Abstract
A hurdle facing DNA vaccine development is the ability to generate strong immune responses systemically and at local immune sites. We report a novel systemically administered DNA vaccination strategy using intramuscular codelivery of CCL27 or CCL28, which elicited elevated peripheral IFN-γ and antigen-specific IgG while driving antigen-specific T-cell secretion of cytokine and antibody production in the gut-associated lymphoid tissue and lung. This strategy resulted in induction of long-lived antibody responses that neutralized influenza A/PR8/34 and protected mice from morbidity and mortality associated with a lethal intranasal viral challenge. This is the first example of the use of CCL27 and CCL28 chemokines as adjuvants to influence a DNA vaccine strategy, suggesting further examination of this approach for manipulation of vaccine-induced immunity impacting both quality and phenotype of responses.
Keywords: DNA vaccines, immune responses, molecular adjuvants, immune modulation
Introduction
DNA-based plasmid vaccines have a number of advantages that make an attractive vaccine platform,1,2 including the fact that this vaccine candidate is a non-live, non-replicating, non-spreading and non-viral platform that to date has an excellent safety profile. Pre-existing immunity or the development of neutralizing antibodies is not an issue for plasmid DNA vaccines, which allow for boosting of a specific immune response with multiple immunizations. Plasmid DNA is also simple to manipulate and produce compared with live vaccines. Furthermore, it is a stable platform that may not require a cold chain for storage. DNA vaccines have been shown to elicit humoral and cellular responses and confer protection in small-animal models and in larger species to some degree.1,3-6
Despite these advantages, enhancing the potency of immune responses generated by DNA vaccines is a critical focus of this platform, because DNA vaccines have shown poor translation from rodent studies to human trials. Some strategies to overcome the limitations in immunogenicity of the platform include gene optimization, heterologous prime-boost strategies, improved delivery techniques, as well as the use of molecular adjuvants to augment DNA vaccine-elicited responses.1,7-11 The inclusion of genes encoding cytokines or chemokines (immune trafficking signals) in a vaccination strategy can alter the magnitude, duration and nature of the immune response and such genes have been tested as vaccine adjuvants.12-17 These strategies have shown promise in improving the efficacy of DNA vaccines in large-animal models such as horses, dogs and pigs, as well as in humans.2,18-22 Currently, several DNA-based veterinary products have been licensed for use.2
One strategy that improves the immunogenicity of DNA vaccines is the use of chemokine molecular adjuvants. The redirection of systemic immune cells in vivo by DNA vaccination using chemokines as part of a DNA-encoded vaccine cocktail has been previously reported.23,24 Chemokines are inflammatory molecules that act primarily as chemoattractants and as activators of lymphocytes through the enhancement of Th1 and Th2 phenotypes.23-27 Thus, modulation of immune responses through the chemokine network is an interesting concept for vaccine design. The ability of mucosally expressed chemokines to act as adjuvants and elicit DNA vaccine-specific immune responses has yet to be shown. CCL27 (cutaneous T cell-attracting chemokine) has a role in the T-lymphocyte biology of the skin, including the chemotaxis of cutaneous memory T cells28,29 and specialized ‘effector’ T helper cells to and/or within epidermal microenvironments,30,31 whereas CCL28 (mucosal-associated epithelial chemokine) is expressed at high levels in the epithelium of various mucosal sites including the small intestine, colon and bronchi.32,33 CCR10, the receptor for chemokines CCL27 and CCL28, is expressed within mucosal and epithelial tissue and helps to recirculate and localize antibody secreting cells,32,34-36 as well as naive, effector and memory T lymphocytes32,33 to mucosal and cutaneous sites. Thus, these chemokines and their cognate receptor, CCR10, are important regulators of mucosal immune responses and T- and B-cell recruitment to specific mucosal sites.
We investigated the importance of the CCR10 chemokine ligands to the resulting immune response to DNA vaccination with influenza A hemagglutinin (HA) plasmid vaccine. We found that codelivery of plasmids expressing CCL27 or CCL28 with a construct encoding influenza A/PR8/34 HA resulted in the induction of elevated levels of antigen-specific T-cell inflammatory cytokines in the periphery as well as in the lung. Furthermore, antigen-specific IgA production in serum or in feces was increased by both adjuvants. After lethal challenge with intranasally delivered PR8 influenza virus, the CCL27- and CCL28-coimmunized mice exhibited improved protection from morbidity and mortality compared with non-adjuvant vaccinated animals. These studies suggest that this strategy has unique mucosal immune-modulating properties, including the ability to enhance antigen-specific IgA responses in vivo, which deserves additional investigation.
Results
Construction and expression of mucosal chemokine plasmids
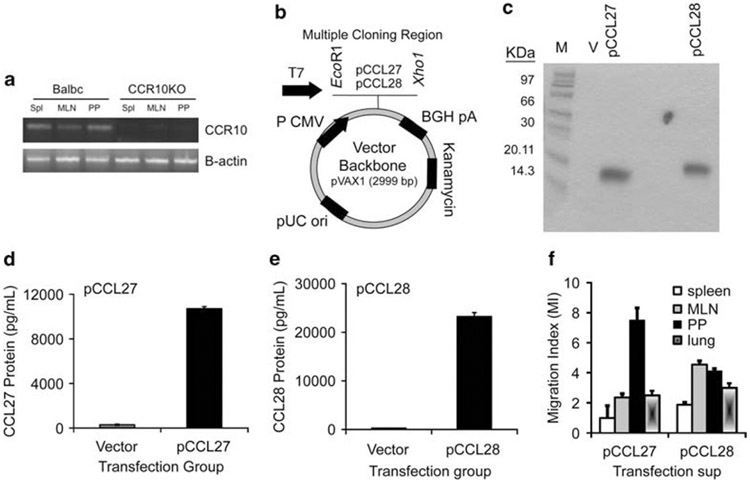
As shown in Figure 1a, CCR10 transcript expression was detected in RNA isolated from lymphocytes from the spleen, Peyer’s patches (PPs) and mesenteric lymph node (MLN) of wild-type BALB/c but not of CCR10 knockout mice,37 suggesting that this receptor could be targeted during an antigen-specific immune response induced after DNA vaccination. To test this hypothesis, plasmid forms of CCL27 (pCCL27) and CCL28 (pCCL28) were constructed and tested for protein expression. The murine forms of CCL27 (363 base pairs) and CCL28 (393 base pairs) genes were generated by PCR using a cDNA template that was synthesized from RNA purified from inflamed ear (CCL27) or mouse small intestine (CCL28). Chemokine cDNA was inserted into the multiple cloning site of the pVAX1 vector (Figure 1b) using the restriction enzymes Ecor1 and Xho1. To confirm that chemokine proteins were being expressed by the constructs, the chemokine plasmids or empty vector (pVAX) was analyzed by in vitro translation assays using specific anti-mouse CCL27 or anti-mouse CCL28 antibodies (Figure 1c). To further confirm expression in vitro, RD cells were transfected with the empty vector (pVAX) control or the chemokine plasmids. Supernatants were collected 48 h after transfection, the levels of secreted CCL27 or CCL28 protein were measured using ELISA and 11 000 pg ml−1 of CCL27 (Figure 1e) and 24 750 pg ml−1 of CCL28 (Figure 1d) were detected in supernatants of transfected cells. Supernatants from pVAX-transfected cells contained minimal levels of either chemokine (~300 pg ml−1). The bioactivity of the plasmid-derived chemokine proteins was confirmed using Transwell chemotaxis assays (Figure 1f). CCR10+ lymphocytes were collected from mucosal tissues (lung lymphocytes, splenocytes, MLN or PP/colonic cryopatch lymphocytes) as designated in Figure 1e and placed in the upper well of a 5-μm Transwell chamber, whereas the bottom well contained supernatants from CCL27- or CCL28-transfected RD cells (Figure 1f). Taken together, these data show that the plasmid expression vectors express specific chemokine proteins that function properly in vitro.
