Abstract
Comparative analysis of chromosomal macrorestriction polymorphism of the two closely related Lactococcus lactis subsp. cremoris strains MG1363 and NCDO763 revealed the presence of a large inversion covering half of the genome. To determine what kind of genetic element could be implicated in this rearrangement, the two inversion junctions of MG1363 and NCDO763 chromosomes were cloned and characterized. Nucleotide sequence analysis showed the presence of one copy of the lactococcal IS905 element in each junction. Each copy of this element contained the same nucleotide mutation that inactivates the putative transposase. Comparison of the sequences surrounding the insertion sequence demonstrated that the large inversion arose from a single-step homologous recombination event between the two defective copies of the IS905 element. The large inversion presumably conferred no selective disadvantage on strain NCDO763 because this rearrangement did not alter the oriC-terC symmetry of the chromosome and the local genetic environment.
During the last decade, the increasing number of available chromosome maps has prompted reconsideration of existing theories about the evolution of bacterial genome organization (7, 16). It is now established that bacteria can be classified into two groups based on genomic stability. The first group contains bacteria in which the overall genomic organization is strongly preserved, as for Escherichia coli (46), Salmonella typhimurium (34), Clostridium perfringens (4), Lactococcus lactis (30), Mycoplasma gallisepticum (56), Halobacterium salinarum (23), and Thermus thermophilus (54), although chromosome rearrangements do occasionally occur. The second group comprises species in which the gene order is highly rearranged, such as Salmonella typhi (37), Rhodobacter capsulatus (44), Helicobacter pylori (26), Bacillus cereus (5), Leptospira interrogans (61), and Pseudomonas aeruginosa (49).
Until recently, it was generally thought that large (≥100-kb) chromosomal inversions were rarely found in natural conditions. However, there is increasing evidence that this kind of genome rearrangement is not exceptional. In addition to the well-known inversion between E. coli K-12 and S. typhimurium LT2 (47), comparison of genomic maps at the interspecies level recently revealed other inversions (12, 33, 36, 38, 41, 61). For example, the chromosomes of S. typhi Ty2 and S. typhimurium LT2 differ by at least three inversions (35). Some large inversions were also found when genome maps comparisons were done at the intraspecies level (Table 1). In one study of 21 P. aeruginosa clone C isolates, as many as nine inversions were observed (50).
TABLE 1.
Large chromosomal inversions within species
| Organism | No. of inversions | Size of inverted region (%) | Inversion junction(s) | Reference |
|---|---|---|---|---|
| Eubacteria (purple) | ||||
| Bordetella pertussis (6 strains) | 2 | 10–25 | ND | 52 |
| Escherichia coli K-12 | ||||
| LN850 and LN1053 | 2 | 40–47 | IS5a | 39 |
| W3110 | 1 | 20 | rrna | 24 |
| BHB2600 | 2 | 2–13 | IS1 and IS5d | 3 |
| 1485IN | 1 | 36 | IS3c | 27 |
| Neisseria gonorrhoeae (6 strains) | 1 | 30 | ND | 21 |
| Salmonella typhi (127 strains) | 20 | ND | rrnb | 37 |
| Pseudomonas aeruginosa clone C (21 strains) | 9 | 19–50 | ND | 50 |
| P. stutzeri (20 strains) | 4 | ND | ND | 22 |
| Eubacteria (gram-positive) | ||||
| Bacillus subtilis | ||||
| 166 | 1 | 45 | Nonhomologousc | 25 |
| 60866 | 1 | 40 | ND | 57 |
| Lactococcus lactis (2 strains) | 1 | 50 | ND | 31 |
| Mycoplasma hominis (5 strains) | 1 | 50 | ND | 28 |
| Eubacteria (spirochetes) | ||||
| Leptopsira interrogans (2 strains) | 1 | 40 | ND | 61 |
| Archaebacteria | ||||
| Halobacterium salinarum (3 strains) | 1 | 10 | ND | 23 |
Determined by Southern hybridization and genetic studies.
Determined by I-CeuI restriction analysis.
Determined by nucleotide sequence analysis.
Determined by restriction analysis.
It has been postulated that ribosomal (rrn) operons or insertion sequences (IS) are the primary targets involved in inversion formation by homologous recombination. However, experimental data have been obtained only for cultured enteric bacteria. In E. coli, the genetic elements implicated in five spontaneous large inversions were characterized. Recombination between rrnD and rrnE operons caused the chromosomal inversion in strain W3110 (24), whereas IS elements were responsible for the remaining inversions (IS3 for strain 1485IN [27], IS5 for strains LN850 and LN1053 [39], and IS5 and IS1 for strain BHB2600 [3]). In S. typhi, rrn loci were implicated in inversions solely on the basis of I-CeuI analysis (37). Only the inversion junctions of strain 1485IN have been characterized by nucleotide sequencing. In gram-positive bacteria, the inversion junctions of only one spontaneous inversion have been characterized at the nucleotide level (25) in Bacillus subtilis 166. This inversion is unlikely to have been caused by homologous recombination and probably occurred during the repair process after γ-ray irradiation. The nature of each of the genetic elements involved in the other inversions described in Table 1 is unknown, except for the inversion in a defined lineage of Neisseria gonorrhoeae that could probably be mediated by homologous recombination between pilE1-opaE and pilS5-opaD regions (21).
L. lactis is a gram-positive mesophilic bacterium extensively used for health and dairy applications (13). The low-resolution chromosome maps of four independent lactococcal strains have been constructed by pulsed-field gel electrophoresis (PFGE). Two strains, DL11 (58) and IL1403 (30), belong to L. lactis subsp. lactis, and the other two, MG1363 (31) and FG2 (9), are L. lactis subsp. cremoris strains. These two subspecies have nucleotide sequences that diverge by 20 to 30% (11), close to the level of divergence between E. coli and S. typhimurium. Comparison of physical maps of the four strains showed an overall conservation of restriction site locations for the two L. lactis subsp. lactis strains but not for the L. lactis subsp. cremoris strain. At the genetic (i.e., gene order) level, different kinds of rearrangements were observed. A large inversion covering almost half of the chromosome (31) was identified by comparing strains of the two different subspecies, and translocation or inversion of four discrete regions had occurred between the two L. lactis subsp. cremoris strains (9). However, as the genome comparisons were made between genetically unrelated strains, it is not possible to follow the genetic events involved in these rearrangements.
