Abstract
The genetics of lactose utilization within the slow-lactose-fermenting Lactococcus lactis strain NCDO2054 was studied with respect to the organization, expression, and evolution of the lac genes. Initially the β-galactosidase gene (lacZ) was cloned by complementation of an Escherichia coli mutant on a 7-kb HpaI fragment. Nucleotide sequence analysis of the complete fragment revealed part of a gal-lac operon, and the genes were characterized by inactivation and complementation analyses and in vitro enzyme activity measurements. The gene order is galK-galT-lacA-lacZ-galE; the gal genes encode enzymes of the Leloir pathway for galactose metabolism, and lacA encodes a galactoside acetyltransferase. The galT and galE genes of L. lactis LM0230 (a lactose plasmid-cured derivative of the fast-lactose-fermenting L. lactis C2) were highly similar at the nucleotide sequence level to their counterparts in strain NCDO2054 and, furthermore, had the same gene order except for the presence of the intervening lacA-lacZ strain NCDO2054. Analysis of mRNA for the gal and lac genes revealed an unusual transcriptional organization for the operon, with a surprisingly large number of transcriptional units. The regulation of the lac genes was further investigated by using fusions consisting of putative promoter fragments and the promoterless β-glucuronidase gene (gusA) from E. coli, which identified three lactose-inducible intergenic promoters in the gal-lac operon. The greater similarity of the lacA and lacZ genes to homologs in gram-negative organisms than to those of gram-positive bacteria, in contrast to the homologies of the gal genes, suggests that the genes within the gal operon of L. lactis NCDO2054 have been recently acquired. Thus, the lacA-lacZ genes appear to have engaged the promoters of the gal operon in order to direct and control their expression.
Lactococci play a vital role in commercial milk fermentations, in which their primary function is to convert lactose to lactic acid. Typical Lactococcus lactis strains were selected as starter cultures because of their rapid fermentation of lactose. They transport lactose via a phosphoenolpyruvate-dependent phosphotransferase system (PTS), and it is subsequently hydrolyzed by phospho-β-galactosidase, giving glucose and galactose-6-phosphate. The galactose-6-phosphate is further metabolized to triose phosphates by the enzymes of the d-tagatose-6-phosphate pathway. It has been suggested that the lactose PTS is essential for rapid homolactic fermentation by starter cultures (29) because the affinity of the lactose PTS for lactose is very high (46) and one lactose molecule is translocated and simultaneously phosphorylated at the expense of one ATP equivalent. The fact that the genes determining these functions are plasmid encoded in many L. lactis strains has aided their characterization with respect to genetic organization and regulation of expression (11, 48).
In contrast to the industrial starter strains, some lactococci ferment lactose slowly and produce a variety of end products, such as acetate, formate, and ethanol, in addition to lactate (13). Although starter lactococci may also convert pyruvate to a variety of end products, for example, by fermentation of galactose alone via the d-tagatose-6-phosphate or Leloir pathway, these pathways are not expressed during lactose fermentation, which is homolactic in most strains (45). Recently a galactose gene cluster which has the gene order galA-galM-galK-galT-galE, encoding a galactose permease, mutarotase, galactokinase, galactose-1-phosphate uridylyltransferase, and UDPglucose 4-epimerase, respectively, has been reported for L. lactis MG1363 (20). L. lactis NCDO2054, which is the best-studied slow lactose fermenter, hydrolyzes lactose via a β-galactosidase and, therefore, metabolizes the galactose moiety of lactose through the Leloir pathway (3). The properties and evolution of the defective nature of lactose metabolism in strain NCDO2054 have intrigued researchers (7, 44). This strain, which can accumulate lactose-6-phosphate to a high intracellular concentration by using an efficient lactose PTS system, contains enzymes of the d-tagatose-6-phosphate pathway (3) but possesses low-level phospho-β-galactosidase activity. Thus, the slow fermentation of lactose has been attributed to this rate-limiting phospho-β-galactosidase activity (7), and it has also been suggested that the high levels of accumulated lactose-6-phosphate might be deleterious to growth of the strain (10). A proton-coupled β-galactoside transport system which has a much higher affinity for galactose and its analogs than for lactose was discovered in NCDO2054 (26); it is probably the same galactose permease described for L. lactis MG1363. However, the precise manner by which lactose is translocated into the cell and made available for hydrolysis by β-galactosidase in order to achieve sufficient metabolism of lactose for growth is still essentially unknown. The presence of an inducible β-galactosidase gene (lacZ) within this Lactococcus strain has attracted additional attention (6, 18) because of the biotechnological potential of the gene and its promoter(s) for the construction of selection and expression vectors.
The present study was undertaken to further our knowledge of the genetics and evolution of lactose metabolism in the unusual L. lactis NCDO2054 strain. Here we describe the location of the lacZ and lacA (galactoside acetyltransferase) genes, associated together with genes encoding enzymes for the Leloir pathway of galactose metabolism. The transcriptional organization and regulation of the lac genes within the gal-lac operon were studied by analysis of mRNA and by using gene fusions consisting of putative promoter fragments and the promoterless β-glucuronidase gene (gusA) from Escherichia coli. The evidence presented suggests that the lacA and lacZ genes have recruited the promoters of the gal operon not simply to drive their expression but to do so in a regulated manner.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
L. lactis NCDO2054 (also known as ATCC 7962) and LM0230 were originally obtained from the American Type Culture Collection, Rockville, Md. E. coli JM109#8, a derivative of JM109 cured of its F′ episome and therefore no longer capable of α-complementation (34), was used for isolation of the β-galactosidase gene. E. coli BZ234, a derivative of C600 containing the F′ episome, was a gift from Tom Bickle, Biozentrum, University of Basel, Switzerland, and was used for routine isolation of plasmid DNA. E. coli MK30-3 was purchased from New England Biolabs, Inc.
L. lactis strains were grown at 30°C in M17 broth (Oxoid, Hampshire, England) containing either glucose, lactose, or galactose at 1% as required. E. coli was routinely grown in Luria-Bertani medium with aeration at 37°C. Antibiotics were added at the following concentrations: ampicillin (Ap) at 100 μg ml−1, chloramphenicol (Cm) at 20 μg ml−1, and erythromycin (Em) at 100 μg ml−1 for E. coli; and Cm at 4 μg ml−1 and Em at 5 μg ml−1 for lactococci. Functional β-galactosidase was detected by the addition of 40 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) ml−1 to Luria-Bertani solid medium, and isopropyl-β-d-thiogalactopyranoside (IPTG), when required, was added at a final concentration of 1 mM. Histochemical screening for gusA-positive clones was performed with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc; Amresco, Solon, Ohio) at a final concentration of 0.5 mM.
Molecular techniques and transformation.
General molecular cloning techniques, restriction enzyme analysis, and transformation of E. coli by CaCl2-induced competence were performed as described by Sambrook et al. (42). Plasmid DNA was isolated from E. coli and L. lactis by using Qiagen columns (Kontron Instruments, Basel, Switzerland); 5 mg of lysozyme ml−1 was added to buffer P1 to aid lysis of the lactococci. Electroporation of L. lactis was performed essentially as described by Wells et al. (52). Total DNA from L. lactis was isolated as described by Hayes et al. (22). Restriction endonucleases, calf intestinal alkaline phosphatase, and T4 DNA ligase were purchased from Boehringer Mannheim and New England Biolabs, Inc., and were used as recommended by the manufacturer. PCR amplification was performed as previously reported (41). DNA sequences of plasmid inserts were determined by the dideoxy-chain termination method (43), using pUC19-derived plasmid DNA as the template, with the M13 universal primer or by primer walking. Custom-made primers were purchased from Microsynth (Balgach, Switzerland). The sequence data were assembled and analyzed by using the Wisconsin package, version 8.0 (Genetics Computer Group [GCG], Madison, Wis.), and amino acid comparisons were performed with the FASTA (36) and GAP (35) programs.
