Abstract
The glucosamine-1-phosphate acetyltransferase activity but not the uridyltransferase activity of the bifunctional GlmU enzyme from Escherichia coli was lost when GlmU was stored in the absence of β-mercaptoethanol or incubated with thiol-specific reagents. The enzyme was protected from inactivation in the presence of its substrate acetyl coenzyme A (acetyl-CoA), suggesting the presence of an essential cysteine residue in or near the active site of the acetyltransferase domain. To ascertain the role of cysteines in the structure and function of the enzyme, site-directed mutagenesis was performed to change each of the four cysteines to alanine, and plasmids were constructed for high-level overproduction and one-step purification of histidine-tagged proteins. Whereas the kinetic parameters of the bifunctional enzyme appeared unaffected by the C296A and C385A mutations, 1,350- and 8-fold decreases of acetyltransferase activity resulted from the C307A and C324A mutations, respectively. The Km values for acetyl-CoA and GlcN-1-P of mutant proteins were not modified, suggesting that none of the cysteines was involved in substrate binding. The uridyltransferase activities of wild-type and mutant GlmU proteins were similar. From these studies, the two cysteines Cys307 and Cys324 appeared important for acetyltransferase activity and seemed to be located in or near the active site.
UDP-N-acetylglucosamine (UDP-GlcNAc), the nucleotide-activated form of N-acetylglucosamine, plays an important role in the biochemistry of all living organisms. In gram-negative bacteria, it is situated at the branch point of the biosynthetic pathways of two essential cell envelope components, peptidoglycan and lipopolysaccharides (24, 28, 31), and it is also required for the formation of the enterobacterial common antigen (15). Conditional-lethal mutants of Escherichia coli altered in the biosynthesis of this essential precursor were characterized by a cell lysis phenotype under restrictive growth conditions (27, 34, 35). The four-step formation of UDP-GlcNAc from fructose-6-P has been now completely elucidated in E. coli (9, 11, 19, 20, 34). It involves the successive actions of GlcN-6-P synthase (GlmS), phosphoglucosamine mutase (GlmM), GlcN-1-P acetyltransferase, and GlcNAc-1-P uridyltransferase (UDP-GlcNAc pyrophosphorylase). We recently showed that the two latter activities were carried by a single 456-amino-acid protein that we named GlmU. The corresponding glmU gene was located just upstream from the GlcN-6-P synthase glmS gene, in the 84-min region of the E. coli chromosome (18, 19, 22, 33).
The bifunctional GlmU enzyme has been purified to homogeneity and shown to exhibit a number of characteristics which suggested that the acetyltransferase and uridyltransferase activities may reside in separate catalytic domains (12, 19). First, the substrates, products, and effectors of the acetyltransferase reaction did not inhibit the uridyltransferase activity, and vice versa. Second, the intermediate GlcNAc-1-P was clearly released from the active acetyltransferase domain prior to transformation by the uridyltransferase domain. Third, portions of the GlmU amino acid sequence exhibiting similarities with that of other previously characterized XDP-sugar pyrophosphorylase and acetyltransferase activities were located in the N-terminal portion and the second third, respectively, of the protein. Finally, the acetyltransferase but not the uridyltransferase was inactivated by thiol-specific reagents, suggesting that a sulfhydryl group(s) may play a role in the catalytic mechanism of GlmU acetyltransferase activity.
The present study is a part of our endeavor to elucidate the structural organization, the substrate binding domains, and the role of specific amino acid residues in the structure and function of the enzyme. The E. coli GlmU enzyme contains four cysteine residues, at positions 296, 307, 324, and 385. To ascertain the role of these cysteines in the structure and function of the two catalytic domains of GlmU, each was replaced by an alanine residue by site-directed mutagenesis. The corresponding mutant proteins, designated C296A, C307A, C324A, and C385A, were purified to near homogeneity and assayed for enzyme activity, sensitivity to thiol reagents, and functional complementation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli JM83 (araΔ[lac-proAB] rpsL thi φ80 dlacZΔM15) (36) and DH5α (supE44 ΔlacU169 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 φ80 dlacZΔM15) (Bethesda Research Laboratories) were used as hosts for plasmids and for preparation of the overproduced GlmU enzymes. Strain UGS83 (JM83 glmU::kan [pGMU]), which carries an inactivated copy of the glmU gene on the chromosome and a wild-type copy of glmU on a plasmid whose replication is thermosensitive, was previously described (18). Strain BMH71-18 mutS, defective in mismatch repair, was used in site-directed mutagenesis experiments (8). 2YT (21) was used as the culture medium, and growth was monitored on the basis of optical density (OD) at 600 nm. Ampicillin, kanamycin, and chloramphenicol were used at 100, 30, and 25 μg ml−1, respectively.
