Abstract
Strain DS988, an Azotobacter vinelandii mutant with a reduced capacity to accumulate poly-β-hydroxybutyrate, was isolated after mini-Tn5 mutagenesis of the UW136 strain. Cloning and nucleotide sequencing of the affected locus revealed a gene homologous to Escherichia coli ptsP which encodes enzyme INtr, a homologue of enzyme I of the phosphoenol pyruvate-sugar phosphotransferase system with an N-terminal domain similar to the N-terminal domain of some NifA proteins. Strain DS988 was unable to grow diazotrophically with 10 mM glucose as a carbon source. Diazotrophic growth on alternative carbon sources such as gluconate was only slightly affected. Glucose uptake, as well as glucose kinase and glucose-6-phosphate-dehydrogenase activities that lead to the synthesis of gluconate-6-phosphate, were not affected by the ptsP mutation. The inability of DS988 to grow diazotrophically in 10 mM glucose was overcome by supplying ammonium or other sources of fixed nitrogen. Acetylene reduction activity but not transcription of the nitrogenase structural gene nifH was shown to be impaired in strain DS988 when it was incubated in 10 mM glucose. The diazotrophic growth defect of DS988 was restored either by increasing the glucose concentration to above 20 mM or by lowering the oxygen concentration. These data suggest that a mutation in ptsP leads to a failure in poly-β-hydroxybutyrate metabolism and in the respiratory protection of nitrogenase under carbon-limiting conditions.
Azotobacter vinelandii is an obligate, aerobic, nitrogen-fixing soil bacterium that undergoes differentiation by forming desiccation-resistant cysts and produces the intracellular polyester poly-β-hydroxybutyrate (PHB). Oxygen limitation initiates the synthesis of this polymer (31). Under relaxed oxygen conditions, acetyl-coenzyme A (CoA) is fed into the tricarboxylic acid cycle, and the resultant CoA inhibits the β-ketothiolase activity, which catalyzes the first step of PHB synthesis. Under oxygen limitation and carbon excess, NADPH increases and inhibits citrate synthase and isocitrate dehydrogenase, raising the levels of acetyl-CoA and lowering the CoA levels; thus, the inhibition of the β-ketothiolase by CoA is overcome, allowing synthesis of PHB to proceed (32).
A. vinelandii fixes nitrogen under fully aerobic growth conditions due to protection of its oxygen labile nitrogenase from inactivation. Protection of nitrogenase is achieved by two mechanisms. In the first, called respiratory protection, A. vinelandii can exhibit one of the highest known respiration rates at the expense of a high rate of carbon and energy source consumption, maintaining a low intracellular oxygen concentration. In the second mechanism, called conformational protection, when oxygen stress occurs nitrogenase undergoes a conformational switch to a reversible inactive but protected state, a process mediated by the FeSII protein (26).
A. vinelandii can grow on a wide variety of carbon sources under diazotrophic conditions (38) and transports carbohydrates by an active transport mechanism. Glucose transport is coupled to the oxidation of l-malate via the respiratory chain (3). d-Glucose is metabolized via the Entner-Doudoroff pathway (4, 17, 34).
The phosphoenolpyruvate (PEP):sugar phosphotransferase system (PTS) mediates the uptake and concomitant phosphorylation of many carbohydrates in a number of bacterial genera. Several phosphoryl-transfer proteins catalyze the relay of phosphate from PEP to the incoming sugar. Enzyme I and Hpr, encoded by ptsI and ptsH, respectively, comprise the soluble PTS proteins and transfer phosphate from PEP to all of the sugar-specific phosphoryl-carrier proteins and are called the general or energy coupling PTS proteins. The enzyme II complexes are carbohydrate specific and are composed of three or four domains organized either as individual polypeptides or as fused proteins, at least one of which is localized in the cytoplasmic membrane (19).
The glucose PTS is widely distributed in genera that are obligate or facultative anaerobes, most frequently in those that ferment glucose via the Embden-Meyerhoff-Parnas pathway, but is absent in bacteria that are strictly aerobic, such as Pseudomonas, Alcaligenes, and Azotobacter spp. (27, 28). However, a fructose PTS is present in Pseudomonas and Alcaligenes spp., where a fructose-1-phosphate kinase activity enables these bacteria to metabolize this sugar by the Embden-Meyerhoff-Parnas pathway (28). A fructose-1 kinase activity has not been detected in Azotobacter spp. (1, 33).
The PTS is not only involved in the transport and phosphorylation of carbohydrates, but it also regulates several metabolic processes, such as catabolism of carbon sources (PTS and non-PTS) by the interrelated phenomena of catabolite repression and inducer exclusion (19).
Genes encoding proteins homologous to PTS components that seem to be involved in other aspects of bacterial physiology have been reported in several bacterial species (21–23, 35). Examples of these pts genes include phbH and phbI, which are present in Alcaligenes eutrophus, where they control accumulation of the reserve polymer PHB (21). Other examples of pts genes are ptsN and npr (ptsO) of Escherichia coli and Klebsiella pneumoniae, encoded within the rpoN operon, whose products are homologous to enzyme IIAFru (IIANtr) and Hpr (Npr), respectively, and very likely regulate induction of ς54 (RpoN)-controlled promoters. In E. coli, IIANtr acts to regulate assimilation of nitrogen derived from organic sources (20). In K. pneumoniae, certain ptsN mutations increase the expression of the pnifH, pnifL, and p2glnA, ς54-dependent promoters, whereas ptsO mutations decrease the expression of these promoters, suggesting that unphosphorylated IIANtr negatively regulates ς54 (15). Since RpoN is the alternative ς54 involved in the transcription of genes related to nitrogen metabolism (among other physiological functions), it has been postulated that IIANtr and Npr could jointly function as a carbon-nitrogen coordinator (23). The enzyme I responsible for the phosphorylation of Npr and IIANtr has not been identified. An E. coli gene called ptsP, which encodes an enzyme I homologue called enzyme INtr with an N-terminal domain homologous to the N-terminal domains of some NifA proteins, has been proposed to be the Npr phosphorylating enzyme (25), but no experimental evidence has been provided.
