Abstract
A deletion of the sigB operon was constructed in three genetically distinct Staphylococcus aureus strains, and the phenotypes of the resulting mutants were analyzed. Compared to the corresponding wild-type strains, the ΔsigB mutants showed reduced pigmentation, accelerated sedimentation, and increased sensitivity to hydrogen peroxide during the stationary growth phase. A cytoplasmic protein missing in the ΔsigB mutants was identified as alkaline shock protein 23, and an extracellular protein excreted at higher levels in one of the ΔsigB mutants was identified as staphylococcal thermonuclease. Interestingly, most sigB deletion phenotypes were only seen in S. aureus COL and Newman and not in 8325, which was found to contain an 11-bp deletion in the regulator gene rsbU. Taken together, our results show that ςB is a global regulator which modulates the expression of several virulence factors in S. aureus and that laboratory strain 8325 is a ςB-defective mutant.
The initial event during infection by the human pathogen Staphylococcus aureus is the expression of certain virulence genes. Virulence factors are required for colonization of host tissue and for protection against the host defense. Timely correct expression of the virulence factors is essential for the establishment and maintenance of an infection and represents a highly regulated process (29). As a prerequisite, the microorganism has to recognize and respond to certain signals provided by the host. Such signals could be temperature (increases upon infection), peroxide (released by macrophages), pH shifts, or the presence or absence of specific carbon or energy sources (24).
In bacteria, alternative sigma factors of RNA polymerase are known to play a crucial role in regulating gene expression upon major changes in the environment. We recently identified the alternative sigma factor ςB in S. aureus 8325 and showed that ςB is induced during stationary phase and upon heat shock (20). The corresponding sigma factor in B. subtilis is known to be itself target of a complex regulatory network, which controls gene expression in response to certain stress and stationary-phase-specific signals (14). It has recently been shown that S. aureus ςB also has sigma factor activity in vitro and that transcription of the global regulator Sar in S. aureus is at least partially controlled by ςB (8). Since the Sar protein represents a global regulator involved in the expression of virulence genes (2, 4), it is tempting to speculate that ςB is directly or indirectly involved in the regulation of virulence genes. To test this hypothesis, we constructed a sigB deletion in several staphylococcal backgrounds and analyzed the phenotype of these mutants. Here we report that deletion of sigB caused a drastic phenotype in two of the three backgrounds tested and revealed a natural ςB defect in strain 8325. Compared to strain COL, strain 8325 has an 11-bp deletion in the gene encoding the ςB regulator RsbU (20, 36), which we suggest is the reason for the ςB defect.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture conditions, and general methods.
The bacterial strains and plasmids used in this study are listed in Table 1. S. aureus cells were routinely grown in Luria-Bertani (LB) medium at 37°C. Antibiotics were used at the following concentrations: for Escherichia coli, ampicillin at 100 μg ml−1; for S. aureus, erythromycin at 10 μg ml−1 and tetracycline at 10 μg ml−1. All DNA manipulations and handling of E. coli were performed in accordance with standard protocols (31). Manipulations with S. aureus were done as described earlier (20).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotypea | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH10B | F− φ80dlacZΔM15 recA1 | Gibco/BRL, Gaithersburg, Md. |
| S. carnosus TM300 | 13 | |
| S. aureus | ||
| RN4220 | NCTC 8325-4-r (restriction mutant) | 19 |
| 8325 | NCTC 8325 | |
| COL | High-Mcr clinical isolate | |
| Newman | High level of clumping factor | 9 |
| 8325 ΔsigB | Deletion of sigB operon, Emr | This study |
| COL ΔsigB | Deletion of sigB operon, Emr | This study |
| Newman ΔsigB | Deletion of sigB operon, Emr | This study |
| Plasmids | ||
| pBLSK(+) | Apr | Stratagene, La Jolla, Calif. |
| pIKET | tetK and ermB in pBLSK+; Apr in E. coli; Tcr Emr in S. aureus | This study |
| pTX15 | Tcr PXyl; staphylococcal origin of replication | 28 |
| pIK58 | 1.1-kb NsiI and 1.7-kb HindIII S. aureus fragments in pIKET | This study |
| pIK64 | 800-bp sigB fragment in pTX15 | This study |
Abbreviations are as follows: Apr, ampicillin resistant; Emr, erythromycin resistant; Mcr, methicillin resistant; Tcr, tetracyclin resistant.
Construction of plasmid pIK58, used to create a sigB deletion mutant.
