Abstract
Antibody biotinylation is a process of attaching biotin molecules to antibodies by chemically modifying specific functional groups on the antibodies without altering their antigen recognition specificity. Biotin, a small vitamin, forms a strong and specific interaction with the protein streptavidin, resulting in a stable biotin-streptavidin (biotin-STV) complex. This biotin-STV interaction is widely exploited in various biotechnological applications, including biosensors. Biosensors are analytical devices that employ biological recognition elements, such as antibodies, enzymes, or nucleic acids, to detect and quantify target analytes in a sample. Antibodies are commonly used as recognition elements in biosensors due to their high specificity and affinity. In this study, the antibody anti-Bovine Serum Albumin (αBSA) has been biotinylated at different antibody:biotin ratios, and the stability of this labeling over time has been investigated. Furthermore, the sensitivity of the biosensor for detecting the Bovine Serum Albumin (BSA) protein has been compared using the biotinylated antibody and the non-biotinylated form, showing a four-fold improvement in detection. This system was also compared with the Enzyme-Linked ImmunoSorbent Assay (ELISA) technique. The advantages of using biotinylated antibodies in biosensors include increased stability and reproducibility of the biorecognition layer, as well as flexibility in sensor design, as different biotinylated antibodies can be utilized for diverse target analytes without altering the sensor's architecture.
Keywords: Biotinylation, Optical biosensor, Sensitivity, Antibodies
1. Introduction
Due to the growing development of society, the advances made in the field of health have given fundamental importance to in vitro detection methods worldwide. Good detection in a diagnosis is the first step to getting an effective treatment, and in this sense, new analytical devices must have certain requirements such as sensitivity and specificity, which are crucial aspects. Some other characteristics that they must have include portability, ease of use, speed, and cost-effectiveness [1].
Biosensors have all these characteristics and provide detection devices with numerous advantages over traditional laboratory diagnostic methods. These advantages include sensitivity, speed, and rapid measurement, as well as their cost-effectiveness and ease of use [1,2].
In recent decades, the development of biosensors has been increasing, both due to clinical needs in the detection of an analyte and other areas, such as environmental applications. Specifically, in the last two years, due to the pandemic that has occurred worldwide, the development of biosensors for SARS-CoV-2 (COVID-19) has been remarkable [3,4].
A biosensor is an analytical device that consists of a biological detection element (molecules for specific recognition) and a physical transducer, which can be optical, mass-based, or electrochemical, among others. The interaction between them aims to transform the obtained biological signal into a measurable response that can provide quantitative or semi-quantitative information, thanks to biological recognition [2,3,5].
Within the characteristics of biosensors, it is important for them to be specific and highly sensitive. Specificity depends on the immobilized biorreceptors on the biosensor's surface, and sensitivity depends on how optimized the immobilization process is [6]. Bioreceptors can include enzymes, aptamers, whole cells, or antibodies, among others. Among these, antibodies have a great advantage due to their selectivity and sensitivity [1,2].
The use of antibodies facilitates the detection and quantification of the protein of interest in biological samples, but for this, it is necessary for the antibodies to be immobilized on the surface in the correct orientation so that the biosensor has the highest possible recognition capacity. Among the procedures described for the immobilization of antibodies, we find physical adsorption, plasma treatment, covalent bonding, and affinity techniques, such as the biotin-streptavidin (biotin-STV) system [5,6].
This biotin-STV system is based on the interaction of biotin (vitamin B7 with a molecular weight of 244.31 g/mol) [7] with STV (a homotetrameric protein derived from the bacterium Streptomyces avidinii with a molecular weight of 56 g/mol). STV is capable of binding four biotin molecules, amplifying the signal, and increasing detection sensitivity [8,9]. This system is widely used today in many immunoassays and for diagnostic purposes because it exhibits the highest non-covalent affinity known, with a Ka = 1015 M−1 [[9], [10], [11]], allowing the development of highly specific biosensors for protein identification and quantification [11]. Furthermore, this interaction shows high stability as it is resistant to extreme pH, temperature, and other denaturing agents, as well as enzymatic degradation, thereby expanding the conditions under which immunoassays can be performed highlighting its versatility as one of its advantages [[7], [8], [9]].
