Abstract
Proline dipeptidase (prolidase) was purified from cell extracts of the proteolytic, hyperthermophilic archaeon Pyrococcus furiosus by multistep chromatography. The enzyme is a homodimer (39.4 kDa per subunit) and as purified contains one cobalt atom per subunit. Its catalytic activity also required the addition of Co2+ ions (Kd, 0.24 mM), indicating that the enzyme has a second metal ion binding site. Co2+ could be replaced by Mn2+ (resulting in a 25% decrease in activity) but not by Mg2+, Ca2+, Fe2+, Zn2+, Cu2+, or Ni2+. The prolidase exhibited a narrow substrate specificity and hydrolyzed only dipeptides with proline at the C terminus and a nonpolar amino acid (Met, Leu, Val, Phe, or Ala) at the N terminus. Optimal prolidase activity with Met-Pro as the substrate occurred at a pH of 7.0 and a temperature of 100°C. The N-terminal amino acid sequence of the purified prolidase was used to identify in the P. furiosus genome database a putative prolidase-encoding gene with a product corresponding to 349 amino acids. This gene was expressed in Escherichia coli and the recombinant protein was purified. Its properties, including molecular mass, metal ion dependence, pH and temperature optima, substrate specificity, and thermostability, were indistinguishable from those of the native prolidase from P. furiosus. Furthermore, the Km values for the substrate Met-Pro were comparable for the native and recombinant forms, although the recombinant enzyme exhibited a twofold greater Vmax value than the native protein. The amino acid sequence of P. furiosus prolidase has significant similarity with those of prolidases from mesophilic organisms, but the enzyme differs from them in its substrate specificity, thermostability, metal dependency, and response to inhibitors. The P. furiosus enzyme appears to be the second Co-containing member (after methionine aminopeptidase) of the binuclear N-terminal exopeptidase family.
Pyrococcus furiosus is a fermentative archaeon which grows optimally at temperatures near 100°C (26). Like many heterotrophic hyperthermophiles, it utilizes proteins and peptides as growth substrates and produces organic acids, CO2, and H2. Several enzymes involved in the catabolism of amino acids have been purified from P. furiosus (2), including aminotransferases (3), glutamate dehydrogenase (34), 2-keto acid oxidoreductases (31, 37, 38), and acetyl coenzyme A synthetases (39). In addition, this organism produces perhaps a dozen or more proteolytic-type enzymes, which are assumed to generate small peptides from the protein-based growth substrates (6, 8, 21, 23, 45). So far, three proteases have been characterized from P. furiosus. These are a membrane-associated serine protease (48), an intracellular protease with trypsin- and chymotrypsin-like activities (28, 29), and an intracellular endopeptidase that cleaves at prolyl residues (30). In addition, two proteases have been purified from other members of the family Thermococcales, including a thiol protease from an unclassified species of Pyrococcus (40) and a serine protease from Thermococcus stetteri (33). To date, however, there have been no reports on the properties of amino acid-yielding peptidases from P. furiosus or related species. In order to further understand the pathways of peptide metabolism in these organisms, we examined P. furiosus for dipeptidase activities. Cell extracts contained very high concentrations of prolidase, a proline-specific dipeptidase, and the characterization of this enzyme is reported herein.
Since prolyl residues confer a conformational constraint on a peptide chain due to the cyclic nature of its pyrrolidine side group, only a few proteases are known that are able to hydrolyze bonds adjacent to proline (22, 49). These enzymes include (i) proline-specific endopeptidase, which hydrolyzes peptides on the carboxyl side of prolyl residues located internally within a polypeptide (-X--Pro-/-X-); (ii) prolyl aminopeptidase, which cleaves the bond between any N-terminal amino acid and a penultimate prolyl residue (NH2-X-/-Pro--X-) in peptides of various lengths; (iii) proline iminopeptidase, which catalyzes cleavage of unsubstituted N-terminal prolyl residues from dipeptides, tripeptides, and polypeptides (Pro-/-X-); (iv) proline specific C-terminal exopeptidase (-X--Pro-/-X-COOH), which releases an amino acid from the C terminus of a peptide with a penultimate proline residue; and (v) prolidase, which only cleaves dipeptides with proline at the C terminus (NH2-X-/-Pro-COOH). These proline-specific enzymes are thought to participate, in concert with other endo- and exopeptidases, in the terminal degradation of intracellular proteins and may also function in the recycling of proline.
Prolidase (iminodipeptidase, EC 3.4.3.7) is widespread in nature and has been isolated from different mammalian tissues (12, 24, 44) as well as from bacteria such as species of Lactobacillus (10, 25) and Xanthomonas (46). While the physiological role of prolidase in bacteria is unclear, a deficiency of this enzyme in humans results in abnormalities of the skin and other collagenous tissues (43). Prolidase also has biotechnological applications. For example, it has a potential use in the dairy industry as a cheese-ripening agent (9) since proline release from proline-containing peptides in cheese reduces bitterness. In addition, it was recently reported (18) that prolidase and an enzyme termed organophosphorus acid anhydrolase (OPAA, EC 3.1.8.1) appear to be one and the same. OPAA hydrolyzes highly toxic, organophosphorus, acetylcholinesterase inhibitors, which include various chemical warfare agents and pesticides. Such enzymes have been characterized from various species of Pseudomonas, Flavobacterium, and Alteromonas and also from eucaryotes (5, 17, 36). The sequence of the OPAA from Alteromonas sp. strain JD6.5 shows similarity to that of human prolidase and, like the latter enzyme, OPAA catalyzes the Mn2+-dependent hydrolysis of X-Pro dipeptides but not of Y-Pro-X tripeptides (18). It seems reasonable to conclude, therefore, that the natural function of OPAA involves peptide metabolism rather than detoxification. Conversely, previously characterized prolidases may be found to be biotechnologically relevant in detoxification strategies.
Prolidase has yet to be characterized from an archaeon or a hyperthermophile. It was therefore of some interest to determine the properties of this enzyme from P. furiosus. In addition, the gene encoding P. furiosus prolidase was cloned and expressed in Escherichia coli, and this allowed a biochemical comparison to be made between the native form (from P. furiosus) and the recombinant form.
MATERIALS AND METHODS
Growth of microorganisms.
P. furiosus (DSM 3638) was grown at 95°C in a 500-liter fermentor with maltose as the carbon source as described previously (13). E. coli BL21(λDE3) (F− ompT [lon] hsdS) was grown in a 100-liter fermentor at 37°C in Luria-Bertani medium supplemented with ampicillin (100 μg/ml) as needed.
Enzyme assay.
