Abstract
New discoveries in drugs and drug delivery systems are focused on identifying and delivering a pharmacologically effective agent, potentially targeting a specific molecular component. However, current drug discovery and therapeutic delivery approaches do not necessarily exploit the complex regulatory network of an indispensable microbiota that has been engineered through evolutionary processes in humans or has been altered by environmental exposure or diseases. The human microbiome, in all its complexity, plays an integral role in the maintenance of host functions such as metabolism and immunity. However, dysregulation in this intricate ecosystem has been linked with a variety of diseases, ranging from inflammatory bowel disease to cancer. Therapeutics and bacteria have an undeniable effect on each other and understanding the interplay between microbes and drugs could lead to new therapies, or to changes in how existing drugs are delivered. In addition, targeting the human microbiome using engineered therapeutics has the potential to address global health challenges. Here, we present the challenges and cutting-edge developments in microbiome-immune cell interactions and outline novel targeting strategies to advance drug discovery and therapeutics, which are defining a new era of personalized and precision medicine.
Graphical Abstract
Quote: “Every Time We Have a Chance to Get Ahead, [Microbes] Move the Finish Line.” – Hidden Figures 2017.
1. Introduction
A diverse variety of microbes, colonizing by trillions, can be found throughout the human body in locations such as the gastrointestinal tract, respiratory tract, skin, and reproductive system. The communities of bacteria and other microorganisms create a localized microbiome that plays an essential role in health and disease [1]. The microbiome is not just a passive bystander, but it actively impacts multiple host functions, including circadian rhythm, nutritional responses, metabolism, and immunity [2]. Specifically, the microbiome plays multiple critical roles in the programming and development of major components of the host’s innate and adaptive immune system. Initially, the high-throughput DNA sequencing technology involved clustering reads based on bacterial and archaeal 16S ribosomal RNA amplicon sequences, followed by whole metagenomic and meta transcriptomic sequencing [3,4], and more recently multiplexed spatial imaging [5] endeavors are defining the function, spatial localization, and real-time activity of microbiomes and revealing interactions between microbial metabolism and host development.
The microbiome composition can differ markedly between individuals and can fluctuate within individuals based on changes in diet, exposure to new environments, and acute or chronic changes linked to disease. As demonstrated by the Human Microbiome Project, an individual’s early breastfeeding exposure, gender, and other demographic factors play a major role in defining communities of individuals with similar microbiomes [6]. Distinct body habitats have spatially diverse and niche-specific microbiomes present [7], which can lead to diseases and uniquely impact the performance of drugs and drug delivery systems. A perturbation of the microbial community which overcomes the tolerance capabilities of the body, termed dysbiosis [8], and the expansion of specific microbial species over others, are implicated in the development of multiple diseases [2], including inflammatory bowel disease (IBDs) [9]; autoimmunity and allergic disorders [10]; Alzheimer’s [11]; depression [12]; and cancers [13,14]. The gut microbiome is instrumental to the energy metabolism and immune response in the host and perturbations in beneficial host-microbe interactions cause aberrant activation of immune cells, consequently leading to diseases and metabolic disorders that involve immunity, including ulcerative colitis and Crohn’s disease [15–18], obesity [19–21], diabetes [22,23], and non-alcoholic fatty liver disease [24], among others. Comparing the microbiomes of healthy individuals to diseased individuals shows that individuals facing acute or chronic illness have less diversity of microorganisms and are more susceptible to infection by foreign microbe populations, known as pathogens, due to the stunted capabilities of their immune systems [25–29].
Therapeutics and microbiome have a bi-directional impact on each other. In a recent drug screen [30], nearly one-fourth of more than 1,000 drugs, marketed for various conditions but not sold as antibiotics, had antibacterial effects when tested against 40 strains of human gut bacteria. In another study, 176 out of 271 drugs incubated with 76 human gut microbes from diverse clades were metabolized markedly to reduce the drug level by more than 20% [31]. By combining high-throughput genetic analyses with mass spectrometry, the study highlighted the causal links of the microbiota with interpersonal variability in microbiomes to interpersonal differences in drug metabolism [31]. These findings have implications for drug discovery, development, and deployment across a spectrum of disease conditions [31,32].
This review describes the human microbiome and its association with diseases and discusses strategies to target immune-microbiome interactions for drug discovery, screening, and development of novel therapeutics, including the modulation of microbial composition, as well as technologies for the improved study of drug-microbe-host interactions. Finally, we examine new directions for the field and discuss the implications these findings will have for medicine and global health moving forward.
2. The microbiome and disease
2.1. Human microbiome biology and pathophysiology
The human microbiota (or microbiome) consists of bacteria, archaea, viruses, and fungi which colonize and symbiotically reside at various interfaces throughout the body, and it is defined by the genomic expression of these co-residing species [33,34]. These epigenetic expressions and microorganism population densities are not only characteristic of specific organ systems but greatly vary between healthy and diseased states [35–37]. Two main organ systems whose microbiomes are heavily influenced by external factors are the respiratory tract (Fig. 1A) and the digestive tract (Fig. 1B), where the mucosal linings are constantly exposed to bacteria from the host’s surrounding environment.
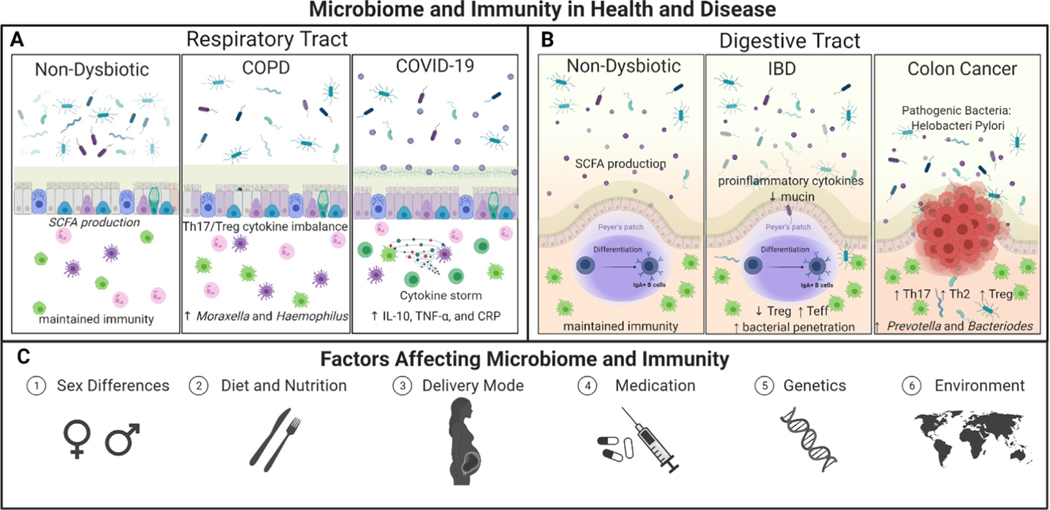
Fig. 1.
Landscape of Microbiome and Immunity. There is a dysregulation of the microbiome populations under health and diseased state conditions, especially in the A) respiratory tract [38–43] and B) the digestive tract [44–46]. C) Various factors influence the microbiome composition and the immune responses in humans, including sex, diet and nutrition, delivery mode, medications such as antibiotics, genetics, and the environment.
The complex compositions of respiratory and digestive tract microbiomes begin from birth, where initial microbial exposure and colonization stem from the rupture of amniotic membranes and passage through the birth canal [47–49]. The childbirth delivery mode plays a critical role in the establishment of the early microbiome composition and the child’s immune system, especially in the first year of development [50–52]. The delivery mode of birth, such as natural birth versus Cesarean section (C-section), and post-natal treatment affects the enrichment of certain species over others. Mainly, infants delivered by C-section do not receive their mother’s commensal genera, such as Bacteroides, which are key microbes for immune development, are replaced by opportunistic strains characteristic of the hospital’s environment [53,54]. Not only do C-sections lack positive microbes associated with neonatal brain, nervous system, and immune system development [55–57], but infants born through C-section have a higher likelihood of developing respiratory infections [52,58,59], allergies [60–64], asthma [65–67], and other metabolic, inflammatory, and immunological diseases [68–71]. This is followed by additional exposure of microbial species and bioregulation due to unique factors such as environment, breastfeeding, and nutritional choices [72–75].
The microbiome also plays a role in the link between human epigenetics and health. Epigenetics is defined as the study of phenotypic changes due to gene expression changes due to the mechanisms such as DNA methylation and histone acetylation that alter the packing of DNA and subsequently its ability to be transcribed and expressed [76]. These changes are regulated by other identified mechanisms including microbial metabolite crosstalk, diet, exposure to antibiotics, and environmental conditions (pH, oxygen, and temperature) that modulate expression [77]. One key attribute of many pathologies (including cancer, cardiac failure, chronic inflammation, and neurodegenerative diseases) is the overexpression of histone deacetylase (HDAC) which induces tighter packing of DNA on the histone protein and reduces the expression of the genetic code [78,79]. However, many gut bacteria produce HDAC inhibiting metabolites, like butyric and valeric acids, making microbiota vital in the body’s ability to counter the initiation and progression of diseases [80]. This is especially important for understanding the link between the microbiota-gut-brain axis, where disturbances in the microbiomes composition or function can lead to both gastroenterological and neurological disorders [81].
Multitudes of factors, like sex differences, diet and nutrition, delivery mode, medication usage, genetics, and environment, play a role in the development of an individuals’ microbiomes and can impact how an individual responds to drugs and therapies (Fig. 1C). There is also an observed heterogeneity between the genes of the same microbial species over time in a single individual. This is because bacteria can adapt to changing environmental conditions through a process called horizontal gene transfer (HGT), where genes encoded in the DNA or RNA of one microbe can be directly transferred to another microbe, leading to the addition or replacement of genes in the receiving microbe’s genome [82]. This can alter the relationship a bacterium has with its host or allow it to obtain a competitive advantage against other microorganisms in the environment [83,84]. Mechanisms of HGT include transformation (DNA uptake from lysed cells), transduction (DNA introduction from a phage), conjugation (DNA transfer from conjugative pili), and through outer membrane vesicles [83]. Sequencing gut bacterial genomes from different populations worldwide has revealed that HGT frequently occurs in the gut microbiome of individual persons and even more so for those in industrialized populations [85]. From a clinical perspective, HGT poses certain for drugs that interact with the microbiome extensively, such as antibiotics. The gut microbiome can adapt by acquiring mutations for antibiotic resistance genes, which can then be and then transferred to other local microbes through HGT. The collection of all intrinsic and mobile resistant genes is known as the “resistome” [86]. Studies have shown that individuals with a prevalent “resistome” are highly unresponsive to antibiotic treatment [87]. The emergence of these multidrug-resistant bacteria poses a threat for many in receiving life-saving treatments and must be key in considering treatment modalities moving forward.
The “germ theory of disease”, initially recognized in science and medicine through the work of the renowned “fathers of microbiology”, Robert Koch and Louis Pasteur, stipulates the potential of microorganisms to be disease-causing agents [88,89]. While their contributions elucidated the existence, correlation to specific diseases, and harmful health impacts of certain pathogenic microorganisms, there has been a long-standing tendency to generalize most external microorganisms as “germs”. However, in recent years there has been a paradigm shift in medicine recognizing the positive impacts of many microbial symbionts. For example, one study has shown that gardening is great for mental health not only as a recreational activity, but through an individual’s exposure to Mycobacterium vaccae, a bacterium found in the soil [90]. Mycobacterium vaccae has been shown to stimulate the host’s serotonin production, leading to a range of health benefits, especially for individuals who suffer from stress-related psychiatric diseases [91]. It has also been utilized by researchers in many therapeutic developments for stress-resilience [92], cognitive function [93], asthma [94], allergies [95], and tuberculosis [96].
In similar attempts to manipulate microbial populations through therapeutic means, supplementing prebiotics and probiotics into food have demonstrated positive impacts for gastrointestinal health. Prebiotics are defined as dietary components and chemicals that promote the growth of certain residing gut microbes over others; in contrast, the term probiotics refers to the direct delivery of bacteria cells to the host for integration into the microbiome [97]. Identification of probiotic bacteria has allowed for progress in the treatment of dysbiosis-linked diseases. For example, the Lactobacillus species have both health-promoting and protective properties against metabolic diseases [98].
Microorganisms are not only being increasingly appreciated for the dynamic roles they play in maintenance and homeostasis but are at the brink of therapeutic translation with more than 40 clinical trials seeking to obtain FDA (Food and Drug Administration) approval for targeted, therapeutic application [99]. There are various drug administration routes, often classified through the location at which the drug is administered and chosen carefully based on convenience, compliance, and the drug’s pharmacokinetics and pharmacodynamic profile [100]. With the convenience and cost-effective nature of oral administration, there is no surprise why this route is the most utilized in the field. For these drug formulations, the primary site of drug absorption is usually the small intestine, where several microbes reside, and the bioavailability of the medication is influenced by the amount of drug absorbed across the intestinal epithelium. [100] Yet, while manipulating microbiota for therapeutics may be promising in theory, the complex workings of the human body and microbiome are not mutually exclusive studies have demonstrated several confounding variables, including nutrition may impact their efficacy and long-term benefits.
2.2. Challenges surrounding microbiome therapeutics
The therapeutic manipulation of gut microbiome-associated diseases remains challenging. A ten-year assessment indicates that ~30–50% of patients with IBD do not respond to current anti-inflammatory treatments, necessitating the need for new therapies [101]. For example, the delivery of bilirubin, an immunosuppressant, a potential therapeutic biomolecule for IBD has shown protective effects in experimental colitis [102]; however, its hydrophobic nature has limited its delivery efficiency and use in clinical translation. Another major challenge in microbiome research is a limited understanding of how to address the growing global phenomenon that is antimicrobial resistance. Although several global actions have been taken to address antimicrobial resistance (AMR), such as the World Health Organization’s 2015 endorsement of the Global Action Plan (GAP) on AMR [103], it remains an area of concern for international coordinated action [104–107].
Critical work by Dr. Jeffrey Gordon has established the importance of understanding the microbiome in the lens of immunity, drug delivery, and nutritional sciences and the impact this knowledge can have on global health, particularly with malnutrition [108–110]. Severe acute malnutrition among children has been addressed in developing countries through standardized treatment protocols involving nutritional intervention [108–110]. However, there has long been a lack of understanding on how this intervention affects the gut microbiome composition, and how that correlates with therapeutic outcomes. One study conducted by Gordon and colleagues on a cohort of Bangladeshi children identified that children with a characterized immature or underdeveloped core microbiota were more likely to be malnourished, even amongst twins and triplets included for consideration of genetic and environmental factors [111]. This study established that early gut microbiome composition correlates to the development of malnourishment.
Continued work from Gordon and colleagues has applied this knowledge to the development of carefully designed nutritional formulations for modulation of the gut microbiome [108]. Their revised formulation, called microbiota-directed complimentary food 2 (MDCF-2), when compared to the standard supplementary food in a one-month trial on children between 12 and 18 months of age, was able to alter the microbiota composition of aged-matched healthy children and indicated improved health status of malnourished children as shown through plasma protein markers [108] Similarly, to use the complimentary food approach to address and modulate microbiota composition in obesity, the group generated snack prototypes using plant fibers from different sustainable sources and were able to identify fiber specific alterations in the microbiome linked to changes plasma proteomes which indicate altered physiology [112]. A limitation to this study however is that it was performed on gnotobiotic mice, meaning it still lacks much work to translate to humans. These studies have contributed largely to the field, but additional investments into these efforts are direly needed, especially for improving the nutritional status of the global community [112].
Given that the gut microbiome is highly modulated by diet, another study aimed to determine how dietary intervention with plant-based fiber and fermented foods influence both the microbiome and immune system of healthy adults [113]. After a 17-week, randomized study using multi-omics such as proteomic and metagenomic sequencing alongside immune profiling, Wastyk et al identified that the two diets influenced the gut microbiota composition and host immune system distinctly. The consumption of fermented foods decreased levels of inflammation in participants, gathered through immune profiling of signaling proteins, cytokine levels, and immune cell composition in blood. In both diets, there was a high correlation between immune cell frequency and the short-chain fatty acids (SCFAs) found in the stool, specifically with an increase in B cell frequency observed with an increase in the SCFA, butyrate. The increase in microbiome diversity observed in the healthy participants from this study also correlated to the decreases in inflammation-linked signals and activity. Finding these links between diet and the immune status of individuals can potentially be used in therapeutic guidance and development.
Historically, it has been challenging to obtain enough subjects from different ethnic and geographic backgrounds in the study of microbiome compositional differences and responses. The inclusion of broader ranges of subjects in future microbiome research studies, such as those from underdeveloped countries and minority ethnicities/races, could help to gain a deeper understanding of the human microbiome in the lens of global health disparities and epidemiology [114]. Inclusion of individuals from non-Westernized countries is especially important for microbiome science given that recent literature points to there being extensive numbers of unexplored human microbiome species mainly harbored by individuals from these areas of the world [115]. With a more complete picture of the human microbiome comes the ability for us to use this information for improved drug design and delivery and more accurate tailoring of therapeutics.