The results reported here show the presence of a ptsP gene in A. vinelandii and provide evidence suggesting that its product is involved in the regulation of PHB metabolism and in the respiratory protection of nitrogenase under carbon-limiting conditions.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The A. vinelandii strains used in this study were UW136 (5) and MV101 (36). E. coli DH5α was used for the isolation and maintenance of plasmids. E. coli SM10 (λpir)/pUT–mini-Tn5–lacZ (7), was used as the donor strain for Tn5 mutagenesis.
A. vinelandii cells were grown in Burk’s medium supplemented with 2% sucrose (BS) or other carbon sources as indicated. Liquid cultures were carried out in 125-ml flasks, containing 25 ml of medium, on a rotatory shaker at 250 rpm and 30°C. In all growth experiments, the inoculum was grown 30 h in BS, washed twice with Burk’s buffer, and then transferred to the indicated medium. Growth is reported as the optical density at 600 nm. Identification of mutant DS988 was carried out on peptone-yeast (PY)-rich medium supplemented with 2% sucrose. The same medium, containing 2% of the indicated carbon source instead of sucrose, was used for some PHB determinations. The antibiotics and concentrations (in micrograms per milliliter) used were as follows: nalidixic acid, 20; kanamycin, 3; rifamicin, 10; tetracycline, 10; and spectinomycin, 50.
Transposon mutagenesis.
Random transposon mutagenesis of UW136 was carried out as described, with a pUT derivative containing the mini-Tn5 lacZ2 transposon (7).
Bacterial matings.
An A. vinelandii library constructed on pCP13 (13) was mobilized from E. coli DH5α to DS988 mutant in a triparental mating by using the helper plasmid pRK2013 (9) and selection on BS supplemented with kanamycin and tetracycline.
Enzyme assays.
Crude extracts for enzyme determination were prepared as follows. Cultures were centrifuged, and the harvested cells were washed twice with the appropriate buffer. The cells were resuspended in 30 mM Tris-HCl buffer (pH 8.2) for glucose kinase and glucose-6-phosphate (P) dehydrogenase determinations, or in 100 mM Tris-HCl (pH 7.88) for β-ketothiolase determinations, with subsequent cell disintegration by ultrasonic treatment at 5°C. Sonicated cell suspensions were centrifuged at 14,000 g for 10 min. For glucose kinase and glucose-6-P dehydrogenase determinations, the supernatants were centrifuged for 1.5 h at 192,000 g.
β-Ketothiolase enzyme was assayed by its thiolysis activity as described by Senior and Dawes (32). The glucose kinase activity was assayed by coupling glucose-6-phosphate dehydrogenase and then measuring the reduction of NADP at 340 nm according to the method of Angell et al. (2). For glucose-6-phosphate dehydrogenase, the reduction of NADP was measured at 340 nm by the method described by Lessie and Vander Wyk (12). Nitrogenase activity was determined in whole cells by the acetylene reduction assay, as described by Bishop et al. (6). β-Galactosidase activity was determined as described by Miller (16).
Glucose uptake.
Glucose transport was measured in cells incubated for 3 h in Burk’s medium with 11 mM glucose as the carbon source. The cells were washed twice with cold Burk’s buffer, resuspended at a concentration of 0.4 mg (dry weight) ml−1, and incubated in a reaction mixture containing 0.5 mM [14C]glucose (1 mCi mmol−1) in Burk’s buffer. Then 0.5-ml samples were removed at 0, 3, 6, 9, 12, and 15 min; the samples were filtered through Millipore HA filters (0.45-μm pore size) and washed once with Burk’s buffer containing 100 mM unlabeled glucose and twice with cold Burk’s buffer. The dried filters were counted in a liquid scintillation counter.
Determination of PHB.
The PHB content of the bacteria was determined by the spectrophotometric method of Law and Slepecky (11).
DNA manipulations.
Standard procedures for restriction endonuclease digestion, agarose gel electrophoresis, purification of DNA from agarose, and DNA ligation were carried out as described by Sambrook et al. (29). DNA sequences were determined by the dideoxy-chain termination method of Sanger et al. (30).
Construction of strain DS989.
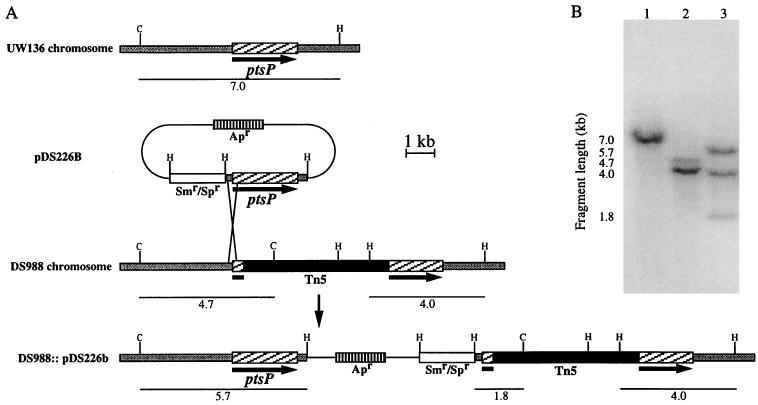
A 3.3-kb BglII DNA fragment containing the 5′ region of the ptsP gene from A. vinelandii UW136 was cloned into plasmid pUC19 to produce pDS20. A 2-kb SmaI fragment containing a tetracycline-resistant gene (Tc) from plasmid pHP45Ω-Tc (8) was inserted into the unique XhoI site present within the ptsP gene in pDS20 to create a ptsP::Tc mutation within the codon for amino acid residue 224 of enzyme INtr. The resultant plasmid pDS20A (Fig. 1), which is unable to replicate in A. vinelandii, was introduced by transformation into strain MV101, a UW136 derivative carrying a nifH::lacZ gene fusion (36). One tetracycline-resistant transformant which was less opaque than MV101, strain DS989, was chosen for further analysis. The substitution of the intact ptsP gene with the ptsP::Tc mutation on the chromosome of the DS989 mutant was confirmed by Southern blotting (data not shown).
