Abstract
The genome of the halophilic archaeon Haloarcula marismortui contains two rRNA operons designated rrnA and rrnB. Genomic clones of the two operons and their flanking regions have been sequenced, and primary transcripts and processing intermediates derived from each operon have been characterized. The 16S, 23S, and 5S genes from the two operons were found to differ at 74 of 1,472 positions, 39 of 2,922 positions, and 2 of 122 positions, respectively. This degree of sequence divergence for multicopy (paralogous) rRNA genes was 10- to 50-fold or more higher than anticipated. The two operons exhibit other profound differences that include (i) the presence in rrnA and the absence in rrnB of tRNAAla and tRNACys genes in the intergenic and distal regions, respectively, (ii) divergent 5′ flanking sequences, and (iii) distinct pathways for processing and maturation of 16S rRNA. Processing and maturation of 16S and 23S rRNA from rrnA operon transcripts and of 23S rRNA from rrnB operon transcripts follow the canonical halophilic pathway, whereas maturation of 16S rRNA from rrnB operon transcripts follows an unusual and different pathway that is apparently devoid of any 5′ processing intermediate.
During ribosome biogenesis, mature rRNAs are endonucleolytically excised from a larger precursor transcript, modified, folded into a complex three-dimensional structure, trimmed, and assembled along with a complete complement of ribosomal proteins into mature subunits. With few exceptions, the major small- and large-subunit rRNA genes are linked within the genome of an organism and cotranscribed in order to maintain control and coordination of these complex processes. Because of the high metabolic demand for protein synthesis, rRNA operons are often multicopy genes, with all copies being identical or nearly identical in both coding and transcribed flanking sequences (1, 13, 24). Any sequence heterogeneity arising from mutations within one of the operons is either eliminated or homogenized to other operons by recombination and gene conversion type processes (11, 19). As a consequence of these processes, rRNA coding and flanking sequences are highly uniform within a species and immune to replacement during infrequent lateral transfer of genetic material across the species boundary.
Although rRNA sequence uniformity is the rule, there are at least two cases where heterogeneity within multicopy (paralogous) rrn genes is substantial. In the eucaryotic parasite Plasmodium berghei, two types of paralogous small-subunit rRNA genes are present (18). These differ by substitution or deletion-insertion at about 5% of the nucleotide positions. The two types of genes appear to be preferentially expressed, respectively, in the blood and insect stages in the life cycle of the parasite.
The second example is the halophilic archaeon Haloarcula marismortui. This organism contains two defined rrn operons in its genome (23). Alignment of the two paralogous 16S sequences has revealed substitutions at 74 of the 1,472 nucleotide (nt) positions (i.e., 5% sequence divergence) (24). All of the substitutions occurred at phylogenetically variable positions, and none affected nucleotide positions that are known to be important in ribosome function. The pattern of nucleotide substitution within the 16S alignment differed in at least one important respect from that expected of independently evolving orthologous 16S rRNA genes: the substitutions were not distributed throughout the 16S gene alignment but rather were concentrated within limited regions of the gene. For example, two-thirds of the nucleotide sequence differences (51 of 74) were located within a 300-nt region (between positions 520 and 820) of the 16S sequences, whereas, for example, only two differences were observed in the adjacent region between positions 220 and 520. In diverging orthologous genes, substitutions occur frequently at many positions that are more or less evenly spread between positions 220 and 820 of the 16S sequence (24, 32). Under standard laboratory conditions of growth, both of the H. marismortui rrn operons appear to be expressed (24). Despite the differences, both of these sequences reside within the halophilic archaeal cluster in small-subunit rRNA phylogenies.
If the situation of sequence heterogeneity within paralogous 16S genes is more common than presently imagined, it could complicate rRNA-based phylogenetic analysis and cause problems with the interpretation of biodiversity as detected by most PCR-based methods (27, 32). Moreover, this situation raises fundamental questions about the composition of genomes and uniformity within the protein synthesis apparatus. How much sequence variability can be accommodated within rRNA without compromising efficiency and accuracy? What roles do the conserved rRNA flanking sequences play in the biogenesis of ribosomes? To appreciate better this complex situation and to understand more thoroughly the potential impact, we have completely sequenced the rrnA and rrnB operons from the genome of H. marismortui and analyzed the in vivo intermediates that accumulate during the processing and maturation of the rrnA and rrnB operon transcripts. The features distinguishing the two operons that have been revealed include (i) sequence heterogeneity within 16S, 23S, and 5S genes; (ii) the presence or absence of spacer and distal tRNA genes; (iii) divergent 5′ flanking sequences; and (iv) distinct pathways for processing and maturation of 16S rRNAs.
MATERIALS AND METHODS
H. marismortui was grown at 37 to 42°C in rich medium (26). The two rrn operons designated rrnA and rrnB were previously cloned as an 8.0-kb HindIII-ClaI fragment and as a 10-kbp HindIII fragment cloned into the vector pBR322, respectively. For some DNA sequence determinations, various fragments were subcloned into pGEM3 or pGEM7 vectors. Sequences were generated with the universal T7 or SP6 primers or with sequence-specific internal primers with the T7 Sequenase system. In some cases, deletions were generated within the inserts of pGEM vectors by using exonuclease III (20). The 7 deaza-2′-deoxyguanosine-5′-triphosphate analog of dGTP was used to help resolve sequence compressions resulting from secondary structure and the high GC content of the template.
Total cellular RNA was isolated by the boiling-sodium dodecyl sulfate lysis method, and the recovered RNA was used for both S1 nuclease protection and primer extension analysis. These methods have been described in detail (7). For S1 protection experiments, 1 to 5 μg of total RNA was hybridized to either 5′- or 3′-end-labeled DNA probes (about 0.05 pmol). The DNA fragments were obtained by restriction enzyme digestion of complete or partial clones of the rrnA or rrnB operons followed by fragment purification in acrylamide gels. Probe DNA fragments were 5′ end labeled with polynucleotide kinase and [γ-32P]ATP or 3′ end labeled with the Klenow fragment of DNA polymerase I and the appropriate [α-32P]deoxynucleoside triphosphate. The hybridization temperature (between 52 and 58°C) was empirically determined for each fragment such that rehybridization of the probe DNA was minimized without interfering with the formation of RNA-DNA hybrids. The molecular length standards employed were the fragments of pBR322 generated by digestion with MspI and 3′ end labeled with the Klenow fragment of DNA polymerase and [α-32P]dCTP. In some cases, size ladders were also generated by using end-labeled DNA fragments in the A and A+G Maxam Gilbert sequencing reactions. Estimates of the length of protection products are accurate to within a few nucleotides and in some instances have been confirmed by primer extension analysis. The protection products often exhibit heterogeneous ends. This heterogeneity could result from either heterogeneity in the RNA ends that enter into the formation of RNA-DNA hybrids or from under- or overdigestion at the ends of the hybrids by the S1 nuclease.
RESULTS
Operon structures.