The prolidase activity assay used was based on the amount of proline liberated from the hydrolysis of dipeptides that contain proline at the C terminus. The proline concentration was determined by a modification of the colorimetric ninhydrin method of Yaron and Mlynar (51). The ninhydrin reagent was prepared by the addition of ninhydrin (3.0% [wt/vol]) to a mixture of 60% (vol/vol) glacial acetic acid and 40% (vol/vol) phosphoric acid followed by a 30-min incubation at 70°C (51). The assay mixture (500 μl) for prolidase contained 50 mM MOPS (3-[N-morpholino]propanesulfonic acid) buffer (pH 7.0), 4 mM Met-Pro (substrate), and 1.2 mM CoCl2 and was incubated at 100°C for 5 min. The reaction was initiated by addition of the enzyme or extract. The mixture was incubated at 100°C for a further 10 min, and the reaction was stopped by the addition of glacial acetic acid (500 μl) followed by the ninhydrin reagent (500 μl). After heating at 100°C for 10 min, the solution was cooled to 23°C and the absorption at 515 nm was determined with an extinction coefficient of 4,570 M−1 · cm−1 for the ninhydrin-proline complex. One unit of prolidase activity is defined as the amount of enzyme that liberates one micromole of proline per minute under these assay conditions.
Purification of P. furiosus prolidase.
Prolidase was purified from P. furiosus under anaerobic conditions at 23°C. Frozen cells (500 g [wet weight]) were thawed in 1,800 ml of 50 mM Tris-HCl buffer (pH 8.0) containing lysozyme (1 mg/ml) and DNase (10 μg/ml) and were lysed by incubation at 37°C for 2 h followed by sonication (Branson 8200 sonicator) for 1 h. A cell extract was obtained by ultracentrifugation at 50,000 × g for 2 h. The supernatant (1,800 ml) was loaded onto a column (10 by 14 cm) of DEAE Fast Flow (Pharmacia, Piscataway, N.J.) equilibrated with 50 mM Tris (pH 8.0) containing 10% (vol/vol) glycerol. The column was eluted at a flow rate of 10 ml/min with a 10-liter linear gradient of 0 to 1.0 M NaCl in the same Tris-glycerol buffer. Prolidase activity was detected as 0.25 to 0.40 M NaCl was applied to the column. The active fractions were combined (1,500 ml), and solid ammonium sulfate was added to a final concentration of 1.5 M. This solution was applied to a column (3.5 by 10 cm) of phenyl Sepharose (Pharmacia) equilibrated with Tris-glycerol buffer containing 1.5 M ammonium sulfate. The column was eluted with a gradient (1 liter) from 1.5 to 0 M ammonium sulfate in the Tris-glycerol buffer at a flow rate of 7 ml/min. Prolidase eluted as 0.45 to 0.78 M ammonium sulfate was applied to the column. The prolidase-containing fractions (250 ml) were concentrated to a volume of 7 ml by ultrafiltration (PM-30 membrane filter; Amicon, Beverly, Mass.) and applied to a column (3.5 by 60 cm) of Superdex-200 (Pharmacia) equilibrated with 50 mM Tris (pH 8.0) containing 0.5 M NaCl at a flow rate of 0.5 ml/min. The active fractions from the Superdex 200 column were applied to a column of HiTrap-Q (1.6 by 2.5 cm; Pharmacia) equilibrated with 50 mM Tris (pH 8.0), and the enzyme was eluted with a gradient (100 ml) from 0 to 0.5 M NaCl in the same buffer at a flow rate of 4 ml/min. Fractions containing prolidase activity (10 ml) eluted as 0.29 to 0.40 M NaCl was applied and were stored at −80°C until being required.
Cloning and expression of the prolidase-encoding gene.
Recombinant P. furiosus prolidase was obtained by PCR amplification of the P. furiosus prolidase gene and subsequent cloning of this gene into the T7-polymerase-driven expression vector pET-21b (Novagen, Milwaukee, Wis.). For the PCR amplification of the prolidase gene, two primers were designed. Primer 1 (ATAGGATCCGGTGAGGAGGTTGTATGAAAGAAAGACTTGAA; Stratagene, La Jolla, Calif.) contained an engineered BamHI site and spans from −21 to +6 on the coding strand. Primer 2 (ATAGGATCCGGTGAGGAGGTTGTATGAAAGAAAGAC; Stratagene) had an engineered NotI site and corresponds to sequence ranging from +1511 to +1541 on the noncoding strand. PCR amplification was performed with native P. furiosus DNA polymerase and a Robocycler 40 (Stratagene) programmed for 39 cycles, each cycle consisting of denaturation at 95°C for 5 min, annealing at 52°C for 2 min, and extension at 72°C for 5 min. The resultant 1.5-kb prolidase gene was first subcloned into the blunt end SrfI site in the vector pCR-Script (Stratagene) to yield plasmid pProl. The prolidase insert DNA contained in plasmid pProl was then sequenced to ensure that no mutations were present in the gene. The prolidase gene was then excised from plasmid pProl by restriction digest with the enzymes BamHI and NotI (Stratagene) and cloned into the BamHI and NotI sites in expression vector pET-21b, resulting in plasmid pET-Prol.
For expression of recombinant prolidase, plasmid pET-Prol was transformed into E. coli BL21(λDE3), which has isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression of T7-RNA polymerase. Prolidase was produced in a culture of BL21(λDE3)-pET-Prol grown in a 100-liter fermentor at 37°C. Expression of the plasmid borne prolidase gene was induced with the addition of IPTG (1 mM) when the culture reached an optical density of 1.0. The induced culture was incubated for 4 h prior to the harvesting of the cells.
Purification of recombinant prolidase.
Recombinant prolidase was purified in three steps. IPTG-induced BL21(λDE3)-pET-Prol cells (10 g [wet weight]) were suspended in 10 ml of 50 mM Tris-HCl, pH 8.0, containing benzamidine HCl (0.5 mg/ml). The cell suspension was passed through a French pressure cell (20,000 lb/in2) twice. The lysed extract was centrifuged at 39,000 × g for 1 h to remove any cellular debris, and the supernatant was diluted to 300 ml with 50 mM Tris-HCl, pH 8.0. Solid ammonium sulfate was slowly added with stirring to a final concentration of 1.5 M, and the solution was applied to a column (3.5 cm by 10 cm) of phenyl Sepharose (Pharmacia) equilibrated with the same buffer at a flow rate of 7 ml/min. The bound protein was eluted with a gradient (1,000 ml) from 1.5 to 0 M ammonium sulfate in 50 mM Tris-HCl, and the recombinant prolidase was eluted as 0.67 to 1.0 M ammonium sulfate was applied. The prolidase-containing fractions were incubated at 100°C for 2.5 h, and denatured E. coli proteins were removed by centrifugation at 27,000 × g for 30 min. The supernatant was diluted threefold with 50 mM Tris-HCl, pH 8.0, as it was applied to a column of HiTrap-Q (1.6 by 2.5 cm; Pharmacia) equilibrated with 50 mM Tris-HCl, pH 8.0. A gradient (100 ml) from 0 to 0.5 M NaCl in the same buffer was applied to the column. The prolidase eluted between 0.25 and 0.37 M NaCl and was stored at −80°C until being required.
Other methods.
