Abstract
Studies of the spirochete Borrelia burgdorferi have been hindered by the scarcity of genetic tools that can be used in these bacteria. For the first time, a method has been developed by which heterologous DNA (DNA without a naturally occurring B. burgdorferi homolog) can be introduced into and persistently maintained by B. burgdorferi. This technique uses integration of circular DNA into the bacterial genome via a single-crossover event. The ability to transform B. burgdorferi with heterologous DNA will now permit a wide range of experiments on the biology of these bacteria and their involvement in the many facets of Lyme disease.
Lyme disease is caused by Borrelia burgdorferi and other closely related spirochetes (3). Spread to mammals through the bites of infected ixodid ticks, it is the most commonly reported arthropod-borne disease affecting humans in the United States, and the number of cases is increasing yearly (4). The disease can manifest in many tissues and organs, including the skin, musculoskeletal, cardiac, and neurologic systems. Furthermore, the causative bacteria can persist for long periods in the mammalian body despite an active immune response directed against bacterial proteins (26, 28).
First reported in 1982 (3), B. burgdorferi has since been the focus of extensive research, yet little is known about how these bacteria cause the many aspects of Lyme disease or how they persistently infect mammals. Infected humans and other mammals produce antibodies directed against a number of B. burgdorferi proteins (1, 5, 7, 8, 10, 24, 29, 35), which indicates that these proteins are produced by the bacteria during mammalian infection. Several of the identified antigens appear to be located on the outer surfaces of the bacteria, suggesting that these proteins may facilitate interactions between the spirochetes and their environments. Synthesis of the B. burgdorferi OspC protein is induced upon tick feeding (25), which suggests that this protein is involved in transmission between the tick and mammalian hosts. Despite these and other studies, the actual functions of most identified Lyme disease antigens remain unknown.
Protein functions in many other bacteria have been studied by using recombinant genetic tools. For example, specific mutant bacteria can be created and studied for altered phenotypes. Protein function is then confirmed if the bacteria are restored to the wild-type phenotype by reintroducing the wild-type gene encoding that protein (the “molecular Koch’s postulates” [18]). Gene expression patterns can also be determined through the use of transcriptional and translational fusions with reporter genes. Such studies are currently impossible with B. burgdorferi, as there are no known methods by which heterologous DNA (DNA without a naturally occurring B. burgdorferi homolog) can be introduced into and stably maintained by these bacteria. A recent study described the transient expression levels of a reporter gene under the transcriptional control of B. burgdorferi promoters, but these fusion constructs could not be maintained by the spirochetes (27). Stable transformation with reporter fusions will be critical for measuring gene expression during the B. burgdorferi infection cycle.
Previous studies have demonstrated that DNA can be introduced into B. burgdorferi and integrate into the genomes of the recipient cells by homologous recombination. Mutant forms of the gyrB gene that encode a subunit of DNA gyrase that is resistant to the antibiotic coumermycin have been developed (23). DNA carrying these mutant gyrB genes can be introduced into B. burgdorferi by electroporation and will recombine with the chromosomal gyrB gene to produce coumermycin-resistant bacteria (20–22). In an earlier study, one of these mutant alleles, gyrBr, was cloned into a segment of a native B. burgdorferi plasmid, cp26, and the recombinant DNA was used to transform B. burgdorferi by electroporation. Coumermycin-resistant colonies were identified in which the gyrBr gene had inserted via allelic exchange with the flanking DNA into the resident cp26 (15). This technique has since been used to disrupt the B. burgdorferi ospC, guaB, and oppAIV genes (2, 32).
An additional result of these studies was the demonstration that coumermycin resistance is a selectable phenotype in merodiploid bacteria. A problem faced when developing a novel genetic system is that one cannot be certain that a foreign marker gene will be expressed at levels adequate to confer a selectable phenotype. Consistent with this, the gyrBr gene is the only selectable marker that has been successfully used for gene disruptions by allelic exchange (2, 15, 32), while attempts to transform B. burgdorferi with other antibiotic resistance markers have not been successful (33).