Figure 1.
Expression of bioactive chemokines from plasmid-encoded chemokine. (a) Reverse transcriptase (RT)-PCR analysis of lymphocytes from the spleen (Spl), mesenteric lymph node (MLN) or Peyer’s patch (PP) from wild-type or CCR10 knockout BALB/c mice. (b) Schematic representation of chemokine expression plasmids. All genes were cloned into the multiple cloning region of the expression vector pVAX (kanamycin resistance) using the restriction enzyme sites, EcoR1 and Xho1. (c) Expression of pCCL27 and pCCL28 was confirmed by a T7-coupled transcription/translation reticulocyte lysate system (TNT). The gel shows size markers (designated M, lane 1), pVAX background control (V, lane 2), 10.9 kDa pCCL27 protein (lane 3) and 12.6 kDa pCCL28 protein (lane 4). (d and e) Expression of bioactive chemokine protein translated from the plasmid forms of CCL27 (d) and CCL28 (e). ELISA was carried out using supernatants from pCCL27- or pCCL28-transfected RD cells (pg ml−1). For each chemokine, vector background control is included (gray bar) vs pCCL27 or pCCL28 (black bar). Data are shown as pg ml−1 of chemokine protein±s.d. of triplicate wells. (f) Migration index of splenocytes, MLN, PP/cryopatch cells and lung cells in response to secreted CCL27- or CCL28-containing supernatants from transfected RD cells.
In vivo bioactivity of chemokine proteins is generated from plasmid CCL27 or CCL28
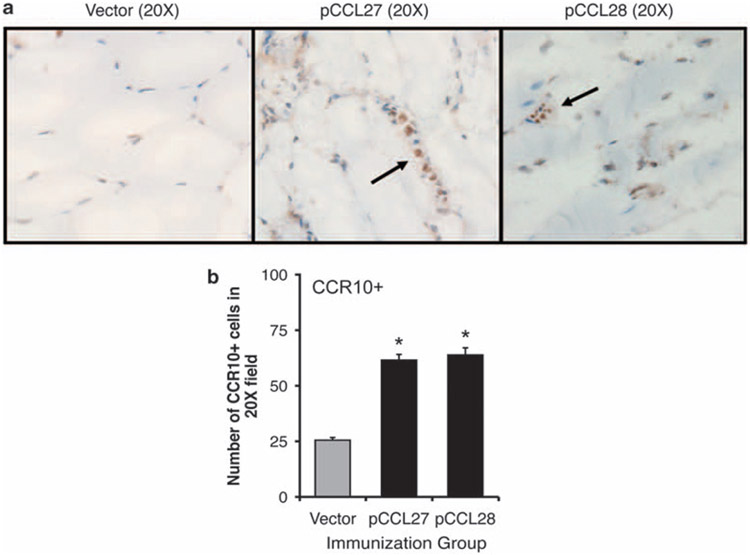
To determine whether intramuscular delivery of pCCL27 or pCCL28 would result in the recruitment of cognate chemokine receptor (that is, CCR10) positive cells to the site of vaccine inoculation, we injected 100 mg of either pVAX empty vector, pCCL27 or pCCL28 constructs in the quadriceps of BALB/c mice (n=4 per group). As described in Materials and methods, injected muscles were excised on day 7 post injection, embedded in OCT medium, snap frozen, serial sectioned and incubated with an anti-mouse CCR10 antibody to determine whether cellular infiltrates of injected muscles expressed CCR10 (Figure 2a). The number of CCR10+ cells was determined by counting DAB (3,3′-Diaminobenzidine) stain-positive cells from six different 20× fields per group. These counts were averaged and plotted as the total number of CCR10+ cells per 20× field (Figure 2b). Intramuscular administration of the chemokine-expressing plasmids resulted in a greater than twofold increase in the recruitment of CCR10+ inflammatory cells when either pCCL27 (Figure 2b, that is, 61±6.7 cells per field) or pCCL28 (Figure 2b, that is, 63±3.3 cells per field) was injected compared with vector backbone alone (Figure 2b, that is, 25±3.2 cells per field).
Figure 2.
Induction of infiltration, after intramuscular injection, by cognate receptor positive cells (a and b). Infiltration of CCR10-positive cells induced following intramuscular injection of 100 μg of pCCL27 and pCCL28. (a) Immunohistochemical staining of quadriceps sections 7 days post immunization with 100 μg of vector backbone control (left panel) or pCCL27 or pCCL28. The number of infiltrating cells was determined by a visual count of six independent 20× fields from the experimental slides viewed randomly under a microscope. The total number of infiltrating CCR10+ cells was averaged and shown±s.d. for vector (gray bar) and pCCL27 or pCCL28 injection (black). (*P<0.05 for statistical comparisons of injections with either pCCL27 or pCCL28 vs vector control.)
Codelivery of pCCL27 or pCCL28 enhances influenza A/PR8/34 HA-specific cytokine secretion from T lymphocytes in the spleen and lung
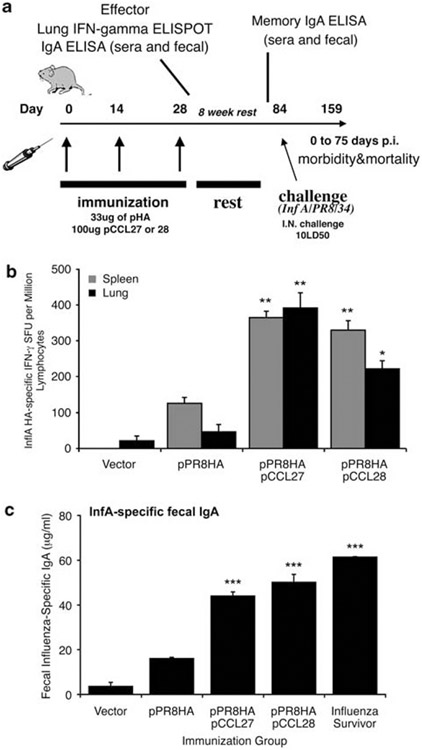
The ability of the plasmid-encoded CCR10 chemokine ligands to modulate antigen-specific T-cell function in a relevant challenge model was examined next. Influenza A/PR8/34 HA plasmid was used as the antigen because vaccine-induced HA-specific T cell and antibody responses have been reported to impact influenza A challenge in the lungs of mice. The role of CCR10 ligands in influenza infection was unknown. As shown in Figure 3a, BALB/c female mice were coimmunized intramuscularly three times with 133 μg of control vector, 33 μg of the hemagglutinin gene from influenza A/PR8/34 (pPR8HA) plus 100 μg of pVAX backbone control DNA, or 33 μg of the pPR8HA in combination with either 100 μg of pCCL27 or of pCCL28. Each injection was separated by a 2-week interval. Both chemokine adjuvants enhanced HA-specific IFN-γ in the spleen compared with vaccination with antigenic plasmid alone (Figure 3b). Coimmunization with CCL27 (393±43 SFC, P<0.001) or CCL28 (222±23 SFC, P<0.005) enhanced the number of HA-specific lung T lymphocytes that secreted IFN-γ several fold compared with pPR8HA immunization alone (47±9 SFC).