Molecular weights were estimated by gel filtration with a column (1 by 27 cm) of Superdex 200 (Pharmacia LKB) with amylase (200,000), alcohol dehydrogenase (150,000), and bovine serum albumin (66,000) as standard proteins. Sodium dodecyl sulfate (SDS)-gel electrophoresis was performed using 12% polyacrylamide by the method of Laemmli (35). Protein concentrations were determined by the Bradford method (11) with bovine serum albumin as the standard. To determine metal content, exogenous metal ions were removed from the prolidase by either dialyzing (membrane cutoff, 8 kDa) the sample against 100 volumes of 50 mM N-(2-hydroxyethyl)piperazine-N′-3-propanesulfonic acid (EPPS), pH 8.0, containing 0.5 M NaCl or by gel filtration with a Superdex 200 column. A complete metal analysis (31 elements) was obtained by plasma emission spectroscopy with a Jarrel Ash Plasma Comp 750 instrument at the Chemical Analysis Laboratory of the University of Georgia. The NH2-terminal sequences of the native and recombinant prolidases were determined by using an Applied Biosystems Model 477 sequencer in the Molecular Genetics Instrumentation Facility (MGIF) of the University of Georgia. Samples were electroblotted onto polyvinylidene difluoride protein-sequencing membranes (Stratagene) from SDS-electrophoresis gels by using a Bio-Rad electroblotting system. Electroblotting was carried out in 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) buffer, pH 11.0, containing methanol (10% [vol/vol]) for 1 h at 50 V. Both strands of the P. furiosus prolidase gene present in plasmid pET-Prol were sequenced in their entirety by the MGIF of the University of Georgia. DNA sequence was analyzed using the computer software programs Genetics Computer Group (University of Wisconsin, Madison) and MacVector (International Biotechnologies, Inc., New Haven, Conn.).
Nucleotide sequence accession number.
The DNA sequence of the prolidase gene is available from GenBank under accession no. AF060010.
RESULTS
Purification of P. furiosus prolidase.
Extracts of P. furiosus cells grown with maltose as the primary carbon source contained high prolidase activity (approximately 2.3 U/mg at 100°C) with the dipeptide Met-Pro as the substrate. For comparison, this specific activity is more than 100-fold higher than the prolidase activity found in cell extracts of Lactobacillus casei grown on casein (25) and 200-fold higher than that in cell extracts of Xanthomonas maltophilia grown in nutrient broth (46). The prolidase of P. furiosus appeared not to be regulated, as the specific activities of extracts of cells grown with yeast extract (5.0 g/liter) and maltose (1.0 g/liter), with yeast extract (5.0 g/liter), peptone (5.0 g/liter) and maltose (1.0 g/liter), or with yeast extract (5.0 g/liter) and maltose (5.0 g/liter) were similar (2.5 ± 0.3 U/mg). Since maltose-grown cells are routinely used in our laboratory to purify various O2-sensitive, oxidoreductase-type enzymes (see reference 2 for an example) these cells were also used for prolidase purification. In addition, the procedure was carried out under anaerobic conditions, not because the prolidase was sensitive to O2, but to allow the purification of enzymes that are from the same batch of P. furiosus cells.
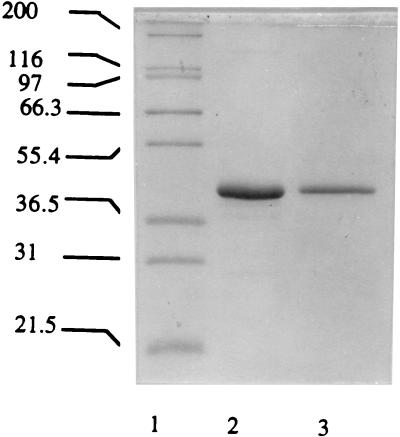
Prolidase activity was not detected in the culture supernatant during either log- or stationary-phase growth of P. furiosus or in the membrane fraction of a cell extract. The activity was present only in the soluble fraction, indicating that the enzyme is a cytoplasmic protein. The results of a typical purification are summarized in Table 1. The enzyme was purified 274-fold with a yield of 4% and a specific activity of approximately 630 U/mg. It therefore constitutes approximately 0.36% of total cytoplasmic protein. When the purified prolidase was treated with SDS sample buffer at 80°C for 10 min prior to electrophoresis, it migrated as a single band corresponding to a molecular mass of 51 kDa. However, when treated at 100°C for 30 min, the protein band migrated with a molecular mass of 42 kDa (see Fig. 1). Presumably, the former conditions result in a partially denatured protein which is retarded in the electrophoretic gel. The prolidase which was eluted from a gel filtration column corresponded to a molecular mass of 100 ± 10 kDa. This result, together with the electrophoretic data, suggests that the enzyme is a homodimer.
TABLE 1.
Purification of prolidase from P. furiosus
| Step | Activitya (U) | Amt of protein (mg) | Sp act (U/mg) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|
| Cell extract | 48,400 | 21,060 | 2.3 | 1 | 100 |
| DEAE Sepharose | 25,600 | 4,500 | 5.7 | 2.5 | 53 |
| Phenyl Sepharose | 10,400 | 855 | 12 | 5.3 | 21 |
| Superdex 200 | 2,180 | 14 | 153 | 66.4 | 4 |
| HiTrap-Q | 1,890 | 3 | 630 | 274 | 4 |
Activity was measured with Met-Pro (4 mM) as the substrate.
FIG. 1.
SDS–12% polyacrylamide gel electrophoresis of purified N- and R-prol. Lane 1, molecular mass markers (kilodaltons): myosin (200), β-galactosidase (116), phosphorylase b (97), bovine serum albumin (66.3), glutamic dehydrogenase (55.4), lactate dehydrogenase (36.5), carbonic anhydrase (31); lane 2, R-prol; lane 3, N-prol.
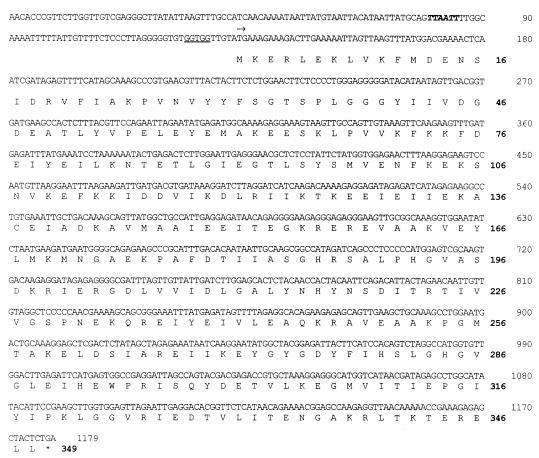
The N-terminal amino acid sequence of the native prolidase was MKERLEKLVKFMDEN. This sequence was used to search the genomic sequence database of P. furiosus, which is nearing completion by the use of multiplex sequencing methods (19). A gene was located whose translated N-terminal region matched exactly the sequence obtained from the enzyme. It consisted of 1,047 bp and encoded a protein of 349 residues with a calculated molecular mass of 39.4 kDa (Fig. 2). The latter value is slightly lower than that (42 kDa) obtained from the SDS-gel analysis, suggesting that the protein is not completely denatured under the conditions used. The enzyme also appears to exhibit nonideal behavior when subjected to gel filtration, since the molecular mass estimated by that method (100 kDa) is higher than that expected (78.8 kDa) for a homodimeric protein. A mass of 39.4 kDa for the prolidase subunit was used in all calculations.