In other bacterial systems, fragments of heterologous DNA have often been introduced on small, independently replicating plasmids. However, many plasmids have limited host ranges and are unlikely to be stably maintained in B. burgdorferi. While some plasmids have wide host ranges, the evolutionary distance between the spirochetes and other eubacteria (12, 36) raises the possibility that even “broad-host-range” plasmids may be unable to replicate and segregate efficiently in B. burgdorferi. To date, there are no known plasmids from other bacteria that can replicate in B. burgdorferi (our unpublished results, this work, and reference 27). A technique is now presented by which heterologous plasmids can be maintained in B. burgdorferi by integration of circular DNA into the bacterial genome via a single-crossover event, thus eliminating the requirement for independent DNA replication and segregation.
MATERIALS AND METHODS
Bacteria.
A culture of B. burgdorferi isolate B31 that has been continuously cultivated for several years in laboratory medium (22) was used for all transformations. A mutant of isolate B31, B31-NGR (15), was the source of the gyrBr gene used in the DNA constructs described below. B. burgdorferi were grown at 35°C in solid Barbour-Stoener-Kelly (BSK) medium (9, 15) or in liquid BSK-H medium (Sigma, St. Louis, Mo.) supplemented with 6% rabbit serum (Sigma).
Recombinant DNA.
Plasmid DNAs were purified from 100-ml Escherichia coli cultures by using Qiagen midi-purification kits (Qiagen, Chatsworth, Calif.) or from 1-ml cultures by a crude boiling-lysis method (19). All restriction endonucleases, T4 DNA ligase, and appropriate buffers were obtained from New England Biolabs (Beverly, Mass.).
As we described earlier (15), the gyrBr gene, including the presumed gyrB promoter, was PCR amplified from B31-NGR chromosomal DNA by using oligonucleotides that introduced a BglII recognition site at the 5′ end and a BclI site at the 3′ end and then cloned into the TA vector pCR2.1 (Invitrogen, San Diego, Calif.). The DNA fragment containing the gyrBr gene was cut from this plasmid with BglII and BclI, separated by agarose gel electrophoresis, and purified with a Qiaex II kit (Qiagen). The gyrBr fragment was ligated into the BglII site of pOK12 (34), and the ligation mix was used to transform E. coli InvαF′ (Invitrogen) by selecting for kanamycin resistance. Plasmids were purified from 1-ml cultures, and insertion of gyrBr into pOK12 was assessed by digestion with restriction endonucleases. DNA sequencing confirmed the insertion and indicated the orientation of gyrBr. One such plasmid, pBLS500, was used thereafter.
A fragment of DNA encoding the B. burgdorferi oppAV gene (2) was subsequently cloned into pBLS500. The oppAV gene was amplified from purified B. burgdorferi B31 plasmid DNA by using the oligonucleotide primers listed in Table 1 and cloned into pCR2.1 (Invitrogen). The oppAV insert was removed from the recombinant plasmid by cleavage at the flanking EcoRI sites and cloned into the EcoRI site of pBLS500 to generate pJLB5.
TABLE 1.
Oligonucleotides used in this work
| Use | Primer no.a | Oligonucleotide | Sequence (5′ to 3′) |
|---|---|---|---|
| Screening of pBLS500 integrants | 1 | pOK12-Fwd2 | TGGGTAACGCCAGGGTTTTC |
| 2 | 246R | CTCTTCATGAATATCGGTAGG | |
| Screening of pJLB5 integrants | pOK12-Rev2 | TGTGGAATTGTGAGCGGATAAC | |
| 1696F | CTTCAGAGATATAAAGGGCTTGGG | ||
| oppAV cloning | oppA.73 | GCGTCTTTTTAAGCCTTTTGCC | |
| oppA.66 | TACTACAAAAACTAGGGTG |
The relative positions and orientations of primers 1 and 2 are indicated in Fig. 2A.
Transformation of B. burgdorferi.
Recombinant plasmids were introduced into B. burgdorferi B31 via electroporation by the previously described procedure (15, 20). Approximately 7 μg of DNA dissolved in 1 μl of distilled water was used in each transformation. Bacteria were resuspended in 5 ml of liquid medium and incubated overnight at 35°C. Aliquots (100 μl) of the transformed culture were each plated in solid BSK medium containing 0.5 μg of coumermycin A1 (Sigma) per ml and incubated at 35°C in a 1% CO2 environment.
Screening of transformed B. burgdorferi.
