SUMMARY/ABSTRACT
Delays in the identification of acute kidney injury (AKI) in hospitalized patients are a major barrier to the development of effective interventions to treat AKI. A recent study by Tomasev and colleagues at DeepMind described a model that achieved a state-of-the-art performance in predicting AKI up to 48 hours in advance.1 Because this model was trained in a population of US Veterans that was 94% male, questions have arisen about its reproducibility and generalizability. In this study, we aimed to reproduce key aspects of this model, trained and evaluated it in a similar population of US Veterans, and evaluated its generalizability in a large academic hospital setting. We found that the model performed worse in predicting AKI in females in both populations, with miscalibration in lower stages of AKI and worse discrimination (a lower area under the curve) in higher stages of AKI. We demonstrate that while this discrepancy in performance can be largely corrected in non-Veterans by updating the original model using data from a sex-balanced academic hospital cohort, the worse model performance persists in Veterans. Our study sheds light on the importance of reproducing artificial intelligence studies, and on the complexity of discrepancies in model performance in subgroups that cannot be explained simply on the basis of sample size.
INTRODUCTION
Delays in the identification of acute kidney injury (AKI) in hospitalized patients are a major barrier to the development of effective interventions to treat AKI.2 By the time changes in typical kidney function biomarkers—serum creatinine and blood urea nitrogen—are detected, damage that is not readily reversed is often already established. This is underscored by recent evidence that automated alerts generated upon AKI onset appear to be ineffective at changing the trajectory of AKI.3 This has led to multiple efforts to develop early warning system scores that predict the onset of AKI with sufficient lead times to support potential interventions.4–8 The most promising of these efforts was recent work by Tomasev and colleagues from DeepMind describing a state-of-the-art model for the continuous prediction of AKI.1 Developed and validated using data from 703,782 US Veterans, the primary recurrent neural network model described in the paper achieved an area under the receiver operating characteristic curve (AUC) of 92.1% when predicting AKI in the next 48 hours. This study was notable for several reasons, including its large sample size, high AUC, and a longer lead time, all of which made this model a clear outlier as compared to previous studies.
Despite its promise, the model has not been implemented within the Veterans Affairs (VA) health system. In this respect, this model represents an example of the “AI chasm,” a term used to describe high-performing AI models that fail to reach the bedside due to challenges involved in real-world implementation.9 The model described in this study is also not publicly available, meaning that it cannot be readily reproduced and evaluated in other clinical settings despite knowledge of the underlying methods and software.10,11 This lack of computational reproducibility among complex AI models in healthcare is a well-recognized barrier to sustaining progress in clinical AI applications.12–16 Because the model was developed in a Veteran population that is 94% male, concerns have also arisen about the generalizability of this model to females,17 and to non-VA contexts where practice patterns may differ. Recent work in medical imaging demonstrates that models trained in predominantly male populations fail to perform well in females.18 This was suggested by the DeepMind study where a lower AKI episode-level sensitivity was observed in females as compared to males (44.8% vs. 56.0%, respectively).
To address these concerns, we sought to evaluate the reproducibility and generalizability of the DeepMind AKI model. Drawing on electronic health record (EHR) data from 278,813 US Veterans, we reproduced key aspects of the DeepMind AKI model, including their methods for data pre-processing, feature selection, transformation of hospitalization data to 6-hour person-period intervals, and outcome definitions. We further assessed the model’s generalizability in a large academic center using data from another 165,359 hospitalizations. Finding that the model performs worse in females, we evaluated an approach to updating the model to correct for this disparity. Both our reconstructed model and the corrected model are publicly available.
RESULTS
Cohort Characteristics
We identified 278,813 VA hospitalizations (from 118 VA hospitals) and 165,359 University of Michigan (UM) hospitalizations meeting inclusion and exclusion criteria. Only the first hospitalization was included for VA patients, whereas all eligible hospitalizations were included for 97,506 UM patients. As compared with UM, patients with VA hospitalizations were more likely to be male (94% vs. 50%), older (mean 70 vs. 57), Black (20% vs. 11%), and to have diabetes (36% vs. 29%). On the other hand, UM patients were more likely to have normal baseline kidney function (baseline eGFR ≥ 60 mL/min/1.73 m2, 81% vs. 73%) and a longer length of stay (mean 6.6 vs. 5.3 days), leading to more 6-hour periods per patient (18 vs. 15) calculated over a maximum of 7 days of hospitalization (Table 1).
Table 1.
Characteristics of the VA and UM cohorts
| Cohort | VA | UM | ||||
|---|---|---|---|---|---|---|
| Training | Validation | Test | Training | Validation | Test | |
| Characteristic | N = 178,453 | N = 44,614 | N = 55,746 | N = 33,077(19,501 patients) | N = 33,034(19,501 patients) | N = 99,248(58,504 patients) |
| Age (years) | 68.8 (13.1) | 68.9 (13.1) | 68.8 (13.1) | 57.3 (18.2) | 57.2 (18.2) | 56.8 (18.2) |
| Sex | ||||||
| Female | 10,083 (5.7%) | 2,557 (5.7%) | 3,119 (5.6%) | 16,381 (49.5%) | 16,605 (50.3%) | 49,280 (49.7%) |
| Male | 168,370 (94.3%) | 42,057 (94.3%) | 52,627 (94.4%) | 16,696 (50.5%) | 16,429 (49.7%) | 49,968 (50.3%) |
| Race | ||||||
| African American | 35,171 (19.7%) | 8,791 (19.7%) | 10,990 (19.7%) | 3,361 (10.2%) | 3,794 (11.5%) | 11,036 (11.1%) |
| Caucasian | 120,962 (67.8%) | 30,308 (67.9%) | 37,835 (67.9%) | 27,594 (83.4%) | 27,277 (82.6%) | 82,164 (82.8%) |
| Other | 15,038 (8.4%) | 3,742 (8.4%) | 4,696 (8.4%) | 1,733 (5.2%) | 1,621 (4.9%) | 4,899 (4.9%) |
| Unknown | 7,282 (4.1%) | 1,773 (4.0%) | 2,225 (4.0%) | 389 (1.2%) | 342 (1.0%) | 1,149 (1.2%) |
| Baseline BMI | 29.6 (6.7) | 29.6 (6.6) | 29.5 (6.6) | 28.8 (6.7) | 28.8 (6.9) | 28.9 (6.8) |
| Unknown | 12,519 (7.0%) | 3,033 (6.8%) | 3,808 (6.8%) | 1,898 (5.7%) | 1,944 (5.9%) | 5,921 (6.0%) |
| Baseline serum creatinine (mg/dL) | 1.1 (0.4) | 1.1 (0.4) | 1.1 (0.4) | 1.0 (0.4) | 1.0 (0.4) | 1.0 (0.4) |
| Baseline eGFR* (mL/min/1.73 m2) | ||||||
| ≥ 60 | 131,108 (73.5%) | 32,836 (73.6%) | 40,874 (73.3%) | 27,016 (81.7%) | 26,813 (81.2%) | 80,530 (81.1%) |
| 45–59 | 27,100 (15.2%) | 6,761 (15.2%) | 8,651 (15.5%) | 3,192 (9.7%) | 3,339 (10.1%) | 9,894 (10.0%) |
| 30–44 | 14,686 (8.2%) | 3,631 (8.1%) | 4,481 (8.0%) | 1,957 (5.9%) | 1,945 (5.9%) | 5,988 (6.0%) |
| 15–29 | 5,470 (3.1%) | 1,365 (3.1%) | 1,706 (3.1%) | 872 (2.6%) | 905 (2.7%) | 2,727 (2.7%) |
| < 15 | 89 (0.0%) | 21 (0.0%) | 34 (0.1%) | 40 (0.1%) | 32 (0.1%) | 109 (0.1%) |
| Baseline diabetes | 64,844 (36.3%) | 16,174 (36.3%) | 20,143 (36.1%) | 9,707 (29.3%) | 9,558 (28.9%) | 28,922 (29.1%) |
| Baseline congestive heart failure | 25,905 (14.5%) | 6,443 (14.4%) | 8,019 (14.4%) | 8,396 (25.4%) | 8,747 (26.5%) | 26,180 (26.4%) |
| Baseline liver disease | 16,672 (9.3%) | 4,214 (9.4%) | 5,349 (9.6%) | 6,469 (19.6%) | 6,541 (19.8%) | 19,606 (19.8%) |
| Surgical service | 41,673 (23.4%) | 10,367 (23.2%) | 13,035 (23.4%) | 5,773 (17.5%) | 5,666 (17.2%) | 16,815 (16.9%) |
| Admitted to ICU | 13,075 (7.3%) | 3,346 (7.5%) | 4,180 (7.5%) | 2,753 (8.3%) | 2,917 (8.8%) | 8,167 (8.2%) |
| Length of stay (days) | 5.3 (12.2) | 5.3 (12.5) | 5.4 (14.2) | 6.6 (7.8) | 6.7 (8.6) | 6.6 (8.3) |
| Number of 6-hour windows ** | 15.1 (9.0) | 15.1 (8.9) | 15.1 (9.0) | 18.4 (8.6) | 18.3 (8.7) | 18.3 (8.7) |
Statistics presented: mean (SD); n (%)
Calculated based on CKD-EPI Creatinine Equation (2021)
Calculated based on a maximum of 7-day hospitalization stay
The incidence of AKI differed in two cohorts and in individual sex groups (Extended Table 1). Among patients without AKI on presentation to the hospital, 10.4% (25,978/250,103) developed AKI during their hospitalization at the VA, whereas at UM, AKI occurred in 16.1% (26,529/164,774) of hospitalizations. Male patients were more likely to experience AKI than females in both cohorts (10.6% vs. 5.6% at the VA, and 18.4% vs. 13.8% at UM).
