FlowJo software (Treestar).
Statistical analysis
All statistical analysis was performed using GraphPad Prism version 6.0 (GraphPad Software). Differences were considered significant if P < 0.05. The Mann–Whitney test was employed for comparing central tendencies of two groups for ADCC response and Nab titers analysis. Antibody titers are reported as GMT (geometric mean titer) with 95% CI. Linear regression was used for the correlation analysis between anti-IgG antibody titers and ADCC response. Kruskal–Wallis test was involved to compare differences between different time points for T cell response.
Role of the funding source
The funder of the study had no role in the study design, data collection, data analysis and data interpretation. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
I-I-I plasma without detectable BA.5 Nabs can facilitate the clearance of BA.5 infection in hACE2-K18 mice within a certain period
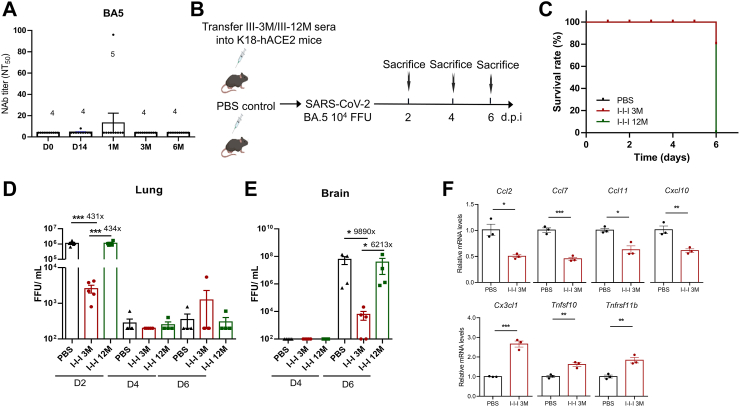
We initially measured the levels of BA.5 Nabs and found that even 14 days after the third inactivated vaccination, BA.5 Nabs remained undetectable (Fig. 1A). To investigate whether other immune factors, except Nabs, can provide protection against BA.5 infection and determine the duration of this protection, we intravenously transferred 200 μL of plasma collected 3 months (I-I-I 3 M group) and 12 months (I-I-I 12 M group) after the booster shot into naive K18-hACE2 mice. Five hours later, the mice were intranasally challenged with BA.5 (Fig. 1B). Surprisingly, 80% of mice in the I-I-I 3 M group survived at day 6 (D6) post-challenge, while all mice in the I-I-I 12 M and PBS groups deceased (Fig. 1C). To investigate whether the protective effect was attributed to direct viral clearance or the reduction of inflammation, we tested the virus titer and cytokine levels in the lungs and brains at indicated time points. The results revealed that the virus titer in the lung of I-I-I 3 M group at day 2 (D2) was 434-fold and 431-fold lower than that in the I-I-I 12 M and PBS groups, respectively (Fig. 1D). Additionally, at D6 post challenge, the virus titer in the brains of the I-I-I 3 M group was significantly lower than that of the I-I-I 12 M (6213-fold) and PBS groups (9890-fold) (Fig. 1E). Moreover, the I-I-I 3 M group exhibited higher levels of antiviral cytokines such as Tnfrsf11b, Tnfsf10, and Cx3cl1, along with lower levels of inflammatory factors, including Ccl2, Ccl7, Ccl11, and Cxcl10 compared to the PBS group (Fig. 1F). Our results clearly demonstrate that I-I-I plasma can induce significant in vivo antiviral protection, rather than adverse effects. However, the protective effect is limited by its duration and can last for at least 3 months.
Fig. 1.
The impact of I-I-I plasma collected at 3 M and 12 M after the third dose of inactivated vaccine on BA.5 infection in K18-hACE2mice. (A) Longitudinal profile of neutralizing antibody titers against Omicron BA.5 measured using conventional virus neutralization test for I-I-I plasma (n = 10). (B) Overview of the mice study design. Plasma collected from individuals at I-I-I 3 M and I-I-I 12 M was intravenously injected into K18-hACE2 mice, while PBS was used as a control. Then, the mice were intranasally challenged with BA.5 and sacrificed at 2, 4, and 6 d.p.i (n = 4 for I-I-I 12 M group and PBS group per timepoint, n = 5 for I-I-I 3 M group per timepoint). (C) Survival rate of challenged mice in I-I-I 3 M, I-I-I 12 M, and PBS groups. (D) Virus titers in the lungs monitored at 2, 4, and 6 d.p.i. (E) Virus titers in the brain monitored at 4 and 6 d.p.i. (F) Selected cytokines and chemokines differentially regulated in I-I-I 3 M and PBS groups at 2 d.p.i. The Mann–Whitney test was employed for comparing central tendencies of every two groups, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
ADCC response induced by I-I-I can originate from both anti-S and anti-N antibodies and lasts longer than 6 months
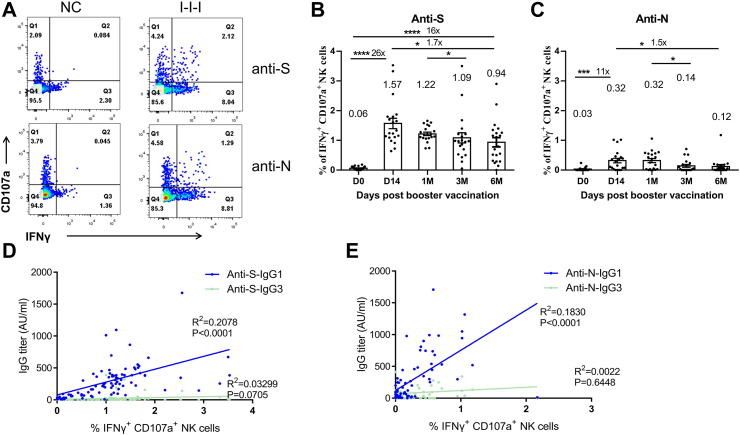
We confirmed the presence of humoral immune effectors other than BA.5 Nabs (Fig. 1) and proceeded to investigate whether ADCC contributes to the observed protective effect and its duration. We conducted ADCC assays based on anti-S and anti-N antibodies at different time points (n = 20 per time point). Both anti-S and anti-N antibodies were found to induce high levels of ADCC responses (Fig. 2A). The ADCC response induced by anti-S antibodies was observed on day 14 (D14) after the booster vaccination and remained relatively high even 6 months (6 M) after the booster shot (Fig. 2B). The ADCC response induced by anti-N antibodies peaked at D14 after the booster shot and remained at the same level for 1 month (1 M), but significantly reduced to 0.12% (95% CI 0–0.24%) of IFN-γ+ CD107a+ NK cells at 6 M (Fig. 2C).