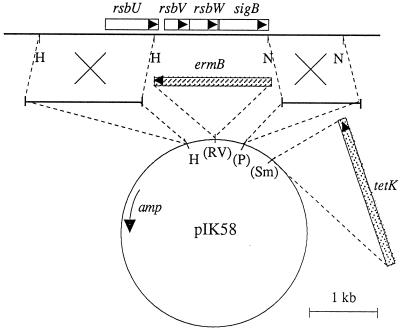
In a first step, we constructed the suicide vector pIKET by introducing appropriate antibiotic resistance cassettes into pBLSK(+), namely, the blunted 1.75-kb AvaI fragment with the ermB gene from transposon Tn551 in the EcoRV site of pBLSK(+), as well as the blunted 2.3-kb HindIII fragment with the tetK gene from pT181 in the SmaI site. This plasmid cannot replicate in S. aureus, and erythromycin resistance can only be rescued by integration of this plasmid into the chromosome. To provide sites for homologous recombination, we cloned the S. aureus 1.1-kb NsiI fragment (downstream region of sigB) in the PstI site of the vector and the 1.7-kb rsbU′-containing HindIII fragment in the HindIII site, leading to plasmid pIK58 (Fig. 1). Fifteen micrograms of this plasmid was used to transform S. aureus RN4220 by electroporation with erythromycin for selection.
FIG. 1.
Physical map of the sigB operon of S. aureus and the construction of a sigB deletion using suicide plasmid pIK58. Open reading frames are depicted as rectangles with arrows indicating their orientation. The genes for ampicillin resistance (amp), erythromycin resistance (ermB), and tetracycline resistance (tetK) are indicated. Recognition sites for restriction enzymes are designated as follows: H, HindIII; N, NsiI; P, PstI; RV, EcoRV; Sm, SmaI. Sites in parentheses were destroyed during cloning. The bold line corresponds to S. aureus chromosomal DNA, the thin line corresponds to pBLSK(+) DNA, and the dashed line shows the cloning procedures. The crosses indicate sites of homologous recombination.
Lipase assay.
To assay lipase activity, strains were grown for 12 h at 37°C in LB medium. Different dilutions of the culture supernatants were tested for lipase activity by monitoring hydrolysis of p-nitrophenyl-caprylate at 405 nm as described elsewhere (27). Calculation of lipase activity was adjusted for cell density and expressed as a percentage of that of the respective wild-type parent. Since the levels of lipase production differed substantially among the three backgrounds tested, dilutions of the supernatants were used to achieve a linear reaction in each case. Supernatants of strain 8325 were diluted 20-fold, those of strain Newman were diluted 4-fold, and those of strain COL were used without dilution; the wild type and mutants were always diluted equally. Lipase activity was also monitored on LB agar plates containing 1% Tween 20 and 1% xylose.
Peroxide susceptibility testing.
For disk assays, bacteria were plated on LB agar and a disk soaked with 10 μl of a 3% H2O2 solution was placed on the surface. Plates were incubated at 37°C for 48 h, and inhibition zones were compared. MICs and MBCs of H2O2 were determined by broth microdilution by using the National Committee for Clinical Laboratory Standards protocol (26) with serial dilutions of H2O2 (2.2 M to 0.125 mM). Microtiter plates were incubated for 48 h at 37°C.
SDS-gel electrophoresis of protein, protein blotting, and N-terminal protein sequencing.
Cellular or excreted proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis by using standard protocols (31). For analysis of excreted proteins, culture supernatants were concentrated about 10- to 15-fold by using Microcon 10 spin columns (Amicon Inc., Beverly, Mass.). Blotting of the proteins onto polyvinylidene difluoride membranes was done as previously described (1), and N-terminal sequencing of the respective proteins was done by P. Hunziker in the Laboratory of Biochemistry, University of Zürich.
Construction of pIK64 for complementation of ΔsigB strains.
For complementation assays, we used vector pTX15, which contains the xylose-inducible promoter PXyl and a tetracycline resistance determinant (28). We first amplified the entire sigB gene by PCR and subcloned it into pBLSK(+). The correct sequence of this fragment was confirmed by plasmid sequencing. We then subcloned the sigB-containing BamHI/EcoRI fragment into the BamHI/EcoRI sites of pTX15 downstream of PXyl, leading to plasmid pIK64. Since pTX15 only contains a staphylococcal origin of replication, we directly transformed pIK64 into S. carnosus TM300 by using protoplast transformation as previously described (13). pIK64 isolated from S. carnosus was then used to transform S. aureus RN4220. From there, the plasmid was moved into wild-type and ΔsigB mutant S. aureus by phage transduction as previously described (20). In those strains, PXyl and, therefore, expression of sigB were induced by adding 1% xylose to the medium. pIK64 could not be used for complementation of strain COL, since this strain naturally contains a tetracycline resistance-encoding plasmid.
RESULTS AND DISCUSSION
Construction of different sigB deletion mutants.
