ABSTRACT
Human leukocyte antigen (HLA)-DPB1 antigens are mismatched in approximately 70% of allogeneic hematopoietic stem cell transplantations (allo-HSCT) from HLA 10/10 matched unrelated donors. HLA-DP-mismatched transplantation was shown to be associated with an increase in acute graft-versus-host disease (GVHD) and a decreased risk of leukemia relapse due to the graft-versus-leukemia (GVL) effect. Immunotherapy targeting mismatched HLA-DP is considered reasonable to treat leukemia following allo-HCT if performed under non-inflammatory conditions. Therefore, we isolated CD4+ T cell clones that recognize mismatched HLA-DPB1 from healthy volunteer donors and generated T cell receptor (TCR)-gene-modified T cells for future clinical applications. Detailed analysis of TCR-T cells expressing TCR from candidate clone #17 demonstrated specificity to myeloid and monocytic leukemia cell lines that even expressed low levels of targeted HLA-DP. However, they did not react to non-hematopoietic cell lines with a substantial level of targeted HLA-DP expression, suggesting that the TCR recognized antigenic peptide is only present in some hematopoietic cells. This study demonstrated that induction of T cells specific for HLA-DP, consisting of hematopoietic cell lineage-derived peptide and redirection of T cells with cloned TCR cDNA by gene transfer, is feasible when using careful specificity analysis.
Key Words: TCR-T, GVHD, leukemia, allogeneic transplantation
INTRODUCTION
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is an established treatment for refractory hematological malignancies.1,2 Allo-HSCT is based on genetic differences between donors and patients that form allogeneic antigens and induce various immune responses.3-6 Infused donor lymphocytes may lead to life-threatening graft-versus-host disease (GVHD); however, graft-versus leukemia/lymphoma (GVL) can induce a strong anti-tumor effect.3,4 Although mortality and morbidity caused by high-dose chemotherapy, infections, and organ damage have been decreased owing to improvements in appropriate management, the survival rate after transplantation remains at approximately 50% over the last two decades, mainly due to the unchanged rate of recurrent malignancies, especially in patients with advanced disease.7-9 Therefore, more effective strategies are needed to prevent relapse and establish treatment methods.10
Transplantation from Human leukocyte antigen (HLA)-A, -B, -C, -DRB1, and-DQB1-matched unrelated donors (10 loci/10 loci) represents the first choice for patients.11 However, recent studies showed that HLA-DPB1 antigen mismatch occurs in approximately 70% of unrelated allo-HSCT cases, and several HLA-DPB1 mismatch combinations decrease the risk of leukemia relapse, and may also increase the risk of GVHD.12-14 As HLA-DPB1 expression is restricted to hematopoietic cells and to a lesser extent, inflamed non-hematopoietic tissues,15 it is unlikely that donor T cells specific for the mismatched HLA-DP allele will induce uncontrollable severe GVHD, suggesting that mismatched HLA-DP is a promising therapeutic target for leukemia relapse following allo-HSCT. Several reports have demonstrated that CD4+ HLA-DP-restricted T cell clones isolated from patients receiving unrelated allo-HSCT or healthy donors, exert anti-leukemic activities.15-19
Recently, chimeric antigen receptor T cells have been introduced into clinics with great success for the treatment of relapsed and refractory CD19-positive leukemia and lymphoma as genetically engineered T cell therapies. However, the number of promising targetable cell surface antigens is limited, ie, in B lymphocyte malignancies.20 The other form of genetically engineered T cells is T-cell receptor (TCR)-modified T cells; however, there have been no clinically approved products to date. Given that TCR recognizes the major histocompatibility antigen and its bound peptides derived from intracellular proteins, the number of targetable antigens is much higher.21,22 In this study, by taking advantage of the features of TCR-T cells, we sought to generate CD4+ T cell clones specific for allogeneic HLA-DP-bound antigens to treat hematological malignancies that relapse following HLA-DP-mismatched allo-HSCT, because CD4+ cytotoxic T cells are indispensable effector T cells in cancer immunity.23 In addition, we focused on HLA-DP alleles such as HLA-DPB1*09:01, whose frequency is relatively high in the Japanese population24 to cover patients in this ethnic group. Finally, we demonstrated that introduction of the inducible HLA-DP expression system could help isolate promising T cell clones with sufficient TCR affinity.
MATERIALS AND METHODS
Sample collection and cell culture
Peripheral blood samples were obtained from healthy donors after obtaining written informed consent. Peripheral blood mononuclear cells were isolated by Ficoll separation and used for preparation of responder T cell and B-lymphoblastoid cell lines. They were frozen and stored at −80 °C until use. Primary leukemia cells were obtained from patients diagnosed with acute leukemia after obtaining written informed consent. Genomic DNA was purified from samples and subjected to HLA typing (HLA Laboratory, Kyoto, Japan). The studies were approved by the Ethics Committee of the Nagoya University Graduate School of Medicine (approval numbers:2018-0246 and 2019-0291) in accordance with the ethical guidelines established by the Japanese Ministry of Health, Labour, and Welfare.
The human acute myeloid leukemia cell lines HL-60; human chronic myeloid leukemia-originated cell lines K562 and MEG-01; human lymphoid leukemia/lymphoma cell lines U266, NALM6, U-937, Raji, and Jurkat; and human cervical adenocarcinoma cell line HeLa were obtained from American Type Culture Collection (ATCC, USA). The human acute myeloid leukemia cell lines MOLM-13, KG-1, HYT-1, and Kasumi-1 were obtained from the Japanese Collection of Research Bioresources Cell Bank, whereas the HYT-1 and THP-1 cell lines were provided by the RIKEN BRC through the National BioResource Project of the MEXT/AMED, Japan. The exact phenotypes of the cell lines are summarized in Table 1. The cell lines were maintained in advanced RPMI 1640 supplemented with penicillin/streptomycin/L-glutamine (PSG) and 2% fetal bovine serum (FBS) (all from Thermo Fisher Scientific, USA), except for the HYT-1 cell line, which was maintained in media supplemented with 10% FBS and 10 ng/mL of granulocyte macrophage colony stimulating factor (PeproTech, USA). HeLa cells (ATCC) were transduced with HLA-DMA, HLA-DMB, and invariant chain (hereafter called HeLa/DMI), and maintained in Iscove’s Modified Dulbecco’s Medium supplemented with PSG and 10% FBS. Epstein-Barr virus-transformed B-lymphoblastoid cell lines were established via infection of B95-8 supernatant with 2 × 106 peripheral blood mononuclear cells and maintained in advanced RPMI 1640 supplemented with PSG and 15% FBS. These cell lines were infected with a modified pLZRS-Rfa retrovirus plasmid (Addgene, USA) encoding HLA-DP α and β cDNA, followed by IRES and truncated nerve growth factor receptor (ΔNGFR) using a Galv-transduced retrovirus packaging cell line (Phoenix-GP, ATCC) and Retronectin (Takara Bio), and finally, positively selected using anti-NGFR antibody and magnetic-activated cell sorting (MACS) system (Miltenyi Biotec, Germany).
Table 1.
| Cell line | Disease of tissue origin | Lineage and comments |
| THP-1 | AML (FAB M5) | Monocytic |
| U-937 | Histiocytic lymphoma | Monocytic, t(10;11) |
| HL-60 | AML (FAB M2) | Promyelocytic |
| MOLM-13 | AML (FAB M5a) | Monocytic, ins(11;9) |
| KG-1 | AML (erythroleukemia) | Myeloblast |
| Kasumi-1 | AML (FAB M2) | Myeloblast, t(8;21) |
| HYT-1 | AML (secondary) | Monocytic |
| MEG-01 | CML, blast crisis | Megakaryoblast, t(9;22) |
| K562 | CML, blast crisis | Erythroid-Myeloid precursor |
| U266 | Myeloma | Plasma cell |
| NALM6 | ALL (pre-B) | B lymphocyte |
| Raji | Burkitt’s lymphoma | B lymphocyte |
| Jurkat | ALL | T lymphocyte |
| HeLa | Cervical adenocarcinoma | Epithelial cell |
ALL: acute lymphoid leukemia
AML: acute myeloid leukemia
CML: chronic myeloid leukemia
FAB: French-American-British classification
HLA-DR and -DP expression in cell lines and primary leukemia cells was examined with anti-HLA-DR (clone L243, BD Biosciences, USA) and anti-HLA-DP (clone B7/21, BD Biosciences), respectively, using LSRFortessa X-20 flow cytometry and FlowJo software (Beckton Dickinson, USA). If necessary, HLA-DP was induced in primary leukemia cells by treatment with interferon-gamma (IFN-γ; 200 JRU/mL, Imunomax-γ, Shionogi) and/or tissue necrosis factor-alpha (TNF-α; 10 ng/mL, Peprotech, USA) for 48 h.