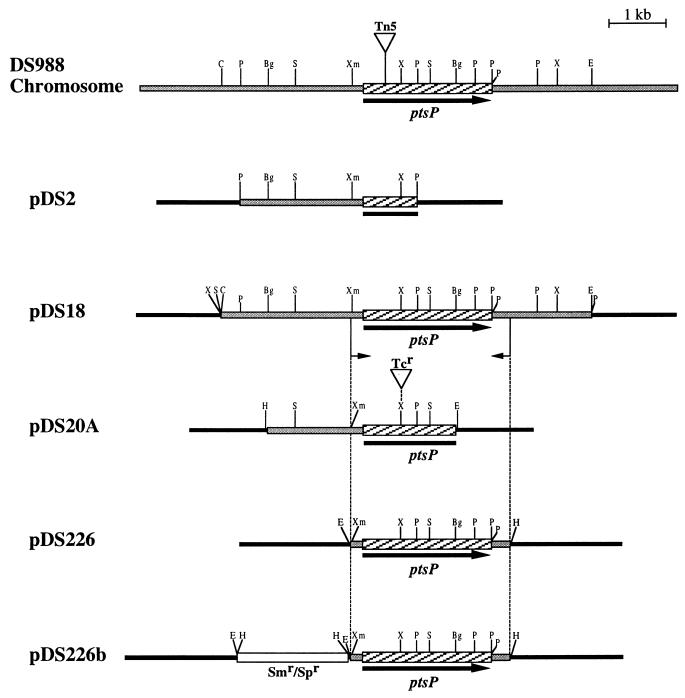
FIG. 1.
Physical map of the A. vinelandii ptsP region and plasmids constructed in this work. The ptsP gene is represented by the arrow. The transposon insertion site is indicated. Vector sequences are represented by black bars. Restriction site abbreviations: Bg, BglII; C, ClaI; E, EcoRI; H, HindIII; P, PstI; S, SalI; X, XhoI; Xm, XmnI.
Construction of plasmids pDS226 and pDS226b.
Oligonucleotides 5′-AGCAGAAGTGGTTCTCCTGC-3′ and 5′-GCATGACCCGCTCGAAGTCTT-3′ and plasmid pDS18, containing the ptsP gene in a 6.4-kb EcoRI-ClaI fragment (Fig. 1), were used to clone the ptsP gene by PCR. The resultant 2.8-kb fragment was cloned into the unique SmaI site of plasmid pUCP20 (37), producing plasmid pDS226 (Fig. 1). An EcoRI-HindIII fragment containing the ptsP gene from pDS226 was cloned into pBR329. The resultant plasmid was used to introduce an Ω-spectinomycin cassette (8) into the EcoRI site to produce plasmid pDS226b, which is shown in Fig. 1.
Nucleotide sequence accession number.
The nucleotide sequence of the ptsP gene reported here has been deposited in the EMBL GenBank and DDBJ Nucleotide Sequence Databases under accession number Y14681.
RESULTS
Isolation of strain DS988.
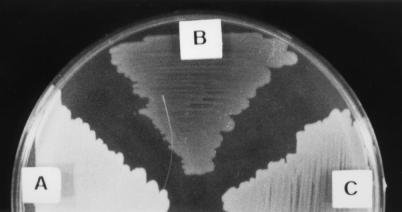
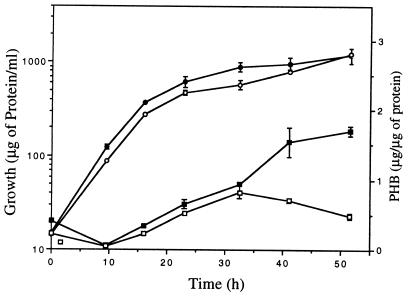
In an attempt to isolate mutants affected in the production of PHB in A. vinelandii, we carried out random mini-Tn5 mutagenesis of strain UW136. This mutagenesis produced strain DS988, which is less opaque than UW136 when grown for 4 days in PY medium plates supplemented with 2% sucrose (Fig. 2). This phenotype is due to a decreased PHB accumulation, since under this condition strain UW136 produced 936 ± 23 μg of PHB per mg of protein, whereas strain DS988 produced 83 ± 46 μg. A kinetic analysis of PHB accumulation during growth of strains UW136 and DS988 in liquid PY-sucrose is shown in Fig. 3. PHB accumulation started in the prestationary phase in both UW136 and DS988; in DS988 it stopped at 32 h, whereas in UW136 it continued during the stationary phase. In UW136 the level of PHB accumulation was 1.8-fold higher in liquid than in solid PY-sucrose cultures. The differences in PHB accumulation between UW136 and DS988 were 11-fold in solid cultures and 3.5-fold in liquid cultures. The ketothiolase activity, which catalyzes the first enzymatic step in PHB biosynthesis, was determined in UW136 and DS988 incubated during 48 h in PY-sucrose liquid cultures; this activity was about 40% of that present in the wild-type strain UW136 (Table 1).
FIG. 2.
Opacity phenotypes of A. vinelandii UW136 (A), DS988 (B), and DS988::pDS226b (C) grown on PY sucrose plates during 5 days.
FIG. 3.
Growth (circles) and PHB accumulation (squares) kinetics by UW136 (solid symbols) and DS988 (open symbols) strains.
TABLE 1.
Effect of ptsP mutation on different enzymatic activitiesa
| Strain | Activity
|
|||||
|---|---|---|---|---|---|---|
| Glucose uptake (nmol min−1 mg−1) | Glucose kinase (nmol min−1 mg−1) | Glucose-6-phosphate dehydrogenase (nmol min−1 mg−1) | Nitrogenase on glucose (nmol h−1 g−1) | Nitrogenase on gluconate (nmol h−1 g−1) | β-Ketothiolase (nmol h−1 g−1)b | |
| UW136 | 4.01 ± 0.56 | 19.7 ± 2 | 311.2 ± 10.7 | 1,096 ± 50 | 9,097 ± 260 | 2,980 ± 138 |
| DS988 | 5.02 ± 0.31 | 18.8 ± 3.2 | 256.1 ± 48.0 | NDc | 4,200 ± 612 | 1,223 ± 36 |
Cells were grown for 20 h on BS, washed with the same medium without a carbon source, and incubated for 4 h on Burk’s medium supplemented with 11 mM glucose or gluconate. Values are the means of three determinations.
