Abstract
A salt-sensitive mutant of Synechocystis sp. strain PCC 6803 defective in the synthesis of the compatible solute glucosylglycerol (GG) was used to search for the gene encoding GG-phosphate synthase (GGPS), the key enzyme in GG synthesis. Cloning and sequencing of the mutated region and the corresponding wild-type region revealed that a deletion of about 13 kb occurred in the genome of mutant 11. This deletion affected at least 10 open reading frames, among them regions coding for proteins showing similarities to trehalose (otsA homolog)- and glycerol-3-phosphate-synthesizing enzymes. After construction and characterization of mutants defective in these genes, it became obvious that an otsA homolog (sll1566) (T. Kaneko et al., DNA Res. 3:109–136, 1996) encodes GGPS, since only the mutant affected in sll1566 showed salt sensitivity combined with a complete absence of GG accumulation. Furthermore, the overexpression of sll1566 in Escherichia coli led to the appearance of GGPS activity in the heterologous host. The overexpressed protein did not show the salt dependence that is characteristic for the GGPS in crude protein extracts of Synechocystis.
The accumulation of compatible solutes represents an important aspect of cellular adaptation to high-salt or high-osmotic-pressure environments. Salt-treated cells are able to accumulate these low-molecular-mass substances in high amounts, since they are hydrophilic, show no net charge, and are compatible with cellular metabolism. Osmoprotective compounds fulfil two different functions: (i) decrease of intracellular water potential to prevent water loss, and (ii) prevention of denaturation of macromolecules in a cellular environment of changed ionic composition and decreased water potential (10). Based on the principal osmoprotective compound accumulated, three salt-tolerant groups of cyanobacteria have been distinguished (25). Strains with lower salt tolerance accumulate the disaccharide sucrose or trehalose, strains with moderate halotolerance synthesize the heteroside glucosylglycerol (GG), and halophilic strains contain the quaternary ammonium compounds glycinebetaine or glutamatebetaine.
The accumulation of GG was characterized by using the moderately halotolerant strain Synechocystis sp. strain PCC 6803. Synechocystis synthesizes mainly GG and traces of sucrose, which enable it to tolerate up to 1.2 M NaCl (24). The biosynthetic pathway begins with ADP-glucose and glycerol-3-phosphate (G3P), which are used by the GG-phosphate synthase (GGPS), and proceeds via the intermediate GG-phosphate (GGP), which is dephosphorylated to GG by the GG-phosphate phosphatase (GGPP) (12). The enzyme activities were found to depend on enhanced salt concentrations in the assays. Furthermore, the GGPS and GGPP activities seem to be reversibly affected by a salt-regulated process. These enzymes could be activated in cells grown in low-salt medium simply by adding salt during the protein extraction and, conversely, could be inhibited in extracts from cells adapted to high-salt medium by omitting salt from the homogenization and assay buffers (12). Beside NaCl, other salts were also found to be effective with regards to the activation of GG-synthesizing enzymes in Synechocystis extracts (15).
Genes encoding proteins involved in cyanobacterial salt adaptation were selected by using the enhanced content of their mRNAs in salt-treated cells (4) or by characterization of salt-sensitive mutants obtained after random cartridge mutagenesis (11). By use of the second strategy, 18 mutants of Synechocystis that were unable to tolerate salt concentrations higher than 550 mM NaCl, which is less than 50% of the salt resistance limit of the wild type (WT), were selected. The salt-sensitive mutants could be divided into two groups: (i) nine mutants that were affected in the synthesis of GG; (ii) nine mutants that showed no change in GG synthesis. The GG-defective mutants showed a larger decrease in their remaining salt tolerance than mutants of the second group. After physiological and genetic characterization of one mutant, which accumulates mainly the intermediate GGP (13), stpA could be identified as the GGPP-encoding gene. In addition to the direct regulation of GGPP enzyme activity by salt, it could be shown that the expression of the stpA gene is also salt stimulated (14).
In the present study, we describe the gene encoding GGPS, the key enzyme in GG synthesis. After cloning and partial sequencing of the mutated site from a GG-defective mutant, it was possible to define the affected region on the chromosome of Synechocystis, since its genome has recently been completely sequenced (17). Open reading frames (ORFs) located in the mutated region were separately mutated, and the mutants were screened for changes in salt tolerance and GG synthesis. By use of this strategy, an otsA homolog could be identified as the GGPS-encoding gene. After overexpression of this gene, GGPS activity was found in Escherichia coli. We propose the designation ggpS for the gene encoding GGPS.
MATERIALS AND METHODS
Strains and culture conditions.
A derivative of the Synechocystis sp. strain PCC 6803 with enhanced transforming capacity, which was used in all experiments, was obtained from S. Shestakov (Moscow State University, Moscow, Russia). Axenic cells were cultured on plates at 30°C under constant illumination by using the mineral medium C (18). Transformants were initially selected on medium containing 10 μg of kanamycin · ml−1 (Sigma) or 0.5 μg of streptomycin · ml−1, while the segregation of clones and cultivation of mutants was performed at 50 μg of kanamycin · ml−1 or 2 μg of streptomycin · ml−1 (11). For the physiological characterization, axenic cultures of the cyanobacteria were grown photoautotrophically in batch cultures as described previously (8). The E. coli strain TG1 was used for routine DNA manipulations (26).
DNA manipulations.