Coumermycin-resistant B. burgdorferi colonies were individually stabbed with toothpicks, and bacteria were transferred to PCR tubes. The PCR amplification solution contained a pair of oligonucleotide primers specific to the introduced recombinant plasmid (Table 1). Reaction conditions consisted of 25 cycles, with each cycle consisting of 30 s at 94°C, 30 s at 50°C, and 1 min at 65°C in a GeneAmp 9600 (Perkin-Elmer, Norwalk, Conn.) with 96-well PCR plates. A blank spot on the plate and solutions alone served as negative controls, and either purified pBLS500 or pJLB5 was used as a positive control. PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining.
Southern blotting.
Potential recombinant B. burgdorferi clones were grown to a density of approximately 108 bacteria per ml in 100 ml of liquid medium, and total bacterial DNA was purified as previously described (16). DNA was digested with restriction endonucleases, separated by pulsed-field agarose gel electrophoresis (30), and transferred to a nylon membrane (ICN, Irvine, Calif.). Membranes with DNA from bacteria transformed with pBLS500 were sequentially hybridized with the purified DNA fragment containing gyrBr described above and with purified pOK12. Membranes from bacteria transformed with pJLB5 were sequentially hybridized with the gyrBr probe and with a previously described oppAV-specific probe (2). Each probe was individually labeled with [α-32P]dATP (Du Pont, Boston, Mass.) by random priming (Life Technologies, Gaithersburg, Md.) and incubated with membranes at 55°C (16). Membranes were washed in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 55°C, and hybridized probes were detected by autoradiography. Radiolabeled probe was stripped from the membrane by washing with boiling water, and probe removal was confirmed by autoradiography before the membrane was rehybridized with a second probe.
Rescue of integrated DNA.
Approximately 125 ng of total DNA from B. burgdorferi transformant clone KS5 or AB1 was subjected to BamHI digestion, followed by phenol extraction and ethanol precipitation. The digested DNA was resuspended in 1× T4 ligase buffer and T4 DNA ligase. The self-ligated DNA was used to transform E. coli XL-1 Blue (Stratagene, La Jolla, Calif.), and the bacteria were plated on Luria-Bertani medium (19) containing 40 μg of kanamycin per ml. Equivalent amounts of B. burgdorferi transformant DNA that had not been treated with either BamHI or T4 DNA ligase were also used to transform E. coli XL-1 Blue.
RESULTS
Construction of a vector and transformation of B. burgdorferi.
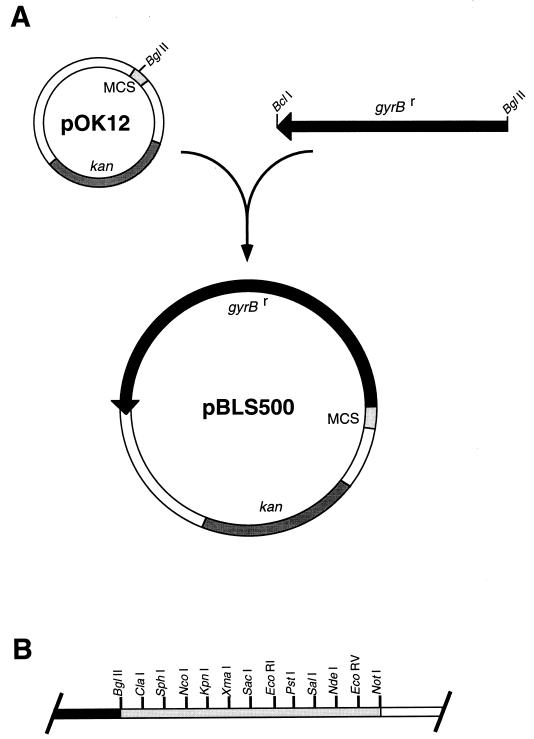
The gyrBr gene was previously amplified by PCR, using oligonucleotide primers that introduced BglII-compatible restriction sites at both ends (15). The 2-kb gyrBr fragment was cloned into the BglII site of pOK12, a 2-kb, low-copy-number E. coli plasmid that confers resistance to kanamycin (34) to produce a recombinant plasmid, pBLS500 (Fig. 1A). Most of the restriction endonuclease cleavage sites present in the pOK12 multiple cloning site (MCS) are unique in pBLS500 (Fig. 1B). The gyrBr gene contains BamHI, HindIII, and MluI recognition sites (data not shown; 15) which are also present in the pOK12 MCS, so these enzymes are unsuitable for cloning additional DNA fragments into pBLS500.