Immunoassays based on this technique require a prior biotinylation process of the antibodies, which involves the attachment of biotin molecules through enzymatic or chemical processes [8], being able to bind to different functional groups such as amines or carboxyls [12]. Additionally, this binding usually does not affect the biological activity or physical characteristics of the molecule [9]. One of the chemical methods of biotinylation involves the use of N-hydroxysuccinimide (NHS) esters of biotin, due to its simplicity as it only involves the mixing of this compound, which, when activated, efficiently reacts with primary amines of the lysine residues to form stable amide bonds [6,13,14].
It is essential that these antibodies are immobilized with good orientation on the surface of biosensors, and for this, there are different methods. The most conventional ones often result in poor utilization of the surface due to the way antibodies are organized. Therefore, once biotinylated, they can be anchored to the surface through interaction with STV, increasing the effective surface area as this reaction provides a flexible linkage between the surface and the immunoglobulin [7,12]. The sensitivity of the sensor is dependent on the orientation of the antibodies in the immobilization process [6].
The application of this system based on biotin-streptavidin interaction is widely distributed in the field of biosensors not only for the immobilization of antibodies but also, for example, in the electrochemical detection of proteases as biomarkers, which are an important therapeutic target in different diseases [15], in the detection of human papillomavirus [16], or in the biofunctionalization of quantum dots to detect dengue markers [17]. Also, among the most recent biosensors that have used this system is the detection of viral DNA in breast cancer [18] or of salmonella bacteria in milk [19].
Optical label-free biosensors are a type of biosensor with advantages over the label process. Our research group has developed a biosensor called Biophotonic Sensing Cells (BICELLs) and are based on two Fabry-Perot interferometers previously reported [[20], [21], [22]], whose interferometric signal is obtained using the previously validated Interferometric Optical Detection Method (IODM) (Fig. 1) [[23], [24]]. This detection method is based on the Increase Relative Optical Power (IROP%) previously described [[21], [24]].
Fig. 1.
Schematic of the detection system. In the upper part of the figure, it is possible to appreciate the KIT used in these tests and a detail of the cell. On the left side is the reading platform used. The lower part of the image shows a schematic of how this biosensor works.
The objective of this work is the establishment of a biotinylating protocol to increase the recognition sensitivity and amplify the signal of the employed optical biosensor. This is a novelty since this system has never before been used as a sensitivity enhancement associated with the IODM and the IROP (%) signal unit. This is an important step in the improvement of this type of biosensor and the method in terms of sensitivity and detection limit in order to expand its applications. For this purpose, antibodies were biotinylated with different ratios of biotin NHS ester, this biotinylation was quantified, and a stability study was conducted under different storage conditions using the Enzyme-Linked ImmunoSorbent Assay (ELISA) technique. Finally, the detection level of the biosensor was compared using the biotin-STV system and direct immobilization of antibody using oxygen plasma, quantifying the Bovine Serum Albumin (BSA) protein.
2. Materials and methods
2.1. αBSA biotination protocol
The anti-BSA (αBSA) antibody biotinylation (Sigma-Aldrich, St. Louis, MO, USA)) was carried out using the compound EZ-Link Sulfo–NHS–Biotin (ThermoFisher Scientific, Waltham, MA USA). For this, 100 μL of the antibody at a concentration of 6.2 mg/mL was mixed with different molar ratios of 10 mM biotin, specifically testing the ratios of 1:10, 1:20, and 1:50 to adjust the number of incorporated biotins. This mixture was incubated for 40 min at room temperature (RT) and then left for 1 h at 4 °C with agitation.
Next, to remove unbound biotins, the mixture was passed through Zeba Spin Desalting Columns with a molecular weight cut-off of 7 kDa (ThermoFisher Scientific). The biotinylated antibody obtained (αBSA-biotin) after passing through the column had a concentration of 0.62 mg/mL.
