Abstract
The increasing abundance of extended spectrum (β-lactamase (ESBL) genes in E. coli, and other commensal and pathogenic bacteria, endangers the utility of third or more recent generation cephalosporins, which are major tools for fighting deadly infections. The role of domestic animals in the transmission of ESBL carrying bacteria has been recognized, especially in low- and middle-income countries, however the horizontal gene transfer of these genes is difficult to assess. Here we investigate blaCTX-M gene diversity (and flanking nucleotide sequences) in E. coli from chicken and humans, in an Ecuadorian rural community and from chickens in another location in Ecuador. The blaCTX-M associated sequences in isolates from humans and chickens in the same remote community showed greater similarity than those found in E. coli in a chicken industrial operation 200 km away. Our study may provide evidence of blaCTX-M transfer between chickens and humans in the community.
Keywords: E. coli; Antimicrobial resistance; Third-generation cephalosporin resistance (TGCR); bla CTX-M ; ISEcp1, spacer sequence; Ecuador; Chickens
1. Introduction
Antimicrobial resistance is recognized as one of the top 10 global public health threats facing humanity [1–3], with an estimated 700,000 annual deaths due to antibiotic-resistant infections, and predictions of 10 million deaths by 2050 [1,3]. Every year, reports on multi-resistant microorganisms increase in different countries, indicating that we are heading into a post-antimicrobial era in which common infections and minor wounds can cause death [4]. Different prevention and control measures have been proposed to mitigate this problem but with minimal success [1,2,5].
Most antimicrobials are administered to animals raised for food, and antimicrobial use in animals is projected to increase to over 100,000 tons by 2030, as global demand for animal protein increases [6]. In Ecuador and in other low- and middle-income countries (LMICs), increased small-scale farming has resulted in greater use of antibiotics as growth promoters (and prophylactics), selecting antimicrobial-resistant bacteria which colonize different hosts and contribute to the dissemination of antimicrobial genes [7,8,9,10].
One of the main antimicrobial resistance genes codes for extended spectrum β-lactamases (ESBLs) which are a rapidly evolving group of enzymes able to hydrolyze third generation cephalosporins, one of the most widely used antibiotics in hospitals and intensive care units [11]. These genes are carried on mobile genetic elements (MGE) which facilitate their spread between bacterial species belonging to different animal microbiomes [12].
Escherichia coli is considered one of the most serious threats in the antimicrobial resistance crisis [13] and it belongs to a type of intestinal bacteria that transfers the most amongst different animal hosts [14]. The blaCTX-M most widely distributed ESBL gene, even though it started to disseminate in E. coli (from the environmental bacteria Kluyvera spp.) since the 1990s [15–17]. The blaCTX-M genes have displaced other ESBLs genes in Enterobacteriaceae, maybe due to highly active MGEs (transposons and plasmids) and successful bacterial clones [15,18,19]. Currently, there are more than 234 blaCTX-M allelic variants in E. coli [20] and only a small number are successful.
In E. coli, the dissemination of blaCTX-M genes occurs clonally and through horizontal gene transfer (HGT). Evidence of antimicrobial-resistant bacteria transmission, from domestic animals to humans, is difficult to obtain; clonal transmission between domestic animals and humans requires the search for genetically identical isolates from a large diversity of E. coli lineages [9,10]. Evidence of HGT requires finding identical plasmids, with the complication that plasmids are diverse, plasmid DNA could be constantly rearranged [21], and resistance genes are usually in highly active transposons (moving back and forth among plasmids, from plasmid to chromosome, or to other MGEs) [22–24,25].
Some reports showing different blaCTX-M allelic variants in E. coli isolates from humans and domestic animals, concluded that role of domestic animals in the current antimicrobial crisis may have been over-estimated [26,27]. However, other studies using isolates with closer spatial-temporal relationships, showed similar allelic variants in isolates from humans and domestic animals [9,10]. Here we analyzed the distribution of blaCTX-M allelic variants and flanking sequences (in conjugable plasmids) from human and chicken isolates in the same community from 2010 to 2017 and compared with similar blaCTX-M allelic variants in E. coli isolated from a chicken industrial operation 200 km away, during overlapping periods.
2. Materials and methods
2.1. Isolates
We analyzed 107 3rth generation cephalosporin resistant E. coli isolates (from a total of 4,518 E. coli isolates analyzed): 38 (35.5 %) from humans (in a rural community) and 69 (64.5 %) from chickens of which 26 (37.7 %) from the same rural community and 43 (62.3 %) were from an industrial operation (Table 2). The isolates were obtained from previous studies (from humans and chickens) in rural communities in northern Coastal Ecuador during 2017 [7,8], in 2009 [28], and 2010–2013 [29] and from chickens from an industrial operation (200 km apart from the remote community) during 2010. In these studies, E. coli was isolated from fecal samples by streaking on MacConkey Agar and incubating for 24 h at 37 °C. Lactose fermenting colonies were tested for β-glucuronidase production, as described previously [7,8]. Resistance profiles were assessed using the Kirby-Bauer disk diffusion method following Clinical and Laboratory Standards Institute (CLSI) recommendations. E. coli ATCC 25922, Staphylococcus aureus ATCC 25923, and Pseudomonas aeruginosa ATCC 27853 were used as controls for antimicrobial susceptibility test. All E. coli were stored frozen at −80 °C until analyzed. Out of 4,518 E. coli isolates, 107 with phenotypic third-generation cephalosporin-resistance (TGCR) were identified. All the protocols were approved by Universidad San Francisco de Quito’s Bioethics committee and University of Michigan’s IRB.