β-Ketothiolase activity was determined from cells grown for 48 h in liquid PY-sucrose medium.
ND, not detected.
Cloning and DNA sequence.
An A. vinelandii cosmid gene library was introduced by conjugation into strain DS988. Two cosmid clones pMS620 and pMS3405 that restored the opacity in strain DS988 were identified.
An 8.2-kb PstI fragment containing the 5.0-kb mini-Tn5 from DS988 mutant was cloned into pBluescript. The resultant plasmid, pDS2t, hybridized to a 3.2-kb PstI fragment of UW136 DNA and to cosmids pMS620 and pMS3405 harboring the wild-type opacity-complementing region.
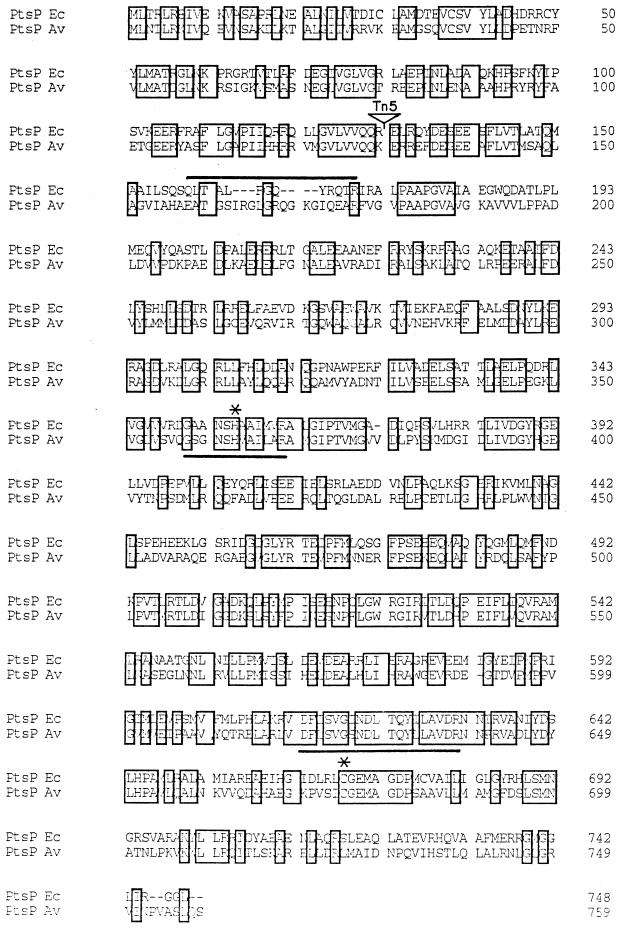
The 3.2-kb PtsI and 6.4-kb EcoRI-ClaI restriction fragments from plasmid pMS620 that hybridized with plasmid pDS2t were cloned. A restriction map of the resultant plasmids pDS2 and pDS18 is shown in Fig. 1. The DNA sequence of a 3-kb region in these plasmids revealed the presence of one open reading frame (ORF) encoding a polypeptide of 759 amino acid residues (Fig. 4), with a calculated molecular weight of 83,640. The exact location of the Tn5 mutation was determined by nucleotide sequencing across the transposon insertion junction and was found to lie within codons 130 and 131 of this ORF (Fig. 4). A database search with the amino acid sequence of this ORF established a high degree of similarity with enzyme I proteins of the PEP PTS and, specifically, an overall 43% identity with enzyme INtr encoded by the ptsP gene of E. coli (25). Accordingly, this ORF was designated ptsP. The A. vinelandii ptsP gene product has an N-terminal domain of 160 amino acids, showing a high degree of identity with the N-terminal domain of the NifA nitrogen fixation regulators of Azospirillum lipoferum, Azospirillum brasiliense, and Herbaspirillum seropedicae, as well as AnfA and VnfA of A. vinelandii (SwissProt accession numbers P54929, P30667, P27713, P12626, and P12627, respectively). As in the E. coli enzyme INtr, in the A. vinelandii protein a putative Q linker is present at the boundaries of the N-terminal and C-terminal domains (Fig. 4, amino acids 158 to 177). The phosphorylation site signature of PEP-utilizing enzymes G-[GA]-x-[TN]-x-H-[STA]-[STAV]-[LIVM](2)-[STAV]-R (Prosite name PS00370) is present at positions 358 to 369 (GSGNSHVAILAR), with the histidyl residue involved in the phosphorylation at position 363. This signature does not correspond exactly to the reported signature for this motif in positions 359 ([GA] > S) and 364 ([STA] > V). We propose the following modification for the signature of the phosphorylation site in PEP-utilizing enzymes: G-[GAS]-x-[TN]-x-H-[STAV](2)-[LIVM](2)-[STAV]-R, since only proteins belonging to this family were identified in release 34 of the SwissProt data bank with this modified signature.
FIG. 4.
Alignment of the deduced amino acid sequences of PtsP from E. coli (Ec) and A. vinelandii (Av). Identical residues are boxed. Histidine involved in phosphorylation and the active site cysteine are marked by asterisks. The signatures of PEP-utilizing enzymes are underlined. The Q linker is overlined. The arrow denotes the position of the Tn5 insertion.
A second signature, one typical of proteins of the family of PEP-utilizing enzymes, [DES]-x-[LIVMF]-2-[LIVMF]-G-[ST]- N-D-[LIVM]-x-Q-[LIVMFYG]-[STALIV]-[LIVMF]-[GAS]- x(2)-R (Prosite name PS00742), is present within amino acids 620 to 638 (DFLSVGSNDLTQYLLAVDR) of A. vinelandii enzyme INtr. A cysteinyl residue, proposed to be implicated in the active site and conserved in all members of this family (24), is present at position 675.
