Abstract
Natural products are a valuable source of pharmaceuticals, providing a majority of the small-molecule drugs in use today. However, their production through organic synthesis or in heterologous hosts can be difficult and time-consuming. Therefore, to allow for easier screening and production of natural products, we demonstrated the use of a cell-free protein synthesis system to partially assemble natural products in vitro using S-Adenosyl Methionine (SAM)-dependent methyltransferase enzyme reactions. The tea caffeine synthase, TCS1, was utilized to synthesize caffeine within a cell-free protein synthesis system. Cell-free systems also provide the benefit of allowing the use of substrates that would normally be toxic in a cellular environment to synthesize novel products. However, TCS1 is unable to utilize a compound like S-adenosyl ethionine as a cofactor to create ethylated caffeine analogs. The automation and reduced metabolic engineering requirements of cell-free protein synthesis systems, in combination with other synthesis methods, may enable the more efficient generation of new compounds.
Graphical Abstract 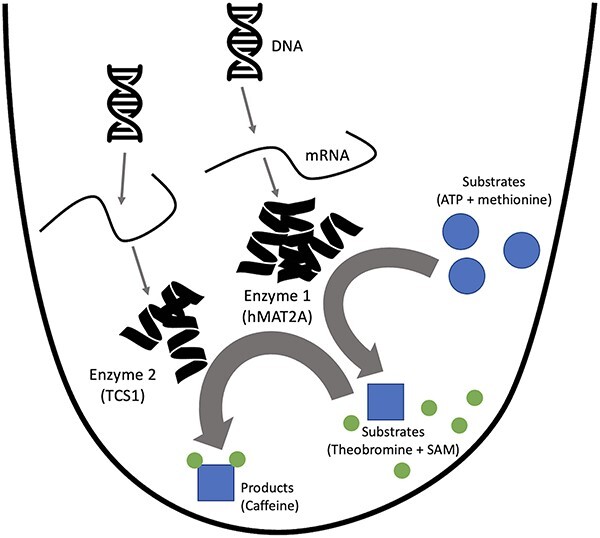
Keywords: cell-free, natural products, biosynthesis, methyltransferases
1. Introduction
Natural products are an essential and highly varied category of small molecules, defined as being produced naturally by some organisms, and these compounds are extremely important to the pharmaceutical industry (1, 2). The majority of all small-molecule testing in the pharmaceutical industry makes use of natural products, and they are considered to be the most productive source of new drugs (2, 3). Despite that fact, many of these companies decommissioned their small-molecule discovery programs in favor of combinatorial chemistry due to its ability to create large libraries which can be efficiently screened for useful biological activity (4). Unfortunately, these new programs failed to create many new useful compounds, with only a few out of millions having any degree of desirable activity (5).
The reason pharmaceutical companies seek out natural products is because of their inherent biological activities. Natural products can be, however, quite large and complex. For instance, two large, valuable groups of natural products are based on polyketides, such as erythromycin, and nonribosomal peptides, such as daptomycin (6). Additionally, Taxol, a diterpene that is perhaps the most important anti-cancer therapeutic in history, is a natural product derived from yew tree bark (7). Taxol is used for the treatment of lung, breast, prostate, bladder, ovarian, esophageal and many other types of cancer and is credited with saving countless lives since it was approved for use in 1993 (8). Current methods of producing Taxol include biosynthesis of precursors in cell culture followed by organic synthesis to complete the process (9). Additionally, many of these compounds involve a methylation step in their production, increasing the value of understanding the activity of methyltransferases (MTases) in cell-free protein synthesis (CFPS) systems (10).
For a biosynthetic approach, the relevant enzymes and pathways must be identified. The traditional method for producing enzymes from bacteria involves transforming cells with a plasmid containing the gene encoding the desired product and then growing a large number of those cells (11). However, recent advances in technology have made it possible to synthesize proteins in vitro using cellular machinery in an open system (12). CFPS involves a single-tube reaction that goes from plasmid to protein (12).
CFPS is a system that allows for the production of mRNA and proteins from plasmids by utilizing cellular lysates containing the necessary machinery for transcription and translation, such as ribosomes and T7 RNA polymerase, while nucleoside triphosphates (NTPs) are used as energy sources for the process. T7 polymerase is utilized for specificity with plasmids containing the corresponding promoter sequence (13). The most common system used in CFPS is an Escherichia coli-based system, but wheat germ–based systems have also been utilized to synthesize a wider range of eukaryotic proteins (14). Additionally, yields can be improved through the introduction of a feeding buffer containing NTPs and other substrates, as well as through the use of continuous flow–based systems that continually replenish substrate (15).
There are several advantages to using CFPS, as opposed to in vivo production. One major advantage is the capability to produce and use toxic compounds, due to the lack of cells that would be killed by cytotoxic effects, thus expanding the range of compounds that can be produced. Another benefit is that the process can be fully automated from the introduction of the plasmid into the system to the production of the protein (16). Additionally, the need for metabolic engineering is greatly reduced because the metabolic load can be essentially ignored. Furthermore, the speed of reactions in these systems is significantly faster, with the entire process of protein/product production taking only 1 day (17).
In another method that can be employed involving CFPS, enzymes of interest are overexpressed in E. coli, followed by the preparation of a CFPS extract from these cultures, which is then utilized to produce a desired product (18). A more streamlined approach would be to execute the enzyme expression and synthesis reaction concurrently within a CFPS reaction. This method offers significant advantages for prototyping applications, including reducing the number of steps required, enhanced flexibility by permitting changes to the CFPS without re-engineering the host organism and improved cost-efficiency as it allows the utilization of a singular cellular extract instead of producing a new one for each enzyme of interest that will be studied.