FIG. 1.
Characteristics of pBLS500. (A) Construction of pBLS500. kan is the kanamycin resistance gene. (B) Unique restriction endonuclease cleavage sites within the MCS of pBLS500. Although the pOK12 MCS contains BamHI, HindIII, and MluI recognition sites, additional sites are present in the gyrBr gene, making these restriction endonucleases unsuitable for cloning DNA fragments into pBLS500.
pBLS500 was introduced into B. burgdorferi isolate B31 under standard electroporation conditions (15, 20) and plated in solid medium containing coumermycin. As a control, another aliquot of bacteria was treated identically but without the addition of DNA into the electroporation cuvette. After 10 days of incubation, many coumermycin-resistant colonies were observed from the culture that had been transformed with pBLS500. Approximately 1,500 coumermycin-resistant colonies were obtained per μg of pBLS500 DNA. No colonies were observed in the plates of mock-transformed bacteria during this time period.
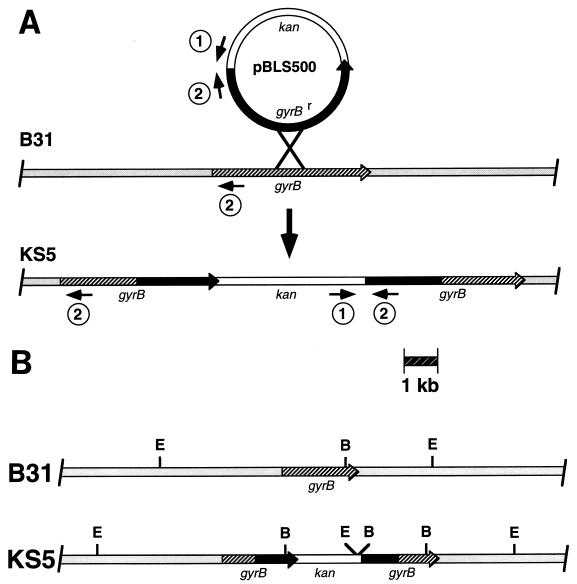
Coumermycin-resistant colonies could arise from bacteria that stably integrated pBLS500 into the chromosome (Fig. 2) or from bacteria that had incorporated only the gyrBr gene into the chromosomal gyrB locus via a double-crossover event. The bacteria might also resolve the integration of pBLS500 by deleting the intervening DNA through recombination between the direct duplicating repeats. Antibiotic-resistant colonies were therefore screened for pBLS500 integration by PCR amplification with oligonucleotide primers that produce a DNA fragment only from bacteria that contain pBLS500 DNA (Table 1 and Fig. 2A). The oligonucleotides used in this initial screening direct the synthesis of an approximately 500-bp PCR fragment from pBLS500 but do not direct synthesis of PCR fragments from wild-type B. burgdorferi B31 (Fig. 2A and 3). Of 250 coumermycin-resistant colonies tested, 3 were found to contain pBLS500 DNA (a frequency of 1.2%). One of these bacterial clones, KS5, was chosen for additional characterization.
FIG. 2.
(A) Diagram of the integration of pBLS500 via a single-crossover event into the gyrB locus of B31 to produce KS5. Locations of oligonucleotide primers used in the identification of pBLS500 integrants by PCR are indicated by the small numbered arrows. Note that oligonucleotide primer number 2 can hybridize with the gyrB genes of both B31 and KS5 but cannot produce a PCR product from B31. The two gyrB genes of KS5 have not been sequenced, so it is unknown which allele contains the coumermycin resistance-conferring mutations. (B) Restriction map of the chromosomes of B31 and KS5 near the gyrB locus. Abbreviations: B, BamHI; E, EcoRI.
FIG. 3.