Fig. 2.
Long-lasting ADCC activation in I-I-I plasma against S and N proteins. Longitudinal plasma samples from I-I-I individuals were utilized for the ADCC assay. (A) The ADCC response against S and N proteins was detected in I-I-I plasma. Kinetics of ADCC response against S (B) and N proteins (C) over time (n = 20 per time point). Correlation analysis between anti-S ADCC (D) or anti-N ADCC (E) and IgG 1 and IgG 3 titers. The Mann–Whitney test was employed for comparing central tendencies of every two groups, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
To assess which IgG subclass induced by I-I-I contributes to ADCC, we measured the levels IgG1 and IgG3 and analyzed the correlations between IgG titers and ADCC responses. Our results revealed that both the anti-S and anti-N ADCC responses were significantly associated with IgG1 titers, while the association with IgG3 titers was less strong (Fig. 2D and E).
I-I-I induced virus-specific CD4+ T cell can proliferate efficiently upon in vitro peptide pool stimulation even at 12 months post-vaccination
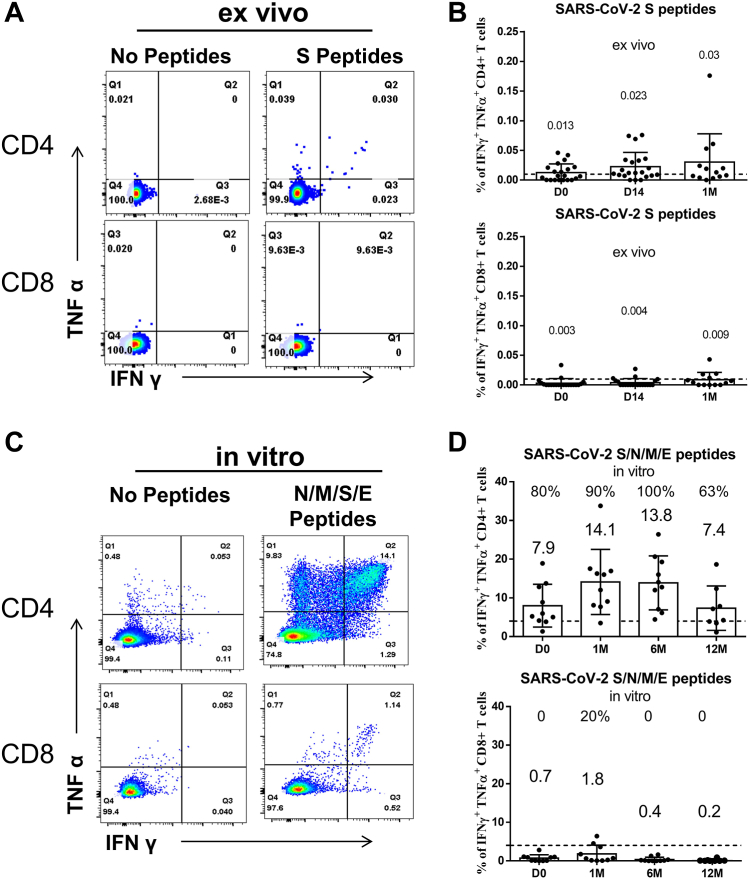
In addition to antibody response, T cell immunity also plays an important antiviral role in SARS-CoV-2.20, 21, 22, 23 We observed that I-I-I can induce a considerable virus-specific CD4+ T cell response, but the levels of these CD4+ T cells by ex vivo assay were not high, even at D14 (0.023% of IFNγ+TNFα+ CD4+ T cells, n = 21) and 1 M (0.03% of IFNγ+TNFα+ CD4+ T cells, n = 13) after vaccination (Fig. 3A and B). To investigate the longevity of these low virus-specific CD4+ T cells and their ability to efficiently proliferate upon encountering the antigen again, such as during infection, we measured the N/M/S/E-specific T cell response by in vitro assay. The results revealed that N/M/S/E-specific CD4+ cells can proliferate in 100% of I-I-I 6 M (n = 10) and 63% of I-I-I 12 M (n = 8) individuals (Fig. 3C and D). However, the expansion capacity, as measured by the percentage of IFNγ+TNFα+ CD4+ T cells, dropped from 13.8% to 7.4% (upper panel, Fig. 3D). Notably, we confirmed that I-I-I cannot efficiently induce a substantial virus-specific CD8+ T cell response, as detected by both ex vivo assay (lower panel, Fig. 3B) and in vitro assay (lower panel, Fig. 3D).
Fig. 3.
Proliferation capacity of virus-specific CD4+T cells lasting 12 months post the third dose in I-I-I individuals. (A) Representative dot plots showing IFNγ and TNFα expression in CD4+ and CD8+ T cells after S peptide ex vivo stimulation at D14 post booster vaccination. (B) The percentage of IFNγ+TNFα+CD4+ T cell (upper panel) and IFNγ+TNFα+CD8+ T cell (lower panel) response at D0, D14, and 1 M of I-I-I vaccination by ex vivo S peptide pool stimulation (n = 21 at D0 and D14, n = 13 at 1 M). (C) Representative dot plots showing IFNγ and TNFα expression in CD4+ and CD8+ T cells at D14 post-booster vaccination upon in vitro N/M/S/E peptide pool stimulation. (D) The percentage of IFNγ+TNFα+CD4+ T cell (upper panel) and IFNγ+TNFα+CD8+ T cell (lower panel) response at different time points after the third dose of inactivated vaccine by in vitro N/M/S/E peptide pool stimulation (n = 10 at D0, 1 M, and 6 M, n = 8 at 12 M).