To verify biotinylation, yield, and differences between ratios, it was quantified using the Pierce Biotin Quantitation Kit (ThermoFisher Scientific). For this, 160 μL of Phosphate Buffer Saline (PBS) (Sigma-Aldrich) were mixed with 10 μL of the HABA (4′-hydroxyazobenzene-2-carboxylic acid)-Avidin compound, and after 15 min, the absorbance was read at 500 nm (A500 HABA/Avidin). Then, 20 μL of biotinylated antibody was added to each of the ratios in triplicate, and as a positive control, biotinylated Horseradish Peroxidase (HRP) enzyme was used. After 15 min, the absorbance at 500 nm was read again (A500 HABA/Avidin/biotin sample). Using these data, the number of biotins bound to the antibody was calculated using the HABA Calculator available on Thermofisher Scientific website (https://www.thermofisher.com/es/es/home/life-science/protein-biology/protein-labeling-crosslinking/biotinylation/biotin-quantitation-kits/haba-calculator.html).
2.2. Verification ELISA assays
ELISA plates (ThermoFisher Scientific) were coated with 50 μL of BSA (Sigma-Aldrich) at different concentrations from 3.2 × 10−4 μg/mL to 5 μg/mL in 1/5 dilutions in carbonate buffer at pH 9.6 (Sigma-Aldrich) overnight at 4 °C and then blocked with casein hydrolysate 1× (1 h at RT). After three washes with PBS-Tween 0.05 % (Sigma-Aldrich) plates were incubated with 50 μL of rabbit polyclonal antibody against BSA (1:1500, Sigma-Aldrich) for 1 h at RT as positive control and 50 μL αBSA-biotin at 1:5000 in the same conditions. After washing, plates were incubated with anti-Rabbit-HRP (αRabbit-HRP) conjugate antibody (1:12,000, Sigma-Aldrich) in the wells incubated with the αBSA, and with STV-HRP at 0.2 mg/mL for wells with the αBSA-biotin for 1 h at RT and washed three times. Finally, 3,3′,5,5′ tetrametilbenzidina (TMB) substrate was added into each well and color development monitored with an ELISA plate reader (Tecan Magellan F50) at 450 nm. Measurements were made in triplicates.
2.3. ELISA stability studies
The biotinylated αBSA at different ratios was stored in a refrigerator at 4 °C and in a freezer at −20 °C to study the stability of the biotinylation process and compare which is the best storage method. For this purpose, ELISAs were performed at 15, 30, and 90 days after being stored under both conditions, following the same procedure described earlier.
2.4. Biosensor tests with and without biotin-STV system
The assays were performed in 65-cell KITs. Each of the sensing cells, called BICELLs [[20], [21], [22]] [[20], [21], [22]] [[20], [21], [22]], has a diameter of 200 μm and is based on a SiO2 layer and a thin layer of SU-8 resin. To carry out the immobilizations, the surface is activated using O2 plasma for 45 s at 40 W, and then the αBSA antibody is incubated at a concentration of 0.62 μg/mL (1.5 μL/cell) for 3 h at 37 °C in a humid chamber, followed by overnight incubation at 4 °C. In parallel, the immobilization of STV is also performed at a concentration of 500 μg/mL (1.5 μL/cell) under the same conditions. Subsequently, a washing step is performed with H2O miliQ (H2OmQ), followed by drying with filtered compressed air. In the KIT incubated with the antibody, BSA protein is incubated at different concentrations (ranging from 1000 μg/mL to 3.9 μg/mL) with a volume of 1.5 μL and left for 2 h at 37 °C in a humid chamber. For the KIT incubated with STV, the αBSA-biotin antibody at a ratio of 20 and a concentration of 0.62 μg/mL (1.5 μL/cell) is then incubated for 2 h at 37 °C in a humid chamber. Finally, the same concentration range as the KIT without STV is incubated. After this final incubation, the KITs are washed with H2OmQ and dried with filtered compressed air. After each immobilization step, the KITs were read on the readout platform to obtain IROP (%) measurements, which are well established and widely referenced [4,21,24].