Cloning of the β-galactosidase gene of L. lactis NCDO2054.
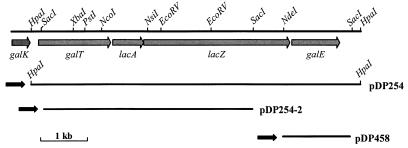
Chromosomal DNA, digested to completion with HpaI, was ligated to pUC19 (54) that had been linearized with SmaI and treated with calf intestinal alkaline phosphatase. The ligation mixture was transformed into E. coli JM109#8, and clones expressing β-galactosidase activity, as evidenced by the formation of blue colonies on Luria-Bertani agar plates supplemented with Ap, X-Gal, and IPTG, were selected. Restriction enzyme analysis of plasmid DNA from several blue colonies revealed common 7-kb HpaI inserts which were in the same orientation relative to the lacZ promoter of pUC19 (Fig. 1). The restriction map constructed for a representative of these plasmids, designated pDP254, was used to locate sites for cloning the DNA upstream of the 5′ HpaI site for nucleotide sequence analysis (Fig. 1).
FIG. 1.
The genetic organization of genes within the gal-lac operon on the chromosome of L. lactis NCDO2054. Relevant restriction sites used in cloning are indicated, and the extent and direction of transcription of the genes are illustrated with shaded arrows. DNA inserts in plasmid constructions pDPD254, pDP254-2, and pDP458, which were used for the isolation and characterization of the genes, and the direction of the lacZ promoter of pUC19 within the plasmids are indicated below.
Directed gene disruption of lacZ and galT.
To disrupt lacZ, the 4.3-kb SacI fragment of pDP254 was cloned into SacI-digested vector pJDC9 (5). The resulting construct, pDP254-2 (Fig. 1), was digested with EcoRV, which removed a 1,007-bp fragment internal to the lacZ gene, and self-religation produced pDP418, which contains an in-frame deletion in the gene (ΔlacZ). This pJDC9-derived plasmid, which cannot replicate in lactococci, was used to transform L. lactis NCDO2054. Em-resistant transformants which harbored a chromosomal copy of pDP418 were obtained that were either blue or white (50:50 ratio) on X-Gal plates. A β-galactosidase-positive and a β-galactosidase-negative transformant were cultivated for approximately 100 generations in the absence of Em. Only the β-galactosidase-negative transformant resulted in Em-sensitive, white colonies when the cultures were screened for the loss of Em resistance and for β-galactosidase activity on X-Gal plates. Several white colonies were examined by PCR amplification of the lacZ region and by Southern hybridization, which confirmed the deletion. Following these experiments, a single strain, designated LL139, that contained a single copy of the ΔlacZ gene was selected.
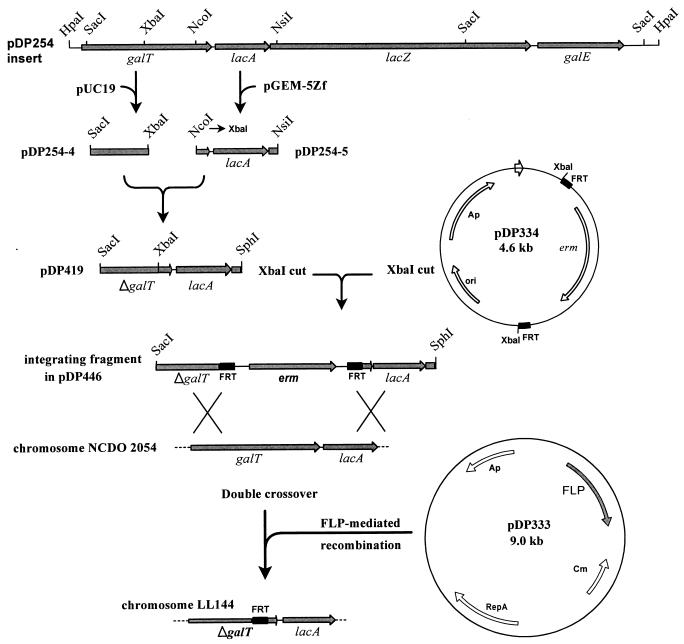
To inactivate the galT gene, a strategy involving the site-specific recombinase (FLP) encoded by the Saccharomyces cerevisiae 2μ circle plasmid, which catalyzes recombination between two target sequences (FRTs), was employed (21). When the FRTs are present as direct copies, FLP-mediated recombination leads to deletion of the intervening DNA, leaving a single copy of an FRT. The disruption of the galT gene was achieved by inserting the erm gene of pAMβ1, which was flanked by direct copies of the S. cerevisiae FRT, into a plasmid-encoded ΔgalT gene. Following integration of this linear structure into the L. lactis NCDO2054 genome via a double crossover, the erm gene was removed by the S. cerevisiae recombinase, resulting in a 580-bp deletion in galT and a single copy of an FRT in the resultant strain, designated L. lactis LL144. The construction of the deletion within the galT gene is described in detail in the legend to Fig. 2.
FIG. 2.
Schematic representation of the construction of the deletion in the galT gene in the gal-lac operon of L. lactis NCDO2054. The 650-bp SacI-XbaI fragment from pDP254, which contains the 5′ portion of galT, was cloned into pUC19, resulting in pDP254-4. The 931-bp NcoI-NsiI fragment, containing the 3′ part of galT from pDP254, was cloned into pGEM-5Zf (Promega), and the NcoI site in the construct was converted to an XbaI site by using the oligonucleotide 5′-CATGAATCTAGATT-3′, generating pDP254-5. The XbaI-NsiI fragment from pDP254-5 was then combined with the SacI-XbaI fragment by ligation into XbaI- and NsiI-digested pDP254-4, which produced a 580-bp deletion in the galT gene (ΔgalT) in plasmid pDP419. Plasmid pUC-EM was constructed by cloning the erythromycin resistance gene (erm) of pAMβ1, obtained as a 1.7-kb AvaI-HindIII fragment from pVA838 (31), into pUC19. The FRT of the site-specific FLP produced by S. cerevisiae was synthesized by using oligonucleotides based on the published DNA sequence (1). The two oligonucleotides FRT1 and FRT2 (Table 1), which were equipped with 3′ overhangs of appropriate recognition sites, were annealed together and ligated into pGEM-5Zf previously digested with the same enzymes, generating the plasmid pGEM-FRT. The restriction sites flanking the FRTs in pGEM-FRT were converted by using a series of linkers to sites which were suitable for cloning on either side of the erm gene in pUC-EM, resulting in plasmid pDP334 (39). Plasmid pDP446 was constructed by cloning the erm gene, flanked by the FRTs from pDP334, into the XbaI site of pDP419. The linear SacI-SphI fragment of pDP446 was transformed into L. lactis NCDO2054, and Em-resistant transformants were obtained. Plasmid pDP333, from which the S. cerevisiae 2μ circle FLP is expressed (40), was subsequently transformed into the latter Em-resistant strains, and the transformants were selected on plates containing Cm. When these transformants were replica plated onto media with and without Em, approximately 90% of the colonies were found to have lost their Em resistance, indicating FLP-mediated excision of the erm gene. A strain designated LL144 was characterized by PCR and Southern hybridization to confirm the presence of the ΔgalT gene and a single copy of an FRT. The temperature-sensitive pDP333 plasmid was cured from LL144 by passaging the culture at 35°C and screening for the loss of Cm resistance.