Construction of plasmids.
Standard procedures for molecular cloning (6, 26) and E. coli cell transformation (5) were used. First, a plasmid suitable for overproduction of wild-type GlmU was constructed as follows. PCR primers were designed to incorporate a BspLU11I site (in bold) 5′ to the initiation codon (underlined) of glmU (33), 5′-GACGCACATGTTGAATAATGCTATGAGC-3′ (primer A), and a PstI site (in bold) 3′ to the gene after the stop codon, 5′-AGCCCTGCAGAATCACTTTTTCTTTACCGG-3′ (primer B). The DNA fragment was amplified from the E. coli chromosome, treated with BspLU11I and PstI, and ligated between the compatible NcoI and PstI sites of vector pTrc99A (Pharmacia). The resulting plasmid, pFP1, allowed expression of the wild-type glmU gene under the control of the strong isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible trc promoter. Two plasmid vectors, pTrcHis30 and pTrcHis60, were constructed for expression of N- and C-terminal His6-tagged enzymes. In these vectors, the polylinker of pTrc99A was replaced with those of pQE30 and pQE60 vectors (Qiagen), respectively. For the construction of pTrcHis30, two oligonucleotides, primer C (5′-CATGCATCACCATCACCATCACG-3′) and primer D (5′-GATCCGTGATGGTGATGGTGATG-3′), were phosphorylated, hybridized together, and inserted between the NcoI and BamHI sites of pTrc99A vector. For the construction of pTrcHis60, two oligonucleotides, primer E (5′-CATGGGAGGATCCAGATCTCATCACCATCACCATCACTA-3′) and primer F (5′-AGCTTAGTGATGGTGATGGTGATGAGATCTGGATCCTCC-3′), were phosphorylated, hybridized together, and inserted between the NcoI and HindIII sites of pTrc99A. For expression of GlmU under a C-terminal His6-tagged form, the glmU gene was amplified with primer A (see above) and primer G (5′-GCCAAGATCTCTTTTTCTTTACCGGACGACG-3′). The resulting fragment was cut with BspLU11I and BglII (in bold) and inserted between the compatible NcoI and BglII sites of pTrcHis60, generating plasmid pFP2. For expression of GlmU under an N-terminal His6-tagged form, the glmU gene was amplified with primer H (5′-GGACGGGATCCTTGAATAATGCTATGAGCGTAGTGA-3′) and primer B (see above). The resulting fragment was cut with BamHI (in bold) and PstI and inserted between the corresponding sites of pTrcHis30, generating plasmid pFP3.
Site-directed mutagenesis.
Plasmids for high-level expression of mutant GlmU enzymes were constructed by using a Transformer TM site-directed mutagenesis kit (Clontech), based on the method of Deng and Nickoloff (8). The sequences of the oligonucleotide primers chosen were 5′-TTGGCACCGGTGCCGTGATTAAAAACAGCG-3′ (C296A, primer I), 5′-GTGATTGGCGATGATGCCGAAATCAGTCCG-3′ (C307A, primer J), 5′-AATCTGGCAGCGGCCGCTACCATTGGCCCGTTT-3′ (C324A, primer K), and 5′-GCGGGAACCATTACCG CCAACTACGATGGTGCG-3′ (C385A, primer L), for replacement of cysteines by alanine residues (codons in bold) at the indicated positions, and 5′-TTGGTGCGGACATCTCGGTAG-3′ (selection primer M) for suppression of the unique EcoRV site lying within the lacIq gene of the target plasmid pFP1 (no change in the amino acid sequence of LacI). Mutagenesis (8) was performed as recommended by the manufacturer, and DNA sequencing of plasmids resistant to EcoRV digestion showed that most of them also carried the expected mutation in the glmU gene. The pFP1 derivative plasmids allowing expression of C296A, C307A, C324A, and C385A mutant enzymes were named pFP1-C296A, pFP1-C307A, pFP1-C324A, and pFP1-C385A, respectively. For expression of the N-terminal His6-tagged mutant enzymes, the 0.8-kb DraIII-PstI fragment from glmU containing the four cysteine codons was deleted from pFP3 and replaced by the corresponding mutated fragment prepared from pFP1-C296A, pFP1-C307A, pFP1-C324A, or pFP1-C385A, yielding plasmid pFP3-C296A, pFP3-C307A, pFP3-C324A, or pFP3-C385A, respectively.
Preparation of crude extracts and enzyme purification.