Growth and PHB accumulation on different carbon sources.
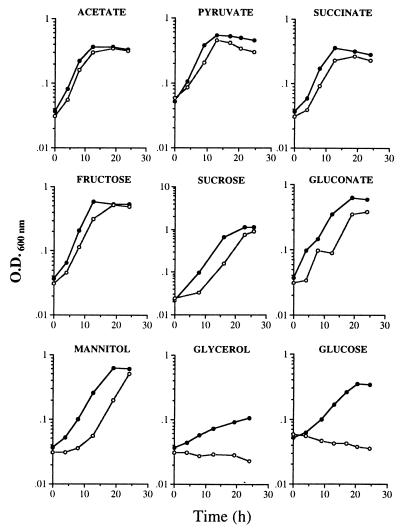
The pts genes are involved in the transport and phosphorylation of sugars, as well as in the regulation of the assimilation of carbon sources (19). Although A. vinelandii does not transport glucose by this system, we tested the ability of strain DS988 to grow diazotrophically in nitrogen-free Burk’s medium with different carbon sources, including sugars and intermediates of the tricarboxylic acid cycle. Strain DS988 grew in all of the carbon sources tested, except on 10 mM glucose or 50 mM glycerol (Fig. 5). A longer lag phase was observed in DS988 grown on sucrose, gluconate, or mannitol.
FIG. 5.
Growth of A. vinelandii strains UW136 (•) and DS988 (○) on Burk’s medium supplemented with different carbon sources. The cultures were pregrown on BS. The concentration for all carbon sources tested was 10 mM, except for acetate (20 mM) and glycerol (50 mM). The data are representative of three different experiments.
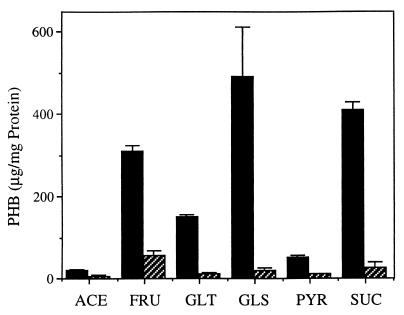
We also determined the effect of the carbon source on the accumulation of PHB by DS988. Strain DS988 was grown diazotrophically on solid Burk’s medium supplemented with 2% fructose, gluconate, glucose, or pyruvate as the sole carbon source. As seen in Fig. 6, PHB accumulation substantially diminished in all of the carbon sources tested. Similar results were obtained when the strain was grown in solid PY-rich medium supplemented with the above-mentioned carbon sources (data not shown).
FIG. 6.
PHB accumulation by A. vinelandii UW136 (solid bars) and DS988 (striped bars) grown on solid Burk’s medium supplemented with 2% different carbon sources: ACE, acetate; FRU, fructose; GLT, gluconate; GLS, glucose; PYR, pyruvate; and SUC, sucrose.
Uptake, phosphorylation, and catabolism of glucose is not affected by the ptsP mutation.
In A. vinelandii the Entner-Doudoroff pathway is the major route for glucose catabolism (4). This, along with the fact that growth on gluconate was not impaired, lead us to hypothesize that either glucose uptake or the first two steps in the Entner-Doudoroff pathway leading to the formation of gluconate-6-phosphate could be affected by the ptsP mutation. Table 1 shows that this is not the case, since both glucose uptake and the glucose kinase and glucose-6-phosphate dehydrogenase activities in DS988 are similar to those of the wild type.
The ptsP mutation affects nitrogen fixation.
Since glucose catabolism seems not to be affected in strain DS988 and since growth inhibition on glucose is observed in nitrogen-free Burk’s medium, the possibility that the ptsP::Tn5 mutation affected nitrogen fixation was tested. Nitrogenase activity, determined by acetylene reduction assays, was not detected in strain DS988 when incubated in Burk’s medium supplemented with 10 mM glucose and was reduced by about 50% in the same medium supplemented with 10 mM gluconate (Table 1). In the wild-type strain UW136 this activity was found to be 9 times lower in Burk’s medium supplemented with 10 mM glucose than in the same medium supplemented with 10 mM gluconate (Table 1). Furthermore, growth on 10 mM glucose was restored by the addition of 10 mM fixed nitrogen source such as ammonium chloride, alanine, asparagine, glutamate, glutamine, or urea (data not shown).
To determine whether the effect of the ptsP mutation on nitrogen fixation was at the level of transcription of the nif structural genes, we constructed, as described in Materials and Methods, strain DS989 (an MV101 derivative, with a ptsP::Tc mutation, which is in turn a UW136 derivative carrying a nifH::lacZ gene fusion). β-Galactosidase activities, determined after 4 h of incubation on Burk’s glucose (inducing condition) and Burk’s glucose supplemented with ammonium chloride (noninducing condition), were similar in strains MV101 and DS989 (Table 2). Thus, no effect on transcription of the nifH gene by the ptsP mutation, as measured by galactosidase activity, was observed.
TABLE 2.
Effect of ptsP mutation on transcription of nifHa
| Strain | Genotype | β-Galactosidase sp act (Miller units) at:
|
||
|---|---|---|---|---|
| 0 h | 4 h
|
|||
| Without ammonium | With ammonium | |||
| MV101 | ptsP (wild type) | 135.5 ± 11.2 | 1,392.9 ± 125 | 82.4 ± 1 |
| DS989 | ptsP | 154.4 ± 51.8 | 1,219.0 ± 111 | 79.2 ± 7 |
Cells were grown for 20 h on BS and 10 mM urea, washed with Burk’s buffer, and incubated for 4 h on Burk’s medium supplemented with 11 mM glucose with or without 10 mM ammonium chloride. Values are the means of three determinations.
Glucose concentrations above 20 mM or oxygen limitation restore diazotrophic growth.