CFPS systems have numerous applications in the synthesis of pharmaceutical biologics (19), but recently, an increasing number of high-value natural products have been synthesized using a CFPS system (20). As part of the path to realizing these advantages, a CFPS-based system has been used to perform the function of the first two of five modules involved in the synthesis of the nonribosomal peptide gramicidin S, a valuable antibiotic (21). A variety of other small-molecule natural products have also been synthesized through modern CFPS research, including chlorogenic acid (22), limonine (23), citronellal (24) and 2,3-butanediol (25). Cell-free systems have also been used to incorporate novel molecules, such as nonstandard amino acids into a system in ways that would be more difficult in vivo (26, 27), including pyrrolysine-based noncanonical amino acids (28). There have even been examples of total synthesis of a nonribosomal peptide natural product in a CFPS system (29). However, the potential to utilize these systems to synthesize natural products from plasmids and precursors in a single reaction has not been fully explored, for example, the use of SAM-dependent MTases to perform modifications and potentially generate novel products. We extend this research by working to produce caffeine using a CFPS system with the SAM-dependent MTase TCS1 (tea caffeine synthase).
To further show the utility of CFPS systems in natural product synthesis, we chose methylation, a common reaction involved in the synthesis of many natural products, and particular methylation performed by SAM-dependent MTases. This is a broad category of enzymes that are involved in the synthesis of many natural products through the transfer of methyl groups to various substrates. Additionally, some SAM-dependent MTases can transfer alkyl groups in the presence of SAM analogs, providing potential for future work with the application of alkylation activity of some SAM-dependent MTases in the presence of SAM analogs (30, 31). However, the use of SAM analogs in cells can be challenging due to their toxicity, but in a CFPS system, the activity of these enzymes could be more easily harnessed without the concerns of toxicity. For example, ethionine and S-adenosyl ethionine (AdoEt) has been shown to be toxic via the mechanism of interfering with natural methylation processes. The toxicity is caused when promiscuous enzymes within the cell are able to utilize the AdoEt to perform ethylation reactions rather than the expected methylation reactions (32, 33).
We chose the synthesis of caffeine as an example of a common and useful natural product that is created by SAM-dependent MTases. There are already established methods for caffeine purification and identification using liquid chromatography-mass spectrometry (LC-MS), and the enzyme TCS1 has previously been expressed in E. coli and found to express well and be soluble (34). These factors make caffeine an ideal candidate for further exploration and demonstration of CFPS systems for product synthesis.
Caffeine can be described as a tri-methylated form of xanthine and has the alternative name 1,3,7-trimethyl xanthine, which fits with how the compound is synthesized in nature. Typically, caffeine is made in multiple methylation steps going from xanthine or xanthosine to a mono-methylated xanthine, then a di-methylated xanthine and finally to caffeine (35, 36). In this study, we are using the tea caffeine synthase enzyme, TCS1, which performs two different methylation steps in the production of caffeine in tea plants, going from 7-methylxanthine to theobromine, and then theobromine to caffeine (37). We will be utilizing the enzyme’s activity in methylating theobromine to caffeine in a CFPS system. Additionally, while many SAM-dependent MTases are thought to be promiscuous and accept varied alkyl group donors (38), the promiscuity of TCS1 is still unexplored, so we will attempt to utilize TCS1 with the cofactor AdoEt to synthesize a novel, ethylated form of caffeine (Figure 1).
Figure 1.
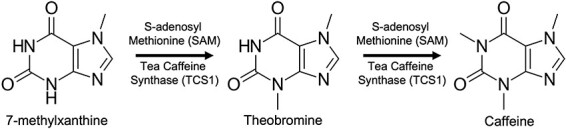
TCS1 caffeine production pathway in this study. TCS1 produces caffeine by the methylation of 7-methylxanthine to theobromine and then to caffeine.
2. Materials and methods
2.1. Plasmid preparation
The plasmid containing the TCS1 gene was made by performing a codon optimization on the tea caffeine synthase gene from Camellia sinensis so that it would be suitable for expression efficiently in E. coli with the Integrated DNA Technologies (IDT) codon optimization tool. This optimized gene was then ordered as a gBlock from IDT and inserted into the pNIC28-Bsa4 plasmid through the NEBuilder homology–based assembly method (39). A plasmid map of the pNIC28-Bsa4 plasmid can be found in the supporting information. This plasmid was then transformed into the DH5α E. coli strain to generate more plasmid, which was then used in the CFPS system to synthesize the TCS1 protein.
2.2. CFPS and purification
CFPS was performed using a Bioneer ExiProgen system using E. coli cellular lysate and Bioneer’s provided master mix of reagents. The reaction was performed in an ExiProgen EC-Tagfree Protein Synthesis Kit K-7320 (Bioneer, Daejeon, Republic of Korea) which includes tobacco etch virus (TEV) protease to cleave the TEV site which was used to attach the 6xHIS tag to the TCS1. The reaction was performed at 30°C using the standard ExiProgen-Tagfree protocol as recommended by Bioneer for a period of 30 h. The pNic28-Bsa4-TCS1 plasmid was diluted to a final concentration of 1.5 ng/μl in a total CFPS reaction volume of 750 μl. The protein was then purified with nickel nitrilotriacetic acid (Ni-NTA) beads, and the mass of the protein was checked on an SDS-PAGE gel. Protein concentration was quantified via a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, USA) by measuring absorbance at 280 nm after placing 1.0 μl of the sample onto the pedestal and using the instrument’s software to calculate based on the protein’s specific extinction coefficient.
2.3. Caffeine synthesis using purified TCS1 from CFPS system
To verify the function of the TCS1, TCS1 was synthesized using a CFPS system, purified using the ExiProgen-Tagfree protocol, and the 6xHIS tag was cleaved by TEV protease. This TCS1 protein was then used to perform an in vitro reaction with TCS1 in a pH 8.0 0.1 M Tris buffer, along with 200 μM MgCl2, 50 μM SAM and 200 μM theobromine. The products were then extracted from this system by performing a chloroform extraction, and the products were checked using LC-MS to verify the presence of caffeine.