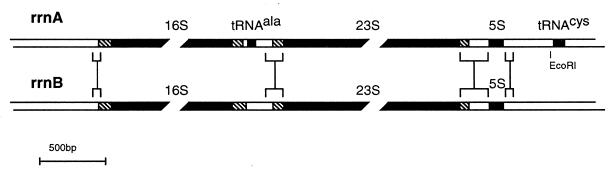
Southern hybridization indicated that the genome of H. marismortui contained two defined rRNA operons. These were cloned as an 8-kbp HindIII-ClaI fragment and as a 10-kbp HindIII-HindIII fragment and were designated rrnA and rrnB, respectively (23). The nucleotide sequences of the rrnA and rrnB operons and their 5′ and 3′ flanking sequences were determined (accession numbers AF034619 and AF034620). The rrnA operon contains 16S, 23S, and 5S genes, as well as a tRNAAla gene in the intergenic spacer and a tRNACys gene in the 3′ distal position (Fig. 1). The rrnB operon contains 16S, 23S, and 5S genes but lacks both the intergenic and the distal tRNA genes. The absence of the distal tRNACys gene from the 3 kbp of unsequenced region distal to the rrnB operon was confirmed by Southern hybridization with a tRNACys-specific oligonucleotide. A subclone from the rrnA operon containing the tRNACys gene exhibited strong hybridization to the oligonucleotide probe, whereas the entire rrnB-containing 10-kbp clone exhibited no hybridization signal (data not shown).
FIG. 1.
Structure of rrnA and rrnB operons of H. marismortui. The chromosomal structures of the rrnA and rrnB operons are indicated. The 16S, 23S, 5S, tRNAAla, and tRNACys genes are represented by solid boxes, the processing inverted repeats are indicated by hatched boxes, and the other flanking and spacer sequences are indicated by open boxes. Flanking and spacer regions longer than 50 nt and greater than 90% identical in sequences between rrnA and rrnB are indicated by brackets. All gene sequences are greater than 95% identical (see Table 1). The complete nucleotide sequences of rrnA and rrnB have been deposited in GenBank (rrnA, AF034619; rrnB, AF034620).
Gene sequences.
Analysis and comparison indicated that the 16S, 23S, and 5S genes from the rrnA and rrnB operons contain an abnormally high number of nucleotide sequence differences. This was first noted for the 1,472-nt 16S genes where substitutions were observed at 74 positions (24). Similarly, for the 2,922-nt 23S genes substitutions were observed at 39 positions, and for the 122-nt 5S genes substitutions were observed at 2 positions (Table 1). As in the rrnA and rrnB 16S genes, differences in the 23S and 5S sequences occur at positions that are phylogenetically variable, and many of the differences are compensatory, affecting both components of a Watson-Crick base pair. None of the substitutions affect positions known to be important in 50S subunit function.
TABLE 1.
Sequence identity of H. marismortui rRNA genes
| Gene comparison | Length (nt) | No. of substitutions | % Identity |
|---|---|---|---|
| 16S (rrnA and rrnB) | 1,472 | 74 | 95.0 |
| 23S (rrnA and rrnB) | 2,922 | 39 | 98.7 |
| 5S (rrnA and rrnB) | 122 | 2 | 98.3 |
A previous study described the 23S and 5S gene sequence from H. marismortui (2). The sequence was apparently obtained by a chromosome walk starting from a fragment containing an rrnA-like 5S gene. For simplicity, this sequence has been designated rrnC. In reality, rrnC may be a composite of the rrnA and rrnB operon sequences that was generated either in vivo through recombination or in vitro through the cloning of overlapping fragments from the two operons. Support for this supposition is based on the following. First, the 5′ portion of the rrnC 23S gene is more similar to the rrnB 23S sequence, the central portion is a composite of rrnA and rrnB, and the 3′ portion is very similar to rrnA. Second, the rrnA and rrnC 23S-5S intergenic spacers, 5S genes, and 5S distal sequences are identical. Third, there were 7 nt substitutions unique to the rrnC 23S sequence, whereas there were 15 nt substitutions unique to the rrnB 23S sequence and 22 substitutions unique to the rrnA 23S sequence. The rrnC 23S gene is only 2,917 nt in length and contains single-nucleotide deletions at positions 680, 1280, 1641, 2417, and 2357 relative to the rrnA and -B 23S sequences. Some of these features suggest that rrnC may be a composite operon sequence rather than an independently evolving entity.
Inverted repeats surrounding the 16S and 23S genes.
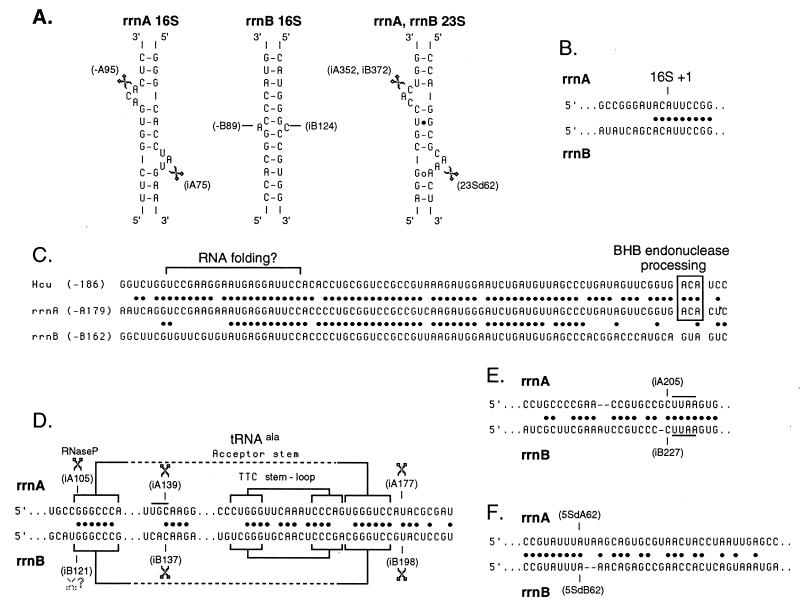
Further analysis of the primary sequences indicated that in both operons, the 16S and 23S genes were surrounded by long inverted repeats (Fig. 1). These repeats are believed to form helical structures in the primary rRNA operon transcripts and to contain the recognition motifs for excision of pre-16S and pre-23S (3, 6, 15). In most archaea, this motif consists of two three-base loops on opposite strands of the helix and separated by four nucleotide base pairs. The motif is believed to be recognized and cleaved between the second and third bases of the two respective loops by a recently characterized bulge-helix-bulge (BHB) endonuclease (15, 21, 30). The concerted cleavages liberate the pre-16S or pre-23S rRNA from the primary transcript. The BHB motif was present within both the 16S and the 23S repeats of rrnA and the 23S repeat of rrnB (Fig. 2A). It was noticeably absent from the 16S repeat of rrnB; this implies that a distinct pathway is used to excise and process 16S rRNA from rrnB operon transcripts.
FIG. 2.