E. coli cells (DH5α or JM83 glmU::kan) carrying plasmids described in this work were grown at 37°C in 2YT-ampicillin medium (500-ml cultures). When the OD of the culture reached 0.1, IPTG was added at a final concentration of 1 mM, and growth was continued for 3 h (final OD = 1). Cells were harvested and washed with 40 ml of cold 20 mM potassium phosphate buffer (pH 7.2) containing 0.5 mM MgCl2 and 0.1% β-mercaptoethanol. The cell pellet was suspended in 5 ml of the same buffer supplemented with a mixture of protease inhibitors: 1 μM leupeptin, 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, and 20 μg of trypsin inhibitor ml−1. Cells were disrupted by sonication in the cold, and the resulting suspension was centrifuged at 4°C for 30 min at 200,000 × g. The supernatant was dialyzed against 100 volumes of the same buffer. Quantitation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of proteins were performed as described earlier (4, 16).
A previously described procedure (18, 19) was used for large-scale preparation of the wild-type GlmU enzyme (18 liters of culture, yielding 700 mg of pure enzyme). Lower amounts of the different His6-tagged enzymes were prepared (500-ml cultures, yielding 5 to 10 mg of enzyme). For the latter preparations, a one-step purification procedure was carried out under native conditions, basically according to the protocol of the manufacturer (Qiagen): binding of His6-GlmU on Ni2+-nitrilotriacetate-agarose (Ni2+-NTA) and washing with 50 mM sodium phosphate buffer (pH 6), containing 0.3 M NaCl, 0.1% β-mercaptoethanol, 20 mM imidazole, and 10% glycerol to remove impurities; elution of His6-GlmU with imidazole (50 to 350 mM) added to washing buffer; and dialysis of His6-GlmU eluate against 20 mM potassium phosphate buffer (pH 7.2) containing 0.1% β-mercaptoethanol and 10% glycerol. The His6-tagged GlmU enzymes prepared in this manner were in all cases 90% pure, as estimated by SDS-PAGE (data not shown).
Enzymatic assays.
Assays for both activities of GlmU were performed as described previously (18, 19). Appropriate dilutions of the enzyme were performed in 20 mM potassium phosphate buffer (pH 7.2) containing 1 mg of bovine serum albumin ml−1 0.5 mM MgCl2, and 0.1% (14 mM) β-mercaptoethanol. One unit of enzyme activity was defined as the amount which catalyzed the synthesis of 1 μmol of product in 1 min. As required for assays in the presence of thiol reagents, the reducing agent was removed from the enzyme by repeated concentrations and dilutions on Centricon-10 membranes (Amicon) at 4°C (no loss of enzyme activity occurred during this procedure). Enzyme was incubated with N-ethylmaleimide (NEM), 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB), 2-nitro-5-thiocyanobenzoic acid (NTCB), p-hydroxymercuribenzoic acid (pHMB), or iodoacetamide in 20 mM potassium phosphate buffer (pH 7.2) prior to assay at 37°C. When required, substrates or β-mercaptoethanol were added in the incubation mixture 30 s before addition of the thiol reagent.
RESULTS AND DISCUSSION
We previously demonstrated that the acetyltransferase activity of GlmU was rapidly lost during chromatographic procedures or incubations in which a thiol-protective agent such as β-mercaptoethanol was omitted or present at concentrations lower than 10 mM (19). For the present study, we made a fresh large-scale preparation of pure GlmU to reinvestigate some kinetic properties of the enzyme and initiate crystallization experiments. Interestingly, a considerable increase in enzyme activity was observed when the entire purification procedure was performed in the presence of a very high concentration of β-mercaptoethanol (14 mM) and 10% glycerol and when appropriate dilutions prior to enzymatic assays were done in phosphate buffer supplemented with 1 mg of bovine serum albumin ml−1. As shown in Table 1, the kcat observed for the acetyltransferase activity of the pure wild-type enzyme was 1,500 s−1, a value 15- to 20-fold higher than those previously reported (12, 19). The uridyltransferase activity of the enzyme preparation was also increased by a similar factor, and the kcat value determined was 350 s−1, compared with 12 to 44 s−1 in previous studies (7, 12, 18).
TABLE 1.
Kinetic parameters of wild-type and mutant GlmU enzymes
| Enzymea |
Km (mM)b
|
kcat (s−1)b
|
||
|---|---|---|---|---|
| GlcN-1-P | AcetylCoA | Acetyl- transferase | Uridyl- transferase | |
| Wild-type GlmU | 0.15 | 0.6 | 1,500 | 350 |
| GlmU-His6 | ND | ND | 120 | 250 |
| His6-GlmU | 0.2 | 0.2 | 1,350 | 330 |
| His6-GlmU C296A | 0.07 | 0.2 | 1,350 | 300 |
| His6-GlmU C307A | 0.25 | 0.09 | 1 | 290 |
| His6-GlmU C324A | 0.12 | 0.14 | 160 | 270 |
| His6-GlmU C385A | 0.2 | 0.25 | 1,450 | 310 |
GlmU His6 and His6-GlmU represent C-terminal and N-terminal, respectively, His6-tagged forms of GlmU.