To investigate the lactococcal genome plasticity, the genomes of L. lactis subsp. cremoris NCDO763 (also called ML3) and MG1363 (19) were compared. Both strains are derivatives of strain NCDO712 (10) and are the lactococcal strains studied most extensively for genetic analysis and molecular biology. Strains NCDO763 and MG1363 differ by only two genome rearrangements: a 30-kb deletion and a large inversion (unpublished data). In this study, we report the identification of this large chromosomal inversion, cloning of the four inversion junctions (designated MG1363-inv1 and -inv2 for strain MG1363 and NCDO763-inv1 and -inv2 for strain NCDO763), and nucleotide characterization of the genetic element involved in the rearrangement.
MATERIALS AND METHODS
Bacterial strains and plasmids.
L. lactis subsp. cremoris NCDO763 (10) was grown in M17 (55), and MG1363 (19) was grown in GM17 (M17 broth containing 0.5% glucose). E. coli DH5α (Life Technologies, Gaithersburg, Md.) was grown in LB broth. Erythromycin was used at concentrations of 150 μg/ml for E. coli and 5 μg/ml for L. lactis. Plasmids are listed in Table 2.
TABLE 2.
Plasmids used
| Name | Description | Reference |
|---|---|---|
| pRC1 | Emr, 3.5-kb cloning vector constructed from pBluescript KSII (+) | 29 |
| pCL52 | Emr, 2.4-kb cloning vector constructed from pBluescript KSII (+) | Unpublished data |
| pCL102 | 5.5-kb HindIII fragment from strain NCDO763 in pRC1 | This work |
| pCL132 | 3.2-kb HindIII fragment from strain NCDO763 in pCL52 | This work |
| pCL133 | Same as pCL132 but inserted in opposite orientation | This work |
| pCL134 | 9-kb EcoRI fragment from strain NCDO763 in pCL52 | This work |
| pCL136 | 1.7-kb EcoRV fragment from pCL134 in pCL52 | This work |
| pCL137 | Same as pCL136 but inserted in opposite orientation | This work |
| pCL138 | 1.4-kb PCR product from strain NCDO763 in pCL52 | This work |
| pCL139 | Same as pCL138 but inserted in opposite orientation | This work |
| pCL200 | 5.4-kb SacI fragment from strain MG1363 in pCL52 | This work |
| pCL201 | 9-kb AccI fragment from strain MG1363 in pCL52 | This work |
| pCL203 | 1.7-kb EcoRV fragment from pCL200 in pCL52 | This work |
| pCL204 | Same as pCL203 but inserted in opposite orientation | This work |
| pCL206 | 1.5-kb BstXI fragment from pCL201 in pCL52 | This work |
| pCL207 | Same as pCL206 but inserted in opposite orientation | This work |
| pCL208 | 1.5-kb AflIII-BstXI fragment from pCL201 in pCL52 | This work |
| pCL209 | Same as pCL206 but inserted in opposite orientation | This work |
| pCL210 | 1-kb EcoRV-SacI fragment from pCL200 in pCL52 | This work |
| pCL211 | 1-kb EcoRV fragment from pCL200 in pCL52 | This work |
| pCL212 | Same as pCL211 but inserted in opposite orientation | This work |
DNA manipulation.
Lactococcal chromosome DNA was extracted as follows. Cells were grown overnight in M17 or GM17 broth containing 50 mM dl-threonine; 1 ml of cell suspension was centrifuged for 1 min (13,000 rpm), and the cell pellet was suspended in 0.75 ml of SET buffer (20% sucrose, 20 mM EDTA, 100 mM Tris [pH 8]) containing lysozyme (5 mg/ml). The mixture was incubated at 37°C for 1 h. Then 100 μl of 10% sodium dodecyl sulfate and 20 μl of proteinase K (25 mg/ml) were added, and incubation was continued at 60°C for 15 min. An equal volume of a phenol-trichloroethane-isoamyl alcohol (25/24/1) was added and mixed by vigorous shaking, and the mixture was centrifuged for 10 min (13,000 rpm). The aqueous layer was removed and reextracted with an equal volume of trichloroethane-isoamyl alcohol (24/1). Nucleic acids were precipitated with 2.5 volumes of ethanol after adjustments to 0.3 M NaCl and collected by centrifugation for 10 min (13,000 rpm). The DNA pellet was washed with 1 ml of 70% ethanol, dried for 5 min in a SpeedVac (Savant), and dissolved in 100 μl of ultrapure water (MilliQ; Millipore, Bedford, Mass.) containing RNase A (10 μg/ml). For PFGE analysis, high-molecular-weight chromosomal DNA of L. lactis was prepared from agarose-embedded cells as described previously (31). Plasmid DNA from E. coli was isolated by using a Qiaprep spin kit (small scale) or Qiagen plasmid midi kit (large scale) as instructed by the manufacturer (Qiagen GmBH, Hilden, Germany). Enzymatic modification of DNA (restriction, ligation, etc.) was performed as instructed by the manufacturers. DNA was introduced in E. coli DH5α by electrotransformation in a Gene Pulser (Bio-Rad, Hercules, Calif.) as recommended by the manufacturer.
PCR amplification and cloning of the NCDO763-inv2 region.
A primer specific for MG1363-inv1 (5′-TTTATTCCTCCAACCTATTA-3′) and another specific for MG1363-inv2 (5′-TAGAAATACGGTCCTGGTCA-3′) were used to PCR amplify (25 cycles of 95°C for 30 s, 48°C for 120 s, and 72°C for 120 s, followed by a final polymerization step at 72°C for 4 min) the NCDO763-inv2 region. The 1.4-kb amplified DNA fragment was purified and cloned into EcoRV-digested plasmid pCL52.
Electrophoresis and Southern hybridization.
Classical 1% agarose gels were made up in 0.1 M TBE (1 M TBE is 1 M Tris base, 1 M boric acid, and 20 mM EDTA). PFGE was performed in a contour-clamped homogeneous electric field system (Pulsaphor Plus; LKB-Pharmacia) in 0.05 M TBE as previously described (32). Standard size markers were phage λ DNA concatemers, obtained as described by Waterbury and Lane (59), and a 1-kb DNA ladder (Life Technologies). Southern hybridization of dried agarose gels was performed as previously described (31). Signals were detected with the BAS1000 bioimaging-analyzer system (Fuji Photo Film Co., Tokyo, Japan) and analyzed with TINA version 2.07c software (Raytest Isotopenβgeräte GmBH).
DNA sequencing and nucleotide analysis.
Automatic double-stranded DNA sequencing was carried out (Génome Express SA, Grenoble, France) for both strands, using universal, reverse, and appropriate synthetic primers. Sequence data were assembled and analyzed with Clone Manager version 4.1 (Scientific & Educational Softwares) and DNAsis version 7.00. DNA and protein sequence similarity searches were carried out with FASTA (45).
Nucleotide sequence accession numbers.