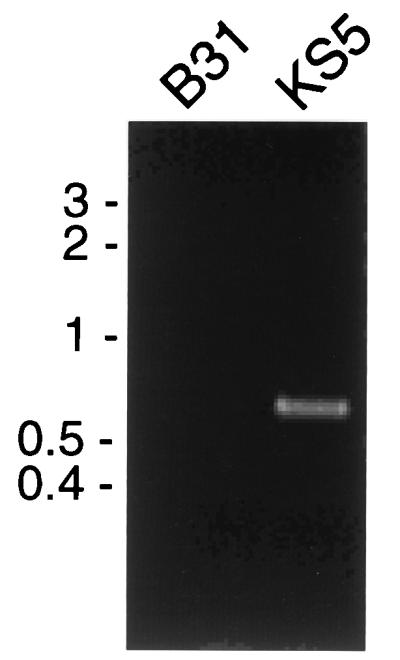
Products obtained by PCR amplification of total DNA from B31 and KS5 using oligonucleotide primers 1 and 2. Reaction products were separated by agarose gel electrophoresis and stained with ethidium bromide. The positions of molecular size markers (in kilobases) are indicated to the left of the gel.
Analysis of transformed B. burgdorferi.
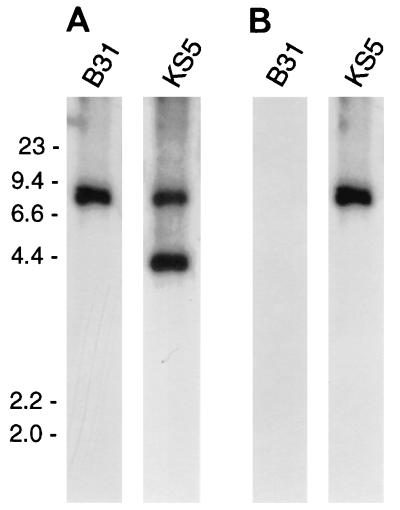
KS5 was further analyzed to confirm that pBLS500 had integrated into the chromosomal gyrB locus as expected. Total genomic DNA (chromosomes and plasmids) was isolated from B31 and KS5, digested with EcoRI, Southern blotted, and sequentially incubated with radiolabeled probes. A probe specific for the B. burgdorferi gyrB gene hybridized with a single B31 EcoRI DNA fragment of the expected size (Fig. 2B and 4A) and with two KS5 EcoRI DNA fragments, as expected (Fig. 2B and 4A). A probe specific for the non-gyrBr region of pBLS500 hybridized to the 7.5-kb EcoRI fragment of KS5 (Fig. 4B). Similar results were obtained from Southern blot analyses of B31 and KS5 DNA digested with other restriction endonucleases (data not shown), confirming that pBLS500 had integrated into the chromosome of KS5 at the gyrB locus.
FIG. 4.
Southern blots of B31 and KS5 DNAs cleaved by EcoRI and hybridized with a probe specific for the B. burgdorferi gyrB gene (A) or with labeled pOK12 (B). The positions of molecular size markers (in kilobases) are indicated to the left of the Southern blots.
KS5 was next analyzed to determine whether the heterologous, integrated DNA could be rescued. The pBLS500 gene encoding kanamycin resistance (kan) and the genes required for replication and segregation in E. coli are flanked by BamHI sites in KS5 (Fig. 2B). Total genomic DNA from KS5 was digested with BamHI, self-ligated, and used to transform E. coli, with selection for kanamycin resistance. Resistant E. coli arose at a frequency of 550 colonies per μg of total KS5 DNA. Six kanamycin-resistant E. coli clones were picked at random, and each was found to contain a plasmid that was identical in size and restriction endonuclease cleavage pattern to the corresponding BamHI fragment of pBLS500 (data not shown). Therefore, neither the pBLS500 replication genes nor the kanamycin resistance gene were detectably altered while integrated into the KS5 chromosome. For a control, E. coli was also transformed with KS5 DNA that had been neither cut nor ligated. No kanamycin-resistant E. coli colonies were obtained from this experiment, indicating that there are no detectable episomal copies of pBLS500 in KS5.
The pOK12-derived kan gene does not confer kanamycin resistance to B. burgdorferi KS5.
pBLS500 contains a gene that confers kanamycin resistance to E. coli; experiments were performed to examine whether pBLS500 also permits B. burgdorferi to grow in the presence of this antibiotic. Approximately fivefold-fewer colonies were formed when wild-type B31 was plated in medium containing 20 μg of kanamycin per ml than in medium without antibiotic. No colonies arose in medium containing 40 μg/ml or higher concentrations of the antibiotic. Similar results were obtained when KS5 was plated in medium containing kanamycin. Additionally, B31 was subjected to electroporation with pBLS500 under the same conditions previously used to generate KS5, except the bacteria were plated in solid medium containing 40 μg of kanamycin per ml rather than coumermycin. No colonies arose from these transformation experiments. We conclude that the pBLS500 kan gene, which encodes an aminoglycoside 3′-phosphotransferase (11, 34), does not confer detectable antibiotic resistance to KS5.