Figure 3.
pCCL27 and pCCL28 immune adjuvant-induced IgA immunity specific against influenza A/PR8/34 mouse model of mucosal lung infection. (a) Schedule for influenza A (PR8) immunization, immune analysis and lethal mucosal challenge. Mice (n=14) were immunized as previously described with 33 μg of hemagglutinin (HA) or in combination with our mucosal chemokines. Effector immune responses were measured 1 week after the third immunization (n=4 mice sacked), and long-lived responses were examined 8 weeks after the final boost (n=10, pre-challenge). (b) Mucosal chemokines enhanced the secretion of IFN-γ influenza-specific A/PR8/34 by lung and splenic T cells (n=4). Lung lymphocytes (black bar) or splenocytes (gray bar) were harvested from immunized mice and used in an IFN-γ ELISpot assay stimulated with either medium control or the CD8+ T cell epitope peptide encoding influenza A/PR8/34 (H1N1) hemagglutinin (IYSTVASSL amino acid 518–526). Values depicted in panel b are resulting values after medium is subtracted. (c) Mucosal chemokine coimmunization augments mucosal-specific IgA in fecal extracts. Pooled fecal pellets (n=10) were analyzed by ELISA for the presence of influenza A/PR8/34 hemagglutinin-specific secretory IgA. *P<0.05, **P<0.01, ***P<0.001 for statistical comparisons of chemokine/pPR8HA groups with pPR8HA antigenic vector alone.
Coimmunization with mucosal chemokines and pPR8HA induces humoral immunity specific against influenza A/PR8/34 HA
At 1 week after the final vaccination, sera or fecal extracts were collected and processed to measure influenza A-specific immunoglobulin in ELISA (Figure 3c, Table 1). As shown in Table 1, coimmunization with either pCCL27 (262 μg ml−1±24.6, P<0.01) or pCCL28 (190 μg ml−1±13, P<0.05) resulted in the significant enhancement of serum IgG levels, resulting in a 2.7-fold or 1.97-fold enhancement of HA-specific IgG, respectively. However, a similar elevation of HA-specific IgG was not detected in fecal extracts (anti-HA IgG levels ranging from 402 to 461 μg ml−1 in all experimental groups). IgA levels from vaccinated mice were also determined. Immunization with the antigenic plasmid (33 μg pPR8HA) resulted in low levels of influenza A-specific IgA in mouse serum (0.37 μg ml−1±0.08) (Table 1). However, coimmunization with pCCL27 or pCCL28 resulted in serum IgA levels of 17.26 and 20.1 μg ml−1, respectively, compared with only 0.37 μg ml−1 elicited by pPR8HA, thus resulting in up to a 54-fold enhancement in influenza-specific IgA (Table 1).
Table 1.
Comparison of influenza A-specific serum and fecal IgG and IgA titers elicited in response to chemokine immunoadjuvant plasmid
| Immunization group | Sera IgG (μg ml−1) |
Fold increase | Sera IgA (μg ml−1) |
Fold increase | Fecal IgG (μg ml−1) |
Fold increase | Fecal IgA (μg ml−1) |
Fold increase |
Sera IgA to IgG |
Fecal IgA to IgG |
|---|---|---|---|---|---|---|---|---|---|---|
| Vector | 45.33 (±8.33) | 0.12 (±0.013) | 461 (±44.89) | 3.63 (±1.04) | ||||||
| pPR8HA | 96.15 (±13.4) | 0.37 (±0.084) | 1.00 | 402 (±29.7) | 1.00 | 16.07 (±0.326) | 1.00 | 1–378 | 1–25 | |
| pPR8HA/pCCL27 | 262.13 (±24.6, P<0.01) | 2.72 | 17.257 (±2.308, P<0.001) | 46.64 | 407 (±32.59) | 1.01 | 44 (±1.027, P=0.001) | 2.74 | 1–20 | 1–9 |
| pPR8HA/pCCL28 | 189.72 (± 13.06, P<0.05) | 1.97 | 20.06 (±4.84, P<0.01) | 54.22 | 456 (±39.74) | 1.13 | 50.133 (±7.032, P<001) | 3.12 | 1–9 | 1–9 |
In fecal extracts, levels of IgA specific for influenza were 16.1 μg ml−1 in the pPR8HA group (Figure 3b). Coimmunization with pCCL27 resulted in higher amounts of secreted influenza-specific IgA (a 2.7-fold enhancement or 44 μg ml−1), whereas the pCCL28-immunized group secreted 50.1 μg ml−1 IgA, a threefold enhancement over pPR8HA immunization in the absence of adjuvant (Figure 3b). Importantly, the levels of influenza-specific IgA elicited by coimmunization with the CCR10 ligands were similar to those detected in the fecal samples from mice that had been infected with and survived an influenza challenge (Figure 3b). Although the level of antigen-specific IgG detected in immunized mice was higher than that of the antigen-specific IgA, the ratio of IgA to IgG in serum and fecal samples is biased toward more HA-specific IgA after chemokine adjuvant immunization. The most potent effect on improving IgA production was induced by pCCL28 (1–9 for both serum and fecal samples) and pCCL27 (1–20 for serum or 1–9 for fecal samples) when compared with the antigenic plasmid group (1–378 serum and 1–25 fecal samples).
Influenza challenge studies
To test whether the chemokine-induced enhancement of influenza A-specific lung lymphocyte cytokine and IgA secretion reaches protective physiological levels, influenza A/PR8/34 lethal intranasal challenge was carried out. Previous studies in our laboratory determined that a 10 LD50 influenza A/PR8/34 challenge in BALB/c mice results in a lethal infection in 100% of naive animals; in contrast, the same challenge after vaccination with 33-μg pPR8HA plasmid results in >80% lethal infection, whereas mice vaccinated with a 50-μg pPR8HA plasmid dose survive 90% of the time.4 Therefore, we choose the 33-μg vaccine dose for the control comparison group.
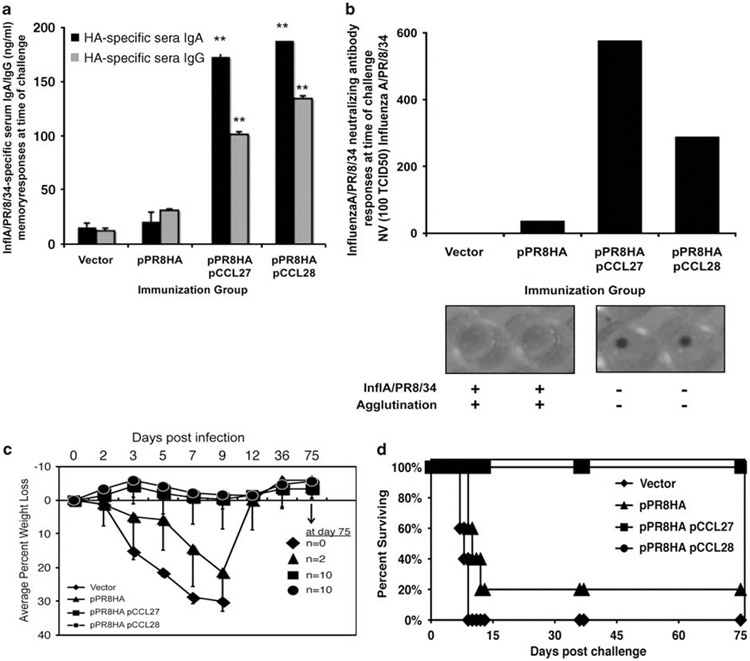
Before challenge and after an 8-week rest post immunization with 33 μg of pPR8HA±the chemokine immune adjuvants, serum was harvested from immunized mice, and long-lived influenza A-specific IgG and IgA levels were measured by ELISA (Figure 4a). Analysis of individual animal serum samples was also carried out after a neutralization assay (six replicate wells each) (Figure 4b). The serum dilution at which 2000TCID50 (50% tissue culture infectious dose) per ml of influenza A/PR8/34 was neutralized by the immune sera is depicted for each study group (Figure 4b). A representative well showing CRBC (chicken red blood cell) agglutination (influenza A/PR8/34, ‘fuzzy’ well) for vector and antigenic plasmid immunization alone, as well as representative wells showing CRBC pooling (well contains influenza A/PR8/34 neutralized virus) for pCCL27 or pCCL28, is shown in Figure 4b. The serum dilution at which 2000TCID50 per ml of influenza A/PR8/34 can be neutralized by pPR8HA plasmid alone is 1–36, whereas that for the pCCL27-adjuvanted mice is 1–576 and that for the pCCL28-adjuvanted mice is 1–288, showing the ability of the CCR10 ligand adjuvants to enhance humoral immunity.
