Abstract
The GcvA protein is required for both glycine-mediated activation and purine-mediated repression of the gcvTHP operon. Random and site-directed PCR mutagenesis was used to create nucleotide changes in gcvA to identify residues of the protein involved in activation, repression, and DNA binding. Single amino acid substitutions at L30 and F31 cause a defect in activation of a gcvT-lacZ fusion but have no effect on repression or DNA binding. Single amino acid substitutions at V32 and S38 cause the loss of binding of GcvA to DNA. A deletion of the carboxy-terminal 14 amino acids of GcvA results in the loss of purine-mediated repression and, consequently, a constitutive activation of a gcvT-lacZ fusion. The results of this study partially define regions of GcvA involved in activation, repression, and DNA binding and demonstrate that these functions of GcvA are genetically separable.
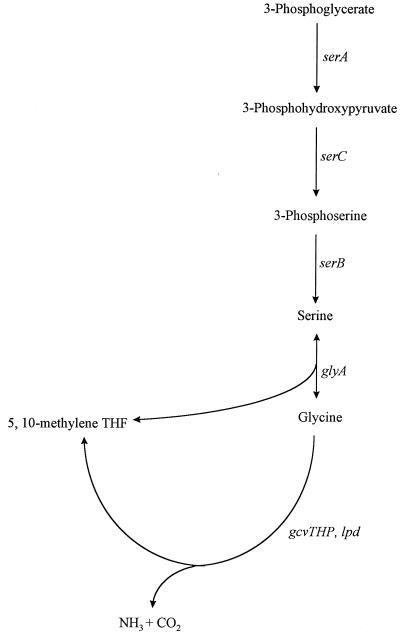
The glycine cleavage (GCV) enzyme system in Escherichia coli is a glycine-inducible, purine-repressible metabolic pathway that catalyzes the oxidative cleavage of glycine to CO2 and NH3 and transfers a one-carbon (C-1) methylene unit to tetrahydrofolate (9) (Fig. 1). The C-1 units are used by the cell in the biosynthesis of purines, methionine, thymine, and other cellular components (13). Three of the proteins of the GCV enzyme complex are encoded by the gcvTHP operon, which maps at 62.5 min on the E. coli chromosome (17, 23). The fourth protein, encoded by the lpd gene, is not part of the gcv operon and maps at min 2.5 (26). Serine hydroxymethyltransferase (glyA gene product) catalyzes the conversion of serine into glycine and the transfer of a C-1 methylene unit to tetrahydrofolate, providing another source of C-1 unit production (13, 15) (Fig. 1). Approximately 15% of all carbon atoms assimilated by E. coli from glucose enter the serine-glycine pathway, making this pathway of central importance in cell physiology (16).
FIG. 1.
The serine-glycine pathway in E. coli. The genes and the gene products are as follows: serA, 3-phosphoglycerate dehydrogenase; serC, 3-phosphoserine aminotransferase; serB, 3-phosphoserine phosphatase; glyA, serine hydroxymethyltransferase; and gcv, glycine cleavage enzyme system. THF, tetrahydrofolate.
Regulation of the gcv operon is complex and is known to involve at least four regulatory proteins. Three of these proteins, Lrp, PurR, and GcvA, have been shown to act directly at the gcv promoter. The leucine responsive regulatory protein (Lrp) is a global regulator in E. coli and is known to activate or repress many genes involved in amino acid metabolism (1, 14). In an lrp mutant, a gcvT-lacZ fusion shows low, noninducible β-galactosidase synthesis (24). In vitro studies have identified multiple Lrp binding sites in the gcv control region (24), and Lrp binding has been shown to bend this region about 90 degrees (unpublished data). However, further studies are required to determine if Lrp’s role in the activation of gcv is structural or if interactions with RNA polymerase (RNAP) or other regulatory proteins occur.
PurR is a global regulator in E. coli involved in negative regulation of many purine and pyrimidine biosynthesis genes (2, 10, 18). PurR, in the presence of exogenous purines, represses gcv expression in vivo twofold and in vitro binds the gcv control region from base pair (bp) −3 to +17 relative to the transcriptional start site of gcvT, the first gene of the gcv operon (30).
GcvA plays a dual role in the regulation of the gcv operon. This protein mediates a six- to sevenfold activation of gcv expression in vivo in the presence of glycine and a fivefold, PurR-independent repression in the presence of purines (30, 31). The GcvA-mediated purine repression of gcv is relieved when gcvA is overexpressed even in the presence of exogenous purines (6). GcvA has a molecular mass of approximately 34 kDa (29) and currently it is unknown what order of multimer GcvA forms (i.e., dimer, tetramer, etc.). DNase I protection studies identified three GcvA binding sites in the gcv control region. There are two adjacent sites beginning at bp −214 and extending to bp −271 and one site overlapping the −35 promoter region, from bp −69 to −34 (32). Binding of GcvA to all three sites in the gcv control region is required for purine-mediated repression of the gcv operon, whereas binding of GcvA to the two upstream sites is required for glycine-mediated induction. GcvA also binds to an extended region in the gcvA control region from bp −28 to +20 relative to the transcription initiation site, negatively autoregulating its own expression (32).