Important sequence motifs located in the flanking and spacer regions of rrnA and rrnB. (A) Small portions of the helical structures of the processing inverted repeats surrounding the 16S and 23S genes in rrnA and rrnB are indicated. The BHB motif is noticeably absent from the rrnB 16S helix. The sites of cleavage detected within the bulges on the 5′ side (left) and 3′ side (right) of each helix are indicated by scissors, and the position of the nucleotide 3′ to the scissile phosphate is indicated. Where negative, numbers correspond to 5′ flanking sequences, with the first nucleotide of 16S rRNA being 1 for the rrnA and rrnB regions, respectively. i, 16S-23S intergenic spacer positions with the first nucleotide after 16S being iA1 or iB1; 23Sd, 23S-5S intergenic spacer with the first nucleotide after 23S being 23Sd1. The 23S-5S intergenic spacer is identical in rrnA and rrnB. (B) The point of sequence convergence at the junction between the 5′ flanking sequences and the beginning of the 16S genes in the rrnA and rrnB operons is indicated. The dots indicate identical nucleotides. (C) A 5′ flanking sequence element conserved between rrnA, rrnB, and the rrn operons of other genera of halophilic archaea are aligned. The numbers in parentheses are the first nucleotide positions of the sequences shown. Dots indicate conserved nucleotides in pairwise comparisons. The 3-nt bulge located in the 5′ portion of the 16S processing helix in rrnA and the single rrn operon of H. cutirubrum is boxed. A conserved sequence proposed to play a role in folding of a conserved pseudoknot within 16S rRNA is indicated (9). (D) 16S-23S intergenic sequence around the tRNAAla in rrnA and the corresponding region of rrnB. Endonucleolytic cleavages occur at or near the indicated positions at the 3′ end of the tRNAAla and tRNAAla-like sequences. Regions of secondary structure within tRNAAla are indicated by brackets; the potential for similar secondary structures in rrnB is also indicated. Dots represent identical nucleotides. The 3′ transcript end detected in the anticodon loop of the tRNAAla structure (position iA139) is indicated; the anticodon sequence is overlined. The position of a corresponding 3′ end in rrnB (position iB137) is also indicated. Finally, a 5′ end resulting from cleavage by RNase P at the 5′ end of tRNAAla is indicated (position iA105). No corresponding end was observed in rrnB (position iB121). (E) The sequence at the point of convergence within the 16S-23S intergenic space between rrnA and rrnB is shown. The UUAA sequence (overlined or underlined) immediately 3′ to the point of perfect convergence represents the archaeal TATA box for the putative intergenic promoters in rrnA and rrnB. The dots represent identical nucleotides. (F) The sequence at the point of divergence in the 5S 3′ flanking region is shown. 5Sd represents the 5S 3′ flanking sequence with the first nucleotide beyond 5S being 5Sd1. The dots represent identical nucleotides.
Transcription initiation and precursor processing in the 5′ flanking regions of rrnA and rrnB.
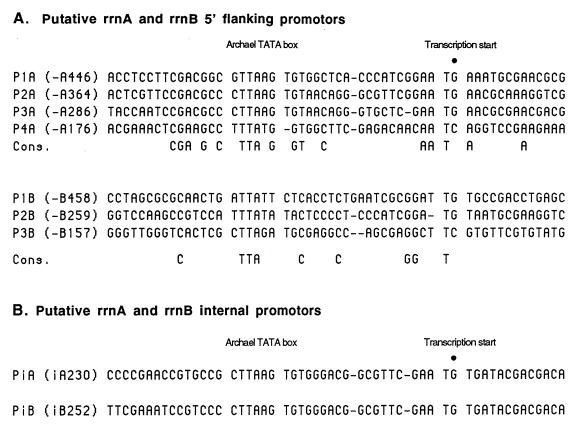
The 5′ flanking regions of the rrnA and rrnB operons contain multiple elements exhibiting a high degree of sequence similarity interspersed within otherwise unrelated sequences. All but one of these conserved elements were identified as putative promoters based on two criteria: sequence similarity to each other and to other well-characterized halophile rrn promoters (5, 6) and (in all but one instance) the presence of a 5′ transcript end mapping to a position at or near the anticipated dinucleotide initiation sites. The rrnA operon contains four promoter-like elements, whereas the rrnB operon contains three such elements (Fig. 3A). Each element possesses a conserved archaeal TATA box centered 22 to 24 nt upstream of the initiation site, which was usually G within a conserved TG dinucleotide. Under standard laboratory culture conditions, both operons are actively transcribed.
FIG. 3.
Alignment of promoter-like sequences from the 5′ flanking and intergenic spacer regions of rrnA and rrnB. The nucleotide sequences of the 5′ flanking promoter-like motifs from rrnA and rrnB (A) and the intergenic promoter-like motifs from rrnA and rrnB (B) are indicated. The sequences are aligned at the archaeal TATA box element and at the dinucleotide transcription initiation site (dot; usually G). The promoter designations and the nucleotide positions of the putative transcription start sites are indicated on the left.
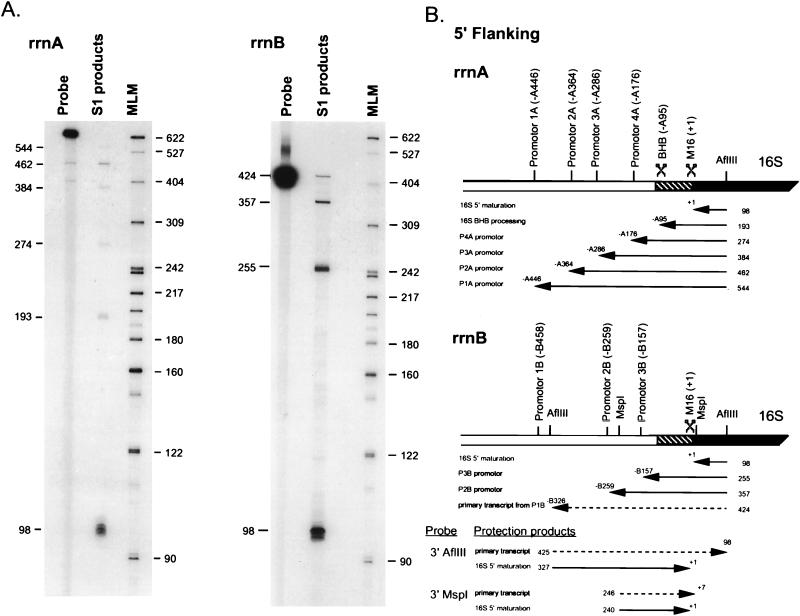
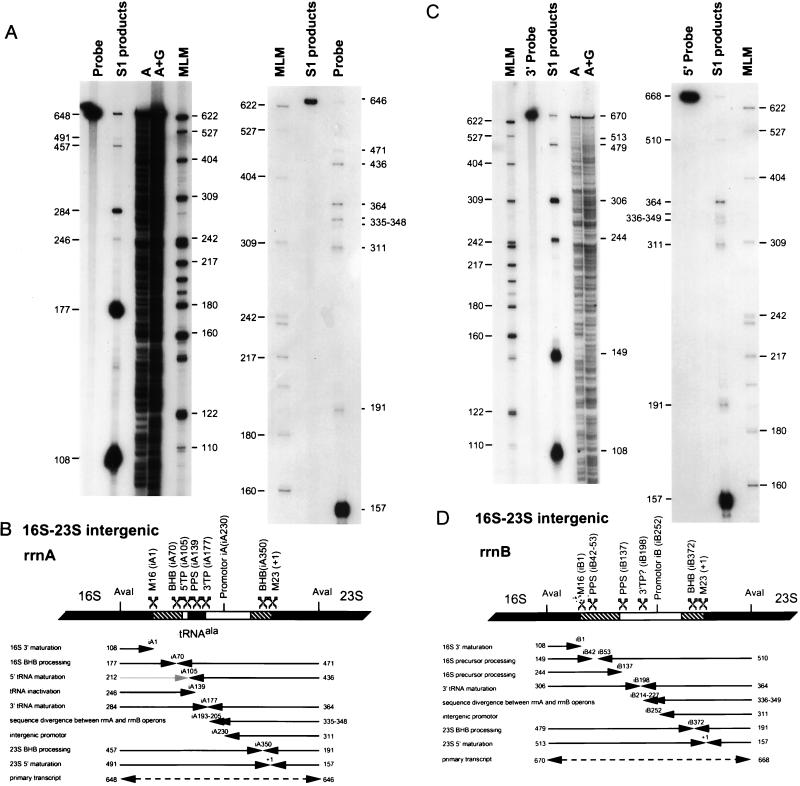
Transcripts derived from the 5′ flanking regions of the rrnA and rrnB operons were detected by S1 nuclease protection assays by using 5′-end-labeled DNA probes specific for each operon (Fig. 4). With the rrnA operon-specific probe, 5′ transcript ends were located at or near position 1 of the 16S rRNA gene and at positions −A95, −A176, −A286, −A364, and −A446 in the 5′ flanking regions (Fig. 4A). The rrnA DNA probe hybridizes mature 16S rRNA derived from both rrnA and rrnB but only hybridizes precursor transcripts from rrnA. This is because the point of sequence divergence between the two operons is at the junction between the 5′ flanking region and the start of the 16S gene (see Fig. 2B). The 5′ end of the product mapping to position −A95 was presumably derived by protection with trailer products generated by endonucleolytic cleavage by the BHB enzyme within the 3-nt bulge on the 5′ side of the 16S rRNA processing stem. The structure of the putative cleavage site is depicted in Fig. 2A. The products with 5′ ends at positions −A446, −A364, −A286, and −A176 correspond to transcripts initiated at the P1A, P2A, P3A, and P4A promoters, respectively.