2.4. Caffeine synthesis and extraction from CFPS system
To perform the caffeine synthesis within the CFPS system, the substrate theobromine (final concentration 2 mM) and the cofactor SAM (final concentration 5 mM) were added to the CFPS system along with the plasmid containing the gene for TCS1. After the protein was synthesized, a chloroform extraction was performed on the CFPS system to remove the caffeine from the system in a form that was more easily analyzed. Chloroform was added to a part of the system and mixed with the aqueous solution. Then, the chloroform layer was drawn off using a glass pipette, and the chloroform was evaporated to leave the caffeine in a purer form for analysis with LC-MS.
2.5. Liquid chromatography mass spectrometry protocol
To verify the production of caffeine from the TCS1, LC-MS was used. The analysis was achieved by liquid chromatography-ultraviolet-mass spectrometry analysis on an Agilent Technologies 6120 Quadrupole LC-MS (with UV detector). Agilent Eclipse Plus C18 column (4.6 × 100 mm) was used for metabolites separation with a linear gradient of 5–95% acetonitrile (v/v) over 15 min in H2O (0.1% formic acid, v/v) at a flow rate of 0.5 ml/min. Extracted ion chromatograms are shown because we are primarily interested in only the species at the expected m/z value of our products and substrates, and it allows us to see our species of interest more clearly even though our extraction protocol left some impurities of other ions in the solution.
2.6. hMAT2a protein synthesis and purification
To obtain the human methionine adenosyltransferase (hMAT2a) used to synthesize the SAM, the pET28a-hMAT2A plasmid was transformed into Rosetta (DE3) E. coli cells. The cells were then grown in a 15 ml flask in Terrific Broth media before being induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. The cells were then incubated overnight at 25°C for 18 h before being spun down in a centrifuge until a pellet was formed. Then, 5 ml of lysis buffer (20 mM hydroxyethylpiperazine ethane sulfonic acid, 300 mM NaCl, pH 7.5 Tris) was added to the pellet, and it was sonicated for until the lysate was clear and the cells were fully lysed. The lysate was then centrifuged for 40 min at 5000 rpm. The lysate was then passed through a Ni-NTA resin and incubated for 30 min at 4°C to bind. The column was then washed two times to removed unwanted compounds. Next, the protein was eluted by the addition to elution buffer containing imidazole to the column. Then, the protein samples were checked on an SDS-PAGE gel to confirm their identity.
3. Results
CFPS systems have immense potential as controlled systems for the synthesis of small molecules. To actualize that potential, we show the production of a natural product within a CFPS system. These synthetic systems, together with the provision of substrates, have the potential to synthesize products more quickly for rapid prototyping and testing natural products. To demonstrate the ability of a CFPS system to produce caffeine, first we demonstrated the production of TCS1 within a CFPS system and then the system was further used to synthesize caffeine, as described later.
3.1. TCS1 protein synthesis within a CFPS system
First, we synthesized the TCS1 enzyme using the CFPS system. The Bioneer Tagfree Protein Synthesis system is a cell-free production system based on a crude E. coli extract. It contains NTPs, amino acids and energy sources required for protein expression, in addition to the E. coli extract (which provides necessary molecular machinery such as ribosomes). We added our plasmid containing the TCS1 gene into the reaction mixture and incubated at 30°C before purifying the protein using Ni-NTA resin. After the protein was purified, we confirmed the production of the protein using an SDS-PAGE gel to check the mass. The apparent mass of the protein matched the expected mass for TCS1 and appeared to be relatively pure, as shown on the SDS-PAGE gel (Figure 2). This indicates that the CFPS system was successful in synthesizing the TCS1 protein. We observed a slightly higher concentration from the positive control Aequorea jellyfish green fluorescent protein (AcGFP) compared to the other proteins, with a total yield of approximately 1 mg of protein for the AcGFP and approximately 0.5 mg for each of the test proteins.
Figure 2.
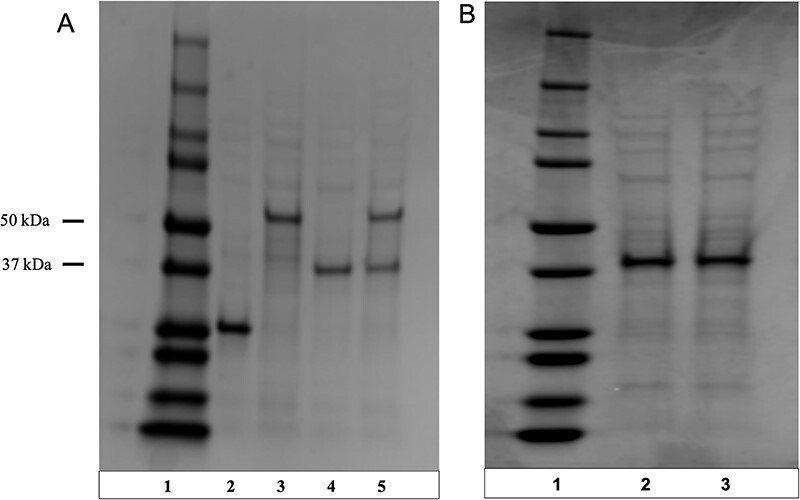
Production of TCS1 and GUD1 (guanine deaminase from Saccharomyces cerevisiae) in a CFPS system with and without substrates present on Coomassie brilliant blue stained SDS-PAGE gels. (A) Production of GUD1, a guanine deaminase which can convert guanine to xanthine in the early stages of caffeine biosynthesis, and TCS1 in a CFPS system. An SDS-PAGE gel shows the production of guanine deaminase (GUD1) and the tea caffeine synthase, TCS1, from the CFPS system. Lane 1 is the ladder. Lane 2 is an AcGFP positive control. Lane 3 is GUD1 (guanine deaminase from S. cerevisiae) with a molecular weight (MW) of ∼57 kDa. Lane 4 is TCS1 with an MW of ∼43 kDa. Lane 5 is GUD1 and TCS1 produced together. This demonstrated the capability of the system to synthesize multiple enzymes at once without major issues. (B) Lane 1 is the ladder. Lane 2 is TCS1 with an MW of ∼43 kDa without theobromine or SAM present in the system. Lane 2 is TCS1 with an MW of ∼43 kDa with both theobromine or SAM present in the system. There was no notable difference in protein production after the introduction of the substrates, so they did not disrupt protein production.