Vaccinated individuals with BA.5 infection showed a significantly higher level of BA.5 Nabs with evident immune imprinting compared to the unvaccinated individuals
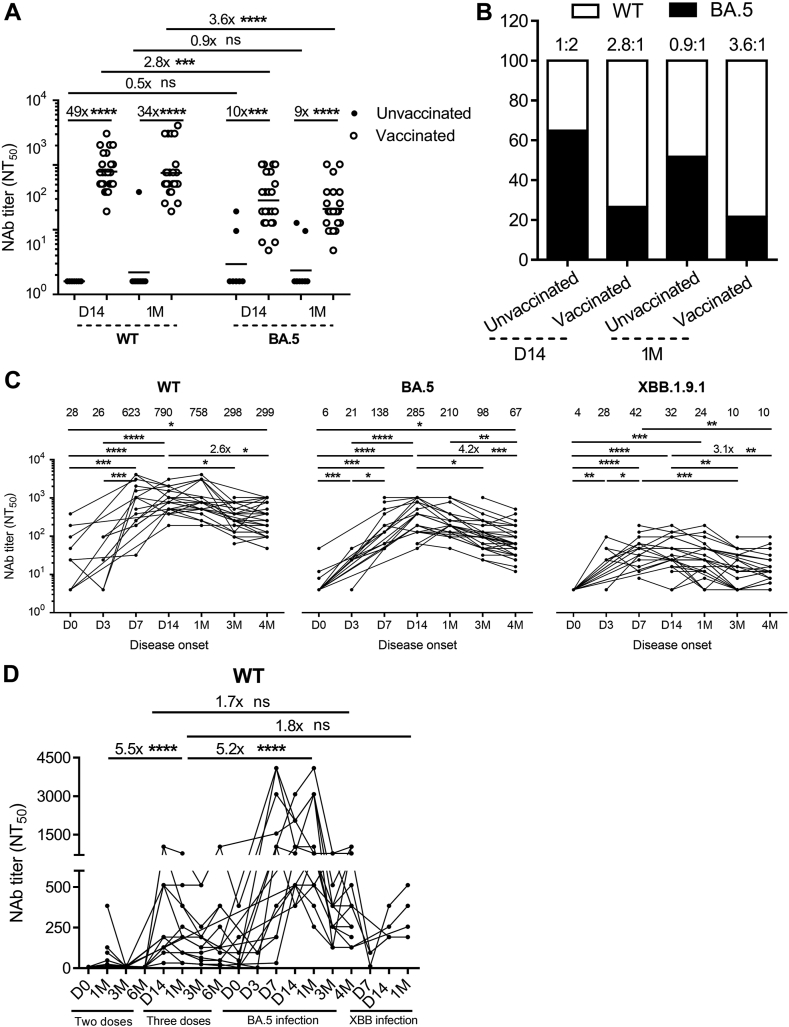
To investigate whether memory B cells were robustly recalled in the vaccinated individuals upon BA.5 infection, we compared the Nabs between 33 vaccinated and 17 unvaccinated participants at D14 and 1 M post BA.5 infection. Our data showed that the vaccinated individuals induced 49-fold and 34-fold higher levels of WT Nabs at D14 (n = 24) and 1 M (n = 23), respectively, and 10-fold and 9-fold higher levels of BA.5 Nabs at D14 and 1 M compared to the unvaccinated individuals (Fig. 4A), demonstrating a strong positive effect caused by vaccination. Interestingly, the vaccinated individuals exhibited a higher ratio between WT and BA.5 Nabs (2.8:1) at D14 than the unvaccinated individuals (1:2) (Fig. 4B). This suggests that the preexisting B cells secreting WT Nabs were preferably recalled in the vaccinated individuals, while the first-time BA.5 infection in unvaccinated individuals occurred prior to inducing Nabs against BA.5, resulting in a different immune imprinting between vaccinated and unvaccinated individuals.
Fig. 4.
BA.5 infection in vaccinated individuals generates higher level of BA.5 Nabs with obvious immune imprinting than unvaccinated individuals. (A) Comparison of Nabs titers against WT and BA.5 between BA.5 infection in unvaccinated and vaccinated individuals (n = 7 at D14 in the unvaccinated individuals, n = 24 at D14 in the vaccinated individuals, n = 10 at 1 M in the unvaccinated individuals, n = 23 at 1 M in the vaccinated individuals). (B) The ratios of Nabs between WT and BA.5 were analyzed in unvaccinated and vaccinated individuals. (C) Nabs titers against WT, BA.5, and XBB.1.9.1 at D0, D3, D7, D14, 1 M, 3 M, and 4 M post-BA.5 infection (n = 11 at D0, n = 7 at D3, n = 16 at D7, n = 24 at D14, n = 23 at 1 M, n = 30 at 3 M, n = 32 at 4 M). (D) Longitudinal analysis of 16 individuals to investigate the evolution of host humoral immunity from two doses of inactivated vaccine to three doses vaccination, to BA.5 infection, and then to XBB reinfection for WT Nabs. The Mann–Whitney test was employed for comparing central tendencies of every two groups, ns, not significant, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
To study the duration of protective Nabs induced by hybrid infection (vaccination-BA.5 infection) against potential emerging variants, such as XBB, we monitored WT, BA.5, and XBB.1.9.1 Nabs at various time points for the vaccinated group. Our data demonstrated strong Nabs against WT, BA.5, and XBB.1.9.1 induced by BA.5 infection, starting from D7 (n = 16), peaking at D14 (n = 24), and significantly declining at 3 M (n = 30) and 4 M (n = 32) (Fig. 4C). The Nabs against WT decreased from 790 (95% CI 684–1284) at D14 to 299 at 4 M, BA.5 decreased from 285 (95% CI 274–572) at D14 to 67 at 4 M, XBB.1.9.1 decreased from 32 at D14 to 10 (95% CI 9–24) with 28% (9/32) individuals had GMT ≤4 at 4 M (Fig. 4C). Our results indicate that while there are still higher Nabs to protect against WT and BA.5 at 4 M, there are individuals at risk of XBB infection at 4 M (Fig. 5).