The considerable variation in the microbiota between individuals, the limited understanding of the role played by microbiome across tissues and organs, and variation in drug metabolism impose a significant challenge to the next phase of research in this area. Technological advancements have enabled the discovery of the bidirectional effects of the microbiome on pharmaceuticals and therapeutics, however obtaining a deeper understanding of the interactions between microbes, immunity, and medicine is critical for the identification of novel therapeutic targets and improvements on current treatment modalities. The dysbiosis of the microbiome in the gut has been seen to have some level of cross talk on other organs, such as the lung through a proposed gut-lung axis [116]. Like the gut, the lining of the lungs also produces mucus that protects the body from the external environment that it is exposed to during breathing. The microbiota that resides within the lung plays a role in promoting immune tolerance [117]. Given the complexities of the microbiome-immune microenvironment in the gut and lungs, it is of vital importance for advancing personalized medicine to evaluate and effectively model how the microbiota affects relevant pathophysiological systems and employ methods to target immune-microbiome interactions.
3. The microbiome and immunity
The body’s immune system orchestrates multiple regulatory functions and protective responses against pathogens in the external environment around us. These immune responses can be classified either as innate, being the first and fastest line of defense in the body, or adaptive, which constitutes the building of long-term, memory responses. The respiratory and digestive tracts are among the key mucosal immunity sites, which also have an active microbiome composition that shapes the local and systemic immune response. In fact, the microbiome can cause immunomodulation, such as an activation or suppression of immune function [118–120]. Hence, an in-depth understanding of the interplay between the immune system and microbiome is necessary for developing effective pharmaceuticals and other therapeutics.
3.1. Innate immunity in the respiratory and digestive tracts
Innate immunity in the respiratory and digestive tracts is mediated through tissue-resident and circulating immune cells, such as macrophages, Natural Killer (NK) cells, dendritic cells (DCs), and innate lymphoid cells (ILCs) [121,122]. These cells are essential in recognizing and reacting to respired allergens, pathogens, and toxins in the respiratory and digestive tracts. The respiratory tract is generally categorized into the upper respiratory tract (nose and pharynx) and the lower respiratory tract (larynx, trachea, bronchi, lungs). The most common bacterial species of the respiratory tract include Streptococcus pyogenes, Streptococcus viridians, Streptococcus pneumoniae, Corynebacterium diptheriae, Staphylococcus aureus, Haemophilus influenza, Mycobacterium tuberculosis, Bordetella pertussis, Klebsiella spp., Neisseria meningitides, Mycoplasma pneumonia, and Pseudomonas aeruginosa, among others [123]. In addition, the lungs may also harbor viruses (Adenovirus, Rhinovirus, Influenza virus, Epstein Baar virus, Measles virus, etc.) and fungal species (Aspergillus spp., Candida albicans, Candida immitis, Candida neoformans, etc.) [124]. The epithelial cells of the airway play a critical role as the first line of innate immune sensors. In performing defense functions, the airway epithelium and submucosa regulate barrier tightness, secrete mucus and antimicrobials, and communicate with immune cell subsets through the production of cytokines, chemokines, and growth factors [125]. The airway epithelium can also sense pathogens through pattern recognition receptors (PRR) such as Toll-like receptors (TLR), Retinoic acid-inducible gene I (RIG)-I-like receptors (RLR), protease-activated receptors (PAR), and Nucleotide-binding and oligomerization domain (NOD)-like receptors [125].
Innate immunity in the gut includes mechanisms of immune regulation like that observed in the lungs. However, there are some identified differences between the lung and gut microbiome in terms of bacterial compositions and mechanisms of immunity. For example, the bacteria Helicobacter pylori shapes the population of local group 2 innate lymphoid cells (ILC2), the dominant ILCs in the stomach that are dependent on the microbiota and are highly responsive to IL-7-induced proliferation, whose production is instigated by the stomach microbiota [126]. ILC2s can also help in the production of IgA by secreting IL-5, which modulates stomach microbial composition [126]. Apart from the production of proinflammatory molecules and ICL2 signaling, the cellular compartments of the innate immune system, including dendritic cells (DCs) and macrophages, also play an important role in combating pathogens during the innate immune response in the gut [127]. Here, the macrophages and DC cells can begin to process antigen and present the antigen to naïve T cells, initiating the adaptive immune response.
3.2. Adaptive immunity in the respiratory and digestive tracts
In the adaptive immune response, naive T cells undergo a series of events including activation, proliferation, and differentiation until they obtain the effector T cell phenotype that allows them to mediate the immune response against the pathogen. T cells are directly involved in the killing of the infected cells and the release of regulatory and proinflammatory mediators necessary for clearance of infection. The humoral immune response mediated by B cells makes up the other part of the adaptive immune responses, where the end goal is the production of neutralizing antibodies that allow for the specific targeting and clearance of the pathogen. Several aspects of activation of humoral immune response using drug delivery systems are reviewed elsewhere by us [128].
Adaptive immune responses in the gut and lungs often occur across the multiple mucosal surfaces present in these anatomical locations, including the upper respiratory and gastrointestinal tract. In these sites, there is often both an innate and adaptive response underplay. Inductive mucosal sites are referred to as the mucosa-associated lymphoid tissues (MALT), which include the gut-associated lymphoid tissues (GALT), Bronchus-Associated Lymphoid Tissue (BALT), and the nasopharyngeal-associated lymphoid tissue (NALT) subcategories [129]. Since these areas are susceptible to infections, the body must be prepared with an appropriate immune cell lineup to fight off potential invaders. In each of these locations, there is a unique establishment of communication and crosstalk between microbes and immune cells, and disturbances in this microbial homeostasis can alter immune responses [130,131].
Mucus, a naturally produced biofluid, contains mucins, salts, lipids, proteins, and cellular signals that vary based on anatomical location and the function of the corresponding organ system [132]. Since mucus is constantly being created, degraded, and cleared along the mucosal linings, different areas vary in thickness, viscoelasticity, and performance of the mucus with maintaining and affecting microbiota [133]. In the gut, goblet and specialized mucous cells synthesize and secrete mucus to protect the secondary lymphoid organs (SLOs) within the MALT from exposure to the external environment [132]. SLOs are areas of immune lymphocyte cell activation and can be observed in the MALT as small clusters of B cells, T cells, and dendritic cells called Peyer’s Patches. Stromal cells in Peyer’s Patches, known as fibroblastic reticular cells (FRCs), play a key role in priming the intestinal microenvironment, especially during organogenesis [134]. During the development of Peyer’s Patches, FRC lineages converge during organogenesis. These specific cell types govern intestinal immunity, including immune responses to SARS-CoV-2 antigen and other immunogens [134,135].
The MALT is covered and protected by a mixture of different specialized epithelial cells that form crypts and villi structures along the intestine [129]. Key signals and cells such as EMC3 and LGR5 + intestinal stem cells allow for maintenance and intestinal homeostasis [136,137]. Within this epithelial lining are microfold (M) cells that allow for transport of luminal antigen toward the Peyer’s Patches, allowing for immune activation, production of antibody-secreting cells and plasmablasts, and regulation of immunoglobulin A (IgA) in the gut [138,139]. Although studies have been conducted and have identified patterns among microbiota and immune dysfunction, the knowledge of the specific microbiota members and the mechanism in which they cause these effects is still limited [140]. The microbiome can imprint a class of T cells in the MALT called mucosal-associate invariant T cells (MAIT) which recognize microbial metabolites presented by MHC class I [141]. Studies by Biram et al have also identified key mechanisms which differ in the Peyer’s Patches compared to the draining lymph node and spleen [142]. They not only identified the sites in the Peyer’s Patch where B cell selection occurred and the cellular events that occurred before germinal center formation but also showed how the B cell receptor (BCR) regulated infiltration of B cells specific to the antigen in the germinal centers (B cell maturation process [128,143–145] and impeded clonal expansion in the subepithelial dome [142]. Another key finding with Peyer’s Patches includes data that suggests that Gut-associated germinal centers undergo clonal selection and antigen-driven maturation in the absence of infection or immunization that is tunable via the presence and diversity of microbiota [146].
Other key environmental factors, like dietary fiber metabolites and exposure to microbiota over time since birth, also regulate lymphoid cell responses and affect resistance to immunopathology [147,148]. Morphologically, PPs have B cell niches, a subepithelial dome (SED) made up of a mixed cell cocktail of B and T cells, stromal cells, macrophages, and dendritic cells, as well as a follicle associated epithelium (FAE) [149]. The FAE consists of specialized intestinal epithelial cells and microfold (M) cells that allow for the transport of luminal antigen and bacteria [149]. When exposed to antigen and T cell-dependent activation, the PP’s naive B cells can become activated, form a germinal center (GCs) and undergo proliferation, somatic hypermutation (SHM), and affinity-based selection before differentiation into memory B cells [150]. The process of somatic hypermutation is initiated by the presence of activation-induced cytidine deaminase (AID) [151]. Somatic hypermutation diversifies the V region of B cell genes through the introduction of point mutations that are selected for providing greater affinity for the antigen [151]. Class switch recombination, in contrast, does not affect the V region but increases the functional diversity of immunoglobulins by replacing the Cμ region in the immunoglobulin gene, which is first expressed with another heavy-chain C region to produce IgG, IgA, or IgE antibodies [151]. T cell cytokines and receptors, like CD40L, play an important role in B cell stimulation, activation, and class switching to different antibody types [151]. These processes and responses have a crucial role in host defense against pathogens whose surface antigens cannot elicit peptide-specific T-cell responses. Under conditions where a pathogen is detected by lymphoid follicles, germinal centers form and respond.
The vast complexity of B cell and T cell activation and function in response is characteristic of the microbiome’s homeostatic conditions and is dependent on the form of associated inflammation or disease state. Maintaining homeostasis is critical for an individual to remain healthy, and there are several ways in which adaptive immune cells contribute to stabilizing the microbiota or that are indicative of dysbiosis. In a positive case, the body’s detection of the receptors TLR2-, MyD88, and PI3K-from bacterial lysates all resulted in the increased proliferation of B cells producing IL-10, which both inhibits T cell activation and the release of proinflammatory cytokines by macrophages [152,153]. The resulting effect of this induction stabilized the mucosal homeostasis and relieved T cell-mediated colitis. In the case of Inflammatory Bowel Disease (IBS), the transfer of an IBS microbiota into a murine germ-free gut resulted in an increased number of T helper cells producing IL-2 (Th2) and IL-17 (Th17), which are critical for T cell growth factor and linking T cell activation to neutrophil innate immunity, respectively [154,155]. There was also a decrease in RORγt + regulatory T cells (Treg), which help maintain tolerance to disturbances in the microbiota. Thus, from the increase in inflammatory response from the Th cells and loss of tolerance correlated to the loss of Treg cells, investigators were able to better understand the pathogenesis of IBS related to the microbiota.
Understanding how the introduction of microbes induces specific immune responses is especially important for predicting how a patient may contract a disorder through exposure to specific classes of microbes, or even how to develop preventative healthcare measures to entirely avoid these pathologies. Physical triggers are especially important to consider. The induction for Th17 cells, for example, is dependent on whether microbes can adhere to the intestinal wall within the gut. A study by Atarashi et al. demonstrated how Th17 cells were induced only when microbes had the ability to adhere to the epithelial cells lining the intestinal wall, whereas adhesion-defective microbes did not yield a Th17 induction [156]. There are also studies focused on how immune cell populations change over the lifetime of a tumor. Yu et al demonstrated with murine models that during tumorgenesis a high prevalence of CD8 + IFN + T cells within a tissue that develops a tumor and the other neighboring tissues, results in a greater likelihood of developing multiple tumors in subsequent stages of tumor metastasis [157]. Despite the important findings we can gain by studying the impacts of adaptive immunity on microbiota in healthy and diseased states, it is important to remember that microbiota composition is correlated to an individual’s geographic population and can have a tremendous impact on an individual’s responses to different bacterial infections, as demonstrated by Porras et al [158].
4. Bidirectional impacts of microbiome and therapeutics
The human microbiome not only can affect host metabolic functions but also has powerful capabilities in the modulation of pharmacotherapies since microbes affect the pharmacokinetics and pharmacodynamics of drugs [159]. This unique balancing act between therapeutic drugs, the microbiome, and the targeted host has been complemented with the birth of Pharmacomicrobiomics [159]. There is a bidirectional impact of the microbiome on therapeutics and of therapeutics on the microbiome, although the mechanisms of these interactions are still not well characterized, especially when considering the role of immune cells and processes in the mix.
Several population studies have been conducted to evaluate the effect of common drugs on the microbiome and have found specific perturbations in those taking the identified drugs, either via a decrease or increase in certain taxa [160]. These bidirectional changes illustrate how commonly used drugs play a role in bacterial strains and population dynamics. Proton pump inhibitors are a class of drugs that are commonly used in the treatment of Gastroesophageal reflux disease (GERD), peptic ulcers, and Helicobacter pylori (H. pylori) infection. These drugs alter specific taxa in the gut microbiome and can lead to decreased colonization resistance and the development of enteric infections, including Clostridium Difficile infections [160,161]. A population-based study found that those using Paracetamol and opioids had a higher abundance of Streptococcaceae, which plays a role in metabolic disease and function [162]. Selective serotonin reuptake inhibitors (SSRIs), which are commonly used for depression were negatively associated with Turicibacteraceae abundance [163]. Also, inhaled anticholinergic inhaled medications have been negatively associated with Ruminococcaceae and Peptococcaceae abundance and alpha composition which suggests that non-oral drug administration might also indirectly influence the gut microbiota [162]. The question of whether non-antibiotic drugs can also have some properties that affect microbial populations remains. One major study conducted to address this was able to assess over 1000 commonly used drugs that were identified as non-antibiotics. From this study, more than 24% of the drugs inhibited the growth of at least one bacterial strain in vitro [30].
Two different modes of action have been proposed to explain how drug usage influences gut microbial composition [160]. First, that drugs can facilitate the translocation of the microbiome from other sites in the body to the gut, thereby inducing dysbiosis. Second, that drugs directly affect the growth of bacteria due to interactions with the microenvironment. This second mode points toward the decrease of certain bacteria in a similar mechanism. Aside from the impacts that the microbiome has on drug metabolism and vice versa, there is significant potential for bioengineering approaches that use microorganisms as a therapeutic. For example, one study reported the successful use of a bacteriophage to decrease mortality by the alcoholic fatty liver through selective removal of Enterococcus faecalis bacteria, which is responsible for producing cytolysins that lead to deterioration of patients with alcoholic hepatitis [164]. However, the microbiome can potentially function through a third mechanism – modulation of the immune response.
Recently, a paradigm shift is seen in associating the interactions between the microbiome and cancer immune therapeutics in the development of anti-tumor efficacy. Various studies have shown that the gut microbiome may immunomodulate antitumor innate and adaptive immune responses. Paulos et al showed, using mouse models, that microbial translocation can augment the function of adoptively transferred self/tumor-specific CD8 + T cells through toll-like receptor (TLR4) signaling [165]. Sivan et al. showed that commensal Bifidobacterium can promote antitumor response to anti–PD-L1 checkpoint inhibitors [166]. They compared murine melanoma tumors in mice harboring distinct commensal microbiota and established differences in antitumor immunity. These differences were eliminated upon cohousing the mice or after fecal transfer. Lida et al further showed that gut microbiome dysbiosis using germ-free mice or antibiotics impaired the response of subcutaneous tumors to CpG-oligonucleotide immunotherapy and platinum chemotherapy [167]. These effects were through the modulation of tumor microenvironment where disruption of microbiome resulted in a poor response from tumor-infiltrating myeloid-derived cells, lower cytokine production and tumor necrosis after CpG-oligonucleotide treatment.
However, the relevance of the microbiome in anti-cancer therapy was only recently established in humans. Gopalakrishnan, V., et al. reported that the gut microbiome modulates response to anti–programmed cell death 1 protein (PD-1) immunotherapy in melanoma patients [14], thus, looking into manners in which maintenance of healthy gut flora could help in optimizing the efficacy of cancer immunotherapy treatments. Analysis of fecal microbiome samples of patients that responded to anti-PD1 therapy versus non-responders revealed significantly higher alpha composition, the relative abundance of Ruminococcaceae bacteria, and functional differences in gut bacteria in responding patients, including enrichment of anabolic pathways. In another study, Routy et al. showed that low levels of the bacterium Akkermansia muciniphila due to antibiotic consumption was associated with poor response to anti-PD-1 inhibitors in patients with epithelial lung and kidney cancers [168].