Table 2.
Characteristics of E. coli isolates from 94 human and chicken fecal isolates, including year sample was collected, the variant number for the blaCTX-M- gene, spacer [number of base pairs between the blaCTX-M- gene and the downstream transposon], Insertion Sequence [IS], and the gene that is downstream from the blaCTX-M- gene. The sequences are ordered according to the type of blaCTX-M- variant.
| Source | Number isolates | Year | blaCTX-M- variant | Upstream Spacer | IS [transposon] | Down-stream Sequences |
|---|---|---|---|---|---|---|
| Chicken Community | 1 | 2017 | 15 | 127 bp | ISEcp1 | N/A |
| Chicken Community | 1 | 2017 | 15 | 127 bp | ISEcp1 | ORF477 |
| Chicken Community | 11 | 2017 | 55 | 127 bp | ISEcp1 | ORF477 |
| Chicken Community | 4 | 2017 | 64 | 45 bp | ISEcp1 | N/A |
| Chicken Community | 3 | 2017 | 64 | 45 bp | ISEcp1 | ORF477 |
| Chicken Community | 4 | 2017 | 65 | 42 bp | ISEcp1 | N/A |
| Chicken industrial | 28 | 2010 | 2 | N/A | N/A | N/A |
| Chicken industrial | 5 | 2010 | 8 | N/A | N/A | N/A |
| Chicken industrial | 1 | 2010 | 12 | 48 bp | ISEcp1 | N/A |
| Chicken industrial | 1 | 2010 | 12 | 48 bp | ISEcp1 | ORF477 |
| Chicken industrial | 4 | 2010 | 14 | 42 bp | ISEcp1 | N/A |
| Human Community | 2 | 2010–2013 | 15 | 48 bp | ISEcp1 | N/A |
| Human Community | 3 | 2010–2013 | 2 | N/A | N/A | N/A |
| Human Community | 4 | 2010–2013 | 9 | 42 bp | ISEcp1 | N/A |
| Human Community | 1 | 2010–2013 | 137/15 | 42 bp | ISEcp1 | N/A |
| Human Community | 2 | 2010–2013 | 14 | 42 bp | ISEcp1 | N/A |
| Human Community | 1 | 2010–2013 | 14 | N/A | ISEcp1 | N/A |
| Human Community | 1 | 2010–2013 | 15 | N/A | ISEcp1 | N/A |
| Human Community | 1 | 2010–2013 | 64 | 48 bp | ISEcp1 | N/A |
| Human Community | 1 | 2010–2013 | 65 | 42 bp | ISEcp1 | N/A |
| Human Community | 1 | 2017 | 1 | 80 bp | ISEcp1 | N/A |
| Human Community | 1 | 2017 | 1 | N/A | N/A | N/A |
| Human Community | 1 | 2017 | 9 | 42 bp | ISEcp1 | N/A |
| Human Community | 3 | 2017 | 15 | 48 bp | ISEcp1 | N/A |
| Human Community | 1 | 2017 | 15 | 48 bp | ISEcp1 | ORF477 |
| Human Community | 1 | 2017 | 15 | N/A | ISEcp1 | N/A |
| Human Community | 1 | 2017 | 15 | N/A | N/A | N/A |
| Human Community | 4 | 2017 | 27 | 42 bp | ISEcp1 | N/A |
| Human Community | 1 | 2017 | 55 | 127 bp | ISEcp1 | N/A |
| Human Community | 1 | 2017 | 64 | 45 bp | ISEcp1 | ORF477 |
2.2. Conjugation experiments
Conjugation experiments were conducted to identify the blaCTX-M- genes that had the potential to be horizontally transferred by plasmids. Conjugation experiments were performed in Lysogeny Broth (LB) with E. coli J53Azr (resistant to sodium azide and susceptible to cefotaxime) as the recipient, and all of our E. coli TGCR isolates as donors [30]. Donor and recipient isolates were cultured in tubes with 5 ml of LB incubated for 12 h at 37 °C, to get cells in a logarithmic growth phase. A 0.5 ml-aliquot of each tube was transferred to 4 ml of fresh LB and incubated for 16 h at 37 °C without shaking. Transconjugants were selected using selective culture media made of Trypticase soy agar (TSA) supplied with sodium azide (200 μg/ml) and cefotaxime (1 mg/ml). All transconjugants were stored frozen at −80 °C.
2.3. Polymerase chain reaction (PCR) and DNA sequencing
Genetic materials from all 107 E. coli TGCR isolates were extracted using DNAzol® (Invitrogen™, USA) following manufacturer protocol and recommendations. A conventional PCR was performed to identify samples carrying blaCTX-M- gene (PCR1); to identify the sequences upstream from the blaCTX-M- gene (PCR2), and to identify sequences downstream from the blaCTX-M- (PCR3).