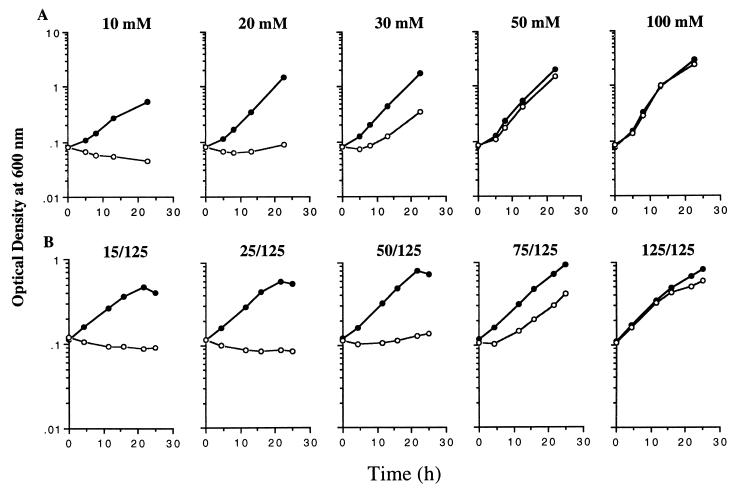
In A. vinelandii cultures, carbon limitation increased the oxygen sensitivity of nitrogenase (10). Since the growth-deficient phenotype of the DS988 mutant was observed on the carbon sources that gave the lowest growth rates (glucose and glycerol), we hypothesized that the effect on nitrogenase activity could be attributed to a defective nitrogenase protection. Diazotrophic growth of strain DS988 was restored when the glucose concentration in the medium was increased to more than 20 mM (Fig. 7A). In a similar way, the lag on gluconate was overcome by increasing its concentration in the medium (data not shown). Growth of DS988 on 10 mM glucose was also restored when the aeration of the culture was lowered by increasing the volume of medium in the flasks (Fig. 7B). These results are consistent with the hypothesis of a failure in the respiratory protection of nitrogenase in DS988.
FIG. 7.
Effect of glucose (A) or oxygen (B) concentration on the growth of A. vinelandii strains UW136 (•) and DS988 (○). Numbers in panel B indicate the volumes of medium used in 125-ml flasks. The data are representative of two different experiments.
The PHB and nitrogen fixation-deficient phenotypes are caused by the ptsP::Tn5 mutation.
Cosmids pMS620 and pMS3405 restored the wild-type colony opacity and the ability to grow on glucose with N2 (data not shown), suggesting that the phenotypes described are caused by the ptsP::Tn5 mutation. Plasmid pDS226 (Fig. 1), which is able to replicate in A. vinelandii and carries the ptsP gene flanked by 215 bp upstream of the ATG start codon and 322 bp downstream of the stop codon, failed to complement the opacity and nitrogen fixation phenotypes of strain DS988, suggesting that the promoter transcribing ptsP is not present in this plasmid or that the phenotype in DS988 is due to a polar effect on genes downstream of ptsP. The fragments containing the ptsP gene, as well as a spectinomycin gene, were cloned into plasmid pBR329; the resultant plasmid pDS226b (Fig. 1), which is unable to replicate in A. vinelandii, was transformed into DS988 for integration into the chromosome. Two types of ampicillin- and spectinomycin-resistant transformants were selected: those that showed the wild-type colony opacity (DS988::pDS226b, Fig. 2) and the ability to grow on N2 in Burk’s medium with 10 mM glucose and those with the DS988 opacity phenotype that were unable to grow on N2 in Burk’s medium with 10 mM glucose. Southern blot analysis (Fig. 8) showed that integration of pDS226b in a transformant (DS988::pDS226b) with the wild-type phenotype occurred between the ptsP promoter region and the Tn5 insertion, thus allowing the wild-type ptsP gene to be transcribed from its own promoter (Fig. 8B). These data confirm that the PHB and nitrogen fixation phenotypes are due to the ptsP mutation and not to a polar effect.
FIG. 8.
Integration of plasmid pDS226b into the chromosome of strain DS988. (A) Schematic representation of the pDS226b integration into the DS988 chromosome. (B) Southern blot hybridization of total genomic DNA from UW136 (lane 1), DS988 (lane 2), and DS988::pDS226b (lane 3) digested with ClaI and HindIII with ptsP as probe. The hybridizing fragments in panel B are as indicated in panel A.
DISCUSSION
Characterization of a mutant affected in its ability to accumulate PHB allowed us to establish the presence in A. vinelandii of a ptsP gene encoding an enzyme INtr homologue that has been recently described in E. coli. Since A. vinelandii has been reported to lack an active PEP:glucose phosphotransferase (28) and to transport glucose by another mechanism (3), the possibility that this enzyme INtr could participate in the transport of other carbohydrates such as fructose, as is the case in Pseudomonas spp. (28), was raised. However, the ptsP mutation did not markedly affect fructose utilization (Fig. 5), a finding in agreement with the absence in A. vinelandii of fructose-1-phosphate kinase activity (1); this is usually associated with PTS-dependent fructose transport in aerobic bacteria (28). Because the assimilation of other carbohydrates was also unaffected, it is unlikely that the A. vinelandii enzyme INtr participates in the transport of carbohydrates or in the regulation of its catabolism.
It has been suggested that in E. coli, enzyme INtr participates in a phosphate relay with Npr and IIANtr proteins (25). In fact, Npr can be a phosphate acceptor from PTS enzyme I-P, although this phosphorylation is less efficient than that of Hpr, and Npr-P is able to transfer the phosphate to IIANtr in vitro (20). It has also been demonstrated that IIANtr of K. pneumoniae can be a substrate of Hpr-P and that an npr mutation diminishes transcription of ς54-dependent promoters in this bacterium, implying that unphosphorylated IIANtr negatively regulates ς54. Enzyme INtr is present in A. vinelandii (this study), and although the presence of npr and ptsN homologues in A. vinelandii has not been demonstrated, sequence analysis of the rpoN region suggested the presence of these genes in this bacterium (14). We show here that although a ptsP mutation negatively affects nitrogenase activity, it does not affect transcription from the nifH ς54-dependent promoter, implying that in A. vinelandii, the enzyme INtr does not participate in the control of this ς54-dependent promoter.