While the model was trained on all windows (including those in which AKI had already occurred), the model was evaluated on only those windows in which patients had not yet achieved the outcome. At the 6-hour window level, the incidence of new-onset AKI in the test set was 3.53% at the VA and 3.76% at UM (Table 2).
Table 2.
Model performance (AUC) of the original VA model at VA and UM, by outcome stage, by sex
| Outcome | VA Test AUC (95% Cl) | UM Test AUC (95% Cl) | ||||
|---|---|---|---|---|---|---|
| All | Female | Male | All | Female | Male | |
| Multiclass | ||||||
| Original VA model | 0.9742(0.9718, 0.9770) | 0.9691(0.9413, 0.9846) | 0.9744(0.9721, 0.9777) | 0.8685(0.8644, 0.8726) | 0.8689(0.8612, 0.8738) | 0.8680(0.8620, 0.8740) |
| AKI-1+ | ||||||
| Incidence | 3.53%(23,957/678,516) | 2.00%(745/37,226) | 3.62(23,212/641,290) | 3.76%(58,382/1,551,354) | 3.18%(24,585/772,665) | 4.34%(33,797/778,689) |
| Original VA model | 0.8196(0.8168, 0.8223) | 0.7943(0.7770, 0.8116) | 0.8194(0.8166, 0.8222) | 0.8469(0.8453, 0.8484) | 0.8477(0.8453, 0.8501) | 0.8439(0.8419, 0.846) |
| AKI-2+ | ||||||
| Incidence | 0.41 %(3,277/806,465) | 0.34%(139/40,855) | 0.41 %(3,138/765,610) | 0.86%(15,076/1,753,474) | 0.75%(6,472/857,809) | 0.96%(8,604/895,665) |
| Original VA model | 0.7741(0.7656, 0.7825) | 0.7636(0.7191, 0.8080) | 0.7749(0.7663, 0.7835) | 0.6550(0.6494, 0.6606) | 0.6504(0.6419, 0.6590) | 0.6622(0.6549, 0.6695) |
| AKI-3+ | ||||||
| Incidence | 0.22%(1,775/821,316) | 0.21 %(88/41,443) | 0.22%(1,687/779,873) | 0.39%(6,976/1,790,447) | 0.32%(2,780/875,621) | 0.46%(4,196/914,826) |
| Original VA model | 0.8341(0.8248, 0.8433) | 0.7111(0.6520, 0.7703) | 0.8393(0.8300, 0.8486) | 0.7981(0.7919, 0.8044) | 0.7627(0.7518, 0.7737) | 0.8271(0.8198, 0.8345) |
| AKI-3D | ||||||
| Incidence | 0.11 %(940/839,964) | 0.15%(61/42,071) | 0.11 %(879/797,893) | 0.08%(1,412/1,817,604) | 0.07%(586/887,574) | 0.09%(826/930,030) |
| Original VA model | 0.9497(0.9429, 0.9565) | 0.8927(0.8251, 0.9602) | 0.9537(0.9487, 0.9588) | 0.9558(0.9507, 0.9609) | 0.9560(0.9480, 0.9641) | 0.9550(0.9483, 0.9618) |
Reproducibility of the DeepMind AKI Model
Among eligible 6-hour windows (those in which the outcome had not already occurred), our gradient-boosted decision tree (GBDT) model predicted any AKI in the next 48 hours with an AUC of 82.0% (95% CI 81.7%, 82.2%) in the VA test set, which was lower than DeepMind’s observed AUC for a similar GBDT of 88.9% (95% CI 88.6%, 89.2%). The rationale behind our selection of a GBDT model is provided in the Methods. The model’s AUCs for AKI stages 2+, 3+, and 3D were 77.4%, 83.4%, and 95.0%, respectively (full details in Table 2). The performance substantially varied between VA hospitals in the test set, with AUCs ranging from 61.5% to 98.5%, suggesting that even a high-performing model may not generalize across all VA sites (Extended Figure 1). Overall, the model was well-calibrated for all levels of AKI (Extended Figure 2a).
However, the model overestimated the risk of AKI 1+ in females as compared to males (Extended Figure 2a). The model also had worse discrimination in females when predicting AKI 3+ and 3D, with AUCs of 71.1% for AKI 3+ (as compared to 83.9% in males) and 89.3% for AKI 3D (as compared to 95.4% in males).
Generalizability of the AKI Model at UM
Among eligible windows, the GBDT model predicted any AKI in the next 48 hours in the UM test set with an AUC of 84.7% (95% CI 84.5%, 84.8%), which was higher than the AUC of 82.0% we observed in the VA test set. In the UM test set, the model’s AUCs for AKI stages 2+, 3+, and 3D were 65.5%, 79.8%, and 95.6%, respectively (full details in Table 2). While the model appeared to generalize well overall, there was a marked difference in AUC for stage 2+ AKI (65.5% at UM vs. 77.4% at the VA).
The model generally overestimated the risk of AKI at all stages, and this finding was worse in females as compared to males (Extended Figure 2b). Also, similar to our finding in the VA test set, the model performed worse in females when predicting AKI stage 3+, with an AUC of 76.3% in females as compared to 82.7% in males (Table 2)
Understanding the Reasons for Differential Performance by Sex
Because the VA population consists of 94% males, one potential reason for the worse performance observed in females is the relatively small number of females who progressed to AKI stage 3+ (n = 94 in entire VA cohort, Extended Table 1). If the worse performance in females was primarily attributable to the lower number of events, then updating the model using data from a sex-balanced population should improve the model’s performance in females. Thus, starting with our 160-tree GBDT model, we continued to further train it using the UM training cohort (in which 50% are females), with early stopping determined based on the UM validation cohort (as described in the Methods). This process added 10 trees to the original model, and we refer to this updated model as the “extended model” to highlight that this 170-tree model contains the original 160 trees within it, and is thus an extension of the original model.
Remarkably, this small extension to the original model improved the performance in the UM test set both overall and between sexes (Table 3). Whereas the original model had poorly predicted AKI 2+ at UM (AUC 65.5%), the extended model performs much better on the UM test set (AUC 81.8%). At AKI stage 3+, where the original model exhibited the largest difference between females and males (AUC 76.3% vs. 82.7%), the performance was much more similar in the extended model (AUC 85.5% for females and 88.6% for males). The overall calibration was also better in the UM test set (Extended Figure 3). While this mechanism of updating a base model in a local population is a promising approach to correcting issues related to model generalizability, the small sample of females used to train the original model does not entirely explain the differential performance by sex. When the extended model was re-evaluated on the VA test set, its performance was worse in females, with an AUC for AKI 3+ of 69.1% in females as compared to 82.8% in males (Extended Table 2).
Table 3.
Model performance (AUC) of the extended VA model at UM, by outcome stage, by sex
| Outcome | UM Test AUC (95% CI) | ||
|---|---|---|---|
| All | Female | Male | |
| Multiclass | |||
| Extended VA model | 0.8780(0.8749, 0.8826) | 0.8757(0.8697, 0.8813) | 0.8795(0.8752, 0.8850) |
| AKI-1+ | |||
| Extended VA model | 0.8523(0.8508, 0.8538) | 0.8535(0.8512, 0.8559) | 0.8490(0.8470, 0.8510) |
| AKI-2+ | |||
| Extended VA model | 0.8181(0.8138, 0.8224) | 0.8135(0.8070, 0.8200) | 0.8236(0.8179, 0.8292) |
| AKI-3+ | |||
| Extended VA model | 0.8722(0.8666, 0.8778) | 0.8554(0.8461, 0.8647) | 0.8858(0.8790, 0.8927) |
| AKI-3D | |||
| Extended VA model | 0.9346(0.9258, 0.9433) | 0.9402(0.9271, 0.9532) | 0.9297(0.9178, 0.9415) |
The extended model’s worse performance in the VA population may be attributable to differences in care patterns between females and males at the VA, or due to differences in female patient characteristics at the VA and UM. As compared to females at the VA, females at UM were older (UM: 58.4 [SD 19.1]; VA: 55.2 years [SD 14.6]), less diverse (UM: 81.4% White; VA: 58.8% White;), and were more likely to have baseline chronic kidney disease (eGFR < 30 at UM: 3.1%; VA: 2.0%) and congestive heart failure (UM: 23.2%; VA: 6.5%), but had a similar body mass index (UM: 29.1 [SD 7.4]; VA: 30.7 [SD 7.4]) and diabetes mellitus (UM: 26.2%; VA: 24.0%).
Differences in model performance were not observed between racial groups (Extended Tables 3 and 4), potentially because the VA population includes a relatively high proportion of Black patients.