Establishment of allo-HLA-DP-specific clones
Responder CD45RA+ CD4+ T cells were isolated from donor peripheral blood mononuclear cells by MACS separation as follows: CD4+ cells were first isolated using a REAlease CD4 MicroBead kit, followed by positive isolation with the anti-CD45RA antibody (all from Miltenyi Biotec).
CD4+ CD45RA+ T cells (1 × 106) were cocultured with 100 Gy-irradiated K562 cells (an erythron-myeloid leukemia cell line that lost whole HLA expression25) stably transduced with CD86 (hereafter called K86) expressing allogeneic HLA-DP α and β cDNA of interest at a ratio of 5:1 in the presence of 20 U/mL interleukin (IL)-2 (Imunase, Shionogi) and restimulated after 2 weeks. The cultures were fed 2–3 times a week. Allogeneic HLA-DP-specific responses were evaluated based on the expression of the activation marker CD154 on CD4+ T cells.26 The MAC-sorted CD154 positive cells were cloned by limiting dilution in the presence of feeder cells (5 × 104 irradiated pooled peripheral blood mononuclear cells and 1 × 104 irradiated B-lymphoblastoid cell lines), 30 ng/mL OKT3 (Takara), and 50 U/mL IL-2 in 96-well round-bottom plates for 14 d. Growing cells were further expanded and subjected to more functional studies.
Tetracycline-inducible HLA-DP expression system
To examine the reactivity of individual T-cell clones toward cell lines expressing decreased levels of cognate HLA-DP molecules, a tetracycline-inducible system was used. The HLA-DPB1*09:01-P2A-HLA-A1*02:01 fragment was inserted into the pRetroX-TetOne 3G vector (Takara Bio) and transduced into Phoenix-GP/Galv packaging cells. The supernatant was used to infect K86 cells, as described above. Finally, single-cell cloning was performed to establish K86 cells that were best controlled for HLA-DP expression using a graded concentration of doxycycline (DOX) (Takara Bio).
Generation of genetically-modified T cells
TCR-α and TCR-β gene sequences from selected T cell clones were determined by RT-PCR using multiplex primers as reported.27,28 To increase gene expression, whole TCR sequences were codon optimized, and the constant regions were murinized as previously reported.29 The TCRα and TCRβ genes were cloned into the retroviral vector, pSplice-TCR-BC1 (Takara Bio).30 The vector was transduced into the GP13 packaging cell line (ATCC) and the culture supernatant was collected after 48 h, and infected to 3-day activated T cells with CD3/28 beads (Thermo Fisher Scientific) in the presence of 50 U/mL IL-2 in a Retronectin (Takara Bio)-coated plate by “Spinfection” according to the manufacturer’s instruction. Resultant TCR-T cells were analyzed for transduction efficiency five days after transduction with PE-labeled anti-TCRVβ18 antibody (#IM2049, Beckman Coulter, USA).
Enzyme-linked immunosorbent assays
Aliquots of T cell clones or TCR-T cells (1 × 105) were co-incubated with various tumor cells (1 × 105) forcibly expressing the HLA-DP restriction molecules for 6 h. For blocking studies, the target cells were pretreated with either control mouse IgG (Wako), anti-HLA-DR (BioLegend, USA), -DP (Cosmo Bio), or -DQ (Novus Bio, USA) antibody at a final concentration of 10 μg/mL for 1 h at room temperature, and subsequently cocultured with effector TCR-T cells for 6 h. TNF-α and IFN-γ production in the supernatant was evaluated using conventional sandwich enzyme-linked immunosorbent assays with a pair of relevant antibodies against TNF-α and IFN-γ (all from R&D Systems, USA) and 3,3′,5,5′-tetramethylbenzidine colorimetric reagents (Sigma-Aldrich, USA). Absorbance at 650 nm was measured on a plate reader using Cytation (Agilent Technologies, USA).
Cytotoxicity assays
For cellular cytotoxicity assays, target cells were first labeled with CytoTell Red 650 (AAT Bioquest, USA) and co-incubated with TCR-T cells at graded effector/target (E/T) ratios. After 48 h, all cells were collected and stained with Fixable Viability Dye eFluor450 (Thermo Fisher). Thereafter, live and dead cells were counted using LSRFortessa X-20 flow cytometer and FlowJo software.
Intracellular cytokine staining
T cell clones and TCR-T cells were evaluated for cytokine production at the single-cell level. Responder T cells were cocultured with target cells for 4–6 h. To stop cytokine secretion, Brefeldin A (BD Bioscience, USA) was added to cultures for the last 2 h. Thereafter, cultured cells were stained first with cell surface antibodies FITC-CD3, APC-Cy7-CD4 (BioLegend, USA), and APC-CD107a (clone H4A3) if necessary, for 15 min at 4 °C, followed by Fixable Viability Dye eFluor450 for 15 min at 4 °C. Finally, cells were stained for intracellularly trapped cytokines with PE-IFN-γ (clone B27), APC-TNF-α (clone MAb11) and AF488-granzyme B (clone CB9) using a Cytofix/Cytoperm kit (BD Biosciences), according to the manufacturer’s instructions. Stained cells were analyzed using LSRFortessa X-20 and FlowJo software.
Statistical analyses
The paired t-test was used to assess the differences between the groups. The differences were considered statistically significant at P <0.05.
RESULTS
Induction of allo-reactive T cells upon stimulation with mismatched HLA-DP-expressing K86 cells
The schematic of the experimental procedure is shown in Figure 1A. As it has been shown to be that allo-reactive T cells are mainly present in the naïve T cell fraction in healthy subjects,31 we decided to use CD45RA+ CD4+ T cells as responders. Therefore, CD45RA+ CD4+ T cells from two healthy donors who do not possess the HLA-DPB1*09:01 allele were stimulated with irradiated K86 cells expressing HLA-DPB1*09:01-P2A-HLA-A1*02:01 (hereafter referred to as K86/B0901), as described in the Methods section. The specific T cell response was evaluated via the elevated expression of CD154 on CD4+ T cells26 (Fig. 1B), indicating that HLA-DPB1*09:01-reactive T cells were indeed induced during culture. CD154+ T cells were sorted, cloned by limiting dilution, and subsequently expanded. The propagated T cell clones showed various degrees of IFN-γ production in response to K86/B0901 via enzyme-linked immunosorbent assay; however, only a small portion of T cell clones showed substantial IFN-γ production (15/97, 15.5% in Donor #1-derived clones, 10/90, 11.1% in Donor #2; Fig. 1C). More than half of these T cell clones showed reactivity to both K86/B0901 and HeLa/DMI/B0901 (a non-hematopoietic cell line forced to express HLA-DPB1*09:01-P2A-HLA-A1*02:01), as shown in Fig. 1D. Others (2/15 in donor #1-drived clones; 4/10 in donor #2-derived clones) showed reactivity against K86/B0901 only. As the goal of this study was to generate T cells reactive to allogeneic HLA-DP expressed on hematopoietic cells, and not non-hematopoietic cells, for the development of TCR-T cells useful for treating relapsed leukemia without inducing GVHD, we selected clones that showed selective reactivity against K86/B0901. A representative reaction pattern for clone #17 is shown in Fig. 1E.