However, it was possible that the addition of the SAM or theobromine substrates may have interfered with the protein synthesis, so we also tested expression in the CFPS system with all substrates and cofactors needed for caffeine synthesis present, in which we found that there was no significant change in protein production when the substrates were added to the system, with a similar final yield of 0.5 mg of TCS1 both with and without the substrates (Figure 2B). We then performed caffeine production assays to demonstrate the enzyme’s activity.
3.2. Caffeine production activity by purified TCS1
We assayed the methylation activity of TCS1 by taking the purified enzyme from the CFPS system and performed a methylation reaction with both theophylline and theobromine substrates. When the TCS1 protein synthesized with the CFPS system was incubated with theophylline and SAM, there were no significant peaks observable by LC-MS other than the substrates (Figure 3B). However, when TCS1 was incubated with theobromine and SAM, a peak was observed at the same retention time and mass as the pure caffeine standard (Figure 3C). This suggests that the TCS1 produced in the CFPS system was functional and capable of synthesizing caffeine.
Figure 3.
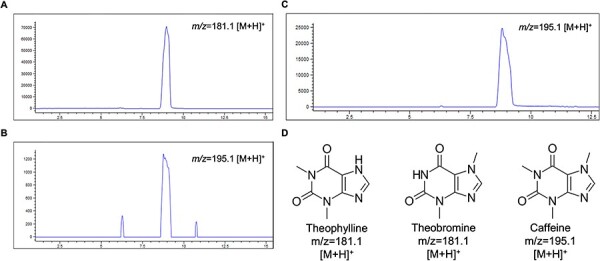
Production of caffeine from theophylline and theobromine substrates using purified TCS1. LC-MS chromatograms show the presence of product or reactant after chloroform extraction from the reaction mixture. The Y-axis is intensity in arbitrary units, and the X-axis is retention time on the column in minutes. (A) Mass filter analyses of the theophylline substrate (m/z = 181.1 [M + H]+) This demonstrates that the theophylline is present and can be detected via LC-MS, so the issue with the theophylline reactions is not caused by lack of substrate. (B) Mass filter analyses of the enzymatic reaction of TCS1 and substrate theophylline. The final product substrate caffeine (m/z = 195.1 [M + H]+) can be detected in the reaction, but the intensity here is close to the level of noise, showing very little, if any, product was made. The additional two peaks seem to be noise, which is visible on the chromatogram due to the extremely low concentration of caffeine produced from the theophylline substrate. This indicates that theophylline does not seem to be a suitable substrate for caffeine production via TCS1. (C) With a theobromine substrate mass filter, m/z = 195.1 [M + H]+ is used to detect caffeine, and the product is clearly produced with a detectable yield due to the clear peak at the expected mass. (D) The structures of theophylline, theobromine and caffeine are shown along with their expected m/z in LC-MS chromatograms.
Our findings contradict previous research that suggested theophylline may be a suitable substrate for caffeine production using TCS1 (34), as we observed that it resulted in extremely low caffeine production in our experimental conditions. In contrast, theobromine was effective and resulted in easily visible caffeine peaks when checked with LC-MS as seen in Figure 3. While theophylline was easily detectable via LC-MS (Figure 3A), it resulted in very low caffeine production close to the level of noise (Figure 3B).
3.3. Caffeine synthesis by TCS1 within a crude CFPS system
To evaluate the ability of TCS1 to produce caffeine within the CFPS system without purification, we added SAM and theobromine substrates to the other compounds required for the CFPS system. After incubation, the resulting solution was analyzed by LC-MS, which showed several peaks due to the presence of various compounds within the CFPS system (Figure 4A). However, we observed an additional peak with an m/z of 195.1 [M + H]+ and the same retention time as the pure caffeine standard (Figure 4), indicating the presence of caffeine in the solution. Although the quantification of caffeine was limited due to the chloroform extraction process, the presence of caffeine in the final solution demonstrates the simultaneous production of TCS1 and the synthesis of caffeine within the CFPS system. Due to the inefficient extraction, we estimate our final concentration of caffeine to be 10 μM in the total reaction volume of 750 ml, based on a comparison mass spectra obtained and the known caffeine standard, which would be a lower bound on the yield of the reaction.
Figure 4.
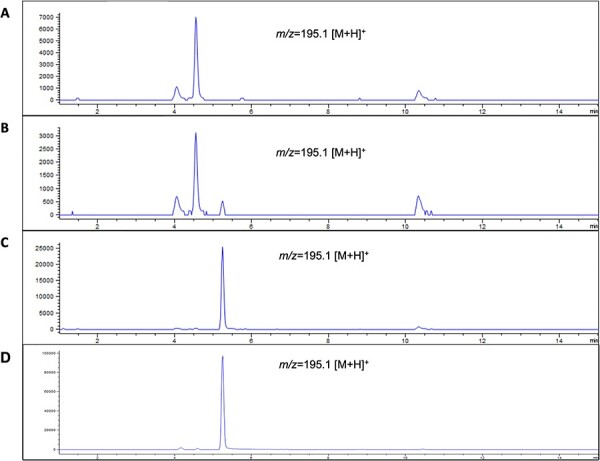
Production of caffeine within a CFPS system. LC-MS chromatograms showing the presence of product or reactant after chloroform extraction from the reaction mixture. The Y-axis is intensity in arbitrary units, and the X-axis is retention time on the column in minutes. Each chromatogram is shown with m/z = 195.1 [M + H]+which would show the presence of caffeine. (A) A negative control chromatogram is shown which is the result of a chloroform extraction of the CFPS system producing TCS1 without the presence of theobromine substrate added. (B) The experimental chromatogram which shows the result of a chloroform extraction on a CFPS system that had theobromine added and simultaneously produced TCS1. (C) A positive control chromatogram that shows the results of concentrated caffeine after a chloroform extraction to show the location of the caffeine peak. (D) A caffeine standard chromatogram to show the expected, characteristic peak for caffeine.