The GenBank accession numbers for MG1363-inv1 and MG1363-inv2 sequences are AJ223960 and AJ223961, respectively. The accession numbers for NCDO763-inv1 and NCDO763-inv2 sequences are AJ223962 and AJ223963, respectively.
RESULTS
Characterization of the large chromosomal inversion in strain NCDO763.
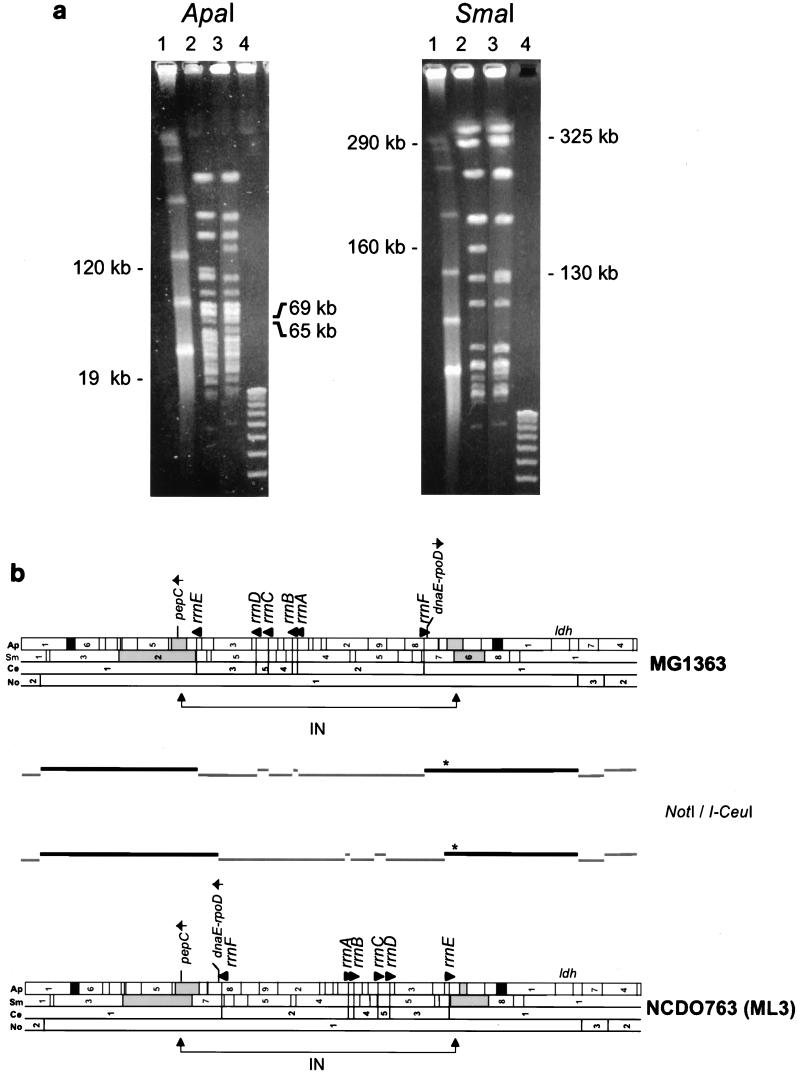
Comparative analysis of ApaI and SmaI fingerprints of the two genomes (Fig. 1a) revealed that the 69- and 65-kb ApaI fragments of MG1363 were replaced by two fragments of 120 and 19 kb and that the 325- and 130-kb SmaI fragments of MG1363 were replaced by two fragments of 290 and 160 kb in NCDO763. This restriction polymorphism did not change the overall size of either genome, as the sums of the fragment pairs were the same (134 kb in MG1363 and 139 kb in NCDO763 for ApaI; 455 kb and 450 kb for SmaI). The simplest explanation for this rearrangement was a chromosomal inversion between the two chromosomes, because a single inversion event does not change the total size of the genome but changes the size of two restriction fragments in each strain (Fig. 1b). Various hybridization probes were used to match the ApaI and SmaI fragments of MG1363 with their counterparts in NCDO763. Despite complex hybridization pattern, the 19-kb ApaI fragment used as the probe strongly hybridized to the 130-kb SmaI and 69-kb ApaI fragments of MG1363 and the 160-kb SmaI and 19-kb ApaI fragments of NCDO763 (Fig. 2b). The 325-kb SmaI and 65-kb ApaI fragments of MG1363 were associated with the 290-kb SmaI and the 120-kb ApaI fragments of NCDO763, using the pepC gene as a probe (data not shown).
FIG. 1.
(a) PFGE of chromosomal DNA from strains MG1363 and NCDO763 digested with restriction enzymes ApaI and SmaI. Lanes: 1, lambda concatemer size standards (each band is a 48.5-kb multimer); 2, NCDO763 DNA; 3, MG1363 DNA; 4, 1-kb DNA ladder. The sizes of fragments correlated with the large chromosomal inversion are indicated beside each gel. Electrophoresis was carried out at 10 V/cm for 13 h at 13°C, with pulse times of 6 s for ApaI and 8 s for SmaI. (b) Physical maps of MG1363 and NCDO763 chromosomes. Fragments are numbered as in reference 31. Arrows indicate the 5′→3′ orientation of the gene. Abbreviations: Ap, ApaI; Ce, I-CeuI; No, NotI; Sm, SmaI; IN, inversion region. The locations of fragments generated by a NotI/I-CeuI double digestion are shown between the maps. Black lines indicate fragments the size of which has changed; asterisks indicate fragments hybridizing to the ldh probe.
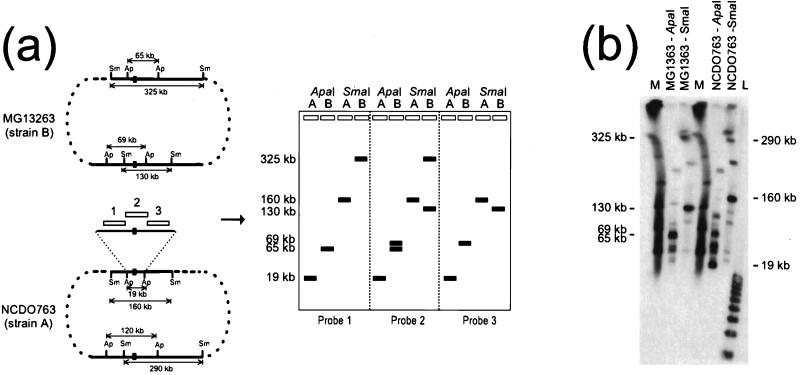
FIG. 2.