FIG. 2.
The 1,047-bp gene encoding P. furiosus prolidase and the deduced amino acid sequence (348 amino acids) are shown. A putative TATA box is indicated in boldface. The ribosomal binding site is underlined, and the translation start site is marked by an arrow.
Purification of recombinant P. furiosus prolidase.
The production of prolidase protein was successfully induced in E. coli cells in the presence of IPTG with maximal induction (as determined by prolidase activity) after a 4-h period of induction at 37°C (data not shown). P. furiosus prolidase activity could be identified in cell extracts of E. coli both by high-temperature (100°C) enzyme assays, which eliminated the host cell prolidase activity, as well as by the appearance of a protein band corresponding to the size of prolidase (42 kDa) after SDS-gel analysis of cell extracts (data not shown). The specific activity of the prolidase in the recombinant E. coli cells was approximately 90 U/mg, which is about 40-fold higher than that present in cell extracts of P. furiosus (under the same assay conditions when Met-Pro was used as a substrate at 100°C). The results of a typical purification of the recombinant prolidase from a cell extract of E. coli BL21(λDE3)-pET-Prol are summarized in Table 2. The enzyme was purified in three steps with a specific activity increase of about 15-fold and a recovery of 54%. It constituted approximately 6.7% of the total cellular protein. However, the specific activity of the purified recombinant form (∼ 1,300 U/mg) was about twofold greater than that of the native enzyme from P. furiosus. The reasons for this are unclear at present (see below). Nevertheless, the recombinant prolidase was indistinguishable from the native protein from P. furiosus when analyzed by SDS-gel electrophoresis, and N-terminal amino acid sequence analysis showed that it contained the same first 15 residues (MKERLEKLVKFMDEN). It should also be noted that the nucleotide sequence of the gene encoding the prolidase was identical to that obtained from the genomic database.
TABLE 2.
Purification of recombinant P. furiosus prolidase from E. coli
| Step | Activitya (U) | Amt of protein (mg) | Sp act (U/mg) | Purification (fold) | Recovery (%) |
|---|---|---|---|---|---|
| Cell extract | 45,400 | 498 | 91 | 1.0 | 100 |
| Phenyl Sepharose | 31,800 | 105 | 302 | 3.3 | 70 |
| Heat treatment | 27,200 | 24 | 1,130 | 12 | 60 |
| HiTrap-Q | 24,400 | 18 | 1,360 | 15 | 54 |
Activity was measured with Met-Pro (4 mM) as the substrate.
Physical properties of native and recombinant prolidases.
Both the native prolidase purified from P. furiosus (hereafter referred to as N-prol) and the recombinant prolidase obtained from E. coli (hereafter referred to as R-prol) were analyzed for metals. The only ones present in significant amounts (>0.1 g-atom/39.4-kDa subunit) were cobalt and zinc. Both R-prol and N-prol contained 1.0 ± 0.3 (mean ± standard deviation) g-atoms of Co/mol of subunit (data from four and eight different prolidase samples, respectively). These data indicate that the difference in the specific activities of N- and R-prol under standard assay conditions was not due to a difference in Co content. R-prol was also purified as described above but with all buffers containing 1 mM CoCl2. The resulting enzyme, after gel filtration (Superdex 200) to remove exogenous Co, also contained 1.0 (± 0.2) g-atoms of Co/subunit. However, this and all other prolidase preparations tested were inactive unless Co2+ ions were added to the assay medium, suggesting that both the recombinant and native forms of the enzyme require occupancy of a second (or more) Co2+ site per subunit for activity and that the two sites have very different affinities (see below). Chemical analysis of R-prol and N-prol also revealed the presence of significant but variable amounts of zinc (1.5 ± 1.0 and 2.0 ± 0.5 g-atoms/subunit from 4 and 12 determinations, respectively). However, there was no correlation between the zinc content and the specific activity of an enzyme preparation (whether R- or N-prol), indicating that the zinc is nonspecifically bound. This was confirmed by treating R-prol (containing 2.0 ± 0.1 g-atoms of Zn/subunit) with EDTA (5 mM in 50 mM EPPS buffer, pH 8.0) for 1 h at 23°C, followed by gel filtration. The resulting enzyme contained only 0.3 ± 0.1 g-atoms of Zn/subunit, but its specific activity under standard assay conditions was not affected by the chelation treatment.
N-prol was very thermostable, with no loss in activity when a sample (0.3 mg/ml in 100 mM MOPS, pH 7.0) was incubated in a sealed vial for 12 h at 100°C. However, the stability was dependent upon protein concentration, as the same enzyme preparation at a concentration of 0.003 mg/ml lost 50% of its activity after a 4-h incubation at that temperature. R-prol was apparently less thermostable than the native form but also exhibited a concentration-dependent response. The time required for a 50% loss in the activity (t50%) of R-prol at 100°C at a concentration of 0.3 or 0.003 mg/ml was 3 or 1 h, respectively. With R-prol at a concentration of 1.5 mg/ml, the optimal pH for stability was 7.0, with t50% values at 100°C decreasing from 7.8 to 0.7, 1.3, 6.4 and 4.5 h at pH 2.0 (100 mM glycine-HCl), 4.5 (100 mM sodium acetate), 8.0 (100 mM EPPS), and 10.0 (100 mM CAPS), respectively. Hence, the enzyme was much more stable under alkaline conditions than it was in acidic media. Addition of Co2+ ions (1 mM) to either enzyme form during the various heat treatments did not affect the results. These data also demonstrate that both the recombinant and native forms of the prolidase are stable under the routine assay conditions (10 min at 100°C).
Catalytic properties of native and recombinant prolidases.
