Fig. 3 |. The structure of the T2T-Y centromere.
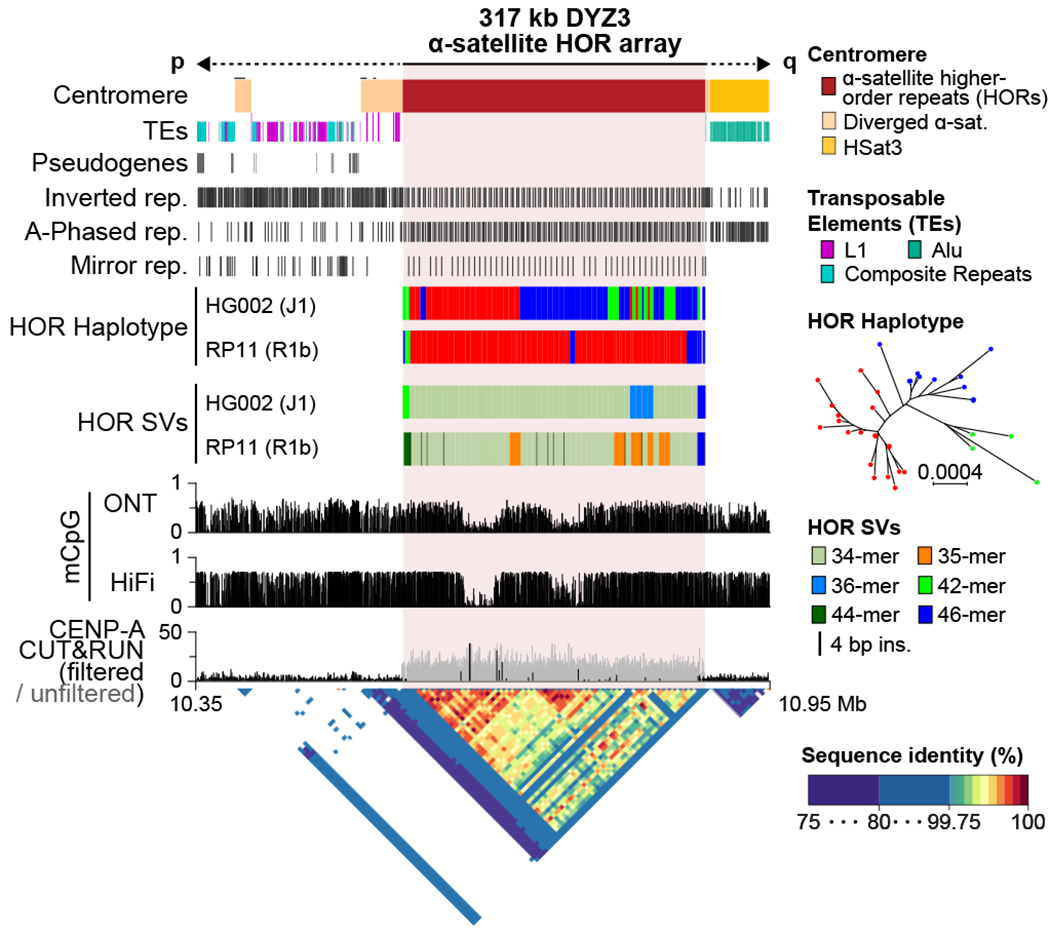
No TEs were found within the DYZ3 array, while L1s (upstream) and Alus (downstream) were found within the diverged alpha satellites (drawn taller than the other Tes). A periodic non-B DNA motif pattern is shown within the HOR array. The HG002-Y (T2T-Y) HOR haplotypes and SVs reveal a different long-range structure and organization compared to a previously assembled centromere from RP11-Y47. Three major HOR haplotypes were identified in HG002-Y based on their phylogenetic distance (red, blue, and green). RP11-Y has no 36-mer variants, but does have a number of 35-mers containing internal duplications. Histograms show the fraction of methylated CpG sites called by both ONT and HiFi, with two hypo-methylated centromeric dip regions (CDR) supported by CENP-A binding signal from CUT&RUN50. A StainedGlass dotplot illustrates high similarity within the HOR array (99.5–100%).