To create a sigB deletion mutant by homologous recombination, we constructed pBLSK(+)-based suicide plasmid pIK58. This plasmid contained two S. aureus DNA fragments flanking the region to be deleted, as well as appropriate antibiotic resistance markers for selection (Fig. 1). We transformed pIK58 into S. aureus RN4220 and obtained approximately 100 erythromycin-resistant colonies. Since no plasmid could be isolated from those transformants, pIK58 must have integrated into the chromosome through at least a single crossover in one of the two homologous regions. The colonies were replica plated onto tetracycline, and three colonies were tetracycline sensitive. This suggested that in these clones a double crossover and, therefore, deletion of the sigB region had occurred (Fig. 1). By using phage transduction and selection for erythromycin resistance, the deletion was transferred to three genetically different backgrounds: S. aureus 8325, COL, and Newman. S. aureus 8325 represents a standard laboratory strain, whereas strain COL is a highly methicillin-resistant clinical isolate. Strain Newman is characterized by a high level of clumping factor and is therefore often used in clumping or adhesion assays (9). After transduction, we isolated erythromycin-resistant and tetracycline-sensitive clones in all three backgrounds. Southern blot hybridization using a sigB-specific probe confirmed that sigB was deleted in all three strains (data not shown). We characterized the phenotype of those mutants as follows.
Reduced pigmentation.
The first obvious phenotype of the sigB deletion mutants was their color on agar plates. Whereas colonies of the wild-type parents of strains COL and Newman produced an orange pigment after 24 h of incubation, the colonies of the respective sigB mutants were unpigmented. In contrast, both wild-type strain 8325 and the ΔsigB mutant were unpigmented. Two major pigments are produced in S. aureus during stationary phase; the yellow carotenoid 4,4′-diaponeurosporene is converted to the orange pigment staphyloxanthin after prolonged cultivation (34). The genes responsible for the biosynthesis of these pigments have been characterized (34, 35). By homology, we were able to identify a ςB-dependent promoter consensus sequence upstream of the staphyloxanthin biosynthesis operon and thus propose that the conversion of 4,4′-diaponeurosporene to staphyloxanthin during late stationary phase is ςB dependent in S. aureus. Interestingly, carotenoid synthesis in Streptomyces setonii is dependent on the ςB homologue CrtS (17).
Increased sedimentation and aggregation.
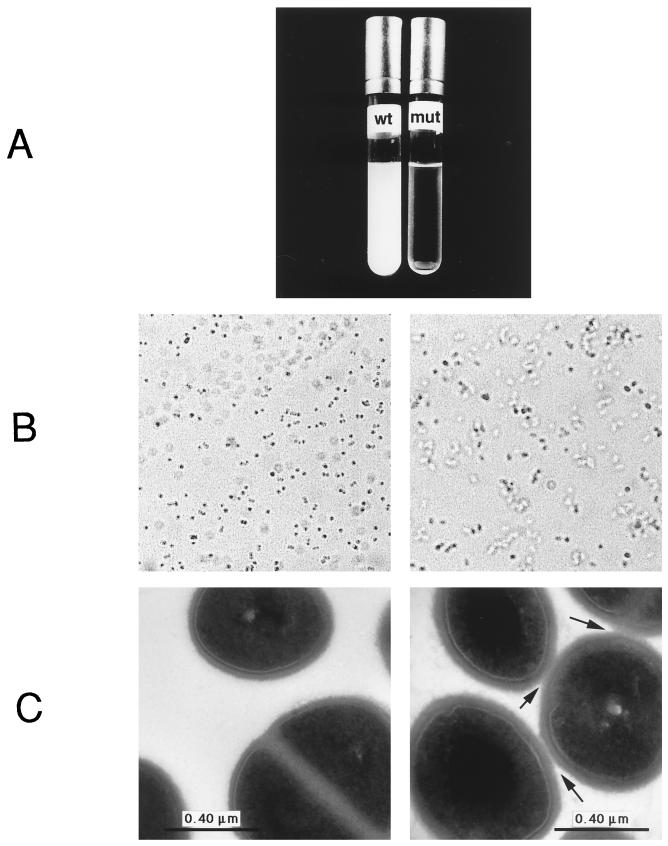
The growth rates of all of the strains tested were unaffected by the sigB deletion, but strain Newman ΔsigB exhibited accelerated sedimentation when a culture grown overnight was left without agitation. In contrast to the wild-type parent, almost all cells in the mutant culture were completely sedimented after less than 1 h (Fig. 2A). Light microscopic analysis revealed that Newman ΔsigB cells were clustered while wild-type cells were mostly separated (Fig. 2B). Electron microscopy confirmed close cell contact in strain Newman ΔsigB (Fig. 2C). Close cell contact can be due to either incomplete separation after cell division or secondary interaction of cell surface proteins, and the correct expression and cellular localization of cell surface proteins is essential for successful colonization of surfaces by S. aureus (11, 12). However, the increased cell aggregation of the ΔsigB mutant was observed only in strain Newman and not in strain COL or 8325. Strain Newman is known to produce high levels of clumping factor, a cell surface-associated fibrinogen receptor (23). It is possible that ςB modulates expression of the clumping factor or of other adhesins, which might lead to the observed cell aggregation also in cultures.