Fig. 1.
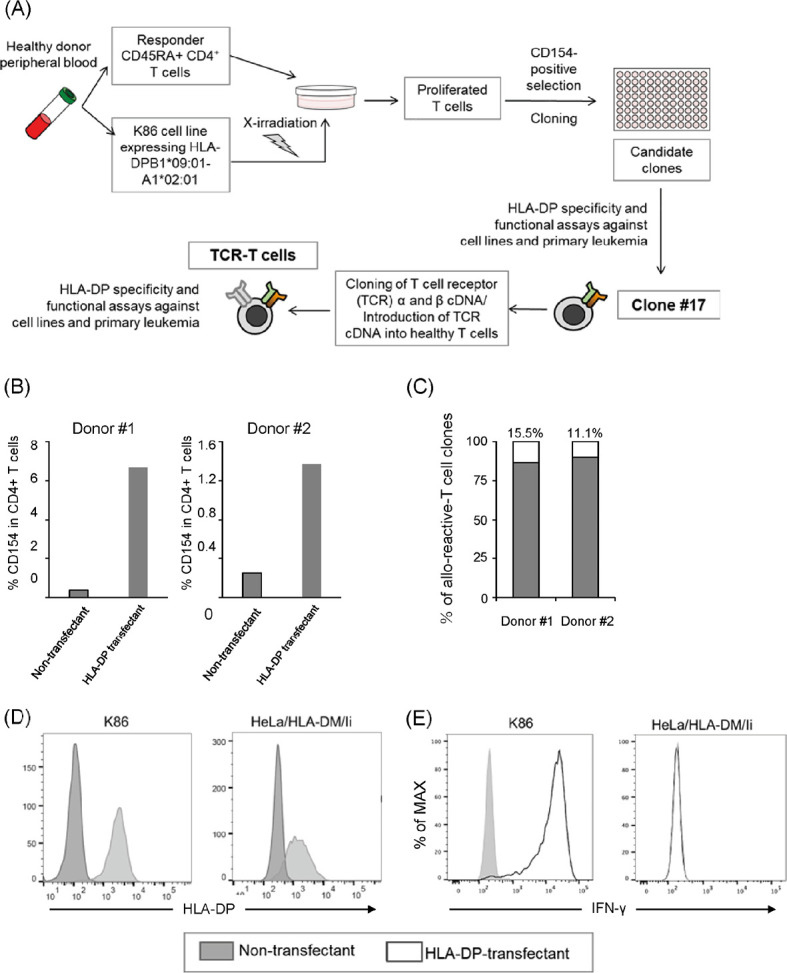
Induction of allo-reactive T cells upon stimulation with mismatched HLA-DP-expressing K86 cells
Fig. 1A: Schematic of experimental procedure from bulk culture to TCR-T cell preparation.
Fig. 1B: In vitro T cell lines were stimulated by CD86-transduced K562 (K86) cells expressing HLA-DPB1*09:01. After 6 h, CD154 expression on CD4+ T cells were examined.
Fig. 1C:Allo-reactivity of the expanded T cell clones assessed via IFN-γ enzyme-linked immunosorbent assay. Black bar = non-reactive T cell clones, white bar = reactive T cell clone.
Fig. 1D: Expression level of HLA-DP in K86 or HeLa cells transduced with HLA-DM and invariant chain (HeLa/DMI) cells with/without transduction with a retroviral vector encoding HLA-DPB1*09:01-P2A-HLA-DPA1*02:01.
Fig. 1E:Clone #17 was cultured with K86 or HeLa/DMI cells with/without HLA-DPB1*09:01-P2A-HLA-DPA1*02:01 transduction for 6 h. IFN-γ production was examined via intracellular staining.
Study acronyms are explained in Abbreviations to main body.
Reactivity of clone #17 against a panel of tumor cell lines
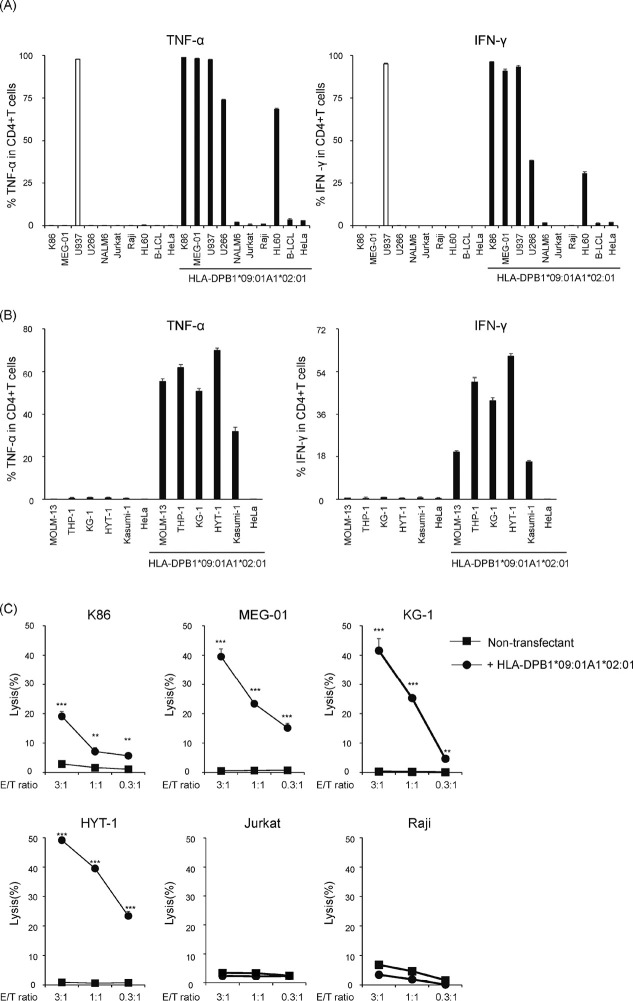
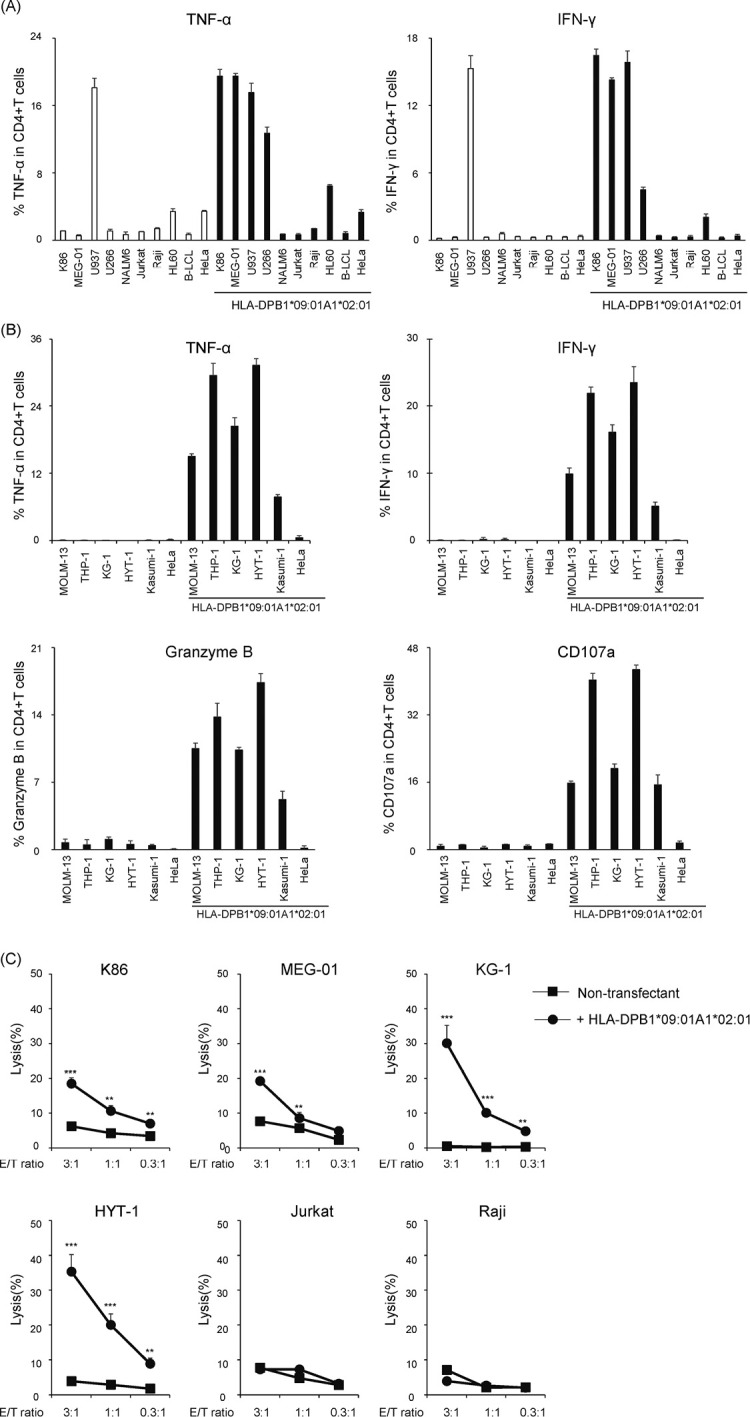
To investigate the reactivity of B0901-specific T cell clone #17, an aliquot of clone #17 was stimulated with various tumor cell lines (all confirmed negative for the HLA-DPB1*09:01 allele) transduced with B0901 and examined for intracellular production of TNF-α and IFN-γ (Fig. 2A and 2B). Flow cytometric analysis demonstrated that clone #17 produced both TNF-α and IFN-γ against leukemia cell lines of myeloid origin such as K86, KG-1, Kasumi-1, HL60, HYT-1, THP-1, MOLM-13, myeloma-derived U266, and MEG-01 (established from a patient with megakaryoblastic transformation of chronic myeloid leukemia) in a B0901-restricted manner, and not against other tumor cell lines. U937 cells of histiocytic lymphoma origin that do not express HLA class II32 were recognized in the absence of HLA-DP expression by flow cytometry, suggesting that the observed recognition was non-specific. These data indicate that the T cell clone might recognize antigens that are limited to certain hematopoietic lineages. Furthermore, we conducted cytotoxicity assays using CytoTell Red 650 labeling. The ability of T cell clones to lyse tumor cells was assessed using graded E/T ratios (Fig. 2C). Consistent with the pattern of TNF-α and IFN-γ production, clone #17 exhibited cytotoxicity against KG-1, HYT-1, K86, and MEGO1, and not against Jurkat and Raji cell lines derived from the lymphoid lineage (Fig. 2B). These results demonstrated that clone #17 could specifically mediate T cell effector functions mainly against myeloid and monoblastic/monocytic leukemia cells and potentially myeloma cells and megakaryoblastic leukemia cells, but not in other tumor cell lines.
Fig. 2.
Recognition of various tumor cell lines using clone #17
Fig. 2A:Clone #17 was co-incubated with various tumor cell lines with/without HLA-DPB1*09:01-P2A-HLA-DPA1*02:01 for 6 h. TNF-α and IFN-γ production was examined via intracellular staining.