The GcvR protein is involved in repression of gcv expression in minimal medium by an unknown mechanism; this repression is enhanced by the addition of purines to the growth medium and is antagonized by the addition of glycine (6). Inactivation of gcvR results in a loss of repression in all three media, and overexpression of gcvR results in enhanced repression in all three media. However, GcvR-mediated repression is dependent on a functional GcvA protein; gcvA mutants no longer show GcvR-mediated purine repression, and the levels of gcvT-lacZ expression in a gcvA mutant overexpressing gcvR on a multicopy plasmid are no lower than in the gcvA gcvR double mutant (6). GcvR has not been shown to bind to the gcv promoter or directly interact with other proteins, and the molecular mechanism of repression is unknown.
The GcvA protein belongs to the LysR-type family of transcriptional regulators (29). Although the LysR family of proteins is large, few of the proteins have been analyzed at the molecular level. This study focuses on identifying the functional residues of GcvA and describes mutations in the gcvA coding region that partially define activation, repression, and DNA binding regions. The results are consistent with a previously proposed model for gcv regulation where Lrp binds to the gcv control region, bending the DNA to position other regulatory proteins into proximity to RNAP so possible protein-protein interactions can occur to either activate or repress gcv operon expression (25).
MATERIALS AND METHODS
Strains and plasmids.
Genotypes of strains and plasmids used in this study are listed in Table 1.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Source or reference |
|---|---|---|
| Strainsa | ||
| GS162 | wt | This laboratory |
| GS986 | gcvA1 purR::Tn10 | This laboratory |
| GS998 | gcvA1 | This laboratory |
| GS1039 | gcvA1 serA25 purR::Tn10 | This laboratory |
| Plasmids | ||
| pGS311 | Single copy plasmid, Apr, derived from plasmid pDF41b | This laboratory |
| pGS335 | 1.3-kb EcoRI-HindIII gcvA fragment in pGS272 (pACYC177 derivative), Apr | This study |
| pGS341 | 1.3-kb EcoRI-HindIII gcvA fragment from pGS335 in pGS311, Apr | This study |
| pGS440 | PCR-derived mutant EcoRI-HindIII gcvA fragment in pGS311, Apr | This study |
| pGS441 | gcvAF31L in pGS341, Apr | This study |
| pGS442 | gcvAF31A in pGS341, Apr | This study |
| pGS443 | gcvAL30A in pGS341, Apr | This study |
| pGS444 | gcvAV32A in pGS341, Apr | This study |
| pGS445 | gcvAS38P in pGS341, Apr | This study |
| pGS472 | ΔgcvA944 in pGS311, Apr | This study |
| pGS473 | His-tagged gcvA in pKK223-3, Apr | This study |
All strains are thi pheA905 ΔlacU169 araD129 rpsL150 and were lysogenized with a λ phage carrying the gcvT-lacZ translational gene fusion.
Plasmid pDF41 (8) was obtained from S. R. Kushner.
Media.
The complex medium used was Luria-Bertani broth (LB) (12). The defined medium used was Vogel and Bonner minimal salts (28) supplemented with 0.4% glucose (glucose medium [GM]) or lactose (lactose medium [LM]) and supplemented with 50 μg of phenylalanine/ml and 1 μg of vitamin B1/ml, since all of the strains used carry the pheA905 and thi mutations. When necessary, additions were made at the following concentrations: phenylethyl-β-d-thiogalactoside (TPEG), 2 mM; inosine, 50 μg/ml; glycine, 300 μg/ml; serine, 200 μg/ml; ampicillin (AP), 50, 100, or 200 μg/ml depending on the plasmid copy number.
Enzyme assays.
β-Galactosidase assays were performed as described by Miller (12). All results are averages from two or more assays, with each sample done in triplicate. All standard deviations were within 14% of the mean. Protein concentrations were determined by using the BIO-RAD Protein Assay Kit II (Bio-Rad, Richmond, Calif.).
DNA manipulation.
Isolation of plasmid DNA, restriction digestions, ligations, DNA sequencing, and plasmid transformations were performed as previously described (19).
Random mutagenesis.
Mutagenesis of gcvA was performed according to the PCR mutagenesis protocol described by Zhou et al. (33). PCR products were obtained by using primers complementary to template DNA outside of the gcvA insert that are upstream of a unique EcoRI cloning site and downstream of a unique HindIII cloning site. The PCR products from several independent mutagenesis reactions were collected, digested with EcoRI and HindIII, purified from a low-melting-point agarose gel, ligated into the EcoRI-HindIII sites of the single copy plasmid pGS311, and transformed into strain GS1039λgcvT-lacZ.
Site-directed mutagenesis.
Site-directed mutagenesis of gcvA was performed in accordance with a PCR “megaprimer” mutagenesis protocol (20). Changes were introduced through internal downstream primers complementary to gcvA except at the position of the desired bp change. PCR products were generated by using an upstream primer which was complementary to vector DNA outside of the gcvA insert and which included a unique EcoRI cloning site. These PCR products were then used as the upstream megaprimers in another round of PCR synthesis, again with the same DNA template. The downstream primer was complementary to an internal segment of gcvA distal to the mutagenic primer and an internal MluI site. PCR products were cut with EcoRI and MluI restriction enzymes, purified from a low-melting-point agarose gel, and ligated into EcoRI- and MluI-digested single copy wild-type (wt) gcvA plasmid pGS341. The resulting plasmids (pGS441, pGS442, pGS443, pGS444, and pGS445) (Table 1) each contained specific bp mutations that were verified by DNA sequence analysis.
Deletion mutagenesis.
A deletion of the carboxy-terminal (C-ter) 42 bp of gcvA was constructed by PCR. The downstream primer was complementary to a region internal to gcvA and included an artificial translation termination codon and a unique HindIII cloning site. The upstream primer was complementary to the template DNA outside of the gcvA insert and included a unique EcoRI cloning site. The PCR products were digested with EcoRI and HindIII, purified from a low-melting-point agarose gel, and ligated into an EcoRI- and HindIII-digested single copy plasmid, pGS311. The resulting plasmid was designated pGS472.