(a) General strategy for identification of a restriction fragment containing an inversion endpoint. Right, schematic representation of the MG1363 and NCDO763 chromosomes. Open rectangles indicate fragments used as probes. Inversion junctions are indicated by black boxes. Restriction fragments are not drawn to scale. Abbreviations: Ap, ApaI; Sm, SmaI. Left, expected hybridization patterns of PFGE-separated ApaI and SmaI fragments of NCDO763 (A) and MG1363 (B) chromosomes with the various probes. (b) Hybridization of PFGE-separated MG1363 and NCDO763 DNA, using the 19-kb ApaI fragment as a probe. A pulse time of 8 s was used. Lane M, lambda concatemer size standards; lane L, 1-kb DNA ladder. Hybridization was performed in the presence of 32P-labeled lambda and the 1-kb ladder.
The chromosomal inversion could also be demonstrated by comparison of the restriction patterns generated with I-CeuI, an endonuclease that recognizes a consensus sequence in the 23S rRNA gene of bacteria (40), followed by hybridization using the ldh gene as the probe. This gene has been accurately mapped to the MG1363 chromosome (31). The NotI/I-CeuI double-digested fragments detected were of a size similar to that expected in the case of an inversion (Fig. 1b).
Cloning the inversion junctions of the NCDO763 and MG1363 chromosomes.
The strategy used for cloning the NCDO763-inv1 region is summarized Fig. 2a. As depicted in this schematic, any restriction fragment located near the left side of an inversion junction (probe 1) should hybridize with the 19-kb ApaI and 160-kb SmaI fragments of the NCDO763 chromosome and with the 65-kb ApaI and the 325-kb SmaI fragments of the MG1363 chromosome. A restriction fragment located close to the right side of the junction (Fig. 2a, probe 3) should hybridize with the 19-kb ApaI and 160-kb SmaI fragments of the NCDO763 chromosome and with the 69-kb ApaI and 130-kb SmaI fragments of the MG1363 chromosome. However, a fragment carrying the inversion junction (Fig. 2a, probe 2) should give a single hybridization signal with the NCDO763 chromosome (19-kb ApaI and 160-kb SmaI fragments) and two with the MG1363 chromosome (65- and 69-kb ApaI fragments and 130- and 325-kb SmaI fragments).
Assuming that the 19-kb ApaI fragment of strain NCDO763 as a probe would generate a probe 2-type hybridization pattern, this fragment was used to probe PFGE-separated SmaI and ApaI fragments of the MG1363 and NCDO763 genomes. The hybridization pattern was more complex than expected (Fig. 2b), suggesting that the 19-kb ApaI fragment contained one or several elements repeated along both chromosomes at a high copy number. The unexpected absence of strong hybridization signals with the 325-kb MG1363 and 290-kb NCDO763 SmaI fragments could be explained only by a location of the inversion junction near one of the two ApaI sites. The above-mentioned strategy was then modified for a two-step cloning procedure. A genomic library of HindIII-digested DNA from strain NCDO763 was constructed and probed with the 19-kb ApaI fragment. Among the positive clones isolated, two HindIII fragments of 3.2 and 5.5 kb (pCL132 and pCL102, respectively [Table 2 and Fig. 3]) generated hybridization patterns consistent with their locations on either side of the inversion endpoint (data not shown). No other HindIII fragment containing the inversion endpoint could be cloned. A second NCDO763 genomic library of EcoRI fragments was then constructed and probed with pCL132 and pCL102. Both fragments hybridized with a 9-kb EcoRI fragment (pCL134). A restriction map of pCL134 was constructed (Fig. 3, NCDO763-inv1), and hybridization experiments using various subfragments of pCL134 as probes showed that the inversion endpoint was localized into a 1.7-kb EcoRV fragment (pCL136) and corresponded to the repeated element (data not shown) responsible for the complex hybridization pattern observed with the 19-kb ApaI fragment as probe (Fig. 2b).
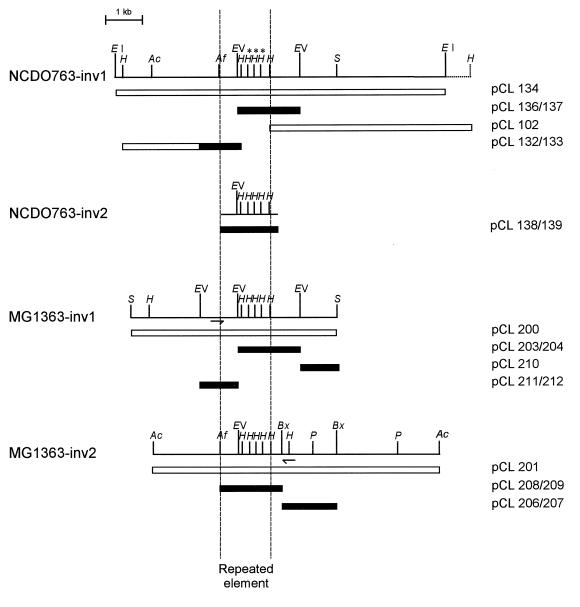
FIG. 3.
Restriction maps of the four regions surrounding the inversion junctions. The various restriction fragments cloned are indicated by open rectangles, and the corresponding plasmids are indicated. Black boxes show the DNA regions that were sequenced. Arrows indicate positions of the primers used for PCR amplification of the NCDO763-inv2 region. Abbreviations: EI, EcoRI; H, HindIII; Ac, AccI; Af, AflIII; EV, EcoRV; S, SacI; Bx, BstXI; P, PstI.
The two inversion endpoints of the MG1363 chromosome were cloned with two monocopy regions of pCL134 fragment, located on either side of the repeated element, as the probe. The first endpoint was obtained by isolating a 5.4-kb SacI fragment (pCL200) that hybridized to probe pCL102, and the second corresponded to a 9-kb AccI fragment (pCL201) that hybridized to a subfragment of pCL132. Restriction maps of these two fragments were constructed (Fig. 3, MG1363-inv1 and MG1363-inv2), and hybridization analysis demonstrated that both regions contained the same repeated element as carried by pCL134 fragment. The second inversion endpoint of the NCDO763 chromosome (Fig. 3, NCDO763-inv2) was obtained by PCR amplification using one primer located to the right of the repeated element of the MG1363-inv2 region and the second located to the left of the repeated element of the MG1363-inv1 region. Hybridization analysis using the amplification product (pCL138) as the probe indicated that the same repeated element as found in pCL134, pCL200, and pCL201 was also present in pCL138 (data not shown).
Sequence analysis of the four inversion endpoints.