DISCUSSION
In our study, we were able to partially reproduce the results reported by the DeepMind team in their development and validation of an AKI model in a population of US Veterans drawing from over 100 VA hospitals. We observed an AUC for predicting any AKI in the next 48 hours of 82.0% in a national VA cohort, which was lower than the AUC of 88.9% for a similar GBDT described in the DeepMind paper. At lower stages of AKI, we found the model to be miscalibrated in females, which aligns with the DeepMind team’s finding of a lower sensitivity in females as compared to males. However, we also uncovered a lower AUC in females as compared to males in higher stages of AUC, a difference that was not evaluated in the DeepMind study. This difference persists when the VA-trained model is transported to a large academic hospital. While further training on a sex-balanced cohort improved the discrepancy in model performance at the academic hospital, it worsened the disparity in model performance at the VA, suggesting that the lower performance in females is related to reasons other than simply a low number of events at the VA.
Our finding that a modeling strategy relying on only VA data results in worse performance in females is troubling. Had the differences been attributable solely to the small sample size of females, these differences should have been correctable by updating the model using information from a sex-balanced cohort as was present at our academic hospital. However, updating the model actually worsened this disparity at the VA, which suggests that other factors such as practice patterns or patient characteristics for females treated at the VA may account for this difference in the VA context.
Our work has limitations that may affect our findings. While we replicated many aspects of the DeepMind study, including similar inclusion and exclusion criteria, a similar modeling strategy, and the inclusion of many of the same predictors (see Supplemental Table 1), we were unable to include International Classification of Diseases, Ninth Revision (ICD-9) codes and clinical note headings as predictors in our model due to computational constraints within the VA computing environment, a limitation not faced by the DeepMind team due to their use of a de-identified dataset in a proprietary computing environment. Billing codes also undergo periodic updates, which can result in models becoming outdated. By the time the DeepMind study was published, ICD-9 codes had been replaced with ICD-10 codes, and the implementation of ICD-11 codes is already underway.19 ICD-9 codes were known to be an important component of the DeepMind AKI model. For example, “malignant neoplasm of [the] kidney” was reported as one of the top features in the original study,1 possibly because this billing code foreshadows an imminent nephrectomy or renal artery embolization.
Our work also has important implications. While sex and gender inequalities in healthcare machine learning models have long been suspected, we provide definitive evidence that this phenomenon can and does occur, and that it is complex, not simply explained away by a low sample size. We also show promising results that some of these differences attributable to models trained in imbalanced populations can be mitigated through further training on a balanced population, which means that base models trained in a large population may be capable of being fine-tuned through a relatively simple mechanism in tree ensemble models. In the interest of promoting transparency, we have made our original and extended models publicly available.
METHODS
Study Cohorts
Our study used data from two cohorts: a national VA cohort drawing on data from 118 VA hospitals, and a University of Michigan (UM) cohort.
National VA cohort.
We collected clinical data on all adult patients admitted at a VA hospital between October 1, 2016 to September 30, 2017. Starting with a cohort of 280,985 US Veterans hospitalized between October 1, 2016 and September 30, 2017, we excluded patients who did not have a creatinine checked during their stay (defined in the next section Data Collection and Processing), had pre-existing end stage renal disease (ESRD), and those who had a baseline creatinine >4.0 mg/dL (because they may have had pre-existing AKI stage 3). Only the first hospitalization for each patient was included in the analysis. The final VA cohort consisted of 278,813 patients, which was randomly divided into training (64%), validation (16%) and test (20%) sets at the patient level.
UM cohort.
We collected clinical data from all adult patients admitted to UM from January 1, 2016 to December 31, 2020 were included. The same exclusion criteria used in the VA cohort were applied to the UM cohort, though all hospitalizations (not only the first) were included. The final UM cohort consisted of 165,359 hospitalizations. Anticipating the need for updating of the VA model at UM, we randomly selected 60% of hospitalizations (sampled at the patient level) for the test set, and set aside the remaining 40% for model updating, which was divided equally into a training (20%) and validation (20%) set.
Predictor Variables
We collected both fixed predictors (i.e., baseline variables) and time-varying predictors (i.e., variables measured on a repeated basis during a hospitalization) in both cohorts.
Fixed predictors included age, height, weight, body mass index (BMI), 17 comorbidities, admission to a surgical service, intensive care unit (ICU) status, baseline serum creatinine (sCr), all of which were captured at the time of admission. Age was top-coded at 89 years. Baseline height and weight were calculated as the mean value from the 3 years preceding admission for VA patients, and the most recent value within the past year for UM patients. If no recent value was identified for UM patients, the first inpatient measurement was used. Height and weight measurements were converted into inches and pounds, respectively, and extreme values were removed. Baseline BMI was calculated using the baseline height and baseline weight. Comorbidities were calculated by Charlson Comorbidity Index using one-year data prior to admission for VA patients and from the current encounter for UM patents.20 Baseline sCr was determined by the following order of preference: (1) mean outpatient sCr between 7–365 days prior to admission, (2) within 7 days prior to admission, and (3) first inpatient sCr test for VA patients or first documented sCr value within 24 hours of admission for UM patients.
Time-varying predictors consisted of inpatient vital signs, laboratory test results and administration of medications. Twenty-six laboratory testing components (serum albumin, alkaline phosphatase, alanine aminotransferase, aspartate transaminase, total and direct bilirubin, blood urea nitrogen, serum calcium, carbon dioxide, serum chloride, serum glucose, high-density lipoprotein cholesterol, hematocrit, hemoglobin A1c, hemoglobin, international normalized ratio, low-density lipoprotein cholesterol, microalbumin-to-creatinine ratio, serum phosphate, platelet count, serum potassium, serum creatinine, serum sodium, total cholesterol, triglyceride, and total white blood cell count) were selected due to universal use across different health systems. Eight vital signs (inpatient weight, systolic blood pressure, diastolic blood pressure, respiratory rate, temperature, pulse, blood oxygen level and central venous pressure) were pulled regardless of the frequency of measurement. Administration of medications was examined for eleven drug classes (aminoglycosides, sympathomimetics, beta blockers, alpha blockers, calcium channel blockers, antilipemic agents, loop diuretics, angiotensin-converting enzyme inhibitors, angiotensin II Inhibitors, non-ionic contrast media, and nonsalicylate antirheumatic non-steroidal anti-inflammatory drugs) as opposed to individual medications.
Data Preprocessing and Feature Engineering
Physiologically infeasible values (e.g., due to a laboratory error) were excluded. Microalbumin-to-creatinine ratios were set to 0 when values were reported only in a text field based on the observation that the text fields reported such values as being below the detectable range. Data elements were time-stamped using the time when values became available to the EHR (i.e., the observation time). The description of variables, the associated units, and valid ranges are shown in Supplemental Table 1.
After extracting the fixed and time-varying predictors, we captured patient states at 6-hour intervals beginning with the time of admission for each patient in a manner similar to the DeepMind AKI study. Patient states were captured up until the final creatinine value, discharge, or death, and truncated at 7 days of hospitalization due to computational constraints. For each 6-hour interval, summary statistics (length, minimum, mean, median, maximum) were calculated for the preceding 48 hours divided into 6-hour windows for vital signs and laboratory test results. Using these summary statistics, additional variables were created based on clinical relevance: the ratio of the most recent maximum sCr to baseline sCr, the difference between the most recent maximum sCr and baseline sCr, and the ratio of most recent maximum BUN to most recent maximum sCr. These three sCr-based predictors, time (hours) from admission, current AKI stage, plus the summary statistics of temporal predictors in the given windowed lookback period, together with the fixed predictors, were used as the full set of 1,467 predictors. The preparation of predictors at VA and UM followed the same procedures with the only exception for CVP predictors. CVP information is not available at UM. Hence, CVP-based predictors were manually added to the predictor set and were all set to missing. The number of administered medications was calculated for the preceding week (7 days) divided into 24-hour sliding windows. More details can be found in Supplementary Table 1. A visual representation of the feature engineering process is shown in Figure 1.
Figure 1. Representation of the Electronic Health Record Data for the Proposed Model.