Average of at least two independent experiments. Standard deviations were less than 10%. ND, not determined.
Inactivation of GlmU acetyltransferase by thiol-specific reagents.
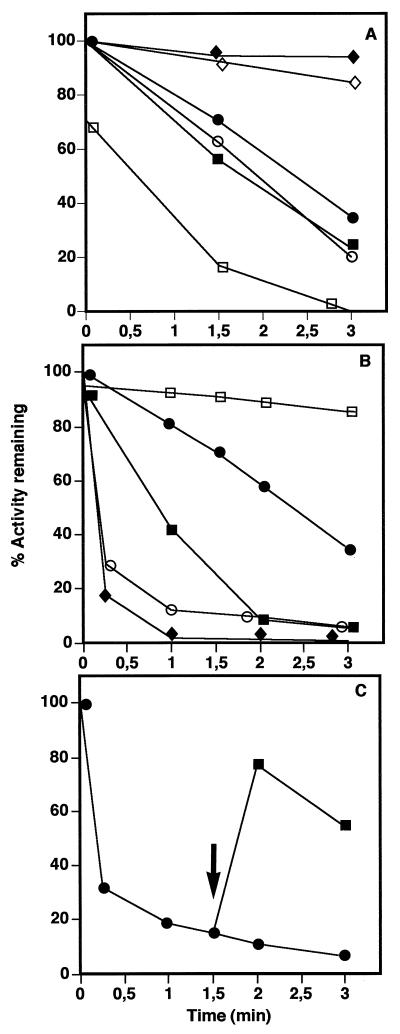
The sensitivity of this new enzyme preparation toward spontaneous inactivation in the absence of a thiol-reducing agent or inactivation by thiol-specific reagents was investigated. A significant loss of acetyltransferase activity was observed when the enzyme was incubated in the absence of β-mercaptoethanol (Fig. 1A). This activity was also readily inactivated within a few minutes by low concentrations of all reagents tested: NEM, DTNB, NTCB, pHMB, and iodoacetamide (Fig. 1B). In each case, the inactivation was time and dose dependent. Furthermore, we showed that inactivation by DTNB was a reversible process, as the activity of the enzyme could be almost quantitatively recovered after addition of 0.1% β-mercaptoethanol (Fig. 1C). The presence of either the substrate acetylcoenzyme A (acetyl-CoA) or β-mercaptoethanol protected the enzyme from spontaneous or reagent-induced inactivation. As observed previously (19), the acetyltransferase was inhibited by its reaction product GlcNAc-1-P. However, neither this compound nor GlcN-1-P, the other substrate of the acetyltransferase, nor UTP, the substrate of the uridyltransferase, protected against inactivation (Fig. 1A). All of these results suggested the presence of cysteine residue(s) in or near the active site of the acetyltransferase. The uridyltransferase activity of GlmU was comparatively very stable and completely insensitive to all the thiol-specific reagents described above (data not shown), indicating that the aforementioned cysteine residue(s) was not involved in the second activity of the bifunctional enzyme.
FIG. 1.
Inactivation of wild-type GlmU acetyltransferase by thiol-specific reagents. (A) Wild-type GlmU was stripped of β-mercaptoethanol and incubated at 37°C in 20 mM potassium phosphate buffer in the absence (•) or presence of 14 mM β-mercaptoethanol (◊), 1.6 mM acetyl-CoA (⧫), 4 mM GlcN-1-P (○), 2.7 mM GlcNAc-1-P (□), or 1 mM UTP (■). At the indicated times, aliquots (1.8 ng of protein) were removed and the residual acetyltransferase activity was measured. The activity at t = 0 was 30% lower in the presence of GlcNAc-1-P because this reaction product inhibits the enzyme (19). (B) Wild-type GlmU was stripped of β-mercaptoethanol and incubated at 37°C in the absence (•) or presence of various thiol-specific reagents: NEM at 50 μM (■), DTNB at 5 μM (○), and NTCB at 50 μM (⧫). Samples (1.8 ng of protein) were removed at the indicated times, and the residual acetyltransferase activity was measured. Curves obtained with iodoacetamide at 100 μM and pHMB at 2.5 μM could be superimposed on that obtained with NEM at 50 μM. □, enzyme incubated with 1.6 mM acetyl-CoA for 30 s prior to addition of NEM at a final concentration of 50 μM. (C) Wild-type GlmU was stripped of β-mercaptoethanol and incubated at 37°C in 20 mM potassium phosphate buffer containing 5 μM DTNB. At t = 1.5 min (arrow), the mixture was divided in two parts, and β-mercaptoethanol was added to one at a final concentration of 14 mM. Samples (1.8 ng of protein) were removed at different times, and the residual acetyltransferase activity was measured. Symbols: •, no addition; ■, addition of β-mercaptoethanol at t = 1.5 min. All experiments were performed at least in duplicate, and standard deviations were less than 10%.