The point at which nitrogenase activity is affected in strain DS988 seems to be a failure in the respiratory protection under carbon-limiting conditions, where oxygen-consuming respiratory protection is restricted. We provided evidence supporting this proposal, since increasing the glucose concentration above 10 mM reestablished the diazotrophic growth of the mutant. It has been proposed that the nitrogenase complex is more sensitive to oxygen inactivation upon energy (carbon) starvation due to a reduced flux of electrons to the complex (10). By lowering the oxygen concentration, the nitrogenase inhibition in DS988 was also overcome.
Upon energy starvation, the conformational protection mediated by the FeSII protein temporarily protects nitrogenase from inactivation and subsequent degradation (18); therefore, it is also possible that the conformational protection by FeSII is affected in DS988. It would be interesting to test a fesII mutant for growth on BS 10 mM glucose. These interpretations imply that glucose is a poor carbon source for A. vinelandii. The doubling time of strain UW136 on glucose under diazotrophic conditions is significantly longer than that observed when grown on gluconate or fructose at the same molar concentrations (Fig. 5). In addition, the high-energy-demanding nitrogenase activity was found to be nine times lower on glucose than that on gluconate (Table 1), and it has been shown that there is a relationship between nitrogenase activity and the supply of carbon source (10). These data are consistent with glucose being an energy-limiting carbon source.
The effect of the ptsP mutation on nitrogenase activity could also be a consequence of the reduced capacity for PHB accumulation. It has been suggested that PHB participates in the regulation of the intracellular oxygen environment by providing a readily oxidized carbon source that could increase the oxidative activity in the absence of exogenous substrate, thus facilitating the respiratory protection of nitrogenase (31).
The control point at which PHB accumulation is affected in DS988 is not known; involvement of PTS homologous proteins in the regulation of PHB metabolism has been reported in A. eutrophus, where mutations in either ptsI (phbI) or ptsH (phbH) genes (encoding enzyme I and Hpr homologues) caused the polymer content to decrease more rapidly than in the wild type. In addition, the opacity of the mutants decreased further after prolonged incubation (21). Thus, the decrease in PHB content seems to be due to PHB degradation. Our data do not rule out the possibility that, in A. vinelandii, the decrease in PHB content caused by the ptsP mutation is due to degradation. However, we observed neither a decrease in opacity after prolonged incubation nor a drastic decrease in PHB. Furthermore, the lower β-ketothiolase activity observed in the DS988 cells (Table 1) implies that it may be due to a decrease in its synthesis. The reduction of PHB is higher than the reduction in the ketothiolase activity; this could be explained by the presence of two ketothiolase activities in A. vinelandii (30a).
ACKNOWLEDGMENTS
This work was supported by grant IN212096 from DGAPA UNAM. D. Segura thanks CONACyT and PADEP-UNAM for financial support during work on his Ph.D. degree.
We thank S. Moreno, J. Guzmán, and Oswaldo Lopez for technical support and G. Soberón, L. Servin, and F. Bastarrachea for reviewing the manuscript.
REFERENCES
- 1.Anderson A J, Hacking A J, Dawes E A. Alternative pathways for the biosynthesis of alginate from fructose and glucose in Pseudomonas mendocina and Azotobacter vinelandii. J Gen Microbiol. 1987;133:1045–1052. [Google Scholar]
- 2.Angell S, Schwarz E, Bibb M J. The glucose kinase gene of Streptomyces coelicolor A3 (2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol Microbiol. 1992;6:2833–2844. doi: 10.1111/j.1365-2958.1992.tb01463.x. [DOI] [PubMed] [Google Scholar]
- 3.Barnes E M. Respiration-coupled glucose transport in membrane vesicles from Azotobacter vinelandii. Arch Biochem Biophys. 1972;152:795–799. doi: 10.1016/0003-9861(72)90275-5. [DOI] [PubMed] [Google Scholar]
- 4.Beale J M, Foster J L. Carbohydrate fluxes into alginate biosynthesis in Azotobacter vinelandii NCIB 8789: NMR investigations of the triose pools. Biochemistry. 1996;35:4492–4501. doi: 10.1021/bi951922v. [DOI] [PubMed] [Google Scholar]
- 5.Bishop P E, Brill W J. Genetic analysis of Azotobacter vinelandii mutant strains unable to fix nitrogen. J Bacteriol. 1977;130:954–956. doi: 10.1128/jb.130.2.954-956.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop P E, Jarlenski D M, Hetherington D R. Evidence for an alternative nitrogen fixation system in Azotobacter vinelandii. Proc Natl Acad Sci USA. 1980;77:7342–7246. doi: 10.1073/pnas.77.12.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Lorenzo V, Herrero M, Jakubzik V, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 9.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhla J, Oelze J. Dependence of nitrogenase switch-off upon oxygen stress on the nitrogenase activity in Azotobacter vinelandii. J Bacteriol. 1988;170:5325–5329. doi: 10.1128/jb.170.11.5325-5329.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Law J H, Slepecky R A. Assay of poly-β-hydroxybutyric acid. J Bacteriol. 1961;82:33–36. doi: 10.1128/jb.82.1.33-36.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lessie T G, Vander Wyk J C. Multiple forms of Pseudomonas multivorans glucose-6-phosphate and 6-phosphogluconate dehydrogenases. J Bacteriol. 1972;110:1107–1117. doi: 10.1128/jb.110.3.1107-1117.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-Salazar J M, Moreno S, Nájera R, Boucher C, Espín G, Soberón-Chávez G, Deretic V. Characterization of the genes coding for the putative sigma factor AlgU and its regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their role in alginate biosynthesis. J Bacteriol. 1996;178:1800–1808. doi: 10.1128/jb.178.7.1800-1808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merrick M J, Gibbins J, Toukdarian A. The nucleotide sequence of the sigma factor gene ntrA (rpoN) of Azotobacter vinelandii: analysis of conserved sequences in NtrA proteins. Mol Gen Genet. 1987;210:323–330. doi: 10.1007/BF00325701. [DOI] [PubMed] [Google Scholar]