Plasmids pCL136, pCL137, pCL138, pCL139, pCL203, pCL204, pCL206, pCL207, pCL208, pCL209, pCL210, pCL211, and pCL212 and parts of pCL132 and pCL133 (Fig. 3 and Table 2) were sequenced to identify the repeated element involved in the large inversion between the two chromosomes. Computer-assisted analysis of the MG1363-inv1 (3,638 bp), MG1363-inv2 (2,942 bp), and NCDO-inv1 (2,896 bp) regions predicted five open reading frames (ORFs) surrounding a 1,313-bp IS element present in each region (Fig. 4). The nucleotide sequence of the IS element was 99.9% identical to that of the previously characterized lactococcal insertion sequence IS905a and 100% identical to that of IS905b. The only difference between IS905a and IS905b is the substitution of a C for a T at position 693 of the published sequence, converting a glutamine codon to a stop codon and truncating about half of the putative transposase (14). A potential transcription terminator with a stem of 20 nucleotides and a loop of 5 nucleotides (ΔG° = −41.2 kcal/mol) was detected 296 nucleotides beyond ORF 2 of the MG1363-inv1 region. The stem had four T residues at its 3′ end, followed by three more T residues that are not part of the stem. This structure is typical of rho-independent transcription termination.
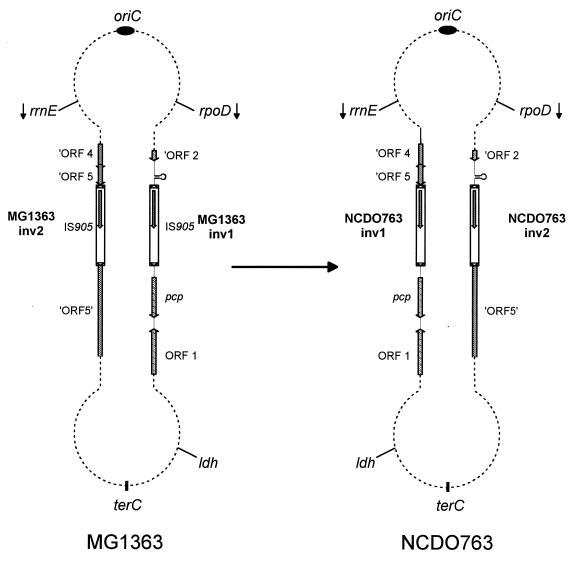
FIG. 4.
Structures of the MG1363 and NCDO763 chromosomes. The genetic organization of the regions surrounding the inversion junctions was deduced from sequence analysis. MG1363-inv1, MG1363-inv2, NCDO763-inv1, and NCDO763-inv2 are not drawn to scale of the chromosome. Arrows indicate the direction of gene transcription. The location of terC is hypothetical.
Computer-based comparison of the deduced amino acid sequences of the five ORFs against nonredundant protein sequence databases was performed. The C-terminal part of ORF 2 (53 amino acids) of the MG1363-inv1 region was 45.3% identical to the C-terminal part of the ceo gene product from the nisin-sucrose transposon Tn5306 of L. lactis. The ceo gene encodes an N5-(carboxyethyl)ornithine synthase (EC 1.5.1.24) involved in the synthesis of N-(CA)-amino acids (15). The C-terminal part of ORF 1 (244 amino acids) was 33.7% identical to the yhaA gene product of B. subtilis. Based on protein domain analysis (8), the ORF 1 and yhaA gene products could be related to the aminoacylase (EC 3.5.1.14) enzyme family (51). The 215-amino-acid product of the ORF located just downstream from IS905 was 58.6% identical to the pcp gene product of Streptococcus pyogenes. The pcp gene encodes a pyrrolidone carboxyl peptidase (PCP; EC 3.4.19.1) which removes N-terminal pyroglutamic acid residues from polypeptides (6). As E. coli K-12 strains are deficient in PCP activity (42), it was possible to phenotypically test the pyrrolidonyl peptidase activity of the MG1363-inv1 region by using a simple colorimetric assay (43). When pCL102 (containing the entire pcp gene) was introduced into E. coli DH5α, the erythromycin-resistant transformants were PCP+, indicating that the lactococcal pcp gene was functional. The products of ORFs 4 and 5 of the MG1363-inv2 region were 27.7 and 29.2% identical to those of ORFs 4 and 5 of Lactobacillus casei bacteriophage A2 (18). The protein encoded by ORF 5 was the putative large subunit of phage terminase, a protein that mediates the specific interaction between the prohead of the virus and its DNA. Sequence similarity comparisons for ORF 5 of the MG1363-inv2 region were based on the in-frame fusion of the two parts of ORF 5, after removal of the nucleotide sequence corresponding to the entire IS905 element, including one of the direct repeats.
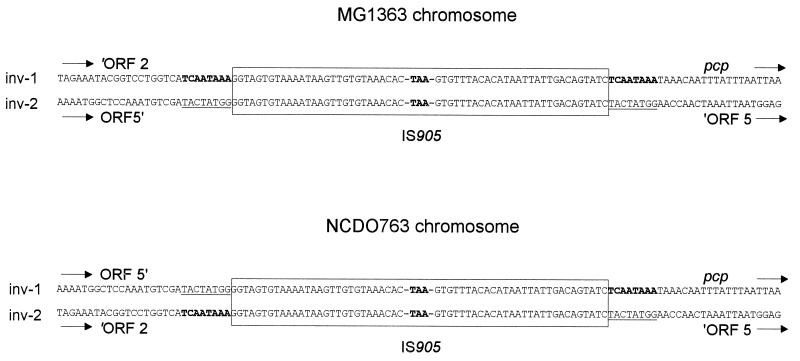
The sequences upstream from the IS905 element of MG1363-inv1 and NCDO763-inv2 regions were identical (Fig. 5). Sequence identity was also observed for regions downstream from the IS905 elements of MG1363-inv1 and NCDO763-inv1. No nucleotide divergence other than the reciprocal exchange downstream from the IS elements was detected between MG1363 and NCDO763 chromosomes. Therefore, the large chromosomal inversion occurred by homologous recombination between the two inversely oriented copies of IS905.
FIG. 5.
Nucleotide sequences of part of the IS905 elements and their flanking regions for the two chromosomes. IS905 is boxed, and only the sequences of both 28-bp indirect repeats and the stop codon generated by the C→T mutation are shown. Direct repeats resulting from the IS905 insertion are indicated in bold in the MG1363-inv1 sequence and underlined in the MG1363-inv2 sequence.