Figure 4.
CCR10 ligands induce long-lived IgG, IgA and neutralizing antibody and protect mice against influenza A/PR8/34 lethal mucosal challenge. (a) Influenza-specific long-lived IgG and IgA titers were determined at time of challenge. Individual serum samples were harvested from mice from each immunization group (n=10 mice per group) and (ng ml−1) levels of pPR8HA-specific IgG (gray bar) and IgA (black bar) were determined by ELISA using a recombinant standard of known concentration. [**P<0.01, for comparisons between chemokine plasmid adjuvanted groups (n=10 mice per experimental group vs n=10 pPR8HA alone]. (b) Coimmunization with CCL27 and CCL28 chemokines elicits anti-influenza A/PR8/34 neutralizing antibodies. Serial fourfold dilutions of individual serum samples from immunized mouse groups were incubated with influenza A/PR8/34 (100 TCID50 virus per well), and results from the chicken erythrocyte sedimentation assay are shown as the serum dilution at which 2000TCID50/ml of influenza A/PR8/34 can be neutralized. Data represent individual mouse sera (n=10) in six replicates per dilution for each immunized mouse group. Representative wells of neutralized virus (influenza A/PR8/34 neutralized wells, pool of erythrocytes at bottom of well) and non-neutralized virus (influenza A/PR8/34 positive in well, agglutination or fuzzy well is present) are shown for each immunization group in panel b. (c and d) Systemic coimmunization with mucosal chemokines protects mice from influenza A/PR8/34 intranasal challenge. Average percent weight loss (morbidity) and death (survival) in groups of mice immunized and challenged. Vector-immunized mice (diamonds) lost weight rapidly (c) and died (n=10) by day 9 post infection (d). Mice that were immunized with pPR8HA (triangles) alone lost an average 18% weight (c) by day 10, whereas 80% died (d) by day 10 post challenge. Mice immunized with CCR10 ligands, pCCL27 and pCCL28, were protected from weight loss (squares and circles, respectively, c) and were protected from mortality (d) in the days after challenge. Using log-rank analysis, pVAX vs pR8HA (P=0.0005), pVAX vs CCL27 (P<0.0001) and pVAX vs CCL28 (P<0.0001); pR8HA vs pR8HA/CCL27 (P=0.0003) and pR8HA vs pR8HA/CCL28 (P=0.0003).
Lethal influenza A intranasal challenge
Three immunizations with pCCL27 or pCCL28 in combination with pPR8HA were carried out on groups of 10 mice each, with each injection separated by 2 weeks (immunization schedule shown in Figure 3a). After an 8-week rest, the mice were challenged intranasally under anesthesia, as described in Materials and methods. After challenge, we observed that vector-immunized animals lost a significant amount of weight by day 5 and 100% of those mice succumbed to infection by day 9 post challenge (Figures 4c and d). In addition, mice immunized with a suboptimal dose of pPR8HA alone lost weight by day 9 p.i., and only 20% of the vaccinated animals survived to day 12 p.i. Animals that received adjuvant vaccines containing pCCL27 or pCCL28 exhibited better protection from weight loss and overall complete survival protection from challenge (Figures 4c and d). These data illustrate the ability of CCR10 ligand adjuvants to enhance protective immunity in this lethal mucosal challenge system.
Discussion
In combination with antigenic plasmids, genetic adjuvants encoding cytokines or chemokines represent a means of modifying the magnitude, duration and nature of the immune response generated in vivo.26,27,38-40 Few studies have measured the effects of genetic adjuvants encoding chemokines on induction of systemic and local immunity and protection, and specifically, none has examined the effects of the interesting CCR10 ligands, CCL27 and CCL28. Expression of CCR10 and its ligands within mucosal and epithelial tissue at the gastrointestinal and lung bronchial sites has been reported to have an important role in the trafficking of specific immune cells to these sites of pathogen exposure.28-31,33,41 Specifically, CCR10, in response to signals delivered by CCL28 and CCL27, contributes to the broader localization of IgA antibody-secreting cells to the small intestine, colon and bronchi.32,33 CCR10 is also expressed on memory CD4+/CLA+ T cells, a marker that is found on populations of skin-homing circulating lymphocytes found to be enriched in cutaneous inflammatory sites as well as in the oral mucosa.28 It is well established that activation or differentiation of T cells can alter the expression of specific chemokine receptors and thereby increase their sensitivity or responsiveness to cognate chemokine ligands.42-44 In addition, chemokines and their receptors can be upregulated during inflammatory stimulation, leading to preferential recruitment of effector lymphocytes to specific tissue microenvironments.45,46
We analyzed a systemically administered DNA vaccination strategy characterized by intramuscular plasmid vaccination with codelivery of CCL27 or CCL28 plasmid adjuvants. We observed that these adjuvants exhibited both systemic and mucosal effects. For example, we observed elevated peripheral IFN-γ production from T cells and enhancement of antigen-specific IgG from B cells systemically. We also observed coordinate enhancement of antigen-specific T-cell secretion of cytokine and antibody production in the gut-associated lymphoid tissue and lung. Moreover, the induction of long-lived antibody responses that can neutralize influenza A/PR8/34 and protect mice from morbidity and mortality associated with a lethal intranasal viral challenge was observed. Plasmid-derived CCL27 and CCL28 elicited antigen-specific humoral and cellular immune responses at important mucosal sites as determined by increased antigen-specific production of IgA in fecal extracts, and increased antigen-specific IFN-γ secretion from T cells collected from the lung. In addition, it is important to note that the ratio of IgA to IgG in sera was also positively skewed to a greater degree in chemokine-adjuvant vaccinated mice. The clear distinction in IgA-modulating activity was evident in fecal IgA measurements as well as in serum IgA assays, suggesting that these adjuvants seem particularly useful in the enhancement of generation of antigen-specific IgA.
Further studies analyzing the immunobiology of the plasmid adjuvants, CCL27 and CCL28, are warranted to further understand their effects on induction of DNA vaccine-induced antigen-specific mucosal immunity. The mucosal tissues are one of the main interfaces between environmental antigens and the immune system and are comprised of a large and highly specialized innate and adaptive arm that protects the mucosal surface against potential pathogens and harbors chronic infections, contributing almost 80% of all lymphocytes that are accumulated in the mucosa-associated lymphoid tissue. Therefore, the development of effective mucosal chemokine DNA vaccine adjuvants that can be delivered parenterally has implications for prophylactic and therapeutic immunization strategies that require both peripheral and mucosal physiologically relevant antigen-specific immunity. Importantly, infections of the lung may be particularly interesting to target with this unique strategy.