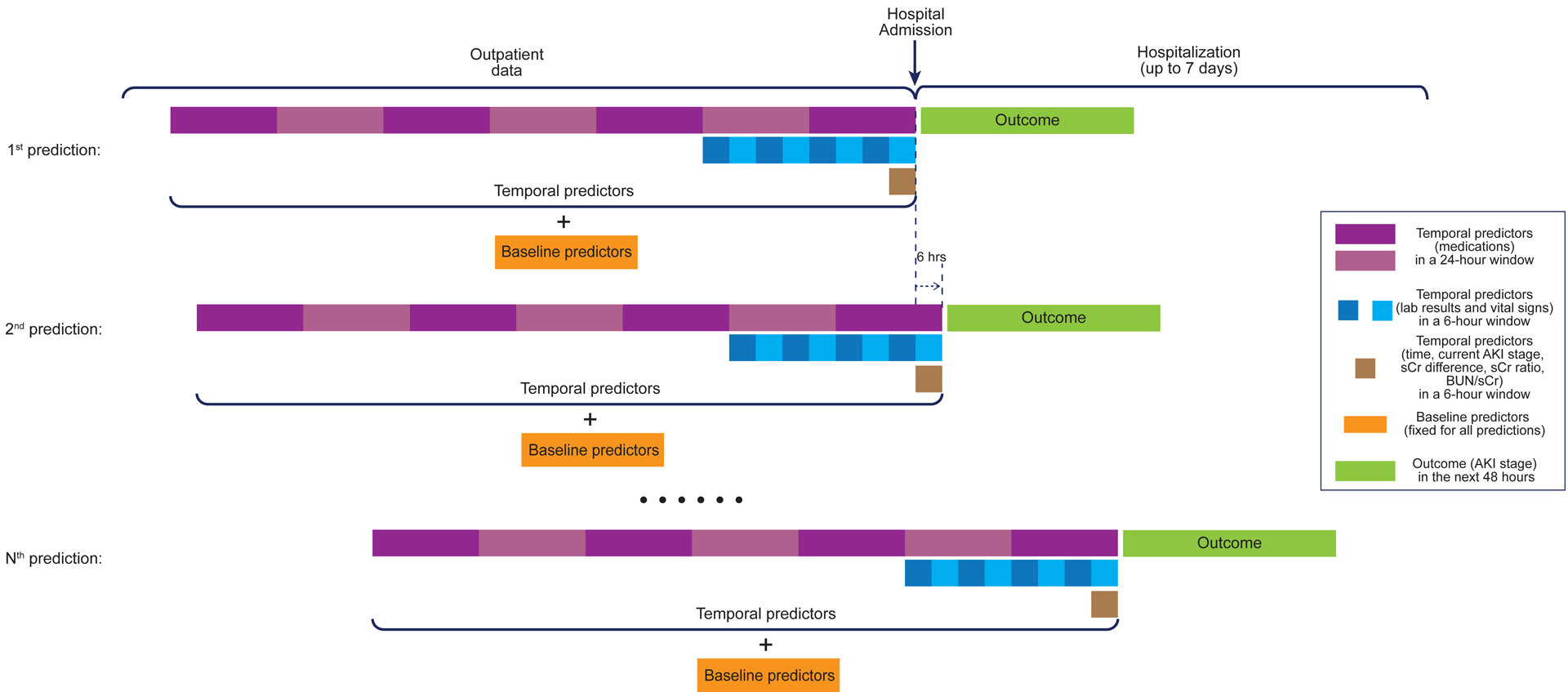
Representation of the electronic health record (EHR) data for our model. EHR data available for each hospitalization were prepared to make an acute kidney injury (AKI) risk prediction every six hours from the time of hospital admission and up to 7 days from admission. For each prediction, baseline predictors (orange blocks) and temporal predictors (purple blocks: medications; blue blocks: lab results and vital signs; brown blocks: time, current AKI stage, serum creatinine [sCr] difference from baseline sCr, sCr ratio to baseline sCr, and BUN to sCr ratio) were prepared and used together to estimate the outcome (AKI stage) in the next 48 hours (green blocks).
Outcome Definition
AKI was defined and staged for severity according to the Kidney Disease: Improving Global Outcomes (KDIGO) international guidelines.21 The outcome was calculated on a rolling basis at 6-hour intervals by comparing the maximum sCr value in the 48-hour prediction window with the baseline sCr. Stage 1 AKI was defined as a sCr level increase ≥ 0.3 mg/dL, but less than twice the baseline sCr or an increase of 1.5 times baseline. Stage 2 AKI reflected an increase of 2 to 3 times the baseline, and stage 3 AKI was an sCr level increase greater than 3 times baseline or an increase to ≥4.0 mg/dL. Stage 3D was determined based on the need for dialysis, where the time of first dialysis was determined based on diagnosis, procedure, and clinic stop codes during hospitalization at VA, and using procedure codes at UM. Thus, at every 6-hour interval at which patient states were captured, outcomes were defined as one of five classes based on the 48-hour prediction window: no AKI, AKI stage 1, AKI stage 2, AKI stage 3, and AKI stage 3D. While models were trained using this multinomial outcome, results reported by AKI stages were grouped according to level of severity. For example, AKI stage 1+ is used to refer to any AKI stage, and AKI stage 2+ refers to AKI stage 2 or greater (including stages 3 and 3D).
Model Development
In the original study, Tomasev et. al. selected a “simple” recurrent neural network (RNN) as their primary model, which achieved an AUC of 92.1% in their test set. Tomasev and colleagues also evaluated 11 other neural network architectures, 2 tree ensembles, and a logistic regression model, all of which performed better than prior studies. For example, the gradient boosted trees (GBDT) achieved an AUC of 88.9%, which still represents state-of-the-art performance. While Tomasev et. al. had access to a de-identified dataset, which allowed them to use DeepMind’s computing infrastructure to train deep learning models, our team was restricted to using the VA’s VINCI platform, which lacks the graphical processing units needed to efficiently train deep learning models. Thus, we opted to reproduce the GBDT model from the Tomasev study.
The GBDT model was trained on the VA training set to predict AKI stage in the next 24 hours as a multinomial outcome (i.e. “No AKI”, “AKI stage 1”, “AKI stage 2”, “AKI stage 3”, “AKI stage 3D”) using 1,467 predictors at each 6-hour step with a maximum of 1000 trees and a maximum depth of 5. The VA validation set was used to determine the need for early stopping based on an improvement in log loss lower than 0.0005 on 5 consecutive rounds based on a moving average calculated after every 10 trees. The categorical predictors were reordered by the mean response of each level for more efficient training. Internally, a separate one-versus-all tree was trained for each outcome class. Using a learning rate of 0.1, the trained VA AKI model stopped training at 160 trees (internally represented as 160 trees per class). Lower learning rates (0.01 and 0.001) produced more trees (because more trees were needed to achieve convergence) but achieved similar results (i.e., AUC), so will not be presented here.
During model training, optimal binary splits were determined by minimizing the error using non-missing data. After a variable split was determined, missing values for that variable were assigned to the direction minimizing the error. When generating predictions, missing values followed the assigned direction.
Model Evaluation
The performance of the GBDT model was evaluated in both the VA test set and the UM test set. The model discrimination was assessed by using the area under the receiver operating characteristic (AUC). The AUC was reported both as a multinomial outcome using Hand and Till’s method,22 and as a series of binary AUCs where at-risk individuals were evaluated on their risk of progression to a higher AKI stage. For example, patients without any AKI to date were evaluated on their risk of developing any AKI (i.e., stage 1 or greater), and patients with no AKI or AKI stage 1 or evaluated on their risk of developing AKI stage 2 or greater, and so on. The 95% confidence intervals were generated using 200 bootstrap resamples for the multiclass AUCs and DeLong’s method for binary AUCs.23 Our primary finding is the AUC calculated when treating each prediction independently in its ability to predict AKI in the next 48 hours, which is closely comparable to the way AUC was calculated in the DeepMind study. To aid with interpretability, we also report hospitalization-level AUCs, which use the maximum predicted probability for each binary outcome over the course of the hospitalization to assess the quality of the predictions at the hospitalization-level (after excluding predictions made after the outcome has occurred). The rationale for this approach has been previously described.24,25 We also evaluated model calibration by comparing deciles of predicted probabilities with observed risk.
Because the make-up of the VA population is different from other hospitals (e.g., 94% male), we examined model performance across sexes and racial groups.
Updating the Model with UM Data
Given prior concerns that models trained at the VA may not generalize to broader populations, we updated the VA model using a UM training/validation set that was set aside prior to model evaluation (as described previously in Study Cohorts). Starting with the original 160-tree GBDT model trained only in the VA population, we continued to train it using the UM training set, with a similar early stopping strategy based on a lack of log loss improvement of 0.0001 after 5 consecutive rounds in the UM validation set. This updated model (which we refer to as the “extended model” to indicate that it includes a portion of the original VA model) added 10 additional trees on top of the original 160 trees, resulting in a total of 170 trees. The updated model was then evaluated in both the UM and VA test sets.
Variable Importance
We assessed variable importance using each variable’s squared influence within the GBDT algorithm aggregated over the tree ensemble.26 Variable importance for the original and extended model are provided in Extended Figure 4.
Software
All data processing and analyses were performed using R 4.0.5 at the VA and R 3.6.1 at UM.27 Transformation of time-series data was performed using the Grammar of Prediction (gpmodels) R package.28 h2o version 3.32.1.3 was used to fit the GBDT model.29 We did not use XGBoost (which was used in the DeepMind study) because while h2o and XGBoost achieve comparable performance for their respective GBDT implementations, h2o’s implementation is more memory-efficient,30 which was a requirement when using the VA’s VINCI computing platform.
Extended Data
Extended Figure 1. Model performance (AUC) of the original VA model at each VA hospital.