The experiments in the CFPS system using TCS1 provide further evidence for the potential use of CFPS systems as a versatile platform for natural product synthesis. Additionally, this finding reveals that CFPS systems can be utilized for single-solution reactions that enable the simultaneous synthesis and utilization of an enzyme for product synthesis, thus eliminating the need for enzyme purification. This simple and efficient process has significant implications for streamlining natural product synthesis in a variety of settings. This simplified system also enables the use of a simple system of mass-balance differential equations for modeling the enzyme activity along with the concentrations of substrates and products over time (see supplemental Figures S2–S5).
3.4. Production of caffeine through coupled reactions of TCS1 and hMAT2A and investigations into the specificity of TCS1 for SAM over AdoEt
One particularly useful advantage of cell-free systems over cellular systems is their resilience against compounds that are toxic to cells but which have a mechanism of toxicity that does not directly interfere with the limited reactions that occur within a CFPS system. This provides the prospect of utilizing CFPS systems in conjunction with substrates that would normally be toxic to cells to synthesize products that could not normally be synthesized in cells. For instance, the use of AdoEt or other alkyl analogs of SAM in a cell-free system may enable the synthesis of a range of alkylated products that would otherwise be impossible to produce in cells. This opens up new possibilities for the creation of novel products based on natural compounds that are difficult to synthesize in cellular systems.
To investigate the potential of TCS1 to be used in a CFPS system to synthesize novel products as part of a coupled reaction, we performed a coupled reaction of TCS1 with hMAT2a (human methionine adenosyltransferase) producing SAM and the ethyl analog of SAM, AdoEt. Due to the low efficiency of our caffeine extraction, we tested the activity of purified TCS1 produced from the CFPS system with the pure SAM, SAM produced through a coupled reaction with hMAT2a and AdoEt produced through a coupled reaction with hMAT2a. We confirmed both the SAM and AdoEt production via LC-MS, and the hMAT2a was able to synthesize both compounds (Figure S7). We observed that TCS1 can perform a coupled methylation reaction with hMAT2a producing SAM; however, the enzyme does not seem to produce any product when it is provided with AdoEt (Figure 5). This indicates that while the enzyme is able to work in concert with hMAT2a, it is not able to utilize the AdoEt provided to it as a substrate.
Figure 5.
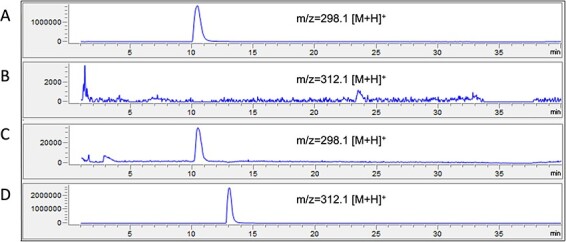
Attempted production of caffeine and ethylated theobromine from theobromine and SAM/AdoEt using purified TCS1. LC-MS chromatograms show the presence of product or reactant after chloroform extraction from the reaction mixture. The Y-axis is intensity in arbitrary units, and the X-axis is retention time on the column in minutes. (A) Mass filter analyses of the caffeine product (m/z = 195.1 [M + H]+) from a reaction mixture with purified TCS1, theobromine and pure SAM. (B) Mass filter analyses of the caffeine product (m/z = 195.1 [M + H]+) from a reaction mixture with purified TCS1, theobromine, purified hMAT2a, adenosine triphosphate (ATP) and methionine. (C) Mass filter analyses of the ethylated theobromine product (m/z = 210.1 [M + H]+) from a reaction mixture with purified TCS1, theobromine, purified hMAT2a, ATP and methionine. The low intensities here represent a lack of product formed, as expected for a negative control. (D) Mass filter analyses of the ethylated theobromine product (m/z = 210.1 [M + H]+) from a reaction mixture with purified TCS1, theobromine, purified hMAT2a, ATP and ethionine. The low intensities here represent a lack of product formed despite presence of AdoEt and theobromine.
4. Discussion
The prospect of utilizing CFPS systems as a platform for natural product synthesis is very exciting, as it has the potential to provide several benefits over traditional methods. As demonstrated by our caffeine synthesis, it is possible to utilize a single-step mixture of a plasmid containing the gene of interest, substrate and cofactors within the CFPS, eliminating the need for multiple steps of protein synthesis and separate reaction runs. This simplified process could be useful for rapid prototyping, allowing for the synthesis of small amounts of various compounds for screening Furthermore, while we focused on the methylation of theobromine to caffeine in this study, it is likely that other SAM-dependent methylation reactions may behave similarly in CFPS systems, making these systems suitable for a wide range of methylation reactions.
The main advantage of this approach lies in its inherent simplicity and flexibility. Traditional methods for producing natural products involve extracting them from their natural sources or synthesizing them using organic chemistry techniques. Both these methods can be highly time-consuming, labor-intensive and technically challenging, limiting their scalability and cost-effectiveness. Conversely, the CFPS system circumvents these problems by creating an environment where biosynthesis of compounds, such as caffeine, can occur in vitro. This controlled environment provides researchers with more flexibility to modulate conditions and carry out experiments, eliminating the time and effort required for maintaining and modifying whole organisms.