DISCUSSION
This study has established for the first time that a spontaneous chromosomal inversion can be mediated by homologous recombination between two inversely oriented copies of an IS element in gram-positive bacteria. This genetic event led to a large inversion of half of the chromosome, including the six rrn operons. An inversion involving the same chromosomal regions was detected when the chromosome maps of two lactococcal strains of different subspecies were compared (31). As the putative replication origin (oriC) of the lactococcal chromosome was recently located near the rrnA operon (60), these two inversions did not affect the distance of genes from oriC. It is noteworthy that most naturally occurring inversions include either the origin or the terminus of replication (terC) and do not disrupt the oriC-terC symmetry. It was recently showed that chromosome length variations of natural isolates of E. coli are distributed symmetrically with respect to the replication origin and terminus (2). Moreover, inversions restoring the chromosomal balance between oriC and terC have been found in S. typhi and S. paratyphi A (35, 36). Altogether, these observations are consistent with there being selective forces maintaining chromosome symmetry (17, 48), with only inversions that do not change the diametrically opposed locations of oriC and terC, or that restore this symmetry following other rearrangements, being selectively fixed in natural conditions.
Any genetic event, including transposition, site-specific recombination, double-strand break repair, and homologous recombination, may account for the occurrence of large chromosomal inversions in natural conditions. However, inversion junctions have been identified in only a few cases, and so it is not known whether a particular type of genetic event is preferentially associated with inversions. Nucleotide sequence analysis of the four chromosomal regions of strains MG1363 and NCDO763 explained the mechanism that generated the inversion in these strains. In contrast to the E. coli 1485IN inversion, in which recombination between the two IS3 elements was accompanied by a deletion of more than 50 kb (27) and probably associated with an IS3 transposition event, inversion between strains MG1363 and NCDO763 required only a single homologous recombination event between the two copies of the IS905 element.
Transposition of the IS905 element generates an 8-bp duplication at the target site (14). Sequence analysis of the chromosomal regions at the vicinity of the IS element revealed the presence of these two direct repeats on either side of the IS905 element in strain MG1363 (5′-TCAATAAA-3′ for the MG1363-inv1 region and 5′-TACTATGG-3′ for the MG1363-inv2 region). Therefore, strain MG1363 has the parental chromosome structure and strain NCDO763 has the genome structure generated by the inversion. The presence of these different direct repeats unambiguously demonstrates that the two copies of IS905 were inserted into the MG1363 chromosome via independent transposition events, despite the C→T transition that removed half of the transposase. As it is highly improbable that the same mutation arose independently in both IS905 copies after transposition, it is likely that (i) translational readthrough could occur at the stop codon (20), generating a low level of functional transposase, (ii) transposition could generate defective copies of the element at relatively high frequency, or (iii) transposase proteins present into the cell could catalyze the transposition of nonfunctional elements. To our knowledge, there is no precedent for any of these patterns of behavior among prokaryotic IS elements. Thus, there may be a new mode of transposition regulation involved here, in addition to those already described (reviewed in reference 1).
The genetic organization of the MG1363 chromosomal sites where the two IS905 elements are inserted is consistent with the direct-selection hypothesis, which postulates that genomic IS elements can occasionally become fixed in a bacterial population through the beneficial mutations they cause (53). Transposition of the IS905 element should not disturb the expression of the surrounding genes in the MG1363-inv1 region, because this element is integrated just downstream from the putative rho-independent transcription terminator of ORF 2, and the presence of a perfect prokaryotic promoter −35 consensus (5′-TTGACA-3′) reading outward 5 bp from the IR-R terminus is likely to create a new hybrid promoter for the pcp gene. Transposition of the IS905 element into the MG1363-inv2 region should confer a selective advantage on the strain, because it disrupts a bacteriophage gene that is essential for the lytic cycle.
The large chromosomal inversion described herein should not have phenotypic effects on the fitness of strain NCDO763, in comparison to strain MG1363, because it did not change the symmetry of the chromosome and therefore should not confer a selective disadvantage for replication of the NCDO763 chromosome. In addition, the inversion did not affect the local genetic environment, such as pcp gene expression in strain NCDO763. However, sensitive competition experiments are required to confirm that the inversion is neutral for cell fitness.
ACKNOWLEDGMENTS
We thank M. Chandler and J. Casadesus for helpful discussions and A. Edelman for reading the manuscript.
This work was supported by grants from the Centre National de la Recherche Scientifique (UPR9007), from the Région Midi-Pyrénées (RECH 9609795), and from the UE-BIOTECH Programme (CT 96-0498). N. Campo was supported by a fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche.
REFERENCES
- 1.Arber W. The generation of variation in bacterial genomes. J Mol Evol. 1995;40:7–12. [Google Scholar]
- 2.Bergthorson U, Ochman H. Distribution of chromosome length variation in natural isolates of Escherichia coli. Mol Biol Evol. 1998;15:6–16. doi: 10.1093/oxfordjournals.molbev.a025847. [DOI] [PubMed] [Google Scholar]
- 3.Birkenbihl R P, Vielmetter W. Cosmid-derived map of E. coli strain BHB2600 in comparison to the map of strain W3110. Nucleic Acids Res. 1989;17:5057–5069. doi: 10.1093/nar/17.13.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canard B, Saint-Joanis B, Cole S T. Genomic diversity and organization of virulence genes in the pathogenic anaerobe Clostridium perfringens. Mol Microbiol. 1992;6:1421–1429. doi: 10.1111/j.1365-2958.1992.tb00862.x. [DOI] [PubMed] [Google Scholar]