Protein purification.
Plasmid pGS473, which overexpresses GcvA, was constructed by PCR. The upstream primer was complementary to gcvA overlapping the translation initiation codon and included an artificial Shine-Dalgarno sequence and a unique EcoRI cloning site. The downstream primer was complementary to a region overlapping the gcvA translation termination codon and included an artificial string of six histidine codons, an artificial translation termination codon, and a unique HindIII cloning site. The resulting PCR product was cut with EcoRI and HindIII and ligated into the expression vector pKK223-3 (Pharmacia Biotech, Piscataway, N.J.) immediately downstream of the inducible tac promoter. Plasmids for overproduction of mutant GcvA proteins were also constructed by PCR. The upstream primer used was as described above, the downstream primer was complementary to an internal segment of gcvA distal to a unique MluI restriction site, and the DNA template used contained specific bp changes in gcvA. The resulting PCR products were cut with EcoRI and MluI and subcloned into EcoRI- and MluI-digested pGS473. Each plasmid, including the wt, was sequenced to ensure that no additional PCR-induced mutations in gcvA had occurred.
Each plasmid was transformed into strain XL1 Blue (New England Biolabs, Beverly, Mass.) and streaked for purity on LB agar plus 200 μg of AP/ml. A single colony was picked and grown in 1 ml of LB broth plus AP at 37°C till mid-log phase. Cells were harvested by centrifugation, resuspended in 10 ml of LB plus AP, and grown at 37°C till mid-log phase. Cells were collected, resuspended in 10 ml of LB broth plus AP and 1 mM isopropyl β-d-thiogalactopyranoside, and grown for an additional 3 h at 37°C. Cells were collected, frozen overnight at −70°C, resuspended in 2 ml of Na-phosphate buffer (50 mM Na-phosphate, 500 mM NaCl) (Qiagen, Chatsworth, Calif.) plus 1 μg of lysozyme/ml, and incubated on ice for 1 h. The cellular suspension was sonicated on ice until it was viscous and clear. Membrane and cytoplasmic fractions were separated through centrifugation, and the supernatant was brought to 33% ammonium sulfate saturation. Precipitated protein was collected by centrifugation and resuspended in 1 ml of TEG buffer (50 mM Tris HCl [pH 7.9], 0.5 mM EDTA, 5% glycerol) (27). A total of 100 μl of Ni2+-nitrilotriacetic acid (NTA) agarose binding resin (Qiagen) was washed three times with TEG buffer before application of the protein sample. Protein was bound to the Ni2+-NTA agarose binding resin for 30 min at 4°C with gentle shaking. The resin was collected by centrifugation at 4°C and then was washed three times with TEG buffer to remove unbound protein. Protein was eluted by stepwise addition of TEG buffer plus imidizole from 75 to 600 mM. The GcvA protein was eluted from 100 to 300 mM imidizole. GcvA protein was visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by staining with Coomassie brilliant blue R (1.25 g of Coomassie brilliant blue dye, 50 ml of acetic acid, 250 ml of ethanol, 250 ml of distilled H2O).
Gel mobility shift assay.
Gel mobility shift (GMS) assays were based on the methods of Fried and Crothers (4) and Garner and Revzin (5). A 759-bp fragment of the gcv control region (nucleotides −466 to +293) was 32P-labeled at a unique EcoRI site by using T4 polynucleotide kinase. The GMS assay was performed by incubating the labeled DNA (<0.75 nM DNA per reaction) in DNA binding buffer (5 mM Tris HCl [pH 7.5], 25 mM KCl, 0.5 mM EDTA, 2.5% glycerol, 0.5 mM dithiothreitol) plus 125 μg of bovine serum albumin/ml for 5 min at 37°C, and then adding 2-μl volumes of twofold serial dilutions of GcvA protein diluted in DNA binding buffer to each reaction (final volume, 20 μl). After an additional 15 min at 37°C, 1 μl of loading buffer (0.1% xylene cyanol, 50% glycerol) was added, and the samples were loaded onto a nondenaturing, 5% polyacrylamide–3% glycine gel.
DNase I protection assay.
The DNase I protection assay is a modified version of the method of Schmitz and Galas (22). The 32P-labeled 759-bp fragment described above was used as a template. The labeled DNA was incubated at 37°C for 5 min in 16-μl reaction mixtures containing DNA binding buffer plus 125 μg of bovine serum albumin/ml, and then 2-μl volumes of twofold serial dilutions of GcvA protein diluted in DNA binding buffer were added and incubated for an additional 15 min. A total of 2 μl of DNase I (1.4 μl of a 1-U/μl solution of DNase I in 19 mM Na-acetate–32 mM CaCl2) was added for 30 s and the reactions were stopped by addition of 5 μl of stop solution (3 M ammonium acetate, 33 μg of sonicated calf thymus DNA/ml, 0.17 M EDTA). The samples were precipitated with ethanol, and the pellets were resuspended in DNA sequencing loading buffer (0.1 M NaOH, 5 M urea, 1 mM EDTA, 0.05% xylene cyanol-bromophenol blue). Samples were loaded onto a 5% polyacrylamide–7 M urea sequencing gel alongside the Maxam and Gilbert (11) A + G and C + T sequencing reactions performed on the same 759-bp 32P-labeled fragment.