FIG. 4.
Analysis of 5′ transcript ends located in the 5′ flanking regions of rrnA and rrnB. Two respective AflIII restriction fragments specific for rrnA and rrnB were 5′ end labeled on the (−) strand at position 98 within the 16S sequence and used as probes against total cellular RNA in S1 nuclease protection assays. (A) The protection products were separated on denaturing polyacrylamide gels. The sizes (in nucleotides) of protection products and molecular length markers (MLM) are indicated beside the autoradiograms. (B) A diagram of rrnA and rrnB is shown. Each protection product is represented by an arrow with the length in nucleotides given on the right and the position of the 5′ transcript end given above the leftward pointing arrowheads. The positions of promoter-like sequence motifs and precursor processing sites are indicated. The 16S gene is a solid box, the 5′ portion of the processing repeat is a hatched box, and the remaining 5′ flanking region is unfilled. The dashed arrow in rrnB represents full-length protection of the probe by transcripts that were presumed to have been initiated at the upstream P1B promoter. At the bottom of panel B, the results of nuclease protection assays with 3′-end-labeled rrnB probes are illustrated (autoradiograms not shown). As above, the dashed arrows represent full-length protection of the probe, and the solid arrows represent partial-length protection of the probe. The length of each product is indicated on the left end and the position of the 3′ transcript end is indicated on the right end of each arrow (see text for details).
With the rrnB operon probe, 5′ transcript ends were located at or near position 1 of 16S rRNA and positions −B157 and −B259 (Fig. 4B). Also apparent was a product corresponding to full-length protection of the DNA probe. The transcripts that protected the entire probe were presumably derived from the putative upstream P1B promoter (position −B458), whereas the two shorter products were derived by protection with transcripts initiated at the P2B and P3B promoters, respectively (Fig. 3A). No other abundant 5′-end protection products were observed, suggesting that there was no endonucleolytic processing within the 5′ flanking region of the rrnB transcript. As mentioned above, the rrnB 16S processing helix noticeably lacks the recognition motif used by the BHB processing endonuclease to excise a precursor 16S rRNA from the primary transcript (Fig. 2A). To confirm the absence of endonucleolytic processing in the 5′ flanking region of rrnB transcripts, the 424-nt AflIII fragment was 3′ end labeled on the minus strand at position −B327 and used as a probe in the nuclease protection assay. Only transcripts derived from rrnB protect this probe. Two protection products were observed. The first corresponds to full-length protection of the probe by rrnB transcripts initiated at the upstream P1B promoter. The second product was about 327 nt long and resulted from partial protection of the probe by an RNA intermediate with a 3′ end at or near position 1 of 16S rRNA (data not shown but see Fig. 4B). This result seems to indicate that the 5′ region of rrnB operon transcripts are subject to a single endonucleolytic cleavage that separates and releases the 5′ end of 16S rRNA from the intact 5′ leader. This result was confirmed by using a 245-nt MspI fragment from rrnB, 3′ end labeled at position −B240, as a probe (Fig. 4B).
Another somewhat longer and more highly conserved motif is also present in the 5′ flanking regions of both the rrnA and the rrnB operons and at a similar position in the rrn operons of other genera of halophilic archaea. Between rrnA and rrnB, the conserved motif is 52 nt long and 92% identical in sequence (Fig. 2C). When compared to the motifs in other halophilic archaea (e.g., Halobacterium cutirubrum), similarity to the rrnA motif extends a further 11 nt in the 5′ direction and a further 16 nt in the 3′ direction (6). The 3′ end of the rrnA and H. cutirubrum motifs correspond to the site of the BHB endonuclease cleavage in the 5′ portion of the 16S processing helix. The 5′ end of the motifs has been proposed to play a role in folding of a universally conserved pseudoknot within 16S rRNA (9). Other sequences within this motif may be involved in other as yet uncharacterized aspects of 16S rRNA processing, folding, or maturation. Outside of this motif and the multiple promoter elements, no other sequence similarity was observed between the 5′ flanking regions of the rrnA and rrnB operons.
Intergenic spacer regions in rrnA and rrnB.
The 16S-23S intergenic spacer regions of rrnA and rrnB are 383 and 405 nt, respectively. The two spacers are identical in sequence over the 3′-distal 183 nt but show only fragmentary similarity over the remaining 5′ proximal regions (Fig. 1). The rrnA operon contains a tRNAAla gene within the 5′ portion of the spacer, whereas the rrnB operon appears to contain only a partial remnant of a tRNAAla-like gene at this position (see below).
Transcripts and processing intermediates derived from the 16S-23S intergenic spaces of the two operons were detected by S1 nuclease protection assays by using both 5′- and 3′-end-labeled DNA probes specific for each operon (Fig. 5). In regions of perfect or near-perfect sequence identity, transcripts from both operons hybridize to either end-labeled probe. These regions of coincident hybridization include the 3′ end of the 16S rRNA (for 3′-end-labeled probes) or the terminal 183 nt of the spacer and the 5′ end of 23S rRNA (for 5′-end-labeled probes). With the 3′-end-labeled DNA probe from rrnA, transcript 3′ ends were located at or near positions iA1, iA70, iA139, iA177, iA350, and position 1 of 23S rRNA. Also apparent was a product of full-length probe corresponding to protection by unprocessed transcripts from the rrnA operon. With the same rrnA DNA probe labeled at the 5′ end, transcript 5′ ends were located at or near positions iA70, iA105, iA177, iA193 to iA205, iA230, and iA350 and position 1 of 23S rRNA. Again, full-length protection of the probe by an unprocessed rrnA operon transcript was also observed (Fig. 5A and B).
FIG. 5.