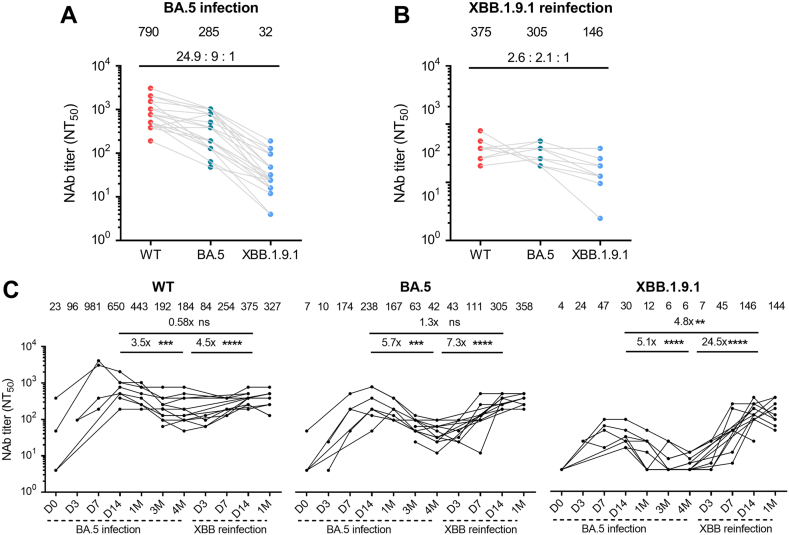
Fig. 5.
Enhancement of Nabs against XBB.1.9.1 and shifting of immune imprinting after XBB reinfection. (A) Examination of immune imprinting after BA.5 infection. The Nab titers against WT, BA.5, and XBB.1.9.1 at D14 post infection were measured, and the ratios compared to XBB.1.9.1 were analyzed (n = 24). (B) To identify the capacity of combating immune imprinting of XBB reinfection, the ratios of Nabs between WT, BA.5, and XBB.1.9.1 were monitored at D14 post XBB reinfection (n = 10). (C) Kinetic analysis of Nabs against WT, BA.5, and XBB.1.9.1 for 12 reinfected individuals to investigate the changes in humoral immunity from BA.5 infection to XBB reinfection. The Mann–Whitney test was employed for comparing central tendencies of every two groups, ns, not significant; ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
To investigate the evolution of host humoral immunity from two doses of vaccine to three doses, to BA.5 infection, and then to XBB reinfection, we compared hybrid immunity with vaccination immunity (Fig. 4D) based on a 2-year longitudinal analysis of WT Nabs (n = 16). At 1 M post-vaccination or infection, we found that Nabs against WT induced by XBB.1.9.1 reinfection and BA.5 infection were 1.8-fold and 5.2-fold higher, respectively, than three-dose vaccination, while two-dose vaccination induced 5.5-fold lower than three doses (Fig. 4D). Regarding the duration of Nabs, compared to D14 post-BA.5 infection, Nabs against WT maintained the same level at 1 M and decreased to 44% at 4 M (Fig. 4D). Importantly, Nabs titers against WT at 4 M post-breakthrough infection were indeed 1.7-fold higher than those at D14 post three-dose vaccination (Fig. 4D), indicating that BA.5 breakthrough infection induced stronger and longer humoral immunity than single vaccinations.
XBB reinfection elevated Nabs against XBB.1.9.1 and shifted immune imprinting
At 4 M post-BA.5 infection, the level of XBB.1.9.1 Nabs remained very low (Fig. 4C). We then continuously followed up the cohort for XBB.1.9.1 reinfection. One participant withdrew from the study after one month of sampling, so we did not test whether she was re-infected with XBB.1.9.1. As expected, after a longitudinal follow-up of the 32 vaccinated-infected individuals, we found that 67% (6/9) individuals of GMT ≤4 were reinfected with XBB.1.9.1 and in total 63% (20/32) of individuals were not reinfected with XBB.1.9.1 at 5 M post BA.5 infection. The ratio between WT, BA.5, and XBB.1.9.1 Nabs at D14 after BA.5 infection (24.9:9:1, n = 24) and after XBB reinfection (2.6:2.1:1, n = 10) exhibited a clear difference in the immune imprinting pattern (Fig. 5A and B). This difference in the Nabs ratio suggests that the antibody response shifted from predominantly targeting WT influenced by the previous vaccine in BA.5 infection to favoring XBB.1.9.1 influenced by the vaccination-BA.5-XBB reinfection (Fig. 5B). Comparing the Nab titers at D14 post-XBB reinfection to that post-BA.5 infection, our results demonstrated a 0.58-fold increase for WT Nabs, a 1.3-fold increase for BA.5 Nabs, and a substantial 4.8-fold increase for XBB Nabs. The shift in immune imprinting was further supported by the Nab titer increase from 4 M after BA.5 infection to D14 after XBB reinfection. XBB.1.9.1 Nabs titer increased 24.5-fold, which was considerably higher than the increases observed for WT (4.5-fold) and BA.5 (7.3-fold) (Fig. 5C).