Enzyme assays.
In preparation for spectrophotometric assays, cells growing exponentially were harvested at an optical density at 600 nm (OD600) of 1.0 and washed in 20 mM phosphate buffer (pH 6.5) containing 10 mM MgCl2 and 1 mM dithioerythritol. The cells were disrupted by shaking at 4°C with glass beads (105 μm-diameter; Sigma) in a Bead Beater (Biospec Products) for three cycles of 3-min treatments, with 1-min intervals in between on ice. Cellular debris was removed by a 15-min centrifugation at 13,000 × g at 4°C, the extracts were stored on ice, and all enzyme assays were completed within 4 h after preparation of the extracts. β-Galactosidase and galactose-1-phosphate uridylyltransferase were assayed at 30 and 25°C, respectively, by standard protocols (24, 32). UDPglucose 4-epimerase activity was determined by the method of Wilson and Hogness (53) with the modifications of Poolman et al. (38). Toluene-treated cells were prepared and assayed for β-galactosidase activity by the method of Miller (32). For the determination of β-glucuronidase activity, cell extracts were prepared as described for spectrophotometric assays, except that GUS buffer (50 mM NaHPO4, pH 7.0; 10 mM β-mercaptoethanol; 1 mM EDTA; 0.1% Triton X-100) was used, and the assay was performed as previously described (8). Protein concentrations were estimated by using the Bio-Rad protein assay reagent (4).
RNA isolation, Northern hybridization, and primer extension analysis.
L. lactis NCDO2054 was grown in glucose-, lactose-, or galactose-containing M17 medium to an OD600 of 0.7. The cells were immediately harvested, and their RNA was extracted by using Macaloid (Rheox) and glass beads (<100 μm diameter; Sigma) as previously described (27). The RNA was resuspended in diethylpyrocarbonate-treated sterile water and stored at −80°C. The RNA obtained was of high purity (A260/A280, >2.0) and showed no degradation of the major 23S and 16S rRNA species. For analysis of transcripts, equal quantities of RNA were glyoxalated and fractionated by using a 1% agarose gel (49). A part of the gel, containing the RNA ladder (0.24 to 9.5 kb; Gibco BRL), was stained with ethidium bromide as recommended by the manufacturer. Transfer of the RNA to a nylon membrane (GeneScreen Plus; DuPont), hybridization, and analysis were carried out according to the manufacturer’s instructions.
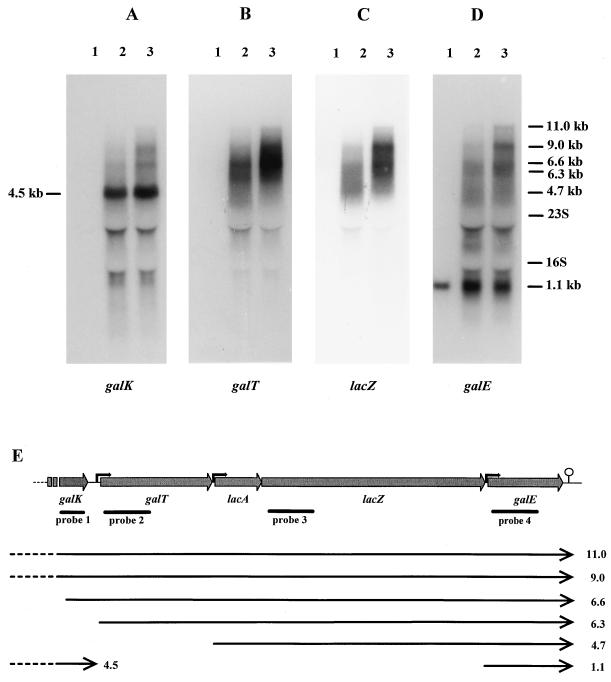
The positions of the DNA probes used in the Northern hybridizations are shown in Fig. 3E. The probes (350 bp for galK and 600 bp for the others) for the galK, galT, lacZ, and galE genes were made by PCR with primer pairs GALT5-GALT10, GALT2-GALT3, LACZ1-LACZ2, and GALE2-GALE3, respectively (Table 1). The PCR fragments were labeled with [α-32P]dCTP by using a Random Primed DNA labeling kit (Boehringer Mannheim) and purified on NICK columns (Pharmacia) before use.
FIG. 3.
Northern blot analysis of gal-lac operon expression from L. lactis NCDO2054 grown on medium containing glucose (lane 1), lactose (lane 2), or galactose (lane 3) and hybridized with probes specific for galK (A), galT (B), lacZ (C), and galE (D). The positions of the 23S (2.9-kb) and 16S (1.5-kb) rRNAs and the estimated sizes of the gal-lac-specific transcripts are indicated. (E) Illustration of the transcriptional units of the gal-lac operon of L. lactis NCDO2054. The positions of potential promoters (black arrows) and the transcriptional terminator ( ) are indicated. The transcripts are represented as lines with arrows, and their estimated lengths (in kilobases) are indicated at the right. Lines labeled probe 1, probe 2, probe 3, and probe 4 show the extents of the DNA probes used in Northern analysis of the mRNA of the operon.
TABLE 1.
Primers cited in this study
| Primer | Sequence |
|---|---|
| FRT1 | CTTTCGGAAACGCTTTGAAGTTCCTATTCCGAAGTTCCTATT |
| CTCTAGAAAGTATAGGAACTTCAGAGCGCTTTTGAAATGCA | |
| FRT2 | TTTCAAAAGCGCTCTGAAGTTCCTATACTTTCTAGAGAATAG |
| GAACTTCGGAATAGGAACTTCAAAGCGTTTCCGAAAGGGCC | |
| GALT2 | TGATGAGCTCTATCACAG |
| GALT3 | ATTGATCTAGAAATCCCA |
| LACZ1 | TTGATGTACTAGAGCGTA |
| LACZ2 | AAGTCTAACAAGTAGTTA |
| GALE2 | TCTTGGTGGAGCAGGATA |
| GALE3 | TCATCACCATAAATAGTA |
| GALT4 | CGGGATCCAAATTGGTTCTGGTTCAACA |
| GALT6 | AATAAGAATTCGAATTGATTGATAAATTGAC |
| GALT5 | CGGGATCCTCATGCCGTTTATGAAAATA |
| GALT10 | TGAACCAGAACCAATTT |
| GALE4 | CTTCGAATTCATCCTGCTCCACCAAGAACT |
| GALE5 | TGACGGATCCCATATGGGTGTAGGTGGAGA |
| LACA1 | GCGGATCCAGAAGTATTTGAACAGGTGT |
| LACA2 | CTGAATTCTGAACTCGTTTGTAGCTTTC |
| 4701 | TTGCATTGACTCACCCAC |
| 4702 | GGCAAGTTTATAGTGATG |
| GUS-AS | GGGTTGGGGTTTCTACAGGACGTA |
| GUS2-AS | GGACGTAACATAAGGGACTCCTCA |
| GALE.PE | CATCGTAACCACGTTTAAGAAGCA |
Primer extension of the galE promoter was performed by annealing 1 pmol of primer GALE.PE or GUS2-AS (Table 1), which are complementary to the galE mRNA and gus mRNA of pCRN20, respectively, to 20 μg of RNA followed by cDNA synthesis as previously described (49). Primer-extended products were electrophoresed on a sequencing gel together with the products of plasmid sequencing reactions obtained with the same primers.