RESULTS
Isolation of a GcvA positive-control mutant.
GcvA functions to both activate and repress gcv expression (30, 31). The first objective of this study was to identify single amino acid substitutions in GcvA that result in the loss of transcriptional activation of the gcv promoter but not the loss of DNA binding or repression. The following selection strategy was used to identify such positive-control (PC) mutants. Strain GS1039 is a serA gcvA double mutant that carries a λgcvT-lacZ gene fusion. The double mutant is a serine auxotroph and cannot grow on a plate containing GM supplemented with glycine because there are insufficient C-1 units available for the conversion of glycine into serine via the serine hydroxymethyltransferase reaction (Fig. 1). If wt gcvA is supplied in trans, growth of the double mutant on a GM plate supplemented with glycine resumes because GcvA, in the presence of glycine, activates expression of the genes encoding the GCV enzyme system, resulting in cleavage of glycine and the production of C-1 units needed for serine production. However, the double mutant does not grow on an LM plate supplemented with serine, inosine, and TPEG (an inhibitor of β-galactosidase) when wt gcvA is supplied in trans because GcvA, in the presence of inosine, represses the λgcvT-lacZ gene fusion, resulting in insufficient β-galactosidase activity for growth on lactose. Lysogen GS1039 was transformed with a plasmid pool carrying PCR-induced random base pair changes in gcvA and plated on LB agar (AP was added to all selection and scoring media). Colonies were patched onto an LM plate supplemented with serine, inosine, and TPEG and a GM plate supplemented with glycine. Our selection assumed that transformants containing mutations in gcvA that decreased the GcvA activator function but not the repressor function would fail to grow on either scoring plate. Mutations that inactivate the repressor function of GcvA allow growth on the LM plate because the λgcvT-lacZ gene fusion is no longer repressed and therefore there is sufficient β-galactosidase activity for growth on this medium. By using this selection several hundred transformants were screened for gcvA mutations. One transformant that did not grow at all on either type of scoring medium was isolated and the plasmid carrying the putative gcvA activation-deficient mutation was isolated and designated pGS440.
To demonstrate that plasmid pGS440 carried the potential PC mutation in gcvA, this plasmid and the single copy wt gcvA plasmid were used to transform strain GS986. GS986 is a λ gcvT-lacZ lysogen that carries chromosomal mutations in gcvA and purR. Transformants were grown in GM alone and GM supplemented with either glycine or inosine and assayed for β-galactosidase activity. The GcvA protein encoded on plasmid pGS440 was unable to activate the gcvT-lacZ fusion in response to glycine compared to the level of activation by the wt GcvA protein encoded on plasmid pGS341 (Table 2). However, the mutant GcvA protein still retained the ability to repress gcvT-lacZ expression in the presence of inosine (Table 2). Previous work showed that binding of GcvA to three sites in the gcv control region is required for repression of a gcvT-lacZ fusion (32). Therefore, it was assumed that the GcvA protein encoded on plasmid pGS440 could still bind to DNA.
TABLE 2.
Effects of amino acid substitutions and deletions in GcvA on gcvT-lacZ expression
| Plasmida | gcvA allele | β-Galactosidase activity in GM withb:
|
Fold activityc | Fold repressiond | ||
|---|---|---|---|---|---|---|
| No supple- ment | Glycine | Inosine | ||||
| None | gcvA1 | 104 | 115 | 103 | 1 | 1 |
| pGS341 | wt gcvA | 160 | 826 | 22 | 5.2 | 7.3 |
| pGS440 | Mutant gcvA | 86 | 113 | 18 | 1.3 | 4.8 |
| pGS441 | gcvAF31L | 86 | 124 | 11 | 1.4 | 7.8 |
| pGS442 | gcvAF31A | 71 | 161 | 7 | 2.3 | 10 |
| pGS443 | gcvAL30A | 135 | 522 | 23 | 3.9 | 5.9 |
| pGS444 | gcvAV32A | 104 | 114 | 102 | 1.1 | 1 |
| pGS445 | gcvAS38P | 113 | 103 | 114 | 1 | 1 |
| pGS472 | ΔgcvA944 | 1,048 | 1,151 | 871 | 1 | 1.2 |
All plasmids were assayed in strain GS986λgcvT-lacZ (purR::Tn10 gcvA1).
Cells were grown in GM with the indicated supplements and assayed for β-galactosidase activity. Activity is expressed in Miller units (14).
Data are differences in activity (fold activation) between transformants grown in GM and GM supplemented with glycine.
Data are differences in activity (fold repression) between transformants grown in GM and GM supplemented with inosine.
Identification of the amino acid responsible for the GcvA PC phenotype.
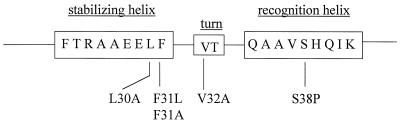
The gcvA gene from plasmid pGS440 was sequenced and five base pair changes were observed. One change, from a T to a C, converted codon 31 from TTT (phenylalanine) to CTT (leucine). Amino acid 31 (aa31) lies in the putative helix-turn-helix (H-T-H) domain of GcvA (Fig. 2) (29) and because of the position of this mutation, we hypothesized that this codon change was most likely responsible for the inability of GcvA to activate the gcvT-lacZ fusion. To confirm this hypothesis, site-directed mutagenesis was used to change codon 31 from a phenylalanine (Phe) to a leucine (Leu) (F31L). The mutation was subcloned into a single copy plasmid, and the plasmid was designated pGS441. This plasmid was transformed into the lysogen GS986, and the transformant was grown in GM and GM supplemented with either glycine or inosine and assayed for β-galactosidase activity. The F31L protein activated and repressed β-galactosidase to levels that were essentially identical to those found with the original transformant (Table 2), indicating that the Leu substitution at aa31 was responsible for the activator-deficient phenotype of the GcvA protein. The gcvAF31L allele was also shown to be recessive, in trans, to a single copy of wt gcvA (data not shown).