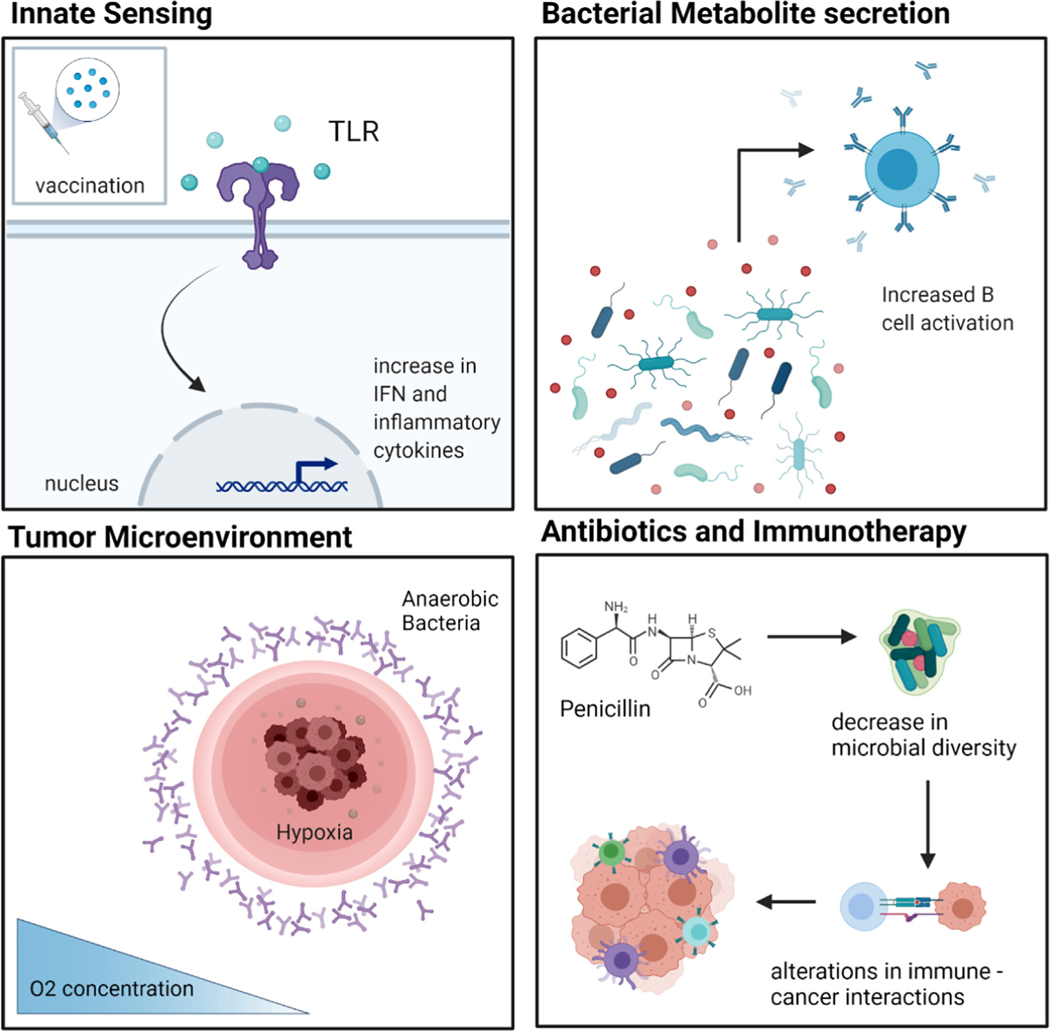
Gut microbes can also contribute to drug efficacy and safety by enzymatically transforming drug structure and altering drug bioavailability, bioactivity, or toxicity [169]. One example is the transformation of the cardiac drug digoxin into the cardioinactive metabolite, dihydrodigoxin, by Eggerthella lenta (E. lenta) residing in the gut microbiome [170]. There are a few proposed ways in which the microbiome interfaces and plays a role in cancer and vaccination, as highlighted in Fig. 2. The presence of bacteria allows for inherent innate sensing mechanisms and the metabolites secreted by the microbiota could also directly affect B cell activation and maturation in the gut. In addition, tumor microenvironments have been shown to have hypoxic conditions which could directly affect anaerobic bacteria populations to thrive in these localized tumor microenvironments (Fig. 2). Under medication treatment with antibiotics and immunotherapies, changes in microbial composition could directly affect or alter cancer cell-immune cell interactions (Fig. 2).
Fig. 2.
Microbiome immune interactions in cancer, infectious disease, and vaccination. Mechanisms by which innate sensing, metabolite secretion, tumor microenvironments and the use of antibiotics combined with immunotherapies can affect the gut microbiome, immune cell response and native tissue.
5. Microbiome and immune targeting strategies
Given the large role that the microbiome plays in immunity and vice versa, it is important to implement proper targeting strategies in the development of drug delivery systems. With more research being conducted on these key topics, including immunomodulation via microbial composition, pre and probiotic engineering, and targeting formulations, there could be a significant improvement in the treatment of disease.
5.1. Immunomodulation via microbial composition
Microbial composition and colonization play a role in pathology and disease state modulation of the immune system. Given the power that these microorganismal communities play, several strategies have been implemented to immunomodulate via control of the growth and presence of certain microbiota. Approaches such as fecal microbiota transplants (FMT) in which the microbiota from healthy donors are processed and transplanted into diseased patients have shown potential to treat Clostridioides difficile infection, IBD, cancer, colitis, and cirrhosis via immunomodulation [171]. Early-stage observations from clinical trials have shown promising results, including how this transplantation reduces gut microbial antibiotic resistance gene transfer and improves responsiveness to anti-PD1 immunotherapy in melanoma patients [172–174]. A Phase 1 clinical trial was conducted to assess the safety and feasibility of FMT followed by anti–PD-1 immunotherapy reinduction in 10 patients with anti–PD-1–refractory metastatic melanoma [172]. After the FMT and anti-PD-1 Therapy, PET-CT imaging followed outcomes on metastatic lesions visualized as black areas concentrated in the leg and groin [172]. For one patient, there was an observed increase in lesion sizes and number but after Day 67, complete resolution of all lesions was demonstrated via additional imaging [172]. Immunohistochemistry staining also succeeded in highlighting a clear increase in CD68 + cell infiltration in the gut lamina propria, an increase in the abundance of intra-tumoral CD8 + T cells, and visualization of tumor necrosis [172].
However, there are some limitations and challenges in the advancing of FMT as commonplace therapies. There is currently limited knowledge on the dose and composition of FMR necessary for effectiveness. This is likely attributed to the complexity and limitations on what is known to constitute a “healthy”, model microbiome. In addition, several studies must be undertaken to ensure reproducibility, safety, regulation, and scalability will be optimal for clinical translation and adaptation. Though FMT has shown promising and exciting results, FMT therapeutic treatment does not demonstrate itself to be the best or most sustainable option for clinical practice, especially until there are well established standard protocols and techniques.
5.2. Synthetic biology and nanotechnology approaches
Other exciting advancements include synthetic biology examples of engineered microbes and nanotechnology. Synthetic biology is used to tune a bacteria’s response cues, leading to targeted diagnostic readout [175–177]. In one case, murine-specific Escherichia coli was modified with a genetic toggle for the lacZ operon, where exposure to anhydrotetracycline (ATC) induces permanent operon expression [178]. The engineered bacteria were then placed within a murine in vivo model [178]. This study demonstrates, the utility of engineered bacteria for diagnostic activation in antibiotic resistance studies in addition to the need to validate in vitro models to in vivo responses which varied dramatically in this study and has been an ongoing concern for developing biomimetic ex vivo models. In another case, bacteria were engineered to protect commensal microbiota from dysbiosis induced by antibiotic treatment [179]. A strain of the Lactococcus lactis bacteria was engineered to degrade broad spectrum β-lactam antibiotics by programming them to produce the β–lactamase enzyme [179]. In this study, Cubillos-Ruiz et al ensured that their engineered live bacteria therapeutic was not capable of undergoing horizontal gene transfer via genetic engineering mechanisms, and when administered to mice treated with ampicillin, the group was able to show conservation of the gut microbiota while maintaining serum antibiotic levels required for treatment [179]. Bacteria have been recently engineered to therapeutically secrete protein tumor necrosis factor α (TNF-α) in response to highly controllable heat stimulation, which resulted in an inhibition of breast cancer tumor growth in mice in vivo [180]. The use of engineered bacteria for therapeutic modulation and protection of the microbiome provides new windows of opportunity for clinical translation in a broad variety of illnesses and could begin to address the spreading problem that is antibiotic resistance and dysbiosis.
In application, this is important for the development and translation of new vaccines, which face considerable challenges with the natural variation microbiota variation between patients and the poor understanding of how vaccines perform under these altered conditions [181]. Work from Pulendran et. al highlights that mice which fail to recognize flagella on gut bacteria resulted in poor germinal center formation [182]. This means that the TLR mediated sensing of flagellin plays a vital role in promoting plasma cell differentiation and the decimation of the gut microbiome in mice using antibiotics also leads to inadequate vaccine outcomes [182]. TLR5 in specific is a key player in the inflammatory response to pathogen expressing flagellum when they bypass the epithelium, the gut‘s the first layer of defense. Depletion of the microbiome in mice from antibiotic use can lead to unfavorable and ineffective outcomes in vaccination.
One clinical trial explored how antibiotic-induced perturbations in the gut microbiome affected the efficiency of an administered vaccine against H1N1 influenza [184]. Evidence points to the importance of microbiome composition with vaccination, high-lighting that lack of vaccination and exposure to flu strains critically affect neutralization and IgG/IgA binding response against H1N1 in antibiotic altered microbiome states [184]. Altered gut microbiomes also can enhance innate immunity and the expression of genes that produce key inflammatory response signals, such as transcription factor activating protein 1 (AP-1) and nuclear receptor 4A1 (NR4A1) [184]. Treatment with antibiotics may also alter seasonal influenza vaccine recipients’ blood metabolome in part via the alteration of bile and lithocholic acids, which likely also affect inflammation and vaccine responses [128]. Antibiotic treatments have potential to be used for enhancement of inflammatory signatures, dendritic cell activation and manipulation toward desired metabolic trajectories. Hence, the gut microbiome is a critical factor to be considered as we move forward in the vaccine development landscape.
Fundamental investigations into poor immune responses to vaccines have driven investigation into how one can engineer vaccines to have a more favorable immune response. By engineering a nanogel vaccine that specifically target toll like receptor 2 (TLR2), a protein on mammalian cell membranes which begins signaling of proinflammatory cytokines in response to bacterial lipopeptides, traditionally unfavorable immune responses in mice were able to be improved as well as the vaccine trafficking and germinal center response [185]. TLR5−/− mice with altered microbiomes resulted in metabolic syndrome and diminished germinal center response to the nanovaccines [185]. The nanogel vaccines also changed gut microbiome composition which suggests a feedback loop with immune responses. Chronic treatment with antibiotics in this study showed poor vaccine responses as well, likely due to interleukin-6 expression levels in the mice. A promising highlight, however, is that low immune responses were rescued by the pyridine-poly (hydroxyethyl methacrylate) (Pyr-pHEMA) nanogel vaccine [185]. This study is the first to underscore the advantages of material-based nanovaccines, which offers immunomodulation for cases of gut inflammatory diseases as well as more robust vaccine response. Hence, there is a space for materials in the context of drug delivery as materials themselves could offer immunomodulatory properties and bias immune response toward more robust clinical outcomes in patients with underlying conditions linked to gut microbiome dysfunction and dysbiosis. Nanotechnology approaches have also been widely proposed in the case of intestinal pathogens that contribute to cancer. One such microorganism is H. pylori, which has been associated with increased risk and prevalence of gut cancers. Knowing the role that these microbes play in cancer allows for smart microbe-inspired technologies and materials as well as the use of these bacteria as targets for future therapeutics [186].
5.3. Prebiotics and probiotic therapeutics
Identification of potential prebiotics and probiotics, as well as novel methods of delivery to patients, brings further insight into vaccine efficacy in relation to diet, microbiota stability, and healthy versus diseased microenvironments. Prebiotics, such as fructo-oligosaccharides and galacto-oligosaccharides, are non-digestible carbohydrates, commonly known as fiber, that are metabolized by the resident microbiota to support their health and growth [187]. Alternatively, probiotics refer to the microorganisms which may yield health benefits for the host in controlled amounts [188]. When prebiotics and probiotics are combined, these are known as Synbiotics [189]. Prebiotics may be delivered either through a patient’s diet or formulated into an orally administered supplement. Since the polysaccharide inulin has been demonstrated to increase the efficacy of immune checkpoint blocker (ICB) therapy against anti-programmed cell death protein-1 (α-PD-1), Han et al formulated a “colon retentive” oral gel which was able to support the host-resident microbiota and improve the T cell immunity of microbiota administered through ICB therapy [190]. In another recent study, Luccia et al worked to improve the immune response of malnourished patients to an oral cholera toxin vaccine through a synbiotic approach [191]. This involved transplanting the microbiota of Bangladeshi children with severe acute malnutrition (SAM) into a murine gut, then administering a specially formulated prebiotic-enhanced diet and exposure to supportive probiotics through a shared cage strategy [191]. With this strategy they demonstrated the success of combining prebiotic and probiotic therapies and their success as adjuvants for modulating the immune response, which may be extended to a variety of other vaccine targets. However, it is important to note that the current understanding of what foods can be considered prebiotics and whether they are “beneficial” is generally relative and variably defined across the field. More so, recent arguments could extend the definition of prebiotics beyond fiber-containing foods to peptides and lipids which have benefits for colon-resident microbiota [192].
Beyond simply integrating naturally beneficial probiotics into a patient’s microbiota as a means of treatment, there are a variety of cases where engineered bacteria have been successfully tailored for applications such as anti-cancer therapeutics. Engineering controllable probiotics can be promising in other applications like preventing drug-resistant bacterial infection [193] as well as in addressing colon cancer [194]. Bacteria over millennia have evolved and adapted to be excellent carriers and survivors in the body. Hence, it is of no surprise that biologists have wondered whether these bacteria can become “living machines,” hacked and reprogrammed for the benefit of human health and medical therapeutics. Chowdhury et al. have further developed this idea by developing bacteria with programmed circuitry that can hone into tumor microenvironments, produce nanobodies, and release them in a localized manner after lysis. Here, the programmed bacteria produced anti-CD47 nanobodies which blocked the anti-phagocytic CD47 receptor that is expressed across various cancer cell lines [195]. This prevents the inhibition of phagocytic cells in the tumor microenvironment while also boosting immune response and honing due to the presence of lysed bacteria, causing an overall increase in the activation and efficacy of this treatment in cancer. Further work has applied this technology to poorly immunogenic or “cold” cancers [196]. This same strategy, if well designed, could also be applied in other health applications either as a standalone therapeutic option or in combination with other pre-established treatment modalities.
Just as microbial composition can be modulated with multiple engineering strategies, the mucus along the gut and lungs also can be engineered to produce more favorable immune responses and outcomes [197]. Similarly, certain beneficial bacteria can be used as bioagents for therapeutic and diagnostic purposes. Advances in nanotechnology and materials science have allowed for the fabrication of protective coatings and new delivery mechanisms for bacteria-based therapies. This provides more control over therapeutics. For example, bacteria can be engineered to be reactivated on demand using a triggerable, pH-responsive nanocoating [198]. This allows for the bacteria to be engineered to remain inactive until after gastric emptying, which allows for improved oral availability and treatment efficacy. Applications of bacteria clinically in the past have shown low treatment efficiency, which is associated with a rapid clearance and inflammatory side effects caused by the immunogenicity of bacteria. To combat the challenge of unfavorable immunogenicity, therapeutic bacteria can be coated with biological membranes that mimic the host mammalian cells. Studies conducted by Cao et al. have shown that by membrane coating probiotic bacteria there was low inflammatory response and accumulation, slow elimination by macrophages, and unaltered bioactivity which is promising for improving treatment efficacies while avoiding side effects [199].
Nanoparticles can also be engineered to target, activate and deliver siRNA to macrophages that can as a result significantly improve their ability to combat bacterial infections [200,201]. As shown by Kim et al, a fusogenic nanoparticle construct with the macrophage targeting peptide, CRV [202], efficiently delivers siRNA payload against the Irf5 gene [200]. This porous silicon nanoparticle formulation inhibited proinflammatory signaling pathways and allowed for targeted and high payload delivery [200]. In mice, a 480% gene silencing in activated macrophages was observed in vivo as well as improved response to therapeutics in both Gram-positive and Gram-negative models of bacterial infection in the muscle and lungs [200]. Lung infections, like pneumonia and cystic fibrosis (CF), present major challenges clinically with appropriate treatment modalities. Though antibiotic drugs have been the traditional treatment for these infections, many have investigated another promising therapeutic agent: phages. They especially show potential in addressing difficult to treat bacterial infections because the mechanism of action for phage induced lysis of bacteria is separate from that of antibiotics. This feature of phage therapy indicates that the complex treatment of multidrug resistant bacteria can be addressed. Engineered polymeric microparticles have been proposed as a drug delivery vehicle for getting active phages into the deep lung via dry powder inhalation for the lysis of bacteria [203]. These microparticles, composed of PLGA, when tested showed a reduction of Pseudomonas aeruginosa bacterial infection and mortality in mice, indicating a promising and potentially clinically translatable option for addressing intractable lung infections [203]. In the case of CF, it is not well known how fungi affects bacteria in the lungs. Scedosporium and Lomentospora species of filamentous fungi that are typically found in CF lungs and present peptidorhamnomannans (PRMs) in their cell wall that affect adhesion and cell-host interactions [204]. One group tested isolated PRMs from these species of fungi and found that they possess antibacterial and anti-biofilm effects, highlighting them as a potential target and future therapeutic for treatment of CF [204]. In specific, PRM isolated from L. prolificans, S. aurantiacum, S. boydii, and S. apiospermum were found to be especially effective against B. cepacia and MRSA [204]. However, P. aeruginosa and E. coli showed resistance to all PRM, meaning that this potential targeting and treatment option could be limited in its range of application [204].