Construction of plasmids for promoter analysis.
The regions preceding the galT, lacA, and galE genes were amplified by PCR with primers GALT4 and GALT6, LACA1 and LACA2, and GALE3 and GALE4, respectively (Table 1). The BamHI and EcoRI sites incorporated in the primer sequences allowed the fragments to be cloned in the correct orientation in front of the β-glucuronidase gene (gusA) of pNZ273 (37). The ligation products were electrotransformed into L. lactis NCDO2054, generating pCRN24, pCRN21, and pCRN20, harboring the galT, lacA, and galE potential promoter regions, respectively. The sequences of the inserts in the plasmids were confirmed by nucleotide sequence analysis with the GUS-AS primer (Table 1).
Nucleotide sequence accession number.
The GenBank accession numbers for the nucleotide sequences reported for L. lactis NCDO2054 and LM0230 are AF082008 and AF082009, respectively.
RESULTS
Nucleotide sequence and organization of the L. lactis NCDO2054 gal-lac region.
Initially the lacZ gene of L. lactis NCDO2054 was cloned on an estimated 7-kb HpaI fragment in pUC19(Fig. 1) by α-complementation of E. coli JM109#8 in the presence of IPTG, a gratuitous inducer of the E. coli lacZ promoter. In the absence of IPTG, α-complementation was no longer observed, suggesting that a functional promoter of L. lactis lacZ might not be present in the insert. The nucleotide sequence of a 7,172-bp region within the HpaI fragment was determined, revealing the presence of four complete open reading frames (ORFs) and one partial ORF, which are initially designated here based on their homology to previously characterized genes (Fig. 1). The lacZ gene was 3,071 bp in length, and although there were four possible translational initiation sites in frame, the most likely was that positioned 6 bp downstream of a putative ribosome-binding site (RBS). Just 11 bp upstream of lacZ and oriented in the same direction was a 784-bp lacA gene encoding a galactoside acetyltransferase. A putative RBS preceded the start codon of lacA by 8 bp. Upstream of lacA, separated by 40 bp, was a 1,479-bp galactose-1-phosphate uridylyltransferase gene (galT) with a potential RBS 6 bp upstream. The short intergenic regions between galT, lacA, and lacZ suggested that these genes might be cotranscribed. The partial sequence (373 bp) of an ORF encoding the galactokinase gene (galK) was found 173 bp upstream of galT. Downstream of lacZ, separated by 51 bp, was a 981-bp UDPgalactose 4-epimerase gene (galE) gene, and this was followed by a stem-loop structure, with a free energy value of −12.7 kcal mol−1, which could function as a transcriptional terminator (Fig. 4D).
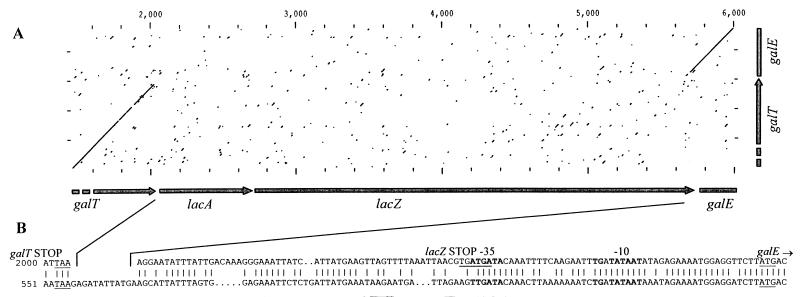
FIG. 4.
(A) Nucleotide sequence of the galK-galT intergenic region and the deduced C- and N-terminal amino acid sequences of the galK and galT genes. (B) Nucleotide sequence of the galT-lacA intergenic region and the deduced C- and N-terminal amino acid sequences of the galT and lacA genes. (C) Nucleotide sequence of the lacZ-galE intergenic region and the deduced C- and N-terminal amino acid sequences of the lacZ and galE genes. (D) Sequence of the potential transcriptional terminator following the galE gene. Translational stop (★) and start (underlined) sequences and potential promoter sequences (boldface) are indicated. The transcriptional start site of the galE promoter is indicated as +1. The precise DNA fragments inserted in the gusA reporter plasmid pNZ273 lie between the arrows (⇒ ⇐). Inverted repeats capable of forming stem-loop structures are indicated by arrowheads under the sequence. Catabolite-responsive element-like (cre-like) sequences are underlined.
Homology values for the predicted amino acid sequences of the genes are presented in Table 2. The deduced sequences of galT and galE and the partial sequence of galK showed the highest degrees of similarity (56 to 71%) to sequences from gram-positive microorganisms (33, 38), especially other lactic acid bacteria. In contrast, the predicted amino acid sequence of the intervening lacZ gene exhibited the greatest homology to gram-negative homologs. A similarity of 60% to the β-galactosidase of E. coli (25) was found, while approximately 40% homology to β-galactosidases of lactic acid bacteria was exhibited. The lacA gene, when compared to related gram-negative genes, showed 54% homology to the galactoside acetyltransferase of E. coli (23) and 46% homology to lacA of Lactobacillus delbrueckii subsp. bulgaricus (28). Although another lacA gene has been discovered in the gram-positive bacterium Streptococcus bovis, its nucleotide sequence has not been reported (15).
TABLE 2.
Homologies of the predicted proteins of the lac-gal genes of L. lactis NCDO2054
| L. lactis gene | Homologous protein from microorganism: | % Identity | % Similarity |
|---|---|---|---|
| galKa | Lactobacillus helveticus | 58 | 64 |
| S. thermophilus | 59 | 64 | |
| E. coli | 41 | 52 | |
| galT | Lactobacillus helveticus | 50 | 57 |
| Bacillus subtilis | 47 | 56 | |
| E. coli | —b | — | |
| lacA | E. coli | 44 | 54 |
| Lactobacillus delbrueckii subsp. bulgaricus | 39 | 46 | |
| lacZ | E. coli | 52 | 60 |
| Lactobacillus delbrueckii subsp. bulgaricus | 32 | 40 | |
| S. thermophilus | 32 | 42 | |
| galE | S. thermophilus | 61 | 71 |
| E. coli | 39 | 57 |
Note that homology values are for the partial sequence of galK.
—, no significant homology evident.
The GC contents of the galT, lacA, lacZ, and galE genes were all in the range 34 to 38%, which is similar to the average 38% GC content of L. lactis. The codon usages of the gal and lac genes were compared to a codon frequency table for L. lactis (47), using the method described by Grantham et al. (16). The D2 (distance squared) values for lacA, lacZ, and galE were all relatively high, i.e., 2.889, 2.074, and 2.454, respectively, and only the codon usage pattern of the galT gene, which gave a D2 value of 1.186, would be considered close to the typical pattern of L. lactis.
Induction of enzyme activities for lactose and galactose metabolism in L. lactis.
During growth on M17-glucose, galactose-1-phosphate uridylyltransferase activity could not be detected, but this activity was induced over 200-fold on lactose medium and over 300-fold on galactose medium (Table 3). A low level of β-galactosidase was present in glucose-grown cultures, and this activity was also induced approximately 300-fold in lactose- or galactose-containing medium. However, a substantial level of UDPglucose 4-epimerase activity was already produced in L. lactis NCDO2054 growing on glucose, and this increased 10-fold in lactose- or galactose-containing medium. The similar levels of induction observed for the uridylyltransferase and β-galactosidase activities suggest that at least the galT and lacZ genes, and probably the intervening lacA gene, are coregulated.
TABLE 3.