Integration of additional DNA into B. burgdorferi.
One goal of these studies will be to complement mutant genes through the introduction of wild-type alleles. To determine whether this will be possible via the plasmid integration technique, a 2-kb fragment of B. burgdorferi DNA containing the oppAV gene (2) was cloned into pBLS500. This plasmid, pJLB5, was introduced into B31 by electroporation, and bacteria were plated in medium containing coumermycin. A total of 276 antibiotic-resistant colonies were screened by PCR for pJLB5 integration with the oligonucleotide primers listed in Table 1, and one integrant was identified. This recombinant clone, AB1, was grown further without coumermycin selection, and extracted DNA was analyzed by Southern blotting with probes specific for oppAV and gyrBr, both of which confirmed that pJLB5 was integrated into the chromosomal gyrB locus (data not shown). As described above for KS5, the integrated pJLB5 DNA was rescued from purified AB1 genomic DNA by digestion with BamHI and self-ligation. It can be concluded, therefore, that at least 6 kb of DNA can be incorporated into and maintained by B. burgdorferi by the plasmid integration method of transformation.
DISCUSSION
By utilizing site-directed integration of circular DNA, for the first time, a method has been developed by which heterologous DNA can be maintained in B. burgdorferi. At least 6 kb of DNA can be introduced into B. burgdorferi by this technique. Furthermore, DNA rescued from the transformed bacteria remained functionally and structurally intact. Plasmid pJLB5 contains an additional 2 kb of B. burgdorferi DNA (the oppAV gene) that could result in homologous recombination, but approximately equivalent transformation efficiencies were obtained when using either pBLS500 or pJLB5 (3:250 versus 1:276, respectively). A possible explanation is that pJLB5 is 50% larger than pBLS500 and is not as efficiently taken up by the bacteria. One microgram of pBLS500 also contains more molecules than a similar weight of pJLB5, which will also affect the determination of transformation efficiency.
To avoid the requirement for independent plasmid replication and segregation, we introduced foreign DNA that can integrate into the B. burgdorferi genome. This technique uses a recombinant plasmid carrying a DNA fragment that is also located in the genome of the bacterium to be transformed. The introduced circular DNA can integrate into the genomic copy of the DNA via a single-crossover event, resulting in a duplication of the target DNA that flanks the remainder of the introduced DNA. This technique has been widely used to introduce heterologous DNA into gram-positive cocci and, recently, Neisseria gonorrhoeae (6, 13, 14, 31). The B. burgdorferi gyrBr gene has an advantage in that it can serve as both the selectable marker and as the DNA segment for targeting homologous recombination with the chromosomal gyrB.
Clearly, the present method of circular plasmid integration can be improved by the use of selectable markers other than the coumermycin resistance gyrBr gene. Additional markers will eliminate the need to screen colonies for DNA integration, permit the integration of DNA at sites other than the gyrB locus, and allow the introduction of additional DNA into previously transformed bacteria. The mutant DNA gyrase in bacteria containing the gyrBr gene may also alter DNA supercoiling, which could affect expression of other genes. Identification of additional selectable markers will also be a valuable first step in constructing independently replicating shuttle vectors for B. burgdorferi. Recombinant DNAs based on pBLS500 can be useful in identifying alternative selectable markers for use in B. burgdorferi. An antibiotic resistance gene can be cloned into pBLS500 and transformed into B. burgdorferi, and integrants can be selected by plating in medium containing coumermycin. Integrants can then be assessed for the ability to grow in the presence of the appropriate antibiotic. The kan gene carried by pBLS500 does not confer kanamycin resistance to B. burgdorferi, but other aminoglycoside 3′-phosphotransferase genes with different promoters or codon usages may be functional in these bacteria. Aminoglycoside resistance can also be conferred by enzymes with aminoglycoside-N-acetyltransferase or aminoglycoside-O-adenyltransferase activity (17).