5.4. Microbiome targeted medicines
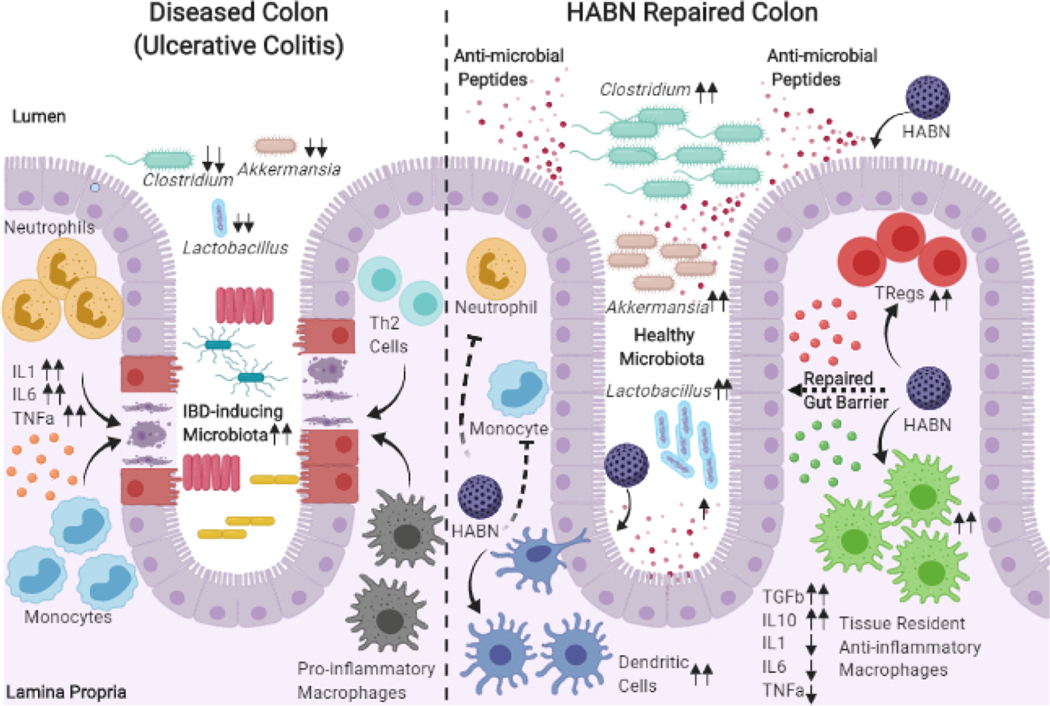
Formulation-based engineering approaches are important for the development of locally targeted treatments that promote microbial replenishment and repair. In cases like inflammatory bowel disease (IBD), there has historically been a limitation on anti-inflammatory treatment efficacy, meaning that there is a need for novel therapeutic options. One such option of interest is the immunosuppressive catabolite and antioxidant, bilirubin, which was once considered by the medical community as a futile by-product of the body [205]. Over time, this paradigm shift on bilirubin has attracted several researchers in considering this molecule in therapeutic development. A limitation in the delivery of bilirubin is its high hydrophobicity, which causes increased binding to plasma albumin and limits its clinical translatability [206]. However, researchers have taken approaches to modify and improve the delivery of bilirubin. Specifically, Lee et. al has showed that hyaluronic acid-bilirubin nanoparticles (HABNs) were able to encapsulate bilirubin aqueously despite being water-insoluble, these HABNs are successful in replenishing the gut commensal microbiome, regulating immune response and restoring the gut barrier in murine inflammatory colitis (Fig. 3) [207]. This advancement in the delivery of a previously troublesome chemical target highlights the importance of using advanced chemistries and materials engineering to improve drug delivery. Moving forward, techniques and approaches such as this can not only open doors for delivering other insoluble molecules of interest but also will likely lead toward a future where nanomedicines are personalized to specific microbial signatures in disease and personalized immune-modulating nanomedicine treatments.
Fig. 3.
HABN repair of the Colon. Visual representation of HABN approach by Lee et. al allows for delivery of insoluble bilirubin and repair of the colon and gut microbiome [207]. Adapted with permission from Singh [208].
We have previously discussed in Section 3 the variety of ways in which innate and adaptive immunity plays, as well as physical mechanisms within the microbiota, lead to healthy and diseased states. By formulating targeted drugs to promote or inhibit surface receptor interactions is a key approach to not only reducing inflammatory responses, but also in resolving the underlying disorders [209]. One example of this is Enterome Biosciences’ development of a fimbrial adhesion inhibitor treatment known as Sibofimloc, which blocks the adhesion of invasive Escherichia coli, and has demonstrated inflammation reduction in patients suffering from active Crohn’s Disease [210].
Outside of targeting microbiota, there are many companies working on drugs that manipulate the microbiota’s metabolizing capabilities to treat a variety of other disorders, especially concerning the gut-brain and gut-lung axes. Dr. Sarkis Mazmanian and his colleagues have previously demonstrated that patients with neuro-divergent and neurodegenerative conditions, such as autism spectrum disorder (ASD), schizophrenia, and Parkinson’s Disease, have different microbiota compositions compared to neurotypical controls [211,212]. Most importantly, they have highlighted how microbial metabolite products directly affect nervous system communications, the potential for drug delivery via the gut to bypass the blood–brain barrier, and have even initiated Phase 1b/2a clinical trials for an oral therapeutic designed to regulate anxiety in ASD patients [213].
Critical work is also underway in developing therapeutics to modulate the gut-lung axis, with high clinical importance following the outbreak of Sars-CoV-2 (COVID-19). With the outbreak of the Sars-CoV-2 pandemic, numerous research groups have led investigations to understand how the gut-lung axis facilitates the course of Sars-CoV-2 infection and subsequent healing, as detailed in a review by de Oliveira et al [214]. These investigations range from studies analyzing the changes in pulmonary and gut microbiota compositions, impacts on the immune system’s ability to clear Sars-CoV-2, related symptoms of the disease in both the pulmonary and digestive tracts, and the beneficial effects of probiotic administration as a means for treating this disease.
Probiotic therapies for pulmonary microbiota have also been extended beyond infectious disease applications to better treat chronic illnesses and metastatic lung cancer. Noci et al compared the responses of probiotic and antibiotic aerosolized therapies in countering immunosuppression in the lungs, which is often linked to the lungs’ high susceptibility as a secondary organ site for tumor metastasis from other organ systems [215]. Here, they demonstrated antibiotic treatment led to a decrease regulatory T cell prevalence while probiotic treatment led to maturation of antigen presenting cells (APCs) responsible for immunosurveillance, ultimately leading to the body’s ability to effectively launch an immune response against metastatic cancers infiltration to the lung. Similarly, others have used nebulized combinations of antioxidants and antibiotics for delivery in treating idiopathic pulmonary fibrosis [216].
While there is promise for therapeutic treatments that function through microbiota targeting and manipulation, there are still many challenges to overcome. These challenges include a low correlation in microbial composition changes between patients in response to these drugs and the need for more advanced biosensors and 3D culture platforms, as well as regulations and best practice policies set forth by the FDA [217]. In the next section, we will explore how these tools are being developed to address this wide array of challenges.
6. Advancements and challenges in therapeutic screening tools
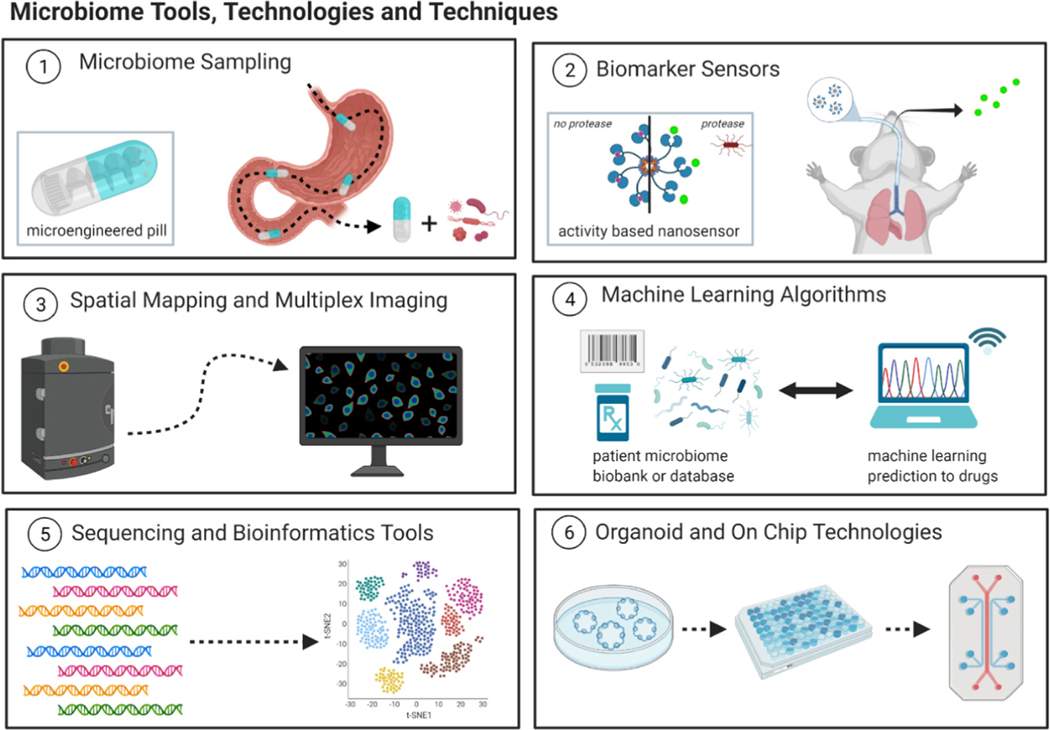
The development of quality tools for the study of microbiome has enabled a better understanding of the clinical importance of this microbiome in health. However, there are still limitations with the tools currently in use for studying the microbiome, especially in that there is not sufficient use of technological advancements for analyzing how it plays a role in therapeutic efficacy. Though there are still many challenges in tools for sampling and analyzing, we propose that certain pushes and applications of technologies could aid in addressing these issues moving forward. Certain technologies in specific have already shown much promise in addressing these limitations, such as the use of spatial mapping, machine learning, and organoid-on-chip technologies (Fig. 4).
Fig. 4.
Microbiome Tools and Technologies of the Future. Several advances in microbiome analysis tools and technologies have allowed for new potential ways of looking into the ways in which pharmaceutical drugs interact with the immune system the microbiome and overall human health.
6.1. Microbiome sampling products and biosensors
Moving forward with microbiome research and dysbiosis, it is important to be able to non-invasively sample and test the microbiome for studies. Advancements by multiple groups have proposed the use of smart ingestible materials to accomplish in vivo sampling of the gut microbiome [218,219]. Having a non-invasive, site differential platform such as this to obtain samples more easily would be beneficial for the microbiome research community and would foster more identified microbial species in pathogenesis and disease progression. Increased attempts to do just this have occurred throughout the past decade. For example, studies ranging from microelectromechanical systems (MEMS) technology approaches to develop a remote-controlled drug delivery to a microbiome sampling capsule system for the goal of diagnosing and treating gastrointestinal diseases have been explored [220–222]. Subsequent studies in the field have continued to make strides in the engineering of products to non-invasively sample the microbiome for further analysis and diagnosis, including some hydrogel materials-based approaches [223,224]. However, despite these considerable efforts there are still roadblocks in the use of microbiome sampling products, mainly related to having accurate representation of the microbial community. These roadblocks are even more apparent when it comes to sampling the microbiome of the respiratory tract, especially since there is a difference in biomass and accessibility to the lower respiratory tract for sampling [225–227]. Hence, there is an urgent need to continue refining these devices for optimal, non-invasive sampling of the microbiome across various regions of the digestive and respiratory tract.
A platform for sampling microbiota could also provide insight on a specific patient’s intestinal health and can allow for physicians to make more informed decisions on the best treatment options for each patient based on their unique microbiome signature and compare to the patterns seen data patients with similar signatures and diseases. With the recent advances and pushes toward personalized medicine, it is clear how a highly thorough, predictive technology could help the next generations of physicians and drug delivery specialists. There is also much promise in the development of infection and microbiome activity-based diagnostics moving forward. The microbiome often produces enzymes and metabolites which are essential for survival. These enzymes and markers can be used as biomarkers for clinically and pharmacologically relevant applications such as monitoring and evaluating therapeutic treatment regimens and global surveying of the microbiome [228]. One enzyme of interest is Bile salt hydrolase (BSH), which is essential for metabolite processing in the intestinal microbiome. A promising biotechnology being developed is a bioluminescent probe which monitors the activity of bile salt hydrolase, which could be beneficial in understanding how BSH modulating drugs play a role in therapeutic intervention [229]. In this study, Khodakivskyi et al demonstrate how these metabolic probes can be used to identify prebiotics that increase levels of this beneficial BSH and linked BSH activity and inflammatory bowel disease [229].
For respiratory tract biosensing, pivotal work by Chan et al has developed protease-sensitive breath biomarkers for the lung [230]. These nano sensors are activity based, meaning that exposure to extracellular proteases such as those present in respiratory disease states, cleave the conjugated peptide substrates and release volatile organic compound reporters which can be exhaled and detected via mass spectrometry [230]. This technology could potentially be used for microbiome specific detection in the lung across a variety of disease states and conditions. These and other biosensor approaches are promising for microbiome monitoring and sensing; however, they are still limitations of these biosensor approaches, including various parameters (sensitivity, ease of use, etc.) that must be refined before translation into clinical practice [231,232].
6.2. Spatial-Omics and multiplexed imaging
Mass data obtained from novel and advanced technologies in gene sequencing and high throughput analysis of the microbiome have highlighted that the communities and species involved are highly diverse. However, much remains to be done within this area. For one, there are limited tools that give insight on the way that these complex communities are organized in across the space and over time, which is clearly important for understanding the crosstalk and interactions which are occurring between microbes and the host. Strides have been taken to address this, as seen by seminal work from Shi et. al. Using high phylogenetic resolution microbiome mapping by fluorescence in situ hybridization (HiPR-FISH) technology, they were able to use binary encoding, spectral imaging and machine learning algorithms to create complex maps of microbial species [5]. This approach allowed for the unique labelling of more than a thousand different isolates and as well as insight into the spatial organization of microbes at the single-cell resolution level.
Recent work by Feng et al has also applied multiplexed imaging techniques to perform in situ imaging of both bacteria and tumor tissue using their designed prokaryotic and eukaryotic hybrid probes for in situ imaging (PEHPSI) system [233]. This study observed that there is an enrichment of gram-negative bacteria in the breast cancer tissue microenvironment and a significant correlation between these bacteria and CD8 + T cell presence in HER2 + breast cancer subtype, providing insightful information on microbiome-immune cell interactions in cancer [233]. Given that it is crucial to understand tumor microenvironment - microbiome interactions, being able to co-detect cancer subtypes alongside bacterial communities provides spatiotemporal information which can help further facilitate our understanding of the mechanisms and how microbial populations contribute to disease pathogenesis and treatment.
Moving forward, this technology can be broadly applied to not only visualize interplay between microbiota and host cells but also to evaluate drugs in context of spatiotemporal distribution and therapeutic response [234]. Antibiotic resistance continues to be a grand challenge in the field and often this innate ability for microbes to evolve and adapt against drugs can be related to horizontal transfer, the carrying over of genes across bacterial species. In specific with antibiotic resistance, these genes which are conserved and shared contribute to a “resistome” and allow for better survival when put up against antibiotic drugs [83]. Hence, it is important to build technologies and robust techniques which can follow this mechanism of horizontal transfer moving forward and perhaps design the next generation of drugs to outsmart microbes in this battle seen with antibiotic resistance. These analysis techniques can also be used to inform drug design and development and potentially even lower the costs of therapeutics currently seen in the market. Collection of microbiome data across various individuals can help identify how certain microbial species could be used in combination with already existing drug formulations. In addition, by having longitudinal studies of individual patients one can observe over time the impact of microbial composition with clinical outcomes.
6.3. Machine learning and Sequencing/Bioinformatics tools
Advanced artificial intelligence techniques such as machine learning algorithms have been applied to the development of microbiome targeting and therapeutics [235]. Machine learning is broadly defined as the process of fitting computational models to data, often ones which are highly complex and mutli-faceted [236]. This can be used for predicting outcomes based off training to a specific dataset and holds much promise when it comes to the mass information one obtains from biological –omics sequencing. Approaches to integrate these learning algorithms to the microbiome include those which aim to predict biomolecular protein–protein interactions between microbiota and the host [237]. Over the years, there has been a steady development of both reference-based prediction tools, like Mangosteen and MIMOSA, as well as machine learning prediction approaches, like Melonnpan [238,239]. These approaches have been used for predicting metabolite profiles given microbiome sequencing data. When comparing the reference based computational approach to machine learning approaches, it has been highlighted that machine learning was able to be trained to predict metabolites most accurately [238,239]. Within the broad machine learning umbrella lie more specific learning methods such as deep learning, artificial neural, and random forest non-linear models, all which have been applied to the microbiome space [236,240–246]. Although machine learning has had a significant impact on biological research and is expected to continue doing so, it also does have its limitations such as natural variance in biological data and potentially not having sufficient data for prediction or understanding complex and undefined areas of biology. However, with growing amounts of detailed data from patients with known outcomes and profiles, machine learning can potentially be used in the future to inform industry and healthcare professionals on decision making on therapeutic interventions.
The future breakthroughs in microbial macromolecules and metabolite sequencing, advanced bioinformatics software tools and development of higher resolution imaging modalities that highlight the microenvironment conditions and microbe-cell interactions will likely be of high impact and interest for microbiome research [247–249]. Microbiome related –omics data processing and bioinformatics tools provide essential examples of how these approaches can further inform and answer key scientific questions about the microbiome, therapeutics and clinical outcomes. These technological advancements will allow for faster discovery of “- omic” differences between species, environmental factors, and anatomic locations, such as immune follicles. Some advances in this understanding are currently underway. For example, complete genome sequencing of murine Peyer’s patches-derived Lactobacillus taiwanensis CLG01 strain points toward this being a potential probiotic with antibacterial and immunomodulatory systemic characteristics [250]. In addition, deeper understanding of the infamous p53 gene, which is an important coding signal in cancer, has been seen to change from tumor suppressive expression to oncogenic depending on the signals and interactions with the gut microbiome based partially on ChIP-seq data analysis and 16S rRNA sequencing [251]. Proteomics and other data analysis techniques can also help in characterization of human tissue and specifically, the use of this type of data analysis in organoid models to study different diseases [252,253].