Enzyme activity measurements
| Strain | Carbohydrate in growth medium | Induction with galactosea | Activity
ofb:
|
||
|---|---|---|---|---|---|
| Galactose-1-phosphate uridylyltransferase | β-Galactosidase | UDPgalactose 4-epimerase | |||
| NCDO2054 | Glucose | No | —c | 3.8 ± 0.2 | 788.5 |
| Lactose | No | 225.9 ± 9.5 | 1,305.4 ± 122.6 | 8,735.2 | |
| Galactose | No | 350.2 ± 34.9 | 1,008.2 ± 80.5 | 7,801.1 | |
| Glucose | Yes | 178.8 | 441.5 | NDd | |
| LL144ΔgalT | Glucose | No | — | 69.3 ± 5.1 | ND |
| Glucose | Yes | — | 283.0 | ND | |
| LL139ΔlacZ | Glucose | No | — | — | ND |
| Galactose | No | 317.8 ± 37.9 | — | ND | |
To check for induction of enzyme activity, 20-ml cultures grown for 16 h in M17-glucose were washed and resuspended in 50 ml of fresh M17 broth containing 1% galactose and incubated at 30°C. Aliquots were removed after 3 h and assayed for galactose-1-phosphate uridyltransferase and β-galactosidase activities.
In nanomoles per minute per milligram of total protein.
—, no measurable enzyme activity; i.e. <0.2 nmol per min per mg.
ND, not determined.
Confirmation of galT, lacZ, and galE gene functions.
L. lactis LL144, which has a deletion in the galT gene, could no longer grow in galactose-containing M17 medium as expected, and no galactose-1-phosphate uridylyltransferase activity could be measured in LL144 even when exponentially growing cells were inoculated into M17-galactose to allow for induction (Table 3). Strain LL144 also could no longer grow in M17-lactose. Measurement of β-galactosidase activity indicated that there was an 18-fold increase in the level detected in glucose-grown cells of strain LL144 compared to that of strain NCDO2054. β-Galactosidase activity remained inducible when glucose-grown cells were inoculated into M17-galactose, as was the case for strain NCDO2054 (Table 3).
Inactivation of the lacZ gene resulted in the loss of the ability of the mutant L. lactis LL139 to grow in M17-lactose and the loss of detectable β-galactosidase activity in cells grown on glucose or galactose. The ΔlacZ disruption did not affect the ability of LL139 to grow on galactose as a carbon source, since this was an in-frame deletion which should not influence the transcription of the following cistron and, furthermore, the galE gene downstream of lacZ is preceded by its own promoter (see below).
Several attempts to disrupt the galE gene by methods similar to those described above failed to produce an L. lactis NCDO2054 strain carrying a deletion in galE. Instead, the function of the gene was confirmed by complementation of the galE mutation in E. coli MK30-3. The 1.4-kb NdeI-SacI fragment of pDP254 harboring galE (Fig. 1) was cloned into the corresponding sites in pGEM-5Zf. The gene was subsequently removed as a SacI-PstI fragment and placed under the control of the lacZ promoter of pUC19 to generate pDP458. Strain MK30-3 transformed with pDP458 could grow on IPTG-supplemented minimal medium with galactose as the sole carbon source, whereas MK30-3 containing pUC19 alone could not grow.
Transcriptional analysis of the gal-lac genes.
To determine the transcriptional organization and the nature of induction of the gal-lac genes, total RNA from glucose-, lactose-, and galactose-grown L. lactis NCDO2054 cells was analyzed by Northern hybridization (Fig. 3). When probes for the galT and lacZ genes were used, surprisingly, several large transcripts whose synthesis was induced during growth on lactose and galactose were visible (Fig. 3B and C). The transcripts which were more intense and sharper in the galactose-grown than in the lactose-grown cultures were estimated to be 11.0, 9.0, 6.6, and 6.3 kb. In addition, a weak signal of approximately 4.7 kb appeared to be present with the lacZ probe for lactose-grown cultures (Fig. 3C). An identical pattern of four large, weak transcripts was observed with the galE probe, but there was also a small band of 1.1 kb present under all growth conditions (Fig. 3D). This 1.1-kb mRNA corresponded to the size of the galE gene, indicating that galE was also transcribed alone from its own promoter and, furthermore, providing evidence of the functionality of the transcriptional terminator downstream of galE. Hybridization with the galK-specific probe revealed three long, weak transcripts of approximately 11.0, 9.0, and 6.6 kb in size which were also detected with the galT, lacZ, and galE probes, but the smaller, 6.3-kb mRNA was not detected (Fig. 3A). An extra, strong transcript, estimated to be 4.5 kb, was observed only with the galK probe in the lactose- and galactose-grown cultures.
When one assumes that the potential terminator following galE is functional, and since the mRNA required for a single transcript for the galT, lacA, lacZ, and galE genes must be at least 6.3 kb, the presence of identically sized transcripts for galT, lacZ, and galE, four of which were estimated to be larger than or equal to 6.3 kb, indicated either that these genes were cotranscribed from several promoters located upstream of galT or that mRNA processing of one or more of the larger transcripts might be occurring. It is noteworthy that the 9.0- and 11.0-kb transcripts containing the entire galKT-lacAZ-galE operon originate upstream of galK.
Characterization of galT, lacA, and galE putative promoters regions.
The regions preceding the galT (Fig. 4A), lacA (Fig. 4B), and galE (Fig. 4C) genes were cloned in front of the gusA reporter gene (resulting in plasmids pCRN24, pCRN21, and pCRN20, respectively) in order to locate and investigate the inducibility of promoters which might be responsible for the smaller transcripts of the gal-lac operon. The extents of the cloned fragments in the plasmids are indicated in Fig. 4, and the results of the study are presented in Table 4. On medium containing glucose, lactose, or galactose, colonies of L. lactis NCDO2054 harboring pCRN20 developed a strong blue color following overnight incubation, indicating the presence of a functional promoter preceding the galE gene. Growth of L. lactis NCDO2054(pCRN24) transformants resulted in blue colonies on X-Gluc plates supplemented with lactose after 48 h of incubation and in pale blue colonies on galactose medium after 72 h, while no color development was observed with glucose. Thus, the region immediately upstream of the galT gene contains a weak promoter which is induced by lactose and, to a much lesser extent, by galactose. The L. lactis NCDO2054(pCRN21) colonies developed a pale blue color on X-Gluc plates containing lactose after 72 h of incubation, but no reaction was detected for glucose- or galactose-containing medium, indicating the presence of a very weak lactose-inducible promoter upstream of the lacA gene. The galT and lacA promoters were both repressed by glucose, since colonies of L. lactis harboring either pCRN24 or pCRN21 failed to develop a blue color when grown in the presence of lactose plus glucose.
TABLE 4.
Development of blue color on plates containing X-Gluc, and β-glucuronidase activity detected for L. lactis strains harboring gal-lac promoter fusion plasmids
| Plasmid | Putative promoter | Growth medium carbohydrate | Color development
ata:
|
β-Glucuronidase activityb | |
|---|---|---|---|---|---|
| 48 h | 72 h | ||||
| pCRN24 | galT | Glucose | − | − | |
| Lactose | + | ++ | NDc | ||
| Galactose | − | + | |||
| Glucose-lactose | − | − | |||
| pCRN21 | lacA | Glucose | − | − | |
| Lactose | − | + | ND | ||
| Galactose | − | − | |||
| Glucose-lactose | − | − | |||
| pCRN20 | galE | Glucose | ++++ | 459 | |
| Lactose | ++++ | 935 | |||
| Galactose | ++++ | 492 | |||
−, no blue color; increasing numbers of + signs indicate more-intense blue color.