Materials and methods
RNA isolation and reverse transcriptase-PCR analysis of murine CCR10 expression
For analysis of CCR10 expression, RNA was extracted from the spleen, MLN, or PP from BALB/c or CCR10KO mice37 (kindly provided by Dr Olivier Morteau and Dr Craig Gerard of Harvard Medical School, Perlmutter Laboratory and Department of Pediatrics). PP were obtained from the duodenum and ileum (small intestine), pooled and disrupted using scissors, tweezers and steel cloth mesh. To obtain a single cell suspension, the lungs, spleens and MLNs from each experimental group were disrupted using a Seward Stomacher 80 (Metrohn, Riverview, FL, USA), put through a 40 μm cell strainer, washed with medium, pelleted and incubated for 5–10 min at room temperature in ACK lysing buffer resulting in the lysis of red blood cells. Using this protocol, more than 95% viable cells are routinely harvested from tissues. Total RNA was extracted from cells isolated from various tissues using the Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA, USA). RNA was eluted in water and RNA concentrations were measured by spectrophotometry. Reverse transcriptase-PCR was performed using the First Strand cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA) using oligo dt (12–18) primers and superscript II to generate cDNA strands (Invitrogen). For PCR, the sense primer used for amplification of wild-type CCR10 was designed and carried out as described by Morteau et al.37 Briefly, primers are specific for the region upstream from the site of insertion of the targeting construct (ATG codon): 5′-CGGAGAAACCCTTGTAGCCAG-3′. The antisense primer was designed 215 bp downstream from the sense primer: 5′-GGCCAAGACTAGGCCATTGCC-3′. The PCR was carried out using a Veriti Thermal Cycler (Applied Biosystems, Foster City, CA, USA) in which conditions were run as reported.37 Specifically, RNA was reverse transcribed for 30 min at 50 °C, and the reverse transcriptase was inactivated by 15-min incubation at 95 °C. The amplification profile was 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s, followed by a 10-min extension at 72 °C. Sense and antisense primers were also designed for the amplification of β-actin (350 bp) for loading controls.
DNA plasmids
For the antigenic plasmids, DNA vaccine constructs expressing influenza A strain PR8 HA protein (pPR8HA) were prepared as previously described.4,6,27,47 The chemokines CCL27 and CCL28 (sequence from Gene Bank, Accession number NM011336 and NM020279, respectively) were cloned from RNA extracted from murine ear skin (CCL27) or murine small intestine (CCL28). The PCR was carried out using a Veriti Thermal Cycler (Applied Biosystems) using the following primers and amplification conditions:
The primers contain the following bases and include flanking restriction sites (EcoR1 and Xho1).
pCCL27 sense: GCCCCCGAATTCGCCGCCATGATGCAGCAAGCAGGGCTCACA
pCCL27 antisense: ATCGGGCTCGAGTTAGTTTTGCTGTTGGGGGTTTG
pCCL28 sense: GCCCCCGAATTCGCCGCCACCATGCAGCAAGCAGGGCTCACA
pCCL28 antisense: ATCGGGCTCGAGCTAACGAGAGGCTTCGTGCCT
PCR conditions for all of the reactions were as follows: 1 cycle at 97 °C for 3 min, 30 cycles at 94 °C for 1 min, 55 °C for 1.15 min and 72 °C for 1.15 min, followed by a final extension at 72 °C for 10 min. The 363 (CCL27) and 393 (CCL28) base pair PCR products were, respectively, ligated into the pVAX1 cloning vector (Invitrogen) after restriction enzyme digestion using EcoR1 and Xho1 (New England Biolabs, Ipswich, MA, USA), which were designed into the PCR primers and are in the multiple cloning region of the vector. All positive clones were verified by sequence analysis.
Plasmid transfection and expression analysis
Expression levels of the plasmid construct were tested after transient transfection of RD cells (ATCC). Cells were plated in 60×15 mm tissue culture dishes at a density of 2.5×105 cells per well in complete Dulbecco’s modified Eagle’s medium plus 10% fetal bovine serum (HyClone, Thermo Scientific, Waltham, MA; GIBCO, Carlsbad, CA, USA) and allowed to adhere overnight. The next day cells were transfected with CCL27 or CCL28 plasmid (3 μg per well) using FuGENE 6 Transfection Reagent (Roche Diagnostics, Indianapolis, IN, USA) according to the protocol provided by the manufacturer. After 48 h, cell supernatants were harvested and analyzed for the presence of murine CCL27 or CCL28 protein by commercial ELISA kits (R&D Systems, Minneapolis, MN, USA).
Chemotaxis assays
Chemotaxis assays were performed using 5-μm Costar Transwell plates (Corning Life Sciences, Lowell, MA, USA). Splenocytes, MLN, PP/cryopatches and lung lymphocytes were isolated from naive BALB/c mice and were placed in the upper wells (2×106 per well), whereas bottom wells contained medium alone or medium from 48-h harvested supernatants from cell lines transfected as above with pCCL27 or pCCL28. After incubation at 37 °C for 2 h, migrated cells in the lower chamber from multiple replicate wells (generally four to five wells per condition) were collected and counted. The data are graphed as a migration index, which is the fold increase in the number of cells migrated in experimental wells divided by the number of cells migrated in background, medium control wells. The chemotaxis experiments were carried out over three independent trials. Data generated from the independent experiments and replicate wells were averaged and shown in Figure 1.
In vitro translation assay
The TNT T7 Coupled Reticulocyte Lysate System (Promega, Madison, WI, USA) and [35S] methionine were used to create labeled CCL27 and CCL28 protein products. pVAX vector alone (negative control), pCCL27 or pCCL28, and [35S] methionine were added to the reaction mixture according to the instructions supplied by the manufacturer. The reaction was carried out at 30 °C for 1 h. Labeled proteins were detected using 5 μg purified monoclonal anti-CCL27 or anti-CCL28 (R&D Systems) at 4 °C with rotation overnight in RIPA buffer. Approximately 5 mg of protein G–sepharose beads (Amersham Biosciences, Piscataway, NJ, USA) (50 μl of 100 mg ml−1 stock) were added to each reaction, and the samples were incubated at 4 °C, with rotation, for 2 h. The beads were washed thrice with binding buffer containing high salt and bovine serum albumin and finally suspended in 2× sample buffer. The protein complexes were eluted from sepharose beads by boiling for 5 min and were electrophoresed on a 12% SDS-polyacrylamide gel electrophoresis gel (Cambrex Corp., East Rutherford, NJ, USA). The gel was fixed, treated with amplifying solution (Amersham Biosciences) and dried for 2 h in a Bio-Rad gel dryer (Bio-Rad, Hercules, CA, USA). The dried gel was exposed to X-ray film at −80 °C overnight and developed using a Kodak automatic developer.
Plasmid immunization and mice
The quadriceps muscles of 6- to 8-week-old female BALB/c mice (The Jackson Laboratory, Bar Harbor, ME, USA) were injected with plasmid DNA formulations containing combinations of vector backbone (pVAX), chemokine adjuvant plasmids (100 μg) and antigenic plasmids (33 μg of pPR8HA, an HA containing plasmid) as described in the figure legends and earlier publications.4,48 Formulations contained 0.25% bupivacaine HCL (Sigma-Aldrich, St Louis, MO, USA) in isotonic citrate buffer. Coadministration of various gene plasmids involved mixing DNA plasmids so that each experimental group contained the same concentration of DNA (vector backbone added to make all groups equal DNA concentrations) before injection in a final volume of 100 μl. All DNA was generated using endofree plasmid maxiprep kit (Qiagen). All animals were housed in a temperature-controlled, light-cycled facility at the University of Pennsylvania, and their care was under the guidelines of the National Institutes of Health and the University of Pennsylvania.
Histology of muscle biopsies from immunized animals
Injected murine quadriceps muscles were excised 7 days after immunization and frozen immediately in OCT medium in a dry ice and methanol bath. Frozen muscles were cut into approximately 1000 serial sections of 5-μm thickness, air dried and fixed for 10 min in 100% acetone. Every other serial section was stained with hematoxylin and eosin before dehydration, mounting and examination for the presence and extent of cellular inflammatory infiltrates. Once the injection site was identified by cellular inflammation (hematoxylin and eosin stain), additional sections corresponding to the injection site were used for immunohistochemical analysis as follows: acetone-fixed 5-μm sections were treated with 0.5% hydrogen peroxide in PBS for 15 min to quench endogenous peroxidases. The sections were washed with PBS, and free biotin was blocked using the avidin+biotin blocking system (Vector Laboratories, Burlingame, CA, USA). Sections were then incubated with the primary antibodies at room temperature for 1 h. For chemokine receptor detection, monoclonal antibodies to mouse CCR10 were used (Abcam, Cambridge, MA, USA), and costained with secondary antibody labeled with biotin. Isotype control antibodies were also used (Pharmingen, BD Biosciences, San Jose, CA, USA). After incubation, the slides were washed thrice with PBS and then developed with the Vectastain Elite ABC kit and Vector DAB substrate (Vector Laboratories). After substrate development, the sections were washed in water and counter-stained with hematoxylin. Slides were rinsed in water, and coverslips were mounted using Histomount (Invitrogen). Immunohistochemical support was provided by Children’s Hospital of Philadelphia, Department of Pathology, Philadelphia, Pennsylvania. The assessment of infiltrating cells was done randomly and blinded by at least two separate investigators. The experiment was repeated thrice. We observed similar results in vaccination studies targeting the tibialis anterior muscle, a small muscle routinely used for immunizations.