Fig. 2B:A series of myeloid and monocytic cell lines was further tested as shown above.
Fig. 2C: Cytotoxicity of clone #17 was evaluated by the decrease of fluorescence-labeled target at graded E/T ratio. Percent lysis is indicated as the mean ±SD and a representative result of at least two independent experiments is shown. Statistical significance was determined using two-tailed Student’s t-test. (**P <0.01, ***P <0.001).
Study acronyms are explained in Abbreviations to main body.
Reactivity of clone #17 against primary leukemia cells
Next, we tested the reactivity of clone #17 against primary leukemia blasts obtained from patients at diagnosis. Of the 15 samples collected, three patients diagnosed with acute myeloid leukemia were identified to possess HLA-DPA1*02:01 and HLA-DPB1*09:01 alleles (Table 2). They were thawed and checked for viability (>90% for all three samples). Then, they were pre-treated with or without a cocktail of IFN-γ and TNF-α for 48 h and tested for HLA-DR and HLA-DP expression. As shown in Fig. 3A, all cells were positive for HLA-DR expression, and HLA-DP was weakly expressed. Following cytokine stimulation, two of the three leukemia samples upregulated HLA-DP expression, whereas one sample (Leuk-11) decreased its expression. Then, these leukemia samples, K86 expressing HLA-DPA1*01:03-P2A-HLA-DPB1*04:02 (negative control) and K86/B0901 (positive control) were cocultured with clone #17 overnight and tested for intracellular cytokine production. Neither INF-γ nor TNF-α was produced against K86 expressing HLA- DPA1*01:03-P2A-HLA-DPB1*04:02, whereas K86/B0901 induced notable IFN-γ and TNF-α production from clone #17. Regarding stimulation with primary leukemia cells, TNF-α production was observed irrespective of pre-treatment, while almost no IFN-γ production was observed in all cocultures (Fig. 3B). Finally, the treatment of leukemia cells with epigenetic modifiers such as histone deacetylase inhibitor33 or DNA methylation inhibitors such as trichostatin A or 5-aza treatment34 to facilitate antigen presentation by HLA molecules did not increase susceptibility to clone #17 (data not shown). These observations indicated 1) that HLA-DP expression was not constitutive and could be induced in the presence of INF-γ, and 2) clone #17 recognized certain antigens expressed in myeloid leukemia cell lines that were not significantly expressed in at least three primary myeloid leukemia cells available and tested in this study.
Table 2.
| UPN | Diagnosis (FAB) | Source | %Blast | %CD34 | HLA-DPB1* |
| 02 | AML (M2) | BM | 76 | 92.9 | 05:01, 09:01 |
| 11 | AML (M0) | BM | 91 | 96.3 | 02:01, 09:01 |
| 15 | AML (M2) | BM | 64 | 83.6 | 05:01, 09:01 |
BM: bone marrow
FAB: French-American-British classification
NA: not applicable
UPN: unique patient number
Fig. 3.
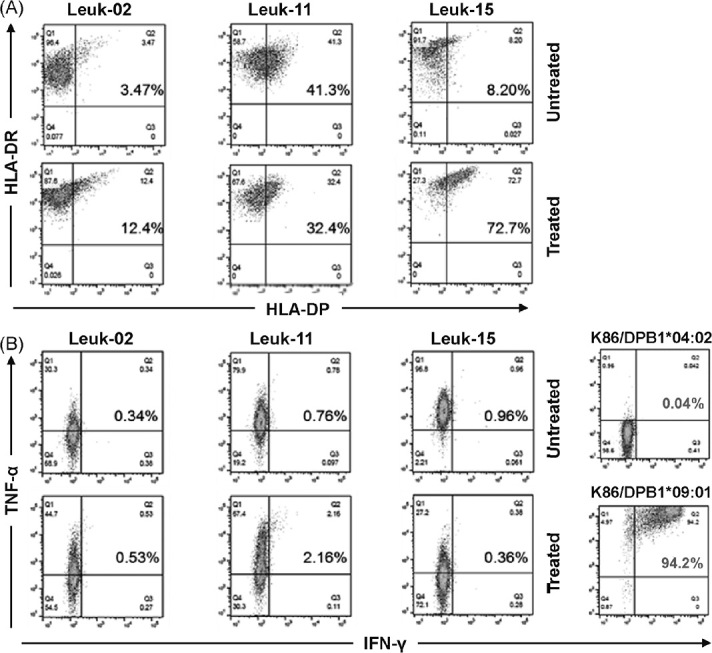
Cytokine production of clone #17 against primary leukemia cells
Fig. 3A: Expression of HLA-DR and HLA-DP on three primary myeloid leukemia cells with or without TNF-α and IFN-γ pre-treatment for 48 h.