In micromoles per minute per milligram of protein.
ND, not detectable.
β-Glucuronidase activity was approximately twofold higher for L. lactis NCDO2054(pCRN20) grown on lactose than the level achieved in medium with glucose or galactose. Therefore, although galE is constitutively expressed from the galE promoter on glucose- and galactose-containing-media, this promoter is weakly induced by lactose. Since X-Gluc is an extremely sensitive substrate for β-glucuronidase activity, the functional galT and lacA promoters are obviously very weak. Thus, despite the use of very concentrated cell extracts, the spectrophotometric-assay substrate (para-nitro-β-d-glucuronic acid) for β-glucuronidase activity was not sensitive enough to measure the strengths of these promoters.
To map the transcriptional start sites of these three putative promoters, primer extension was performed with total RNA from NCDO2054. A primer-extended product, 5 bp downstream of the galE gene −10 region, was reproducibly obtained from mRNA of glucose-grown cells (Fig. 4C). The same start point was obtained for NCDO2054 containing the promoter cloned in pCRN20. The galT and lacA regions also contain functional promoters, but no reproducible extended products were detected with the mRNA of lactose- or galactose-grown cells. The fact that the galT and lacA genes are present on several transcripts, of which the larger mRNAs are less abundant, could explain the difficulties of obtaining a clearcut result. The experiments were repeated with NCDO2054 harboring the recombinant pCRN21 or pCRN24, but still no signals were detected by primer extension.
Analysis of the insertion or deletion point of lacA-lacZ in the gal-lac operon.
The lacA and lacZ genes of L. lactis NCDO2054 exhibit substantial homology to homologs from gram-negative organisms, while the flanking galT and galE genes are most similar to related gram-positive genes. This suggests that the lacA and lacZ genes may have been recently acquired by the gal-lac operon of strain NCDO2054. Since dairy starter L. lactis strains do not possess a β-galactosidase gene but have the enzymatic potential to ferment galactose via the Leloir pathway, it was interesting to determine if a related gal operon was present in a rapid-lactose-fermenting L. lactis strain and, if so, to determine the putative insertion point of the lacA and lacZ genes. L. lactis LM0230 is a strain derived from the fast lactose fermenter L. lactis C2 which has been cured of its lactose plasmid (12). Using a set of primers, 4701 and 4702 (Table 4), which were homologous to the galT and galE genes of strain NCDO2054, an 848-bp fragment was amplified from the chromosome of L. lactis LM0230. Sequence analysis indeed revealed that the 3′ and 5′ ends of ORFs were very similar at the nucleotide sequence level to the galT and galE genes, respectively, of strain NCDO2054 (Fig. 5A). The interspersed lacA and lacZ genes of NCDO2054 were missing in strain LM0230. Instead, LM0230 contained a 110-bp intergenic region which, upon introduction of gaps, exhibited homology to the 3′ end of the lacZ gene of strain NCDO2054. Nevertheless, the promoter regions preceding the galE gene of both strains were conserved and highly homologous (Fig. 5B).
FIG. 5.
(A) Dot plot presentation of nucleotide sequence comparisons between the galT-lacA-lacZ-galE of L. lactis NCDO2054 (horizontal axis) and the 5′ end of galT and 3′ end of galE of L. lactis LM0230 (vertical axis). Numbers above the dot plot indicate nucleotide positions. (B) Nucleotide sequences of the galT-lacA and lacZ-galE intergenic regions of L. lactis NCDO2054 (upper sequence) aligned with the galT-galE intergenic region of L. lactis LM0230 (lower sequence). Identical nucleotides (|) are indicated. The genes encoded by the DNA are shown above the sequence. The translational start and stop sequences are underlined, and potential promoter sequences for galE are indicated in boldface.
DISCUSSION
The present study describes the location and characterization of the lacA and lacZ genes of L. lactis NCDO2054 as a functional part of an operon also encoding the galK, galT, and galE genes and confirms the functions of these genes, which are involved in the metabolism of galactose and lactose. Furthermore, evidence for the recent acquisition within this gal-lac operon of the lac genes, which have engaged the promoters of the original gal operon in order to direct and control their transcription, is provided.
Organization of the gal-lac region.
In many organisms, combinations of the galK, galT, galE, and galM (mutarotase gene) genes, encoding enzymes for the Leloir pathway of galactose metabolism, are clustered or organized in a single operon (11, 20). In E. coli, the gal operon contains galE-galT-galK-galM, in that order, and the genes encoding galactose transport are located elsewhere (50). Recently a galactose gene cluster which has the gene order galA-galM-galK-galT-galE, with galA encoding a galactose permease, was reported for L. lactis MG1363 (20). Similarly, the genes for lactose utilization are usually grouped together; e.g., in E. coli the lacZYA genes constitute an operon, with lacY encoding a lactose permease (2). In the gram-positive organism Streptococcus thermophilus, the lacZ gene is cotranscribed with the gene responsible for lactose transport, lacS, and although the genes encoding enzymes of the Leloir pathway are located just upstream of lacSZ, they are not part of the same operon (9, 38). The nucleotide sequences of the lacA and lacZ genes of L. lactis NCDO2054 have been reported (17, 18), but their proximity to each other and their location within a gal-lac operon have not been described. So far, the organization galKT-lacAZ-galE is unique since it is the only occurrence of lac genes interspersed in an operon with genes encoding enzymes for the Leloir pathway. Despite the considerable similarities of the L. lactis lacA and lacZ genes to their counterparts in E. coli, the lac gene orders of these two bacteria are different.
Functions of galT, lacZ, and galE.
The functions of the galT and lacZ genes were confirmed by deleting internal portions of the genes (Table 3). Surprisingly, the ΔgalT mutation in strain LL144 resulted in the inability to grow on lactose or galactose. In E. coli, the growth of mutants lacking galactose-1-phosphate uridylyltransferase is arrested by accumulation of galactose-1-phosphate (30). Similarly, when LL144 is grown in the presence of galactose or lactose, this same intermediate may be responsible for the observed growth inhibition. There was a slight increase in expression of the downstream lacZ gene in LL144 grown in glucose-containing medium. The remaining copy of the recombinase target site (FRT) in the ΔgalT gene may have created a weak promoter which is not regulated, or, alternatively, the 600-bp deletion within galT may have resulted in a more stable transcript. Either of these possibilities could have enhanced expression of the genes downstream of galT. β-Galactosidase activity could still be induced in LL144 in the presence of galactose, which suggests that alterations in transcription of lacZ are not the obstacle to growth on lactose.
Attempts to confirm the function of galE by gene disruption were unsuccessful, which suggests that the UDPgalactose 4-epimerase enzyme possesses a function(s) other than the metabolism of galactose. Since the galE gene was expressed at a substantial level from its own promoter even in the absence of lactose or galactose, it is not unreasonable to assume that this other function(s) must be essential for viability of L. lactis cells. UDPgalactose 4-epimerase has been demonstrated to be involved in the preparation of carbohydrate residues for incorporation into complex polymers such as exopolysaccharide(s) or lipopolysaccharide(s) for cell wall synthesis (14).
Transcriptional regulation within the gal-lac operon.