Overproduction, purification, and kinetic properties of mutant GlmU enzymes.
The bifunctional GlmU protein comprises 456 amino acids, with four cysteines, at positions 296, 307, 324, and 385, as deduced from the glmU gene sequence (18, 33). Each cysteine was converted to alanine by oligonucleotide-directed mutagenesis of expression plasmid pFP1. pFP1 and its four mutant derivatives (pFP1-C296A, pFP1-C307A, pFP1-C324A, and pFP1-C385A) were transformed into the thermosensitive glmU mutant strain UGS83 (18). All of them were shown to restore growth of UGS83 at the restrictive temperature, even in the absence of IPTG, suggesting that the activities of the mutant proteins expressed under these conditions were enough to ensure normal cell growth. These different strains were induced with IPTG for 3 h, and the crude protein extracts were tested for both activities of GlmU. While strains overproducing wild-type or C296A and C385A mutant proteins contained about the same levels of acetyltransferase (90, 70, and 160 U/mg of protein, respectively), this activity appeared about 1,000- and 10-fold lower in strains overproducing the C307A and C324A mutant proteins (0.07 and 9 U/mg of protein, respectively). It should be noted that expression patterns for these plasmids were similar, the band of overexpressed wild-type or mutant protein representing in each case approximately 10% of total cell proteins, as judged by SDS-PAGE (data not shown). This finding suggested that among the four cysteines, only Cys307 and Cys324 were important for the acetyltransferase activity of GlmU. The levels of uridyltransferase in these extracts were all in the same range (from 30 to 42 U/mg of protein), indicating that none of the cysteine residues was essential for the second activity of GlmU, a finding consistent with the insensitivity of this enzyme to thiol-specific reagents.
Because of the very low level of acetyltransferase activity determined in strains carrying plasmid pFP1-C307A, a risk remained that part of the activity did not correspond to the overexpressed C307A protein but was the result of the activity of some cellular enzyme catalyzing nonspecifically and at a low rate the same reaction. This possibility and the fact that GlmU enzymes represented only 10 to 15% of proteins from crude cell extracts prompted us to purify the different forms of GlmU enzyme. We constructed pFP2, a plasmid similar to pFP1 but expressing the enzyme tagged by a C-terminal amino acid extension consisting of Arg-Ser-His6. The GlmU-His6 protein was greatly overproduced in pFP2-harboring cells upon IPTG induction, and only one chromatographic step on Ni2+-NTA was required for its almost complete purification (data not shown). However, while the uridyltransferase activity of the enzyme was normal, its acetyltransferase activity appeared to be reduced by a factor of 12 (Table 1). The reason for this loss of activity is unknown, but it may relate to involvement of the C-terminal end of the protein in the acetyltransferase activity. The unexpected influence of a C-terminal His tag on the kinetic parameters of an enzyme was also recently reported for the Bacillus licheniformis β-lactamase (17), showing that the addition of a C-terminal His tag to a protein might not always be as neutral as generally assumed. Consequently, the GlmU enzyme with a C-terminal His tag did not appear to be a reliable model for investigating the effects of site-directed mutations on the acetyltransferase activity. Therefore, we constructed a plasmid pFP3, for overproduction of the N-terminally His6-tagged form of GlmU. As shown in Table 1, problems described above were not observed with the purified His6-GlmU enzyme, which carried wild-type levels of both acetyltransferase and uridyltransferase activities. The Km values for both substrates acetyl-CoA and GlcN-1-P were determined and also appeared quite similar to those of the enzyme without a His tag (Table 1). The corresponding mutated plasmids pFP3-C296A, pFP3-C307A, pFP3-C324A, and pFP3-C385A were constructed and used for the overproduction and purification of the four His6-tagged mutant GlmU enzymes. Cultures of 0.5 liter were sufficient to prepare approximately 5 to 10 mg of purified enzymes, each one appearing almost homogeneous, as a predominant 50-kDa band on Coomassie blue-stained SDS-gels (data not shown). As shown in Table 1, the kcat values of the C296A and C385A enzymes did not change extensively relative to the values for the unaltered acetyltransferase. On the other hand, the kcat values of the C307A and C324A enzymes were approximately 0.07 and 12% of the value for the unaltered acetyltransferase, respectively, indicating that these two cysteines (in particular Cys307) were important for activity. This finding was in agreement with data involving crude protein extracts. The Km values for both substrates acetyl-CoA and GlcN-1-P were practically unaffected by these different mutational changes (Table 1), suggesting that none of the cysteine residues was essential for substrate binding. It should be noted that the yields and chromatographic behaviors of the four mutated enzymes were comparable to those of the unaltered GlmU enzyme, suggesting that none of these residues was essential for overall stability of the enzyme. In particular, the behavior of the different mutant enzymes during chromatography on gel filtration columns (Superose 12) was the same as that of the unaltered enzyme and was consistent with a similar trimeric oligomerization state (data not shown), as reported previously for the wild-type enzyme (18).