Isolation of total DNA from Synechocystis was done as described previously (4). All other techniques, such as plasmid isolation, transformation of E. coli, ligation, and restriction analysis (restriction enzymes were obtained from New England Biolabs), were standard methods (26). Labeling of DNA probes with digoxigenin for Southern hybridization was done by using the PCR DIG probe synthesis kit (Boehringer Mannheim). Sequencing was performed by the dideoxy chain termination method with [α-35S]dATP (Amersham Buchler) and the Sequenase 2.0 kit (U.S. Biochemicals). Double-stranded plasmid DNA was isolated by using the QIAprep plasmid kit (Qiagen). The following synthetic primers were specifically used for sequencing the regions flanking the aphII gene: CAGGCCTGGTATGAGTCAGC (Kan5′); ATTTTTATCTTGTGCAATGT (Kan3′) (custom oligonucleotide synthesis by Pharmacia). Computer analysis of DNA and protein sequences was done by using DNASIS/PROSIS and Blast (2) software packages. The plasmid vector pGEM7 (Promega) was used to clone the ORFs of interest after PCR amplification of DNA fragments. The following primers, used for amplification of selected genes, were designed by using the complete genome sequence from Synechocystis sp. strain PCC 6803 (17) and contain added restriction sites (underlined) in a 5′ extension: primer ggpS5, CGGGATCCCGAGGGGGGCAAAAACAATGTCAA; ggpS3, CGGGATCCCTACATTTGGGGGGGCTGTCCC; glpD5, AAAAGAATTCATGCGTAATTTCCCAGAA; glpD3, CTGATACTCGAGTCAGTGGAGACAATAGTC; glpK5, AAAGGATCCATGACAGCAAAACATAAT; glpK3, AAAAGAATTCTCACTGGTCAACGGATAC. Usually, the primers are complementary to the sequences behind and before the start and stop codons (shown in bold letters), respectively, of the corresponding genes. The primer ggpS5 anneals 714 bp upstream of the start codon of the ggpS gene, and the primer ggpSE3′ binds 73 bp downstream of the ggpS gene.
Generation of insertion mutants.
For the generation of mutants in specific ORFs, the aphII gene (conferring kanamycin resistance) from pUC4K (30) or the smr gene cartridge (conferring streptomycin resistance) from pBSL130 (1) was integrated at selected unique restriction sites into the ORFs cloned into E. coli vectors (Table 1). Plasmid DNA of these constructs was isolated from E. coli by using the QIAprep spin plasmid mini kit (Qiagen). About 1 μg of DNA was used for transformation of Synechocystis, and antibiotic-resistant (Kmr Smr) clones were selected (11).
TABLE 1.
Plasmids and Synechocystis sp. strain PCC 6803 mutants used and constructed in this study (see Fig. 1)
| Plasmid or mutant | Descriptiona |
|---|---|
| Plasmids | |
| pGG | pGEM7 containing ggpS (sll1566) as a 2.1-kb BamHI fragment obtained by PCR with ggpS5 and ggpS3 primers; final size, 5.2 kb |
| pGΔG::Km2 | pGG containing an inactivated ggpS gene (aphII gene is inserted in the SnaBI/StuI deletion in the opposite direction of ggpS); final size, 6.1 kb |
| pGΔG::Sm2 | pGG containing an inactivated ggpS gene (smr gene is inserted in the SnaBI/StuI deletion in the opposite direction of ggpS); final size, 6.7 kb |
| pGD | pGEM7 containing glpD (sll1085) as a 1.7-kb EcoRI/XhoI fragment obtained by PCR with glpD5 and glpD3 primers; final size, 4.7 kb |
| pGD::Km2 | pGD containing an inactivated glpD gene (aphII gene is inserted at the unique BlpI site in the opposite direction of glpD); final size, 5.9 kb |
| pGD::Sm2 | pGD containing an inactivated glpD gene (smr gene is inserted at the unique BlpI site in the opposite direction of glpD); final size, 6.7 kb |
| pGK | pGEM7 containing glpK (slr1672) as a 1.5-kb EcoRI/BamHI fragment obtained by PCR with glpK5 and glpK3 primers; final size, 4.5 kb |
| pGK::Km2 | pGK containing an inactivated glpK gene (aphII gene is inserted at the BstEII deletion in the opposite direction of glpK); final size, 5.6 kb |
| pBADGGPS | pBAD/HisB containing ggpS (sll1566) as a 1.6-kb BglII/KpnI fragment obtained by PCR with ggpSE5′ and ggpSE3′ used for ggpS overexpression in E. coli LMG194; final size, 5.7 kb |
| Mutants | |
| ΔGK2 | Salt-sensitive Synechocystis mutant impaired in ggpS (sll1566) obtained after transformation of the WT with pGΔG::Km2 |
| ΔGS2 | Salt-sensitive Synechocystis mutant impaired in ggpS (sll1566) obtained after transformation of the WT with pGΔG::Sm2 |
| DK2 | Salt-tolerant Synechocystis mutant impaired in glpD (sll1085) obtained after transformation of the WT with pGD::Km2 |
| ΔKK2 | Salt-tolerant Synechocystis mutant impaired in glpK (slr1672) obtained after transformation of the WT with pGK::Km2 |
| DS2ΔKK2 | Salt-tolerant Synechocystis mutant impaired in glpD (sll1085) and glpK (slr1672) obtained after transformation of ΔKK2 with pGD::Sm2 |
Salt tolerance of the mutants was tested by the ability to grow in 0.55 M NaCl or to lyse at this salt concentration.