PCR1 was carried out using degenerate primers [31]. PCR2 used the blaCTX-M- gene and ISEcp1 sequences [31]. Both sequences are often in close proximity. PCR3 was designed to obtain complete coding sequences of blaCTX-M- genes and the downstream sequences. We used degenerate primers of blaCTX-M- and orf477 [32]. All amplicons obtained were sequenced. Primers used in this study are listed in Table 1 and the locations of the flanking regions are shown in Fig. 1. Accession numbers of the sequences are in supplemental material, Table S1. Amplicons were sequenced using Sanger’s method at Research Technology Support Facility, Michigan State University.
Table 1.
Sequences of primers used in PCR experiments in this study. PCR experiment described in the text.
| PCR | Primer forward | Primer reverse | Reference |
|---|---|---|---|
| PCR1 | 5′-ATGTGCAGYACCAGTAARGTKATGGC-3′ | 5′-TGGGTRAARTARGTSACCAGAAYCAGCGG-3′ | Fang et al., 2008 |
| PCR2 | 5’-TGCTCTGTGGATAACTTGC-3’ | 5’-GCCATMACYTTACTGGTRCTGCACAT-3’ | Poirel et al., 2003 |
| PCR3A | 5’-GAATACTGATGTAACACGGATTG-3’ | 5’-TCGTTTCGTGGTGCTGAATTT-3’ | Hu et al., 2018 |
| PCR3B | 5’-CGTMTCTTYCAGAATAAGGAATCCC-3’ | 5’-TCGTTTCGTGGTGCTGAATTT-3’ | Hu et al., 2018 |
Fig. 1.
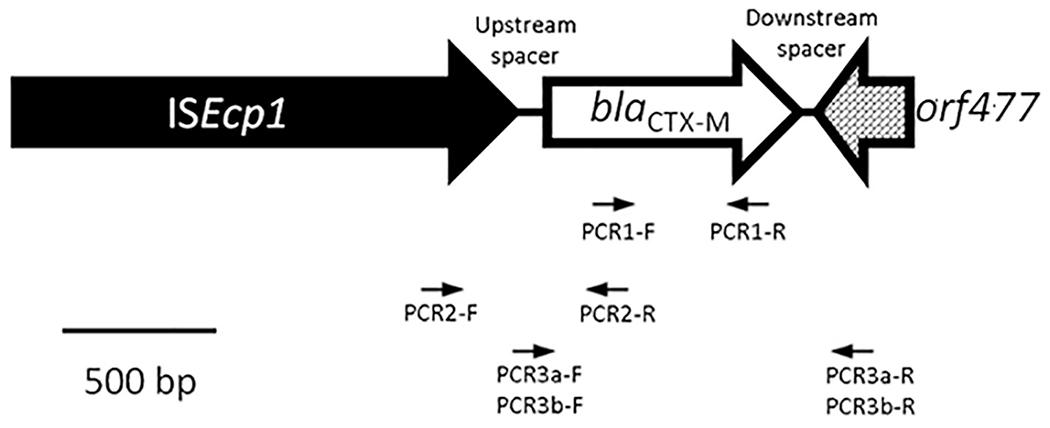
Schematic representation of usual orientation of the blaCTX-M- genes. Here we see the transposon ISEcp1 [Insertion Sequence] upstream from the blaCTX-M- gene, and the orf477 gene in the downstream region. Arrows indicates the primers used in this study.
2.4. blaCTX-M- variant distribution analysis
We assumed that an evidence of gene transfer in the local community were the presence of isolates with: 1) same blaCTX-M- allele variant; 2) same number of nucleotides and same nucleotide sequences between the blaCTX-M- gene and the transposable element; 3) had the same transposon (or Insertion Sequence); and 4) had the same downstream gene (Fig. 1).
2.5. Statistical analysis
We used R statistical software to run principal component analysis (PCA) and ANOVA to evaluate the relationship between blaCTX-M allele variants (from humans’ and chickens’ E. coli) in the community and those from chickens in the industrial operation. We used GraphPrim 9.5 for the graphic representation.
3. Results
3.1. Conjugation experiments
We obtained 102 transconjugants (from isolates carrying blaCTX-M genes); 37 (36.3 %) from humans and 65 (63.7 %) from chickens. The origin of chicken’s isolates was: 40 (61.5 %) were from industrial operation and 25 (38.5 %) were from the rural community (Table 2).
3.2. Nucleotide sequence analysis of blaCTX-M genes
We obtained PCR1 amplicons from all transconjugants, however only 94 produced clean DNA sequences. All blaCTX-M genes found in isolates from chickens in the rural community were also present in isolates from humans of the same community (blaCTX-M-15 blaCTX-M-27, blaCTX-M-55, blaCTX-M-64, blaCTX-M-65); none of these variants were present in isolates from the chicken industrial operation. Isolates from humans in the community shared only 2 variants with chickens in the industrial operation (blaCTX-M-2 and blaCTX-M-14) (Table 2, Fig. 3). The prevalence of some shared variants was different for humans and chickens in the community: blaCTX-M-15 was present in 9 human and 2 chicken isolates, blaCTX-M-55 was present in 1 human and 11 chicken isolates; blaCTX-M-64 was present in 2 human and 7 chicken isolates. The prevalence of the shared blaCTX-M-2 gene was different for isolates from humans and chickens in the industrial operation (3 and 28 respectively). Three variants (blaCTX-M-1,blaCTX-M-9, blaCTX-M-137/15) were present only in human isolates and 2 variants (blaCTX-M-8, blaCTX-M-12) were present only in isolates from chickens in the industrial operation (Table 2).