Method for killing mouse, tissue or sample harvest and cell purification for immune analysis
At end points designated in the legends, the animals were sedated using Avertin (Sigma-Aldrich) and appropriate amounts of blood and fresh fecal pellets (pooled or run individually as designated in the legend) were taken before the animals were killed. After the animals were killed, the spleen, lung, MLN and PP/colonic cryopatches from each mouse were harvested from each experimental group, and tissue was either pooled or run individually, as designated in the figure legends, into a 15-ml conical tube containing R10 medium (RPMI1640 plus 10% fetal bovine serum, 500 μg l−1 penicillin and 500 μg l−1 streptomycin) (GIBCO; Invitrogen). PPs were obtained from the duodenum and ileum (small intestine), pooled and disrupted using scissors, tweezers and steel cloth mesh. The lungs, spleens and MLNs from each experimental group were crushed into a single cell suspension using a Seward Stomacher 80 (Metrohn), put through a 40-μm cell strainer, washed with medium, pelleted and incubated for 5–10 min at room temperature in ACK lysing buffer, resulting in the lysis of red blood cells. All cells were washed, resuspended in medium and counted (cell viability was determined using trypan blue stain) using a hemacytometer.
IFN-γ ELISpot
IFN-γ ELISpot was performed as previously described.4 Briefly, ELISpot Millipore 96-well plates (Millipore, Billerica, MA, USA) were coated with an R&D Systems anti-mouse IFN-γ capture antibody and incubated for 24 h at 4 °C. A total of 2×105 splenocytes from immunized mice (n=4 mice per group) were added to each well of an ELISpot plate and stimulated at 37 °C 5% CO2, in the presence of R10 (negative control), concanavalin A (positive control) or specific peptide (HA) antigens (10 μg ml−1). Cells were stimulated with the CD8+ T-cell epitope peptide encoding influenza A/PR8/34 (H1N1) HA (IYSTVASSL amino acid 518–526) synthesized by IDS (Fountain Hills, AZ, USA). After 24 h of stimulation, the plates were washed and incubated overnight at 4 °C with biotinylated anti-mouse IFN-γ antibody (R&D Systems). The plates were subsequently washed and streptavidin–alkaline phosphatase (R&D Systems) was added to each well and incubated for 2 h at room temperature. The plates were washed, and BCIP (5-bromo-4-chloro-3′ indolyl phosphate p-toluidine salt) and NBT (nitro blue tetrazolium chloride) (R&D Systems) were added to each well. Pooled spleen samples from four mice were used in these experiments because previous experiments in our laboratory determined small variability and consistent responses between BALB/c mice within an experimental group receiving vaccination with pPR8HA constructs.
Processing fecal extract for antibody measurement
Fresh fecal pellets were collected from vaccinated mice and pooled for each experimental group. Stool was dissolved in the following buffer in a specific weight/volume ratio. In total, 1 g of thawed stool was dissolved with 4 ml of PBS pH 7.5 [0.05% Tween 20, soybean trypsin inhibitor (100 μg ml−1), EDTA (0.05 M)+phenyl-methylsulfonylfluoride at 10 mM]. The suspension was incubated for 15 min with frequent vortexing, and the sediment was pelleted by centrifugation at 1200 r.p.m. for 5 min. The fecal supernatant was filtered using a 70-μm strainer and centrifuged again at 16 000 g for 15 min. Supernatants were further diluted in 1% BSA or frozen in 0.1% BSA/0.02% sodium azide.
Analysis of IgG and IgA binding antibodies by ELISA
ELISA was used to determine influenza A/PR8/34 HA-specific IgG and IgA in mouse serum and fecal extract as described.49,50 Blood samples were harvested from mice by retro-orbital bleeds. To measure effector (1 week post final immunization) anti-influenza IgG and IgA responses, serum and feces were collected for analysis by ELISA. The data shown Table 1 are from a representative experiment of six independent experiments. Pooled fecal samples from four mice were used to measure effector phase responses, whereas for the influenza challenge study, mouse serum was obtained from 10 mice per experimental group, processed and run individually in ELISA analysis as designated in Figures 3 and 4 and Table 1. Corning EIA/RIA plates were coated with UV-inactivated influenza A/PR8/34 diluted in PBS (Mediatech, Manassas, VA, USA) at a final volume of 100 μl per well and incubated overnight at 4°C. Plates were washed with PBS/Tween (0.05% Tween 20) thrice and blocked against non-specific binding with 200 μl of blocking buffer/diluent (3% BSA in PBS) for 2 h at room temperature. The plates were washed, and pooled sera or fecal extract from immunized mice was diluted in PBS/1% BSA and was added to wells at a final volume of 100 μl. Samples were added in triplicate, at dilutions from 1 to 25 to 1 to 3200 dilution series to determine end point titer (defined as the lowest sample dilution at which measurements for an experimental group are equal to those for vector-immunized mouse sample OD values) and incubated at room temperature for 2 h or overnight at 4 °C. Bound antibodies were detected with horseradish peroxidase-labeled goat anti-mouse IgG or IgA (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and developed with substrate TMB H2O2 (Sigma-Aldrich). The color reaction was stopped with 2n H2SO4, and the absorbance at 450 nm was read in an EL312 Bio-Kinetics microplate reader (BioTek Instruments, Winooski, VT, USA). The amount of total IgG or IgA in serum or fecal secretions was calculated by interpolating the optical densities on calibration curves using the DeltaSoft II program (BioMetallics, Princeton, NJ, USA).
Virus titer and virus neutralization assay
The virus neutralization was performed in microtiter plates as described previously.51 The influenza A/PR8/34 (H1N1) was diluted in PBS, and 100-μl aliquots were dispensed into wells of flat-bottom microtiter plates (105.6 TCID50 per ml or 100TCID50 virus per well, 2000TCID50 per ml) that contained 50 μl of serum from immunized mice (n=10), positive controls and negative control serum dilutions, with 6–12 replicates per dilution. After 1 h at 37 °C, 50 μl of this mixture was added to a monolayer of MDCK (Madin-Darby canine kidney) epithelial cells, plated at a cell density of 2.5×106 cells per ml in ISCCM–0.1% BSA medium. After 8 h, medium containing trypsin (32 μg ml−1) was added to each well, and the cultures were incubated for an additional 2.5 days. Culture supernatants (25 μl) were tested by hemaglutination assay for the presence of virus. Using round-bottom polystyrene microtiter plates, culture supernatants were mixed with 25 μl of chicken erythrocytes (RBC) (1% in PBSN), and the pattern of RBC sedimentation was recorded after the plates were incubated for 35 min at room temperature. The neutralization titer is defined as the dilution of serum or antibody sample at which infection with 2000TCID50 per ml of influenza A/PR8/34 is neutralized in 50% of the culture.
HA/Flu challenge model
At 8 weeks after the last boost, the cohorts were challenged intranasally with a dose of 10 LD50 (2000TCID50) live influenza A/PR8/34. The animals were temporally anesthetized with anesthetic (Avertin) given by i.p. injection and subsequently processed for influenza A/PR8/34 infection. In total, 30 μl of dosed virus diluted in PBS was applied to the nares of anesthetized mice, resulting in the aspiration of the virus into upper and lower airways. Each day after infection, a decrease in total body weight was measured as a surrogate of morbidity and survival was noted as a measure of mortality (Figure 4). Challenge studies used 10 animals per experimental group and were repeated twice.