FIG. 2.
Comparison of liquid cultures of wild-type (wt) and ΔsigB mutant (mut) forms of S. aureus Newman. (A) Sedimentation of a culture grown overnight is shown after 1 h without shaking. Overnight cultures were also analyzed by light microscope (×1,000 magnification) (B) or by electron microscope (C). In both cases, aggregation of the cells was visible. The arrows indicate areas of close contact between cells.
Decreased susceptibility to H2O2 during stationary phase.
Since H2O2 represents an important stress factor for S. aureus during infection, we tested the H2O2 susceptibility of the ΔsigB mutants in a disk diffusion assay. The diameters of the inhibition zones were the same in the wild type and ΔsigB mutants, yet interestingly, after 48 h of incubation, we repeatedly observed wild-type colonies of strains COL and Newman close to the disk (Fig. 3). Those colonies were never observed with the sigB deletion mutants or with wild-type strain 8325. It was possible that those colonies represented highly H2O2-resistant second-site mutants. However, when they were reassayed for H2O2 susceptibility, they were not highly resistant to H2O2, but the same growth pattern as in the original assay was observed. This implies that the appearance of such highly H2O2-resistant colonies in late stationary phase (48 h) is due to a transient effect, possibly an adaptive response to H2O2. We also examined the MICs and MBCs of H2O2 (Table 2). For the wild-type COL and Newman strains, the MBC was higher than the MIC, whereas for COL ΔsigB, Newman ΔsigB, and wild-type and ΔsigB 8325, the MICs and MBCs were identical. Thus, higher concentrations of H2O2 are required to kill wild-type COL and Newman cells than to inhibit growth, whereas in the respective sigB deletion mutants, as well as in strain 8325, the same concentration of H2O2 which inhibits growth also kills the cells. This is in accordance with the results seen in the zone assay, where after 48 h only wild-type COL and Newman cells could again grow after being initially inhibited. Taken together, these data suggest that in S. aureus growth at high concentrations of peroxide during late stationary phase requires ςB.
FIG. 3.
Comparison of the H2O2 susceptibilities of wild-type (wt) and ΔsigB mutant (mut) strains of S. aureus in a disk diffusion assay after 48 h. The primary inhibition zones of all strains were comparable, but wild-type strain COL and Newman colonies growing close to the central H2O2 disk were visible after 48 h of incubation.
TABLE 2.
Susceptibilities of S. aureus sigB mutant strains and their wild-type parents to H2O2
| Strain | MIC (mM) | MBC (mM) |
|---|---|---|
| 8325 | ||
| Wild type | 0.5 | 0.5 |
| ΔsigB mutant | 0.5 | 0.5 |
| COL | ||
| Wild type | 1 | 4 |
| ΔsigB mutant | 1 | 1 |
| Newman | ||
| Wild type | 0.25 | 4 |
| ΔsigB mutant | 0.25 | 0.25 |
H2O2 is detoxified by the enzyme catalase, which is considered to be an important virulence factor in S. aureus. Catalase protects the cells from the oxidative burst released from host macrophages upon infection, and in S. aureus, a correlation between catalase activity and virulence has been observed (16, 22). In E. coli, expression of the two catalase genes is regulated by the stationary-phase sigma factor ςS (15, 25), and in B. subtilis, one catalase gene has been shown to be regulated by ςB (10). S. aureus also appears to have multiple catalase activities (7), and since the growth of S. aureus at high concentrations of H2O2 in late stationary phase was ςB dependent, we suggest that expression of at least one of the catalases is regulated by ςB. Preliminary data from catalase activity stains of native protein gels with total protein from S. aureus wild-type and ΔsigB strains revealed the existence of at least two bands with catalase activity, of which one was less abundant in the ΔsigB strains (data not shown). However, further biochemical analysis is required to define those activities.
Expression of Asp23 and thermonuclease is affected by the sigB deletion.