Fig. 3.
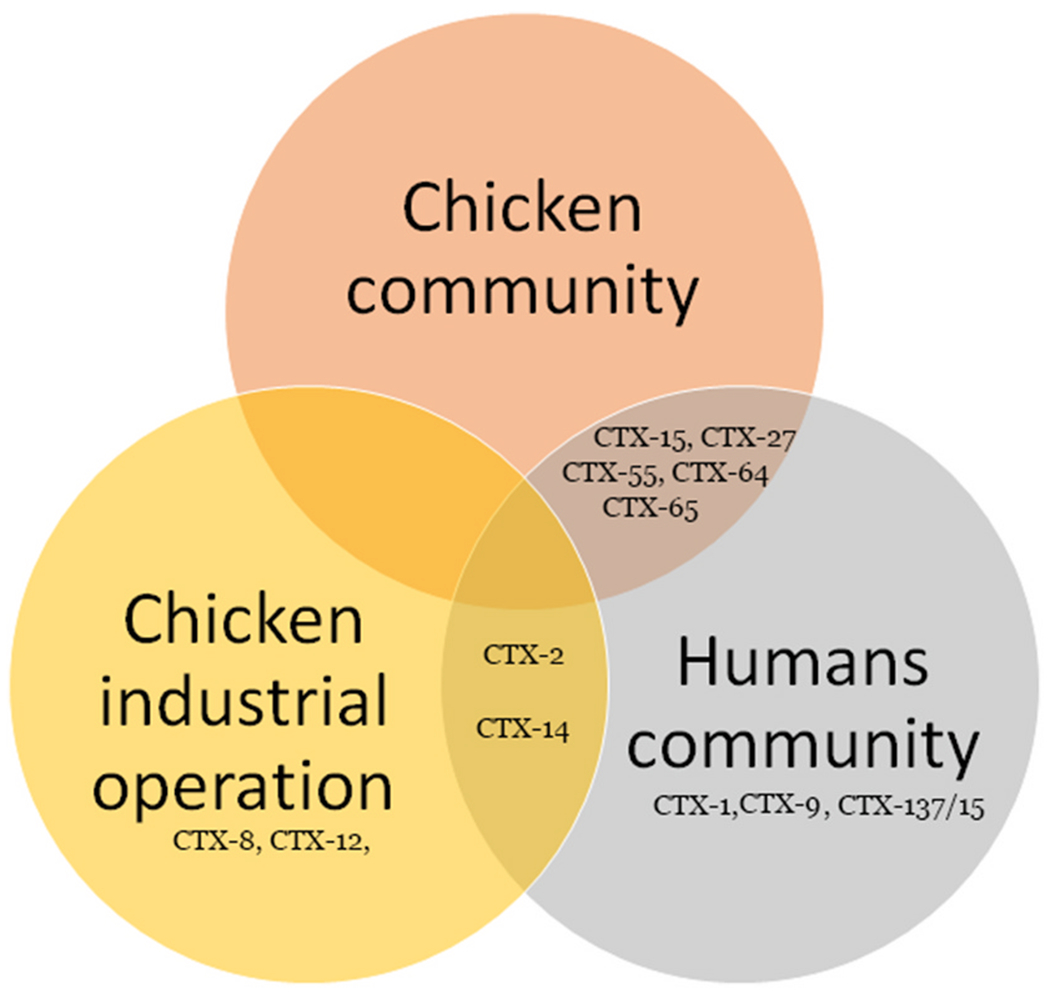
Venn Diagram showing the distribution of blaCTX-M- gene variants found in humans and chickens in different locations.
3.3. Nucleotide sequence analysis of blaCTX-M flanking regions
The transposable element (ISEcp1), and the spacer sequences (between the blaCTX-M and ISEcp1) (Table 2) were detected in 51 blaCTX-M positive isolates. The isolates that have the same blaCTX-M seemed to share the same spacer size except for blaCTX-M-15 (2 isolates had a 127bp spacer, while 6 isolates had a 48bp spacer) and blaCTX-M-65 (1 isolate had 48bp while 8 isolates had 45 bp) (Table 2). In 18 isolates we identified the orf477 downstream blaCTX-M gene, however we did not see an association with any source of samples. We were able to identify a possible recombinant of blaCTX-M-15 and blaCTX-M-137 in one isolate (Fig. 2). All accession numbers for these allelic variants are listed in Table S1.
Fig. 2.