We also observed that theophylline seems to be a very poor substrate for TCS1 when compared to theobromine within a CFPS system, whereas previous studies have indicated that it may be a good substrate for caffeine production (34). It is possible that the conditions within a CFPS system differ in some way to make theobromine a better substrate, but more research is needed to better understand why theobromine performed so much better in this case compared to theophylline.
The objective of employing non-natural SAM analogs is to facilitate the synthesis of various natural product derivatives using pre-characterized enzymes. This approach provides an efficient way to produce an array of compounds, bearing structural similarity to existing natural products, with minor modifications. Subsequently, these novel compounds can be screened for potentially beneficial pharmaceutical properties. This can create a large number of compounds which may be more likely to be biologically relevant than a purely combinatorial chemistry–based approach.
Using a singular substrate and enzyme, while varying the cofactors, it is possible to generate a vast array of compounds with diverse modifications on the substrate. Considering caffeine as an example, it possesses three methylation sites compared to xanthine. If an enzyme could perform methylation, ethylation and propylation on each of these sites, a potential of 33 or 27 possible derivatives could be generated using a single enzyme–substrate pair. The challenge lies in the fact that these alkylation reactions are unsuitable for in vivo procedures due to the toxic nature of the cofactors. As such, the ultimate objective is to establish a system that enables the execution of these alkylation reactions in a single step to enable rapid prototyping of new compounds. Such a system would significantly enhance the speed and efficiency of generating a multitude of novel compounds for further testing and development.
While our method showcases certain benefits, it is not without limitations, particularly when contrasted with traditional natural product synthesis approaches. One prominent issue pertains to our extraction process, which produced an apparently low yield and incorporated unknown compounds from the CFPS system bearing a similar mass to caffeine. Prospective studies could focus on refining this extraction technique to bolster both product purity and yield. There are also a number of alternative extraction methods which could be used both to optimize the final product’s concentration and to more effectively extract other target products. These methods include percolation, reflux extraction and supercritical fluid extraction, which have all been used for the extraction of natural products in the past. (40, 41) Furthermore, the comparatively minor scale of our reactions may present another challenge. Scaling up to produce higher quantities of the product might necessitate the adoption of novel CFPS systems, such as continuous flow systems, to generate increased amounts of the product. Nonetheless, the principal advantage of our system potentially resides in its ability to rapidly prototype new compounds. In this context, the generation of small quantities of a broad spectrum of compounds may outweigh the need for producing large volumes of a single compound.
The concentration achieved in this experiment is low compared to what the yield could be if further optimized; however, it should be sufficient for a number of high-throughput drug screening methods, for example, enzyme-linked immunosorbent assay, surface plasmon resonance and isothermal titration calorimetry. Each of these typically require concentration in the nanomolar to micromolar range to detect the products in question and can be used for high-throughput drug screening (42–44). Therefore, the concentration of 10 µM we have achieved would indeed be suitable for these assays and could be used for high-throughput drug screening. This implies that this method may have applications in generating small amounts of compounds which can then be screened for useful biological activity.
Our results suggest that TCS1 may not be a suitable enzyme for performing ethylation reactions without modification to increase its promiscuity. One potential approach to increasing the promiscuity of TCS1 would be to engineer the enzyme by altering the SAM-binding pocket or other modifications to the active site. This could potentially enable TCS1 to effectively use alkyl SAM analogs in a coupled reaction. Some enzymes have previously been shown to be promiscuous and accept larger SAM analogs, such as the enzymes CouO and RebM (45, 46). Therefore, while it may be possible to synthesize novel products through alkylation with promiscuous MTases in a CFPS system, they must either be naturally promiscuous enzymes, like CouO or RebM as opposed to TCS1, or enzymes that were engineered to increase promiscuity. Further work could be done on exploring other MTases that could be used in a similar system and on engineering less promiscuous enzymes to enhance their ability to use alkyl SAM analogs.
However, it is crucial to note that while this study offers an example of using a CFPS system for product synthesis, challenges remain to be addressed before CFPS platforms can be broadly utilized for large-scale industrial production. Research into optimizing the reaction conditions, such as temperature, pH and nutrient concentrations, is necessary to improve the efficiency and yield of the biosynthesis process, as our yield of caffeine seemed lower than traditional measures. This may indicate that a more important role for CFPS product synthesis could be in the form of rapid prototyping, in which small amounts of many different products are created. When using the multiple possible SAM analogs as alkyl group donors onto a substrate, a variety of derivative products could be created, which can then be screened for useful biological activity. Then, when a useful natural product-like compound is identified, it could be synthesized on a larger scale with more convention product synthesis methods.
Supplementary Material
Acknowledgments
The Thorson Lab generously provided us with the pET28a-hMAT2A plasmid, which was instrumental in our production of SAM and AdoEt in coupled reactions with TCS1. We are grateful for their assistance and support in this project.
Contributor Information
Fanglong Zhao, Department of Chemical and Biomolecular Engineering, Rice University, Houston, TX, USA.
Xue Gao, Department of Chemical and Biomolecular Engineering, Rice University, Houston, TX, USA; Department of Chemistry, Rice University, Houston, TX, USA; Department of Bioengineering, Rice University, Houston, TX, USA.
George N Phillips, Jr., Department of Biosciences, Rice University, Houston, TX, USA; Department of Chemistry, Rice University, Houston, TX, USA.
Supplementary data
Supplementary data are available at SYNBIO Online.
Data availability
All data generated or analyzed in this paper are either included in the paper/supplementary data or can be made available upon reasonable request by the corresponding author.
Material availability
Plasmids and other materials are available at reasonable request from Rice University via a Material Transfer Agreement.
Funding
Gulf Coast Consortia, on the Houston Area Molecular Biophysics Program [T32 GM008280 to A.D.]; National Institutes of Health [R01 GM115261 and R01 CA217255 to G.N.P., R35 GM138207 to X.G.]; Robert A. Welch Foundation [C-1952 to X.G.].
Author contributions
A.D. designed and performed experiments, wrote and edited the manuscript.