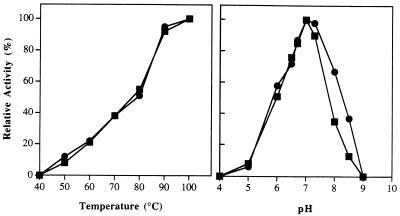
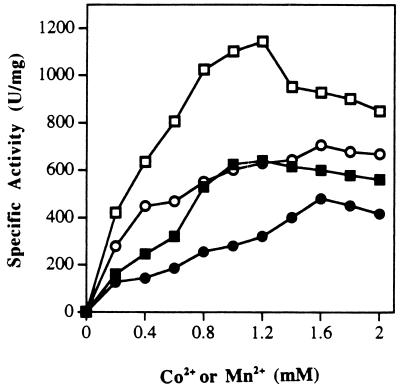
The catalytic activities of N- and R-prol showed virtually identical responses to changes in temperature and pH (Fig. 3). Both showed a pH optimum at 7.0 and a temperature optimum of ≥100°C. Indeed, under the assay conditions used, neither form exhibited detectable activity at temperatures of ≤40°C. From the temperature-dependent data, the calculated activation energies for N- and R-prol are 11.9 ± 67.34 and 10.3 ± 77.86 kcal/mol, respectively. All assay reaction mixtures used for both N- and R-prol included 1.2 mM Co2+, and these ions could not be replaced with other divalent (Mg2+, Ca2+, Fe2+, Zn2+, Cu2+, or Ni2+) or monovalent (Na+ or K+) cations (no activity was detected), with the exception of Mn2+. The effects of Co2+ and Mn2+ concentrations on the activities of N- and R-prol are shown in Fig. 4. The two enzyme forms showed very similar responses, with maximal activities at concentrations of 1.2 mM CoCl2 and 1.6 mM MnCl2, with the latter supporting approximately 75% of the activity of the former. However, both cations caused some inhibition when added above their optimal concentration (Fig. 4). The apparent association constants for Co2+ and Mn2+ were 0.24 and 0.62 mM for N-prol and 0.5 and 0.66 mM for R-prol, respectively. When N-prol was incubated with 1 mM Co2+ ions and then assayed in the absence of the metal, there was no difference in the specific activity (compared to standard conditions where Co2+ ions are included in the assay medium). However, when the sample was preincubated with 1 mM Co2+ and then dialyzed (against 3,000 volumes of 50 mM MOPS buffer, pH 7.0), only 5% of the activity remained. Addition of Co2+ ions (1.0 mM) to the dialyzed enzyme completely restored enzyme activity (assayed in the absence of Co2+ ions), but when EDTA (1 mM) was also added, no activity was detected. It therefore appears that the purified forms of both N- and R-prol contain one tightly bound Co2+ ion per subunit, but that one (or more) additional cation(s), which can be either Co2+ or Mn2+, is required for activity.
FIG. 3.
The effects of pH (A) and temperature (B) on the activities of N-prol (squares) and R-prol (circles). The assay mixtures contained prolidase (0.015 μg), Met-Pro (4 mM), and CoCl2 (1.2 mM). For determination of the effects of pH, the following buffers (each at 50 mM) were used: sodium acetate, pH 5.0; bis-Tris-HCl, pH 6.0; MOPS, pH 7.0; EPPS, pH 8.0; CHES (2-[N-cyclohexylamino]-ethanesulfonic acid), pH 9.0; and CAPS, pH 10.0. The assays were carried out at 100°C. For determination of the effects of temperature, the buffer used was 50 mM MOPS, pH 7.0. An N-prol activity level of 100% corresponds to 600 U/mg while 100% R-prol activity corresponds to 1,250 U/mg (with Met-Pro as substrate and measured at 100°C).
FIG. 4.
The effects of Co2+ and Mn2+ ions on the activities of N- and R-prol. The assay mixtures contained 0.02 μg of N-prol (solid symbols) or R-prol (open symbols), Met-Pro (4 mM), and various concentrations of either CoCl2 (squares) or MnCl2 (circles).
Prolidase was identified by its ability to hydrolyze the dipeptide Met-Pro, and this substrate was used in all routine assays. The activities of N- and R-prol with other peptides are shown in Table 3. Both gave virtually identical results. They only hydrolyzed dipeptides with Pro at the C terminus (not the N terminus), and the nature of the N-terminal residue was critical, with significant activity occurring only with peptides containing nonpolar amino acids. Kinetic analyses were conducted using N-Prol with the five prolyl-containing dipeptides with which it showed significant activity (Table 3). All exhibited normal Michaelis-Menton-type kinetics, and the kinetic constants, shown in Table 4, were calculated from linear double-reciprocal plots. The affinities of the enzyme for Met-Pro and Leu-Pro, the two substrates with which it showed the highest kcat/Km values, are lower than might have been anticipated, but the kcat/Km values for all five dipeptides are not too dissimilar, suggesting perhaps that all are of physiological significance. The affinity of R-prol for Met-Pro is comparable to that of the native enzyme (Table 4) although the kcat value is more than twofold higher, consistent with the results obtained under standard assay conditions. The activities of the native and recombinant forms were not significantly affected when they were treated (at 25°C for 30 min, prior to assaying under standard conditions) with any of the following protease (thiol or serine) inhibitors (each at 1 mM final concentration): iodoacetate, ES-64 (l-transepoxysuccinyl-leucylamido (4-guanido) butane), N-ethyl maleimide, phenyl methane sulfonyl fluoride, or diisopropylphosphofluoride.
TABLE 3.
Substrate specificity of the native and recombinant forms of P. furiosus prolidase
| Substratea | Relative activity (%)b
|
|
|---|---|---|
| N-prol | R-prol | |
| Met-Pro | 100 | 100 |
| Leu-Pro | 75 | 79 |
| Val-Pro | 46 | 10 |
| Phe-Pro | 25 | 24 |
| Ala-Pro | 23 | 17 |
| Lys-Pro | 4 | 10 |
| Gly-Pro | 0 | 1 |
| Pro-Ala | 0 | 0 |
| Pro-Hydroxypro-Pro | 0 | 0 |
| Lys-Trp-Ala-Pro | 1 | 0 |
| Gly-Arg-Gly-Asp-Thr-Pro | 0 | 0 |
| Pro-Pro-Gly-Phe-Ser-Pro | 1 | 0 |
| Pro-Pro-Gly-Phe-Ser-Pro-Phe-Arg | 0 | 0 |
| N-acetyl Pro | 0 | 0 |
All substrates were used at a final concentration of 4 mM.
The rate of hydrolysis is expressed as a percentage of the activity compared to that obtained at 100°C with Met-Pro as the substrate. An activity of 100% corresponds to 600 U/mg for N-prol and 1,350 U/mg for R-prol.
TABLE 4.
Kinetic parameters for substrates of P. furiosus prolidase
| Prolidase | Substratea | Km (mM) | Vmax (μmol/min/mg) | kcatb (s−1) | kcat/Km (mM−1s−1) |
|---|---|---|---|---|---|
| N-prol | Met-Pro | 2.8 | 645 | 271 | 97 |
| Leu-Pro | 3.0 | 645 | 271 | 90 | |
| Val-Pro | 4.2 | 175 | 74 | 18 | |
| Ala-Pro | 8.3 | 250 | 105 | 13 | |
| Phe-Pro | 20.0 | 1,000 | 420 | 21 | |
| R-prol | Met-Pro | 3.3 | 1,250 | 525 | 45 |
All assays were carried out at 100°C in 50 mM MOPS, pH 7.0, containing 1.2 mM CoCl2.
Based on a minimum molecular mass of 39.4 kDa.
DISCUSSION
P. furiosus contains significant intracellular concentrations of prolidase, an enzyme that appears to hydrolyze only dipeptides that contain Pro at the C terminus and a nonpolar residue at the N terminus. This finding is consistent with the proteolytic nature of the organism, although the rather high Km values (3 to 20 mM) determined for the enzyme’s substrates suggest that such dipeptides must be present at significant intracellular concentrations in vivo. The gene encoding the enzyme was successfully expressed in E. coli although, surprisingly, the recombinant form had a higher specific activity than the native prolidase. Why this is the case is unclear since the molecular weight (as judged by SDS-gel electrophoresis), N-terminal amino acid sequence, activation by metal ions (Co2+ and Mn2+), temperature and pH dependence, and substrate specificities of the two enzyme forms were indistinguishable. The fact that the recombinant form was slightly less thermostable than the native protein suggests that it may not be completely folded, and perhaps this additional flexibility leads to enhanced catalytic activity.