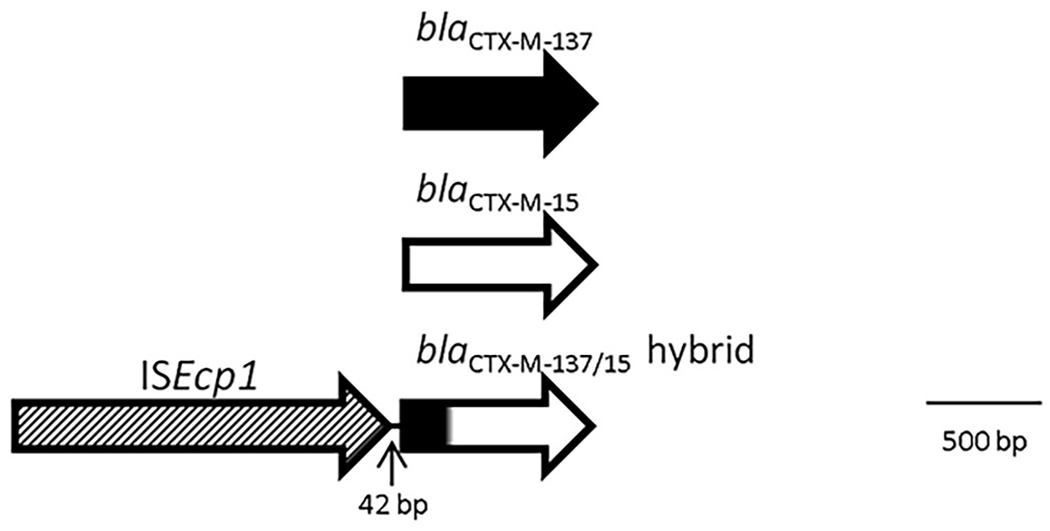
Recombination scheme between blaCTX-M-137 and blaCTX-M-15, sample isolated from human feces, GenBank accession number MZ314224.
We found difference in blaCTX-M genes (and adjacent sequences) associated with different animal species; some blaCTX-M variants (blaCTX-M-1, blaCTx-M-9) were found only in human isolates and not in chickens in the community. Additionally, spacers flanking blaCTX-M-15 in human and chicken isolates (in the same community) were different (Table 2); we also found different proportion of strains carrying same variants: human isolates had more blaCTX-M-15 than chickens in the same community (45 %; n = 9 vs. 8 %; n = 2); chicken isolates had more blaCTX-M-55 than human isolates in same community (45 %; n = 11 vs. 5 %, n = 1).
3.4. Statistical analysis
The ANOVA and Pearson correlation indicate that human isolates (from the community) have larger differences with chickens in the industrial operation than with isolates from the same community (p= < 0.0001) (Fig. S1). Similarly, Principal Component Analysis indicate that location (community or industrial operation) has more influence in variability than animal species (p = < 0.0001) (Fig. S1).
4. Discussion
The results of our study show a difference in the distribution of blaCTX-M variants in E. coli isolates from 2 different locations in Ecuador, from 2010 to 2017. The same blaCTX-M variants and the same flanking sequences (spacer and transposons) found in chicken isolates were also found in human isolates in the community. Contrastingly, no blaCTX-M variants were shared between chicken’s isolates in the community and those from the industrial operation 200 km apart (even in isolates from the same year) (Table 2 and Fig. 3); the ANOVA and PCA were consistent with this observation. We suggest that the distribution of blaCTX-M variants is a supportive evidence for transference of blaCTX-M genes among E. coli from chickens and humans in the same community. Transposable elements (insertion sequences, like those found in our study) are actively moving important antimicrobial resistance genes among plasmids. The HGT of blaCTX-M (and other antimicrobial genes) is complex; these blaCTX-M genes (and flanking sequences) are associated with transposable elements (such as ISEpc1 or IS26) and promote high frequency transposition among different plasmids and chromosomes [33,34,35–38] and the transposition of genes among plasmids increases the possibilities of HGT [34,38]. Transposition also makes it difficult to detect transmission, even with the use of whole genome sequencing [39,40]. Despite our arguments in favor of HGT, it is also possible that the statistically significant association of blaCTX-M variants with locations may be due to the dissemination of one clone or few clonally related strains in the community, however, clonally transmission of blaCTX-M variants is rare in rural communities [39].
We did find 2 variants in isolates from chickens at the industrial operation and humans in the community (blaCTX-M-2, blaCTX-M-14), however, the proportion of isolates carrying the blaCTX-M-2 in the two species was different (15 % in human’s compared to 78 % in chicken’s).
There were differences in the frequencies E. coli (carrying the same blaCTX-M variant and flanking sequences) obtained from chicken and humans (Table 2). These results suggest that transmission of these genes may occur more frequently among strains from the same animal species and less frequently among strains from different animal species; greater interaction with members of the same animal species may contribute to the transference of bacteria and bacterial genes among these bacteria.
Diference in variant distribution overtime was also evident as 31 % of 2017 isolates had blaCTX-M-55 whereas no isolate from 2010 to 2013 had this variant; 21 % of 2017 isolates (from humans and chickens) had blaCTX-M-15 compared with 5 % in isolates obtained in 2010–2013; 21 % of 2017 isolates had blaCTX-M-64 whereas 1.8 % of isolates from 2010 to 2013 had this variant (see Table 2). These results are also in agreement with previous research showing that blaCTX-M variants change overtime in a given location [22].