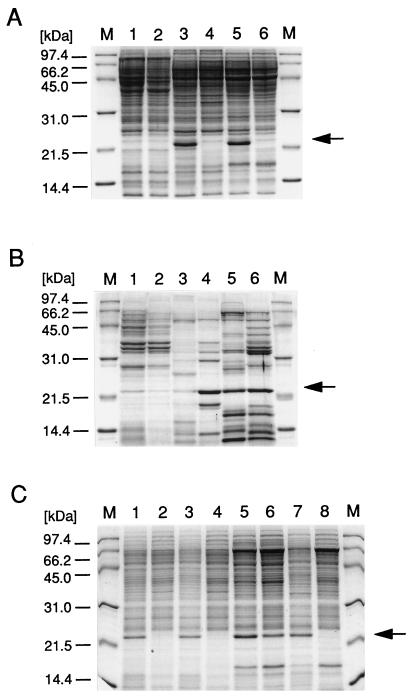
By using SDS-polyacrylamide gel electrophoresis, we compared the total protein expression patterns and the levels of excreted protein of the wild-type and ΔsigB strains (Fig. 4). The patterns of total protein expression of the wild-type and mutant strains were very similar. However, one very abundant protein of about 23 kDa was clearly missing in the ΔsigB mutants of strains COL and Newman and also in wild-type 8325 (arrow in Fig. 4A). N-terminal sequencing of this protein revealed the amino acid sequence MTVDNNKAKQAYDNQ, which shows 100% identity with the first 15 amino acids of alkaline shock protein 23 (Asp23) of S. aureus. The asp23 gene was previously found to encode a 169-amino-acid protein with an unknown function and a molecular mass of 19.2 kDa (21). Although the molecular mass of Asp23 is predicted to be 19.2 kDa, it reveals an apparent molecular mass of 23 kDa on a protein gel. It was furthermore demonstrated that expression of Asp23 is strongly induced upon a pH upshift to 10 (21). The transcriptional start site of asp23 has been mapped (21), and upon examination of the sequence, we found a perfect ςB-dependent promoter consensus sequence, instead of the proposed ςA promoter, at the correct distance from the transcriptional start of the gene. These observations strongly suggest that asp23 is a ςB target in S. aureus. In growth assays, the ΔsigB mutant strains were slightly more sensitive to a pH upshift from 7 to 10 during exponential growth phase (data not shown). However, the function of Asp23 in S. aureus and its possible role in permitting S. aureus to grow at high pHs remain to be determined.
FIG. 4.
SDS-polyacrylamide gel electrophoresis analysis of wild-type and ΔsigB mutant S. aureus strains. In A, about 20 μg of total cellular protein per lane was analyzed; in B, about 5 μg of excreted protein per lane was analyzed. Samples were analyzed as follows: lanes 1 and 2, strain 8325; lanes 3 and 4, strain COL; lanes 5 and 6, strain Newman. Extracts in lanes 1, 3, and 5 were from the wild-type parent, and those in lanes 2, 4, and 6 were from the corresponding ΔsigB mutant. In C, 10 μg of total cellular protein per lane from the following strains was analyzed: lanes 1 and 2, strain 8325 wild type; lanes 3 and 4, strain 8325 ΔsigB; lanes 5 and 6, strain Newman wild type; lanes 7 and 8, strain Newman ΔsigB. Lanes 1, 3, 5, and 7 were complemented with plasmid pIK64. A protein standard was loaded in lanes M. The arrows in A and C indicate the protein identified as Asp23. The arrow in B indicates the protein identified as staphylococcal thermonuclease.
On gels of the excreted proteins of S. aureus, the amount and pattern of excreted proteins varied significantly among wild-type strains 8325, COL, and Newman (Fig. 4B). Comparison of each wild-type parent with the respective ΔsigB mutant revealed three classes of proteins: (i) those which were identical in the wild type and the mutant, (ii) those which were less abundant in the mutant, and (iii) those which were enhanced in the mutant. We determined the N-terminal sequence of a 23-kDa protein from ΔsigB mutant strains COL and Newman (arrow in Fig. 4B) and in both cases obtained the amino acid sequence SQTDNGVNR, which showed 100% identity with amino acids 64 to 72 of the staphylococcal thermonuclease (5, 6, 18). Staphylococcal thermonuclease is an excreted toxin of 231 amino acids with a molecular mass of 25.5 kDa. Amino acids 1 to 63 serve as a signal peptide which is cleaved off after protein export. Therefore, the mature excreted protein begins with amino acid 64, which is exactly where the homology to our protein sequence starts. Thermonuclease was hardly detectable in wild-type and mutant 8325 but highly abundant in both wild-type and ΔsigB mutant strain Newman. Interestingly, in strain COL, thermonuclease was more prominent in the ΔsigB mutant than in the wild type, indicating that in this strain ςB has a negative effect either on protein expression or on export of this protein into the medium. Taken together, our data indicate that deletion of ςB can have both positive and negative effects on certain proteins. It also shows that additional strain-specific factors may exist which modulate ςB-dependent regulation in the respective background.
Increased lipase activity.