Fig. 3B:Production of TNF-α and IFN-γ from clone #17 following cocultivation of leukemia cells and positive/negative control cells assessed via intracellular cytokine staining.
Study acronyms are explained in Abbreviations to main body.
Redirection of specificity of the genetically modified-polyclonal healthy T cells with TCR of clone #17 against a panel of tumor cell lines
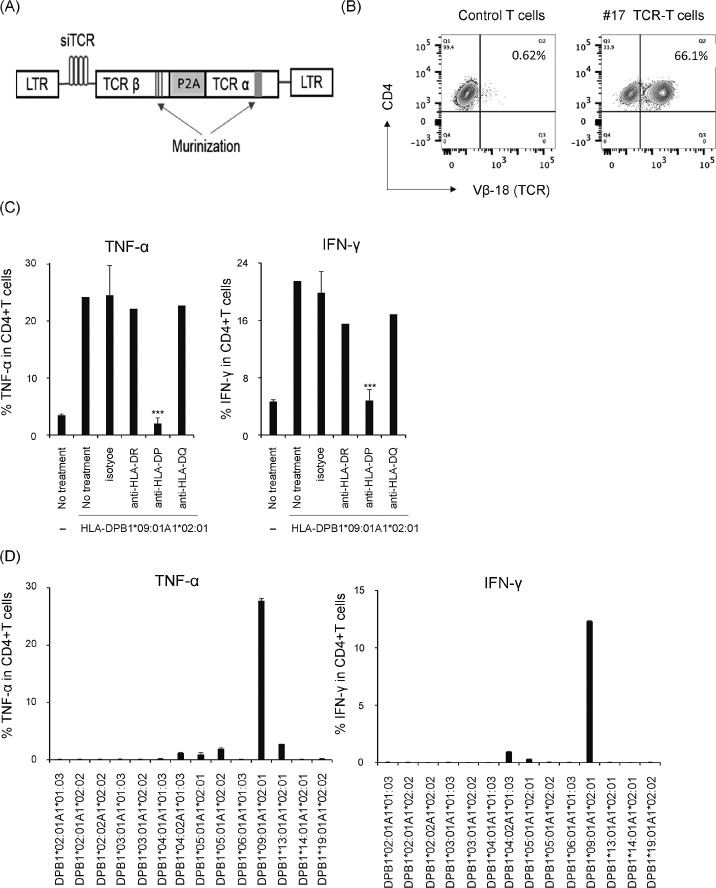
Next, we questioned whether the B0901-specificity of clone #17 could successfully redirect T cells from healthy donors by TCR gene transfer. Hence, cDNAs of both TCR-α and -β chains of clone #17 were PCR-amplified and subjected to direct sequencing. The α chain consisted of TRAV13-1*01 and TRAJ6*01, and the β chain consisted of TRBV18*01, TRBD1*01, and TRBJ2-7*01. The obtained sequences were codon-modified and inserted into the pSplice-TCR-BC1 retroviral vector (Fig. 4A) as described in the “Material and Methods” such that the siRNA transcribed from the vector would not destroy the clone #17 TCR mRNA also transcribed from the vectors. Thereafter, the retroviral supernatant from producer cells was used to generate TCR genetically modified T cells originating from HLA-DPB1*09:01-negative healthy donors. As shown in Fig. 4B, flow cytometric analysis confirmed the expression of the clone #17 on CD4+ T cells using an anti-TCRVβ18 antibody. We first examined whether TCR-T cells recognize only HLA-DP molecules and no other HLA class II molecules (HLA-DR, HLA-DQ) by blocking assays with antibodies against HLA-DR/-DP/-DQ. As shown in Fig. 4C, only the anti-HLA-DP antibody showed a substantial reduction in TNF-α and IFN-γ production. Second, we examined the potential reactivity of TCR-T cells against HLA-DP alleles other than HLA-DPB1*09:01; cross-reaction with other HLA-DP molecules might cause dangerous graft-versus-host reactions when administered in vivo for therapeutic purposes. Hence, TCR-T cells were co-incubated with K86 expressing various HLA-DP alleles that are commonly found in the Japanese population24 and tested for TNF-α and IFN-γ production. As shown in Fig. 4D, TCR-T-cells selectively reacted with K86/B0901. These results indicated that TCR-T cells expressing clone #17 TCR cDNA modified with murinization and siTCR incorporation exhibited stringent specificity against the targeted HLA-A1*02:01/HLA-DPB1*09:01/ molecule. Third, we examined the specificity of TCR-T cells toward the same panel of tumor cell lines as tested with clone #17. In agreement with the recognition pattern of parental clone #17, TCR-T cells produced not only TNF-α and IFN-γ against the same target cells (Fig. 5A) but also additional target cell lines of myeloid or monocyte/monoblast origin (Figure 5B, upper panels). In addition, TCR-T cells expressed granzyme B and CD107a (a hallmark of degranulation) against the additional target cells (Figure 5B, lower panels) and induced lysis in four of the six cell lines that were tested (Fig. 5C). These results indicated that TCR-T cells recognized leukemia cells that neither lost specificity to the peptide antigen/HLA-DP complex nor acquired undesired new reactivity due to chimera formation between transduced TCR chains and endogenous TCR chains.
Fig. 4.
Specificity of TCR-T cells with clone #17 TCR
Fig. 4A: Structure of retroviral vector. siRNA, shRNA specific to endogenous TCR mRNA.
Fig. 4B:Expression of transduced TCR on cultured CD4+ T cells assessed with anti-TCR-Vβ18 antibody.
Fig. 4C: TCR-T cells were co-cultured with K86 cells expressing HLA-DPB1*09:01 with the indicated blocking antibodies for 6 h. TNF-α and IFN-γ production was examined via intracellular staining.
Fig. 4D:K86 cells were individually transduced with the retroviral vector encoding various HLA-DPA1 and -DPB1 combinations, which are frequently observed in the Japanese population. They were cocultured with TCR-T cells and cytokine production was evaluated as above. Data were processed as in Fig. 2.
Study acronyms are explained in Abbreviations to main body.
Fig. 5.
Recognition of various tumor cell lines using TCR-T cells with clone #17 TCR
Fig. 5A and 5B:TCR-T cells with #17 TCR were cocultured as shown in the legend of Fig. 2 and assessed for cytokine production.
Fig. 5C:Cytotoxicity of the TCR-T cells was evaluated by the decrease of fluorescently labeled target at the graded E/T ratio.
Study acronyms are explained in Abbreviations to main body.
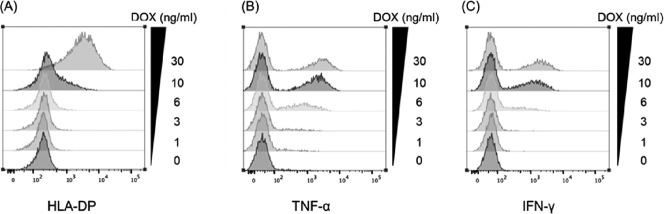
TCR-T cells can recognize target expressing HLA-DP low level
Finally, we examined whether the failure of clone #17 to recognize primary leukemia cells might be caused by the low expression level of HLA-DP on these target cells. To test this, we generated K86 cells whose surface HLA-DPB1*09:01 expression was controlled by doxycycline concentration using a tetracycline-inducible system. After 48 h of induction with a graded concentration of DOX, some of the K86 cells were stained for HLA-DP expression and the remaining K86 cells were cocultured with TCR-T cells overnight. Thereafter, production of TNF-α and IFN-γ was evaluated by intracellular cytokine staining followed by flow cytometry. As shown in Fig. 6A, HLA-DP expression started to be weakly positive at a DOX concentration of 6 ng/mL. At this level, a substantial portion of TCR-T cells produced TNF-α (Fig. 6B) and a lower amount of IFN-γ (Fig. 6C). At a DOX concentration of 10 ng/mL, both TNF-α and IFN-γ were produced by as many TCR-T cells as was observed at a concentration of 30 ng/mL. These data indicated that TCR-T cells express a high-affinity TCR that can engage with an extremely limited amount of cell surface HLA-DP expression.