F.Z. performed LC-MS experiments and revised the manuscript.
X.G. supervised analysis experiments and revised the manuscript.
G.N.P. designed experiments, wrote and edited the manuscript.
Conflict of interest statement.
There are no conflicts of interest declared by the authors.
References
- 1. Koehn F.E. and Carter G.T. (2005) The evolving role of natural products in drug discovery. Nat. Rev: Drug Discov., 4, 206–220. [DOI] [PubMed] [Google Scholar]
- 2. Li J.W.-H. and Vederas J.C. (2009) Drug discovery and natural products: end of an era or an endless frontier?. Science, 325, 161–165. [DOI] [PubMed] [Google Scholar]
- 3. Harvey A.L. (2008) Natural products in drug discovery. Drug Discov. Today, 13, 894–901. [DOI] [PubMed] [Google Scholar]
- 4. Kodadek T. (2011) The rise, fall and reinvention of combinatorial chemistry. Chem. Commun., 47, 9757–9763. [DOI] [PubMed] [Google Scholar]
- 5. Kubinyi H. (2003) Drug research: myths, hype and reality. Nat. Rev. Drug Discov., 2, 665–668. [DOI] [PubMed] [Google Scholar]
- 6. Baltz R.H., Miao V. and Wrigley S.K. (2005) Natural products to drugs: daptomycin and related lipopeptide antibiotics. Nat. Prod. Rep., 22, 717–741. [DOI] [PubMed] [Google Scholar]
- 7. Heinig U. and Jennewein S. (2009) Taxol: a complex diterpenoid natural product with an evolutionarily obscure origin. Afr. J. Biotechnol., 8, 1370–1385. [Google Scholar]
- 8. Saville M.W., Lietzau J., Pluda J.M., Wilson W.H., Humphrey R.W., Feigel E., Steinberg S.M., Broder S., Yarchoan R., Odom J. et al. (1995) Treatment of HIV-associated Kaposi’s sarcoma with paclitaxel. Lancet, 346, 26–28. [DOI] [PubMed] [Google Scholar]
- 9. McElroy C., Jennewein S. (2018) Taxol® biosynthesis and production: from forests to fermenters. In: Schwab W, Lange BM, Wüst M (eds). Biotechnology of Natural Products. Springer International Publishing, Cham, pp. 145–185. [Google Scholar]
- 10. Zhang C., Sultan S.A., T R. and Chen X. (2021) Biotechnological applications of S-adenosyl-methionine-dependent methyltransferases for natural products biosynthesis and diversification. Bioresour. Bioprocess., 8, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lichty J.J., Malecki J.L., Agnew H.D., Michelson-Horowitz D.J. and Tan S. (2005) Comparison of affinity tags for protein purification. Protein Expr. Purif., 41, 98–105. [DOI] [PubMed] [Google Scholar]
- 12. Carlson E.D., Gan R., Hodgman C.E. and Jewett M.C. (2012) Cell-free protein synthesis: applications come of age. Biotechnol. Adv., 30, 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kwon Y.-C. and Jewett M.C. (2015) High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci. Rep., 5, 8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aceti D.J., Bingman C.A., Wrobel R.L., Frederick R.O., Makino S., Nichols K.W., Sahu S.C., Bergeman L.F., Blommel P.G., Cornilescu C.C. et al. (2015) Expression platforms for producing eukaryotic proteins: a comparison of E. coli cell-based and wheat germ cell-free synthesis, affinity and solubility tags, and cloning strategies. J. Struct. Funct. Genomics, 16, 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spirin A.S. (2004) High-throughput cell-free systems for synthesis of functionally active proteins. Trends Biotechnol., 22, 538–545. [DOI] [PubMed] [Google Scholar]
- 16. Beebe E.T., Makino S., Nozawa A., Matsubara Y., Frederick R.O., Primm J.G., Goren M.A. and Fox B.G. (2011) Robotic large-scale application of wheat cell-free translation to structural studies including membrane proteins. New Biotechnol., 28, 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gregorio N.E., Levine M.Z. and Oza J.P. (2019) A user’s guide to cell-free protein synthesis. Methods Protoc., 2, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dudley Q.M., Anderson K.C. and Jewett M.C. (2016) Cell-free mixing of Escherichia coli crude extracts to prototype and rationally engineer high-titer mevalonate synthesis. ACS Synth. Biol., 5, 1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chiba C.H., Knirsch M.C., Azzoni A.R., Moreira A.R. and Stephano M.A. (2021) Cell-free protein synthesis: advances on production process for biopharmaceuticals and immunobiological products. Biotechniques, 70, 126–133. [DOI] [PubMed] [Google Scholar]
- 20. Ji X., Liu W.-Q. and Li J. (2022) Recent advances in applying cell-free systems for high-value and complex natural product biosynthesis. Curr. Opin. Microbiol., 67, 102142. [DOI] [PubMed] [Google Scholar]
- 21. Goering A.W., Li J., McClure R.A., Thomson R.J., Jewett M.C. and Kelleher N.L. (2017) In vitro reconstruction of nonribosomal peptide biosynthesis directly from DNA using cell-free protein synthesis. ACS Synth. Biol., 6, 39–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Niu F.-X., Yan Z.-B., Huang Y.-B. and Liu J.-Z. (2021) Cell-free biosynthesis of chlorogenic acid using a mixture of chassis cell extracts and purified spy-cyclized enzymes. J. Agric. Food Chem., 69, 7938–7947. [DOI] [PubMed] [Google Scholar]
- 23. Dudley Q.M., Karim A.S., Nash C.J. and Jewett M.C. (2020) In vitro prototyping of limonene biosynthesis using cell-free protein synthesis. Metab. Eng., 61, 251–260. [DOI] [PubMed] [Google Scholar]