2.5. Correlation between IODM and ELISA
To compare the sensitivity obtained in the biosensor using the biotin-STV system, a sandwich-type ELISA was performed. For this, the αBSA antibody (Thermofisher Scientific) was immobilized on the plate at a 1:1000 dilution in carbonate buffer (50 μL/well) and left overnight at 4 °C. After three washes with 0.05 % PBS-Tween (200 μL/well per wash), the surface was blocked with 1× casein hydrolysate (200 μL/well) for 1 h at RT. The same wash was performed, and BSA protein at different concentrations (ranging from 15 μg/mL to 0.03 μg/mL) (100 μL/well) was incubated for 2 h at RT. Three washes were performed, and αBSA-biotin (Sigma-Aldrich) at a 1:1500 dilution (50 μL/well) was incubated for 2 h at RT. Finally, after the washes, streptavidin-HRP at 0.2 μg/mL (100 μL/well) was incubated for 1 h at RT. It was developed with TMB substrate (75 μL/well) and the reaction was stopped with 2 N HCl (75 μL/well). The obtained color was measured at 450 nm.
3. Results
3.1. Biotinylated antibody quantitation
Once the αBSA antibody was biotinylated with different molar excesses of biotin (10, 20, and 50), it was quantified using the Pierce Biotin Quantitation Kit. The HABA reagent in this kit allows for an estimation of the molar ratio of biotin per protein because when the biotinylated sample is mixed with the HABA-Avidin compound, biotin competes with HABA for binding sites on avidin. Due to its high affinity, biotin displaces HABA, resulting in a proportional decrease in absorbance at 500 nm. In this way, the absorbance of the reagent at 500 nm was measured before and after mixing it with the αBSA-biotin samples at different ratios. The results are shown in Table 1.
Table 1.
Results obtained using the Pierce Biotin Quantitation Kit.
| Positive control | Ratio 10 | Ratio 20 | Ratio 50 | |
|---|---|---|---|---|
| INITIAL ABSORBANCE (500NM) | 0.655 | 0.6841 | 0.6419 | 0.6543 |
| FINAL ABSORBANCE (500NM) | 0.552 | 0.6015 | 0.6001 | 0.6159 |
| ΔABSORBANCE (500NM) | 0.103 | 0.0826 | 0.0418 | 0.0384 |
| MOLECULAR WEIGHT (G/MOL) | 40,000 | 150,000 | 150,000 | 150,000 |
| CONCENTRATION (MG/ML) | 1 | 6.2 | 6.2 | 6.2 |
| BIOTIN MOLECULES PER IGG MOLECULE | 2 | 1 | 6 | 5 |
Based on the results, it can be observed that the 20 ratio has the highest number of biotin molecules per antibody molecule (6 in this case). There is only a one-biotin molecule difference compared to the 50 ratio, but since it is an amplification system, this difference could be significant. On the other hand, the 10 ratio turned out to be a very low ratio, as it only added one biotin molecule per antibody molecule. Similarly, these results help us verify the biotinylation protocol and quantify the biotinylation process. The data acquired is confirmed through the positive control results aligning with the product specification sheets. The reason why fewer biotin molecules were added in the 50 ratio than in the 20 ratio may be due to a competitiveness between these molecules because of the high concentration. It also corresponds to what was expected based on the specifications of the product used.
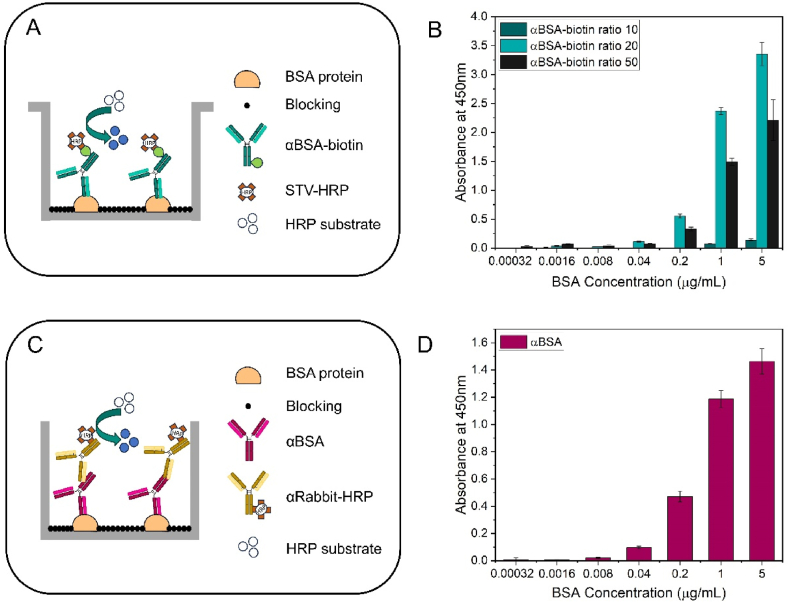
To verify the proper functioning of the biotinylated antibodies and determine which of the three ratios provides the best signal, direct ELISAs were performed (Fig. 2A). As a positive control, another direct ELISA was conducted using the αBSA antibody without biotinylation, recognizing the protein (Fig. 2C). In this way, we also tested the proper functioning of the protein-antibody pair (BSA-αBSA).