Fig. 6.
TCR-T cells can recognize target expressing HLA-DP, even at a low level. K86 cells transduced with the tetracycline-inducible vector encoding HLA-DPA1*02:01-P2A-DPB1*09:01 was manageable with the DOX concentration using the tetracycline-inducible system
Fig. 6A: After 48 h of induction with a graded concentration of DOX, some K86 cells were stained for HLA-DP expression.
Fig. 6B and 6C:The remaining cells were cocultured with TCR-T cells overnight. Thereafter, TNF-α and IFN-γ production was evaluated using intracellular cytokine staining.
Study acronyms are explained in Abbreviations to main body.
Because we found TCR-T cells could recognize target cells with low HLA-DP expression, we tested the reactivity of TCR-T cells against the same primary leukemia cells tested by clone #17 (Fig, 7A and B). In this analysis, we also examined expression of granzyme B and CD107a to evaluate the multifunctionality of TCR-T cells. As shown in Figure 7B, TCR-T cells were found to be able to produce IFN-γ in addition to TNF-α to all three primary leukemia cells, unlike in the case of clone #17. They also expressed CD107a and granzyme B (Fig. 7C), thereby suggesting that TCR-T cells possess multifunctionality.
Fig. 7.
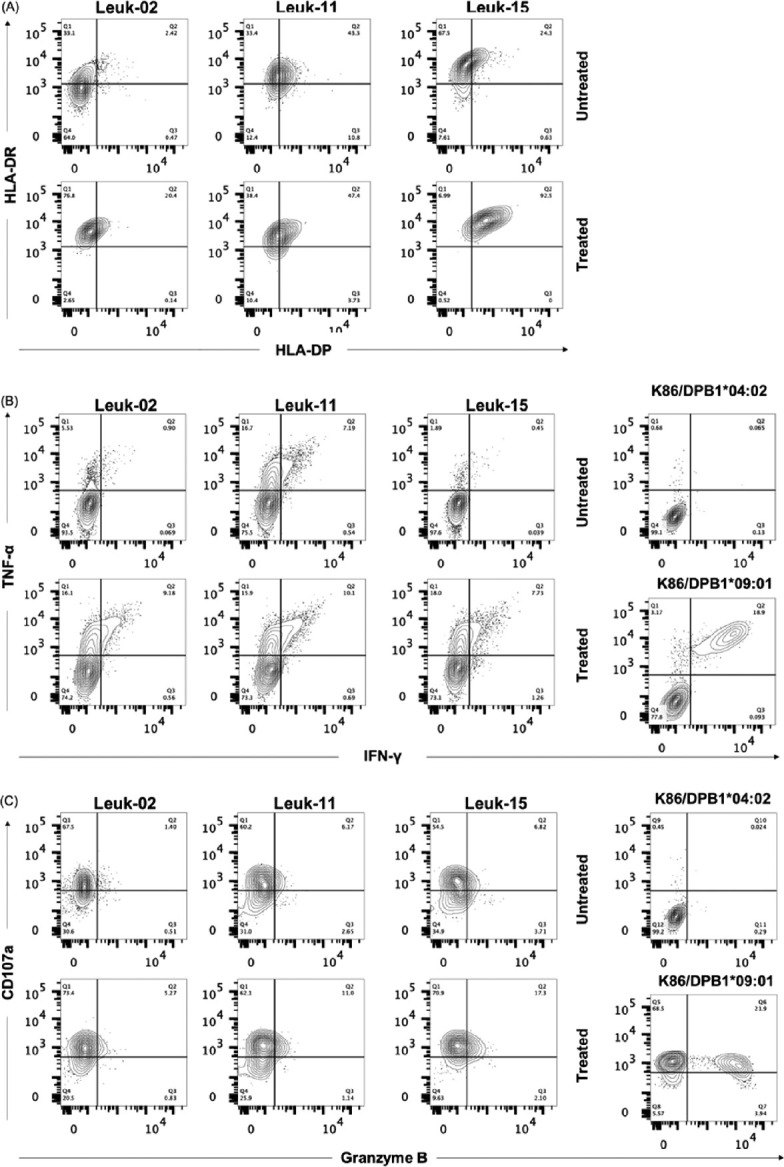
Cytokine production of TCR-T cells against primary leukemia cells
Fig. 7A:Expression of HLA-DR and HLA-DP on three primary myeloid leukemia cells with or without TNF-α and IFN-γ pre-treatment for 48 h.
Fig. 7B:Production of TNF-α and IFN-γ from TCR-T cells following cocultivation of leukemia cells and positive/negative control cells assessed via intracellular cytokine staining.
Fig. 7C:Expression of granzyme B and CD107a by TCR-T cells tested as shown above.
Study acronyms are explained in Abbreviations to main body.
DISCUSSION
In this study, we established HLA-DPB1*09:01-restricted CD4+ T cell clone #17 from HLA-DPB1*09:01-negative healthy donors by repeated stimulation with K562 cells co-transduced with CD86 and HLA-DPA1*02:01/HLA-DPB1*09:01. Thereafter, we demonstrated that redirection of T cells transduced with TCR isolated from clone #17 was feasible and definite, as shown by a detailed specificity analysis. Efficient expression of exogenous TCR might be made possible by incorporation of murinization of TCR constant regions29 and inclusion of siTCR technology,30 although their degree of contribution was not evaluated individually for this particular TCR. Our TCR-T cells specific for HLA-DPB1*09:01 are next to the two HLA-DPB1*03:01/04:01-specific TCR-T cells reported by Klobuch et al.35
Clone #17 produced TNF-α and IFN-γ and lysed the B09:01-transduced leukemia cell lines, such as KG-1 and HYT-1 of the myeloid lineage. Moreover, TCR-T cells carrying clone #17-derived TCR were able to produce cytokines in response to stimulation with K86, expressing an extremely low level of HLA-DP molecules, which was far weaker than that observed in cytokine pre-treated primary leukemia cells. Although clone #17 failed to produce IFN-γ when stimulated with three primary myeloid leukemia cells possessing HLA-DPA1*02:01/HLA-HLA-DPB1*09:01 alleles even after cytokine treatment, TCR-T cells expressing clone #17 TCR did produce IFN-γ and also granzyme B. This could be partly explained by the use of T cells from healthy donors to prepare TCR-T cells, whereas clone #17 was proliferated by multiple expansion cultures, which could result in exhaustion, where it could produce IFN-γ by target cells expressing a high level of HLA-DP such as K86 /DPB1*09:01. Finally, neither clone #17 or TCR-T cells recognized transduced HeLa/DMI/B0901 cells expressing moderate levels of HLA-DP. As HeLa cells have been used as a representative of non-hematopoietic cells together with dermal fibroblasts,16,18,19 our findings suggest that the target antigen recognized by clone #17 and its derivative TCR-T cells are likely to be expressed in hematopoietic cells of the myeloid and monocytic lineage. We could not exclude the possibility that clone #17 target antigen could be overexpressed in immortalized myeloid leukemia cell lines rather than primary myeloid leukemia cells.