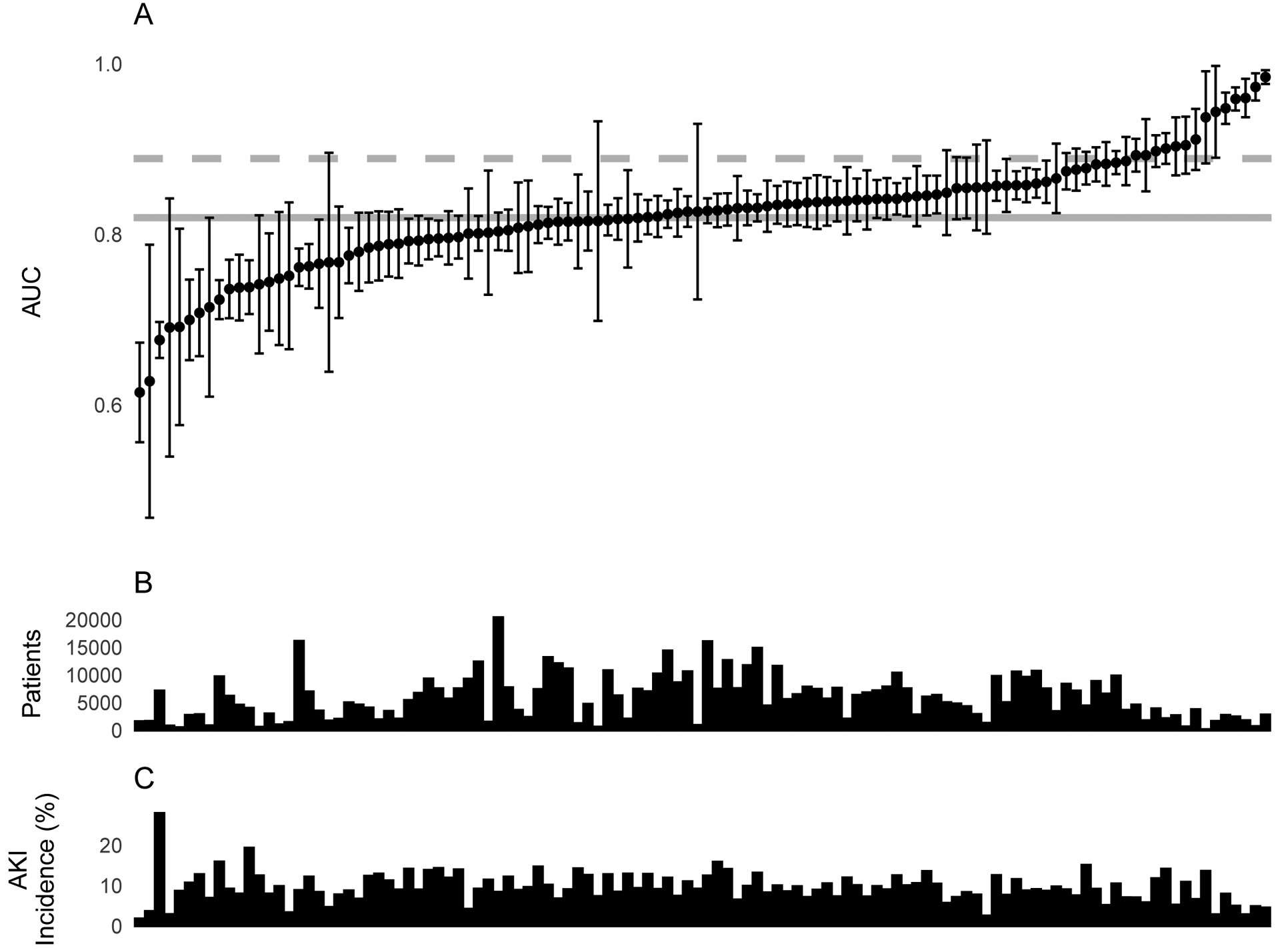
Model performance of the original model at each VA hospital in the test set, along with characteristics of each VA hospital. A. Model performance with respect to area under the curve (AUC) with DeLong 95% CI of the original VA model for predicting AKI-1+ at each VA hospital. B. Number of patients (after excluding those with AKI 1+ at baseline) at each VA hospital. C. Hospitalization-level AKI-1+ incidence in the test set (after excluding those with AKI 1+ at baseline) at each VA hospital. A total of 114 VA hospitals are shown in the figure. The results of the remaining four VA hospitals are not shown due to small cohort sizes.
Extended Figure 2. Calibration of the original VA model a) VA test set b) UM test set.
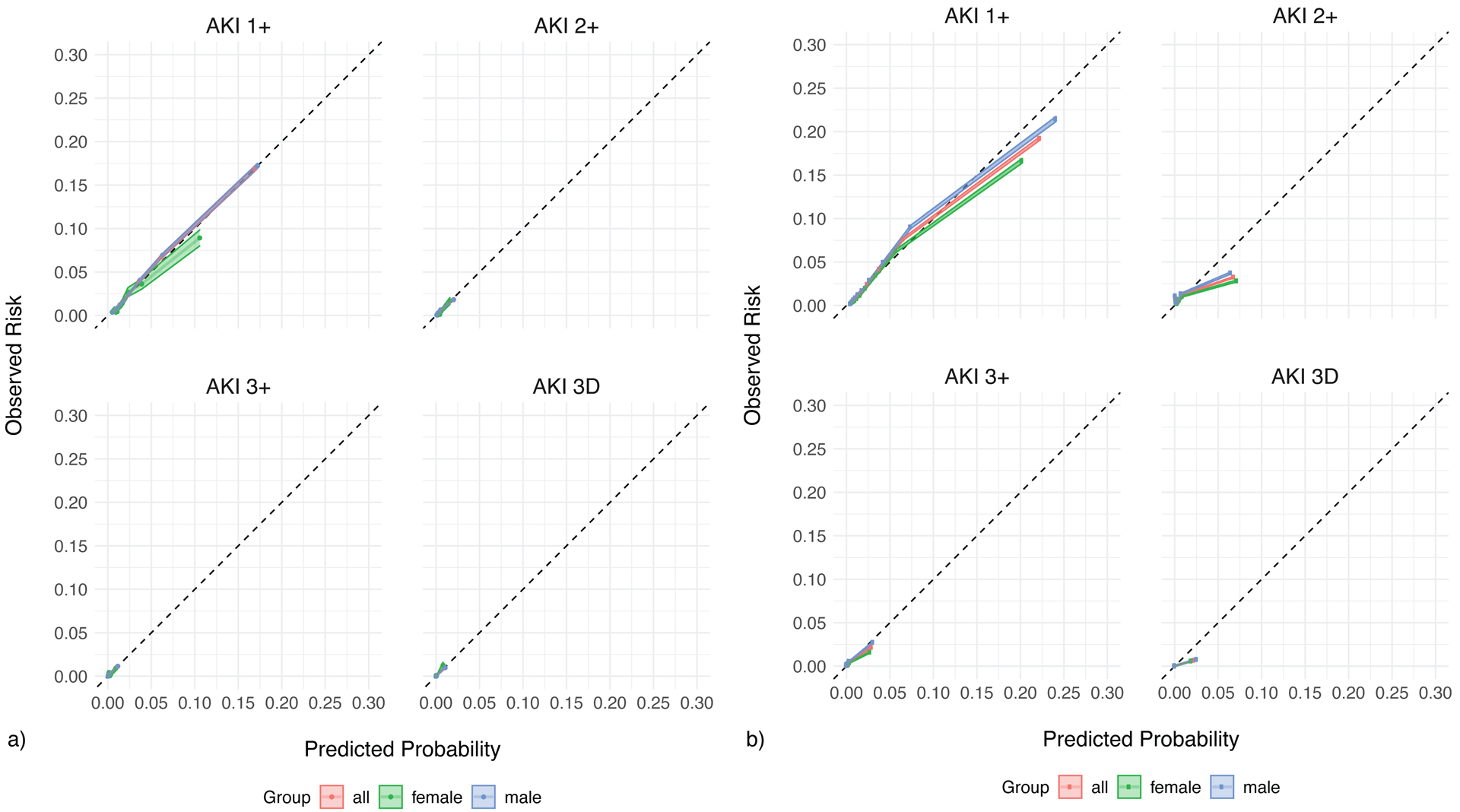
The calibration of the original model on the a) VA test set and b) UM test set. The predicted probabilities (deciles) are plotted against the observed probabilities with 95% confidence intervals. The diagonal line demonstrates the ideal calibration. The model calibration is examined for all patients (red), females only (green), and males only (blue).
Extended Figure 3. Calibration of the extended VA model at UM.