A major obstacle to studying the biology of B. burgdorferi and its role in Lyme disease has been a paucity of genetic tools that can be used in these bacteria. The technique described herein should open the doors to a number of important experiments. Phenotypes associated with known mutations can be confirmed by complementation with wild-type alleles to see if defects are corrected, although most such studies will require other selectable markers in addition to coumermycin resistance. The successful integration of pJLB5, which contains a full-length copy of the oppAV gene, indicates that such experiments will be possible. Additionally, fusions between B. burgdorferi genes and reporter genes encoding easily detectable products will be useful in determining the expression patterns of B. burgdorferi genes during infection and in the transmission cycle between mammalian and arthropod hosts.
ACKNOWLEDGMENTS
We thank Scott Samuels for providing B31-NGR; Tom Schwan, Michael Chausee, and Joseph Hinnebusch for comments on the manuscript; Gary Hettrick and Robert Evans for graphic support; and Kelly Matteson for secretarial assistance.
REFERENCES
- 1.Aguero-Rosenfeld M E, Nowakowski J, McKenna D F, Carbonaro C A, Wormser G P. Serodiagnosis in early Lyme disease. J Clin Microbiol. 1993;31:3090–3095. doi: 10.1128/jcm.31.12.3090-3095.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bono J L, Tilly K, Stevenson B, Hogan D, Rosa P. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology. 1998;144:1033–1044. doi: 10.1099/00221287-144-4-1033. [DOI] [PubMed] [Google Scholar]
- 3.Burgdorfer W, Barbour A G, Hayes S F, Benach J L, Grunwaldt E, Davis J P. Lyme disease—a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control. Lyme disease—United States, 1996. Morbid Mortal Weekly Rep. 1997;46:531–535. [PubMed] [Google Scholar]
- 5.Craft J E, Fischer D K, Shimamoto G T, Steere A C. Antigens of Borrelia burgdorferi recognized during Lyme disease: appearance of a new immunoglobulin M response and expansion of the immunoglobulin G response late in the illness. J Clin Invest. 1986;78:934–939. doi: 10.1172/JCI112683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drake S L, Sandstedt S A, Koomey M. PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol Microbiol. 1997;23:657–668. doi: 10.1046/j.1365-2958.1997.2511618.x. [DOI] [PubMed] [Google Scholar]
- 7.Dressler F, Whalen J A, Reinhardt B N, Steere A C. Western blotting in the serodiagnosis of Lyme disease. J Infect Dis. 1993;167:392–400. doi: 10.1093/infdis/167.2.392. [DOI] [PubMed] [Google Scholar]
- 8.Engstrom S M, Shoop E, Johnson R C. Immunoblot interpretation criteria for serodiagnosis of early Lyme disease. J Clin Microbiol. 1995;33:419–427. doi: 10.1128/jcm.33.2.419-427.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurtti T J, Munderloh U G, Johnson R C, Ahlstrand G G. Colony formation and morphology in Borrelia burgdorferi. J Clin Microbiol. 1987;25:2054–2058. doi: 10.1128/jcm.25.11.2054-2058.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma B, Christen B, Leung D, Vigo-Pelfrey C. Serodiagnosis of Lyme borreliosis by Western immunoblot: reactivity of various significant antibodies against Borrelia burgdorferi. J Clin Microbiol. 1992;30:370–376. doi: 10.1128/jcm.30.2.370-376.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oka A, Sugisaki H, Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981;147:217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- 12.Paster B J, Dewhirst F E, Weisburg W G, Tordoff L A, Fraser G J, Hespell R B, Stanton T B, Zablen L, Mandelco L, Woese C R. Phylogenetic analysis of the spirochetes. J Bacteriol. 1991;173:6101–6109. doi: 10.1128/jb.173.19.6101-6109.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Podbielski A, Spellerberg B, Woischnik M, Pohl B, Lütticken R. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS) Gene. 1996;177:137–147. doi: 10.1016/0378-1119(96)84178-3. [DOI] [PubMed] [Google Scholar]
- 14.Proctor R A. The use of selective mutagenesis to study the pathogenesis of Gram-positive bacterial diseases. J Lab Clin Med. 1992;119:5–10. [PubMed] [Google Scholar]