Protein overexpression.
For overexpression and purification of the GGPS, the pBAD Expression System (Invitrogen), which uses the tightly controlled promoter of the arabinose operon from E. coli, was used. The ggpS ORF was amplified from Synechocystis chromosomal DNA by PCR with the Elongase-Enzyme-Mix (GIBCO, Life Technologies) and the following primers: 5′-GGAAGATCTATGAATTCATCCCTTGTGATC-3′ (ggpSE5′; BglII site underlined, start codon in bold letters) and 5′-CGGGGTACCCCT/AAACTCTAACTTTGG-3′ (ggpSE3′; KpnI site underlined) (custom oligonucleotide synthesis by Pharmacia). The translational start codon immediately behind a BglII site was used to clone the fragment (BglII and KpnI) in frame with the N-terminal polyhistidine tag into pBAD/HisB (Invitrogen). For overexpression, the recombinant plasmid carrying the ggpS gene (pBADGGPS) was transferred into E. coli LMG194 (Invitrogen). The cells were cultured at 37°C in LB medium until the suspension reached an A600 of 0.5. The expression of the protein was induced by the addition of arabinose (0.0002%), and the suspension was incubated for 4 h. The proteins were extracted from E. coli by sonication with homogenization buffer (20 mM Na phosphate buffer, 500 mM NaCl [pH 7.8]). The extract was applied to a nickel chelate column (ProBond; Invitrogen), where the fusion protein was bound. The GGPS protein was eluted from the matrix by passing homogenization buffer with an increasing concentration of imidazole through the column (wash buffer containing 75 mM imidazole; elution buffer containing 300 mM imidazole). To reduce the salt concentration, the eluted protein was dialyzed against 20 mM Tris-maleate buffer (pH 7.8), containing 5 mM MgCl2. The proteins were electrophoretically separated by using 12% polyacrylamide gels (14) or used for enzyme assays.
Physiological characterization.
The content of low-molecular-mass carbohydrates was analyzed by high-performance liquid chromatography (27). The activities of GGPS and GGPP were determined in vitro by using the 14C-labeled substrate G3P (Amersham Buchler) and buffers containing no or enhanced NaCl contents (0 or 324 mM NaCl added). The reaction products were separated by thin-layer chromatography (TLC). The in vitro determination of these enzyme activities has been described in detail previously (12). To complete dephosphorylation of GGP in measurements of the GGPS isolated from E. coli after overexpression of the ggpS gene, 1 U of alkaline phosphatase (Boehringer Mannheim) was directly added to the assay mixtures for an additional 30 min at 30°C. Quantification of radioactive spots was done with a phosphoimager (BAS 1000; Fuji). Photosynthetic oxygen evolution was measured with a Clark-type electrode (8). Growth and cell density were monitored by determining the absorbance of diluted cyanobacterial suspensions at 750 nm (A750) with a spectrophotometer (U2000; Hitachi).
RESULTS
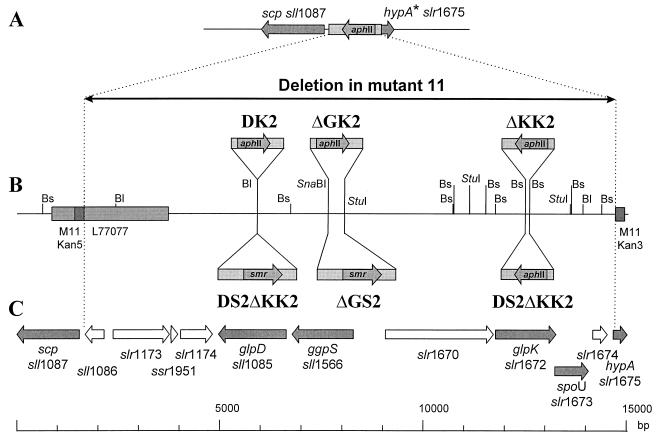
The salt-sensitive mutant 11 was generated by random integration of an aphII gene cartridge and found to be completely defective in GG synthesis (11). To characterize the genetic basis for this defect, the mutated site was cloned (for details of the strategy, see reference 13). The cyanobacterial DNA flanking the 5′ end of the aphII gene cartridge was partly sequenced (sequence M11Kan5) (data not shown) and used as a probe for screening a Synechocystis gene library to obtain the corresponding WT region. From the selected phage clone, a 4.5-kb SalI fragment could be cloned and partly sequenced (the sequence has been deposited in GenBank under accession no. L77077 [hereafter called sequence L77077]). After comparison with the complete genome sequence of Synechocystis (17), it was found that the sequence L77077 corresponds to the region between bp 1941414 and bp 1944348. This sequence contains three ORFs (sll1087, sll1086, slr1173; designation in accordance with work of Kaneko et al. [17]) which were subsequently mutated by integration of an aphII gene. However, none of these single mutants showed any alteration in salt adaptation, indicating that the gene affected in mutant 11 leading to impaired GG synthesis is not harbored on the sequenced fragment (data not shown).