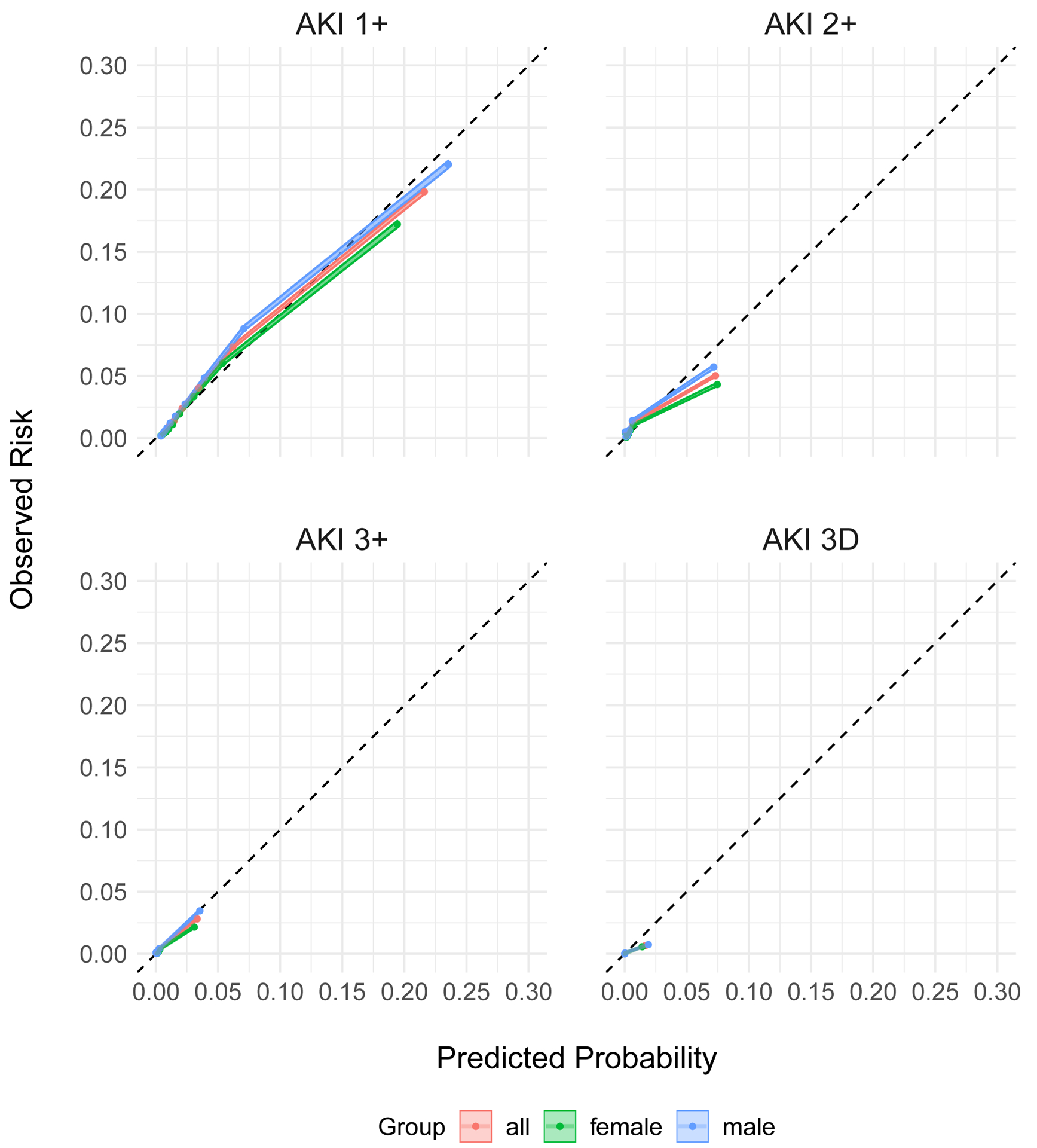
The calibration of the extended model in the UM test set. The predicted probabilities (deciles) are plotted against the observed probabilities with 95% CI. The diagonal line demonstrates the ideal calibration. The model calibration is examined for all patients (red), females only (green), and males only (blue).
Extended Figure 4. Predictor importance plot of the original and extended VA model.
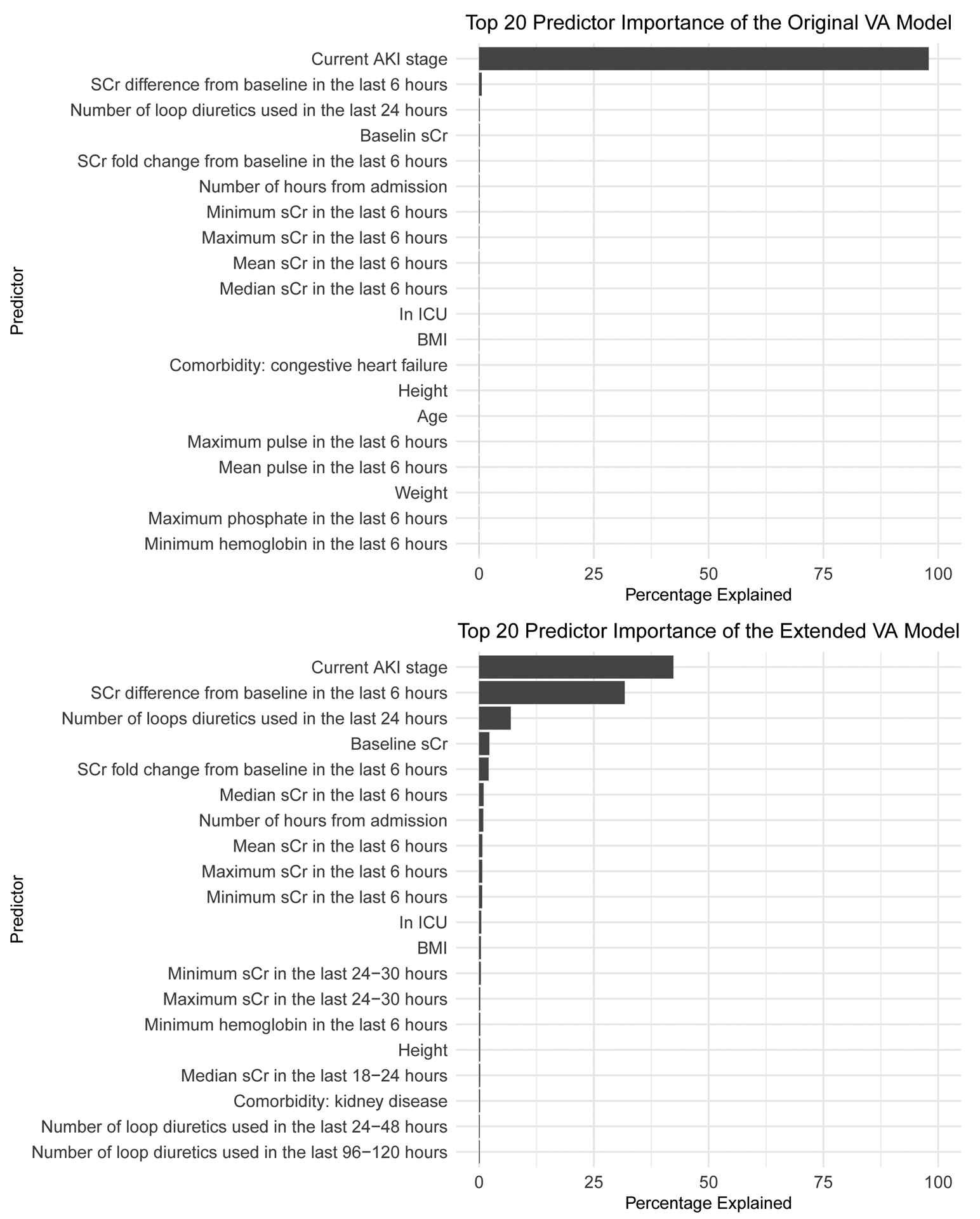
Top 20 important predictors of the original VA model (top) and the extended VA model (bottom). Predictors are ranked by their relative importance and expressed as a percentage.
Extended Table 1.
AKI incidence in the VA and UM cohorts, by acute kidney injury stage, by sex
| Outcome | VA (all) | UM (all) | ||||
|---|---|---|---|---|---|---|
| All | Female | Male | All | Female | Male | |
| Hospitalization level | ||||||
| AKI 1+ | 10.39%(25,978/250,103) | 6.04%(890/14,741) | 10.66%(25,088/235,362) | 16.10%(26,529/164,774) | 13.79%(11,307/82,009) | 18.39%(15,222/82,765) |
| AKI 2+ | 1.52%(4,127/271,850) | 1.11 %(171/15,463) | 1,54%(3,956/256,387) | 3.93%(6,494/165,192) | 3.55%(2,917/82,184) | 4.31%(3,577/83,008) |
| AKI 3+ | 0.82%(2,244/275,030) | 0.60%(94/15,613) | 0.83%(2,150/259,417) | 1.76%(2,914/165,276) | 1.46%(1,198/82,227) | 2.07%(1,716/83,049) |
| AKI 3D | 0.31 %(278,799) | 0.13%(21/15,758) | 0.33%(856/263,041) | 0.23%(388/165,338) | 0.18%(152/82,260) | 0.28%(236/83,078) |
| Outcome | VA (all) | UM (all) | ||||
| All | Female | Male | All | Female | Male | |
| Multiclass predictions, every 6 hours | N = 4,213,375 | N = 215, 923 | N = 3,997,452 | N = 3,033,165 | N = 1,478,583 | N = 1,554,582 |
| No AKI | 3,277,669(77.8) | 185,228(85.8) | 3,092,441(77.4) | 2,723,535 (89.8) | 1,350,169(91.3) | 1,373,366(88.3) |
| AKI-1 | 737,264(17.5) | 22,690(10.5) | 714,574(17.9) | 231,626(7.6) | 94,951(6.4) | 136,675(8.8) |
| AKI-2 | 87,500(2.1) | 3,801(1.8) | 83,699(2.1) | 39,700(1.3) | 18,614(1.3) | 21,086(1.4) |
| AKI-3 | 93,376(2.2) | 3,757(1.7) | 89,619(2.2) | 30,444(1.0) | 11,912(0.8) | 18,532(1.2) |
| AKI-3D | 17,566(0.4) | 447(0.2) | 17,119(0.4) | 7,860(0.3) | 2,937(0.2) | 4,923(0.3) |
| Outcome | VA (all) | UM (all) | ||||
| All | Female | Male | All | Female | Male | |
| Binary predictions for each stage among patients who have not reached that stage, every 6 hours | ||||||
| AKI 1 + | 3.55%(120,255/3,386,277) | 2.22%(4,191/189,054) | 3.63%(116,064/3,197,223) | 3.76%(97,197/2,583,269) | 3.18%(40,973/1,288,611) | 4.34%(56,224/1,294,658) |
| AKI 2+ | 0.41%(16,323/4,029,154) | 0.35%(722/208,549) | 0.41%(15,601/3,820,605) | 0.87% 25,286/2,921,450) | 0.76%(10,922/1,428,256) | 0.96%(14,364/1,493,194) |
| AKI 3+ | 0.21%(8,700/4,109,724) | 0.19%(404/212,042) | 0.21%(8,296/3,897,682) | 0.39%(11,767/2,983,230) | 0.31%(4,566/1,457,716) | 0.47%(7,201/1,525,514) |
| AKI 3D | 0.12%(5,116/4,200,925) | 0.06%(140/215,616) | 0.12%(4,976/3,985,309) | 0.08%(2,367/3,027,672) | 0.06%(925/1,475,646) | 0.09%(1,442/1,549,659) |
Extended Table 2.