We observed more blaCTX-M variant diversity in human isolates than in chicken isolates in the community. Additionally, we found blaCTX-M-64 and blaCTX-M-65 first in humans in the community in 2010–2013 and later in chickens in the community in 2017 (Table 2). It is tempting to speculate that these results indicate that humans may be the source of some of these gene variants found in E. coli from chickens. This transmission may be associated with low sanitary infrastructure in this community [41], allowing the chickens to become colonized with E. coli from humans. Other researchers have presented evidence of ARG transmission between E. coli from humans and domestic animals due to a shared environment, direct contact between humans and domestic animals, or ingestion of food contaminated with E. coli [7,8,9,10].
In Ecuador, blaCTX-M-14, blaCTX-M-15, blaCTX-M-55, and blaCTX-M-65 variants have been reported in clinical isolates [42–44]. Studies in other Ecuadorian rural communities have shown that humans and domestic animals in the same communities share E. coli with blaCTX-M-2, blaCTX-M-14, blaCTX-M-15, blaCTX-M-55, and blaCTX-M-65, suggesting a mechanism of spread (clonal and HGT of blaCTX-M genes) between humans and domestic animals [10].
Previous research has described the close relationship between the ISEcp1 (and other insertion sequences) with blaCTX-M genes [15,45,46]. These insertion sequences are thought to have been involved in the original mobilization blaCTX-M genes from the bacteria Kluyvera spp [15]. Most of the members of the blaCTX-M gene family have been associated with ISEcp1 and five different DNA spacer sequences between the insertion sequence and the blaCTX-M gene [15]. The length of the spacer sequence is often associated with specific a blaCTX-M gene [47] and differences in the location of the ISEcp1 (Table 2) indicate frequent transposable activity, something that has been observed previously [23].
We also detected a possible recombinant of blaCTX-M-137 and blaCTX-M-15 in one isolate, being the first 218 nucleotides form blaCTX-M-137 and the rest from blaCTX-M-15 (Fig. 2). It is the first report of this allelic variant; we could not find other sequences close to it during BLAST search (accession number MZ314224).
The small number of TGCR isolates analyzed is a major limitation of our study. We obtained 3 to 5 E. coli colonies from each culture plate (without cephalosporin selection); this approach allows to obtain strains that are numerically dominant (regardless of the resistance status) in the intestines and feces [48]. Numerically dominant strains may correspond to the strains that disseminate the most in the environment. We present evidence that the combination of epidemiological data (location, time frame, animal species) and DNA sequence data form blaCTX-M genes and flanking sequences could provide evidence of blaCTX-M genes transfer between bacteria in humans and domestic animals inhabiting the same community. We showed that nucleotide sequences of blaCTX-M gene and flanking regions is a very useful tool to study the transmission of these genes. This information is critical to detect sources and trends in the transmission of this ESBL genes in communities.
5. Conclusions
The results of our research suggest that the blaCTX-M- variants are transmitted between chickens and humans in a rural community in Ecuador. We also present indirect evidence of transmission of antimicrobial resistance genes from humans to chickens in a rural community with low sanitary infrastructure. This study supports the notion that domestic animals play an important role in the circulation of ESBL genes.
Supplementary Material
Acknowledgments
This research was funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Healthh, United States of America; grants: K01 AI103544, R01 AI050038, R01AI137679, and R01AI167989. The authors thank Deysi Parrales and Barbara Pibaque for their help with preparation of culture media and Liseth Salinas for her suggestions for the manuscript.
Footnotes
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.medmic.2023.100092.
References
- [1].Dadgostar P. Antimicrobial resistance: implications and costs. Infect Drug Resist 2019. Dec 20;12:3903–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2019. 10.15620/cdc:82532. [DOI] [Google Scholar]
- [3].World Health Organization WHO. New report calls for urgent action to avert antimicrobial resistance crisis. 2019. https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis.
- [4].Reardon S. WHO warns against ‘post-antibiotic’ era. Nature 2014. 10.1038/nature.2014.15135. [DOI] [Google Scholar]