The cyanobacterial DNA fragment flanking the aphII cartridge at the 3′ end in mutant 11 did not hybridize with DNA from the phage clone isolated by using the DNA fragment neighboring the 5′ end. Additionally, the 200 bp of the estimated sequence of the 3′ flanking fragment (sequence M11Kan3) (data not shown) exhibited no similarities to sequence L77077. These were the first indications that, during the integration of the aphII gene into the genome of mutant 11, a large deletion had occurred. Such deletions have already been reported from studies using this mutagenesis strategy for Synechocystis (7, 13). The exact size of the deletion could be estimated by comparison with the complete genome sequence (17). Sequence M11Kan3 was identical to the Synechocystis genome region between bp 1955270 and bp 1955316. Combined with the assigned region for sequence L77077, these comparisons clearly show that in mutant 11, about 13.0 kb of the WT genome, containing at least 10 ORFs, is missing (Fig. 1).
FIG. 1.
Schematic drawing showing the genetic organization, restriction map, and protein-encoding regions of the chromosomal site affected in Synechocystis mutant 11 (A), the corresponding site of the WT of Synechocystis (C) (17), and the insertion sites of the aphII and smr genes in selected sites of the different genes to obtain directed mutants of different ORFs (B). Shaded arrows represent genes encoding putative proteins in Synechocystis; empty arrows represent genes encoding hypothetical proteins in Synechocystis; small arrows labeled with aphII represent inserted aphII gene cassettes; small arrows labeled with smr represent inserted smr gene cassettes; shaded boxes represent regions sequenced in this work and sequences L77077, M11Kan5, and M11Kan3; asterisks indicate partial deleted genes; a long horizontal black arrow represents the deletion that occurred during integration of an aphII gene in mutant 11. Bl, BlpI; Bs, BstEII.
To clarify whether any of these ORFs plays a role in GG synthesis, mutants impaired in only one or two of them, which are most probably responsible for the salt-sensitive phenotype, were constructed (Fig. 1; Table 1). After amplification and cloning of DNA fragments corresponding to single ORFs, antibiotic resistance gene cartridges conferring resistance to kanamycin or streptomycin were introduced into their sequences, leading to genes inactivated by insertion or partial deletion. Constructs showing inserted aphII and smr gene cartridges in an orientation opposite to the transcription direction of the affected genes (Fig. 1) were selected and transformed into Synechocystis WT cells. Since these plasmids do not harbor an origin of replication for Synechocystis, stable antibiotic-resistant clones could be obtained only after exchange of the mutated gene copy with the WT copy on the chromosome by homologous recombination. By using this strategy, single mutants affected in sll1085, sll1566, and slr1672, probably coding for a glycerol-3-phosphate dehydrogenase (GlpD; 51% amino acid identity with human GlpD [19]), a trehalose-phosphate synthase (OtsA) (for similarities, see Table 2), and a glycerol kinase (GlpK; 61% amino acid identity with GlpK from E. coli [22]), respectively, and a double mutant affected in both sll1085 and slr1672 were obtained (Table 1; Fig. 1).
TABLE 2.
Results of amino acid sequence comparisons of Synechocystis protein Sll1566 and several trehalose-phosphate synthasesa
| Gene | Accession no. | Organism | Sequence
|
Refer- ence | |
|---|---|---|---|---|---|
| Total no. of aa | % Iden- tical aa | ||||
| otsA | Q50167 | Mycobacterium leprae | 498 | 48 | 28 |
| otsA | O06353 | Mycobacterium tuberculosis | 500 | 47 | 23 |
| tpsB | Q00217 | Aspergillus niger | 480 | 46 | 31 |
| tpsA | Q00075 | Aspergillus niger | 517 | 44 | 31 |
| tps | Q92410 | Candida albicans | 478 | 44 | 32 |
| tps1 | Q00764 | Saccharomyces cerevisiae | 495 | 44 | 5 |
| ggs1 | Q07158 | Kluyveromyces lactis | 488 | 43 | 20 |
| otsA | P31677 | Escherichia coli | 473 | 42 | 16 |
| tps1 | P40387 | Schizosaccharomyces pombe | 479 | 42 | 6 |
| otsA | P55612 | Rhizobium sp. | 464 | 39 | 9 |
Amino acid sequence comparisons were performed by using BLAST (2). The sequence of Synechocystis protein Sll1566 (GGPS or OtsA) which is 499 amino acids (aa) in length, was compared with those of several trehalose-phosphate synthases.
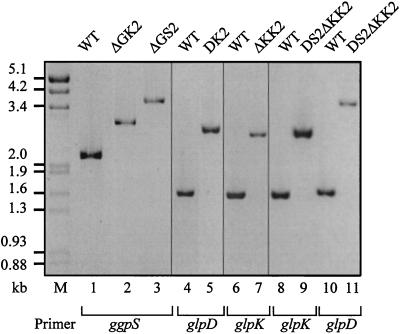
After cultivation for several generations on increasing concentrations of the appropriate antibiotic, chromosomal DNA of selected clones was obtained and compared to the WT DNA by PCR and Southern blot analyses. In all cases, when mutant DNA was used, the PCR analyses showed fragments that were larger than the fragments obtained with WT DNA (Fig. 2). The size differences exactly corresponded to the increases expected by insertion of an aphII gene cassette (1.2 kb) or a smr gene cassette (1.8 kb). In the case of the mutants affected in sll1566 (ΔGK2 and ΔGS2), the size increase was 0.3 kb less (Fig. 2), since before insertion of the antibiotic resistance cartridges about 0.3 kb was deleted from this gene by double digestion with StuI/SnaBI (Fig. 1; Table 1). Furthermore, in all PCRs with mutant DNA, the WT fragment was completely absent (Fig. 2). The results of the PCR analyses were confirmed by Southern hybridization experiments (data not shown). Additionally, the integration of the aphII or smr gene cartridge into the genome of the mutants could be verified by using DNA probes specific for these antibiotic resistance genes. Thus, the plasmid constructs used for transformation of Synechocystis to obtain the mutants were correctly integrated by double crossover and the mutants were completely segregated, since no WT copy remained visible.