Omicron BA.5 breakthrough infection resulted in enhanced virus-specific CD4+ and CD8+ T cell responses with different differentiation patterns
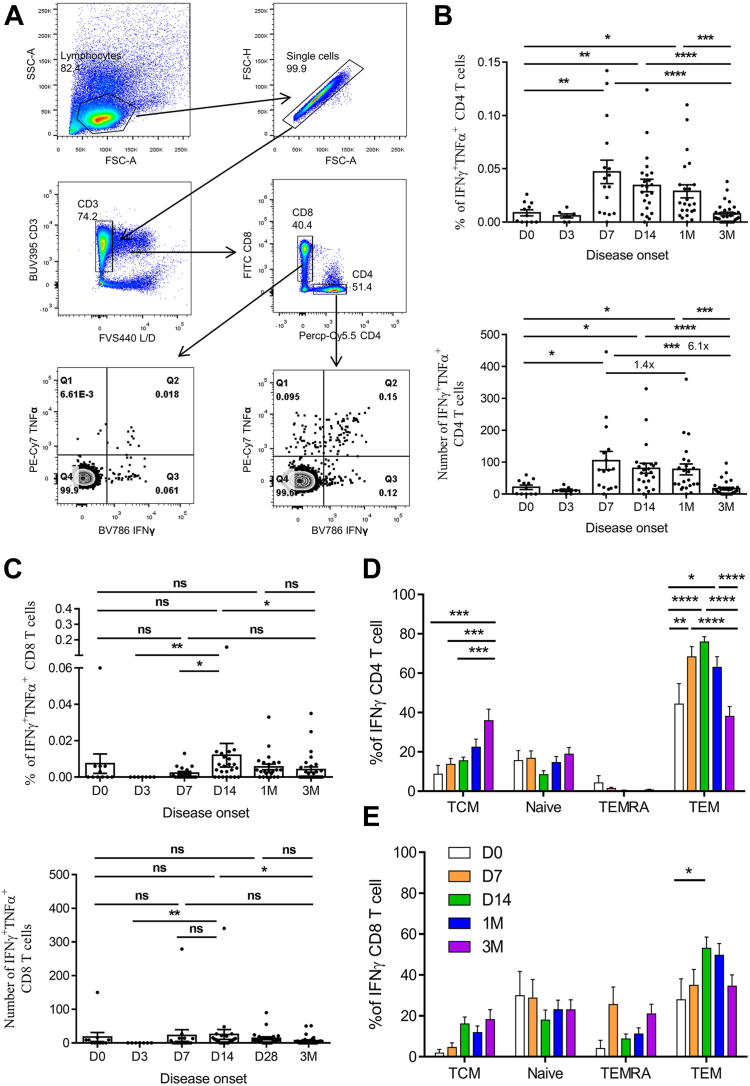
Considering that three doses of inactivated vaccines may not sufficiently induce high levels of virus-specific T cell responses, it is essential to investigate whether BA.5 breakthrough infection can stimulate robust virus-specific T cell responses. To assess this, we measured the levels of IFNγ+TNFα+ CD4+ and CD8+ T cells using peptide pools of S/N/M/E proteins. Our findings revealed that the percentage of S/N/M/E-specific IFNγ+TNFα+ CD4+ T cells peaked at D7, which was significantly higher than at baseline (D0) (0.047, 95% CI 0.023–0.070 vs 0.009, 95% CI 0.002–0.015). This elevated response remained at a high level at 1 M (0.034, 95% CI 0.022–0.046). However, at 3 M post-infection, the percentage of S/N/M/E-specific CD4+ T cells notably decreased to 0.008 (95% CI 0.004–0.011, n = 30) of IFNγ+TNFα+ CD4+ T cells (Fig. 6A and B). Additionally, the number of S/N/M/E-specific CD4+ T cells per million PBMC cells were 105 (95% CI 44–166) at D7 (n = 16), which was 1.2-fold and 5.9-fold higher than those at 1 M (84, 95% CI 46–121, n = 23) and 3 M (18, 95% CI 9–27, n = 30) (Fig. 6B). In contrast, the kinetics of S/N/M/E-specific CD8+ T cells showed a delay compared to the CD4+ response, with the highest level peaking at D14 with 24 (95% CI 1–47) IFNγ+TNFα+ CD8+ T cells per million PBMCs (Fig. 6C).
Fig. 6.