Lipase represents an important virulence factor that is excreted mainly during stationary growth phase and can easily be assayed either on plates or in liquid culture supernatants. On plates, we observed that all of the sigB deletion strains studied produced more lipase than did their respective wild-type parents (data not shown). This effect was confirmed by a quantitative assay of culture supernatants. Interestingly, the levels of lipase expression varied significantly among the three wild-type strains, with strain 8325 showing the highest activity and strain COL showing the lowest activity. Since we focused on the effect of the sigB deletion in each case, the lipase activity of each wild-type strain was set as 100% and the ΔsigB mutant levels were compared with those of the corresponding parents (Table 3). For strains 8325 and COL, lipase production was about 1.5 times as high in the ΔsigB mutant as in the parent, and for strain Newman, mutant lipase activity was increased three- to fourfold. Those values were highly reproducible and were confirmed in four independent assays. Hence, we speculate that lipase production or excretion during stationary growth phase is negatively regulated by ςB or by a ςB-dependent factor.
TABLE 3.
Lipase activities of S. aureus sigB mutant strains and their wild-type parents
| Strain | Avg % activity (SD)a |
|---|---|
| 8325 | |
| Wild type | 100 |
| ΔsigB mutant | 162 (8) |
| COL | |
| Wild type | 100 |
| ΔsigB mutant | 153 (16) |
| Newman | |
| Wild type | 100 |
| ΔsigB mutant | 373 (3) |
The values shown are results of four independent assays.
Complementation of ΔsigB mutants restores pigmentation and Asp23 production.
To prove that the observed phenotypes of the mutants are due to the deletion of sigB itself, we complemented the mutants with plasmid pIK64, carrying the sigB gene under control of the xylose-inducible promoter PXyl. pIK64 was introduced into the wild-type and ΔsigB mutant forms of strains 8325 and Newman but could not be used in strain COL because of the overlapping tetracycline resistance encoded by plasmid pIK64 and a plasmid naturally present in this background. When ΔsigB strains 8325 and Newman containing pIK64 were grown in medium with xylose, the formerly pigmentless strains produced a very intense orange pigment, even during exponential growth phase and not only in late stationary phase. In contrast, without xylose in the medium, no such pigment was produced. These results show that (i) sigB alone restores pigmentation in the pigmentless ΔsigB mutants and (ii) overexpression of plasmid-encoded sigB causes overexpression of the orange pigment staphyloxanthin even much earlier than in the wild-type situation. Therefore, we conclude that the operon for staphyloxanthin biosynthesis is a direct target of ςB. pIK64 even caused orange pigmentation in otherwise pigmentless wild-type 8325, meaning that the missing pigmentation in this strain is not due to a loss or defect of the pigmentation genes but rather to a nonfunctional ςB protein, most likely due to a defective RsbU protein, as described later in the report.
We also analyzed the expression of the protein Asp23 in the complemented mutants (Fig. 4C). Addition of pIK64 and xylose restored Asp23 expression in wild-type 8325, ΔsigB 8325, and ΔsigB Newman (lanes 1, 3, and 7). In wild-type strain Newman, the presence of pIK64 even slightly increased the amount of Asp23 (lane 5). These results support our assumption that asp23 is also a direct target of ςB. Taken together, the complementation data prove that deletion of ςB alone leads to the observed phenotypes in the mutants described.
Deletion of 11 bp of rsbU in strain 8325 and derivatives.
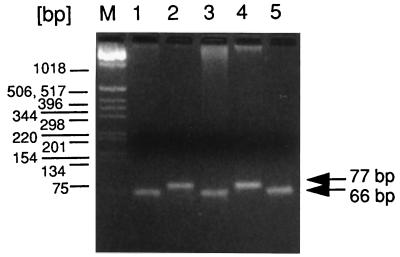
We previously found that strain 8325 contains an 11-bp deletion in the rsbU gene in comparison to the B. subtilis sequence and also in comparison to the sigB sequence in S. aureus COL (20). To test whether this deletion was present in other 8325 derivatives as well, we performed PCR with oligonucleotides flanking the deletion site. PCR should give rise to a 77-bp fragment in the wild type and a 66-bp fragment in a mutant with an 11-bp deletion. Two independent 8325 strains (lanes 1 and 5) and strain RN4220 (an 8325 derivative) (lane 3) had an 11-bp deletion, whereas strains COL and Newman (lanes 2 and 4) had no deletion (Fig. 5).
FIG. 5.
PCR of different S. aureus strains using primers flanking the 11-bp deletion in the rsbU gene. Amplification products were analyzed on a 4% agarose gel as follows: lane 1, strain 8325; lane 2, strain COL; lane 3, strain RN4220, lane 4, strain Newman; lane 5 independent source of strain 8325. A 100-bp DNA ladder was loaded in lane M as a molecular size marker.
In B. subtilis, an intact RsbU phosphatase is required for full ςB activity under stress conditions (32, 33). Therefore, it is to be expected that S. aureus ςB can only be activated under the respective stress conditions in strains with functional RsbU. Consequently, it is plausible that deletion of ςB results in a mutant phenotype only in strains with intact RsbU.