FIG. 2.
Putative H-T-H DNA binding domain of GcvA. The amino acids that are part of the H-T-H are denoted as letters inside the boxes and the amino acid changes made are shown below the boxes. The recognition helix likely interacts with target DNA and the stabilizing helix secures the recognition helix in the proper conformation once bound to the target DNA.
To determine whether the Phe at aa31 of GcvA was required for activation or if the Leu substitution at this position altered protein structure, thus diminishing GcvA-mediated activation, codon 31 was site specifically changed to an alanine (Ala) (Materials and Methods). Ala has no side chain beyond the β-carbon to interfere with protein structure. The single copy plasmid, containing the Ala substitution at codon 31 in gcvA (F31A), was transformed into the lysogen GS986, and the transformant was grown in GM and GM supplemented with either glycine or inosine and assayed for β-galactosidase activity. Although the F31A protein displayed a twofold glycine-mediated activation and more severe purine-mediated repression, the phenotype of F31A is similar to that of F31L and the original activator-deficient gcvA isolate (Table 2), indicating that the Phe at aa31 of GcvA is required for glycine-mediated activation of the gcvT-lacZ fusion but not for inosine-mediated repression. Similar to the gcvAF31L activation-deficient mutant, the gcvAF31A allele was also shown to be recessive, in trans, to a single chromosomal copy of the wt gcvA allele (data not shown).
Roles of aa30 and aa32 of GcvA in regulation of the gcv operon.
Several LysR family member proteins have been shown to interact directly with RNAP to mediate transcriptional regulation (7). Therefore, it is possible that aa31 may be part of a larger contact surface on GcvA that interacts with RNAP to activate gcv expression. To investigate this possibility, aa30 and aa32 were changed from a Leu to an Ala and from a Val to an Ala, respectively, and the mutations were subcloned into a single copy plasmid (Fig. 2) (Materials and Methods). The resulting plasmids, pGS443 and pGS444, were transformed into the lysogen GS986, and the transformants were grown in GM and GM supplemented with either glycine or inosine and assayed for β-galactosidase activity. The L30A protein exhibited a small loss of glycine-mediated activation of the gcvT-lacZ fusion, but no loss of inosine-mediated repression (Table 2), indicating that aa30 is involved in glycine-mediated activation but to a lesser extent than aa31. The ability of the mutant L30A protein to repress the gcvT-lacZ fusion in the presence of inosine indicated that the protein was not altered in its DNA binding ability. The V32A protein no longer activated or repressed the gcvT-lacZ fusion (Table 2), indicating that the amino acid at position 32 was important for both functions. The gcvAL30A and gcvAV32A alleles present on single copy plasmids were recessive, in trans, to a single chromosomal copy of the wt gcvA allele (data not shown).
aa38 of GcvA is involved in DNA binding.
The second objective of this study was to identify amino acids in GcvA that are involved in DNA binding. The GcvA protein was shown to bind to the gcv operon and gcvA control regions (32). We assumed that binding occurred through the putative H-T-H DNA binding domain located in the amino-terminal portion of the GcvA protein (Fig. 2). The Ser at position 38 in the H-T-H domain of the GcvA protein is highly conserved among the H-T-H domains of other LysR family member proteins and is thought to be directly involved in DNA binding (21). To determine if this amino acid is important for GcvA binding to the gcv and gcvA control regions, codon 38 was changed to a proline (Pro), and the gcvA mutation was subcloned into a single copy plasmid designated pGS445 (Materials and Methods). This plasmid was transformed into the lysogen GS986, and the transformant was grown in GM and GM supplemented with either glycine or inosine and β-galactosidase activities were measured. The substitution of a Pro for a Ser at aa38 in GcvA rendered the protein unable to activate or repress the gcvT-lacZ fusion (Table 2). The gcvAS38P allele also showed a trans-dominant phenotype to a chromosomal copy of wt gcvA (data not shown).
The C-ter 14 aa’s of the GcvA protein are required for purine-mediated repression of gcv.
The final objective of this study was to determine which amino acids in GcvA are involved in purine-mediated repression. Deletion analysis is often used to elucidate functional domains of proteins and this approach was taken to locate the repressor domain(s) of GcvA. Plasmid pGS472 carries the ΔgcvA944 allele, where the C-ter 14 aa’s of the corresponding GcvA protein are deleted (Materials and Methods). This plasmid was transformed into the lysogen GS986, and the transformant was grown in GM and GM supplemented with either glycine or inosine and β-galactosidase activities were measured. Deleting the C-ter 14 aa’s of GcvA caused the gcvT-lacZ fusion to be expressed constitutively at a high level under all growth conditions (Table 2), suggesting that the C-ter portion of GcvA is involved in purine-mediated repression of the gcv operon. Because the deletion protein activated the gcvT-lacZ fusion so highly, it was assumed that the protein could bind DNA. A wt chromosomal copy of gcvA was shown to be trans-dominant to the ΔgcvA944 allele (data not shown).
Binding of the mutant GcvA proteins to the gcv control region.