6.4. Development of organoid modelling Technologies:
The modelling and exploration of the functionality of the human microbiome has been aided by the development and establishment of animal models [254]. A humanized animal model of the gut microbiome can be instrumental for mechanistic understanding of microbial activities as well as preclinical development of therapies. In fact, studies have clearly demonstrated that humanized gnotobiotic mouse models have contributed to the advancements in biomedicine, bridging human and animal physiology in health and disease [255]. However, inadequate understanding of the full intricacies of the gut microbiota and lack of standardized protocols in these models place major constraints on translatability of the system. Other modelling techniques, hence, can provide a better understanding and mimicry of the biological processes occurring with the microbiome and immunity.
The rise of organoids, self-organized organ-like cell aggregates that originate from multipotent stem cells, has allowed for advanced levels of biomimicry to be achieved [256,257]. This unique technology has become a promising area of biomedical research which holds potential in both research and medicine. Across the scientific community, organoids look to be the next top model for studying human biology and are a significant step forward for personalized medicine. Significant strides have been taken in understanding and developing robust intestinal and lung organoids [258–265]. Polyethylene glycol (PEG) hydrogels have been used for generating human intestinal organoids for in vivo delivery as a therapeutic [266–268]. Other gut and intestinal organoids have also been used as research tools and models for digestive tract diseases such as inflammatory bowel disease (IBD) [269–273]. Others have sought to bridge the gap of understanding intestinal microbiota function regarding luminal physiology through organoid culture and microbiota supplementation through microneedle injection and high-throughput culture [274].
Recapitulating certain biological and mechanical cues in gut and lung organoids, such as the contractile nature of the tissue, has been an area of interest in the field which has shed light on the importance of signaling pathways and stem cell differentiation [275–277]. When it comes to lung organoids, similar advancements have been seen that allowed for improved modelling of the physiological environment in vitro. For example, lung organoids and models have been used to model lung branch ex vivo morphogenesis and study pharmacokinetics of inhaled drugs [278–280]. However, lung organoids can be limited in their use for personalized medicine in that for certain cases such as in modeling lung cancer, it can become quite difficult to distinguish between tumor and healthy organoid tissues and keep track of the cell states [281].
The development of immune competent organoids has been an advancing scientific pursuit as of late but to a much more limited extent. Intestinal organoids have begun to interface with immune biology to recapitulate drug effects and overall responses in cases like with coronavirus and enterovirus infections [282–285]. Initiatives such as the Organoid Cell Atlas project focuses on characterizing organoids and in vitro cellular systems at the single-cell level to help facilitate the use and manufacturing of obtained single cell and genomics banked data for application in human organoids [286].
Biomaterials are used in the generation of organoids because of their ability to provide a carefully engineered scaffold with cellular cues for the clusters of stem-like cells to grow and begin developing on. Because of this, it is not a surprise that many advancements in materials science and manufacturing have become the foundation for smarter approaches in organoid growth and design. For example, different types of hydrogels have been engineered to support the growth of intestinal organoids and to promote favorable immune responses [287–291]. Different manufacturing advancements in biomaterials have also allowed for more control of tissue shape and spatiotemporal architecture. Technologies such as bioprinting have also allowed for complex organoids to be developed either through using cells and materials or predeveloped organoids as building blocks to creating macroscale tissues ex vivo [292].
There are several limitations of organoids in the context of microbiomes and therapeutics. One such limitation is that few organoid models are capable of directly integrating of the human gut microbiome since cells are still susceptible to bacterial infection in the dish and eventual cellular death. Headway in the gut organoid space has started to address this co-culture issue but still requires an added level of complexity and advancement to truly mimic in vivo conditions. In addition, different classifications of bacteria, such as anaerobes and aerobes, require different parameters for survival in culture in a way like microbiota in vivo. Most established organoid systems have several limitations including reproducing the level of physiological function, heterogeneity, and the ability to monitor readouts such as metabolite production to name a few [257]. However, many of these limitations can now be combatted using rising engineering technologies and approaches, which will hopefully allow for further exploration and acquisition of knowledge in this field.
6.5. Development of Organoid-on-Chip modeling technologies
Biomaterials have also affected the way in which organoids can be put onto other platforms, such as microfluidic chips, to better recapitulate environmental cues. Integrating microfluidics to develop organoid-on-a-chip platforms is advantageous in that they facilitate better nutrient disbursement, gas exchange, and recapitulate 3D tissue architecture and physiology across the body [256,293,294]. Specifically, advances with gut-on-chip microfluidic models have seen great development over the past years, with different platforms aiming to recapitulate important gastrointestinal features such as peristalsis, tissue structure and flow and its induced shear stresses, and even more recently the co-culture with bacterial species [295–298]. However, limitations continue with the ability to culture bacteria alongside these organoid platforms, especially since most are not compatible with the 0.5% luminal oxygen levels required of anaerobic bacteria [299]. Additional roadblocks with using microfluidic organ-on-chip platform continues to be that not all physiological phenomena like vascularization and presence of lymphatics are not easily integrated into the system [300]. Although there are certain limitations of organoid modelling, microfluidic integrated models have shown great potential as a strategy for both understanding disease mechanisms and screening therapeutics [298,301].
Recent breakthroughs in intestinal organoid on-chip technologies now include more control over morphogenesis and tissue architecture. Work from Nikolaev et al has shown how epithelial organoids can be engineered to self-organize into crypts and villi-like structures to develop more physiologically relevant mini-intestines [302]. The work of Dr. Nancy Albritton’s group has also achieved these architectures [303], in addition to devices analyzing factors that impact the microbiome including the mucus deposition, self-sustaining oxygen gradients, and chemical gradients [296,304]. This addition of mechanical and structural cues at the cellular level might seem minor but new findings in transcriptomics point to microfluidic conditions being more favorable for certain cell types, like epithelium, compared to static conditions [305–307].
Similarly, lung-on-chip platforms have advanced recently to model diseases and assess immune responses [308]. With nanomedicine growing in clinical translation, an area of application for these modeling platforms includes using them to monitor the transport, adsorption, and efficacy of various nanoparticles in vitro [309,310]. Most lung-on-chip platforms lack integration of microvasculature networks and the presence of interstitial fibroblasts. To address this, Mejías et al designed a microfabricated device that behaves and mimics the human lung interstitial-airway interface, which after characterization point toward the fabrication being a more pathophysiological relevant model for complex pulmonary disease such as CF [311]. Other lung-on-a-chip have also used stretchable materials like elastin and collagen to create stretchable alveoli to mimic breathing movements with more tunability with thickness, composition, and stiffness [312]. Biomimetic validation of these platforms has only recently been achieved, where these models present a noteworthy potential to model the complex biology of immune cells and microbiota cross-talk. However, when integrating the microbiome into these modeling platforms, it is important to consider how experimental conditions such as the choice of cell culture media, oxygen levels, and microbial composition might influence antibiotic, activity, and immune studies [313].
7. Future Directions:
The key impacts of microbiome research are the development of novel technologies and techniques for identifying immune-microbiome interactions and targets, especially in how these engineered constructs may translate to the formulation and delivery of clinical therapeutics. The potential impacts that this type of research can provide span across multiple areas of medicine in a personalized fashion. In addition to advancements currently being done in synthetic biology, pre/probiotic engineering, nanotechnology, organoids, microfluidics, and 3D printed technologies, a new area which may further pave the way in microbiome research includes work within modelling the human microbiome-gutbrain axis [314–318]. Findings in this field have shown that interactions between gut microbiota and cells such as enteric neurons and local lymphoid cells can regulate blood glucose and shape IgA responses, which opens more potential paths and targets for future therapies [319,320].
The importance of understanding the human microbiome echoes through society to this date, especially as we monitor and reflect upon the COVID-19 pandemic that has induced serious global health concerns. The COVID-19 causative virus, Severe Acute Respiratory Coronavirus 2 (SARS-CoV-2), has been shown to infect human gut enterocytes that express ACE2 [321–323] and alter the respiratory and gut microbiota of infected individuals [324–327]. The severity of COVID-19 is also linked with the microbiome and could be tied to disease prognosis [41,325,328]. Though some of these studies remain limited in their patient size and disease time-line, the findings highlight an interesting and under-recognized perspective on COVID-19 immunity and the microbiome. These findings hold powerful clinical implications and potential solutions, such as using short-chain fatty acids like butyrate for clinical diagnosis and precision intervention techniques and prebiotics to modulate and inhibit certain identified bacterial culprits related to worse clinical outcomes [329]. With this disease and beyond, the microbiome can be the missing piece needed to unleash a revolution in personalized medicine.
The medicine of tomorrow will be shaped by the microbiome findings of today. With these key pieces of information and the exemplary engineering and scientific advancements in immune-microbiome interactions, a new era of personalized and precision medicine will be born. One in which both medical treatment and drug design and development will be guided not just by a patient’s medical history but by their distinct microbiomes as well.
Acknowledgments
This work was supported in part by 5R01AI132738-06 and 5R21AI160136-02 grants from the National Institute of Allergy and Infectious Diseases of the US National Institutes of Health and funding by the Wellcome Leap HOPE program awarded to A. S. V.M.J gratefully acknowledges funding support from NIH grant T32-GM08433. A.N.M gratefully acknowledges that this material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-2039655. Abstract Figure and Figs. 1, 2, and 4 were created with Biorender.com.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Ursell LK, Metcalf JL, Parfrey LW, Knight R, Defining the human microbiome, Nutrition reviews 70 (2012) S38–S44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zheng D, Liwinski T, Elinav E, Interaction between microbiota and immunity in health and disease, Cell research 30 (6) (2020) 492–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Forster SC, Kumar N, Anonye BO, Almeida A, Viciani E, Stares MD, Dunn M, Mkandawire TT, Zhu A, Shao Y, Pike LJ, Louie T, Browne HP, Mitchell AL, Neville BA, Finn RD, Lawley TD, A human gut bacterial genome and culture collection for improved metagenomic analyses, Nat Biotechnol 37 (2) (2019) 186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, Brady A, Creasy HH, McCracken C, Giglio MG, McDonald D, Franzosa EA, Knight R, White O, Huttenhower C, Strains, functions and dynamics in the expanded Human Microbiome Project, Nature 550 (7674) (2017) 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Shi H, Shi Q, Grodner B, Lenz JS, Zipfel WR, Brito IL, De Vlaminck I, Highly multiplexed spatial mapping of microbial communities, Nature 588 (7839) (2020) 676–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ding T, Schloss PD, Dynamics and associations of microbial community types across the human body, Nature 509 (7500) (2014) 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]., Nature 486 (7402) (2012) 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E, Dysbiosis and the immune system, Nat Rev Immunol 17 (4) (2017) 219–232. [DOI] [PubMed] [Google Scholar]
- [9].Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham KS, Brislawn CJ, Casero D, Courtney H, Gonzalez A, Graeber TG, Hall AB, Lake K, Landers CJ, Mallick H, Plichta DR, Prasad M, Rahnavard G, Sauk J, Shungin D, Vázquez-Baeza Y, White RA, Braun J, Denson LA, Jansson JK, Knight R, Kugathasan S, McGovern DPB, Petrosino JF, Stappenbeck TS, Winter HS, Clish CB, Franzosa EA, Vlamakis H, Xavier RJ, Huttenhower C, Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases, Nature 569 (7758) (2019) 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Choi S-C, Brown J, Gong M, Ge Y, Zadeh M, Li W, Croker BP, Michailidis G, Garrett TJ, Mohamadzadeh M, Morel L, Gut microbiota dysbiosis and altered tryptophan catabolism contribute to autoimmunity in lupus-susceptible mice, Science translational medicine 12 (551) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang X, Sun G, Feng T, Zhang J, Huang X, Wang T, Xie Z, Chu X, Yang J, Wang H, Chang S, Gong Y, Ruan L, Zhang G, Yan S, Lian W, Du C, Yang D, Zhang Q, Lin F, Liu J, Zhang H, Ge C, Xiao S, Ding J, Geng M, Sodium oligomannate therapeutically remodels gut microbiota and suppresses gut bacterial amino acids-shaped neuroinflammation to inhibit Alzheimer’s disease progression, Cell research 29 (10) (2019) 787–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, Schiweck C, Kurilshikov A, Joossens M, Wijmenga C, Claes S, Van Oudenhove L, Zhernakova A, Vieira-Silva S, Raes J, The neuroactive potential of the human gut microbiota in quality of life and depression, Nature microbiology 4 (4) (2019) 623–632. [DOI] [PubMed] [Google Scholar]
- [13].Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA, The microbiome, cancer, and cancer therapy, Nature medicine 25 (3) (2019) 377–388. [DOI] [PubMed] [Google Scholar]
- [14].Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Szczepaniak Sloane R, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Marcelo Riquelme Sanchez E, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA, Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients, Science 359 (6371) (2018) 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Danese S, Fiocchi C, Ulcerative colitis N Engl J Med 365 (18) (2011) 1713–1725. [DOI] [PubMed] [Google Scholar]
- [16].Singh V, Yeoh BS, Carvalho F, Gewirtz AT, Vijay-Kumar M, Proneness of TLR5 deficient mice to develop colitis is microbiota dependent, Gut Microbes 6 (4) (2015) 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Carvalho FA et al. , Interleukin-1beta (IL-1beta) promotes susceptibility of Toll-like receptor 5 (TLR5) deficient mice to colitis, Gut 61 (3) (2012) 373–384. [DOI] [PubMed] [Google Scholar]
- [18].Vijay-Kumar M, et al. , Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest, 2007. 117(12): p. 3909–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Turnbaugh PJ, et al. , An obesity-associated gut microbiome with increased capacity for energy harvest. Nature, 2006. 444(7122): p. 1027–31. [DOI] [PubMed] [Google Scholar]
- [20].Walters WA, Xu Z, Knight R, Meta-analyses of human gut microbes associated with obesity and IBD, FEBS Lett 588 (22) (2014) 4223–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ley RE, Turnbaugh PJ, Klein S, Gordon JI, Microbial ecology: human gut microbes associated with obesity, Nature 444 (7122) (2006) 1022–1023. [DOI] [PubMed] [Google Scholar]
- [22].Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT, Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5, Science 328 (5975) (2010) 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Brugman S et al. , Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes?, Diabetologia 49 (9) (2006) 2105–2108 [DOI] [PubMed] [Google Scholar]
- [24].Dumas M-E, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, Fearnside J, Tatoud R, Blanc V, Lindon JC, Mitchell SC, Holmes E, McCarthy MI, Scott J, Gauguier D, Nicholson JK, Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice, Proc Natl Acad Sci U S A 103 (33) (2006) 12511–12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Manor O, Dai CL, Kornilov SA, Smith B, Price ND, Lovejoy JC, Gibbons SM, Magis AT, Health and disease markers correlate with gut microbiome composition across thousands of people, Nature Communications 11 (1) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Frost F, Kacprowski T, Rühlemann M, Pietzner M, Bang C, Franke A, Nauck M, Völker U, Völzke H, Dörr M, Baumbach J, Sendler M, Schulz C, Mayerle J, Weiss FU, Homuth G, Lerch MM, Long-term instability of the intestinal microbiome is associated with metabolic liver disease, low microbiota diversity, diabetes mellitus and impaired exocrine pancreatic function, Gut 70 (3) (2021) 522–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Johnson K-V-A, Gut microbiome composition and diversity are related to human personality traits, Human Microbiome Journal 15 (2020) 100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Budding A, et al. , An age dependent pharyngeal microbiota signature associated with SARS-CoV-2 infection. Available at SSRN 3582780, 2020. [Google Scholar]