- [5].Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, Colomb-Cotinat M, Kretzschmar ME, Devleesschauwer B, Cecchini M, Ouakrim DA, Oliveira TC, Struelens MJ, Suetens C, Monnet DL, Burden of AMR Collaborative Group. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis 2019. Jan;19(1):56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci U S A 2015. May 5;112(18):5649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hedman HD, Eisenberg JNS, Trueba G, Rivera DLV, Herrera RAZ, Barrazueta JV, Rodriguez GIG, Krawczyk E, Berrocal VJ, Zhang L. Impacts of small-scale chicken farming activity on antimicrobial-resistant Escherichia coli carriage in backyard chickens and children in rural Ecuador. One Health 2019. Nov 7;8:100112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hedman HD, Eisenberg JNS, Vasco KA, Blair CN, Trueba G, Berrocal VJ, Zhang L. High prevalence of extended-spectrum beta-lactamase CTX-M-producing Escherichia coli in small-scale poultry farming in rural Ecuador. Am J Trop Med Hyg 2019. Feb;100(2):374–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Falgenhauer L, Imirzalioglu C, Oppong K, Akenten CW, Hogan B, Krumkamp R, Poppert S, Levermann V, Schwengers O, Sarpong N, Owusu-Dabo E, May J, Eibach D. Detection and characterization of ESBL-producing Escherichia coli from humans and poultry in Ghana. Front Microbiol 2019. Jan 15;9:3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Salinas L, Loayza F, Cárdenas P, Saraiva C, Johnson TJ, Amato H, Graham JP, Trueba G. Environmental spread of extended spectrum beta-lactamase (ESBL) producing Escherichia coli and ESBL genes among children and domestic animals in Ecuador. Environ Health Perspect 2021. Feb;129(2):27007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rawat D, Nair D. Extended-spectrum (β-lactamases in Gram negative bacteria. J Global Infect Dis 2010. Sep;2(3):263–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Falagas ME, Karageorgopoulos DE. Extended-spectrum beta-lactamase-producing organisms. J Hosp Infect 2009. Dec;73(4):345–54. [DOI] [PubMed] [Google Scholar]
- [13].CDC. Antibiotic resistance threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2019. [Google Scholar]
- [14].Moeller AH, Suzuki TA, Phifer-Rixey M, Nachman MW. Transmission modes of the mammalian gut microbiota. Science 2018. Oct 26;362(6413):453–7. [DOI] [PubMed] [Google Scholar]
- [15].Cantón R, González-Alba JM, Galán JC, CTX-M- Enzymes. Origin and diffusion. Front Microbiol 2012. Apr 2;3:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rossolini GM, D’Andrea MM, Mugnaioli C. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin Microbiol Infect 2008. Jan;14(Suppl 1):33–41. [DOI] [PubMed] [Google Scholar]
- [17].Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, Ayala J, Coque TM, Kern-Zdanowicz I, Luzzaro F, Poirel L, Woodford N. CTX-M-: changing the face of ESBLs in Europe. J Antimicrob Chemother 2007. Feb;59(2):165–74. [DOI] [PubMed] [Google Scholar]
- [18].Rupp ME, Fey PD. Extended spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae: considerations for diagnosis, prevention and drug treatment. Drugs 2003;63(4):353–65. [DOI] [PubMed] [Google Scholar]
- [19].Coque TM, Baquero F, Canton R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Euro Surveill 2008. Nov 20;13(47):19044. [PubMed] [Google Scholar]
- [20].National Center for Biotechnology Information NCBI. Reference Gene Catalog. https://www.ncbi.nlm.nih.gov/pathogens/refgene/#blactx-m.
- [21].Brolund A, Rajer F, Giske CG, Melefors Ö, Titelman E, Sandegren L. Dynamics of resistance plasmids in extended-spectrum-(β-lactamase-producing Enterobacteriaceae during Postinfection colonization. Antimicrob Agents Chemother 2019. Mar 27;63(4):e02201–e02218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M- β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother 2017. Aug 1;72(8):2145–55. [DOI] [PubMed] [Google Scholar]
- [23].Hamamoto K, Tokunaga T, Yagi N, Hirai I. Characterization of blaCTX-M-14 transposition from plasmid to chromosome in Escherichia coli experimental strain. Int J Med Microbiol 2020. Feb;310(2):151395. [DOI] [PubMed] [Google Scholar]
- [24].Gómez-Gómez C, Blanco-Picazo P, Brown-Jaque M, Quirós P, Rodríguez-Rubio L, Cerdà-Cuellar M, Muniesa M. Infectious phage particles packaging antibiotic resistance genes found in meat products and chicken feces. Sci Rep 2019. Sep 16;9(1):13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Eckert C, Gautier V, Arlet G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J Antimicrob Chemother 2006. Jan;57(1): 14–23. [DOI] [PubMed] [Google Scholar]
- [26].Day MJ, Hopkins KL, Wareham DW, Toleman MA, Elviss N, Randall L, Teale C, Cleary P, Wiuff C, Doumith M, Ellington MJ, Woodford N, Livermore DM. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: an epidemiological surveillance and typing study. Lancet Infect Dis 2019. Dec;19(12):1325–35. [DOI] [PubMed] [Google Scholar]
- [27].Ludden C, Raven KE, Jamrozy D, Gouliouris T, Blane B, Coll F, de Goffau M, Naydenova P, Horner C, Hernandez-Garcia J, Wood P, Hadjirin N, Radakovic M, Brown NM, Holmes M, Parkhill J, Peacock SJ. One health genomic surveillance of Escherichia coli Demonstrates Distinct lineages and mobile genetic elements in isolates from humans versus Livestock. mBio 2019. Jan 22;10(1):e02693–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Armas-Freire PI, Trueba G, Proaño-Bolaños C, Levy K, Zhang L, Marrs CF, Cevallos W, Eisenberg JN. Unexpected distribution of the fluoroquinolone-resistance gene qnrB in Escherichia coli isolates from different human and poultry origins in Ecuador. Int Microbiol 2015. Jun;18(2):85–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Braykov NP, Eisenberg JN, Grossman M, Zhang L, Vasco K, Cevallos W, Muñoz D, Acevedo A, Moser KA, Marrs CF, Foxman B, Trostle J, Trueba G, Levy K. Antibiotic resistance in animal