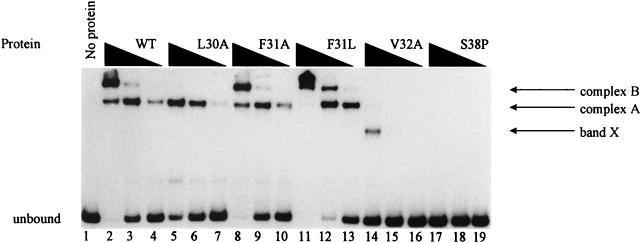
The DNA binding abilities of the mutant GcvA proteins described above were based on previous work which demonstrated that GcvA-mediated repression of the gcv operon requires the binding of GcvA to three sites in the gcv control region (32). Therefore, it was assumed that the F31L, F31A, and L30A GcvA proteins could bind DNA normally based on their ability to repress a gcvT-lacZ fusion. However, the V32A and S38P proteins could not repress or activate a gcvT-lacZ fusion; thus, it was assumed that these proteins could not bind DNA. To verify these assumptions, we used GMS assays to show in vitro whether the mutant GcvA proteins could bind to the gcv control region. The F31A and F31L GcvA proteins could bind to the gcv DNA template as well as or better than the wt GcvA protein (Fig. 3, compare lanes 2 to 4 with lanes 8 to 10 and 11 to 13), indicating that DNA binding had not been significantly altered in these two mutant proteins. The L30A GcvA protein displayed less than a twofold difference in binding affinity for the gcv template compared to that of the wt GcvA protein (Fig. 3, compare lanes 2 to 4 and lanes 5 to 7). Complex B was not seen in the GMS assay at the L30A protein concentration used (Fig. 3, lanes 5 to 7). However, additional GMS assays have shown that complex B does form when the L30A protein concentration is approximately 20 nM (data not shown). The S38P GcvA protein, which caused the loss of both activation and repression of a gcvT-lacZ fusion and was dominant in trans to wt GcvA, could no longer bind to the gcv DNA template (Fig. 3, lanes 17 to 19). The V32A protein, which also caused the loss of both activation and repression of gcvT-lacZ expression (Table 2), required approximately four times the mutant protein to bind the gcv DNA template compared to that of the wt protein (Fig. 3, compare lanes 2 to 4 and lanes 14 to 16). Furthermore, in the V32A protein GMS assay a band of faster mobility was observed which did not occur in the wt GcvA GMS pattern (Fig. 3, compare lanes 2 to 4 and lane 14, see band X).
FIG. 3.
GMS assay for the binding of wt and mutant GcvA proteins to the gcv control region. A 759-bp DNA fragment containing the gcv control region was 32P-labeled and incubated with twofold serial dilutions of purified mutant or wt GcvA protein. The unbound 32P-labeled gcv DNA fragment is indicated. Complexes A and B indicate GcvA bound to sites 2 and 3, and sites 1, 2, and 3 of the gcv control region, respectively (see Fig. 4). Band X is an anomalous band seen only with the GcvA V32A protein. Lane 1, no protein; lanes 2 to 4, 10.0, 5.0, and 2.5 nM wt GcvA, respectively; lanes 5 to 7, 10.0, 5.0, and 2.5 nM L30A GcvA, respectively; lanes 8 to 10, 10.0, 5.0, and 2.5 nM F31A GcvA, respectively; lanes 11 to 13, 10.0, 5.0, and 2.5 nM F31L GcvA, respectively; lanes 14 to 16, 10.0, 5.0, and 2.5 nM V32A GcvA, respectively; lanes 17 to 19, 10.0, 5.0, and 2.5 nM S38P GcvA, respectively.
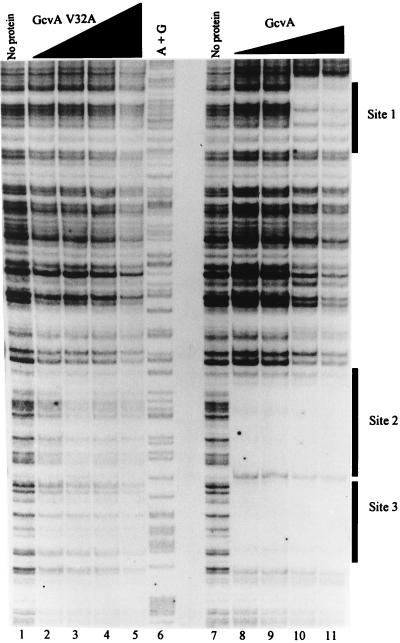
Because the V32A GcvA protein displayed an altered GMS pattern compared to that of the wt GcvA protein, we wanted to determine if the mutant protein was binding to GcvA sites 1, 2, and 3 or if it was binding nonspecifically to the DNA. Therefore, a DNase I protection assay was performed on the gcv promoter region (Materials and Methods) with both the mutant V32A GcvA protein and the wt GcvA protein. The wt GcvA protein protected sites 2 and 3 from DNase I digestion at the lowest protein concentration used (25 nM) (Fig. 4, lane 8) and an approximately fourfold higher concentration (100 nM) was required for protection of site 1 (Fig. 4, lane 10). The V32A protein did not protect sites 1, 2, or 3 from DNase I digestion even as the protein concentration was increased from 50 to 400 nM (Fig. 4, lanes 2 to 5). Thus, although the V32A protein showed weak binding to the gcv control region in the GMS assay, a specific target site was not detected by DNase I footprinting.
FIG. 4.