BA.5 breakthrough infection elevated virus-specific CD4+and CD8+T cell responses with a differentiation pattern. (A) Gating strategy showing IFNγ and TNFα expression in CD4+ and CD8+ T cells after N/M/S/E peptides ex vivo stimulation. The percentages (upper panel) and number/million PBMC (lower panel) of IFNγ+TNFα+CD4+ T cells (B) and IFNγ+TNFα+CD8+ T cells (C) at different time points post BA.5 infection were analyzed (n = 11 at D0, n = 7 at D3, n = 16 at D7, n = 23 at D14 and 1 M, n = 30 at 3 M). (D) Frequency of N/M/S/E-specific memory CD4+ and CD8+ T cell phenotypes at different time points post-BA.5 infection (n = 11 at D0, n = 16 at D7, n = 23 at D14 and 1 M, n = 30 at 3 M). Kruskal–Wallis test was involved to compare differences between different time points for T cell response, ns, not significant; ∗P < 0.05, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
To further investigate the differentiation pattern of virus-specific memory T-cell subsets in BA.5 breakthrough infection, we utilized CD45RA and CCR7 surface markers to subdivide memory T cell subsets into naïve T cells (Tnaïve, CD45RA+CCR7+), central memory T cells (TCM, CD45RA−CCR7+), effector memory T cells (TEM, CD45RA−CCR7−), and CD45RA+ effector memory T cells (TEMRA, CD45RA+CCR7−). Our results suggested that TEM was the dominant subtype of IFNγ+CD4+ T cells. The proportion of IFNγ+CD4+ TEM significantly increased from 44% (95% CI 21–67%) at D0 (n = 11) to 68% (95% CI 57–79%) at D7 (n = 16), reaching a peak of 76% (95% CI 70–81%) at D14 (n = 23) (Fig. 6D). Subsequently, the percentage of IFNγ+CD4+ TEM decreased over time, dropping to 38% (95% CI 28–48%) at 3 M (n = 30) post-infection (Fig. 6D). Interestingly, the level of IFNγ+CD4+ TCM increased to 36% (95% CI 24–48%) at 3 M, which was significantly higher than that at D0, D7, and D14 (Fig. 6D). These results suggest that at early post-infection, virus-specific CD4+ T cells were activated with a TEM phenotype, which transitioned to a TCM phenotype at 3 M post-infection. In contrast, IFNγ+CD8+ T cells exhibited higher proportions of Tnaïve (29%, 95% CI 9–48%) and TEMRA (26%, 95% CI 7–44%) phenotypes at D7. With a significant increase in IFNγ+CD8+ TEM from 28% (95% CI 5–51%) at D0 to 53% (95% CI 42–64%) at D14, the IFNγ+CD8+ Tnaïve and TEMRA decreased to 18% (95% CI 8–28%) and 9% (95% CI 4–14%), respectively, at D14 (Fig. 6E). Subsequently, the proportion of IFNγ+CD8+ TEM decreased to 35% (95% CI 23–46%) at 3 M, and the proportion of IFNγ+CD8+ Tnaïve and TEMRA recovered to 23% (95% CI 13–33%) and 21% (95% CI 11–31%), respectively, at 3 M (Fig. 6E). Moreover, a lower proportion of IFNγ+CD8+ TCM was observed during BA.5 infection (Fig. 6E).
Discussion
In this study, we have provided evidence that I-I-I can offer protection against BA.5 infection, although the effectiveness of this protection depends on the time elapsed since the last dose of vaccination. Our findings shed light on why asymptomatic infections were more commonly observed during the BA.2 outbreak from March to June 2022 than during the BA.5 pandemic outbreak in December 2022 and January 2023. Reports have confirmed that there is still 22% vaccine efficacy against BA.2 infection, but a high 93% efficacy against severe/fatality infection.1 This discrepancy in efficacy is likely due to the fact that during the BA.2 outbreak, it was only around 3 months after receiving the inactivated vaccine, which provided both protective ADCC from binding antibodies and virus-specific CD4+ T cell immunity. In contrast, during the BA.5 pandemic, symptomatic BA.5 infections were more prevalent, at around 85%, possibly because only CD4+ T cell immunity remained effective for combating BA.5 infection, given that it had been 12 months post-vaccination. However, it is important to note that the vaccinated individuals showed much higher WT and BA.5 Nabs compared to the unvaccinated BA.5-infected individuals (Fig. 4A). This indicates that vaccination induced memory B cells secreting BA.5 Nabs were rapidly recalled and stimulated to much higher levels to fight against BA.5 infection. Our kinetic analyses further support these findings, showing a rapid recalling activation of N/M/S/E-specific CD4+ T cell response after BA.5 infection in vaccinated individuals (Fig. 6B). Overall, our data demonstrate that I-I-I can partially protect against BA.5 infection through both binding antibodies and T cell immunity, even in the absence of specific BA.5 Nabs and can also facilitate the rapid generation of BA.5 Nabs.
Our ex vivo assay data indicated that I-I-I induced a low level of virus-specific CD4+ T cell response, but our in vitro data showed that these virus-specific CD4+ T cells were capable of proliferating upon antigen re-encounter. Importantly, this was further confirmed in the BA.5 breakthrough infection, where virus-specific CD4+ T cell responses were rapidly recalled at D7 post-infection, with a significantly higher proportion of TEM phenotype (Fig. 6B and D). Our findings strongly suggest that a quick and differentiated virus-specific CD4+ T cell response plays an important role in combating BA.5 infection, which was consistent with the observation that Delta and Omicron breakthrough infections also rapidly recall spike-specific CD4+ T cells.24 In contrast, we did not detect virus-specific CD8+ T cell responses in our ex vivo or in vitro assays at various time points after I-I-I vaccination (Fig. 3B and D), which was consistent with previous findings that inactivated virus and subunit vaccines are less efficient in inducing CD8+ T cells.3,25 However, post BA.5 infection, virus-specific CD8+ T cell responses peaked at D14 with a higher proportion of Tnaïve and TEMRA phenotypes, showing a slower kinetics compared to CD4+ T cell response (Fig. 6). This suggests that CD8+ T cell response is primarily inducted by BA.5 infection. Based on our findings, after BA.5 breakthrough infection, there will be several immune effectors, including Nabs, ADCC, virus-specific T cell immunity, which can provide protection against future emerging variants. If the variants continue to evolve based on the present circulating strains rather than jumping from an intermediate host to create totally new variants, we speculate that future COVID-19 waves will be smaller and shorter in duration with fewer symptoms.