- 15.Rosa P, Samuels D S, Hogan D, Stevenson B, Casjens S, Tilly K. Directed insertion of a selectable marker into a circular plasmid of Borrelia burgdorferi. J Bacteriol. 1996;178:5946–5953. doi: 10.1128/jb.178.20.5946-5953.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosa P A, Schwan T G. A specific and sensitive assay for the Lyme disease spirochete Borrelia burgdorferi using the polymerase chain reaction. J Infect Dis. 1989;160:1018–1029. doi: 10.1093/infdis/160.6.1018. [DOI] [PubMed] [Google Scholar]
- 17.Rouch D A, Byrne M E, Kong Y C, Skurray R A. The aacA-aphD gentamycin and kanamycin resistance determinant of Tn4001 from Staphylococcus aureus: expression and nucleotide sequence analysis. J Gen Microbiol. 1987;133:3039–3052. doi: 10.1099/00221287-133-11-3039. [DOI] [PubMed] [Google Scholar]
- 18.Salyers A A, Whitt D D. Bacterial pathogenesis: a molecular approach. Washington, D.C: American Society for Microbiology; 1994. [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Samuels D S. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol Biol. 1995;47:253–259. doi: 10.1385/0-89603-310-4:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samuels D S, Garon C F. Oligonucleotide-mediated genetic transformation of Borrelia burgdorferi. Microbiology. 1997;143:519–522. doi: 10.1099/00221287-143-2-519. [DOI] [PubMed] [Google Scholar]
- 22.Samuels D S, Mach K E, Garon C F. Genetic transformation of the Lyme disease agent Borrelia burgdorferi with coumarin-resistant gyrB. J Bacteriol. 1994;176:6045–6049. doi: 10.1128/jb.176.19.6045-6049.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samuels D S, Marconi R T, Huang W M, Garon C F. gyrB mutations in coumermycin A1-resistant Borrelia burgdorferi. J Bacteriol. 1994;176:3072–3075. doi: 10.1128/jb.176.10.3072-3075.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwan T G, Kime K K, Schrumpf M E, Coe J E, Simpson W J. Antibody response in white-footed mice (Peromyscus leucopus) experimentally infected with the Lyme disease spirochete (Borrelia burgdorferi) Infect Immun. 1989;57:3445–3451. doi: 10.1128/iai.57.11.3445-3451.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwan T G, Piesman J, Golde W T, Dolan M C, Rosa P A. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sigal L H. Lyme disease: a review of aspects of its immunology and immunopathogenesis. Annu Rev Immunol. 1997;15:63–92. doi: 10.1146/annurev.immunol.15.1.63. [DOI] [PubMed] [Google Scholar]
- 27.Sohaskey C D, Arnold C, Barbour A G. Analysis of promoters in Borrelia burgdorferi by use of a transiently expressed reporter gene. J Bacteriol. 1997;179:6837–6842. doi: 10.1128/jb.179.21.6837-6842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steere A C. Lyme disease. N Engl J Med. 1989;321:586–596. doi: 10.1056/NEJM198908313210906. [DOI] [PubMed] [Google Scholar]
- 29.Stevenson B, Bono J L, Schwan T G, Rosa P. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect Immun. 1998;66:2648–2654. doi: 10.1128/iai.66.6.2648-2654.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevenson B, Tilly K, Rosa P A. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J Bacteriol. 1996;178:3508–3516. doi: 10.1128/jb.178.12.3508-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao L, LeBlanc D J, Ferretti J J. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene. 1992;120:105–110. doi: 10.1016/0378-1119(92)90016-i. [DOI] [PubMed] [Google Scholar]
- 32.Tilly K, Casjens S, Stevenson B, Bono J L, Samuels D S, Hogan D, Rosa P. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol Microbiol. 1997;25:361–373. doi: 10.1046/j.1365-2958.1997.4711838.x. [DOI] [PubMed] [Google Scholar]
- 33.Tilly, K., A. Elias, and B. Stevenson. Unpublished results.
- 34.Vieira J, Messing J. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene. 1991;100:189–194. doi: 10.1016/0378-1119(91)90365-i. [DOI] [PubMed] [Google Scholar]
- 35.Wilske B, Schierz G, Preac-Mursic V, von Busch K, Kuhbeck R, Pfister H-W, Einhaupl K. Intrathecal production of specific antibodies against Borrelia burgdorferi in patients with lymphocytic meningoradiculitis (Bannwarth’s syndrome) J Infect Dis. 1986;153:304–314. doi: 10.1093/infdis/153.2.304. [DOI] [PubMed] [Google Scholar]
- 36.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]