FIG. 2.
PCR analyses using chromosomal DNA from the WT (lanes 1, 4, 6, 8, and 10) and mutants (ΔGK2 [lane 2], ΔGS2 [lane 3], DK2 [lane 5], ΔKK2 [lane 7], and DS2ΔKK2 [lanes 9 and 11]) (see Table 1) of Synechocystis as a template and primers specific for selected genes ggpS5 and ggpS3 [lanes 1 to 3], glpD5 and glpD3 [lanes 4, 5, 10, and 11], and glpK5 and glpK3 [lanes 6 to 9]) in order to verify the lesions in the chromosomal DNA of the mutants. Lane M, fragment size marker, EcoRI/HindIII-cut λ DNA.
In physiological experiments, the growth of the mutants was compared to that of WT cells under different culture conditions. Under low-salt conditions, no significant differences were observed in cultures on solid or in liquid media. But, differences became obvious after cells were transferred to media containing NaCl concentrations above 500 mM. Under those culture conditions, cells of the mutants impaired in sll1566 (ΔGK2 and ΔGS2) were unable to grow and lysed after a few days, while all mutants impaired in glpD and/or glpK (DK2, ΔKK2, and DS2ΔKK2) were able to adapt to high-salt concentrations (tested up to 684 mM NaCl). The degree of salt sensitivity of mutants ΔGK2 and ΔGS2 resembled the initial salt sensitivity of mutant 11, which was also unable to grow on media with NaCl concentrations above 550 mM (Table 1).
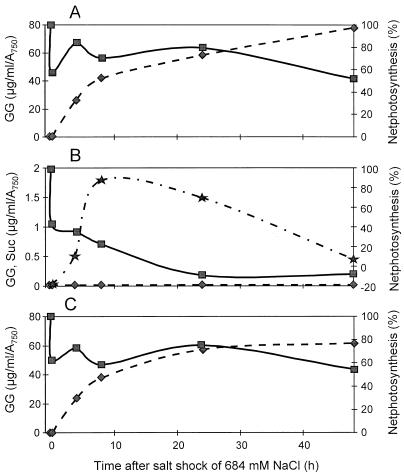
The kinetics of alterations in photosynthesis and accumulation of osmoprotective compounds were investigated after applying a salt shock of 684 mM NaCl to WT and mutant cells (Fig. 3). In all cultures, an immediate inhibition of photosynthesis was observed after salt was added, which is characteristic of salt-shocked cells of Synechocystis (8, 13). In WT cells, as well as mutant cells affected in glpD and/or glpK (only the data for the double mutant DS2ΔKK2 are shown, which are also representative for the corresponding single mutants), photosynthesis recovered after 4 h and became nearly adapted to the high-salt environment after 48 h. On the contrary, net photosynthesis in cells of the ΔGK2 mutant decreased progressively and was completely inhibited after only 24 h (Fig. 3). Nevertheless, the cells of mutant ΔGK2 seemed not to be dead, since they continued to respire for at least 2 additional days.
FIG. 3.
Photosynthetic oxygen evolution (■) and accumulation of GG (⧫) and sucrose (★) after a salt shock of 684 mM NaCl in WT (A), mutant ΔGK2 (B), and mutant DS2ΔKK2 (C) Synechocystis cells. In WT and mutant DS2ΔKK2 cells, only traces of sucrose, which were below the limit for accurate detection by the high-performance liquid chromatography systems, were present.
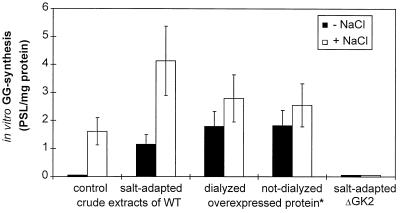
The accumulation of the osmoprotective compound GG started immediately in salt-shocked WT and DS2ΔKK2 mutant cells and reached comparable values 24 to 48 h after the salt shock. But, in salt-treated cells of mutants ΔGK2 and ΔGS2, no traces of GG could be detected. Instead of GG, these cells accumulated increased amounts of sucrose (Fig. 3). In the WT, only traces of sucrose could be detected. Enhanced sucrose accumulation has already been found in cells of another GG-defective mutant of Synechocystis (11). However, compared to the GG content in WT cells, the accumulated amounts of sucrose in the mutants represent less than 10%, which is not sufficient to balance the external salt concentration. Additionally, assays to detect GGPS activity were performed under different test conditions. In crude protein extracts from WT cells, GGPS activity was detected but only in tests where the assay buffer contained 340 mM NaCl. With extracts from the mutants ΔGK2 and ΔGS2, absolutely no GGPS activity was detectable regardless of whether the cells were salt adapted or the assays contained salt (Fig. 4). Therefore, the estimates of GGPS activity confirmed the data on GG accumulation in the cells.