Northern analysis of the gal and lac genes revealed an unusual transcriptional organization for the operon, with a surprisingly large number of transcriptional units, as illustrated in Fig. 3E. The galKT-lacAZ-galE genes were transcribed together as part of the two longest mRNAs, with sizes of 11.0 and 9.0 kb, which both originated well upstream of the galK gene. A communication by Grossiord et al. (19) described a 7.5-kb mRNA transcript of the gal operon of L. lactis MG1363 and determined by primer extension that its 5′ end was just upstream of the galA gene. The occurrence of several smaller transcripts was reported as well and was explained as being due to mRNA processing at the galM-galK and galK-galT junctions. Since the MG1363 gal operon lacks the ca. 3.6-kb lacAZ cistrons, this major 7.5-kb transcript is the equivalent of the 11-kb transcript of NCDO2054. The smaller, 9.0- and 6.6-kb transcripts of NCDO2054 correspond to the respective smaller processed mRNA fragments of MG1363. As observed by Grossiord et al. (19) and ourselves, these transcripts are all induced by galactose and are responsible for the major expression of the operon.
Interestingly, the level of induction of the β-galactosidase activity of NCDO2054 was approximately 30% higher in the lactose-grown cultures than in those grown on galactose (Table 3) (6). Furthermore, Northern blot analysis with the lacZ probe revealed the appearance of a weak signal at approximately 4.7 kb in the lactose-grown cultures only (Fig. 3C). These results strongly suggest the presence of an additional lactose-inducible promoter activity following galT and preceding lacA or lacZ.
In fact, DNA sequence analysis identified within the gal-lac operon three putative promoter sequences that preceded the galT, lacA, and galE genes. These were investigated by transcriptional fusion to the promoterless β-glucuronidase reporter gene (Table 4). All three promoters were induced by lactose, and the galT promoter was also induced by galactose, although to a lesser extent. Both the galT and lacA promoters were repressed by glucose, while the galE promoter was constitutively active on glucose and galactose. A search within the galT and lacA promoter regions located sequences that were homologous to the 14-bp palindromic catabolite-responsive element sequences of catabolite-repressed promoters in gram-positive bacteria (51). Comparison with the consensus sequence (5′-TGWAANCGNTNWCA-3′) showed 12 of 14 and 9 of 14 matches with the potential sequences within the galT and lacA promoter regions, respectively (Fig. 4A and B).
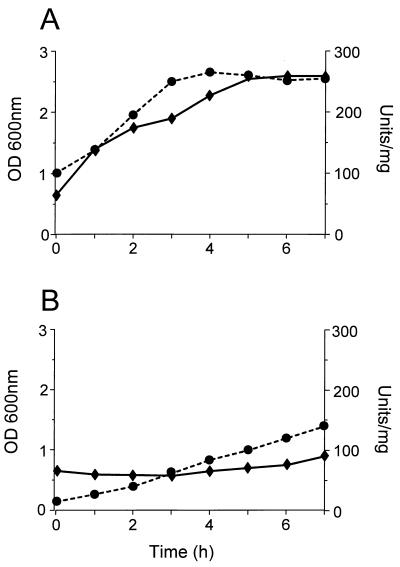
Despite these observations, NCDO2054 has a long lag phase, prior to growth on lactose, during which time β-galactosidase activity is induced far more rapidly during growth on galactose than on lactose and the strain grows substantially slower on lactose (Fig. 6). Therefore, the overall driving force for the transcriptional expression of this gal-lac operon is dependent on induction by galactose rather than lactose. It is noteworthy that the manner by which lactose becomes available for hydrolysis by β-galactosidase within strain NCDO2054 is presently unknown. Thus, it is possible that the form of lactose required for induction of the gal-lac transcripts is a limiting factor within the cell if the growth medium contains lactose as the unique carbon source (e.g., milk).
FIG. 6.
Growth and β-galactosidase activity of L. lactis NCDO2054 in M17 medium containing galactose (A) or lactose (B). A 16-h culture of strain NCDO2054 grown in M17-glucose was pelleted, washed, and resuspended (1:10) in fresh M17 broth supplemented with 0.5% galactose or lactose. At hourly time intervals, the OD600 (—♦—) was measured and a 10-ml sample was removed to assay for β-galactosidase activity. One unit of β-galactosidase enzyme is equivalent to 1 μmol of o-nitrophenol released from o-nitrophenyl-β-d-galactopyranoside (ONPG) per minute. The β-galactosidase activity, in units per milligram of total protein, is indicated (---•---).
Evolution of lacA-lacZ within the gal-lac operon.
The galT and galE genes of fast-lactose-fermenting L. lactis LM0230 were highly similar to their counterparts in strain NCDO2054 at the nucleotide sequence level, and they have the same gene order except for the presence of the intervening lacA-lacZ within strain NCDO2054. The greater degree of similarity of the lacA and lacZ genes to homologs of gram-negative organisms than to those of gram-positive bacteria implies that the genes have likely been recently acquired within the gal operon of NCDO2054 rather than being lost via the generation of a lac gene-free gal operon as in LM0230. This supports the theory that this slow-fermenting strain has arisen from a fast lactose fermenter as opposed to the reverse situation (7). Horizontal gene transfer or insertion sequence-mediated events may have played a role in procurement of these genes by NCDO2054. However, the galT-lacA and lacZ-galE intergenic regions evolved into very compact sequences, leaving behind insufficient traces with which to ascertain the mode of acquisition of the lac genes within the operon.
It is interesting that several promoters are involved in the transcription of the gal-lac genes. The lactose-inducible lacA promoter activity may have resulted from a remnant promoter structure which was linked to the lac genes from their original source. The other promoter activities directing the transcription of the lac genes were probably already present in the original gal operon. However, some of these promoters, especially those preceding the galT gene, may have evolved further to allow lacA-lacZ gene expression levels adequate for the efficient metabolism of lactose in this strain.
ACKNOWLEDGMENTS
We thank the Swiss National Science Foundation for financial support within SPP Food Biotechnology Program no. 5002-044544.
We are very grateful to Gilbert Lamothe for photography and to Harald Brüssow and Ralf Zink for critically reviewing the manuscript. We also thank Willem de Vos, NIZO, Ede, The Netherlands, for the kind gift of pNZ273.