Our 2-year longitudinal study provided a comprehensive view of the host humoral immunity’s evolution from two-dose inactivated vaccines to three doses, BA.5 infection, and then XBB reinfection (Table 3). As expected, we observed that hybrid immunity induced significantly higher Nabs than single vaccinations. A hybrid vigor immunity resulting from a combination of natural immunity and vaccine-generated immunity is observed in several studies.26, 27, 28 It is conceivable that further vaccination in vaccinated-infected individuals will robustly elevate immunity. During BA.5 breakthrough infection, we noticed that the peak value of Nabs against WT was 2.8-fold (790 vs 285) higher than the Nab titer against BA.5 at D14 post-BA.5 infection (Fig. 4C). This can be attributed to immune imprinting induced by WT vaccination, which has been reported in several studies.17,19,29 BA.5, being antigenically distinct from WT,6 primarily recalls memory B cells shared between BA.5 and WT vaccination, which masks the de novo generation of variant-specific B cells, thus hindering the generation of satisfactory humoral immune responses toward BA.5.16,18,30 Thus, BA.5 breakthrough infection induced lower Nab against BA.5 than against WT. Coincidentally, Cao et al. also found that mice receiving a single booster of variant spike protein had significantly lower Nabs against the boosting variant compared to WT,31 indicating the presence of immune imprinting. Interestingly, XBB.1.9.1 reinfection did not reinforce the production of WT Nabs and BA.5 Nabs; instead, it led to more XBB.1.9.1 Nabs than BA.5 infection and single vaccination. The change in Nabs ratio (Fig. 5A and B) confirmed a significant shift in immune imprinting from WT bias to XBB bias due to BA.5-XBB reinfection. Furthermore, the absolute amount of WT Nabs at D14 post XBB reinfection was approximately 47% of those observed in BA.5 infection (Figs. 4C and 5C). Despite the decrease, this level of WT Nabs is still sufficiently high to combat the WT virus for a certain period. These findings immunologically confirm that the WT strain can be excluded from future vaccines, which aligns with the recommendations of the WHO and FDA.
Table 3.
Summary of the immunogenicity for vaccination-BA.5 infection-XBB reinfection.
| Two doses | Three doses | BA.5 infection | XBB re-infection | |
|---|---|---|---|---|
| Nabs against WT | + | ++ | ++++ | ++ |
| Nabs against BA.5 | − | − | +++ | +++ |
| Nabs against XBB.1.9.1 | / | / | + | +++ |
| Virus-specific CD4+ T cell | − | + | +++ | / |
| Virus-specific CD8+ T cell | − | − | ++ | / |
+: positive; ++: significant high level; +++: very high level; ++++: extremely high level; −: undetectable; /: not tested.
Our study has several limitations. Firstly, the sample size for BA.5 breakthrough infections, particularly among the unvaccinated-infected individuals, was relatively small. This limited sample size may restrict the generalizability of the findings. Additionally, we also noted that the between-group comparisons were not controlled for factors such as age, gender and health status, which may have influenced the antibody responses. Therefore, further studies should incorporate large sample sizes and more precise comparative analyses to provide more accurate and comprehensive insights.
Our small-scale but long-term study has provided valuable insights into the dynamic timeline of immune response recruitment under different circumstances, namely vaccination alone and vaccination-infection scenarios. We characterized the strength, differentiation, and duration of different immune effectors and retrospectively summarized the protective mechanisms induced by I-I-I, BA.5 infection, and XBB reinfection. Our data showed the profound impact of vaccination-infection deeply on immune imprinting, which subsequent influences the response to emerging variants. These observations offer crucial guidance for future vaccine design.
Contributors
ZW, NZ, ZZ and JZ designed this study. ZW and TC interpret data and drafted the manuscript. TC, JS and WL conducted the animal experiment. TC, XS, SL, MH, WL, CL, TS, LC, RW, XS, QL, JS, JL and SD performed neutralizing assay and flow cytometry assay. XS, SL and MH conducted ADCC assay. TC, XS, SL and MH collected samples. WL helped to conduct the statistical analysis. NZ, ZZ and JZ made critical revision of the manuscript. All authors read and approved the final manuscript.
Data sharing statement
All the data will be available upon reasonable request to the corresponding author with publication.
Declaration of interests
We declare no conflict of interest.
Acknowledgements
This study was supported by R&D Program of Guangzhou National Laboratory (SRPG23-005), National Key Research and Development Program of China (2022YFC2604104, 2019YFC0810900), S&T Program of Guangzhou Laboratory (SRPG22-006), and National Natural Science Foundation of China (81971485, 82271801, 81970038), Emergency Key Program of Guangzhou Laboratory (EKPG21-30-3), Zhongnanshan Medical Foundation of Guangdong Province (ZNSA-2020013), and State Key Laboratory of Respiratory Disease (J19112006202304).
The authors wish to thank all participating donors.
Contributor Information
Jincun Zhao, Email: zhaojincun@gird.cn.
Zhuxiang Zhao, Email: zhaozhuxiang@126.com.
Nanshan Zhong, Email: nanshan@vip.163.com.
Zhongfang Wang, Email: wangzhongfang@gird.cn.