The selective GVL effect devoid of unwanted GVHD is the key to success in T-cell therapy targeting allo-antigens.2,3 It is well known that pro-inflammatory cytokines, such as TNF-α and IFN-γ, secreted under inflammatory conditions, such as infections, induce HLA class II expression in non-hematopoietic cells. These cells can be targeted by HLA class II (including HLA-DP)-specific T cells which may result in GVHD occurrence.36 The two murinized TCR-T cells reported by Klobuch et al broadly recognized HLA-DPB1*04:01-positive cells irrespective of their origin, and HLA-DPB1*03:01-positive cells in an HLA-DP expression level-dependent manner, respectively.35 They suggested that both TCRs might recognize peptides ubiquitously expressed by hematopoietic and non-hematopoietic cells.35 In contrast, from this perspective, clone #17 and its derivative TCR-T might recognize a peptide selectively expressed in some hematopoietic cells because they did not react to HeLa/DMI cells expressing sufficient levels of HLA-DP for recognition, as estimated by inducible HLA-DP experiments. To address this, it is necessary to identify antigenic peptides. We attempted to perform negative selection-based screening using a pooled K86/HLA-B09:01 cell line infected with the sgRNA CRISPR-Cas9 lentivirus library and clone #17 cells, by assuming that surviving K86 cells from clone #17 killing would have lost genes encoding the cognate antigen or proteins involved in the antigen presentation process. To date, we have not identified the candidate gene. Alternatively, screening of peptide libraries isolated from HLA-DP molecules obtained from K86/B0901 could be another option.37
Finally, it has been reported that HLA-DP expression is associated with GVHD and/or GVL effects after unrelated allo-HSCT.38,39 A high level of expression of the mismatched HLA-DPB1 allele of the patient correlates with the risk of GVHD and GVL effects.38,39 Alternatively, Toffalori et al have shown that patients suffering from leukemia relapse after allo-HSCT developed loss or decrease in HLA class-DR, -DQ, and -DP expression in leukemia cells, caused by epigenetic downregulation of the HLA class II regulator CIITA.40 These findings suggested that high expression of HLA-DP in leukemia cells or generation of HLA-DP-specific T cells with high affinity TCR is key to developing effective T cell-based immunotherapy. Previous reports demonstrated that the use of histone deacetylase inhibitor33 or DNA methylation inhibitors such as trichostatin A or 5-aza treatment,34 or IFN-γ40 increases or restores HLA-DP expression. Thus, the combination of these inhibitors and allogeneic HLA-DP-specific TCR-T cells is a promising therapy for leukemia relapse following allo-HSCT.
In conclusion, we demonstrated that induction of T cells specific for HLA-DP, consisting of hematopoietic cell lineage-derived peptide, and redirection of T cells with cloned TCR cDNA by gene transfer is possible. For clinical applications, further efforts to identify more T cells that can selectively recognize primary leukemia cells with limited HLA-DP expression, and not non-hematopoietic cells, are necessary. A detailed evaluation of TCR affinity with the inducible HLA-DP expression system we devised will help isolate promising T cell clones suitable for clinical application.
ACKNOWLEDGEMENTS
The authors thank all members of the Department of Immunology, Nagoya University Graduate School of Medicine, for technical support and discussions regarding this study. The authors also thank staff of Division for Medical Research Engineering, Nagoya University Graduate School of Medicine for their technical support. This study was supported in part by Grants-in-Aid for Scientific Research (grant nos. 18K08341 and 21K08369 to Y.A.) from the Ministry of Education, Culture, Sports, Science, by AMED under Grant Number JP21ek0510027 (to Y.A.), by Aichi Cancer Research Foundation (to Y.A.) and by the Bristol-Myers Squibb endowed chair in Cancer Biomarker Research (to Y.A.).
CONFLICTS OF INTEREST
Y.A. received honoraria and research funding from Bristol-Myers Squibb. All other authors declare no competing financial interests.
Abbreviations
- Allo-HSCT
allogeneic hematopoietic stem cell transplantation
- DOX
doxycycline
- GVHD
graft-versus host defense
- HLA
human leukocyte antigen
- IFN-γ
interferon-gamma
- TCR
T cell receptor
- TNF-α
tissue necrosis factor-alpha
REFERENCES
- 1.Miller JS, Warren EH, van den Brink MR, et al. NCI First International Workshop on The Biology, Prevention, and Treatment of Relapse After Allogeneic Hematopoietic Stem Cell Transplantation: Report from the Committee on the Biology Underlying Recurrence of Malignant Disease following Allogeneic HSCT: Graft-versus-Tumor/Leukemia Reaction. Biol Blood Marrow Transplant. 2010;16(5):565–586. doi: 10.1016/j.bbmt.2010.02.005. [DOI] [PMC free article] [PubMed]
- 2.Falkenburg JH, Jedema I. Allo-reactive T cells for the treatment of hematological malignancies. Mol Oncol. 2015;9(10):1894–1903. doi: 10.1016/j.molonc.2015.10.014. [DOI] [PMC free article] [PubMed]
- 3.Akatsuka Y, Morishima Y, Kuzushima K, Kodera Y, Takahashi T. Minor histocompatibility antigens as targets for immunotherapy using allogeneic immune reactions. Cancer Sci. 2007;98(8):1139–1146. doi: 10.1111/j.1349-7006.2007.00521.x. [DOI] [PMC free article] [PubMed]
- 4.Wu CJ, Ritz J. Induction of tumor immunity following allogeneic stem cell transplantation. Adv Immunol. 2006;90:133–173. doi: 10.1016/S0065-2776(06)90004-2. [DOI] [PubMed]
- 5.Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373(9674):1550–1561. doi: 10.1016/S0140-6736(09)60237-3. [DOI] [PMC free article] [PubMed]
- 6.Nash RA, Storb R. Graft-versus-host effect after allogeneic hematopoietic stem cell transplantation: GVHD and GVL. Curr Opin Immunol. 1996;8(5):674–680. doi: 10.1016/s0952-7915(96)80085-9. [DOI] [PubMed]
- 7.Duval M, Klein JP, He W, et al. Hematopoietic stem-cell transplantation for acute leukemia in relapse or primary induction failure. J Clin Oncol. 2010;28(23):3730–3738. doi: 10.1200/JCO.2010.28.8852. [DOI] [PMC free article] [PubMed]
- 8.Craddock C, Labopin M, Pillai S, et al. Factors predicting outcome after unrelated donor stem cell transplantation in primary refractory acute myeloid leukaemia. Leukemia. 2011;25(5):808–813. doi: 10.1038/leu.2011.13. [DOI] [PubMed]
- 9.Falkenburg JHF, Jedema I. Graft versus tumor effects and why people relapse. Hematology Am Soc Hematol Educ Program. 2017;2017(1):693–698. doi: 10.1182/asheducation-2017.1.693. [DOI] [PMC free article] [PubMed]
- 10.Loke J, Malladi R, Moss P, Craddock C. The role of allogeneic stem cell transplantation in the management of acute myeloid leukaemia: a triumph of hope and experience. Br J Haematol. 2020;188(1):129–146. doi: 10.1111/bjh.16355. [DOI] [PMC free article] [PubMed]
- 11.Lee SJ, Klein J, Haagenson M, et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood. 2007;110(13):4576–4583. doi: 10.1182/blood-2007-06-097386. [DOI] [PubMed]
- 12.Morishima Y, Kashiwase K, Matsuo K, et al. Biological significance of HLA locus matching in unrelated donor bone marrow transplantation. Blood. 2015;125(7):1189–1197. doi: 10.1182/blood-2014-10-604785. [DOI] [PMC free article] [PubMed]