- [29].Ma ZS, Testing the Anna Karenina principle in human microbiome-associated diseases. Iscience, 2020. 23(4): p. 101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, Brochado AR, Fernandez KC, Dose H, Mori H, Patil KR, Bork P, Typas A, Extensive impact of non-antibiotic drugs on human gut bacteria, Nature 555 (7698) (2018) 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL, Mapping human microbiome drug metabolism by gut bacteria and their genes, Nature 570 (7762) (2019) 462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Javdan B, Lopez JG, Chankhamjon P, Lee Y-C, Hull R, Wu Q, Wang X, Chatterjee S, Donia MS, Personalized Mapping of Drug Metabolism by the Human Gut Microbiome, Cell 181 (7) (2020) 1661–1679.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Peterson J, Garges S, Giovanni M, McInnes P, L.u. Wang JA Schloss, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M, The NIH human microbiome project, Genome research 19 (12) (2009) 2317–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Eloe-Fadrosh EA, Rasko DA, The human microbiome: from symbiosis to pathogenesis, Annual review of medicine 64 (1) (2013) 145–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hilty M, et al. , Disordered microbial communities in asthmatic airways. PloS one, 2010. 5(1): p. e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ramsheh MY, Haldar K, Esteve-Codina A, Purser LF, Richardson M, Müller-Quernheim J, Greulich T, Nowinski A, Barta I, Stendardo M, Boschetto P, Korzybski D, Prasse A, Parr DG, Hohlfeld JM, Döme B, Welte T, Heath S, Gut I, Morrissey JA, Ziegler-Heitbrock L, Barer MR, Singh D, Brightling CE, Lung microbiome composition and bronchial epithelial gene expression in patients with COPD versus healthy individuals: a bacterial 16S rRNA gene sequencing and host transcriptomic analysis. The Lancet, Microbe 2 (7) (2021) e300–e310. [DOI] [PubMed] [Google Scholar]
- [37].Sánchez-Alcoholado L, Ramos-Molina B, Otero A, Laborda-Illanes A, Ordóñez R, Medina JA, Gómez-Millán J, Queipo-Ortuño MI, The Role of the Gut Microbiome in Colorectal Cancer Development and Therapy Response, Cancers 12 (6) (2020) 1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Shilts MH, et al. , Severe COVID-19 Is Associated With an Altered Upper Respiratory Tract Microbiome. Frontiers in cellular and infection microbiology, 2022: p. 1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Paudel KR et al. , Role of lung microbiome in innate immune response associated with chronic lung diseases, Frontiers in medicine 7 (2020) 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cervilha DAB, Ito JT, Lourenço JD, Olivo CR, Saraiva-Romanholo BM, Volpini RA, Oliveira-Junior MC, Mauad T, Martins MA, Tibério IFLC, Vieira RP, Lopes FDTQS, The Th17/Treg cytokine imbalance in chronic obstructive pulmonary disease exacerbation in an animal model of cigarette smoke exposure and lipopolysaccharide challenge association, Scientific reports 9 (1) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yeoh YK, Zuo T, Lui G-Y, Zhang F, Liu Q, YL A. Li A.CK. Chung, Cheung CP, YK E. Tso KSC Fung V. Chan, Ling L, Joynt G, Hui D-C, Chow KM, Ng SSS, Li T-M, WY R. Ng T.CF. Yip, Wong G-H, Chan FKL, Wong CK, Chan PKS, Ng SC, Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19, Gut 70 (4) (2021) 698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Jing X, Xu M, Song D, Yue T, Wang Y, Zhang P, Zhong Y, Zhang M, Lam T-Y, Faria NR, De Clercq E, Li G, Association between inflammatory cytokines and anti-SARS-CoV-2 antibodies in hospitalized patients with COVID-19, Immunity & Ageing 19 (1) (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].del Valle-Mendoza J et al. , Comparison of cytokines levels among COVID-19 patients living at sea level and high altitude, BMC infectious diseases 22 (1) (2022) 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Niccolai E, Russo E, Baldi S, Ricci F, Nannini G, Pedone M, Stingo FC, Taddei A, Ringressi MN, Bechi P, Mengoni A, Fani R, Bacci G, Fagorzi C, Chiellini C, Prisco D, Ramazzotti M, Amedei A, Significant and conflicting correlation of IL-9 with Prevotella and Bacteroides in human colorectal cancer, Frontiers in immunology 11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chen L, Wang J, Gut microbiota and inflammatory bowel disease, WIREs Mechanisms of Disease (2015) e1540. [DOI] [PubMed] [Google Scholar]
- [46].Lee M, Chang EB, Inflammatory bowel diseases (IBD) and the microbiome—searching the crime scene for clues, Gastroenterology 160 (2) (2021) 524–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wampach L, Heintz-Buschart A, Fritz JV, Ramiro-Garcia J, Habier J, Herold M, Narayanasamy S, Kaysen A, Hogan AH, Bindl L, Bottu J, Halder R, Sjöqvist C, May P, Andersson AF, de Beaufort C, Wilmes P, Birth mode is associated with earliest strain-conferred gut microbiome functions and immunostimulatory potential, Nature communications 9 (1) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Korpela K, Costea P, Coelho LP, Kandels-Lewis S, Willemsen G, Boomsma DI, Segata N, Bork P, Selective maternal seeding and environment shape the human gut microbiome, Genome research 28 (4) (2018) 561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S, Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid, Scientific reports 6 (1) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Busi SB, de Nies L, Habier J, Wampach L, Fritz JV, Heintz-Buschart A, May P, Halder R, de Beaufort C, Wilmes P, Persistence of birth mode-dependent effects on gut microbiome composition, immune system stimulation and antimicrobial resistance during the first year of life, ISME Communications 1 (1) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Turroni F, Milani C, Duranti S, Lugli GA, Bernasconi S, Margolles A, Di Pierro F, van Sinderen D, Ventura M, The infant gut microbiome as a microbial organ influencing host well-being, Italian journal of pediatrics 46 (1) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Reyman M et al. , Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life, Nature communications 10 (1) (2019) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, Bokulich NA, Song SJ, Hoashi M, Rivera-Vinas JI, Mendez K, Knight R, Clemente JC, Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer, Nature medicine 22 (3) (2016) 250–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shao Y, Forster SC, Tsaliki E, Vervier K, Strang A, Simpson N, Kumar N, Stares MD, Rodger A, Brocklehurst P, Field N, Lawley TD, Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth, Nature 574 (7776) (2019) 117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Nunez N, Réot L, Menu E, Neonatal immune system ontogeny: the role of maternal microbiota and associated factors. How might the non-human primate model enlighten the path?, Vaccines 9 (6) (2021) 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gerhardt S, Mohajeri MH, Changes of colonic bacterial composition in Parkinson’s disease and other neurodegenerative diseases, Nutrients 10 (6) (2018) 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lu J, Claud EC, Connection between gut microbiome and brain development in preterm infants, Developmental psychobiology 61 (5) (2019) 739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Baumfeld Y, Walfisch A, Wainstock T, Segal I, Sergienko R, Landau D, Sheiner E, Elective cesarean delivery at term and the long-term risk for respiratory morbidity of the offspring, European journal of pediatrics 177 (11) (2018) 1653–1659. [DOI] [PubMed] [Google Scholar]
- [59].de Steenhuijsen Piters WAA, Binkowska J, Bogaert D, Early Life Microbiota and Respiratory Tract Infections, Cell Host & Microbe 28 (2) (2020) 223–232. [DOI] [PubMed] [Google Scholar]
- [60].Zimmermann P et al. , Association between the intestinal microbiota and allergic sensitization, eczema, and asthma: a systematic review, Journal of Allergy and Clinical Immunology 143 (2) (2019) 467–485. [DOI] [PubMed] [Google Scholar]
- [61].Galazzo G et al. , Development of the microbiota and associations with birth mode, diet, and atopic disorders in a longitudinal analysis of stool samples, collected from infancy through early childhood, Gastroenterology 158 (6) (2020) 1584–1596. [DOI] [PubMed] [Google Scholar]
- [62].Stokholm J et al. , Maturation of the gut microbiome and risk of asthma in childhood, Nature Communications 9 (1) (2018) 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chua H-H et al. , Intestinal dysbiosis featuring abundance of Ruminococcus gnavus associates with allergic diseases in infants, Gastroenterology 154 (1) (2018) 154–167. [DOI] [PubMed] [Google Scholar]
- [64].Renz H, Skevaki C, Early life microbial exposures and allergy risks: opportunities for prevention, Nature Reviews Immunology (2020) 1–15. [DOI] [PubMed] [Google Scholar]
- [65].Stokholm J et al. , Delivery mode and gut microbial changes correlate with an increased risk of childhood asthma, Science translational medicine 12 (569) (2020). [DOI] [PubMed] [Google Scholar]
- [66].Sbihi H et al. , Thinking bigger: How early-life environmental exposures shape the gut microbiome and influence the development of asthma and allergic disease, Allergy 74 (11) (2019) 2103–2115. [DOI] [PubMed] [Google Scholar]
- [67].Ronan V, Yeasin R, Claud EC, Childhood Development and the Microbiome—The Intestinal Microbiota in Maintenance of Health and Development of Disease During Childhood Development, Gastroenterology 160 (2) (2021) 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Andersen V et al. , Caesarean Delivery and Risk of Chronic Inflammatory Diseases (Inflammatory Bowel Disease, Rheumatoid Arthritis, Coeliac Disease, and Diabetes Mellitus): A Population Based Registry Study of 2,699,479 Births in Denmark During 1973–2016, Clinical epidemiology 12 (2020) 287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Murata C et al. , Delivery mode-associated gut microbiota in the first 3 months of life in a country with high obesity rates: A descriptive study, Medicine 99 (40) (2020), e22442 e22442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ardic C et al. , Caesarean delivery increases the risk of overweight or obesity in 2-year-old children, Journal of Obstetrics and Gynaecology 41 (3) (2021) 374–379. [DOI] [PubMed] [Google Scholar]
- [71].Lamont RF, Møller Luef B, and Stener Jørgensen J, Childhood inflammatory and metabolic disease following exposure to antibiotics in pregnancy, antenatally, intrapartum and neonatally. F1000Research, 2020. 9: p. F1000 Faculty Rev-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Pannaraj PS et al. , Association between breast milk bacterial communities and establishment and development of the infant gut microbiome, JAMA pediatrics 171 (7) (2017) 647–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].LeMay-Nedjelski L et al. , Examining the relationship between maternal body size, gestational glucose tolerance status, mode of delivery and ethnicity on human milk microbiota at three months post-partum, BMC Microbiology 20 (1) (2020) 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Roggero P et al. , Analysis of immune, microbiota and metabolome maturation in infants in a clinical trial of Lactobacillus paracasei CBA L74-fermented formula, Nature Communications 11 (1) (2020) 2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Brushett S et al. , The Effects of Urbanization on the Infant Gut Microbiota and Health Outcomes. Frontiers, Pediatrics 8 (408) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Allis CD, Jenuwein T, The molecular hallmarks of epigenetic control, Nature Reviews Genetics 17 (8) (2016) 487–500. [DOI] [PubMed] [Google Scholar]
- [77].Romano KA, Rey FE, Is maternal microbial metabolism an early-life determinant of health?, Lab Animal 47 (9) (2018) 239–243 [DOI] [PubMed] [Google Scholar]
- [78].Gallinari P et al. , HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics, Cell research 17 (3) (2007) 195–211. [DOI] [PubMed] [Google Scholar]
- [79].Li G, Tian Y, Zhu W-G, The roles of histone deacetylases and their inhibitors in cancer therapy, Frontiers in Cell and Developmental Biology 8 (2020) 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Yuille S et al. , Human gut bacteria as potent class I histone deacetylase inhibitors in vitro through production of butyric acid and valeric acid, PLoS One 13 (7) (2018) e0201073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Louwies T et al. , The microbiota-gut-brain axis: an emerging role for the epigenome, Experimental Biology and Medicine 245 (2) (2020) 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Keeling PJ, Palmer JD, Horizontal gene transfer in eukaryotic evolution, Nature Reviews Genetics 9 (8) (2008) 605–618. [DOI] [PubMed] [Google Scholar]
- [83].Brito IL, Examining horizontal gene transfer in microbial communities, Nature Reviews Microbiology (2021) 1–12. [DOI] [PubMed] [Google Scholar]
- [84].Lerner A, Matthias T, Aminov R, Potential effects of horizontal gene exchange in the human gut, Frontiers in immunology 8 (2017) 1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Groussin M et al. , Elevated rates of horizontal gene transfer in the industrialized human microbiome, Cell 184 (8) (2021) 2053–2067.e18. [DOI] [PubMed] [Google Scholar]
- [86].Singh S, Verma N, Taneja N, The human gut resistome: Current concepts & future prospects, The Indian journal of medical research 150 (4) (2019) 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].McInnes RS et al. , Horizontal transfer of antibiotic resistance genes in the human gut microbiome, Current Opinion in Microbiology 53 (2020) 35–43. [DOI] [PubMed] [Google Scholar]
- [88].National Research Council (US) Committee to Update Science, M., and Animals., A Theory of Germs. . Science, Medicine, and Animals. 2004, Washington (DC): National Academies Press (US). [Google Scholar]
- [89].Rickles D, In Handbook of the Philosophy of Science, Philosophy of Medicine. 2011: North-Holland. [Google Scholar]
- [90].Lowry CA et al. , Identification of an immune-responsive mesolimbocortical serotonergic system: potential role in regulation of emotional behavior, Neuroscience 146 (2) (2007) 756–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Amoroso M et al. , Rapidly Growing Mycobacterium Species: The Long and Winding Road from Tuberculosis Vaccines to Potent Stress-Resilience Agents, International Journal of Molecular Sciences 22 (23) (2021) 12938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Loupy KM et al. , Comparing the effects of two different strains of mycobacteria, Mycobacterium vaccae NCTC 11659 and M. vaccae ATCC 15483, on stress-resilient behaviors and lipid-immune signaling in rats, Brain, behavior, and immunity 91 (2021) 212–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Matthews DM, Jenks SM, Ingestion of Mycobacterium vaccae decreases anxiety-related behavior and improves learning in mice, Behavioural processes 96 (2013) 27–35. [DOI] [PubMed] [Google Scholar]
- [94].Xiao H et al. , Effects of Mycobacterium vaccae Aerosol Inhalation on Airway Inflammation in Asthma Mouse Model, Journal of Aerosol Medicine and Pulmonary Drug Delivery 34 (6) (2021) 374–382. [DOI] [PubMed] [Google Scholar]
- [95].Strygin A et al. , Mycobacterium vaccae lysate induces anti-allergic immune response in vitro, Bulletin of Experimental Biology and Medicine 170 (2) (2020) 226–229. [DOI] [PubMed] [Google Scholar]
- [96].Xu L et al. , Immunotherapeutical potential of Mycobacterium vaccae on M. tuberculosis infection in mice, Cellular & Molecular Immunology 6 (1) (2009) 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Green M, Arora K, Prakash S, Microbial medicine: Prebiotic and probiotic functional foods to target obesity and metabolic syndrome, International journal of molecular sciences 21 (8) (2020) 2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Azad MAK et al. , Probiotic Species in the Modulation of Gut Microbiota: An Overview, Biomed Res Int 2018 (2018) 9478630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Jimenez M, Langer R, Traverso G, Microbial therapeutics: new opportunities for drug delivery, The Journal of experimental medicine 216 (5) (2019) 1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kim J, and De Jesus O, Medication Routes of Administration, 2021. [PubMed] [Google Scholar]
- [101].Neurath MF, Current and emerging therapeutic targets for IBD, Nat Rev Gastroenterol Hepatol 14 (5) (2017) 269–278. [DOI] [PubMed] [Google Scholar]
- [102].Longhi MS et al. , Bilirubin suppresses Th17 immunity in colitis by upregulating CD39, JCI insight 2 (9) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Munkholm L, Rubin O, The global governance of antimicrobial resistance: a cross-country study of alignment between the global action plan and national action plans, Globalization and health 16 (1) (2020) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Global action plan on antimicrobial resistance, WHO Report; (2015). [Google Scholar]
- [105].W.H. Organization, Monitoring global progress on antimicrobial resistance: tripartite AMR country self-assessment survey (TrACSS), 2019–2020: global analysis report, 2021. [Google Scholar]
- [106].Cassini A et al. , Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis, The Lancet infectious diseases 19 (1) (2019) 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Stewardson AJ et al. , Effect of carbapenem resistance on outcomes of bloodstream infection caused by Enterobacteriaceae in low-income and middle-income countries (PANORAMA): a multinational prospective cohort study, The Lancet Infectious Diseases 19 (6) (2019) 601–610. [DOI] [PubMed] [Google Scholar]
- [108].Chen RY et al. , A microbiota-directed food intervention for undernourished children, New England Journal of Medicine 384 (16) (2021) 1517–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chen RY et al. , Duodenal microbiota in stunted undernourished children with enteropathy, New England Journal of Medicine 383 (4) (2020) 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Gehrig JL et al. , Effects of microbiota-directed foods in gnotobiotic animals and undernourished children, Science 365 (6449) (2019), p. eaau4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Subramanian S et al. , Persistent gut microbiota immaturity in malnourished Bangladeshi children, Nature 510 (7505) (2014) 417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Delannoy-Bruno O et al. , Evaluating microbiome-directed fibre snacks in gnotobiotic mice and humans, Nature 595 (7865) (2021) 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Wastyk HC et al. , Gut-microbiota-targeted diets modulate human immune status, Cell 184 (16) (2021), pp. 4137–4153. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Porras AM, Brito IL, The internationalization of human microbiome research, Current opinion in microbiology 50 (2019) 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Pasolli E et al. , Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle, Cell 176 (3) (2019), pp. 649–662. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Sencio V, Machado MG, Trottein F, The lung–gut axis during viral respiratory infections: The impact of gut dysbiosis on secondary disease outcomes, Mucosal Immunology (2021) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Sommariva M et al. , The lung microbiota: role in maintaining pulmonary immune homeostasis and its implications in cancer development and therapy, Cellular and Molecular Life Sciences 77 (14) (2020) 2739–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Belkaid Y, Hand TW, Role of the microbiota in immunity and inflammation, Cell 157 (1) (2014) 121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Wu HJ, Wu E, The role of gut microbiota in immune homeostasis and autoimmunity, Gut Microbes 3 (1) (2012) 4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Cao Y et al. , Critical Role of Intestinal Microbiota in ATF3-Mediated Gut Immune Homeostasis, J Immunol 205 (3) (2020) 842–852. [DOI] [PubMed] [Google Scholar]
- [121].Ardain A, Marakalala MJ, Leslie A, Tissue-resident innate immunity in the lung, Immunology 159 (3) (2020) 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Thaiss CA et al. , The microbiome and innate immunity, Nature 535 (7610) (2016) 65–74. [DOI] [PubMed] [Google Scholar]