Fig. 2.
A) Description of the protocol followed in the direct ELISA for the BSA detection with the αBSA-biotin. B) Data obtained using the ELISA technique, where absorbance at 450 nm is represented based on the BSA concentration in μg/mL for the signals obtained from the three biotinylated antibodies at different ratios (10, 20, and 50). C) Detailed outline of the protocol followed in the indirect ELISA for the detection of BSA with the non-biotinylated αBSA. D) Representation of the signals obtained in the direct ELISA, as a positive control, of the absorbance versus the concentration of BSA in μg/mL.
Comparing Fig. 2B and D, it can be observed that a higher absorbance signal is obtained when using the biotinylated antibodies, due to the signal amplification that the system undergoes, being almost three times higher. On the other hand, if we compare the signals obtained in the ELISAs performed with αBSA-biotin at the three ratios in Fig. 2B, a higher signal is observed when using the biotinylated antibody at a ratio of 20, which also coincides with the quantification data since it is the antibody with the highest number of attached biotin molecules. Based on these results, the antibody biotinylated at a ratio of 10 was not used for any subsequent experiments since the signal obtained with it in this initial assay is minimal.
3.2. Stability of biotinylated antibody
To test the stability of biotinylation over time and determine the best way to store it, the biotinylated antibody at different ratios was aliquoted and stored under two different storage conditions, at 4 °C and −20 °C. ELISAs were performed at 15, 30, and 90 days. Comparing Fig. 3A and B, as in previous results, it is observed that the signals are higher when using the αBSA antibody biotinylated at a ratio of 20. Additionally, it is observed that the signal decreases over time for both conditions, obtaining a higher signal in both cases when the antibody is stored at −20 °C. Therefore, biotinylation remains stable for 90 days since the protocol was carried out, considering that there is a slight decrease in the signal, and the best way to preserve it is at −20 °C.
Fig. 3.
A) Representation of the absorbance signal at 450 nm versus the concentration of immobilized BSA in μg/mL for the αBSA-biotin antibody at a ratio of 20. B) Depiction of the absorbance signal at 450 nm versus the concentration of immobilized BSA in μg/mL for the αBSA-biotin antibody at a ratio of 50.
3.3. Evaluation of biotinyilated antibody on interferometric biosensor
Once their functionality was tested and the optimal ratio for the biotinylation protocol was selected, we proceeded to perform the experiments on the KITs. For this purpose, 65-cell KITs were used, and their surfaces were activated using an oxygen plasma process. In some KITs, the surface was biofunctionalized with streptavidin, while in others, the αBSA antibody was directly immobilized as control (Fig. 4A). After all the incubation and protein recognition steps, the ΔIROP (%) signals were compared depending on the type of assay. As observed in Fig. 4B, the signals for the detection of BSA protein at different concentrations are higher when the surface has been coated with streptavidin. Therefore, we have greater sensitivity in the sensor, being able to detect up to 3.9 μg/mL of BSA (5.9 ng total) compared to 15.62 μg/mL (23 ng total) detected without using the biotin-STV system. In this case, detection is improved almost fourfold.
Fig. 4.
A) Schematic comparison of antibody immobilization on the sensing surface with and without streptavidin. B) Comparison of the ΔIROP (%) signal obtained based on the detected concentration of BSA in μg/mL when the biosensor surface is biofunctionalized with streptavidin or not. Abbreviations: streptavidin (STV), Increase Relative Optical Power (IROP).