In vivo activity of wild-type and mutant GlmU enzymes.
Replacement of each cysteine with an alanine residue produced GlmU molecules that were still functional, as judged by their ability to support growth under restrictive conditions of a thermosensitive glmU mutant strain. Complementation by the C307A mutant was a priori unexpected, considering its greatly reduced acetyltransferase activity (only 0.1% of the wild-type activity). However, it should be noted that functional complementation was tested with mutated genes present on multicopy plasmids. In particular, we estimated here that gene expression from the pFP1 plasmid in the absence of IPTG was about 10-fold higher than that detected in a wild-type plasmidless strain. Also, we previously reported that the GlmU enzyme was present in great excess in growing E. coli cells, compared to the specific requirements in UDP-GlcNAc molecules of the peptidoglycan and lipopolysaccharide biosynthesis pathways (19). This was particularly evident for the acetyltransferase activity, which is the highest activity of the bifunctional GlmU enzyme. Finally, it was earlier described that the peptidoglycan content of E. coli cells could be decreased by up to 50% without loss of viability (reference 18 and references therein). Taken together, these data suggested that expression of a greatly altered mutant enzyme from a multicopy plasmid could provide enough biosynthetic activity to support normal cell growth and cell integrity. Such an unexpected finding was recently encountered during site-directed mutagenesis of essential residues from E. coli UDP-N-acetylmuramate:l-alanine ligase (3). To determine the effects of the mutational changes introduced into the protein sequence, future work should involve the transfer of the mutated glmU genes in single copy onto the E. coli chromosome.
Function of cysteine residues in the bifunctional GlmU enzyme.
The results obtained in this study allowed several important conclusions regarding the functions of the four cysteine residues in the E. coli GlmU enzyme. The sensitivity of the enzyme to thiol-specific reagents suggested to us that for E. coli GlmU, either the reagent interacted with catalytically functional cysteine residues or alkylation of -SH groups and incorporation of a bulky group (such as an NEM moiety) into the protein elicited gross conformational changes and attendant loss of enzyme activity. It was clear that none of the cysteines was absolutely required for the catalytic mechanism of the enzyme, as none of the mutations resulted in a complete loss of activity; the most dramatic change was in a functional protein with 0.1% residual activity. Only three GlcNAc-1-P uridyltransferases, from E. coli (18), Neisseria gonorrhoeae (29), and Bacillus subtilis (13), were characterized to date. The bifunctionality of the GlmU enzyme was demonstrated only in E. coli (19), but GlmU enzymes from other bacterial species most probably also carry the acetyltransferase activity. None of the four cysteines of the E. coli enzyme appeared strictly conserved in the three sequences (29) and in sequences of other putative GlmU enzymes revealed by systematic sequencing of bacterial genomes (data not shown). Since it was unlikely that cysteine residues would be prerequisite for catalysis by GlmU from E. coli but not other species, we proposed that residues Cys307 and Cys324 were located in or near the active site of the acetyltransferase but were not directly involved in the catalytic process. It was also conceivable that the formation of a covalent complex between the reagents and the cysteines resulted in steric deformation of the tertiary structure of the enzyme, but this was unlikely for several reasons. Reversibility of the inhibition by DTNB with a reducing agent indicated that irreversible changes in enzyme structure had not occurred. Furthermore, the ability of the substrate acetyl-CoA to protect the enzyme against inactivation by thiol-specific reagents was consistent with the reactive cysteine residue(s) being near or in the active site of the acetyltransferase domain.