- [123].Kelly BJ et al. , Composition and dynamics of the respiratory tract microbiome in intubated patients, Microbiome 4 (1) (2016) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Dickson RP et al. , The microbiome and the respiratory tract, Annual review of physiology 78 (2016) 481–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Hartl D et al. , Innate immunity of the lung: from basic mechanisms to translational medicine, Journal of innate immunity 10 (5–6) (2018) 487–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Ohno H, Satoh-Takayama N, Stomach microbiota, Helicobacter pylori, and group 2 innate lymphoid cells, Experimental & Molecular Medicine 52 (9) (2020) 1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Wang C et al. , The Impact of SARS-CoV-2 on the Human Immune System and Microbiome, Infectious Microbes & Diseases 3 (1) (2021) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Singh A, Eliciting B cell immunity against infectious diseases using nanovaccines, Nat Nanotechnol 16 (1) (2021) 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].McGhee JR, Fujihashi K, Inside the mucosal immune system, PLoS Biol 10 (9) (2012) e1001397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Neurath MF, Targeting immune cell circuits and trafficking in inflammatory bowel disease, Nature Immunology 20 (8) (2019) 970–979. [DOI] [PubMed] [Google Scholar]
- [131].Caruso R, Lo BC, Núñez G, Host–microbiota interactions in inflammatory bowel disease, Nature Reviews Immunology 20 (7) (2020) 411–426. [DOI] [PubMed] [Google Scholar]
- [132].Bansil R, Turner BS, The biology of mucus: Composition, synthesis and organization, Advanced drug delivery reviews 124 (2018) 3–15. [DOI] [PubMed] [Google Scholar]
- [133].Wang BX, Wu CM, Ribbeck K, Home, sweet home: how mucus accommodates our microbiota, The FEBS Journal 288 (6) (2021) 1789–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Prados A et al. , Fibroblastic reticular cell lineage convergence in Peyer’s patches governs intestinal immunity, Nature Immunology 22 (4) (2021) 510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Komban RJ et al. , Activated Peyer0 s patch B cells sample antigen directly from M cells in the subepithelial dome, Nature Communications 10 (1) (2019) 2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Huang M et al. , Emc3 maintains intestinal homeostasis by preserving secretory lineages, Mucosal Immunology (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Tan SH et al. , A constant pool of Lgr5+ intestinal stem cells is required for intestinal homeostasis, Cell Reports 34 (4) (2021) 108633. [DOI] [PubMed] [Google Scholar]
- [138].Kobayashi N et al. , The roles of Peyer’s patches and Microfold cells in the gut immune system: relevance to autoimmune diseases, Frontiers in immunology 10 (2019) 2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Beller A et al. , Specific microbiota enhances intestinal IgA levels by inducing TGF-b in T follicular helper cells of Peyer’s patches in mice, European journal of immunology 50 (6) (2020) 783–794. [DOI] [PubMed] [Google Scholar]
- [140].Ahern PP, Maloy KJ, Understanding immune–microbiota interactions in the intestine, Immunology 159 (1) (2020) 4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Constantinides MG et al. , MAIT cells are imprinted by the microbiota in early life and promote tissue repair, Science 366 (6464) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Biram A et al. , BCR affinity differentially regulates colonization of the subepithelial dome and infiltration into germinal centers within Peyer’s patches, Nature Immunology 20 (4) (2019) 482–492. [DOI] [PubMed] [Google Scholar]
- [143].Victora GD, Wilson PC, Germinal center selection and the antibody response to influenza, Cell 163 (3) (2015) 545–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Meyer-Hermann M et al. , A theory of germinal center B cell selection, division, and exit, Cell Rep 2 (1) (2012) 162–174. [DOI] [PubMed] [Google Scholar]
- [145].Mesin L, Ersching J, Victora GD, Germinal Center B Cell Dynamics, Immunity 45 (3) (2016) 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Nowosad CR et al. , Tunable dynamics of B cell selection in gut germinal centres, Nature 588 (7837) (2020) 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Al Nabhani Z et al. , A weaning reaction to microbiota is required for resistance to immunopathologies in the adult, Immunity 50 (5) (2019), pp. 1276–1288. e5. [DOI] [PubMed] [Google Scholar]
- [148].Sepahi A et al. , Dietary fiber metabolites regulate innate lymphoid cell responses, Mucosal immunology 14 (2) (2021) 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Bar-Ephraim YE, Kretzschmar K, Clevers H, Organoids in immunological research, Nature Reviews Immunology 20 (5) (2020) 279–293. [DOI] [PubMed] [Google Scholar]
- [150].Lycke NY, Bemark M, The regulation of gut mucosal IgA B-cell responses: recent developments, Mucosal Immunology 10 (6) (2017) 1361–1374. [DOI] [PubMed] [Google Scholar]
- [151].Murphy K and Weaver C, Janeway’s immunobiology. 2016: Garland science. [Google Scholar]
- [152].Mishima Y et al. , Microbiota maintain colonic homeostasis by activating TLR2/MyD88/PI3K signaling in IL-10–producing regulatory B cells, The Journal of clinical investigation 129 (9) (2019) 3702–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Couper KN, Blount DG, Riley EM, IL-10: the master regulator of immunity to infection, The Journal of Immunology 180 (9) (2008) 5771–5777. [DOI] [PubMed] [Google Scholar]
- [154].Zenobia C, Hajishengallis G, Basic biology and role of interleukin-17 in immunity and inflammation, Periodontology 69 (1) (2000, 2015,) 142–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Britton GJ et al. , Microbiotas from humans with inflammatory bowel disease alter the balance of gut Th17 and RORct+ regulatory T cells and exacerbate colitis in mice, Immunity 50 (1) (2019), pp. 212–224. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Atarashi K et al. , Th17 cell induction by adhesion of microbes to intestinal epithelial cells, Cell 163 (2) (2015) 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Amy IY et al. , Gut microbiota modulate CD8 T cell responses to influence colitis-associated tumorigenesis, Cell reports 31 (1) (2020) 107471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Porras AM et al. , Geographic differences in gut microbiota composition impact susceptibility to enteric infection, Cell Reports 36 (4) (2021) 109457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Aziz RK et al. , Translating pharmacomicrobiomics: Three actionable challenges/prospects in 2020. Omics: a journal of integrative biology 24 (2) (2020) 60–61. [DOI] [PubMed] [Google Scholar]
- [160].Weersma RK, Zhernakova A, Fu J, Interaction between drugs and the gut microbiome, Gut 69 (8) (2020) 1510–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Freedberg DE et al. , Proton pump inhibitors alter specific taxa in the human gastrointestinal microbiome: a crossover trial, Gastroenterology 149 (4) (2015), pp. 883–885. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Jackson MA et al. , Gut microbiota associations with common diseases and prescription medications in a population-based cohort, Nature communications 9 (1) (2018) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Zheng P et al. , Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism, Molecular psychiatry 21 (6) (2016) 786–796. [DOI] [PubMed] [Google Scholar]
- [164].Duan Y et al. , Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease, Nature 575 (7783) (2019) 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Paulos CM et al. , Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling, J Clin Invest 117 (8) (2007) 2197–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Sivan A et al. , Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy, Science 350 (6264) (2015) 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Iida N et al. , Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment, Science 342 (6161) (2013) 967–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Routy B et al. , Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors, Science 359 (6371) (2018) 91–97. [DOI] [PubMed] [Google Scholar]
- [169].Koppel N, Rekdal VM, Balskus EP, Chemical transformation of xenobiotics by the human gut microbiota, Science 356 (6344) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Haiser HJ et al. , Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics, Gut microbes 5 (2) (2014) 233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].D. Goldenberg S and Merrick B, The role of faecal microbiota transplantation: looking beyond Clostridioides difficile infection. Therapeutic advances in infectious disease, 2021. 8: p. 2049936120981526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Baruch EN et al. , Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients, Science 371 (6529) (2021) 602–609. [DOI] [PubMed] [Google Scholar]
- [173].Davar D et al. , Fecal microbiota transplant overcomes resistance to anti–PD-1 therapy in melanoma patients, Science 371 (6529) (2021) 595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Bajaj JS et al. , Fecal Microbiota Transplant in Cirrhosis Reduces Gut Microbial Antibiotic Resistance Genes: Analysis of Two Trials, Hepatology communications 5 (2) (2021) 258–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Manina G, Dhar N, McKinney JD, Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms, Cell host & microbe 17 (1) (2015) 32–46. [DOI] [PubMed] [Google Scholar]
- [176].Claudi B et al. , Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy, Cell 158 (4) (2014) 722–733. [DOI] [PubMed] [Google Scholar]
- [177].Helaine S et al. , Internalization of Salmonella by macrophages induces formation of nonreplicating persisters, Science 343 (6167) (2014) 204–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Certain LK et al. , Using engineered bacteria to characterize infection dynamics and antibiotic effects in vivo, Cell Host & Microbe 22 (3) (2017), pp. 263–268. e4. [DOI] [PubMed] [Google Scholar]
- [179].Cubillos-Ruiz A et al. , An engineered live biotherapeutic for the prevention of antibiotic-induced dysbiosis. Nature, Biomedical Engineering (2022). [DOI] [PubMed] [Google Scholar]
- [180].Li L et al. , Precise Thermal Regulation of Engineered Bacteria Secretion for Breast Cancer Treatment In Vivo, ACS Synthetic Biology 11 (3) (2022) 1167–1177. [DOI] [PubMed] [Google Scholar]
- [181].Pulendran B, Immunology taught by vaccines, Science 366 (6469) (2019) 1074–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Oh JZ et al. , TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination, Immunity 41 (3) (2014) 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].reference
- [184].Hagan T et al. , Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans, Cell 178 (6) (2019) 1313, 1328 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Mosquera MJ et al. , Immunomodulatory nanogels overcome restricted immunity in a murine model of gut microbiome–mediated metabolic syndrome, Science advances 5 (3) (2019), p. eaav9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Song W, Anselmo AC, Huang L, Nanotechnology intervention of the microbiome for cancer therapy, Nature nanotechnology 14 (12) (2019) 1093–1103. [DOI] [PubMed] [Google Scholar]
- [187].Davani-Davari D et al. , Prebiotics: definition, types, sources, mechanisms, and clinical applications, Foods 8 (3) (2019) 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Fijan S, Microorganisms with claimed probiotic properties: an overview of recent literature, International journal of environmental research and public health 11 (5) (2014) 4745–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Probiotics: What You Need To Know. 2019 August 2019. [Google Scholar]
- [190].Han K et al. , Generation of systemic antitumour immunity via the in situ modulation of the gut microbiome by an orally administered inulin gel, Nature biomedical engineering 5 (11) (2021) 1377–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Di Luccia B et al. , Combined prebiotic and microbial intervention improves oral cholera vaccination responses in a mouse model of childhood undernutrition, Cell host & microbe 27 (6) (2020), pp. 899–908. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Katsnelson A, Core Concept: Prebiotics gain prominence but remain poorly defined, Proceedings of the National Academy of Sciences 113 (50) (2016) 14168–14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Tan L et al. , Engineered probiotics biofilm enhances osseointegration via immunoregulation and anti-infection. Science, Advances 6 (46) (2020), p. eaba5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Zheng DW et al. , Prebiotics-Encapsulated Probiotic Spores Regulate Gut Microbiota and Suppress Colon Cancer, Advanced Materials 32 (45) (2020) 2004529. [DOI] [PubMed] [Google Scholar]
- [195].Chowdhury S et al. , Programmable bacteria induce durable tumor regression and systemic antitumor immunity, Nature medicine 25 (7) (2019) 1057–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Gurbatri CR et al. , Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies, Science translational medicine 12 (530) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Werlang C, Cárcarmo-Oyarce G, Ribbeck K, Engineering mucus to study and influence the microbiome, Nature Reviews Materials 4 (2) (2019) 134–145. [Google Scholar]
- [198].Feng P et al. , On-Demand Bacterial Reactivation by Restraining within a Triggerable Nanocoating, Advanced Materials 32 (34) (2020) 2002406. [DOI] [PubMed] [Google Scholar]
- [199].Cao Z et al. , Camouflaging bacteria by wrapping with cell membranes, Nature Communications 10 (1) (2019) 3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Kim B et al. , Fusogenic porous silicon nanoparticles as a broad-spectrum immunotherapy against bacterial infections, Nanoscale horizons (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Kim B et al. , Immunogene therapy with fusogenic nanoparticles modulates macrophage response to Staphylococcus aureus, Nature communications 9 (1) (2018) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Tang T et al. , Tumor-specific macrophage targeting through recognition of retinoid X receptor beta, Journal of Controlled Release 301 (2019) 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Agarwal R et al. , Inhaled bacteriophage-loaded polymeric microparticles ameliorate acute lung infections, Nature biomedical engineering 2 (11) (2018) 841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].de Oliveira EB et al. , Peptidorhamnomannans from Scedosporium and Lomentospora species display microbicidal activity against bacteria commonly present in Cystic Fibrosis patients. Frontiers in Cellular and Infection, Microbiology 10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Vítek L, Bilirubin as a signaling molecule, Medicinal research reviews 40 (4) (2020) 1335–1351. [DOI] [PubMed] [Google Scholar]
- [206].Čvorović J, Passamonti S, Membrane transporters for bilirubin and its conjugates: A systematic review, Frontiers in pharmacology 8 (2017) 887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Lee Y et al. , Hyaluronic acid–bilirubin nanomedicine for targeted modulation of dysregulated intestinal barrier, microbiome and immune responses in colitis, Nature Materials 19 (1) (2020) 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Singh A, Materials modulate immunity and gut microbiome, Nature Materials 19 (1) (2020) 3–4. [DOI] [PubMed] [Google Scholar]
- [209].Cully M, Microbiome therapeutics go small molecule, Nature Reviews Drug Discovery 18 (8) (2019) 569–573. [DOI] [PubMed] [Google Scholar]
- [210].Reinisch W et al. , A SAFETY, PHARMACOKINETIC AND PHARMACODYNAMIC STUDY OF SIBOFIMLOC, A NOVEL FIMH BLOCKER IN PATIENTS WITH ACTIVE CROHN’S DISEASE, Journal of Gastroenterology and Hepatology (2022). [DOI] [PubMed] [Google Scholar]
- [211].Morais LH, Schreiber HL, Mazmanian SK, The gut microbiota–brain axis in behaviour and brain disorders, Nature Reviews Microbiology 19 (4) (2021) 241–255. [DOI] [PubMed] [Google Scholar]
- [212].Needham BD et al. , Plasma and fecal metabolite profiles in autism spectrum disorder, Biological Psychiatry 89 (5) (2021) 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Stewart Campbell A et al. , Safety and target engagement of an oral small-molecule sequestrant in adolescents with autism spectrum disorder: an open-label phase 1b/2a trial, Nature Medicine (2022) 1–7. [DOI] [PubMed] [Google Scholar]
- [214].de Oliveira GLV et al. , Microbiota modulation of the gut-lung axis in COVID-19, Frontiers in Immunology (2021) 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Le Noci V et al. , Modulation of pulmonary microbiota by antibiotic or probiotic aerosol therapy: a strategy to promote immunosurveillance against lung metastases, Cell reports 24 (13) (2018) 3528–3538. [DOI] [PubMed] [Google Scholar]
- [216].Varone F et al. , Evaluation of the lung microbiome as a therapeutic target in the management of idiopathic pulmonary fibrosis: role of antioxidant/antibiotic combination therapy, Eur Rev Med Pharmacol Sci 23 (14) (2019) 6379–6386. [DOI] [PubMed] [Google Scholar]
- [217].Mimee M, Citorik RJ, Lu TK, Microbiome therapeutics—advances and challenges, Advanced drug delivery reviews 105 (2016) 44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Waimin JF et al. , Smart capsule for non-invasive sampling and studying of the gastrointestinal microbiome, RSC Advances 10 (28) (2020) 16313–16322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Nejad HR et al. , Ingestible Osmotic Pill for In-vivo Sampling of Gut Microbiome, BioRxiv (2019) 690982. [Google Scholar]
- [220].Koziolek M et al. , Investigation of pH and temperature profiles in the GI tract of fasted human subjects using the Intellicap® system, Journal of pharmaceutical sciences 104 (9) (2015) 2855–2863. [DOI] [PubMed] [Google Scholar]
- [221].Tang Q et al. , Current sampling methods for gut microbiota: a call for more precise devices, Frontiers in cellular and infection microbiology 10 (2020) 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Rezaei Nejad H et al. , Ingestible osmotic pill for in vivo sampling of gut microbiomes, Advanced Intelligent Systems 1 (5) (2019) 1900053. [Google Scholar]
- [223].Liu X et al. , Magnetic living hydrogels for intestinal localization, retention, and diagnosis, Advanced Functional Materials 31 (27) (2021) 2010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Mettu S et al. , Perspective on constructing cellulose-hydrogel-based gut-like bioreactors for growth and delivery of multiple-strain probiotic bacteria, Journal of Agricultural and Food Chemistry 69 (17) (2021) 4946–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Dickson RP, Approaches to Sampling the Respiratory Microbiome, The Microbiome in Respiratory Disease (2022) 3–19. [Google Scholar]
- [226].Baker JM et al. , Whole lung tissue is the preferred sampling method for amplicon-based characterization of murine lung microbiota, Microbiome 9 (1) (2021) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Saladié M et al. , Microbiomic analysis on low abundant respiratory biomass samples; improved recovery of microbial DNA from bronchoalveolar lavage fluid, Frontiers in microbiology (2020) 2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Chan LW, Advances in activity-based diagnostics for infectious disease and microbiome health. Current Opinion, Biomedical Engineering (2021) 100296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Khodakivskyi PV et al. , Noninvasive imaging and quantification of bile salt hydrolase activity: From bacteria to humans. Science, Advances 7 (6) (2021), p. eaaz9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Chan LW et al. , Engineering synthetic breath biomarkers for respiratory disease, Nature nanotechnology 15 (9) (2020) 792–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Wang J, Childers WS, The Future Potential of Biosensors to Investigate the Gut-Brain Axis, Frontiers in Bioengineering and Biotechnology 9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Sosnowski K, Akarapipad P, Yoon JY, The future of microbiome analysis: Biosensor methods for big data collection and clinical diagnostics, Medical Devices & Sensors 3 (5) (2020) e10085. [Google Scholar]