FIG. 4.
Salt dependence of the specific GGPS activities in crude protein extracts from control and salt-adapted cells (342 mM NaCl) of the WT and mutant ΔGK2 of Synechocystis as well as GGPS protein obtained after overexpression in E. coli. The overexpressed GGPS protein was taken directly after purification (buffer containing 20 mM Na phosphate, 500 mM NaCl, and 300 mM imidazole [pH 7.8]) or after extensive dialyzation (buffer containing 20 mM Tris-maleate and 5 mM MgCl2 [pH 7.8]). Enzyme assays were performed in NaCl-free or NaCl-containing (342 mM NaCl) buffer. Radioactivity of the GG spots was determined with a phosphoimager and is expressed as pixel intensities per area (PSL). Means ± standard deviations of values are shown. An asterisk indicates specific activities of the overexpressed protein that were divided by the factor 1,000.
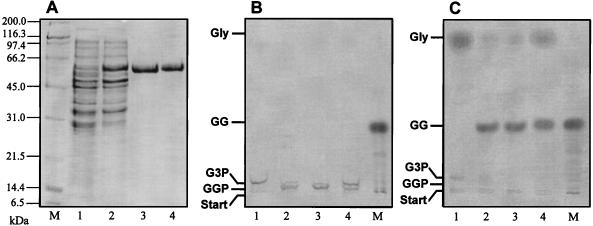
After the sequence encoding Sll1566 was cloned in a plasmid of the pBAD series and expressed in arabinose-treated cells of E. coli LMG194, a protein of about 58 kDa could be purified by using the polyhistidine tag added to the N terminus of the Synechocystis protein (Fig. 5A). This size of the purified protein corresponded very well with the theoretical molecular mass of 56 kDa deduced from the amino acid sequence of Sll1566 (17). Proteins from various steps of the purification were used in GGPS enzyme assays. Crude extracts from E. coli cells containing the plasmid pBADGGPS as well as the purified protein showed the expected enzyme activity, the in vitro formation of GGP from ADP-glucose and G3P, while extracts from cells carrying the control plasmid pBAD/HisB showed no GGP formation (Fig. 5B). The absence of dephosphorylated GG in these assays clearly demonstrates that E. coli does not have GGPP activity and that other phosphatases could not substitute for this activity. After treatment of aliquots of the enzyme assays with alkaline phosphatase, the end product GG was observed (Fig. 5C). The GG spots are somewhat more intensively labeled than the GGP spots, reflecting the better solubility of GG compared to that of GGP in absolute ethanol, which was used to dissolve the dried reaction products to avoid high salt contents in the TLC separations. Enzyme activity with the purified GGPS isolated from E. coli was obtained whether or not the test buffers contained enhanced NaCl concentrations. Furthermore, the dialyzed Sll1566 protein also showed high enzyme activity under low-salt conditions, whereas the enzyme from WT cells was completely inhibited (Fig. 4 and 5). Hence, the activity of the overexpressed protein is independent of the NaCl content in the assays, while the enzyme in crude extracts from Synechocystis needs enhanced salt concentrations for its activity (Fig. 4).
FIG. 5.
Overexpression of GGPS (Sll1566) from Synechocystis in E. coli. (A) Coomassie blue-stained gel from electrophoretic separation of soluble proteins from different fractions of the purification procedure; (B and C) TLC separation of reaction products obtained after determination of GGPS activity by using soluble proteins from different fractions of the purification procedure. The reaction products were directly applied to the TLC plate (B) or pretreated with alkaline phosphatase (C). Lanes: 1, proteins extracted from E. coli harboring the empty pBAD/HisB vector; 2, proteins extracted from E. coli cells harboring the ggpS gene on the pBADGGPS vector; 3, purified GGPS protein with the His tag; 4, purified GGPS protein that was dialyzed overnight against low-salt-containing buffer; M, marker lanes (protein broad-range marker from Bio-Rad [A] and 14C-labeled GG isolated from salt-stressed Synechocystis cells [B and C]).
DISCUSSION
The fragment deleted from the chromosome of mutant 11 contains at least 10 ORFs (Fig. 1). For four ORFs, a putative function can be assigned from sequence comparisons, while the other six ORFs encode hypothetical proteins (17). After reevaluation of the sequence similarities with proteins from the database, at least two different possibilities could explain the defect in GG biosynthesis by the deletion found in mutant 11. First, it is most probable that sll1566, which resembles otsA, encodes GGPS. The amino acid sequence of Sll1566 shows significant homologies to several trehalose-phosphate synthases involved in osmotically induced trehalose synthesis in bacteria and yeasts (Table 2). These enzymes are glucosyltransferases, which are functionally and biochemically closely related to the GGPS. But, in salt-stressed cells of Synechocystis, no trehalose accumulation was found (24, 27), which makes it unlikely that a trehalose-phosphate synthase exists in this strain. Second, it is possible that sll1085 and slr1672 encode enzymes capable of synthesizing G3P (GlpD [glycerol-3-phosphate dehydrogenase] and GlpK [glycerol kinase], respectively) (17), one of the precursors for GG biosynthesis. Their defect could lead to a depletion in G3P, which induces GG deficiency in salt-treated cells.