Kinetic and metal analyses indicated that P. furiosus prolidase (both native and recombinant) has at least two binding sites per subunit for Co2+ ions. One appears to be an integral part of the protein (and not removed by purification or dialysis) while the other(s) has an association constant of ∼0.3 mM and is essential for catalysis. In this regard the P. furiosus enzyme resembles certain members of the broad class of binuclear metallohydrolases represented by the N-terminal exopeptidases, the active sites of which also contain two metal ions that typically differ in their exchange kinetics (50). Prototypical members of this family are bovine leucine aminopeptidase (15) and Aeromonas proteolytica aminopeptidase (20), each of which contains two Zn2+ ions per catalytic unit. The zinc atoms of the A. proteolyticus enzyme can be replaced in vitro with cobalt, and apparently like P. furiosus prolidase, this amino peptidase can be prepared containing just one metal ion per active site (7). The only naturally occurring, cobalt-dependent members of this enzyme class are the methionine aminopeptidases (4), and these contain two Co2+ ions per active site. The crystal structure of the E. coli enzyme (42) shows that the two cobalt ions are coordinated by five amino acid residues (Asp97, Asp108, His171, Glu204, and Glu235), and all five are conserved in the sequences of the other methionine aminopeptidases (16, 41, 47). Interestingly, although the amino acid sequence of P. furiosus prolidase shows no significant similarity with those of methionine aminopeptidases, all five of the cobalt-coordinating residues are conserved in the P. furiosus enzyme (Asp209, Asp220, His280, Glu313, and Glu327) (Fig. 5). Clearly, spectroscopic and structural analyses of the P. furiosus enzyme will be required to determine if it does, in fact, contain a binuclear cobalt site and if the site is analogous to that of the methionine aminopeptidase. Such studies are in progress. In this regard, the ability of P. furiosus prolidase to be activated by Mn2+ ions suggests that an active enzyme form containing a Co-Mn binuclear center should be possible, and this should facilitate the interpretation of spectroscopic data. On the other hand, both R- and N-prol also contained significant amounts of zinc. This appears to be nonspecifically bound, however, as Zn2+ ions did not support enzyme activity (in place of Co2+ or Mn2+ ions) and typical zinc-binding motifs, e.g., HEXXH (32), were not present in the sequence.
FIG. 5.
Alignment of the amino acid sequence of P. furiosus prolidase with other prolidases (Prol), Alteromonas OPAA, and E. coli methionine aminopeptidase (MAP). The GenBank accession numbers for the prolidases other than the one sequenced in this work are as follows: P46545, L. delbrueckii prolidase; P15034, E. coli prolidase; U56398, Alteromonas OPAA; and P07906, E. coli methionine aminopeptidase. Identical residues are designated by the gray shading while similar residues are boxed. The five residues that are ligands to the binuclear cobalt site in the subunit of E. coli methionine aminopeptidase are indicated by asterisks.
Database searches indicated that the amino acid sequence of P. furiosus prolidase showed significant similarity to the sequences of all known prolidases and to a putative prolidase in the genome sequence of the archaeon Methanococcus jannaschii (14). The P. furiosus enzyme showed overall similarities of 69, 61, 58, 56, and 53% with respect to the prolidases from M. jannaschii, Lactobacillus delbrueckii, Haemophilus influenzae, and E. coli and the human prolidase, respectively. In all of these enzymes, there are three extended regions of identity in the C terminus, YFXHXLGHXVGLEVHE (P. furiosus prolidase residues 277 to 292), GMVXTIEPGIY (residues 307 to 317), and GGVRIED (residues 322 to 328). These regions contain three of the five putative Co2+-binding residues mentioned above for the P. furiosus enzyme and presumably form the active-site residues in all of these enzymes. Thus, of the five residues in P. furiosus enzyme that are proposed to bind Co2+ ions, all of them are conserved in the other prolidases (Fig. 5).
The prolidase from P. furiosus represents the first such enzyme to be purified from either an archaeon or a hyperthermophile. All other prolidases are from mesophilic sources (10, 12, 24, 25, 27, 46, 52) and are maximally active at temperatures up to 55°C. As might be expected, the P. furiosus enzyme is by far the most thermostable example of a prolidase, with a temperature optimum above 100°C and no loss of activity after 12 h at this temperature. Indeed, it is one of the most thermostable enzymes known of any type (1). Like the P. furiosus enzyme, the prolidases from X. maltophilia (46) and from eucaryotic sources (guinea pig brain [12], human erythrocytes [24], and bovine intestine [52]) are dimers (although their subunits are larger, 50 to 58 kDa), whereas the enzymes from Lactobacillus lactis (10) and L. casei (25) are monomers (∼42 kDa). So far, however, P. furiosus prolidase is the only one that has been shown to contain cobalt as an integral part of the protein. The enzymes from L. casei, X. maltophilia, and human beings are activated by Mn2+ although their metal contents have not been reported, while it is not known if the prolidases from Aureobacterium esteraromaticum (27), L. lactis, guinea pig brain and bovine intestine contain a metal center or if they are activated by any metal ion. However, the high similarities in the sequences of these enzymes (Fig. 5), and the conservation of the five putative Co-binding residues found in P. furiosus, suggest that they all contain a similar and presumably binuclear metal center, even though the nature of the metal (Mn2+ or Co2+) may not be the same.
On the other hand, there are differences in substrate specificities of the various prolidases. For example, the P. furiosus enzyme is the only one which utilizes only dipeptides with proline at the C (but not the N) terminus. The prolidases of L. lactis and A. esteraromaticum hydrolyze dipeptides with Pro at either the N- or C-terminal position, and the enzymes from L. casei and guinea pig brain efficiently cleave some dipeptides with no prolyl residue. Similarly, the mesophilic prolidases (with the exception of the L. casei enzyme) are inhibited by cysteine protease inhibitors such as N-ethyl maleimide and p-chloromercuribenzoate, suggesting that a reactive cysteine is required for catalysis. However, the P. furiosus enzyme was not inhibited by these reagents. Only one Cys residue is present in this prolidase, and this residue is not conserved in any of the mesophilic prolidases, which contain between 2 (M. jannaschii) and 12 (human) Cys residues per subunit. Thus, it would seem unlikely that a Cys residue is directly involved in the catalytic mechanism of any of these prolidases.