3.4. Comparison IODM technique with ELISA technique
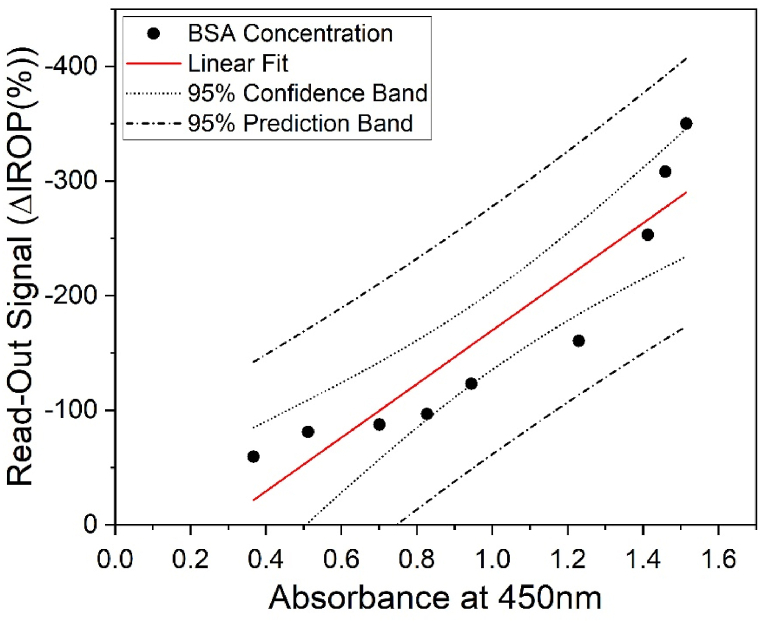
In order to compare the two techniques (ELISA and IODM), a sandwich ELISA was performed, where the ELISA plate was coated with an antibody to detect different concentrations of the BSA protein, similar to the assay conducted in the KITs. In Fig. 5, we can observe the results obtained in this ELISA for a concentration range between 15 μg/mL to 0.03 μg/mL.
Fig. 5.
Comparison between the signals obtained from the biosensor when the surface is coated with streptavidin and the sandwich ELISA for a total amount of BSA in μg.
Due to the significant difference in the volume used for incubation between the ELISA (50 μL) and IODM (1.5 μL), instead of comparing concentrations, we compare the total amount of BSA in μg determined with both techniques. In this way, 100 μL at 15 μg/mL would be equivalent to a total of 1.5 μg of BSA, which corresponds to the first measured concentration in the KIT, which is 1.5 μL at 1000 μg/mL, resulting in a total of 1.5 μg. Dilutions are then performed in a similar manner to obtain the linearity in both cases.
The results obtained through IODM under the same conditions as the ELISA are shown in Fig. 4. The comparison shows that the experimental points corresponding with confidence levels at 95 % approximately.
On the other hand, this technology allows us, thanks to the improvement with the biotin-STV system, to reach detection limits comparable to ELISA. It is important to note that in this way, we are detecting directly without the need for secondary antibodies or chemical developers, which saves steps, time, and reagents in protein concentration determination.
4. Conclusions
In this study, a IODM technique developed in previous works was used to evaluate the improvement in sensitivity of biosensors using the biotin-STV system. To achieve this, a biotinylation protocol was first established with the appropriate ratio for our application, with a ratio of 20 yielding the best results. The stability of antibody biotinylation was determined to be 90 days or potentially longer, and storage conditions were set at −20 °C to ensure better long-term performance.
Furthermore, it was demonstrated that the sensitivity was improved due to better antibody orientation facilitated by the biotin-STV system. This resulted in a more effective sensing surface capable of detecting lower concentrations, achieving an almost fourfold improvement (from 5.9 ng compared to 23 ng total) in sensitivity compared to the biosensor without the streptavidin-coated surface.
Finally, in comparison to ELISA, the biosensor based in interferometric technique exhibited a wider linear range and was capable of reaching the detection limits of the ELISA technique. It offers several advantages, such as direct detection without the use of secondary antibodies or chemical developers, reduced steps in the protocol, time and reagents used. Therefore, this system could be employed to determine different biomarkers since a universal surface is achieved in the biosensor through the immobilization of streptavidin (STV), it becomes possible to biofunctionalize it with the biotinylated antibody of interest for each application.
Funding
This work was supported by the Spanish Ministry of Economy and Competitiveness project, Spain TEC2017-84846-R, Madrid Regional Research Program IND2019/IND-17207 and Ministry of Science and Innovation project, Spain pdc2021-121424-I00.