As mentioned previously, the purified GlmU protein exhibited a number of characteristics which suggested that its two activities reside in separate catalytic domains (12, 19). Portions of the GlmU amino acid sequence exhibiting homologies with sequences of other XDP-sugar pyrophosphorylase and acetylase activities were located in the N-terminal part and second third, respectively, of the protein. Careful examination of the amino acid sequences of the GlmU enzymes showed that all contained incomplete tandem hexapeptide repeats with the consensus sequence (L/I/V)(G/X)XXXX (29). The same feature was observed in the sequences of the UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acyltransferase (LpxD) and UDP-GlcNAc 3-hydroxymyristoyltransferase (LpxA), two enzymes involved in lipopolysaccharide biosynthesis (1, 14, 23), and also in LacA, CysE, and NodL acetyltransferases (10). On this basis, these different enzymes have been suggested to form a single family of acetyl- and acetyltransferase (10, 30, 32). The crystal structures of LpxA and other members of this hexapeptide protein family revealed that the hexapeptide repeat sequence directed folding of a highly unusual structural domain termed a left-handed parallel β helix (LβH) (2, 25). It was suggested that the LβH domain was involved in the subunit interface of these proteins (all were trimeric enzymes, as observed for GlmU) as well as in catalysis (reference 2 and references therein). In the GlmU protein, this particular sequence was located between amino acid residues 250 to 350, in which region cysteines Cys307 and Cys324 from the E. coli enzyme are located. The observation that the mutation or alkylation of these two cysteines resulted in an alteration of the enzyme activity confirmed the role of this particular region in the reaction of acetylation. The absence of a concomitant loss of uridyltransferase activity was consistent with the organization of the enzyme in two distinct functional domains (12, 19). The deleterious effect of a C-terminal His tag on only the acetyltransferase activity of GlmU was also consistent with this model.
ACKNOWLEDGMENTS
This work was supported by a grant from the Centre National de la Recherche Scientifique (URA 1131) and a grant “Biotechnologies” from the Ministère de l’Education Nationale, de la Recherche et de la Technologie (97.C.0177). The financial support by Hoechst Marion Roussel AG to the laboratory and in particular to F.P. is greatly acknowledged.
We kindly thank Dominique Le Beller and Florence Fassy for their continued interest in and encouragement of this work.
REFERENCES
- 1.Anderson M S, Bull H G, Galloway S M, Kelly T M, Mohan S, Radika K, Raetz C R H. UDP-N-acetylglucosamine acyltransferase of Escherichia coli. The first step of endotoxin biosynthesis is thermodynamically unfavorable. J Biol Chem. 1993;268:19858–19865. [PubMed] [Google Scholar]
- 2.Beaman T W, Sugantino M, Roderick S L. Structure of the hexapeptide xenobiotic acetyltransferase from Pseudomonas aeruginosa. Biochemistry. 1998;37:6689–6696. doi: 10.1021/bi980106v. [DOI] [PubMed] [Google Scholar]
- 3.Bouhss A, Mengin-Lecreulx D, Blanot D, van Heijenoort J, Parquet C. Invariant amino acids in the Mur peptide synthetases of bacterial peptidoglycan synthesis and their modification by site-directed mutagenesis in the UDP-MurNAc:l-alanine ligase from Escherichia coli. Biochemistry. 1997;36:11556–11563. doi: 10.1021/bi970797f. [DOI] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principles of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Dagert M, Ehrlich S D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979;6:23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- 6.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor; 1972. [Google Scholar]
- 7.De Luca C, Lansing M, Crescenzi F, Martini I, Shen G-J, O’Regan M, Wong C-H. Overexpression, one-step purification and characterization of UDP-glucose dehydrogenase and UDP-N-acetylglucosamine pyrophosphorylase. Bioorg Med Chem. 1996;4:131–142. doi: 10.1016/0968-0896(95)00159-x. [DOI] [PubMed] [Google Scholar]
- 8.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmids by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 9.Dobrogosz W J. Effect of amino sugars on catabolite repression in Escherichia coli. J Bacteriol. 1968;95:578–584. doi: 10.1128/jb.95.2.578-584.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Downie J A. The nodL gene from Rhizobium leguminosarum is homologous to the acetyltransferases encoded by lacA and cysE. Mol Microbiol. 1989;3:1649–1651. doi: 10.1111/j.1365-2958.1989.tb00150.x. [DOI] [PubMed] [Google Scholar]
- 11.Dutka-Malen S, Mazodier P, Badet B. Molecular cloning and overexpression of the glucosamine synthetase gene from Escherichia coli. Biochimie. 1988;70:287–290. doi: 10.1016/0300-9084(88)90073-9. [DOI] [PubMed] [Google Scholar]
- 12.Gehring A M, Lees W J, Mindiola D J, Walsh C T, Brown E D. Acetyltransfer precedes uridylyltransfer in the formation of UDP-N-acetylglucosamine in separate active sites of the bifunctional GlmU protein of Escherichia coli. Biochemistry. 1996;35:579–585. doi: 10.1021/bi952275a. [DOI] [PubMed] [Google Scholar]