and environmental samples associated with small-scale poultry farming in Northwestern Ecuador. mSphere 2016. Feb 10;1(1):e00021–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang Y, Liao K, Gao H, Wang Q, Wang X, Li H, Wang R, Wang H. Decreased Fitness and Virulence in ST10 Escherichia coli Harboring blaNDM-5 and mcr-1 against a ST4981 strain with blaNDM-5. Front Cell Infect Microbiol 2017. Jun 8;7:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fang H, Ataker F, Hedin G, Dornbusch K. Molecular epidemiology of extended-spectrum beta-lactamases among Escherichia coli isolates collected in a Swedish hospital and its associated health care facilities from 2001 to 2006. J Clin Microbiol 2008. Feb;46(2):707–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hu X, Gou J, Guo X, Cao Z, Li Y, Jiao H, He X, Ren Y, Tian F. Genetic contexts related to the diffusion of plasmid-mediated CTX-M-55 extended-spectrum beta-lactamase isolated from Enterobacteriaceae in China. Ann Clin Microbiol Antimicrob 2018. Mar 23;17(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sheppard AE, Stoesser N, Wilson DJ, Sebra R, Kasarskis A, Anson LW, Giess A, Pankhurst LJ, Vaughan A, Grim CJ, Cox HL, Yeh AJ. Modernising Medical Microbiology (MMM) informatics group, Sifri CD, Walker AS, Peto TE, crook DW, Mathers AJ. Nested Russian Doll-like genetic mobility Drives Rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob Agents Chemother 2016. May 23;60(6):3767–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Che Y, Yang Y, Xu X, Brinda K, Polz MF, Hanage WP, Zhang T. Conjugative plasmids interact with insertion sequences to shape the horizontal transfer of antimicrobial resistance genes. Proc Natl Acad Sci U S A 2021;118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lartigue MF, Poirel L, Aubert D, Nordmann P. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring beta-lactamase gene blaCTX-M- of Kluyvera ascorbata. Antimicrob Agents Chemother 2006. Apr;50(4):1282–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Harmer CJ, Moran RA, Hall RM. Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. mBio 2014. Oct 7;5(5):e01801–e01814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Varani A, He S, Siguier P, et al. The IS6 family, a clinically important group of insertion sequences including IS26. Mobile DNA 2021;12:11. 10.1186/S13100-021-00239-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yao Y, Maddamsetti R, Weiss A, et al. Intra- and interpopulation transposition of mobile genetic elements driven by antibiotic selection. Nat Ecol Evol 2022;6:555–64. [DOI] [PubMed] [Google Scholar]
- [39].Salinas L, Loayza F, Cárdenas P, Saraiva C, Johnson TJ, Amato H, Graham JP, Trueba G. Environmental spread of extended spectrum beta-lactamase (ESBL) producing Escherichia coli and ESBL genes among children and domestic animals in Ecuador. Environ Health Perspect 2021. Feb;129(2):27007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Loayza F, Graham JP, Trueba G. Factors Obscuring the role of E. coli from domestic animals in the global antimicrobial resistance crisis: an evidence-Based Review. Int J Environ Res Publ Health 2020. 28;17(9):3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bhavnani D, Goldstick JE, Cevallos W, Trueba G, Eisenberg JN. Impact of rainfall on diarrheal disease risk associated with unimproved water and sanitation. Am J Trop Med Hyg 2014. Apr;90(4):705–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Delgado DY, Barrigas ZP, Astutillo SG, Jaramillo AP, Ausili A. Detection and molecular characterization of β-lactamase genes in clinical isolates of Gramnegative bacteria in Southern Ecuador. Braz J Infect Dis 2016. Nov-Dec;20(6):627–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Soria Segarra C, Soria Baquero E, Cartelle Gestal M. High prevalence of CTX-M-1 like enzymes in Urinary isolates of Escherichia coli in Guayaquil, Ecuador. Microb Drug Resist 2018. May;24(4):393–402. [DOI] [PubMed] [Google Scholar]
- [44].Barba P, Loaiza K, Mora F, Fors M, Vinueza-Burgos C, Fernández-Moreira E. Multidrug-resistant Escherichia coli isolated from canine faeces in a public park in Quito, Ecuador. J Glob Antimicrob Resist 2019. Sep;18:263–8. [DOI] [PubMed] [Google Scholar]
- [45].Ma L, Siu LK, Lu PL. Effect of spacer sequences between bla(CTX-M) and ISEcp1 on bla(CTX-M-) expression. J Med Microbiol 2011. Dec;60(Pt 12):1787–92. [DOI] [PubMed] [Google Scholar]
- [46].Tian SF, Chu YZ, Chen BY, Nian H, Shang H. ISEcp1 element in association with bla (CTX-M-) genes of E. coli that produce extended-spectrum β-lactamase among the elderly in community settings. Enferm Infecc Microbiol Clín 2011. Dec;29(10):731–4. [DOI] [PubMed] [Google Scholar]
- [47].Poirel L, Kämpfer P, Nordmann P. Chromosome-encoded Ambler class A beta-lactamase of Kluyvera georgiana, a probable progenitor of a subgroup of CTX-M-extended-spectrum beta-lactamases. Antimicrob Agents Chemother 2002. Dec;46(12):4038–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lautenbach E, Bilker WB, Tolomeo P, Maslow JN. Impact of diversity of colonizing strains on strategies for sampling Escherichia coli from fecal specimens. J Clin Microbiol 2008. Sep;46(9):3094–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