The only other enzyme with which the P. furiosus prolidase showed significant sequence similarity was that of OPAA from Alteromonas (51% similarity and 24% identity). This enzyme is capable of hydrolyzing a variety of toxic organophosphorus compounds (17). Although OPAA is a monomeric enzyme, in contrast to dimeric P. furiosus prolidase, its activity is dependent upon Mn2+ ions. Moreover, the five residues proposed to coordinate the binuclear metal center in the P. furiosus enzyme are also conserved in the sequence of OPAA (Fig. 4). Thus, while OPAA has a broad substrate specificity and is capable of hydrolyzing P-F, P-C and P-O bonds (17), it exhibits comparable activity with prolidase-type dipeptides such as Leu-Pro and Ala-Pro. Furthermore, like the P. furiosus enzyme, it does not hydrolyze tri- or tetrapeptides or dipeptides with Pro at the N terminus (18). Clearly, these two enzymes are closely related, and the effectiveness of the P. furiosus enzyme in degrading organophosphorus compounds is currently being explored.
ACKNOWLEDGMENTS
This research was supported by grants from the Office of Industrial Technology of the Department of Energy (SW994-19/RXE-7-17039) and the National Science Foundation (BCS-9632657).
REFERENCES
- 1.Adams M W W, editor. Enzymes and proteins from hyperthermophilic microorganisms. Adv Protein Chem. 1996;48:1–509. [Google Scholar]
- 2.Adams M W W, Kletzin A. Oxidoreductase-type enzymes and redox proteins involved in the fermentative metabolisms of hyperthermophilic archaea. Adv Protein Chem. 1996;48:101–180. doi: 10.1016/s0065-3233(08)60362-9. [DOI] [PubMed] [Google Scholar]
- 3.Andreotti G, Cubellis M V, Nitti G, Sannia G, Mai X, Adams M W W, Marino G. An extremely thermostable aromatic aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim Biophys Acta. 1995;1247:90–96. doi: 10.1016/0167-4838(94)00211-x. [DOI] [PubMed] [Google Scholar]
- 4.Arfin S M, Kendall R L, Hall L, Weaver L H, Stewart A L, Matthews B W, Bradshaw R A. Eukaryotic methionyl aminopeptidases: two classes of cobalt-dependent enzymes. Proc Natl Acad Sci USA. 1995;92:7714–7718. doi: 10.1073/pnas.92.17.7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Attaway H, Nilson J O, Baya A M, Voll M J, White W E, Grimes D J, Colwell R R. Bacterial detoxification of diisopropyl fluorophosphate. Appl Environ Microbiol. 1987;53:1685–1689. doi: 10.1128/aem.53.7.1685-1689.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer M W, Halio S B, Kelly R M. Proteases and glycosyl hydrolases from hyperthermophilic microorganisms. Adv Protein Chem. 1996;48:271–310. doi: 10.1016/s0065-3233(08)60364-2. [DOI] [PubMed] [Google Scholar]
- 7.Bennett B, Holz R C. EPR studies on the mono- and dicobalt (11)-substituted forms of the aminopeptidase from Aeromonas proteolytica. Insight into the catalytic mechanism of dinuclear hydrolases. J Am Chem Soc. 1997;119:1923–1933. [Google Scholar]
- 8.Blumentals I I, Robinson A S, Kelly R M. Characterization of sodium dodecyl sulfate-resistant proteolytic activity in the hyperthermophilic archaebacterium Pyrococcus furiosus. Appl Environ Microbiol. 1990;56:1255–1262. doi: 10.1128/aem.56.7.1992-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bockelmann W. The proteolytic system of starter and non-starter bacteria: components and their importance for cheese ripening. Int Dairy J. 1995;5:977–994. [Google Scholar]
- 10.Booth M, Jennings V, Fhaolain I N, O’Cuinn G. Prolidase activity of Lactobacillus lactis subsp. cremoris AM2: partial purification and characterization. J Dairy Res. 1990;57:245–254. [Google Scholar]
- 11.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 12.Browne P, O’Cuinn G. The purification and characterization of a proline dipeptidase from guinea pig brain. J Biol Chem. 1983;268:6147–6145. [PubMed] [Google Scholar]
- 13.Bryant F O, Adams M W W. Characterization of hydrogenase from the hyperthermophilic archaebacterium, Pyrococcus furiosus. J Biol Chem. 1989;264:5070–5079. [PubMed] [Google Scholar]
- 14.Bult C J, White O, Olsen G J, Zhou L X, Fleischmann R D, Sutton G C, Blake J A, Fitzgerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hana M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 15.Burley S K, David P R, Sweet R M, Taylor A, Lipscomb W N. Structure determination and refinement of bovine lens leucine aminopeptidase and its complex with bestatin. J Mol Biol. 1992;224:113–140. doi: 10.1016/0022-2836(92)90580-d. [DOI] [PubMed] [Google Scholar]
- 16.Chang Y H, Teichert U, Smith J A. Molecular cloning, sequencing, deletion, and overexpression of a methionine aminopeptidase gene from Saccharomyces cerevisiae. J Biol Chem. 1992;267:8007–8011. [PubMed] [Google Scholar]
- 17.Cheng T-C, Harvey S V, Stroup A N. Purification and properties of a highly active organophosphorus acid anhydrolase from Alteromonas undina. Appl Environ Microbiol. 1993;59:3138–3140. doi: 10.1128/aem.59.9.3138-3140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng T C, Harvey S P, Chen G L. Cloning and expression of a gene encoding a bacterial enzyme for decontamination of organophosphorus nerve agents and nucleotide sequence of the enzyme. Appl Environ Microbiol. 1996;62:1636–1641. doi: 10.1128/aem.62.5.1636-1641.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherry J L, Young H H, Disera L J, Ferguson F M, Kimball A W, Dunn D M, Gesteland R F, Weiss R B. Enzyme-linked fluorescent detection for automated multiplex DNA-sequencing. Genomics. 1994;20:68–74. doi: 10.1006/geno.1994.1128. [DOI] [PubMed] [Google Scholar]
- 20.Chevrier B, Schalk C, Dorchymont H, Rondeau J M, Tarnus C, Moras D. Crystal structure of Aeromonas proteolytica aminopeptidase—a prototypical member of the Co-catalyzing zinc enzyme family. Structure. 1994;2:283–291. doi: 10.1016/s0969-2126(00)00030-7. [DOI] [PubMed] [Google Scholar]
- 21.Connaris H, Cowan D A, Sharp R J. Heterogeneity of proteinases from the hyperthermophilic archaeobacterium Pyrococcus furiosus. J Gen Microbiol. 1991;137:1193–1199. [Google Scholar]
- 22.Cunningham D F, O’Connor B. Proline specific peptidases. Biochim Biophys Acta. 1997;1343:160–186. doi: 10.1016/s0167-4838(97)00134-9. [DOI] [PubMed] [Google Scholar]
- 23.Eggen R, Geerling A, Watts J, de Vos W M. Characterization of pyrolysin, a hyperthermoactive serine protease from the archaebacterium Pyrococcus furiosus. FEMS Microbiol Lett. 1990;71:17–20. [Google Scholar]
- 24.Endo F, Tanoue A, Nakai H, Hata A, Indo Y, Titani K, Matsuda I. Primary structure and gene localization of human prolidase. J Biol Chem. 1989;264:4476–4481. [PubMed] [Google Scholar]