Wild-type strain 8325 resembles the COL and Newman ΔsigB mutants with respect to Asp23 expression, H2O2 susceptibility, and pigmentation. Therefore, we suggest that strain 8325 represents a natural ςB-defective mutant most probably due to the 11-bp deletion in the rsbU gene. In contrast, ΔsigB strain 8325 does show a mutant phenotype with respect to lipase production during stationary growth phase, where ςB might activate a negative effector. From work with B. subtilis, it is known that ςB can be activated via two pathways: during exponential phase by an RsbU-dependent pathway and by an RsbU-independent pathway during stationary phase (33). We propose that to control lipase production during stationary phase, ςB does not require stress induction via RsbU but can still be activated during stationary phase in an RsbU-independent way.
In summary, we have demonstrated that a sigB deletion in S. aureus produces a pleiotropic phenotype. Our results suggest that asp23 and the operon for staphyloxanthin biosynthesis are direct targets of ςB in S. aureus. In B. subtilis, ςB is a stationary-phase- and stress-specific sigma factor and several target genes are known to be ςB dependent. However, in many cases, the function of the target genes is unknown, and a sigB mutation has no clear phenotype. Stress survival does also not appear to be impaired in the mutants (14). ςB may not be essential for survival but might give a competitive advantage under specific environmental conditions. In a pathogen like S. aureus, expression of virulence genes is not essential but enables the cells to colonize and survive in human hosts, who serve as a specialized ecological niche. Several of the functions which we have demonstrated to be modulated by ςB in S. aureus, such as peroxide resistance, possibly alkali stress response, cell aggregation, or lipase and thermonuclease production, may play an important role during infection. In addition, expression of the global regulator Sar is ςB dependent (8). The Sar protein is required for expression of the regulator Agr (3), which, in turn, affects the expression of a variety of virulence factors (30). Thus, we suggest that S. aureus ςB is a stress- and stationary-phase-specific global regulator which is directly and indirectly involved in the expression of virulence genes. We predict that the identification of additional ςB-dependent target genes will provide insight into the regulatory pathways controlling the process of infection.
ACKNOWLEDGMENTS
We thank Ursula Hardegger for excellent technical assistance and Brigitte Berger-Bächi for fruitful discussions. We also thank Peter Hunziker, Institute of Biochemistry, University of Zürich, for protein sequencing. This work was done in the laboratory of F. H. Kayser, whose generous support is gratefully acknowledged.
P.G. was supported by NF grant 31-46762.96 of the Schweizerischer Nationalfonds.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R F, Moore D D, Seidmann J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 2.Cheung A L, Koomey J M, Butler C A, Projan S J, Fischetti V A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung A L, Projan S J. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J Bacteriol. 1994;176:4168–4172. doi: 10.1128/jb.176.13.4168-4172.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung A L, Ying P. Regulation of α- and β-hemolysins by the sar locus of Staphylococcus aureus. J Bacteriol. 1994;176:580–585. doi: 10.1128/jb.176.3.580-585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cone J L, Cusumano C L, Taniuchi H, Anfinsen C B. Staphylococcal nuclease (Foggi strain). II. The amino acid sequence. J Biol Chem. 1971;246:3103–3110. [PubMed] [Google Scholar]
- 6.Davis A, Moore I B, Parker D S, Taniuchi H. Nuclease B. A possible precursor of nuclease A, an extracellular nuclease of Staphylococcus aureus. J Biol Chem. 1977;252:6544–6553. [PubMed] [Google Scholar]
- 7.Degteva G K, Malakhova T A. Quantitative indices of the activity and the electrophoretic characteristics of staphylococcal catalase. Zh Mikrobiol Epidemiol Immunobiol. 1979;6:31–34. [PubMed] [Google Scholar]
- 8.Deora R, Tseng T, Misra T K. Alternative transcription factor sigma ςSB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J Bacteriol. 1997;179:6355–6359. doi: 10.1128/jb.179.20.6355-6359.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duthie E S, Lorenz L L. Staphylococcal coagulase: mode of action and antigenicity. J Gen Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