Model performance (AUC) of the extended VA models at VA, by outcome stage, by sex
| Outcome | VA Test AUC(95% CI) | ||
|---|---|---|---|
| All | Female | Male | |
| Multiclass | |||
| Extended VA model | 0.9530(0.9501, 0.9574) | 0.9474(0.9208, 0.9653) | 0.9531(0.9506, 0.9579) |
| AKI-1+ | |||
| Extended VA model | 0.8178(0.8150, 0.8178) | 0.7892(0.7717, 0.8067) | 0.8178(0.8150, 0.8206) |
| AKI-2+ | |||
| Extended VA model | 0.7593(0.7507, 0.7679) | 0.7432(0.6976, 0.7888) | 0.7602(0.7515, 0.7690) |
| AKI-3+ | |||
| Extended VA model | 0.8230(0.8131, 0.8329) | 0.6907(0.6318, 0.7495) | 0.8284(0.8184, 0.8384) |
| AKI-3D | |||
| Extended VA model | 0.9355(0.9261, 0.9450) | 0.8925(0.8252, 0.9599) | 0.9385(0.9298, 0.9472) |
Extended Table 3.
Model performance (AUC) of the original and extended VA models at VA, by outcome stage, by race
| Outcome | VA Test AUC(95% Cl) | ||||
|---|---|---|---|---|---|
| All | Caucasian | African American | Other | Unknown | |
| Multiclass | |||||
| Original VA model | 0.9742(0.9718, 0.9770) | 0.9729(0.9686, 0.9759) | 0.9758(0.9704, 0.9812) | 0.9796(0.9740, 0.9842) | 0.9714(0.9603, 0.9809) |
| Extended VA model | 0.9530(0.9501, 0.9574) | 0.9506(0.9450, 0.9544) | 0.9563(0.9498, 0.9633) | 0.9591(0.9528, 0.9664) | 0.9526(0.9375, 0.9638) |
| AKI-1+ | |||||
| Incidence | 3.53%(23,957/678,516) | 3.47%(16,115/463,936) | 3.78%(4,825/127,634) | 3.43%(2,017/58,830) | 3.56%(1,000/28,116) |
| Original VA model | 0.8196(0.8168, 0.8223) | 0.8217(0.8184, 0.8250) | 0.8109(0.8047, 0.8171) | 0.8174(0.8078, 0.8269) | 0.8277(0.8137, 0.8417) |
| Extended VA model | 0.8178(0.8150, 0.8206) | 0.8196(0.8162, 0.8230) | 0.8109(0.8046, 0.8171) | 0.8145(0.8048, 0.8241) | 0.8264(0.8124, 0.8404) |
| AKI-2+ | |||||
| Incidence | 0.41 %(3,277/806,465) | 0.36%(1,997/548,168) | 0.49%(761/155,339) | 0.57%(393/69,438) | 0.38%(126/33,520) |
| Original VA model | 0.7741(0.7656, 0.7825) | 0.7596(0.7485, 0.7707) | 0.7937(0.7767, 0.8107) | 0.8026(0.7820, 0.8233) | 0.8070(0.7625, 0.8514) |
| Extended VA model | 0.7593(0.7507, 0.7679) | 0.7463(0.7351, 0.7575) | 0.7815(0.7644, 0.7986) | 0.7752(0.7525, 0.7980) | 0.7898(0.7423, 0.8373) |
| AKI-3+ | |||||
| Incidence | 0.22%(1,775/821,316) | 0.19%(1,040/557,294) | 0.27%(424/159,121) | 0.34%(238/70,859) | 0.21%(73/34,042) |
| Original VA model | 0.8341(0.8248, 0.8433) | 0.8189(0.8067, 0.8312) | 0.8486(0.8281, 0.8691) | 0.8706(0.8524, 0.8889) | 0.8451(0.8042, 0.8861) |
| Extended VA model | 0.8230(0.8131, 0.8329) | 0.8103(0.7974, 0.8231) | 0.8375(0.8162, 0.8588) | 0.8442(0.8196, 0.8688) | 0.8418(0.7941, 0.8895) |
| AKI-3D | |||||
| Incidence | 0.11%(940/839,964) | 0.10%(567/568,043) | 0.14%(225/164,387) | 0.12%(88/72,834) | 0.17%(60/34,700) |
| Original VA model | 0.9497(0.9429, 0.9565) | 0.9500(0.9407, 0.9593) | 0.9332(0.9184, 0.9479) | 0.9696(0.9539, 0.9853) | 0.9684(0.9568, 0.9801) |
| Extended VA model | 0.9355(0.9261, 0.9450) | 0.9350(0.9221, 0.9479) | 0.9153(0.8940, 0.9366) | 0.9644(0.9490, 0.9797) | 0.9632(0.9506, 0.9758) |
Extended Table 4.
Model performance (AUC) of the original and extended VA models at UM, by outcome stage, by race
| Outcome | UM Test AUC(95% Cl) | ||||
|---|---|---|---|---|---|
| All | Caucasian | African American | Other | Unknown | |
| Multiclass | |||||
| Original VA model | 0.8685(0.8644, 0.8726) | 0.8697(0.8649, 0.8742) | 0.8625(0.8500, 0.8714) | 0.8689(0.8421, 0.8861) | 0.8576(0.8375, 0.8916) |
| Extended VA model | 0.8780(0.8749, 0.8826) | 0.8799(0.8755, 0.8850) | 0.8733(0.8622, 0.8806) | 0.8759(0.8529, 0.8936) | 0.8565(0.8385, 0.8885) |
| AKI-1+ | |||||
| Incidence | 3.76%(58,382/1,551,354) | 3.71 %(47,489/1,281,347) | 4.22%(7,389/174,932) | 3.49%(2,688/77,092) | 4.54%(816/17,983) |
| Original VA model | 0.8469(0.8453, 0.8484) | 0.8460(0.8443, 0.8477) | 0.8433(0.8390, 0.8476) | 0.8561(0.8491, 0.8631) | 0.8859(0.8762, 0.8957) |
| Extended VA model | 0.8523(0.8508, 0.8538) | 0.8514(0.8497, 0.8531) | 0.8488(0.8446, 0.8530) | 0.8624(0.8555, 0.8693) | 0.8905(0.8811, 0.8999) |
| AKI-2+ | |||||
| Incidence | 0.86%(15,076/1,753,474) | 0.83%(12,064/1,445,319) | 1.01 %(2,033/201,650) | 0.80%(686/86,207) | 1.44%(293/20,298) |
| Original VA model | 0.6550(0.6494, 0.6606) | 0.6519(0.6456, 0.6581) | 0.6535(0.6383, 0.6687) | 0.6680(0.6412, 0.6948) | 0.7646(0.7312, 0.7980) |
| Extended VA model | 0.8181(0.8138, 0.8224) | 0.8158(0.8110, 0.8205) | 0.8215(0.8101, 0.8330) | 0.8406(0.8210, 0.8601) | 0.8342(0.8040, 0.8644) |
| AKI-3+ | |||||
| Incidence | 0.39%(6,976/1,790,447) | 0.37%(5,451/1,475,447) | 0.50%(1,038/206,309) | 0.40%(350/87,868) | 0.66%(137/20,742) |
| Original VA model | 0.7981(0.7919, 0.8044) | 0.7925(0.7853, 0.7998) | 0.8187(0.804, 0.8333) | 0.8063(0.7776, 0.8349) | 0.8585(0.8190, 0.8979) |
| Extended VA model | 0.8722(0.8666, 0.8778) | 0.8763(0.8701, 0.8826) | 0.8518(0.8366, 0.8670) | 0.8626(0.8367, 0.8885) | 0.8980(0.8632, 0.9327) |
| AKI-3D | |||||
| Incidence | 0.08%(1,412/1,817,604) | 0.07%(981/1,496,642) | 0.12%(258/210,914) | 0.15%(134/89,093) | 0.19%(39/20,955) |
| Original VA model | 0.9558(0.9507, 0.9609) | 0.9546(0.9483, 0.9609) | 0.9584(0.9503, 0.9666) | 0.9540(0.9354, 0.9725) | 0.9581(0.9082, 1) |
| Extended VA model | 0.9346(0.9258, 0.9433) | 0.9375(0.9276, 0.9475) | 0.9332(0.9118, 0.9546) | 0.9299(0.9023, 0.9575) | 0.8748(0.7809, 0.9687) |
Extended Table 5.