- 24. Richardson K.N., Black W.B. and Li H. (2020) Aldehyde production in crude lysate- and whole cell-based biotransformation using a noncanonical redox cofactor system. ACS Catal., 10, 8898–8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kay J.E. and Jewett M.C. (2015) Lysate of engineered Escherichia coli supports high-level conversion of glucose to 2,3-butanediol. Metab. Eng., 32, 133–142. [DOI] [PubMed] [Google Scholar]
- 26. Hong S.H., Kwon Y.-C. and Jewett M.C. (2014) Non-standard amino acid incorporation into proteins using Escherichia coli cell-free protein synthesis. Front. Chem., 2, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cui Z., Johnston W.A. and Alexandrov K. (2020) Cell-free approach for non-canonical amino acids incorporation into polypeptides. Front. Bioeng. Biotechnol., 8, 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ranji Charna A., Des Soye B.J., Ntai I., Kelleher N.L. and Jewett M.C. (2022) An efficient cell-free protein synthesis platform for producing proteins with pyrrolysine-based noncanonical amino acids. Biotechnol. J., 17, e2200096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhuang L., Huang S., Liu W.-Q., Karim A.S., Jewett M.C. and Li J. (2020) Total in vitro biosynthesis of the nonribosomal macrolactone peptide valinomycin. Metab. Eng., 60, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang C., Weller R.L., Thorson J.S. and Rajski S.R. (2006) Natural product diversification using a non-natural cofactor analogue of S-adenosyl-L-methionine. J. Am. Chem. Soc., 128, 2760–2761. [DOI] [PubMed] [Google Scholar]
- 31. Zhang J. and Zheng Y.G. (2016) SAM/SAH analogs as versatile tools for SAM-dependent methyltransferases. ACS Chem. Biol., 11, 583–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khupse R., Sarkar A.B. (2023) Ethionine. In: Wexler P. (ed). Reference Module in Biomedical Sciences. Elsevier: Amsterdam, Netherlands, pp. 431–434. [Google Scholar]
- 33. Waterfield C.J., Westmoreland C., Asker D.S., Murdock J.C., George E. and Timbrell J.A. (1998) Ethionine toxicity in vitro: the correlation of data from rat hepatocyte suspensions and monolayers with in vivo observations. Arch. Toxicol., 72, 588–596. [DOI] [PubMed] [Google Scholar]
- 34. Li M., Sun Y., Pan S., Deng W., Yu O. and Zhang Z. (2017) Engineering a novel biosynthetic pathway in Escherichia coli for the production of caffeine. RSC Adv., 7, 56382–56389. [Google Scholar]
- 35. Ogawa M., Herai Y., Koizumi N., Kusano T. and Sano H. (2001) 7-Methylxanthine methyltransferase of coffee plants: gene isolation and enzymatic properties. J. Biol. Chem., 276, 8213–8218. [DOI] [PubMed] [Google Scholar]
- 36. Jin J.-Q., Yao M.-Z., Ma C.-L., Ma J.-Q. and Chen L. (2016) Natural allelic variations of TCS1 play a crucial role in caffeine biosynthesis of tea plant and its related species. Plant Physiol. Biochem., 100, 18–26. [DOI] [PubMed] [Google Scholar]
- 37. Jin L., Bhuiya M.W., Li M., Liu X., Han J., Deng W., Wang M., Yu O., Zhang Z. and Riezman H. (2014) Metabolic engineering of Saccharomyces cerevisiae for caffeine and theobromine production. PloS One, 9, e105368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huber T.D., Clinger J.A., Liu Y., Xu W., Miller M.D., PhillipsG.N. Jr and Thorson J.S. (2020) Methionine adenosyltransferase engineering to enable bioorthogonal platforms for AdoMet-utilizing enzymes. ACS Chem. Biol., 15, 695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Savitsky P., Bray J., Cooper C.D.O., Marsden B.D., Mahajan P., Burgess-Brown N.A. and Gileadi O. (2010) High-throughput production of human proteins for crystallization: the SGC experience. J. Struct. Biol., 172, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Q.-W., Lin L.-G. and Ye W.-C. (2018) Techniques for extraction and isolation of natural products: a comprehensive review. Chin. Med., 13, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Azmir J., Zaidul I.S.M., Rahman M.M., Sharif K.M., Mohamed A., Sahena F., Jahurul M.H.A., Ghafoor K., Norulaini N.A.N. and Omar A.K.M. (2013) Techniques for extraction of bioactive compounds from plant materials: a review. J. Food Eng., 117, 426–436. [Google Scholar]
- 42. Hayashi Y., Matsuda R., Maitani T., Imai K., Nishimura W., Ito K. and Maeda M. (2004) Precision, limit of detection and range of quantitation in competitive ELISA. Analy. Chem., 76, 1295–1301. [DOI] [PubMed] [Google Scholar]
- 43. Vander Meulen K.A., Horowitz S., Trievel R.C. and Butcher S.E. (2016) Measuring the kinetics of molecular association by isothermal titration calorimetry. Methods Enzymol., 567, 181–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ashley J., Piekarska M., Segers C., Trinh L., Rodgers T., Willey R. and Tothill I.E. (2017) An SPR based sensor for allergens detection. Biosens. Bioelectron., 88, 109–113. [DOI] [PubMed] [Google Scholar]
- 45. Singh S., McCoy J.G., Zhang C., Bingman C.A., Phillips G.N. and Thorson J.S. (2008) Structure and mechanism of the rebeccamycin sugar 4′-O-methyltransferase RebM*. J. Biol. Chem., 283, 22628–22636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pavkov-Keller T., Steiner K., Faber M., Tengg M., Schwab H., Gruber-Khadjawi M., Gruber K. and Jeltsch A. (2017) Crystal structure and catalytic mechanism of CouO, a versatile C-methyltransferase from Streptomyces rishiriensis. PLOS One, 12, e0171056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed in this paper are either included in the paper/supplementary data or can be made available upon reasonable request by the corresponding author.