- 10.Engelmann S, Lindner C, Hecker M. Cloning, nucleotide sequence, and regulation of katE encoding a ςB-dependent catalase in Bacillus subtilis. J Bacteriol. 1995;177:5598–5605. doi: 10.1128/jb.177.19.5598-5605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster T J, McDevitt D. Surface-associated proteins of Staphylococcus aureus: their possible roles in virulence. FEMS Microbiol Lett. 1994;118:199–205. doi: 10.1111/j.1574-6968.1994.tb06828.x. [DOI] [PubMed] [Google Scholar]
- 12.Francois P, Vaudaux P, Foster T J, Lew D P. Host-bacteria interactions in foreign body infections. Infect Control Hosp Epidemiol. 1996;17:514–520. doi: 10.1086/647358. [DOI] [PubMed] [Google Scholar]
- 13.Götz F, Schumacher B. Improvements of protoplast transformation in Staphylococcus aureus. FEMS Microbiol Lett. 1987;40:285–288. [Google Scholar]
- 14.Hecker M, Schumann W, Voelker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 15.Ivanova A, Miller C, Glinsky G, Eisenstark A. Role of rpoS (katF) in OxyR-independent regulation of hydroperoxidase I in Escherichia coli. Mol Microbiol. 1994;12:571–578. doi: 10.1111/j.1365-2958.1994.tb01043.x. [DOI] [PubMed] [Google Scholar]
- 16.Kanafani H, Martin S E. Catalase and superoxide dismutase activities in virulent and nonvirulent Staphylococcus aureus isolates. J Clin Microbiol. 1985;21:607–610. doi: 10.1128/jcm.21.4.607-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato F, Hino T, Nakaji A, Tanaka M, Koyama Y. Carotenoid synthesis in Streptomyces setonii ISP5395 is induced by the gene crtS, whose product is similar to a sigma factor. Mol Gen Genet. 1995;247:387–390. doi: 10.1007/BF00293207. [DOI] [PubMed] [Google Scholar]
- 18.Kovacevic S, Veal L E, Hsiung H M, Miller J R. Secretion of staphylococcal nuclease by Bacillus subtilis. J Bacteriol. 1985;162:521–528. doi: 10.1128/jb.162.2.521-528.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreiswirth B N, Löfdahl S, Betley M J, O’Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 20.Kullik I, Giachino P. The alternative sigma factor ςB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch Microbiol. 1997;167:151–159. doi: 10.1007/s002030050428. [DOI] [PubMed] [Google Scholar]
- 21.Kuroda M, Ohta T, Hayashi H. Isolation and the gene cloning of an alkaline shock protein in methicillin resistant Staphylococcus aureus. Biochem Biophys Res Commun. 1995;207:978–984. doi: 10.1006/bbrc.1995.1281. [DOI] [PubMed] [Google Scholar]
- 22.Mandell G L. Catalase, superoxide dismutase, and virulence of Staphylococcus aureus. In vitro and in vivo studies with emphasis on staphylococcal-leucocyte interaction. J Clin Invest. 1975;55:561–566. doi: 10.1172/JCI107963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDevitt D, Francois P, Vaudaux P, Foster T J. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol. 1994;11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 24.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukhopadhyay S, Schellhorn H E. Induction of Escherichia coli hydroperoxidase I by acetate and other weak acids. J Bacteriol. 1994;176:2300–2307. doi: 10.1128/jb.176.8.2300-2307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 27.Peschel A, Augustin J, Kupke T, Stevanovic S, Götz F. Regulation of epidermin biosynthetic genes by EpiQ. Mol Microbiol. 1993;9:31–39. doi: 10.1111/j.1365-2958.1993.tb01666.x. [DOI] [PubMed] [Google Scholar]
- 28.Peschel A, Ottenwälder B, Götz F. Inducible production and cellular location of the epidermin biosynthetic enzyme EpiQ using an improved staphylococcal expression system. FEMS Microbiol Lett. 1996;137:279–284. doi: 10.1111/j.1574-6968.1996.tb08119.x. [DOI] [PubMed] [Google Scholar]
- 29.Projan S J, Novick R P. The molecular basis of pathogenicity. In: Crossley K B, Archer G L, editors. The staphylococci in human diseases. New York, N.Y: Churchill Livingstone; 1997. pp. 55–81. [Google Scholar]
- 30.Recsei P, Kreiswirth B, O’Reilly M, Schlievert P, Gruss A, Novick R P. Regulation of exoprotein gene expression in Staphylococcus aureus by AgaR. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Voelker U, Dufour A, Haldenwang W G. The Bacillus subtilis rsbU gene product is necessary for RsbX-dependent regulation of ςB. J Bacteriol. 1995;177:114–122. doi: 10.1128/jb.177.1.114-122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang W G. Separate mechanisms activate ςB of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol. 1995;177:3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wieland B, Feil C, Gloria-Maerker E, Thumm G, Lechner M, Bravo J-M, Poralla K, Götz F. Genetic and biochemical analyses of the biosynthesis of the yellow carotenoid 4,4′-diaponeurosporene of Staphylococcus aureus. J Bacteriol. 1994;176:7719–7726. doi: 10.1128/jb.176.24.7719-7726.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wieland, K. P., and F. Götz. 1996. Unpublished data.
- 36.Wu S, de Lencastre H, Tomasz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]