Model performance (AUC) of the original and extended VA models at the hospitalization level at VA and UM, by outcome stage, by sex
| Outcome | VA Test AUC(95% Cl) | UM Test AUC(95% Cl) | ||||
|---|---|---|---|---|---|---|
| All | Female | Male | All | Female | Male | |
| AKI-1+ | ||||||
| Incidence | 10.30%(5,158/50,031) | 5.61 %(164/2,925) | 10.60%(4,994/47,106) | 16.10%(15,924/98,887) | 13.94%(6,848/49,112) | 18.23%(9,076/49,775) |
| Original VA model | 0.7729(0.7664, 0.7794) | 0.7700(0.7344, 0.8056) | 0.7708(0.7642, 0.7774) | 0.7227(0.7185, 0.727) | 0.7231(0.7166, 0.7295) | 0.7162(0.7104, 0.7220) |
| Extended VA model | 0.7710(0.7645, 0.7775) | 0.7643(0.7281, 0.8004) | 0.7690(0.7623, 0.7756) | 0.7393(0.7351, 0.7435) | 0.7419(0.7356, 0.7482) | 0.7311(0.7254, 0.7368) |
| AKI-2+ | ||||||
| Incidence | 1,50%(818/54,412) | 1,04%(32/3,069) | 1,53%(786/51,343) | 3.91 %(3,874/99,149) | 3.57%(1,758/49,228) | 4.24%(2,116/49,921) |
| Original VA model | 0.7708(0.7550, 0.7866) | 0.8195(0.7478, 0.8912) | 0.7689(0.7528, 0.7850) | 0.6871(0.6786, 0.6956) | 0.6705(0.6579, 0.6831) | 0.7010(0.6896, 0.7125) |
| Extended VA model | 0.7512(0.7354, 0.7671) | 0.7965(0.7240, 0.8691) | 0.7492(0.7330, 0.7654) | 0.7665(0.7585, 0.7746) | 0.7444(0.7321, 0.7566) | 0.7853(0.7748, 0.7959) |
| AKI-3+ | ||||||
| Incidence | 0.83%(457/54,989) | 0.71 %(22/3,100) | 0.84%(435/51,889) | 1.76%(1,745/99,199) | 1,50%(737/49,258) | 2.02%(1,008/49,941) |
| Original VA model | 0.8215(0.8023, 0.8406) | 0.8093(0.7136, 0.9050) | 0.8223(0.8028, 0.8419) | 0.7668(0.7541, 0.7795) | 0.7128(0.6924, 0.7332) | 0.8055(0.7898, 0.8213) |
| Extended VA model | 0.8159(0.7962, 0.8356) | 0.7913(0.6944, 0.8883) | 0.8171(0.7970, 0.8373) | 0.8287(0.8173, 0.8401) | 0.7832(0.7642, 0.8022) | 0.8613(0.8476, 0.8750) |
| AKI-3D | ||||||
| Incidence | 0.29%(163/55,745) | 0.29%(9/3,119) | 0.29%(154/52,626) | 0.23%(225/99,235) | 0.19%(96/49,276) | 0.26%(129/49,959) |
| Original VA model | 0.9577(0.9448, 0.9706) | 0.9227(0.7766, 1.0) | 0.9593(0.9490, 0.9697) | 0.9675(0.9587, 0.9762) | 0.9696(0.9589, 0.9803) | 0.9662(0.9526, 0.9799) |
| Extended VA model | 0.9455(0.9259, 0.9651) | 0.9231(0.7788, 1.0) | 0.9464(0.9275, 0.9653) | 0.9651(0.9561, 0.9741) | 0.9669(0.9555, 0.9784) | 0.9636(0.9439, 0.9774) |
Supplementary Material
COMPETING INTERESTS:
K.S. receives grant funding from Teva Pharmaceuticals and Blue Cross Blue Shield of Michigan for unrelated work, and serves on an advisory board for Flatiron Health.
This work was supported in part by the Veterans Health Association Innovation Program (contract number 36C10B18C2766).
Footnotes
ADDITIONAL INFORMATION: Supplementary Information is available for this paper.
Data Availability
This study used data from the national Veterans Health Administration’s Corporate Data Warehouse and the University of Michigan. Analyses were performed in secure locations within the VA and UM information systems, respectively. The data in this study are not publicly available because they contain protected health information, and restrictions apply to their use. The models trained in this study are available at https://github.com/ML4LHS/va-aki-model.
MAIN REFERENCES
- 1.Tomašev N et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature 572, 116–119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoste EAJ et al. Global epidemiology and outcomes of acute kidney injury. Nat. Rev. Nephrol 14, 607–625 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Wilson FP et al. Automated, electronic alerts for acute kidney injury: a single-blind, parallel-group, randomised controlled trial. Lancet 385, 1966–1974 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koyner JL, Adhikari R, Edelson DP & Churpek MM Development of a Multicenter Ward-Based AKI Prediction Model. Clin. J. Am. Soc. Nephrol 11, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koyner JL, Carey KA, Edelson DP & Churpek MM The Development of a Machine Learning Inpatient Acute Kidney Injury Prediction Model*. Crit. Care Med 46, 1070 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Peng J-C et al. Development of mortality prediction model in the elderly hospitalized AKI patients. Sci. Rep 11, 1–10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haines RW et al. Acute Kidney Injury in Trauma Patients Admitted to Critical Care: Development and Validation of a Diagnostic Prediction Model. Sci. Rep 8, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motwani SS et al. Development and Validation of a Risk Prediction Model for Acute Kidney Injury After the First Course of Cisplatin. J. Clin. Oncol 36, 682 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCradden MD, Stephenson EA & Anderson JA Clinical research underlies ethical integration of healthcare artificial intelligence. Nat. Med 26, 1325–1326 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Tomašev N et al. Use of deep learning to develop continuous-risk models for adverse event prediction from electronic health records. Nat. Protoc 16, 2765–2787 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Google. https://google/ehr-predictions. GitHub https://github.com/google/ehr-predictions.
- 12.Haibe-Kains B et al. Transparency and reproducibility in artificial intelligence. Nature 586, E14–E16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDermott MBA et al. Reproducibility in machine learning for health research: Still a ways to go. Sci. Transl. Med 13, (2021). [DOI] [PubMed] [Google Scholar]
- 14.Stupple A, Singerman D & Celi LA The reproducibility crisis in the age of digital medicine. npj Digital Medicine 2, 1–3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter RE, Attia ZI, Lopez-Jimenez F & Friedman PA Pragmatic considerations for fostering reproducible research in artificial intelligence. npj Digital Medicine 2, 1–3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh K, Beam AL & Nallamothu BK Machine learning in clinical journals: Moving from inscrutable to informative. Circulation. Cardiovascular qualoity and outcomes vol. 13 e007491 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbins R et al. AI systems are worse at diagnosing disease when training data is skewed by sex. STAT https://www.statnews.com/2020/05/25/ai-systems-training-data-sex-bias/ (2020). [Google Scholar]
- 18.Larrazabal AJ, Nieto N, Peterson V, Milone DH & Ferrante E Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis. Proc. Natl. Acad. Sci. U. S. A 117, 12592–12594 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Classification of Diseases (ICD). https://www.who.int/standards/classifications/classification-of-diseases.
- 20.Sundararajan V et al. New ICD-10 version of the Charlson comorbidity index predicted in-hospital mortality. J. Clin. Epidemiol 57, 1288–1294 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Khwaja A KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin. Pract 120, c179–84 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Hand DJ & Till RJ A Simple Generalisation of the Area Under the ROC Curve for Multiple Class Classification Problems. Mach. Learn 45, 171–186 (2001). [Google Scholar]
- 23.DeLong ER, DeLong DM & Clarke-Pearson DL Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics vol. 44 837 (1988). [PubMed] [Google Scholar]
- 24.Wong A et al. External Validation of a Widely Implemented Proprietary Sepsis Prediction Model in Hospitalized Patients. JAMA Intern. Med 181, 1065–1070 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh K et al. Evaluating a Widely Implemented Proprietary Deterioration Index Model among Hospitalized Patients with COVID-19. Ann. Am. Thorac. Soc 18, 1129–1137 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman JH Greedy Function Approximation: A Gradient Boosting Machine. The Annals of Statistics vol. 29 1189–1232. [Google Scholar]
- 27.Team, R. C. & Others. R: A language and environment for statistical computing. (2013).
- 28.Singh K ML4LHS/gpmodels: A Grammar of Prediction Models. GitHub; https://github.com/ML4LHS/gpmodels. [Google Scholar]
- 29.R Interface for the ‘H2O’ Scalable Machine Learning Platform [R package h2o version 3.36.0.2]. (2022).
- 30.Pafka S szilard/GBM-perf: Performance of various open source GBM implementations. GitHub; https://github.com/szilard/GBM-perf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study used data from the national Veterans Health Administration’s Corporate Data Warehouse and the University of Michigan. Analyses were performed in secure locations within the VA and UM information systems, respectively. The data in this study are not publicly available because they contain protected health information, and restrictions apply to their use. The models trained in this study are available at https://github.com/ML4LHS/va-aki-model.