Analysis of 5′ and 3′ transcript ends located in the 16S-23S intergenic spaces of rrnA and rrnB. Two respective AvaI restriction fragments specific for rrnA and rrnB were 3′ end labeled on the (−) strand at position 1365 within the 16S gene or 5′ end labeled on the (−) strand at position 157 within the 23S gene and used as probes against total cellular RNA in S1 nuclease protection assays. The protection products were separated on denaturing polyacrylamide gels. The size in nucleotides of protection products and molecular length markers (MLM) are indicated beside each autoradiogram. (A) Autoradiograms with the rrnA probe either 3′ labeled (left) or 5′ labeled (right). The lanes marked A and A+G are Maxam Gilbert sequence reactions on the 3′-labeled probe. (B) Diagram illustrating the rrnA 383-nt 16S-23S intergenic space and a summary of the protection products obtained with the 3′-labeled probe (3′ transcript end sites are represented by rightward pointing arrows) and the 5′-labeled probe (5′ transcript end sites are represented by leftward pointing arrows). The estimated length of each product is indicated at the start of the arrow, and the positions of the respective 3′ or 5′ transcript ends within the intergenic spaces are indicated above the arrowhead. Opposing arrowheads represent sites of endonuclease cleavage. The gray arrow represents a product with a 3′ end at position iA105 that was not observed. The overlapping arrowheads at positions iA193 to iA205 represent multiple protection products. The dashed double-headed arrow represents full-length protection of both the 3′- and 5′-labeled probes with unprocessed transcripts from rrnA. The 16S, tRNAAla, and 23S genes are represented as solid boxes, the inverted processing repeats are indicated as hatched boxes, and other intergenic sequences are shown as open boxes. M16, maturation site at the 3′ end of 16S rRNA; BHB, BHB cleavage site; 5′TP, 5′ tRNA processing site (presumably RNase P); PPS, uncharacterized precursor processing site; 3′TP, 3′ tRNA processing site (apparently by an endonuclease; see the text); M23, 5′-end maturation of 23S rRNA (apparently an endonuclease; see the text). (C) Autoradiograms with the rrnB probe either 3′ labeled (left) or 5′ labeled (right). (D) Diagram of the rrnB 405-nt 16S-23S intergenic space. Other details are as described for panel B.
When both a leader 3′ transcript end and a trailer 5′ transcript end map to a position at or near the same nucleotide position, we have inferred that they are generated by a single endonucleolytic cleavage event within the primary transcript. By this criterion, putative endonucleolytic cleavages occur at or near positions iA70, iA177, and iA350 and somewhat surprisingly at position 1 of 23S rRNA. Positions iA70 and iA350 correspond to BHB endonuclease recognition motifs that occur within the 3′ portion of the 16S and the 5′ portion of the 23S processing stems, respectively (Fig. 2A). Detection of an endonuclease cleavage at or very near position 1 of 23S rRNA infers that this cleavage could precede at least in some instances cleavage by the BHB endonuclease in the 23S rRNA processing stem (at position iA350). The final cleavage at or near position iA177 occurs at the 3′ end of the tRNA sequence. The 5′ end of the trailer released by this cleavage has been mapped by primer extension and appears to correspond to the first base following the encoded tRNA sequence (i.e., position iA177; data not shown, but see Fig. 2D). If this primer extension result is reliably detecting a 5′ transcript end and is not the result of a tRNA structure-induced stop at the top of the acceptor stem, it implies that tRNA 3′-end generation is endonucleolytic.
The last 3′ transcript end was mapped to position iA139 within the anticodon loop of the tRNA sequence. Since no corresponding 5′ end was mapped to this position, the 3′ end could have been generated by either exonuclease digestion from a site located 3′ to position iA139 or by endonuclease cleavage at this site to produce a 3′ trailer that has escaped detection by the 5′-end-labeled probe used here. The presence of a 3′ end within the tRNA anticodon loop suggests that excision and maturation of the tRNAAla are substoichiometric. Similar cleavages have been observed within the tRNA sequences in the transcripts from the single-copy rrn operon of H. cutirubrum (4).
There were three protection products with 5′ ends located at positions iA105, iA193 to iA206, and iA230 that were not paired with corresponding 3′ protection products. The first, at iA105, corresponds to the mature 5′ end of tRNAAla. The 5′ end of all tRNAs is believed to be generated by RNase P endonucleolytic cleavage (17, 28). The simplest explanation for the absence of a product with a 3′ end at this position is that cleavage by the BHB endonuclease at position iA76 preceded cleavage by RNase P at the 5′ end of the tRNA gene; as a consequence, only the shorter product with a 3′ end at position iA76 would be visible. The second product representing a heterogeneous collection with 5′ ends between iA193 and iA206 represents the area of sequence convergence between the rrnA and rrnB spacers. These protection products resulted from rrnB operon transcripts hybridizing to the rrnA operon probe and from imprecise trimming by S1 nuclease (Fig. 2E). The third product, with a 5′ end near position iA230, corresponds to a putative internal promoter (Fig. 3B) presumably used to compensate for premature transcription termination within the operon and augment the production of 23S and 5S rRNA. Similar internal promoters have been identified in the rrn operons of other halophilic archaea (22).
Similar experiments were carried out with 3′- and 5′-end-labeled probes from rrnB (Fig. 5C and D). Again, these probes hybridize predominantly rrnB operon transcripts but also some rrnA transcripts in regions of perfect or near-perfect sequence identity. With the 3′-end-labeled probe, transcript 3′ ends were located at or near positions iB1, iB42, iB137, iB198, and iB372 and position 1 of 23S rRNA. With the same 5′-end-labeled probe, transcript 5′ ends were located at or near positions iB53, iB198, iB214 to iB227, iB252, and iB372 and position 1 of 23S rRNA. Again, full-length protection of the 5′ and 3′ labeled probes by the primary rrnB transcript was observed.
By the criteria stated above, there appear to be three endonucleolytic cleavages in the intergenic spacer of rrnB. The first of these, at position iB198, occurs immediately following the tRNA remnant in the rrnB spacer (Fig. 2D). Alignment of this region with the tRNAAla from the rrnA spacer reveals a substantial degree of sequence and structural similarity. It seems likely that a single activity may be responsible for both of these cleavages. The second endonucleolytic cleavage was at position iB372 and corresponded to the BHB endonuclease cleavage in the 5′ portion of the 23S processing stem. The third was at position 1 of 23S rRNA and again indicates that BHB excision of pre-23S rRNA does not always precede the endonucleolytic cleavage near or at the 5′ end of 23S rRNA. Because these last two cleavages occur in a region of sequence identity between rrnA and rrnB, the trailer fragments protecting the 5′-end-labeled probe were from both rrnA and rrnB. In contrast, the 5′ leader fragments protecting the 3′-end-labeled probe were only derived from rrnB.
There were two additional 5′ protection products with ends located at positions iB214 to iB227 and position iB252 that are easily explained. The first heterogeneous group maps to the area where the rrnA and rrnB spacer sequences converge (Fig. 2E). The protection products result from rrnA operon transcripts hybridizing to the rrnB operon probe and imprecise trimming by S1 nuclease. The second product with a 5′ end near position iB252 corresponds to a putative internal promoter (Fig. 3B). As in the rrnA operon, this promoter is apparently utilized to augment production of 23S and 5S rRNA. The 3′ transcript end detected at position iB137 occurs within the tRNA remnant and may be related to the 3′ end detected at position iA139 within the tRNAAla sequence in rrnA (Fig. 2D). There is no obvious explanation for the 3′ transcript end detected at position iB42 or the 5′ transcript end detected at position iB53.