- 13.Petersdorf EW, Malkki M, O’hUigin C, et al. High HLA-DP Expression and Graft-versus-Host Disease. N Engl J Med. 2015;373(7):599–609. doi: 10.1056/NEJMoa1500140. [DOI] [PMC free article] [PubMed]
- 14.Fleischhauer K, Shaw BE. HLA-DP in unrelated hematopoietic cell transplantation revisited: challenges and opportunities. Blood. 2017;130(9):1089–1096. doi: 10.1182/blood-2017-03-742346. [DOI] [PubMed]
- 15.Ibisch C, Gallot G, Vivien R, et al. Recognition of leukemic blasts by HLA-DPB1-specific cytotoxic T cell clones: a perspective for adjuvant immunotherapy post-bone marrow transplantation. Bone Marrow Transplant. 1999;23(11):1153–1159. doi: 10.1038/sj.bmt.1701768. [DOI] [PubMed]
- 16.Rutten CE, van Luxemburg-Heijs SA, Griffioen M, et al. HLA-DP as specific target for cellular immunotherapy in HLA class II-expressing B-cell leukemia. Leukemia. 2008;22(7):1387–1394. doi: 10.1038/leu.2008.90. [DOI] [PubMed]
- 17.Herr W, Eichinger Y, Beshay J, et al. HLA-DPB1 mismatch alleles represent powerful leukemia rejection antigens in CD4 T-cell immunotherapy after allogeneic stem-cell transplantation. Leukemia. 2017;31(2):434–445. doi: 10.1038/leu.2016.210. [DOI] [PubMed]
- 18.Laghmouchi A, Hoogstraten C, van Balen P, Falkenburg JHF, Jedema I. The allogeneic HLA-DP-restricted T-cell repertoire provoked by allogeneic dendritic cells contains T cells that show restricted recognition of hematopoietic cells including primary malignant cells. Haematologica. 2019;104(1):197–206. doi: 10.3324/haematol.2018.193680. [DOI] [PMC free article] [PubMed]
- 19.Rutten CE, van Luxemburg-Heijs SA, Halkes CJ, et al. Patient HLA-DP-specific CD4+ T cells from HLA-DPB1-mismatched donor lymphocyte infusion can induce graft-versus-leukemia reactivity in the presence or absence of graft-versus-host disease. Biol Blood Marrow Transplant. 2013;19(1):40–48. doi: 10.1016/j.bbmt.2012.07.020. [DOI] [PubMed]
- 20.June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC. CAR T cell immunotherapy for human cancer. Science. 2018;359(6382):1361–1365. doi: 10.1126/science.aar6711. [DOI] [PubMed]
- 21.Schmitt TM, Stromnes IM, Chapuis AG, Greenberg PD. New Strategies in Engineering T-cell Receptor Gene-Modified T cells to More Effectively Target Malignancies. Clin Cancer Res. 2015;21(23):5191–5197. doi: 10.1158/1078-0432.CCR-15-0860. [DOI] [PMC free article] [PubMed]
- 22.Zhang J, Wang L. The Emerging World of TCR-T Cell Trials Against Cancer: A Systematic Review. Technol Cancer Res Treat. 2019;18:1533033819831068. doi: 10.1177/1533033819831068. [DOI] [PMC free article] [PubMed]
- 23.Richardson JR, Schöllhorn A, Gouttefangeas C, Schuhmacher J. CD4+ T Cells: Multitasking Cells in the Duty of Cancer Immunotherapy. Cancers(Basel). 2021;13(4):596. doi: 10.3390/cancers13040596. [DOI] [PMC free article] [PubMed]
- 24.Gonzalez-Galarza FF, McCabe A, Santos EJMD, et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020;48(D1):D783-D788. doi: 10.1093/nar/gkz1029. [DOI] [PMC free article] [PubMed]
- 25.Kornbluth J, Spear B, Raab SS, Wilson DB. Evidence for the role of class I and class II HLA antigens in the lytic function of a cloned line of human natural killer cells. J Immunol. 1985;134(2):728–735. doi: 10.4049/jimmunol.134.2.728. [DOI] [PubMed]
- 26.Möller JF, Möller B, Wiedenmann B, Berg T, Schott E. CD154, a marker of antigen-specific stimulation of CD4 T cells, is associated with response to treatment in patients with chronic HCV infection. J Viral Hepat. 2011;18(7):e341-e349. doi: 10.1111/j.1365-2893.2010.01430.x. [DOI] [PubMed]
- 27.Akatsuka Y, Martin EG, Madonik A, Barsoukov AA, Hansen JA. Rapid screening of T-cell receptor (TCR) variable gene usage by multiplex PCR: application for assessment of clonal composition. Tissue Antigens. 1999;53(2):122–134. doi: 10.1034/j.1399-0039.1999.530202.x. [DOI] [PubMed]
- 28.Hamana H, Shitaoka K, Kishi H, Ozawa T, Muraguchi A. A novel, rapid and efficient method of cloning functional antigen-specific T-cell receptors from single human and mouse T-cells. Biochem Biophys Res Commun. 2016;474(4):709–714. doi: 10.1016/j.bbrc.2016.05.015. [DOI] [PubMed]
- 29.Sommermeyer D, Uckert W. Minimal amino acid exchange in human TCR constant regions fosters improved function of TCR gene-modified T cells. J Immunol. 2010;184(11):6223–6231. doi: 10.4049/jimmunol.0902055. [DOI] [PubMed]
- 30.Okamoto S, Mineno J, Ikeda H, et al. Improved expression and reactivity of transduced tumor-specific TCRs in human lymphocytes by specific silencing of endogenous TCR. Cancer Res. 2009;69(23):9003–9011. doi: 10.1158/0008-5472.CAN-09-1450. [DOI] [PubMed]
- 31.Bleakley M, Heimfeld S, Jones LA, et al. Engineering human peripheral blood stem cell grafts that are depleted of naive T cells and retain functional pathogen-specific memory T cells. Biol Blood Marrow Transplant. 2014;20(5):705–716. doi: 10.1016/j.bbmt.2014.01.032. [DOI] [PMC free article] [PubMed]
- 32.Micouin A, Rouillard D, Bauvois B. Induction of macrophagic differentiation and cytokine secretion by IgG1 molecules in human normal monocytes and myelogenous leukemia cells. Leukemia. 1997;11(4):552–560. doi: 10.1038/sj.leu.2400602. [DOI] [PubMed]
- 33.Bae J, Hideshima T, Tai YT, et al. Histone deacetylase (HDAC) inhibitor ACY241 enhances anti-tumor activities of antigen-specific central memory cytotoxic T lymphocytes against multiple myeloma and solid tumors. Leukemia. 2018;32(9):1932–1947. doi: 10.1038/s41375-018-0062-8. [DOI] [PMC free article] [PubMed]
- 34.Suárez-Alvarez B, Rodriguez RM, Calvanese V, et al. Epigenetic mechanisms regulate MHC and antigen processing molecules in human embryonic and induced pluripotent stem cells. PLoS One. 2010;5(4):e10192. doi: 10.1371/journal.pone.0010192. [DOI] [PMC free article] [PubMed]
- 35.Klobuch S, Hammon K, Vatter-Leising S, et al. HLA-DPB1 Reactive T Cell Receptors for Adoptive Immunotherapy in Allogeneic Stem Cell Transplantation. Cells. 2020;9(5):1264. doi: 10.3390/cells9051264. [DOI] [PMC free article] [PubMed]
- 36.Stevanovic S, van Bergen CA, van Luxemburg-Heijs SA, et al. HLA class II upregulation during viral infection leads to HLA-DP-directed graft-versus-host disease after CD4+ donor lymphocyte infusion. Blood. 2013;122(11):1963–1973. doi: 10.1182/blood-2012-12-470872. [DOI] [PubMed]
- 37.Khilji MS, Faridi P, Pinheiro-Machado E, et al. Defective Proinsulin Handling Modulates the MHC I Bound Peptidome and Activates the Inflammasome in beta-Cells. Biomedicines. 2022;10(4):814. doi: 10.3390/biomedicines10040814. [DOI] [PMC free article] [PubMed]
- 38.Petersdorf EW, Bengtsson M, De Santis D, et al. Role of HLA-DP Expression in Graft-Versus-Host Disease After Unrelated Donor Transplantation. J Clin Oncol. 2020;38(24):2712–2718. doi: 10.1200/JCO.20.00265. [DOI] [PMC free article] [PubMed]
- 39.Martin PJ, Levine DM, Storer BE, Sather CL, Spellman SR, Hansen JA. Genetic associations with immune-mediated outcomes after allogeneic hematopoietic cell transplantation. Blood Adv. 2022;6(8):2608–2617. doi: 10.1182/bloodadvances.2021005620. [DOI] [PMC free article] [PubMed]
- 40.Toffalori C, Zito L, Gambacorta V, et al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat Med. 2019;25(4):603–611. doi: 10.1038/s41591-019-0400-z. [DOI] [PubMed]