Protection from DNase I digestion of the gcv control region by wt and V32A GcvA proteins. A 759-bp 32P-labeled DNA fragment containing the gcv control region was incubated with twofold serial dilutions of wt and V32A gcvA proteins (Materials and Methods) and digested with DNase I. The partial digestion products were run on a denaturing 5% polyacrylamide gel adjacent to the Maxam-Gilbert A + G sequencing reactions. Lane 1, no protein; lanes 2 to 5, V32A GcvA at 50, 100, 200, and 400 nM, respectively; lane 6, A + G sequencing reaction; lane 7, no protein; lanes 8 to 11, wt GcvA at 25, 50, 100, and 200 nM, respectively.
DISCUSSION
Direct contact of transcriptional activators with the C-ter domain of the α-subunit of RNAP is required for initiation of transcription at many bacterial promoters (7). One role of these activators in transcription is to recruit RNAP to the promoter. The domains of activator proteins involved in making contact with RNAP can be defined through identification of PC mutants in the activators that block activation but do not affect binding to DNA. We isolated two putative PC mutations in gcvA where the encoded gene products could no longer fully activate gcvT-lacZ expression in response to glycine but could still repress the fusion in response to purines. An Ala substitution at aa31 of GcvA showed that the Phe at this position in wt GcvA provides a critical side chain required for glycine-mediated activation of the gcv operon. The Leu at position 30 was also identified through Ala substitution to play a minor role in transcriptional activation. These two amino acids possibly define a surface-exposed region that interacts with RNAP.
Several LysR family member proteins have been shown to interact with the C-ter domain of the α-subunit of RNAP (7). Thus, it is possible that GcvA may also interact with the α-subunit. We set up a genetic selection to screen for suppressor mutations in the rpoA gene, encoding the α-subunit of RNAP, specific for the F31L PC mutant. However, we have not been successful in isolating such suppressors. It is possible that multiple base pair changes are required to generate the necessary amino acid substitution in the α-subunit that would recognize the mutant F31L GcvA protein or that the change required in rpoA is lethal. We have, however, isolated mutations in the rpoA gene that result in a GcvA-dependent loss of transcriptional activation at the gcv promoter, providing evidence that RNAP and GcvA interact to mediate regulation (7a). If the GcvA protein interacts directly with RNAP from sites 3 and 2, this would be unusual for a sigma 70-activated promoter. Activation generally occurs through a mechanism that involves at least one target site that is located near the promoter such that the activator can touch RNAP in a way that a high level of activation is achieved (3). Although a GcvA binding site occurs from bp −69 to −34, this site functions only in GcvA-mediated repression (32). Although this site is within what might be considered a normal activation location (3), presumably the activator domain of the GcvA protein is unexposed or unable to make an appropriate contact with RNAP from this site.
The serine at position 38 of GcvA is involved in DNA binding. The S38P GcvA protein could not bind to the gcv control region in vivo or in vitro, and the gcvAS38P allele was trans-dominant to the wt gcvA allele. These data suggest that multimerization is occurring between the S38P and wt GcvA proteins and that the mutant protein likely maintains an otherwise native conformation despite the Pro substitution. Initially we hypothesized that amino acid V32 might be involved in activation because it lies adjacent to amino acid F31 and could be part of the same activation region. However, phenotypically the Ala substitution at position 32 caused both the loss of activation and repression of a gcvT-lacZ fusion, and in vitro and in vivo it caused the loss of DNA binding to the gcv control region. If aa32 is involved in DNA binding, we hypothesized that the V32A protein would be dominant in trans to the wt GcvA protein, like the S38P GcvA protein. However, the gcvAV32A allele was recessive to the wt gcvA allele. Therefore, the specific defect of this mutant is not clear from the phenotype. One explanation for the phenotype is that the V32A substitution prevents multimerization with other GcvA monomers. This is consistent with the high level of V32A GcvA protein required in vitro to bind gcv DNA (Fig. 2 and 3). aa32 is in the turn between the two helices of the H-T-H domain (Fig. 2). If the V32A substitution no longer allows multimerization of GcvA, this would suggest that the residues involved in multimerization and the residues involved in activation are adjacent in the GcvA protein. It is also possible that the V32A change has altered the overall protein structure in such a way that the protein monomers can no longer interact with the wt GcvA, thus accounting for the recessive phenotype.
GcvR is also involved in negative regulation of the gcv operon, and the ability of GcvR to repress gcv requires a functional GcvA protein, even when GcvR is overexpressed (6). One model for GcvR involvement in gcv regulation hypothesizes that a GcvA-GcvR heterocomplex might form in response to high purine levels and function as a repressor for the gcv operon. We have shown that deleting the C-ter 14 aa’s of GcvA results in the loss of purine-mediated repression and constitutive expression of a gcvT-lacZ fusion (Table 2). We believe that the C-ter 14 aa’s of GcvA are part of a region that might interact with GcvR to repress the gcv operon. We are using random and site-directed mutagenesis reactions to determine which of the 14 aa’s are required for repression and to further define repressor regions.
The results of this study have allowed us to form a crude map of the functional regions of the GcvA protein. The isolation of additional mutations, coupled with a careful biochemical analysis of the mutant proteins, should define the functional regions of GcvA and further our knowledge of how transcriptional regulators interact with other regulatory proteins and RNAP to control transcription initiation.