Statistical analysis
Data are presented as the mean±s.d. calculated from triplicate wells of pooled lymphocytes from each experimental group. Where appropriate, the statistical difference between immunization groups was assessed using a two-tailed, paired Student’s t-test that yielded a specific P value for each experimental group. Comparisons between samples with a P value <0.05 were considered to be statistically different and therefore significant. A Kaplan–Meier survival analysis was used to depict results for the influenza challenge study, and statistical differences between vaccination groups were determined to be statistically different using a log-rank test.
Acknowledgements
We would like to thank the University of Pennsylvania Department of Pathology Histology Core and Daniel Martinez for their help with immunohistochemical analyses. We would also like to thank Dr Jean D Boyer for her scientific guidance. CCR10 knockout mice were a generous gift from Drs Craig Gerard and Olivier Morteau. Amy Quinn and Carolyn Clarke provided molecular cloning expertise. In addition, we would like to acknowledge Dr Walter Gerhard, Krystyna Mozdzanowska and Darya Zharikova for their helpful comments and expertise on influenza A/PR8/34 mouse studies and neutralization assays. We would like to thank Mr Albert Sylvester, Ms Noshin Kathuria, Philip Choe and Michelle Nater for their help in the mucosal immune assays and analysis. Dr Jiri Mestecky also contributed scientific guidance for the humoral analysis on mucosal tissue and fecal extracts. We would also like to acknowledge Pamela Fried and Diana Winters from Drexel University College of Medicine Academic Publishing Services for their editorial, formatting and journal submission expertise. This work is supported by grants funded through the National Institutes of Health including an F32AI054152 through NIAIDS (MK), an N01A1154 NIAIDS-HVDDT (DBW), an NIH-NIAID-HIVRAD grant (DBW) and a T32-A107632 (KAK).
Footnotes
Conflict of interest
The laboratory notes possible commercial interests associated with this work, which may include Wyeth, VGX, Inovio, BMS, Virxsys, Ichor, Merck, Althea and Aldevron.
References
- 1.Schoenly KA, Weiner DB. HIV-1 vaccine development: recent advances in the CTL platform ‘spotty business’. J Virol 2008; 82: 3166–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat Rev Genet 2008; 9: 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kodihalli S, Kobasa DL, Webster RG. Strategies for inducing protection against avian influenza A virus subtypes with DNA vaccines. Vaccine 2000; 18: 2592–2599. [DOI] [PubMed] [Google Scholar]
- 4.Kutzler MA, Robinson TM, Chattergoon MA, Choo DK, Choo AY, Choe PY et al. Coimmunization with an optimized IL-15 plasmid results in enhanced function and longevity of CD8 T cells that are partially independent of CD4 T cell help. J Immunol 2005; 175: 112–123. [DOI] [PubMed] [Google Scholar]
- 5.Chattergoon MA, Saulino V, Shames JP, Stein J, Montaner LJ, Weiner DB. Co-immunization with plasmid IL-12 generates a strong T-cell memory response in mice. Vaccine 2004; 22: 1744–1750. [DOI] [PubMed] [Google Scholar]
- 6.Chattergoon MA, Kim JJ, Yang JS, Robinson TM, Lee DJ, Dentchev T et al. Targeted antigen delivery to antigen-presenting cells including dendritic cells by engineered Fas-mediated apoptosis. Nat Biotechnol 2000; 18: 974–979. [DOI] [PubMed] [Google Scholar]
- 7.Barouch DH, Letvin NL, Seder RA. The role of cytokine DNAs as vaccine adjuvants for optimizing cellular immune responses. Immunol Rev 2004; 202: 266–274. [DOI] [PubMed] [Google Scholar]
- 8.Lori F, Weiner DB, Calarota SA, Kelly LM, Lisziewicz J. Cytokine-adjuvanted HIV-DNA vaccination strategies. Springer Semin Immunopathol 2006; 28: 231–238. [DOI] [PubMed] [Google Scholar]
- 9.Ulmer JB, Wahren B, Liu MA. Gene-based vaccines: recent technical and clinical advances. Trends Mol Med 2006; 12: 216–222. [DOI] [PubMed] [Google Scholar]
- 10.Fuller DH, Loudon P, Schmaljohn C. Preclinical and clinical progress of particle-mediated DNA vaccines for infectious diseases. Methods 2006; 40: 86–97. [DOI] [PubMed] [Google Scholar]
- 11.Hokey DA, Weiner DB. DNA vaccines for HIV: challenges and opportunities. Springer Semin Immunopathol 2006; 28: 267–279. [DOI] [PubMed] [Google Scholar]
- 12.Lee S, Gierynska M, Eo SK, Kuklin N, Rouse BT. Influence of DNA encoding cytokines on systemic and mucosal immunity following genetic vaccination against herpes simplex virus. Microbes Infect 2003; 5: 571–578. [DOI] [PubMed] [Google Scholar]
- 13.Kaneko H, Bednarek I, Wierzbicki A, Kiszka I, Dmochowski M, Wasik TJ et al. Oral DNA vaccination promotes mucosal and systemic immune responses to HIV envelope glycoprotein. Virology 2000; 267: 8–16. [DOI] [PubMed] [Google Scholar]
- 14.Kusakabe K, Xin KQ, Katoh H, Sumino K, Hagiwara E, Kawamoto S et al. The timing of GM-CSF expression plasmid administration influences the Th1/Th2 response induced by an HIV-1-specific DNA vaccine. J Immunol 2000; 164: 3102–3111. [DOI] [PubMed] [Google Scholar]
- 15.Biragyn A, Belyakov IM, Chow YH, Dimitrov DS, Berzofsky JA, Kwak LW. DNA vaccines encoding human immunodeficiency virus-1 glycoprotein 120 fusions with proinflammatory chemoattractants induce systemic and mucosal immune responses. Blood 2002; 100: 1153–1159. [DOI] [PubMed] [Google Scholar]
- 16.Bagarazzi ML, Boyer JD, Javadian MA, Chattergoon MA, Shah AR, Cohen AD et al. Systemic and mucosal immunity is elicited after both intramuscular and intravaginal delivery of human immunodeficiency virus type 1 DNA plasmid vaccines to pregnant chimpanzees. J Infect Dis 1999; 180: 1351–1355. [DOI] [PubMed] [Google Scholar]
- 17.Wang B, Dang K, Agadjanyan MG, Srikantan V, Li F, Ugen KE et al. Mucosal immunization with a DNA vaccine induces immune responses against HIV-1 at a mucosal site. Vaccine 1997; 15: 821–825. [DOI] [PubMed] [Google Scholar]
- 18.Liu MA, Wahren B, Karlsson Hedestam GB. DNA vaccines: recent developments and future possibilities. Hum Gene Ther 2006; 17: 1051–1061. [DOI] [PubMed] [Google Scholar]
- 19.Dhama K, Mahendran M, Gupta PK, Rai A. DNA vaccines and their applications in veterinary practice: current perspectives. Vet Res Commun 2008; 32: 341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bergman PJ. Canine oral melanoma. Clin Tech Small Anim Pract 2007; 22: 55–60. [DOI] [PubMed] [Google Scholar]
- 21.Martin JE, Pierson TC, Hubka S, Rucker S, Gordon IJ, Enama ME et al. A West Nile virus DNA vaccine induces neutralizing antibody in healthy adults during a phase 1 clinical trial. J Infect Dis 2007; 196: 1732–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu MA, Ulmer JB. Human clinical trials of plasmid DNA vaccines. Adv Genet 2005; 55: 25–40. [DOI] [PubMed] [Google Scholar]