The experiments using the mutants affected in selected ORFs provided clear evidence that sll1566 encodes GGPS, as was assumed as the first and most likely explanation for the defect found in mutant 11. We propose the designation ggpS for this gene. The GGPS shows high amino acid sequence similarities with trehalose-phosphate synthases from heterotrophic bacteria and yeasts (Table 2). By using these sequence data, an evolutionary relationship of the GG and trehalose-synthesizing enzymes can be proposed. In contrast to the situation in E. coli, where the two genes encoding the enzymes involved in trehalose synthesis are organized in one operon (16, 29), in Synechocystis, the genes encoding the enzymes involved in GG synthesis are obviously not transcribed together. The stpA gene encoding GGPP (14), the second enzyme involved in GG synthesis, is situated far from the ggpS gene on the chromosome of Synechocystis (slr0746; bp 3041493 to 3042407) (17). Mutation of only the ggpS gene led to a salt-sensitive phenotype, which is based on a completely impaired GG synthesis. Coupling between a defect in osmolyte synthesis and reduced salt tolerance was also reported for E. coli, where mutants in otsA as well as otsB (encoding trehalose-phosphate phosphatase) were found to be salt or osmosensitive (29). Diminished salt tolerance was found for yeast mutants defective in glycerol and, to a lesser extent, trehalose synthesis (21).
The characterization of the mutants strongly indicated that sll1566 encodes the key enzyme in GG synthesis. This conclusion was finally verified by overexpression of sll1566 in E. coli, leading to the appearance of GGPS activity in the heterologous host. However, GGPS purified from E. coli showed the enzyme activity under low-salt test conditions as well; therefore, the NaCl dependency of GGPS characteristic (12) for this enzyme in crude protein extracts from Synechocystis was lost. The same situation was previously found for GGPP, which was purified from E. coli by using a different expression and purification system (14). Therefore, in Synechocystis cells, an inhibitory mechanism which inhibits the GGPS and GGPP proteins under low-salt conditions should be present. This inhibition is obviously abolished by a NaCl-triggered process in Synechocystis and does not exist in E. coli.
The possibility to delete ggpS, glpD, and glpK from the chromosome of Synechocystis clearly shows that these three genes are not essential for growth under standard conditions. For ggpS, this is not surprising, since GG synthesis is completely absent in cells grown under low-salt conditions. The glpD and glpK genes are also dispensable for cells cultivated in high-salt media. Those cells showed an enhanced content of G3P that is probably needed for GG synthesis (15), but in addition, in control cells G3P has to be synthesized to ensure lipid and other biosyntheses. On the Synechocystis genome, there exists another gene encoding a putative NAD-dependent G3P dehydrogenase (gpsA or slr1755) (17), which obviously expresses sufficient enzyme to produce G3P in the mutants impaired in glpD and glpK. Nevertheless, we cannot rule out that the glpD gene product is significantly involved in G3P production in control and especially salt-stressed cells of the WT. The close proximity to ggpS makes this possibility likely. A comparable situation was reported for Saccharomyces cerevisiae, where one G3P dehydrogenase (GPD1) plays a dominant role in osmoadaptation and another enzyme (GPD2) plays a dominant role in adaptation to anoxic conditions. In single mutants, these enzymes can partly replace each other, while a double mutant was highly osmosensitive and failed to grow under anoxic conditions (3). In future studies, we will analyze how the expression of the ggpS gene is regulated by salt. Furthermore, by using the purified GGPS from E. coli, factors involved in the NaCl-mediated regulation of its enzyme activity will be isolated from Synechocystis.
ACKNOWLEDGMENTS
We thank T. Kaneko, Kazusa DNA Research Institute, Kisarazu, Japan, for sending us sequence data before the complete genome sequence was published and B. Haselkorn, University of Chicago, Chicago, Ill., for critical reading of the manuscript. The excellent technical assistance of B. Brzezinka and I. Dörr is greatly appreciated.
The work at the University of Rostock was supported by a grant of the Deutsche Forschungsgemeinschaft (DFG).