Data availability statement
The authors do not have permission to share data.
CRediT authorship contribution statement
A.M.M. Murillo: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. M. Holgado: Funding acquisition, Investigation, Project administration, Validation, Writing – review & editing, Supervision. M. Laguna: Conceptualization, Funding acquisition, Investigation, Project administration, Supervision, Validation, Writing – review & editing.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Miguel Holgado reports financial support was provided by Spain Ministry of Science and Innovation. Maria Fe Laguna reports financial support was provided by Madrid Regional Research Program. Miguel Holgado reports financial support was provided by Spain Ministry of Economy and Competitiveness. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Long F., Zhu A., Shi H. Recent advances in optical biosensors for environmental monitoring and early warning. Sensors. 2013;13:13928–13948. doi: 10.3390/s131013928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Dorst B., Mehta J., Bekaert K., Rouah-Martin E., De Coen W., Dubruel P., Blust R., Robbens J. Recent advances in recognition elements of food and environmental biosensors: a review. Biosens. Bioelectron. 2010;26:1178–1194. doi: 10.1016/j.bios.2010.07.033. [DOI] [PubMed] [Google Scholar]
- 3.Wu C.C., Chiang Y.H., Chiang H.Y. A label-free electrochemical impedimetric immunosensor with biotinylated-antibody for SARS-CoV-2 nucleoprotein detection in saliva. Biosensors. 2022;12 doi: 10.3390/bios12050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murillo A.M.M., Tomé-Amat J., Ramírez Y., Garrido-Arandia M., Valle L.G., Hernández-Ramírez G., Tramarin L., Herreros P., Santamaría B., Díaz-Perales A., Holgado M. Developing an Optical Interferometric Detection Method based biosensor for detecting specific SARS-CoV-2 immunoglobulins in Serum and Saliva, and their corresponding ELISA correlation. Sensor. Actuator. B Chem. 2021;345 doi: 10.1016/j.snb.2021.130394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thvenot D.R., Toth K., Durst R.A., Wilson G.S. Electrochemical biosensors: recommended definitions and classification (technical report) Pure Appl. Chem. 1999;71:2333–2348. doi: 10.1351/pac199971122333. [DOI] [PubMed] [Google Scholar]
- 6.Welch N.G., Scoble J.A., Muir B.W., Pigram P.J. Orientation and characterization of immobilized antibodies for improved immunoassays. Biointerphases. 2017;12 doi: 10.1116/1.4978435. (Review) [DOI] [PubMed] [Google Scholar]
- 7.Luong J.H.T., Vashist S.K. Chemistry of biotin-streptavidin and the growing concern of an emerging biotin interference in clinical immunoassays. ACS Omega. 2020;5:10–18. doi: 10.1021/acsomega.9b03013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dundas C.M., Demonte D., Park S. Streptavidin-biotin technology: improvements and innovations in chemical and biological applications. Appl. Microbiol. Biotechnol. 2013;97:9343–9353. doi: 10.1007/s00253-013-5232-z. [DOI] [PubMed] [Google Scholar]
- 9.Wilchek M., Bayer E.A. Introduction to avidin-biotin technology. Methods Enzymol. 1990;184:5–13. doi: 10.1016/0076-6879(90)84256-G. [DOI] [PubMed] [Google Scholar]
- 10.Livnah O., Bayer E.A., Wilchek M., Sussman J.L. Three-dimensional structures of avidin and the avidin-biotin complex. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5076–5080. doi: 10.1073/pnas.90.11.5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delgadillo R.F., Mueser T.C., Zaleta-Rivera K., Carnes K.A., González-Valdez J., Parkhurst L.J. 2019. Detailed Characterization of the Solution Kinetics and Thermodynamics of Biotin, Biocytin and HABA Binding to Avidin and Streptavidin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho I.H., Paek E.H., Lee H., Kang J.Y., Kim T.S., Paek S.H. Site-directed biotinylation of antibodies for controlled immobilization on solid surfaces. Anal. Biochem. 2007;365:14–23. doi: 10.1016/j.ab.2007.02.028. [DOI] [PubMed] [Google Scholar]