REFERENCES
- 1.Andrews B J, Proteau G A, Beatty L G, Sadowski P D. The FLP recombinase of the 2μ circle DNA of yeast: interaction with its target sequences. Cell. 1985;40:795–803. doi: 10.1016/0092-8674(85)90339-3. [DOI] [PubMed] [Google Scholar]
- 2.Beckwith J. The lactose operon. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 1444–1452. [Google Scholar]
- 3.Bissette D L, Anderson R L. Lactose and d-galactose metabolism in group N streptococci: presence of enzymes for both the d-galactose 1-phosphate and d-tagatose 6-phosphate pathways. J Bacteriol. 1974;117:318–320. doi: 10.1128/jb.117.1.318-320.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen J-D, Morrison D A. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene. 1988;64:155–164. doi: 10.1016/0378-1119(88)90489-1. [DOI] [PubMed] [Google Scholar]
- 6.Citti J E, Sandine W E, Elliker P R. β-Galactosidase of Streptococcus lactis. J Bacteriol. 1965;89:937–942. doi: 10.1128/jb.89.4.937-942.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crow V L, Thomas T D. Properties of a Streptococcus lactisstrain that ferments lactose slowly. J Bacteriol. 1984;157:28–34. doi: 10.1128/jb.157.1.28-34.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Ruyter P G G A, Kuipers O P, Beerthuyzen M M, van Alen-Boerrigter I, de Vos W M. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J Bacteriol. 1996;178:3434–3439. doi: 10.1128/jb.178.12.3434-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vos W M. Metabolic engineering of sugar catabolism in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:127–146. doi: 10.1007/BF00395934. [DOI] [PubMed] [Google Scholar]
- 10.de Vos W M, Simons G. Molecular cloning of lactose genes in dairy lactic streptococci: the phospho-β-galactosidase and β-galactosidase genes and their expression products. Biochimie. 1988;70:461–473. doi: 10.1016/0300-9084(88)90083-1. [DOI] [PubMed] [Google Scholar]
- 11.de Vos W M, Vaughan E E. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol Rev. 1994;15:217–237. doi: 10.1111/j.1574-6976.1994.tb00136.x. [DOI] [PubMed] [Google Scholar]
- 12.Efstathiou J D, McKay L L. Inorganic salts resistance associated with a lactose-fermenting plasmid in Streptococcus lactis. J Bacteriol. 1977;130:257–265. doi: 10.1128/jb.130.1.257-265.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farrow J A E. Lactose hydrolyzing enzymes in Streptococcus lactis and Streptococcus cremorisand also in some other species of streptococci. J Appl Bacteriol. 1980;49:493–503. doi: 10.1111/j.1365-2672.1980.tb04724.x. [DOI] [PubMed] [Google Scholar]
- 14.Gabriel O. Biosynthesis of sugar residues for glycogen, peptidoglycan, lipopolysaccharide, and related systems. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 1. Washington, D.C: American Society for Microbiology; 1987. pp. 504–511. [Google Scholar]
- 15.Gilbert H J, Hall J. Molecular cloning of Streptococcus bovislactose catabolic genes. J Gen Microbiol. 1987;133:2285–2293. [Google Scholar]
- 16.Grantham R, Gautier C, Gouy M, Jacobzone M, Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981;9:r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffin H G, Gasson M G. The gene (lacA) encoding galactoside acetyltransferase from Lactococcus lactis. Biotechnol Lett. 1994;16:1125–1130. [Google Scholar]
- 18.Griffin H G, MacCormick C A, Gasson M G. Cloning, DNA sequence, and regulation of expression of a gene encoding β-galactosidase from Lactococcus lactis. DNA Sequence J Sequencing Mapping. 1996;6:337–346. doi: 10.3109/10425179609047572. [DOI] [PubMed] [Google Scholar]
- 19.Grossiord B P, Luesink E J, Kuipers O P, de Vos W M. Abstracts of the ASM Conference on Streptococcal Genetics: genetics of the streptococci, enterococci, and lactococci. Washington, D.C: American Society for Microbiology; 1998. Transcriptional analysis and regulation of the genes from the galactose operon of Lactococcus lactis, abstr. 2B-20; p. 72. [Google Scholar]
- 20.Grossiord B P, Vaughan E E, Luesink E, de Vos W M. Genetics of galactose utilization via the Leloir pathway in lactic acid bacteria. Lait. 1998;78:77–84. [Google Scholar]
- 21.Hartley J L, Donelson J E. Nucleotide sequence of the yeast plasmid. Nature. 1980;286:860–864. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- 22.Hayes F, Fitzgerald G F, de Vos W M, Daly C. Molecular organization of the minimal replicon of novel, narrow-host-range, lactococcal plasmid pCI305. Plasmid. 1991;25:16–26. doi: 10.1016/0147-619x(91)90003-f. [DOI] [PubMed] [Google Scholar]
- 23.Hediger M A, Johnson D F, Nierlich D P, Zabin I. DNA sequence of the lactose operon: the lacAgene and the transcriptional terminator region. Proc Natl Acad Sci USA. 1985;82:6414–6418. doi: 10.1073/pnas.82.19.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Isselbacher K J. Uridyl transferase. In: Bergmeyer H V, editor. Methods in enzymatic analysis. I. New York, N.Y: Academic Press, Inc.; 1974. pp. 802–805. [Google Scholar]
- 25.Kalnins A, Otto K, Ruther U, Muller-Hill B. Sequence of the lacZ gene of Escherichia coli. EMBO J. 1983;2:593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kashket E R, Wilson T H. Role of metabolic energy in the transport of β-galactosides by Streptococcus lactis. J Bacteriol. 1972;109:784–789. doi: 10.1128/jb.109.2.784-789.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuipers O P, Beerthuyzen M, Seizen R J, de Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisIgenes for development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 28.Lapierre, L., and J.-E. Germond. Personal communication.
- 29.Lawrence R C, Thomas T D, Terzaghi B E. Reviews of the progress in dairy science: cheese starters. J Dairy Res. 1976;43:141–193. [Google Scholar]
- 30.Lin E C C. Dissimilatory pathways for sugars, polyols, and carboxylates. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 1. Washington, D.C: American Society for Microbiology; 1987. pp. 244–284. [Google Scholar]
- 31.Macrina F L, Tobian J A, Jones K R, Evans R P, Clewell D B. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene. 1982;19:345–353. doi: 10.1016/0378-1119(82)90025-7. [DOI] [PubMed] [Google Scholar]
- 32.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 33.Mollet B, Pilloud N. Galactose utilization in Lactobacillus helveticus: isolation and characterization of the galactokinase (galK) and galactose-1-phosphate uridyl transferase (galT) genes. J Bacteriol. 1991;173:4464–4473. doi: 10.1128/jb.173.14.4464-4473.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgenthaler, P. Unpublished data.
- 35.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 36.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Platteeuw C, Simons G, de Vos W M. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1994;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poolman B, Royer T J, Mainzer S E, Schmidt B F. Carbohydrate utilization in Streptococcus thermophilus: characterization of the genes for aldose 1-epimerase (mutarotase) and UDPglucose 4-epimerase. J Bacteriol. 1990;172:4037–4047. doi: 10.1128/jb.172.7.4037-4047.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pridmore, R. D. Unpublished data.
- 40.Pridmore, R. D., and M. Richard. Unpublished data.
- 41.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Ehlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas T D. Regulation of lactose fermentation in group N streptococci. Appl Environ Microbiol. 1976;32:474–478. doi: 10.1128/aem.32.4.474-478.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas T D, Turner K W, Crow V L. Galactose fermentation by Streptococcus lactis and Streptococcus cremoris: pathways, products, and regulation. J Bacteriol. 1980;144:672–682. doi: 10.1128/jb.144.2.672-682.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson J. Lactose metabolism in Streptococcus lactis: phosphorylation of galactose and glucose moieties in vivo. J Bacteriol. 1979;140:774–785. doi: 10.1128/jb.140.3.774-785.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Guchte M, Kok J, Venema G. Gene expression in Lactococcus lactis. FEMS Microbiol Lett. 1992;88:73–92. doi: 10.1111/j.1574-6968.1992.tb04958.x. [DOI] [PubMed] [Google Scholar]
- 48.van Rooijen R J. Characterization of the Lactococcus lactis lactose genes and regulation of their expression. Ph.D. thesis. Ede, The Netherlands: NIZO; 1993. [Google Scholar]
- 49.van Rooijen R J, de Vos W M. Molecular cloning, transcriptional analysis and nucleotide sequence of lacR, a gene encoding the repressor of the lactose phosphotransferase system of Lactococcus lactis. J Biol Chem. 1990;265:18499–18503. [PubMed] [Google Scholar]
- 50.Weickert M J, Adhya S. The galactose regulon of Escherichia coli. Mol Microbiol. 1993;10:245–251. doi: 10.1111/j.1365-2958.1993.tb01950.x. [DOI] [PubMed] [Google Scholar]
- 51.Weickert M J, Chambliss G H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wells J M, Wilson P M, Le Page R W F. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol. 1993;74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 53.Wilson D B, Hogness D S. Galactokinase and uridine diphosphogalactose 4-epimerase from Escherichia coli. Methods Enzymol. 1966;8:229–240. [Google Scholar]
- 54.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]