23S distal region in rrnA and rrnB.
The 23S 3′ flanking region of rrnA contains a 5S gene and a tRNACys gene, whereas rrnB contains only a 5S gene (Fig. 1). The 23S-5S intergenic spaces in the two operons are 139 nt long, identical in sequence, and designated 23Sd1 to 23Sd139 in a 5′-to-3′ direction (Fig. 1). The 5S genes are 122 nt long and differ in sequence at positions 73 and 106. The 5S distal regions are identical in sequence for the first 61 nt except for a single difference at position 5Sd2; beyond position 5Sd61, the two operons show no significant sequence similarity (Fig. 2F). The 5S-tRNACys intergenic space in rrnA is 407 nt long.
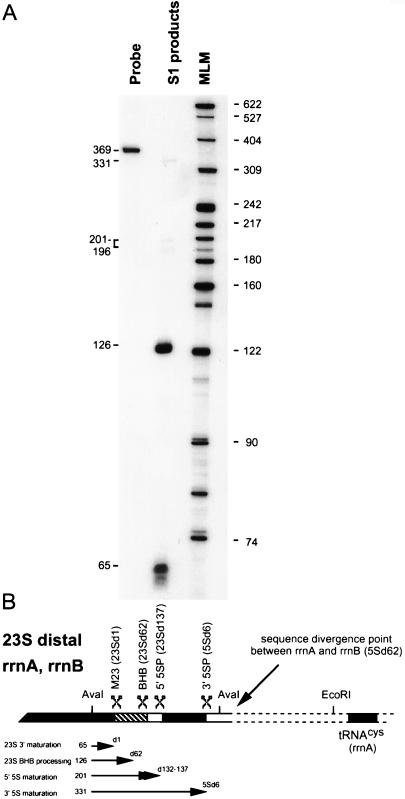
Processing of primary rRNA transcripts in the region distal to the 23S genes was examined by using a 3′-end-labeled 369-nt AvaI fragment that overlapped the last 65 nt of the 23S gene, the 23S-5S intergenic space, the 5S gene, and the first 43 nt of the 5S distal regions (Fig. 6). Transcripts from both operons hybridized efficiently to this probe in spite of the three nucleotide substitutions that exist within and immediately following the 5S sequence. With this probe, 3′ transcript ends were located at or near positions 23Sd1, 23Sd62, 23Sd132 to 23Sd137, and 5Sd6. There was no apparent protection of the entire probe, and the products resulting from cleavages around the 5S sequence were very faint. This indicates that primary transcripts containing the 5S sequence are rapidly and efficiently processed and therefore difficult to detect by nuclease protection assays. The shortest product with an end at 23Sd1 corresponds to the mature 3′ end of 23S rRNA, and the somewhat longer product with an end near position 23Sd62 corresponds to the BHB endonuclease processing site in the 3′ side of the 23S processing stem (Fig. 2A). The other two very faint 3′ ends were located about 4 to 8 nt upstream of the 5S sequence (positions 23Sd132 to 23Sd137) and about 6 nt downstream of the 5S sequence (5Sd6). These locations suggest that the 5S sequence may be excised as a precursor from the primary transcript and subsequently trimmed by a few nucleotides at the 5′ and 3′ ends. In Escherichia coli, pre-5S is excised by RNase E and subsequently trimmed a few nucleotides at the 5′ and 3′ ends (16). An RNase E-like endonuclease activity has been recently identified with H. marismortui and may be responsible for these pre-5S cleavages (14). Although precursor transcripts containing the tRNACys gene have been detected by nuclease protection assays, the results were neither definitive nor informative because of their low abundancy and rapid processing.
FIG. 6.
Analysis of 3′ transcript ends located in 23S-5S intergenic and flanking regions of rrnA and rrnB. Transcripts from both rrnA and rrnB were analyzed by using an AvaI fragment 3′ end labeled on the (−) strand at position 2859 within the 23S gene as a probe in S1 nuclease protection assays. (A) Autoradiogram illustrating protection products with the sizes in nucleotides of protection products (left) and of molecular length markers (MLM; right) indicated. (B) Diagram of the 23S-5S intergenic and 5S 3′ flanking regions. The 23S, 5S, and tRNACys genes are represented by solid boxes, the 23S inverted repeat is indicated by a hatched box, and other intergenic and flanking regions are indicated by open boxes. The dashed region, including the tRNACys gene, is unique to the rrnA operon. Other designations are as described in the legend to Fig. 5.
DISCUSSION
Two rrn operons were initially identified on 20- and 10-kb fragments from the genome of H. marismortui and subsequently cloned as 8.0-kb ClaI-HindIII (rrnA) and 10-kb HindIII (rrnB) fragments (23), respectively. Characterization of these clones indicates that they are surprisingly disparate: (i) the respective 16S, 23S, and 5S genes contain a substantial number of nucleotide sequence differences (Table 1); (ii) except for a short region of sequence conservation (Fig. 2C) and general promoter arrangement, the 5′ flanking regions exhibit little sequence similarity; (iii) processing of 16S rRNA from the rrnA operon transcript follows the canonical archaeal pathway, whereas the processing of 16S rRNA from the rrnB operon occurs by direct endonucleolytic cleavage at the 5′ maturation site; and (iv) the rrnA operon contains tRNAAla and tRNACys genes in the 16S-23S intergenic and 5S distal regions, respectively, whereas the rrnB operon is apparently devoid of functional tRNA genes. Although only qualitative, the results of the S1 nuclease protection experiments reported here and in our earlier work (24) indicate that both operons contribute more or less equally to the production of ribosomes.
Implications for rRNA-based phylogeny and biodiversity.
Based upon sequence similarity alone, it would normally be concluded that the two 16S genes were orthologs derived from distinct species of halophilic archaea. Procaryotic divergence of 16S rRNA proceeds at a rate of about 1% per 50 × 106 years, suggesting that the two 16S sequences (with 5% divergence) were derived from a common ancestor about 250 × 106 years ago. This represents the same degree of 16S sequence divergence that separates E. coli and Serratia marcescens (8, 25). However, there is ample reason to believe that these sequences are not true orthologs. First, both were present in genomic DNA prepared from a culture derived from an isolated single colony. Second, the nucleotide sequence differences are not distributed throughout the 16S gene but rather are concentrated within limited regions; 51 of the 74 substitutions occur between nucleotide positions 520 and 820 of the 16S gene. A much more even distribution of substitutions across the entire sequence was observed for independently evolving orthologous 16S genes from several genera of halophilic archaea (24). Third, although the two 16S sequences are dissimilar, other regions of the rrnA and rrnB operons, including the 3′ portion of the 16S-23S intergenic spacer, the 23S-5S spacer, and the first 62 nt of the 5S 3′ flanking sequence, are virtually identical (Fig. 1). When orthologous halophilic rrn operons are compared, sequence divergence in spacer and flanking regions is more rapid and extensive than is divergence within structural rRNA genes.