References
- 1.Tang L., Wang F.-Z., Rodewald L.E., et al. Real-world effectiveness of primary series and booster doses of inactivated coronavirus disease 2019 vaccine against Omicron BA.2 variant infection in China: a retrospective cohort study. J Infect Dis. 2023;228(3):261–269. doi: 10.1093/infdis/jiad090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X., Wang Y., Hu C., et al. Effectiveness of a booster dose of COVID-19 vaccines during an outbreak of SARS-CoV-2 Omicron BA.2.2 in China: a case-control study. Hum Vaccin Immunother. 2023;19(1) doi: 10.1080/21645515.2023.2194189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao Z., Cui T., Huang M., et al. Heterologous boosting with third dose of coronavirus disease recombinant subunit vaccine increases neutralizing antibodies and T cell immunity against different severe acute respiratory syndrome coronavirus 2 variants. Emerg Microb Infect. 2022;11(1):829–840. doi: 10.1080/22221751.2022.2048969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong J., Liu S., Cui T., et al. Heterologous booster with inhaled adenovirus vector COVID-19 vaccine generated more neutralizing antibodies against different SARS-CoV-2 variants. Emerg Microbes Infect. 2022;11(1):2689–2697. doi: 10.1080/22221751.2022.2132881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao Y., Yisimayi A., Jian F., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608(7923):593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuekprakhon A., Nutalai R., Dijokaite-Guraliuc A., et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185(14):2422–2433.e13. doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Islam M.R., Shahriar M., Bhuiyan M.A. The latest Omicron BA.4 and BA.5 lineages are frowning toward COVID-19 preventive measures: a threat to global public health. Health Sci Rep. 2022;5(6) doi: 10.1002/hsr2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou R., Liu N., Li X., et al. Three-dose vaccination-induced immune responses protect against SARS-CoV-2 Omicron BA.2: a population-based study in Hong Kong. Lancet Reg Health West Pac. 2023;32 doi: 10.1016/j.lanwpc.2022.100660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu A., Wei P., Man M., et al. Antigenic characterization of SARS-CoV-2 Omicron subvariants XBB.1.5, BQ.1, BQ.1.1, BF.7 and BA.2.75.2. Signal Transduct Targeted Ther. 2023;8(1):125. doi: 10.1038/s41392-023-01391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagemann K., Riecken K., Jung J.M., et al. Natural killer cell-mediated ADCC in SARS-CoV-2-infected individuals and vaccine recipients. Eur J Immunol. 2022;52(8):1297–1307. doi: 10.1002/eji.202149470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim J.M.E., Hang S.K., Hariharaputran S., et al. A comparative characterization of SARS-CoV-2-specific T cells induced by mRNA or inactive virus COVID-19 vaccines. Cell Rep Med. 2022;3(11) doi: 10.1016/j.xcrm.2022.100793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andeweg S.P., de Gier B., Eggink D., et al. Protection of COVID-19 vaccination and previous infection against Omicron BA.1, BA.2 and Delta SARS-CoV-2 infections. Nat Commun. 2022;13(1):4738. doi: 10.1038/s41467-022-31838-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powell A.A., Kirsebom F., Stowe J., et al. Protection against symptomatic infection with delta (B.1.617.2) and omicron (B.1.1.529) BA.1 and BA.2 SARS-CoV-2 variants after previous infection and vaccination in adolescents in England, August, 2021–March, 2022: a national, observational, test-negative, case-control study. Lancet Infect Dis. 2023;23(4):435–444. doi: 10.1016/S1473-3099(22)00729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu K.-L., Jiang X.-L., Zhan B.-D., et al. Durability of neutralization against Omicron subvariants after vaccination and breakthrough infection. Cell Rep. 2023;42(2) doi: 10.1016/j.celrep.2023.112075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee W.S., Tan H.-X., Reynaldi A., et al. Durable reprogramming of neutralising antibody responses following breakthrough Omicron infection. medRxiv. 2023 doi: 10.1126/sciadv.adg5301. 2023.02.19.23286159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao Y., Jian F., Wang J., et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature. 2023;614(7948):521–529. doi: 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chemaitelly H., Ayoub H.H., Tang P., et al. Immune imprinting and protection against repeat reinfection with SARS-CoV-2. N Engl J Med. 2022;387(18):1716–1718. doi: 10.1056/NEJMc2211055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park Y.-J., Pinto D., Walls A.C., et al. Imprinted antibody responses against SARS-CoV-2 Omicron sublineages. Science. 2022;378(6620):619–627. doi: 10.1126/science.adc9127. [DOI] [PubMed] [Google Scholar]
- 19.Reynolds C.J., Pade C., Gibbons J.M., et al. Immune boosting by B.1.1.529 (Omicron) depends on previous SARS-CoV-2 exposure. Science. 2022;377(6603) doi: 10.1126/science.abq1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z., Yang X., Zhou Y., et al. COVID-19 severity correlates with weaker T-cell immunity, hypercytokinemia, and lung epithelium injury. Am J Respir Crit Care Med. 2020;202(4):606–610. doi: 10.1164/rccm.202005-1701LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertoletti A., Le Bert N., Tan A.T. SARS-CoV-2-specific T cells in the changing landscape of the COVID-19 pandemic. Immunity. 2022;55(10):1764–1778. doi: 10.1016/j.immuni.2022.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarke A., Potesta M., Varchetta S., et al. Early and polyantigenic CD4 T cell responses correlate with mild disease in acute COVID-19 donors. Int J Mol Sci. 2022;23(13):7155. doi: 10.3390/ijms23137155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandran A., Rosenheim J., Nageswaran G., et al. Rapid synchronous type 1 IFN and virus-specific T cell responses characterize first wave non-severe SARS-CoV-2 infections. Cell Rep Med. 2022;3(3) doi: 10.1016/j.xcrm.2022.100557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koutsakos M., Reynaldi A., Lee W.S., et al. SARS-CoV-2 breakthrough infection induces rapid memory and de novo T cell responses. Immunity. 2023;56(4):879–892.e4. doi: 10.1016/j.immuni.2023.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirai T., Yoshioka Y. Considerations of CD8(+) T cells for optimized vaccine strategies against respiratory viruses. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.918611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds C.J., Pade C., Gibbons J.M., et al. Prior SARS-CoV-2 infection rescues B and T cell responses to variants after first vaccine dose. Science. 2021;372(6549):1418–1423. doi: 10.1126/science.abh1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goel R.R., Apostolidis S.A., Painter M.M., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals after mRNA vaccination. Science Immunol. 2021;6(58) doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bobrovitz N., Ware H., Ma X., et al. Protective effectiveness of previous SARS-CoV-2 infection and hybrid immunity against the omicron variant and severe disease: a systematic review and meta-regression. Lancet Infect Dis. 2023;23(5):556–567. doi: 10.1016/S1473-3099(22)00801-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aydillo T., Rombauts A., Stadlbauer D., et al. Immunological imprinting of the antibody response in COVID-19 patients. Nat Commun. 2021;12(1):3781. doi: 10.1038/s41467-021-23977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis-Gardner M.E., Lai L., Wali B., et al. Neutralization against BA.2.75.2, BQ.1.1, and XBB from mRNA bivalent booster. N Engl J Med. 2023;388(2):183–185. doi: 10.1056/NEJMc2214293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yisimayi A., Song W., Wang J., et al. Repeated Omicron infection alleviates SARS-CoV-2 immune imprinting. bioRxiv. 2023 2023.05.01.538516. [Google Scholar]