- 5.Carlson C R, Grønstad A, Kolstø A B. Physical maps of the genomes of three Bacillus cereus strains. J Bacteriol. 1992;174:3750–3756. doi: 10.1128/jb.174.11.3750-3756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleuziat P, Awadé A C, Robert-Baudouy J. Molecular characterization of pcp, the structural gene encoding the pyrrolidone carboxylyl peptidase from Streptococcus pyogenes. Mol Microbiol. 1992;6:2051–2063. doi: 10.1111/j.1365-2958.1992.tb01378.x. [DOI] [PubMed] [Google Scholar]
- 7.Cole S T, Saint Girons I. Bacterial genomics. FEMS Microbiol Rev. 1994;14:139–160. doi: 10.1111/j.1574-6976.1994.tb00084.x. [DOI] [PubMed] [Google Scholar]
- 8.Corpet F, Gouzy J, Kahn D. The ProDom database of protein domain families. Nucleic Acids Res. 1998;26:323–326. doi: 10.1093/nar/26.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson B E, Kordias N, Dobos M, Hillier A J. Genomic organization of lactic acid bacteria. In: Venema G, Huis in’t Veld J H J, Hugenholtz J, editors. Lactic acid bacteria: genetics, metabolism and applications. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 65–87. [Google Scholar]
- 10.Davies F L, Underwood H M, Gasson M J. The value of plasmid profile for strain identification in lactic streptococci and the relationship between Streptococcus lactis 712, ML3 and C2. J Appl Bacteriol. 1981;51:325–337. [Google Scholar]
- 11.Delorme C, Godon J J, Ehrlich S D, Renault P. Mosaic structure of large regions of the Lactococcus lactis subsp. cremoris chromosome. Microbiology. 1994;140:3053–3060. doi: 10.1099/13500872-140-11-3053. [DOI] [PubMed] [Google Scholar]
- 12.Dempsey J A F, Wallace A B, Cannon J G. The physical map of the chromosome of a serogroup A strain of Neisseria meningitidis shows complex rearrangements relative to the chromosome of the two mapped strains of the closely related species N. gonorrhoeae. J Bacteriol. 1995;177:6390–6400. doi: 10.1128/jb.177.22.6390-6400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Roissart H, Luquet F M. Bactéries lactiques: aspects fondamentaux et technologiques. Uriage, France: Lorica; 1994. [Google Scholar]
- 14.Dodd H M, Horn N, Gasson M J. Characterization of IS905, a new multicopy insertion sequence identified in lactococci. J Bacteriol. 1994;176:3393–3396. doi: 10.1128/jb.176.11.3393-3396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donkersloot J A, Thompson J. Cloning, expression, sequence analysis, and site-directed mutagenesis of the Tn5306-encoded N5-(carboxyethyl)ornithine synthase from Lactococcus lactis K1. J Biol Chem. 1995;270:12226–12234. doi: 10.1074/jbc.270.20.12226. [DOI] [PubMed] [Google Scholar]
- 16.Fonstein M, Haselkorn R. Physical mapping of bacterial genomes. J Bacteriol. 1995;177:3361–3369. doi: 10.1128/jb.177.12.3361-3369.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.François V, Louarn J, Rebollo J E, Louarn J M. Replication termination, nondivisible zones, and structure of the Escherichia coli chromosome. In: Drlica K, Riley M, editors. The bacterial chromosome. Washington, D.C: American Society for Microbiology; 1990. pp. 351–359. [Google Scholar]
- 18.Garcia P, Alonso J C, Suarez J E. Molecular analysis of the cos region of the Lactobacillus casei bacteriophage A2 gene product 3, gp3, specifically binds to its downstream cos region. Mol Microbiol. 1997;23:505–514. doi: 10.1046/j.1365-2958.1997.d01-1863.x. [DOI] [PubMed] [Google Scholar]
- 19.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gesteland R F, Atkins J F. Recoding: dynamic reprogramming of translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 21.Gibbs C P, Meyer T F. Genome plasticity in Neisseria gonorrhoeae. FEMS Microbiol Lett. 1996;145:173–179. doi: 10.1111/j.1574-6968.1996.tb08574.x. [DOI] [PubMed] [Google Scholar]
- 22.Ginard M, Lalucat J, Tümmler B, Römling U. Genome organization of Pseudomonas stutzeri and resulting taxonomic and evolutionary considerations. Int J Syst Bacteriol. 1997;47:132–143. doi: 10.1099/00207713-47-1-132. [DOI] [PubMed] [Google Scholar]
- 23.Hackett N R, Bobovnikova Y, Heyrovska N. Conservation of chromosomal arrangement among three strains of the genetically unstable archaeon Halobacterium salinarium. J Bacteriol. 1994;176:7711–7718. doi: 10.1128/jb.176.24.7711-7718.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill C W, Harnish B W. Inversions between ribosomal RNA genes of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:7069–7072. doi: 10.1073/pnas.78.11.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarvis E D, Chen S, Rudner R. Genetic structure and DNA sequences at junctions involved in the rearrangements of Bacillus subtilis strains carrying the trpE26 mutation. Genetics. 1990;126:785–797. doi: 10.1093/genetics/126.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Q J, Hiratsuka K, Taylor D E. Variability of gene order in different Helicobacter pylori strains contributes to genome diversity. Mol Microbiol. 1996;20:833–842. doi: 10.1111/j.1365-2958.1996.tb02521.x. [DOI] [PubMed] [Google Scholar]
- 27.Komoda Y, Enomoto M, Tominaga A. Large inversion in Escherichia coli K-12 1485IN between inversely oriented IS3 elements near lac and cdd. Genetics. 1991;129:639–645. doi: 10.1093/genetics/129.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ladefoged S A, Christiansen G. Physical and genetic mapping of the genomes of five Mycoplasma hominis strains by pulsed-field gel electrophoresis. J Bacteriol. 1992;174:2199–2207. doi: 10.1128/jb.174.7.2199-2207.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Bourgeois P, Lautier M, Mata M, Ritzenthaler P. New tools for the physical and genetic mapping of Lactococcus strains. Gene. 1992;111:109–114. doi: 10.1016/0378-1119(92)90610-2. [DOI] [PubMed] [Google Scholar]
- 30.Le Bourgeois P, Lautier M, Mata M, Ritzenthaler P. Physical and genetic map of the chromosome of Lactococcus lactis subsp. lactis IL1403. J Bacteriol. 1992;174:6752–6762. doi: 10.1128/jb.174.21.6752-6762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Bourgeois P, Lautier M, van den Berghe L, Gasson M J, Ritzenthaler P. Physical and genetic map of the Lactococcus lactis subsp. cremoris MG1363 chromosome: comparison with that of Lactococcus lactis subsp. lactis IL1403 reveals a large genome inversion. J Bacteriol. 1995;177:2840–2850. doi: 10.1128/jb.177.10.2840-2850.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Bourgeois P, Mata M, Ritzenthaler P. Genome comparison of Lactococcus strains by pulsed-field gel electrophoresis. FEMS Microbiol Lett. 1989;59:65–70. doi: 10.1016/0378-1097(89)90460-6. [DOI] [PubMed] [Google Scholar]