- 25.Fernández-Esplá M D, Martín-Hernández M C, Fox P F. Purification and characterization of a prolidase from Lactobacillus casei subsp. casei IFPL 731. Appl Environ Microbiol. 1997;63:314–316. doi: 10.1128/aem.63.1.314-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiala G, Stetter K O. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch Microbiol. 1986;145:56–61. [Google Scholar]
- 27.Fujii M, Nagaoka Y, Imamura S, Shimizu T. Purification and characterization of a prolidase from Aureobacterium esteraromaticum. Biosci Biotechnol Biochem. 1996;60:1118–1122. doi: 10.1271/bbb.60.1118. [DOI] [PubMed] [Google Scholar]
- 28.Halio S B, Blumentals I I, Short S A, Merrill B M, Kelly R M. Sequence, expression in Escherichia coli, and analysis of the gene encoding a novel intracellular protease (PfpI) from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1996;178:2605–2612. doi: 10.1128/jb.178.9.2605-2612.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halio S B, Bauer M W, Mukund S, Adams M W W, Kelly R M. Purification and characterization of two functional forms of intracellular protease PfpI from the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microbiol. 1997;63:289–295. doi: 10.1128/aem.63.1.289-295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harwood V J, Denson J D, Robinson-Bidle K A, Schreier H J. Overexpression and characterization of a prolyl endopeptidase from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1997;179:3613–3618. doi: 10.1128/jb.179.11.3613-3618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heider J, Mai X, Adams M W W. Characterization of 2-ketoisovalerate ferredoxin oxidoreductase, a new and reversible coenzyme A-dependent enzyme involved in peptide fermentation by hyperthermophilic archaea. J Bacteriol. 1996;178:780–787. doi: 10.1128/jb.178.3.780-787.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hooper N M. Families of zinc metalloproteases. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- 33.Klingeberg M, Galunsky B, Sjoholm C, Kasche V, Antranikian G. Purification and properties of a highly thermostable, sodium dodecyl sulfate-resistant, and stereospecific proteinase from the extremely thermophilic archaeon Thermococcus stetteri. Appl Environ Microbiol. 1995;61:3098–3104. doi: 10.1128/aem.61.8.3098-3104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klump H, DiRuggiero J, Kessel M, Park J B, Adams M W W, Robb F T. Glutamate dehydrogenase from the hyperthermophile Pyrococcus furiosus: thermal denaturation and activation. J Biol Chem. 1992;267:22681–22685. [PubMed] [Google Scholar]
- 35.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Landis W G, DeFrank J J. Enzymatic hydrolysis of toxic organofluorophosphate compounds. Adv Appl Biotechnol Ser. 1990;4:183–201. [Google Scholar]
- 37.Mai X, Adams M W W. Indolepyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. A new enzyme involved in peptide fermentation. J Biol Chem. 1994;269:16726–16732. [PubMed] [Google Scholar]
- 38.Mai X, Adams M W W. Characterization of a fourth type of 2-keto acid-oxidizing enzyme from hyperthermophilic archaea: 2-ketoglutarate ferredoxin oxidoreductase from Thermococcus litoralis. J Bacteriol. 1996;178:5890–5896. doi: 10.1128/jb.178.20.5890-5896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mai X, Adams M W W. Purification and characterization of two reversible and ADP-dependent acetyl coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1996;178:5897–5903. doi: 10.1128/jb.178.20.5897-5903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morikawa M, Izawa Y, Rashid N, Hoaki T, Imanaka T. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl Environ Microbiol. 1994;60:4559–4566. doi: 10.1128/aem.60.12.4559-4566.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura K, Nakamura A, Takamatsu H, Yoshikawa H, Yamane K. Cloning and characterization of a Bacillus subtilis gene homologous to E. coli secY. J Biochem. 1990;107:603–607. doi: 10.1093/oxfordjournals.jbchem.a123093. [DOI] [PubMed] [Google Scholar]
- 42.Roderick S L, Matthews B W. Structure of the cobalt-dependent methionine aminopeptidase from Escherichia coli—a new type of proteolytic enzyme. Biochemistry. 1993;32:3907–3912. doi: 10.1021/bi00066a009. [DOI] [PubMed] [Google Scholar]
- 43.Scriver R C, Smith R J, Phang J M. Disorders of proline and hydroxyproline metabolism. In: Stanbury J B, Wyngaarden J B, Fredrickson D S, Goldstein J L, Brown M S, editors. The metabolic basis of inherited diseases. New York, N.Y: McGraw Hill; 1983. pp. 360–381. [Google Scholar]
- 44.Sjostrom H, Noren O, Josefsson L. Purification and specificity of pig intestinal prolidase. Biochim Biophys Acta. 1973;327:457–470. doi: 10.1016/0005-2744(73)90429-4. [DOI] [PubMed] [Google Scholar]
- 45.Snowden L J, Blumentals I I, Kelly R M. Regulation of intracellular proteolysis in the hyperthermophilic archaebacterium Pyrococcus furiosus. Appl Environ Microbiol. 1992;58:1134–1141. doi: 10.1128/aem.58.4.1134-1141.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suga K, Kabashima T, Ito K, Tsuru D, Okamura H, Kataoka J, Oshimoto T Y. Prolidase from Xanthomonas maltophilia: purification and characterization of the enzyme. Biosci Biotechnol Biochem. 1995;59:2087–2090. doi: 10.1271/bbb.59.2087. [DOI] [PubMed] [Google Scholar]
- 47.Tsunasawa S, Izu Y, Miyagi M, Kato I. Methionine aminopeptidase from the hyperthermophile archaeon Pyrococcus furiosus: molecular cloning and overexpression in Escherichia coli of the gene, and characteristics of the enzyme. J Biochem. 1997;122:843–850. doi: 10.1093/oxfordjournals.jbchem.a021831. [DOI] [PubMed] [Google Scholar]
- 48.Voorhorst W G B, Eggen R I L, Geerling A C M, Platteeuw C, Siezen R J, DeVos W M. Isolation and characterization of the hyperthermostable serine protease, pyrolysin, and its gene from the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1996;271:20426–20431. doi: 10.1074/jbc.271.34.20426. [DOI] [PubMed] [Google Scholar]
- 49.Walter R, Simmons W H, Yoshimoto T. Proline specific endo- and exopeptidases. Mol Cell Biochem. 1980;30:111–127. doi: 10.1007/BF00227927. [DOI] [PubMed] [Google Scholar]
- 50.Wilcox D E. Binuclear metallohydrolases. Chem Rev. 1996;96:2435–2458. doi: 10.1021/cr950043b. [DOI] [PubMed] [Google Scholar]
- 51.Yaron A, Mlynar D. Aminopeptidase-P. Biochem Biophys Res Commun. 1968;32:658–663. doi: 10.1016/0006-291x(68)90289-1. [DOI] [PubMed] [Google Scholar]
- 52.Yoshimoto T, Matsubara F, Kawano E, Tsuru D. Prolidase from bovine intestine: purification and characterization. J Biochem. 1983;94:1889–1896. doi: 10.1093/oxfordjournals.jbchem.a134542. [DOI] [PubMed] [Google Scholar]