- 13.Shioya R., Yamada K., Kido K., Takahashi H., Nozawa A., Kosako H., Sawasaki T. A simple method for labeling proteins and antibodies with biotin using the proximity biotinylation enzyme TurboID. Biochem. Biophys. Res. Commun. 2022;592:54–59. doi: 10.1016/j.bbrc.2021.12.109. [DOI] [PubMed] [Google Scholar]
- 14.Mao S.Y. Chapter 7: biotinylation of antibodies. Methods Mol. Biol. 2010;588:49–52. doi: 10.1007/978-1-59745-324-0. [DOI] [PubMed] [Google Scholar]
- 15.Xia N., Sun Z., Ding F., Wang Y., Sun W., Liu L. Protease biosensor by conversion of a homogeneous assay into a surface-tethered electrochemical analysis based on streptavidin-biotin interactions. ACS Sens. 2021;6:1166–1173. doi: 10.1021/acssensors.0c02415. [DOI] [PubMed] [Google Scholar]
- 16.Lv Q., Wang Y., Su C., Lakshmipriya T., Gopinath S.C.B., Pandian K., Perumal V., Liu Y. Human papilloma virus DNA-biomarker analysis for cervical cancer: signal enhancement by gold nanoparticle-coupled tetravalent streptavidin-biotin strategy. Int. J. Biol. Macromol. 2019;134:354–360. doi: 10.1016/j.ijbiomac.2019.05.044. [DOI] [PubMed] [Google Scholar]
- 17.Tran L., Park S. Highly sensitive detection of dengue biomarker using streptavidin-conjugated quantum dots. Sci. Rep. 2021;11:1–12. doi: 10.1038/s41598-021-94172-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh A., Gopinath S.C.B., Firdous S.M., Ramanathan S. Early detection of viral DNA in breast cancer using fingered aluminium interdigitated electrode modified by Streptavidin-biotin tetravalent complex. J. Indian Chem. Soc. 2022;99 doi: 10.1016/j.jics.2022.100604. [DOI] [Google Scholar]
- 19.Yue X., Sun J., Yang T., Dong Q., Li T., Ding S., Liang X., Feng K., Gao X., Yang M., Huang G., Zhang J. Rapid detection of Salmonella in milk by a nuclear magnetic resonance biosensor based on the streptavidin-biotin system and O-carboxymethyl chitosan target gadolinium probe. J. Dairy Sci. 2021;104:11486–11498. doi: 10.3168/jds.2021-20716. [DOI] [PubMed] [Google Scholar]
- 20.Lavín Á., Casquel R., Sanza F.J., Laguna M.F., Holgado M. Efficient design and optimization of bio-photonic sensing cells (BICELLs) for label free biosensing. Sensor. Actuator. B Chem. 2013;176:753–760. doi: 10.1016/j.snb.2012.09.058. [DOI] [Google Scholar]
- 21.Holgado M., Sanza F.J., López A., Lavín Á., Casquel R., Laguna M.F. Description of an advantageous optical label-free biosensing interferometric read-out method to measure biological species. Sensors. 2014;14:3675–3689. doi: 10.3390/s140203675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanza F.J., Holgado M., Ortega F.J., Casquel R., López-Romero D., Bañuls M.J., Laguna M.F., Barrios C.A., Puchades R., Maquieira A. Bio-Photonic Sensing Cells over transparent substrates for anti-gestrinone antibodies biosensing. Biosens. Bioelectron. 2011;26:4842–4847. doi: 10.1016/j.bios.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Maigler M.V., Holgado M., Laguna M.F., Sanza F.J., Santamaria B., Lavin A., Espinosa R.L. A new device based on interferometric optical detection method for label-free screening of C-reactive protein. IEEE Trans. Instrum. Meas. 2018:1–7. doi: 10.1109/TIM.2018.2876073. [DOI] [Google Scholar]
- 24.Holgado M., Maigler M.V., Santamaría B., Hernandez A.L., Lavín A., Laguna M.F., Sanza F.J., Granados D., Casquel R., Portilla J., Riesgo T. Towards reliable optical label-free point-of-care (PoC) biosensing devices. Sensor. Actuator. B Chem. 2016;236:765–772. doi: 10.1016/j.snb.2016.06.047. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors do not have permission to share data.