REFERENCES
- 1.Alexeyev M F, Shokolenko I N, Croughan T P. Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene. 1995;160:63–67. doi: 10.1016/0378-1119(95)00108-i. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansell R, Granath K, Hohmann S, Thevelein J M, Adler L. The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J. 1997;16:2179–2187. doi: 10.1093/emboj/16.9.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apte S K, Haselkorn R. Cloning of salinity stress-induced genes from the salt-tolerant nitrogen-fixing cyanobacterium Anabaena torulosa. Plant Mol Biol. 1990;15:723–733. doi: 10.1007/BF00016122. [DOI] [PubMed] [Google Scholar]
- 5.Bell W, Klaassen P, Ohnacker M, Boller T, Herweijer M, Schoppink P, Van der Zee P, Wiemken A. Characterization of the 56-kDa subunit of yeast trehalose-6-phosphate synthase and cloning of its gene reveal its identity with the product of CIF1, a regulator of carbon catabolite inactivation. Eur J Biochem. 1992;209:951–959. doi: 10.1111/j.1432-1033.1992.tb17368.x. [DOI] [PubMed] [Google Scholar]
- 6.Blazquez M A, Stucka R, Feldmann H, Gancedo C. Trehalose-6-P synthase is dispensable for growth on glucose but not for spore germination in Schizosaccharomyces pombe. J Bacteriol. 1994;176:3895–3902. doi: 10.1128/jb.176.13.3895-3902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chauvat F, Rouet P, Bottin H, Boussac A. Mutagenesis by random cloning of an Escherichia coli kanamycin resistance gene into the genome of the cyanobacterium Synechocystis sp. PCC 6803: selection of mutants defective in photosynthesis. Mol Gen Genet. 1989;216:51–59. doi: 10.1007/BF00332230. [DOI] [PubMed] [Google Scholar]
- 8.Erdmann N, Fulda S, Hagemann M. Glucosylglycerol accumulation during salt acclimation of two unicellular cyanobacteria. J Gen Microbiol. 1992;138:363–368. [Google Scholar]
- 9.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 10.Galinski E A. Compatible solutes of halophilic eubacteria: molecular principles, water-solute interactions, stress protection. Experientia. 1993;49:487–496. [Google Scholar]
- 11.Hagemann M, Zuther E. Selection and characterization of mutants of the cyanobacterium Synechocystis sp. PCC 6803 unable to tolerate high salt concentrations. Arch Microbiol. 1992;158:429–434. [Google Scholar]
- 12.Hagemann M, Erdmann N. Activation and pathway of glucosylglycerol biosynthesis in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology. 1994;140:1427–1431. [Google Scholar]
- 13.Hagemann M, Richter S, Zuther E, Schoor A. Characterization of a glucosylglycerol-phosphate accumulating, salt-sensitive mutant of the cyanobacterium Synechocystis sp. PCC 6803. Arch Microbiol. 1996;166:83–91. doi: 10.1007/s002030050360. [DOI] [PubMed] [Google Scholar]
- 14.Hagemann M, Schoor A, Jeanjean R, Zuther E, Joset F. The stpA gene from Synechocystis sp. strain PCC 6803 encodes the glucosylglycerol-phosphate phosphatase involved in cyanobacterial osmotic response to salt shock. J Bacteriol. 1997;179:1727–1733. doi: 10.1128/jb.179.5.1727-1733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagemann, M. Unpublished results.
- 16.Kaasen I, McDougall J, Strøm A R. Analysis of the otsBA operon for osmoregulatory trehalose synthesis in Escherichia coli and homology of the OtsA and OtsB proteins to the yeast trehalose-6-phosphate synthase/phosphatase complex. Gene. 1994;145:9–15. doi: 10.1016/0378-1119(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 17.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Nruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 18.Kratz W A, Myers J. Nutrition and growth of several blue-green algae. Am J Bot. 1955;42:282–287. [Google Scholar]
- 19.Lehn D A, Brown L J, Simonson G D, Moran S M, MacDonald M J. The sequence of a human mitochondrial glycerol-3-phosphate dehydrogenase-encoding cDNA. Gene. 1994;150:417–418. doi: 10.1016/0378-1119(94)90469-3. [DOI] [PubMed] [Google Scholar]
- 20.Luyten K, de Koning W, Tesseur I, Ruiz M C, Ramos J, Cobbaert P, Thevelein J M, Hohmann S. Disruption of the Kluyveromyces lactis GGS1 gene causes inability to grow on glucose and fructose and is suppressed by mutations that reduce sugar uptake. Eur J Biochem. 1993;217:701–713. doi: 10.1111/j.1432-1033.1993.tb18296.x. [DOI] [PubMed] [Google Scholar]
- 21.Mager W H, Varela J C S. Osmostress response of the yeast Saccharomyces. Mol Microbiol. 1993;10:253–258. [PubMed] [Google Scholar]
- 22.Muramatsu S, Mizuno T. Nucleotide sequence of the region encompassing the glpKF operon and its upstream region containing a bent DNA sequence of Escherichia coli. Nucleic Acids Res. 1989;17:4378. [PMC free article] [PubMed] [Google Scholar]
- 23.Philipp W J, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom B R, Jacobs W R, Jr, Cole S T. An integrated map of the genome of the tubercle bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed R H, Stewart W D P. Osmotic adjustment and organic solute accumulation in unicellular cyanobacteria from freshwater and marine habitats. Mar Biol. 1985;88:1–9. [Google Scholar]
- 25.Reed R H, Borowitzka L J, Mackay M A, Chudek J A, Foster R, Warr S R C, Moore D J, Stewart W D P. Organic solute accumulation in osmotically stressed cyanobacteria. FEMS Microbiol Rev. 1986;39:51–56. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Schoor A, Erdmann N, Effmert U, Mikkat S. Determination of the cyanobacterial osmolyte glucosylglycerol by high-performance liquid chromatography. J Chromatogr A. 1995;704:89–97. [Google Scholar]
- 28.Smith, D. R., and K. Robison. 1994. Unpublished sequence data.
- 29.Strøm A R, Kaasen I. Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol Microbiol. 1993;8:205–210. doi: 10.1111/j.1365-2958.1993.tb01564.x. [DOI] [PubMed] [Google Scholar]
- 30.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 31.Wolschek M F, Kubicek C P. The filamentous fungus Aspergillus niger contains two “differentially regulated” trehalose-6-phosphate synthase-encoding genes, tpsA and tpsB. J Biol Chem. 1997;272:2729–2735. doi: 10.1074/jbc.272.5.2729. [DOI] [PubMed] [Google Scholar]
- 32.Zaragoza, O., M. A. Blazquez, and C. Gancedo. 1996. Unpublished sequence data.