From the above considerations, we conclude that the rrnA and rrnB operons are both present within the genome of H. marismortui. However, the evolutionary history of the operons cannot be ascertained with any degree of certainty. They may be strict paralogs, i.e., generated by a gene duplication event and never separated by a speciation event. If this were the case, it suggests that the rrnB operon has lost both the tRNAAla and tRNACys genes and developed an alternative pathway for processing 16S rRNA, since these features appear to be ancestral within halophilic rrn operons. Alternatively, the two operons may be ancient orthologs brought back together by lateral transfer to create a chimeric genome. If so, it suggests that recombination or gene conversion-type processes have partially rehomogenized certain parts of the two operons, whereas other parts of the operons have retained their ancestral features. Whatever the correct scenario, it is clear that the different rRNA sequences do not necessarily mean distinct species. Moreover, the potential complications for interpreting phylogeny or biodiversity assessments based upon partial 16S rRNA sequences are evident (25, 27, 32).
Structural considerations.
Essentially all of the ribosomal protein genes from H. marismortui have been cloned and sequenced (29); each gene appears to be unique and have a single copy, and each protein appears to be capable of assembling into particles with either the rrnA or rrnB operon RNAs (24). This implies that the sequence or structural requirements for the RNA-protein interactions are maintained in spite of numerous nucleotide substitutions within or near certain protein binding sites. The 50S ribosomal subunits from H. marismortui have been crystallized for X-ray diffraction analysis and three-dimensional structural characterization (12). If these crystals contain both rrnA and rrnB 23S sequences, it implies that the nucleotide differences have little or no effect on the overall structure of the 50S subunit. The majority of substitutions in both 16S and 23S rRNAs occur in duplex regions and are compensatory. Such compensatory mutations would have little or no effect on the higher-order structure of 23S rRNA.
Maintenance of two disparate rrn operons.
Are there selective forces that contribute to the maintenance of two disparate rrn operons in H. marismortui and, if so, what might they be? In P. berghei, where there are also two types of 18S rRNA designated A and C, it has been suggested that differential expression may play a role in the types of proteins synthesized during the different developmental stages of the complex life cycle (18, 31). The switch from A to C expression occurs through the control of rRNA processing. In gametocytes, precursor transcripts from C-type genes are not processed, and ribosomes containing A-type RNA predominate. In the zygote and the early ookinete, processing of C-type RNA is accelerated and A-type ribosomes are targeted for selective degradation. In H. marismortui, both rrnA and rrnB RNAs are present in approximately equal amounts in ribosomes during cultivation under standard laboratory conditions (24). Moreover, it is interesting that, as in P. berghei, the pathways for processing 16S rRNA from the rrnA and rrnB transcripts are fundamentally different. We have previously suggested that salinity fluctuations in hypersaline environments pose unique challenges to resident halophilic organisms (4, 8). The presence of two distinct ribosome populations may allow H. marismortui to maintain essential protein synthesis during periods of environmental stress (4). Other characterized halophile species contain either a single rrn operon or two identical rrn operons (6, 15).
Processing of precursor transcripts from rrnA and rrnB.
The products observed in S1 nuclease protection assays indicate that there are at least four sites of endonuclease cleavage in the 383-nt 16S-23S spacer of transcripts derived from the rrnA operon. The first was at the BHB motif in the 3′ portion of the 16S processing helix. It is unclear whether maturation at the 3′ end of 16S is an endo- or exonucleolytic event. If it is endonucleolytic, it is a late event and occurs only after endonuclease cleavage either at the BHB motif in the 16S processing stem or at some other downstream site.
The second detectable endonuclease cleavage occurs at the 3′ end of the tRNA. In E. coli, an endonuclease cleaves several nucleotides 3′ to the mature tRNA sequences, and 3′-end maturation is subsequently carried out by a regiment of exonucleases (10). We have mapped the site of endonuclease cleavage to position iA177 by primer extension. If this result is correct (see Results), it suggests that 3′ exonuclease trimming is not involved in H. marismortui. In E. coli, the 3′ CCA end of the tRNA is encoded in the tRNA gene, whereas in archaea the addition of CCA is posttranscriptional and presumably added by a tRNA nucleotidyltransferase (see Fig. 2D). The 5′ end of tRNA is known to be generated by RNase P endonuclease (17, 28); the 3′ end of the leader released by RNase P cleavage was not detected by an S1 protection assay, whereas the 5′ end of the trailer was detected. This implies that RNase P cleavage is a late event and occurs only after the BHB endonucleolytic cleavage in the 3′ portion of the 16S processing stem. The detection of a 3′ end site at position iA139 in the middle of the tRNA suggests the presence of an alternate processing pathway that leads to tRNA degradation. It is unclear whether this product was generated by an exo- or endonuclease.
The third and fourth endonuclease cleavages occurred at the BHB motif in the 5′ portion of the 23S processing helix and near or at the 5′ end of 23S rRNA, respectively. The fact that the leader product of the maturation cleavage was observed indicates that 23S 5′-end maturation can on occasion occur before any of the other cleavages in the intergenic space, including excision of pre-23S from the primary transcript.
Only three endonucleolytic processing events were observed in the 405-nt 16S-23S spacer of transcripts derived from the rrnB operon. These correspond to three of the cleavages within the rrnA spacer: at the 3′ end of the tRNA-like sequence, at the BHB motif in the 5′ portion of the 23S processing helix, and at the 5′ 23S maturation site. Again, it is clear that 23S 5′-end maturation can occur on occasion before any of the other cleavages in the intergenic space.
As illustrated in Fig. 2D, the rrnB intergenic spacer appears to contain a tRNA-like sequence capable of forming a well-defined acceptor and TΨC stem. This sequence is almost certainly not a functional tRNA for several reasons: (i) it lacks the conserved GG dinucleotide in the DHU loop and the UUC trinucleotide in the TΨC loop, and (ii) the DHU and anticodon stems and loops are poorly conserved and organized. Based upon the presence of GU base pairs at position 3 in the acceptor stem, this sequence appears to be the remnant of a tRNAAla gene. Nonetheless, formation of a coaxial stack between the acceptor-like and the TΨC-like stems is apparently sufficient for endonucleolytic cleavage at the 3′ end of the structure at position iB198. Somewhat surprisingly, no products with either 5′ or 3′ ends located near the 5′ end of the tRNA-like structure were detected, although the structure should be a suitable substrate for RNase P. It is possible that RNase P cleavage, if it occurs at all, occurs late, after both upstream and downstream cleavages have taken place. Because of this, the product would not be visible in the S1 nuclease protection assay that was employed. The product with a 3′ end at iB137 within the rrnB tRNA-like sequence was detected. Comparison with the rrnA 3′ end at position iA139 revealed that both ends are located immediately 5′ to the tetranucleotide CAAG; in rrnA, this forms part of the anticodon loop in tRNAAla (Fig. 2D).
Processing and maturation of 16S rRNA from the rrnA operon transcript follow the canonical halophilic archaeal pathway that utilizes the BHB endonuclease to excise pre-16S rRNA. This is followed by maturation at the 5′ and 3′ ends. The rrnB operon transcript lacks the BHB motif in the 16S processing stem. No precursor 5′ ends were detected in the 5′ portion of the 16S processing stem. Instead, the rrnB primary transcript appears to be cleaved directly by a maturation endonuclease at the 5′ end of 16S rRNA. In the intergenic region, a 3′ transcript end at position iB42 and a 5′ transcript end at position iB53 were located in the 3′ portion of the stem. The significance of these ends and their relationship to each other is unknown.
ACKNOWLEDGMENTS
This work was supported by a grant from the Medical Research Council of Canada.
We thank Deidre de Jong-Wong for technical assistance.