- 33.Liu S L, Hessel A, Sanderson K E. The XbaI-Bln I-CeuI genomic cleavage map of Salmonella enteritidis shows an inversion relative to Salmonella typhimurium LT2. Mol Microbiol. 1993;10:655–664. doi: 10.1111/j.1365-2958.1993.tb00937.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu S L, Sanderson K E. I-CeuI reveals conservation of the genome of independent strains of Salmonella typhimurium. J Bacteriol. 1995;177:3355–3357. doi: 10.1128/jb.177.11.3355-3357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu S L, Sanderson K E. Rearrangements in the genome of the bacterium Salmonella typhi. Proc Natl Acad Sci USA. 1995;92:1018–1022. doi: 10.1073/pnas.92.4.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu S L, Sanderson K E. The chromosome of Salmonella paratyphi A is inverted by recombination between rrnH and rrnG. J Bacteriol. 1995;177:6585–6592. doi: 10.1128/jb.177.22.6585-6592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S L, Sanderson K E. Highly plastic chromosomal organization in Salmonella typhi. Proc Natl Acad Sci USA. 1996;93:10303–10308. doi: 10.1073/pnas.93.19.10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Garcia P, St Jean A, Amils R, Charlebois R L. Genomic stability in the archaeae Haloferax volcanii and Haloferax mediterranei. J Bacteriol. 1995;177:1405–1408. doi: 10.1128/jb.177.5.1405-1408.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Louarn J M, Bouché J P, Legendre F, Louarn J, Patte J. Characterization and properties of very large inversions of the E. coli chromosome along the origin-to-terminus axis. Mol Gen Genet. 1985;201:467–476. doi: 10.1007/BF00331341. [DOI] [PubMed] [Google Scholar]
- 40.Marshall P, Lemieux C. Cleavage pattern of the homing endonuclease encoded by the fifth intron in the chloroplast subunit rRNA-encoding gene of Chlamydomonas eugametos. Gene. 1991;104:1241–1245. doi: 10.1016/0378-1119(91)90256-b. [DOI] [PubMed] [Google Scholar]
- 41.Michaux-Charachon S, Bourg G, Jumas-Bilak E, Guigue-Talet P, Allardet-Servent A, O’Callaghan D, Ramuz M. Genome structure and phylogeny in the genus Brucella. J Bacteriol. 1997;179:3244–3249. doi: 10.1128/jb.179.10.3244-3249.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulczyk M, Szewczuk A. Pyrrolidonyl peptidase in bacteria: a new colorimetric test for differentiation of Enterobacteriaceae. J Gen Microbiol. 1970;61:9–13. doi: 10.1099/00221287-61-1-9. [DOI] [PubMed] [Google Scholar]
- 43.Mulczyk M, Szewczuk A. Pyrrolidonyl peptidase activity: a simple test for differentiating staphylococci. J Gen Microbiol. 1972;70:383–384. doi: 10.1099/00221287-70-2-383. [DOI] [PubMed] [Google Scholar]
- 44.Nikolskaya T, Fonstein M, Haselkorn R. Alignment of a 1.2 Mb chromosomal region from three strains of Rhodobacter capsulatus reveals a significantly mosaic structure. Proc Natl Acad Sci USA. 1995;92:10609–10613. doi: 10.1073/pnas.92.23.10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perkins J D, Don Heath J, Sharma B R, Weinstock G M. XbaI and BlnI genomic cleavage maps of Escherichia coli K-12 strain MG1655 and comparative analysis of other strains. J Mol Biol. 1993;232:419–445. doi: 10.1006/jmbi.1993.1401. [DOI] [PubMed] [Google Scholar]
- 47.Riley M, Sanderson K E. Comparative genetics of Escherichia coli and Salmonella typhimurium. In: Drlica K, Riley M, editors. The bacterial chromosome. Washington, D.C: American Society for Microbiology; 1990. pp. 85–95. [Google Scholar]
- 48.Roth J R, Benson N, Galitsky T, Haack K, Lawrence J G, Miesel L. Rearrangements of the bacterial chromosome: formation and applications. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Resnikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 2256–2276. [Google Scholar]
- 49.Römling U, Greipel J, Tümmler B. Gradient of genomic diversity in the Pseudomonas aeruginosa chromosome. Mol Microbiol. 1995;17:323–332. doi: 10.1111/j.1365-2958.1995.mmi_17020323.x. [DOI] [PubMed] [Google Scholar]
- 50.Römling U, Schmidt K D, Tümmler B. Large genome rearrangements discovered by the detailed analysis of 21 Pseudomonas aeruginosa clone C isolates found in environment and disease habitats. J Mol Biol. 1997;271:386–404. doi: 10.1006/jmbi.1997.1186. [DOI] [PubMed] [Google Scholar]
- 51.Sakanyan V, Desmarez L, Legrain C, Charlier D, Mett I, Kochikyan A, Savchenko A, Boyen A, Falmagne P, Pirard A, Glansdorff N. Gene cloning, sequence analysis, purification, and characterization of a thermostable aminoacylase from Bacillus stearothermophilus. Appl Environ Microbiol. 1993;59:3878–3888. doi: 10.1128/aem.59.11.3878-3888.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stibitz S, Yang M S. Genomic fluidity of Bordetella pertussis assessed by a new method for chromosomal mapping. J Bacteriol. 1997;179:5820–5826. doi: 10.1128/jb.179.18.5820-5826.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Syvanen M. Insertion sequences and their evolutionary role. In: de Bruijn F J, Lupski J R, Weinstock G M, editors. Bacterial genomes: physical structure and analysis. New York, N.Y: Chapman & Hall; 1997. pp. 213–220. [Google Scholar]
- 54.Tabata K, Hoshino T. Mapping of 61 genes on the refined physical map of the chromosome of Thermus thermophilus HB27 and comparison of genome organization with that of T. thermophilus HB8. Microbiology. 1996;142:401–410. doi: 10.1099/13500872-142-2-401. [DOI] [PubMed] [Google Scholar]
- 55.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tigges E, Minion F C. Physical map of Mycoplasma gallisepticum. J Bacteriol. 1994;176:4157–4159. doi: 10.1128/jb.176.13.4157-4159.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toda T, Itaya M. I-CeuI recognition sites in the rrn operons of the Bacillus subtilis 168 chromosome: inherent landmark for genome analysis. Microbiology. 1995;141:1937–1945. doi: 10.1099/13500872-141-8-1937. [DOI] [PubMed] [Google Scholar]
- 58.Tulloch D L, Finch L R, Hillier A J, Davidson B E. Physical map of the chromosome of Lactococcus lactis subsp. lactis DL11 and localization of six putative rRNA operons. J Bacteriol. 1991;173:2768–2775. doi: 10.1128/jb.173.9.2768-2775.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waterbury P G, Lane M J. Generation of lambda concatemers for use as pulsed-field electrophoresis size markers. Nucleic Acids Res. 1987;15:3930. doi: 10.1093/nar/15.9.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wind A, Hansen E B. Molecular mechanisms in DNA replication and recombination 1996. Silverthorne, Colo: Keystone Symposia; 1996. Cloning and characterisation of the dnaA region in Lactococcus lactis, abstr. 276; p. 65. [Google Scholar]
- 61.Zuerner R L, Herrmann J L, Saint Girons I. Comparison of genetic maps for two Leptospira interrogans serovars provides evidence for two chromosomes and intraspecies heterogeneity. J Bacteriol. 1993;175:5445–5451. doi: 10.1128/jb.175.17.5445-5451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]