- [233].Feng Z et al. , In situ imaging for tumor microbiome interactions via imaging mass cytometry on single-cell level, Cytometry Part A n/a(n/a) (2022). [DOI] [PubMed] [Google Scholar]
- [234].Liu J et al. , Integration of epidemiologic, pharmacologic, genetic and gut microbiome data in a drug–metabolite atlas, Nature medicine 26 (1) (2020) 110–117. [DOI] [PubMed] [Google Scholar]
- [235].McCoubrey LE et al. , Harnessing machine learning for development of microbiome therapeutics, Gut Microbes 13 (1) (2021) 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Greener JG et al. , A guide to machine learning for biologists, Nature Reviews Molecular Cell Biology 23 (1) (2022) 40–55. [DOI] [PubMed] [Google Scholar]
- [237].Lim H et al. , Artificial intelligence approaches to human-microbiome protein–protein interactions, Current Opinion in Structural Biology 73 (2022) 102328. [DOI] [PubMed] [Google Scholar]
- [238].Yin X et al. , A comparative evaluation of tools to predict metabolite profiles from microbiome sequencing data, Frontiers in microbiology 11 (2020) 3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Mallick H et al. , Predictive metabolomic profiling of microbial communities using amplicon or metagenomic sequences, Nature communications 10 (1) (2019) 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Bhadra P et al. , AmPEP: Sequence-based prediction of antimicrobial peptides using distribution patterns of amino acid properties and random forest, Scientific reports 8 (1) (2018) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Ma Y et al. , Identification of antimicrobial peptides from the human gut microbiome using deep learning, Nature Biotechnology (2022) 1–11. [DOI] [PubMed] [Google Scholar]
- [242].Morton JT et al. , Learning representations of microbe–metabolite interactions, Nature methods 16 (12) (2019) 1306–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].McCoubrey LE et al. , Machine Learning Predicts Drug Metabolism and Bioaccumulation by Intestinal Microbiota, Pharmaceutics 13 (12) (2021) 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Sharma AK et al. , A novel approach for the prediction of species-specific biotransformation of xenobiotic/drug molecules by the human gut microbiota, Scientific reports 7 (1) (2017) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Noecker C et al. , Metabolic model-based integration of microbiome taxonomic and metabolomic profiles elucidates mechanistic links between ecological and metabolic variation, MSystems 1 (1) (2016), e00013 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Rahman SF et al. , Machine learning leveraging genomes from metagenomes identifies influential antibiotic resistance genes in the infant gut microbiome, MSystems 3 (1) (2018), e00123 e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Biteen JS et al. , Tools for the Microbiome: Nano and Beyond, ACS Nano 10 (1) (2016) 6–37. [DOI] [PubMed] [Google Scholar]
- [248].Bolyen E et al. , Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2, Nature Biotechnology 37 (8) (2019) 852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Stuart T et al. , Comprehensive integration of single-cell data, Cell 177 (7) (2019), pp. 1888–1902. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Li X-Y et al. , Complete genome sequencing of Peyer’s patches-derived Lactobacillus taiwanensis CLG01, a potential probiotic with antibacterial and immunomodulatory activity, BMC microbiology 21 (1) (2021) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Kadosh E et al. , The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic, Nature 586 (7827) (2020) 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Kip AM et al. , Proteomics analysis of human intestinal organoids during hypoxia and reoxygenation as a model to study ischemia-reperfusion injury, Cell Death & Disease 12 (1) (2021) 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Ludikhuize MC et al. , Protocol to profile the bioenergetics of organoids using Seahorse, STAR Protocols 2 (1) (2021) 100386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Sharma A, Im SH, Special issue on the human microbiome: from symbiosis to therapy, Exp Mol Med 52 (9) (2020) 1361–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Park JC, Im S-H, Of men in mice: the development and application of a humanized gnotobiotic mouse model for microbiome therapeutics, Experimental & Molecular Medicine 52 (9) (2020) 1383–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Yu F, Hunziker W, Choudhury D, Engineering Microfluidic Organoid-on-a-Chip Platforms. Micromachines 10 (3) (2019) 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Hofer M, Lutolf MP, Engineering organoids, Nature Reviews Materials (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Sato T et al. , Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium, Gastroenterology 141 (5) (2011) 1762–1772. [DOI] [PubMed] [Google Scholar]
- [259].Spence JR et al. , Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro, Nature 470 (7332) (2011) 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Workman MJ et al. , Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system, Nature medicine 23 (1) (2017) 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Tsai Y-H et al. , In vitro patterning of pluripotent stem cell-derived intestine recapitulates in vivo human development, Development 144 (6) (2017) 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Nikolic MZ´ et al. , Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids, Elife 6 (2017) e26575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Sachs N et al. , Long-term expanding human airway organoids for disease modeling, The EMBO journal 38 (4) (2019) e100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Dye BR et al. , A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids, Elife 5 (2016) e19732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Chen Y-W et al. , A three-dimensional model of human lung development and disease from pluripotent stem cells, Nature cell biology 19 (5) (2017) 542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Cruz-Acuña R et al. , PEG-4MAL hydrogels for human organoid generation, culture, and in vivo delivery, Nature protocols 13 (9) (2018) 2102–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Cruz-Acuña R et al. , Synthetic hydrogels for human intestinal organoid generation and colonic wound repair, Nature cell biology 19 (11) (2017) 1326–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Meran L et al. , Engineering transplantable jejunal mucosal grafts using patient-derived organoids from children with intestinal failure, Nature Medicine 26 (10) (2020) 1593–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Angus HC et al. , Intestinal organoids as a tool for inflammatory bowel disease research, Frontiers in medicine 6 (2020) 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].LeSavage BL, Heilshorn SC, Defined matrices bring IBD to 3D, Nature Materials 20 (2) (2021) 124–125. [DOI] [PubMed] [Google Scholar]
- [271].Dheer R, Young VB, Stem-cell-derived models: tools for studying role of microbiota in intestinal homeostasis and disease, Current Opinion in Gastroenterology 37 (1) (2021) 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Park G-S et al. , Emulating Host-Microbiome Ecosystem of Human Gastrointestinal Tract in Vitro, Stem Cell Reviews and Reports 13 (3) (2017) 321–334. [DOI] [PubMed] [Google Scholar]
- [273].Tsuchiya M et al. , Functional analysis of isoflavones using patient-derived human colonic organoids, Biochemical and Biophysical Research Communications 542 (2021) 40–47. [DOI] [PubMed] [Google Scholar]
- [274].Williamson IA et al. , A high-throughput organoid microinjection platform to study gastrointestinal microbiota and luminal physiology, Cellular and molecular gastroenterology and hepatology 6 (3) (2018) 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].DiMarco RL et al. , Engineering of three-dimensional microenvironments to promote contractile behavior in primary intestinal organoids, Integrative Biology 6 (2) (2013) 127–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Merenda A, Fenderico N, Maurice MM, Wnt signaling in 3D: recent advances in the applications of intestinal organoids, Trends in cell biology 30 (1) (2020) 60–73. [DOI] [PubMed] [Google Scholar]
- [277].Rabata A et al. , 3D cell culture models demonstrate a role for FGF and WNT signaling in regulation of lung epithelial cell fate and morphogenesis, Frontiers in cell and developmental biology 8 (2020) 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Vazquez-Armendariz AI, Herold S, From Clones to Buds and Branches: The Use of Lung Organoids to Model Branching Morphogenesis Ex Vivo, Frontiers in Cell and Developmental Biology 9 (2021) 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Van Der Vaart J, Clevers H, Airway organoids as models of human disease, Journal of internal medicine (2020). [DOI] [PubMed] [Google Scholar]
- [280].Barros AS, Costa A, Sarmento B, Building three-dimensional lung models for studying pharmacokinetics of inhaled drugs, Advanced drug delivery reviews (2020). [DOI] [PubMed] [Google Scholar]
- [281].Dijkstra KK et al. , Challenges in Establishing Pure Lung Cancer Organoids Limit Their Utility for Personalized Medicine, Cell Reports 31 (5) (2020) 107588. [DOI] [PubMed] [Google Scholar]
- [282].Zhao X et al. , Human Intestinal Organoids Recapitulate Enteric Infections of Enterovirus and Coronavirus, Stem Cell Reports 16 (3) (2021) 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [283].Krüger J et al. , Drug Inhibition of SARS-CoV-2 Replication in Human Pluripotent Stem Cell-Derived Intestinal Organoids, Cellular and Molecular Gastroenterology and Hepatology 11 (4) (2021) 935–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Tindle C, et al. , Adult stem cell-derived complete lung organoid models emulate lung disease in COVID-19. bioRxiv, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [285].Zhou J et al. , Infection of bat and human intestinal organoids by SARS-CoV-2, Nature medicine 26 (7) (2020) 1077–1083. [DOI] [PubMed] [Google Scholar]
- [286].Bock C et al. , The Organoid Cell Atlas, Nature Biotechnology 39 (1) (2021) 13–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Curvello R et al. , Engineered Plant-Based Nanocellulose Hydrogel for Small Intestinal Organoid Growth, Advanced Science 8 (1) (2021) 2002135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Curvello R et al. , A thermo-responsive collagen-nanocellulose hydrogel for the growth of intestinal organoids, Materials Science and Engineering: C (2021) 112051. [DOI] [PubMed] [Google Scholar]
- [289].Protein-Functionalized Poly(ethylene glycol) Hydrogels as Scaffolds for Monolayer Organoid Culture. Tissue Engineering Part C: Methods, 2021. 27 (1): p. 12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Agarwal T et al. , Recent advances in chemically defined and tunable hydrogel platforms for organoid culture, Bio-Design and Manufacturing (2021) 1–34.33014512 [Google Scholar]
- [291].Graney PL et al. , Organoid Polymer Functionality and Mode of Klebsiella Pneumoniae Membrane Antigen Presentation Regulates Ex Vivo Germinal Center Epigenetics in Young and Aged B Cells, Advanced Functional Materials 30 (48) (2020) 2001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [292].Brassard JA et al. , Recapitulating macro-scale tissue self-organization through organoid bioprinting, Nature Materials 20 (1) (2021) 22–29. [DOI] [PubMed] [Google Scholar]
- [293].Trapecar M et al. , Human physiomimetic model integrating microphysiological systems of the gut, liver, and brain for studies of neurodegenerative diseases. Science, Advances 7 (5) (2021), p. eabd1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [294].Jalili-Firoozinezhad S, Miranda CC, Cabral JMS, Modeling the Human Body on Microfluidic Chips, Trends in Biotechnology (2021). [DOI] [PubMed] [Google Scholar]
- [295].Shin W, Kim HJ, 3D in vitro morphogenesis of human intestinal epithelium in a gut-on-a-chip or a hybrid chip with a cell culture insert, Nature Protocols 17 (3) (2022) 910–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Kim R et al. , An in vitro intestinal platform with a self-sustaining oxygen gradient to study the human gut/microbiome interface, Biofabrication 12 (1) (2019) 015006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Trietsch SJ et al. , Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes, Nature communications 8 (1) (2017) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [298].Kasendra M et al. , Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids, Scientific reports 8 (1) (2018) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [299].Jalili-Firoozinezhad S et al. , A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip, Nature biomedical engineering 3 (7) (2019) 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].Shirure VS,Hughes CCW,George SC,EngineeringVascularizedOrganoid-on-a-Chip Models, Annual Review of Biomedical Engineering 23 (1) (2021) p. null. [DOI] [PubMed] [Google Scholar]
- [301].Mead BE and Karp JM, All models are wrong, but some organoids may be useful. 2019, BioMed Central. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Nikolaev M et al. , Homeostatic mini-intestines through scaffold-guided organoid morphogenesis, Nature 585 (7826) (2020) 574–578. [DOI] [PubMed] [Google Scholar]
- [303].Hinman SS, et al. , In vitro generation of self-renewing human intestinal epithelia over planar and shaped collagen hydrogels. Nature protocols, 2021. 16(1): p. 352–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Wang Y et al. , Building a thick mucus hydrogel layer to improve the physiological relevance of in vitro primary colonic epithelial models. Cellular and Molecular, Gastroenterology and Hepatology 8 (4) (2019), pp. 653–655. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [305].Kulthong K et al. , Transcriptome comparisons of in vitro intestinal epithelia grown under static and microfluidic gut-on-chip conditions with in vivo human epithelia, Scientific reports 11 (1) (2021) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Shin W et al. , Robust Formation of an epithelial layer of human intestinal organoids in a polydimethylsiloxane-based gut-on-a-chip microdevice, Frontiers in medical technology 2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [307].Mead BE, et al. , High-throughput organoid screening enables engineering of intestinal epithelial composition. bioRxiv, 2020. [Google Scholar]
- [308].Saygili E et al. , Human lung-on-chips: Advanced systems for respiratory virus models and assessment of immune response, Biomicrofluidics 15 (2) (2021) 021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Meghani N et al. , Evaluation and live monitoring of pH-responsive HSA-ZnO nanoparticles using a lung-on-a-chip model, Archives of Pharmacal Research 43 (2020) 503–513. [DOI] [PubMed] [Google Scholar]
- [310].Arefi SMA et al. , Simulation of nanoparticle transport and adsorption in a microfluidic lung-on-a-chip device, Biomicrofluidics 14 (4) (2020) 044117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [311].Mejías JC et al. , A 96-well format microvascularized human lung-on-a-chip platform for microphysiological modeling of fibrotic diseases, Lab on a Chip 20 (19) (2020) 3601–3611. [DOI] [PubMed] [Google Scholar]
- [312].Zamprogno P et al. , Second-generation lung-on-a-chip with an array of stretchable alveoli made with a biological membrane, Communications biology 4 (1) (2021) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].Vandeplassche E et al. , Influence of the lung microbiome on antibiotic susceptibility of cystic fibrosis pathogens, European Respiratory Review 28 (152) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Moysidou C-M, Owens RM, Advances in modelling the human microbiome–gut–brain axis in vitro, Biochemical Society Transactions (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Yi W et al. , Effect of temperature stress on gut-brain axis in mice: Regulation of intestinal microbiome and central NLRP3 inflammasomes, Science of The Total Environment 772 (2021) 144568. [DOI] [PubMed] [Google Scholar]
- [316].George AK et al. , Rebuilding Microbiome for Mitigating Traumatic Brain Injury: Importance of Restructuring the Gut-Microbiome-Brain Axis, Molecular Neurobiology (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Bannerman CA et al. , The gut-brain axis and beyond: Microbiome control of spinal cord injury pain in humans and rodents, Neurobiology of Pain 9 (2021) 100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [318].Keogh CE et al. , Myelin as a regulator of development of the microbiota-gut-brain axis, Brain, Behavior, and Immunity 91 (2021) 437–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [319].Muller PA et al. , Microbiota-modulated CART+ enteric neurons autonomously regulate blood glucose, Science 370 (6514) (2020) 314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [320].He J-W et al. , Perspectives on how mucosal immune responses, infections and gut microbiome shape IgA nephropathy and future therapies, Theranostics 10 (25) (2020) 11462–11478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [321].Xu Y et al. , Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding, Nature medicine 26 (4) (2020) 502–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [322].Wu Y et al. , Prolonged presence of SARS-CoV-2 viral RNA in faecal samples, The lancet Gastroenterology & hepatology 5 (5) (2020) 434–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [323].Lamers MM et al. , SARS-CoV-2 productively infects human gut enterocytes, Science 369 (6499) (2020) 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [324].Zuo T et al. , Alterations in gut microbiota of patients with COVID-19 during time of hospitalization, Gastroenterology 159 (3) (2020), pp. 944–955. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [325].Mazzarelli A et al. , 16S rRNA gene sequencing of rectal swab in patients affected by COVID-19, PloS one 16 (2) (2021) e0247041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [326].Tian Y et al. , Gut Microbiota May Not Be Fully Restored in Recovered COVID-19 Patients After 3-Month Recovery. Frontiers, Nutrition 8 (182) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [327].Tao W et al. , Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18, Medicine in Microecology 5 (2020) 100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [328].Chhibber-Goel J, Gopinathan S, Sharma A, Interplay between severities of COVID-19 and the gut microbiome: implications of bacterial co-infections?, Gut Pathogens 13 (1) (2021) 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [329].Xu R et al. , Temporal association between human upper respiratory and gut bacterial microbiomes during the course of COVID-19 in adults, Communications Biology 4 (1) (2021) 240. [DOI] [PMC free article] [PubMed] [Google Scholar]