REFERENCES
- 1.Branlant C A, Krol M A, Machott A, Pouyet J, Ebel J-P, Edwards K, Kossel H. E. coli 23S rRNA heterogeneity. Nucleic Acids Res. 1981;9:4303. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brombach M, Specht T, Erdmann V A, Ulbrich N. Complete nucleotide sequence of a 23S ribosomal RNA gene from Halobacterium marismortui. Nucleic Acids Res. 1989;17:3293. doi: 10.1093/nar/17.8.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chant J, Dennis P P. Archaebacteria: transcription and processing of ribosomal RNA sequences in Halobacterium cutirubrum. EMBO J. 1986;5:1091–1097. doi: 10.1002/j.1460-2075.1986.tb04327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dennis, P. P. Expression of ribosomal RNA operons in halophilic archaea, In A. Oren (ed.), Microbiology and biogeochemistry of hypersaline environments, in press. CRC Press, Inc., Boca Raton, Fla.
- 5.Dennis P P. Multiple promoters for the transcription of the ribosomal RNA gene cluster in Halobacterium cutirubrum. J Mol Biol. 1985;186:456–461. doi: 10.1016/0022-2836(85)90117-2. [DOI] [PubMed] [Google Scholar]
- 6.Dennis P P. The ribosomal RNA operons of halophilic archaebacteria. In: Rodriguez-Valera F, editor. General and applied aspects of halophilic microorganisms. New York, N.Y: Plenum Press; 1991. pp. 251–257. [Google Scholar]
- 7.Dennis P P, Chow J. Analysis of RNA transcripts in archaebacteria. In: Robb F T, Place A R, Sowers K R, Schreier H J, Das Sarma S, Fleischmann E M, editors. Protocols for archaebacterial research. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 163–178. [Google Scholar]
- 8.Dennis P P, Shimmin L C. Evolutionary divergence and salinity-mediated selection in halophilic archaea. Microbiol Mol Biol Rev. 1997;61:90–104. doi: 10.1128/mmbr.61.1.90-104.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dennis P P, Russell A G, Moniz de Sá M. Formation of the 5′ end pseudoknot in small subunit ribosomal RNA: involvement of U3-like sequences. RNA. 1997;3:337–343. [PMC free article] [PubMed] [Google Scholar]
- 10.Deutscher M. Ribonuclease multiplicity diversity and complexity. J Biol Chem. 1993;268:13011–13014. [PubMed] [Google Scholar]
- 11.Dover G A. Molecular drive: a cohesive mode of species evolution. Nature. 1982;299:111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- 12.Evers U, Franceschi F, Boddeker N, Yenath A. Crystallography of halophilic ribosomes: the isolation of an internal ribonucleoprotein complex. Biophys Chem. 1994;50:3–16. doi: 10.1016/0301-4622(94)85015-1. [DOI] [PubMed] [Google Scholar]
- 13.Fleishmann R D, Adams M D, White O, Clayton R A, Kirkness E F, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 14.Franzetti B, Sohlberg B, Zaccai G, von Gabain A. Biochemical and serological evidence for an RNase E-like activity in halophilic Archaea. J Bacteriol. 1997;179:1180–1185. doi: 10.1128/jb.179.4.1180-1185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrett R A, Dalgaard J, Larson N, Kjems J, Mankin A. Archaeal rRNA operons. Trends Biochem Sci. 1991;16:22–26. doi: 10.1016/0968-0004(91)90011-j. [DOI] [PubMed] [Google Scholar]
- 16.Ghora B K, Apirion D. Structural analysis and in vitro processing of p5 rRNA of a 9S RNA molecule isolated from an rne mutant of Escherichia coli. Cell. 1978;15:1055–1066. doi: 10.1016/0092-8674(78)90289-1. [DOI] [PubMed] [Google Scholar]
- 17.Gopalan V, Talbot S J, Altman S. RNA-protein interactions in RNase P. In: Nagai K, Mattaj I, editors. RNA protein interactions. Oxford, England: Oxford University Press; 1995. pp. 103–127. [Google Scholar]
- 18.Gunderson J H, Sogin M L, Wollett G, Hollingdale M, de la Cruz V F, Waters A P, McCutchan T F. Structurally distinct, stage-specific ribosomes occur in Plasmodium. Science. 1987;238:933–937. doi: 10.1126/science.3672135. [DOI] [PubMed] [Google Scholar]
- 19.Hancock J M, Dover G A. Compensatory slippage in the evolution of ribosomal RNA genes. Nucleic Acids Res. 1990;18:5949–5954. doi: 10.1093/nar/18.20.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henikoff S. Exonuclease III generated deletions for DNA sequence analysis. Promega Notes, no. 8. Madison, Wis: Promega Corp.; 1987. pp. 1–3. [Google Scholar]
- 21.Kelman-Leyer K, Armbruster D W, Daniels C J. Characterization of the Haloferax volcanii tRNA intron endonuclease gene reveals a relationship between the archaeal and eucaryal tRNA intron processing system. Cell. 1997;89:839–847. doi: 10.1016/s0092-8674(00)80269-x. [DOI] [PubMed] [Google Scholar]
- 22.Mankin A S, Skripkin E A, Kagramanova V K. A putative internal promoter in the 16S/23S intergenic spacer of the rRNA operon of archaebacteria and eubacteria. FEBS Lett. 1987;219:269–273. doi: 10.1016/0014-5793(87)80233-8. [DOI] [PubMed] [Google Scholar]
- 23.Mevarech M, Hirsch-Twizer S, Goldman S, Jakobson E, Eisenberg H, Dennis P P. Isolation and characterization of the rRNA gene clusters of Halobacterium marismortui. J Bacteriol. 1989;171:3479–3485. doi: 10.1128/jb.171.6.3479-3485.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mylvaganam S, Dennis P P. Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismortui. Genetics. 1992;130:399–410. doi: 10.1093/genetics/130.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochman H, Wilson A. Evolution in bacteria: evidence for a universal rate in cellular genomes. J Mol Evol. 1987;26:74–86. doi: 10.1007/BF02111283. [DOI] [PubMed] [Google Scholar]
- 26.Oren A, Lau P P, Fox G E. The taxonomic status of Halobacterium marismortui from the Dead Sea: a comparison with Halobacterium vallesmortis. Syst Appl Microbiol. 1988;10:251–258. doi: 10.1016/S0723-2020(88)80009-2. [DOI] [PubMed] [Google Scholar]
- 27.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 28.Pace N R, Brown J W. Evolutionary perspective on the structure and function of ribonuclease P, a ribozyme. J Bacteriol. 1995;177:1919–1928. doi: 10.1128/jb.177.8.1919-1928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholzen T, Arndt E. The alpha operon equivalent genome region in the extreme halophilic archaebacterium Haloarcula (Halobacterium) marismortui. J Biol Chem. 1992;267:12123–12130. [PubMed] [Google Scholar]
- 30.Thompson L D, Daniels C J. Recognition of exon intron boundaries by the Halobacterium volcanii tRNA intron endonuclease. J Biol Chem. 1990;265:18104–18111. [PubMed] [Google Scholar]
- 31.Waters A P, Syin C, McCutchan T F. Developmental regulation of state-specific ribosome populations in Plasmodium. Nature. 1989;342:438–440. doi: 10.1038/342438a0. [DOI] [PubMed] [Google Scholar]
- 32.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]