ACKNOWLEDGMENT
This investigation was supported by Public Health Service grant GM26878 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Calvo J M, Matthews R G. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi K Y, Zalkin H. Structural characterization and corepressor binding of the Escherichia coli purine repressor. J Bacteriol. 1992;174:6207–6214. doi: 10.1128/jb.174.19.6207-6214.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fried M, Crothers D M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garner M M, Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981;9:3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghrist A C, Stauffer G V. Characterization of the Escherichia coli gcvR gene encoding a negative regulator of gcv expression. J Bacteriol. 1995;177:4980–4984. doi: 10.1128/jb.177.17.4980-4984.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishihama A. Protein-protein communication within the transcription apparatus. J Bacteriol. 1993;175:2483–2489. doi: 10.1128/jb.175.9.2483-2489.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Jourdan, A., and G. Stauffer. Unpublished data.
- 8.Kahn M, Kolter R, Thomas C, Figurski D, Meyer R, Remaut E, Helinski D R. Plasmid cloning vehicles derived from plasmids ColE1, F, R6K, and RK2. Methods Enzymol. 1979;68:268–280. doi: 10.1016/0076-6879(79)68019-9. [DOI] [PubMed] [Google Scholar]
- 9.Kikuchi G. The glycine cleavage system: composition, reaction mechanism, and physiological significance. Mol Cell Biochem. 1973;1:169–187. doi: 10.1007/BF01659328. [DOI] [PubMed] [Google Scholar]
- 10.Kilstrup M, Meng L M, Neuhard J, Nygaard P. Genetic evidence for a repressor of synthesis of cytosine deaminase and purine biosynthesis enzymes in Escherichia coli. J Bacteriol. 1989;171:2124–2127. doi: 10.1128/jb.171.4.2124-2127.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 12.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 13.Mudd S H, Cantoni G L. Biological transmethylation, methyl-group neogenesis and “one-carbon” metabolic reactions dependent upon tetrahydrofolic acid. In: Florkin M, Stotz E H, editors. Comprehensive biochemistry. Vol. 15. Amsterdam, The Netherlands: Elsevier Publishing Co.; 1964. pp. 1–47. [Google Scholar]
- 14.Newman E B, D’Ari R, Lin R T. The leucine-Lrp regulon in E. coli: a global response in search of a raison d’etre. Cell. 1992;68:617–619. doi: 10.1016/0092-8674(92)90135-y. [DOI] [PubMed] [Google Scholar]
- 15.Pizer L I. Glycine synthesis and metabolism in Escherichia coli. J Bacteriol. 1965;89:1145–1150. doi: 10.1128/jb.89.4.1145-1150.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pizer L I, Potochny M L. Nutritional and regulatory aspects of serine metabolism in Escherichia coli. J Bacteriol. 1964;88:611–619. doi: 10.1128/jb.88.3.611-619.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plamann M D, Rapp W D, Stauffer G V. Escherichia coli K12 mutants defective in the glycine cleavage enzyme system. Mol Gen Genet. 1983;192:15–20. doi: 10.1007/BF00327641. [DOI] [PubMed] [Google Scholar]
- 18.Rolfes R J, Zalkin H. Regulation of Escherichia coli purF. J Biol Chem. 1988;263:19641–19652. [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Sarkar G, Sommer S. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 21.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:596–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 22.Schmitz A, Galas D J. The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res. 1979;6:111–137. doi: 10.1093/nar/6.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stauffer G V, Fogarty S, Stauffer L T. Characterization of the Escherichia coli gcv operon. Gene. 1994;142:17–22. doi: 10.1016/0378-1119(94)90349-2. [DOI] [PubMed] [Google Scholar]
- 24.Stauffer L T, Stauffer G V. Characterization of the gcv control region from Escherichia coli. J Bacteriol. 1994;176:6159–6164. doi: 10.1128/jb.176.20.6159-6164.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stauffer L T, Stauffer George V. Spacing and orientation requirements of the GcvA-binding sites 3 and 2 and the Lrp-binding region for gcvT-lacZ expression in Escherichia coli. Microbiology. 1998;144:1417–1422. doi: 10.1099/00221287-144-5-1417. [DOI] [PubMed] [Google Scholar]
- 26.Steiert P S, Stauffer L T, Stauffer G V. The lpd gene product functions as the L protein in the Escherichia coli glycine cleavage enzyme system. J Bacteriol. 1990;172:6142–6144. doi: 10.1128/jb.172.10.6142-6144.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang H, Severinox K, Goldfarb A, Ebright R H. Rapid RNA polymerase genetics: one-day, no column preparation of reconstituted recombinant Escherichia coli RNA polymerase. Proc Natl Acad Sci USA. 1995;92:4902–4906. doi: 10.1073/pnas.92.11.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 29.Wilson R L, Stauffer G V. DNA sequence and characterization of GcvA, a LysR family regulatory protein for the Escherichia coli glycine cleavage enzyme system. J Bacteriol. 1994;176:2862–2868. doi: 10.1128/jb.176.10.2862-2868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson R L, Stauffer L T, Stauffer G V. Roles of the GcvA and PurR proteins in negative regulation of the Escherichia coli glycine cleavage enzyme system. J Bacteriol. 1993;175:5129–5134. doi: 10.1128/jb.175.16.5129-5134.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson R L, Steiert P S, Stauffer G V. Positive regulation of the Escherichia coli glycine cleavage enzyme system. J Bacteriol. 1993;175:902–904. doi: 10.1128/jb.175.3.902-904.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson R L, Urbanowski M L, Stauffer G V. DNA binding sites of the LysR-type regulator GcvA in the gcv and gcvA control region of Escherichia coli. J Bacteriol. 1995;177:4940–4946. doi: 10.1128/jb.177.17.4940-4946.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Y, Zhang X, Ebright R H. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 1991;19:6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]