- 15.Merrick M J, Taylor M, Saier M H, Reizer J. The role of genes downstream of the n N structural gene rpoN in Klebsiella pneumoniae. In: Tikhonovich I A, Provorov N A, Romanov V I, Newton W E, editors. Nitrogen fixation: fundamentals and application. Amsterdam, The Netherlands: Kluwer Academic Publishers; 1995. pp. 189–194. [Google Scholar]
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 431–435. [Google Scholar]
- 17.Morteson L E, Wilson W. Initial stages in the breakdown of carbohydrates by Azotobacter vinelandii. Arch Biochem Biophys. 1955;53:425–435. doi: 10.1016/0003-9861(54)90423-3. [DOI] [PubMed] [Google Scholar]
- 18.Moshiri F, Kim J W, Fu C, Maier R J. The FeSII protein of Azotobacter vinelandii is not essential for aerobic nitrogen fixation, but confers significant protection to oxygen-mediated inactivation of nitrogenase in vitro and in vivo. Mol Microbiol. 1994;14:101–114. doi: 10.1111/j.1365-2958.1994.tb01270.x. [DOI] [PubMed] [Google Scholar]
- 19.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase system in bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Powell B S, Court D L, Inada T, Nakamura Y, Michotey V, Cui X, Reizer A, Saier M H, Reizer J. Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. J Biol Chem. 1995;279:4822–4839. doi: 10.1074/jbc.270.9.4822. [DOI] [PubMed] [Google Scholar]
- 21.Pries A, Priefert H, Krüger N, Steinbüchel A. Identification and characterization of two Alcaligenes eutrophus gene loci relevant to the poly(β-hydroxybutyric acid)-leaky phenotype which exhibit homology to the ptsH and ptsI of Escherichia coli. J Bacteriol. 1991;173:5843–5853. doi: 10.1128/jb.173.18.5843-5853.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reizer J, Peterkofsky A, Romano A H. Evidence for the presence of heat-stable protein (Hpr) and ATP-dependent Hpr kinase in heterofermentative lactobacilli lacking phosphoenolpyruvate:glucose phosphotransferase activity. Proc Natl Acad Sci USA. 1988;85:2041–2045. doi: 10.1073/pnas.85.7.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reizer J, Reizer A, Saier M H, Jacobson G R. A proposed link between nitrogen and carbon metabolism involving protein phosphorylation in bacteria. Protein Sci. 1992;1:722–726. doi: 10.1002/pro.5560010604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reizer J, Hoischen C, Reizer A, Pham T N, Saier M H. Sequence analyses and evolutionary relationships among the energy-coupling proteins enzyme I and Hpr of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. Protein Sci. 1993;2:506–521. doi: 10.1002/pro.5560020403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reizer J, Reizer A, Merrick M J, Plunkett III G, Rose D J, Saier M H. Novel phosphotransferase-encoding genes revealed by analysis of the Escherichia coli genome: a chimeric gene encoding an enzyme I homologue that possesses a putative sensory transduction domain. Gene. 1996;181:103–108. doi: 10.1016/s0378-1119(96)00481-7. [DOI] [PubMed] [Google Scholar]
- 26.Robson R L, Postgate J R. Oxygen and hydrogen in biological nitrogen fixation. Annu Rev Microbiol. 1980;34:183–207. doi: 10.1146/annurev.mi.34.100180.001151. [DOI] [PubMed] [Google Scholar]
- 27.Romano A H, Eberhard S J, Dingle S L, McDowell T D. Distribution of the phosphoenolpyruvate:glucose phosphotransferase system in bacteria. J Bacteriol. 1970;104:808–813. doi: 10.1128/jb.104.2.808-813.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romano A H, Saier M H. Evolution of the bacterial phosphoenolpyruvate:sugar phosphotransferase system. Section I. Physiological and organismic considerations. In: Mortlock R P, editor. Evolution of metabolic function. Boca Raton, Fla: CRC Press, Inc.; 1992. pp. 143–170. [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Segura, D., E. Vargas, and G. Espín. Unpublished results.
- 31.Senior P J, Beech G A, Ritchie G A, Dawes E A. The role of oxygen limitation in the formation of poly-β-hydroxybutyrate during batch and continuous culture of Azotobacter beijerinckii. Biochem J. 1972;128:1193–1201. doi: 10.1042/bj1281193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Senior P J, Dawes E A. The regulation of poly-β-hydroxybutyrate metabolism in Azotobacter beijerinckii. Biochem J. 1973;134:225–238. doi: 10.1042/bj1340225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stephenson M P, Jackson F A, Dawes E A. Further observations on carbohydrate metabolism and its regulation in Azotobacter beijerinckii. J Gen Microbiol. 1978;109:89–96. [Google Scholar]
- 34.Still G G, Wang C H. Glucose catabolism in Azotobacter vinelandii. Arch Biochem Biophys. 1964;105:126–132. doi: 10.1016/0003-9861(64)90243-7. [DOI] [PubMed] [Google Scholar]
- 35.Titgemeyer F, Walkenhorst J, Cui X, Reizer J, Saier M H. Proteins of the phosphoenolpyruvate:sugar phosphotransferase system in Streptomyces: possible involvement in the regulation of antibiotic production. Res Microbiol. 1994;145:89–92. doi: 10.1016/0923-2508(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 36.Walmsley J, Kennedy C. Temperature-dependent regulation by molybdenum and vanadium of expression of the structural genes encoding three nitrogenases in Azotobacter vinelandii. Appl Environ Microbiol. 1991;57:622–624. doi: 10.1128/aem.57.2.622-624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.West S E H, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Contribution of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence at the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;128:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 38.Wong T Y, Maier R J. H2-dependent mixotrophic growth of N2-fixing Azotobacter vinelandii. J Bacteriol. 1985;163:528–533. doi: 10.1128/jb.163.2.528-533.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]