- 23.McKay PF, Barouch DH, Santra S, Sumida SM, Jackson SS, Gorgone DA et al. Recruitment of different subsets of antigen-presenting cells selectively modulates DNA vaccine-elicited CD4+ and CD8+ T lymphocyte responses. Eur J Immunol 2004; 34: 1011–1020. [DOI] [PubMed] [Google Scholar]
- 24.Kutzler MA, Weiner DB. Developing DNA vaccines that call to dendritic cells. J Clin Invest 2004; 114: 1241–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyer JD, Kim J, Ugen K, Cohen AD, Ahn L, Schumann K et al. HIV-1 DNA vaccines and chemokines. Vaccine 1999; 17 (Suppl 2): S53–S64. [DOI] [PubMed] [Google Scholar]
- 26.Sin J, Kim JJ, Pachuk C, Satishchandran C, Weiner DB. DNA vaccines encoding interleukin-8 and RANTES enhance antigen-specific Th1-type CD4(+) T-cell-mediated protective immunity against herpes simplex virus type 2 in vivo. J Virol 2000; 74: 11173–11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JJ, Yang JS, Dentchev T, Dang K, Weiner DB. Chemokine gene adjuvants can modulate immune responses induced by DNA vaccines. J Interferon Cytokine Res 2000; 20: 487–498. [DOI] [PubMed] [Google Scholar]
- 28.Morales J, Homey B, Vicari AP, Hudak S, Oldham E, Hedrick J et al. CTACK, a skin-associated chemokine that preferentially attracts skin-homing memory T cells. Proc Natl Acad Sci USA 1999; 96: 14470–14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunkel EJ, Butcher EC. Chemokines and the tissue-specific migration of lymphocytes. Immunity 2002; 16: 1–4. [DOI] [PubMed] [Google Scholar]
- 30.Reiss Y, Proudfoot AE, Power CA, Campbell JJ, Butcher EC. CC chemokine receptor (CCR)4 and the CCR10 ligand cutaneous T cell-attracting chemokine (CTACK) in lymphocyte trafficking to inflamed skin. J Exp Med 2001; 194: 1541–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soler D, Humphreys TL, Spinola SM, Campbell JJ. CCR4 versus CCR10 in human cutaneous TH lymphocyte trafficking. Blood 2003; 101: 1677–1682. [DOI] [PubMed] [Google Scholar]
- 32.Lazarus NH, Kunkel EJ, Johnston B, Wilson E, Youngman KR, Butcher EC. A common mucosal chemokine (mucosae-associated epithelial chemokine/CCL28) selectively attracts IgA plasmablasts. J Immunol 2003; 170: 3799–3805. [DOI] [PubMed] [Google Scholar]
- 33.Pan J, Kunkel EJ, Gosslar U, Lazarus N, Langdon P, Broadwell K et al. A novel chemokine ligand for CCR10 and CCR3 expressed by epithelial cells in mucosal tissues. J Immunol 2000; 165: 2943–2949. [DOI] [PubMed] [Google Scholar]
- 34.Feng N, Jaimes MC, Lazarus NH, Monak D, Zhang C, Butcher EC et al. Redundant role of chemokines CCL25/TECK and CCL28/MEC in IgA+ plasmablast recruitment to the intestinal lamina propria after rotavirus infection. J Immunol 2006; 176: 5749–5759. [DOI] [PubMed] [Google Scholar]
- 35.Hieshima K, Kawasaki Y, Hanamoto H, Nakayama T, Nagakubo D, Kanamaru A et al. CC chemokine ligands 25 and 28 play essential roles in intestinal extravasation of IgA antibody-secreting cells. J Immunol 2004; 173: 3668–3675. [DOI] [PubMed] [Google Scholar]
- 36.Nakayama T, Hieshima K, Izawa D, Tatsumi Y, Kanamaru A, Yoshie O. Cutting edge: profile of chemokine receptor expression on human plasma cells accounts for their efficient recruitment to target tissues. J Immunol 2003; 170: 1136–1140. [DOI] [PubMed] [Google Scholar]
- 37.Morteau O, Gerard C, Lu B, Ghiran S, Rits M, Fujiwara Y et al. An indispensable role for the chemokine receptor CCR10 in IgA antibody-secreting cell accumulation. J Immunol 2008; 181: 6309–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calarota SA, Weiner DB. Enhancement of human immunodeficiency virus type 1-DNA vaccine potency through incorporation of T-helper 1 molecular adjuvants. Immunol Rev 2004; 199: 84–99. [DOI] [PubMed] [Google Scholar]
- 39.Shedlock DJ, Weiner DB. DNA vaccination: antigen presentation and the induction of immunity. J Leukoc Biol 2000; 68: 793–806. [PubMed] [Google Scholar]
- 40.Xu R, Megati S, Roopchand V, Luckay A, Masood A, Garcia-Hand D et al. Comparative ability of various plasmid-based cytokines and chemokines to adjuvant the activity of HIV plasmid DNA vaccines. Vaccine 2008; 26: 4819–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Homey B, Alenius H, Muller A, Soto H, Bowman EP, Yuan W et al. CCL27-CCR10 interactions regulate T cell-mediated skin inflammation. Nat Med 2002; 8: 157–165. [DOI] [PubMed] [Google Scholar]
- 42.Loetscher P, Seitz M, Baggiolini M, Moser B. Interleukin-2 regulates CC chemokine receptor expression and chemotactic responsiveness in T lymphocytes. J Exp Med 1996; 184: 569–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sallusto F, Schaerli P, Loetscher P, Schaniel C, Lenig D, Mackay CR et al. Rapid and coordinated switch in chemokine receptor expression during dendritic cell maturation. Eur J Immunol 1998; 28: 2760–2769. [DOI] [PubMed] [Google Scholar]
- 44.Sallusto F, Mackay CR, Lanzavecchia A. Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells. Science 1997; 277: 2005–2007. [DOI] [PubMed] [Google Scholar]
- 45.Olson TS, Ley K. Chemokines and chemokine receptors in leukocyte trafficking. Am J Physiol Regul Integr Comp Physiol 2002; 283: R7–R28. [DOI] [PubMed] [Google Scholar]
- 46.Lukacs NW. Migration of helper T-lymphocyte subsets into inflamed tissues. J Allergy Clin Immunol 2000; 106: S264–S269. [DOI] [PubMed] [Google Scholar]
- 47.Robinson HL, Boyle CA, Feltquate DM, Morin MJ, Santoro JC, Webster RG. DNA immunization for influenza virus: studies using hemagglutinin- and nucleoprotein-expressing DNAs. J Infect Dis 1997; 176 (Suppl 1): S50–S55. [DOI] [PubMed] [Google Scholar]
- 48.Kim JJ, Nottingham LK, Sin JI, Tsai A, Morrison L, Oh J et al. CD8 positive T cells influence antigen-specific immune responses through the expression of chemokines. J Clin Invest 1998; 102: 1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogawa T, Tarkowski A, McGhee ML, Moldoveanu Z, Mestecky J, Hirsch HZ et al. Analysis of human IgG and IgA subclass antibody-secreting cells from localized chronic inflammatory tissue. J Immunol 1989; 142: 1150–1158. [PubMed] [Google Scholar]
- 50.Mestecky J, Jackson S, Moldoveanu Z, Nesbit LR, Kulhavy R, Prince SJ et al. Paucity of antigen-specific IgA responses in sera and external secretions of HIV-type 1-infected individuals. AIDS Res Hum Retroviruses 2004; 20: 972–988. [DOI] [PubMed] [Google Scholar]
- 51.Mozdzanowska K, Feng J, Eid M, Zharikova D, Gerhard W. Enhancement of neutralizing activity of influenza virus-specific antibodies by serum components. Virology 2006; 352: 418–426. [DOI] [PubMed] [Google Scholar]