- 13.Hove-Jensen B. Identification of tms-26 as an allele of the gcaD gene, which encodes N-acetylglucosamine-1-phosphate uridyltransferase in Bacillus subtilis. J Bacteriol. 1992;174:6852–6856. doi: 10.1128/jb.174.21.6852-6856.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly T M, Stachula S A, Raetz C R H, Anderson M S. The firA gene of Escherichia coli encodes UDP-3-O-(R-3-hydroxymyristoyl)-glucosamine N-acetyltransferase, the third step of endotoxin biosynthesis. J Biol Chem. 1993;268:19866–19874. [PubMed] [Google Scholar]
- 15.Kuhn H-M, Meier-Dieter U, Mayer H. ECA, the enterobacterial common antigen. FEMS Microbiol Rev. 1988;54:195–222. doi: 10.1111/j.1574-6968.1988.tb02743.x. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K, Favre M. Maturation of the head of bacteriophage T4. J Mol Biol. 1973;80:575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- 17.Ledent P, Duez C, Vanhove M, Lejeune A, Fonzé E, Charlier P, Rhazi-Filali F, Thamm I, Guillaume G, Samyn B, Devreese B, van Beeumen J, Lamotte-Brasseur J, Frère J-M. Unexpected influence of a C-terminal-fused His-tag on the processing of an enzyme and on the kinetic and folding parameters. FEBS Lett. 1997;413:194–196. doi: 10.1016/s0014-5793(97)00908-3. [DOI] [PubMed] [Google Scholar]
- 18.Mengin-Lecreulx D, van Heijenoort J. Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J Bacteriol. 1993;175:6150–6157. doi: 10.1128/jb.175.19.6150-6157.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mengin-Lecreulx D, van Heijenoort J. Copurification of glucosamine-1-phosphate acetyltransferase and N-acetylglucosamine-1-phosphate uridyltransferase activities of Escherichia coli: characterization of the glmU gene product as a bifunctional enzyme catalyzing two subsequent steps in the pathway for UDP-N-acetylglucosamine synthesis. J Bacteriol. 1994;176:5788–5795. doi: 10.1128/jb.176.18.5788-5795.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mengin-Lecreulx D, van Heijenoort J. Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J Biol Chem. 1996;271:32–39. doi: 10.1074/jbc.271.1.32. [DOI] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 22.Plumbridge J A, Cochet O, Souza J M, Altamirano M M, Calcagno M L, Badet B. Coordinated regulation of amino sugar-synthesizing and -degrading enzymes in Escherichia coli K-12. J Bacteriol. 1993;175:4951–4956. doi: 10.1128/jb.175.16.4951-4956.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raetz C R H. Molecular genetics of membrane phospholipid synthesis. Annu Rev Genet. 1986;20:253–295. doi: 10.1146/annurev.ge.20.120186.001345. [DOI] [PubMed] [Google Scholar]
- 24.Raetz C R H. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1035–1063. [Google Scholar]
- 25.Raetz C R H, Roderick S L. A left-handed parallel β helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Sarvas M. Mutant of Escherichia coli K-12 defective in d-glucosamine biosynthesis. J Bacteriol. 1971;105:467–471. doi: 10.1128/jb.105.2.467-471.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevenson G, Neal B, Liu D, Hobbs M, Packer N H, Batley M, Redmond J W, Lindquist L, Reeves P. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J Bacteriol. 1994;176:4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ullrich J, van Putten J. Identification of the gonococcal glmU gene encoding the enzyme N-acetylglucosamine-1-phosphate uridyltransferase involved in the synthesis of UDP-GlcNAc. J Bacteriol. 1995;177:6902–6909. doi: 10.1128/jb.177.23.6902-6909.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaara M. Eight bacterial proteins, including UDP-N-acetylglucosamine acyltransferase (LpxA) and three other transferases of Escherichia coli, consist of a six-residue periodicity theme. FEMS Microbiol Lett. 1992;97:249–254. doi: 10.1016/0378-1097(92)90344-n. [DOI] [PubMed] [Google Scholar]
- 31.Van Heijenoort J. Murein synthesis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1025–1034. [Google Scholar]
- 32.Vuorio R, Hirvas L, Vaara M. The Ssc protein of enteric bacteria has significant homology to the acyltransferase LpxA of lipid A biosynthesis, and to three acetyltransferases. FEBS Lett. 1991;292:90–94. doi: 10.1016/0014-5793(91)80841-p. [DOI] [PubMed] [Google Scholar]
- 33.Walker J E, Gay N J, Saraste M, Eberle A N. DNA sequence around the Escherichia coli unc operon. Completion of the sequence of a 17 kilobase segment containing asnA, oriC, unc, glmS and phoS. Biochem J. 1984;224:799–815. doi: 10.1042/bj2240799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White R J. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem J. 1968;106:847–858. doi: 10.1042/bj1060847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu H C, Wu T C. Isolation and characterization of a glucosamine-requiring mutant of Escherichia coli K-12 defective in glucosamine-6-phosphate synthetase. J Bacteriol. 1971;105:455–466. doi: 10.1128/jb.105.2.455-466.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]