Abstract
Rhizobial capsular polysaccharides (RKPs) play an important role in the development of a nitrogen-fixing symbiosis with the plant host and in Sinorhizobium meliloti AK631 functional rkpABCDEF genes are required for the production of RKPs. After cloning the rkpF gene, we overexpressed and purified the derived protein product (RkpF) in Escherichia coli. Like acyl carrier protein (ACP), the RkpF protein can be labeled in vivo with radioactive β-alanine added to the growth medium. If homogeneous RkpF protein is incubated with radiolabeled coenzyme A in the presence of purified holo-ACP synthase from E. coli, an in vitro transfer of 4′-phosphopantetheine to the RkpF protein can be observed. The conversion from apo-RkpF protein to holo-RkpF protein seems to go along with a major conformational change of the protein structure, because the holo-RkpF protein runs significantly faster on native polyacrylamide gel electrophoresis than the apo-RkpF protein. Electrospray mass spectrometric analysis reveals a mass of 9,585 for the apo-RkpF protein and a mass of 9,927 for the holo-RkpF protein. Our data show that RkpF is a novel ACP.
The interaction of bacteria of the genera Rhizobium, Sinorhizobium, Azorhizobium, and Bradyrhizobium, collectively called rhizobia, with the roots of leguminous host plants leads to the formation of a new organ, the nodule, in which atmospheric nitrogen is fixed. During the early events of nodulation, plant flavonoids induce the nod genes, which are rhizobial genes essential for nodulation. Most of the nod genes are involved in the production of lipo-chitin oligosaccharide signal molecules, which are able to induce nodule primordia on their respective host plants (31). The later events of nodule development leading to a mature, nitrogen-fixing nodule are less well understood. For the infection of host plants that form indeterminate nodules, the production of extracellular polysaccharides (EPSs) by the microsymbiont is generally thought to be an essential requirement. However, an exoB-deficient mutant of Sinorhizobium (formerly Rhizobium) meliloti 41, S. meliloti AK631, is unable to produce EPSs but still forms mature, nitrogen-fixing nodules (22). In S. meliloti AK631, a novel acidic polysaccharide that is rich in 3-deoxy-d-manno-2-octulosonic acid (Kdo) but different from classical lipopolysaccharide (24) can take over the functional role of EPSs with regard to the formation of nitrogen-fixing nodules in an exo mutant. This novel polysaccharide is structurally analogous to group II K antigens (capsular polysaccharides) of Escherichia coli. The production of such K-like antigens in Sinorhizobium (rhizobial capsular polysaccharides [RKPs]) can be modulated by certain flavonoids and by host root extract (25). Interestingly, the presence of RKPs is required for the induction of plant genes from the flavonoid biosynthetic pathway in alfalfa leaves (1), suggesting that in addition to lipo-chitin oligosaccharides, the RKPs might be another class of compounds functioning as rhizobial signals for the host plant.
For the production of RKPs in S. meliloti, the gene rkpZ (formerly lpsZ) and genes of the fix-23 region are required (26). Because rkpZ is normally absent in S. meliloti 1021 derivatives, such strains do not form capsular polysaccharides and therefore have an absolute requirement for exopolysaccharides. The fix-23 region consists of four complementation units, and complementation unit I was found to contain six genes (rkpABCDEF) coding for protein products that show a considerable degree of similarity and organization similar to those of the rat fatty acid synthase multifunctional enzyme domains (15, 19, 23). Complementation units II, III, and IV consist of four additional genes (rkpGHIJ), with one of the gene products (RkpG) sharing similarity with acyltransferases (15). This prediction of function in fatty acid biosynthesis and transfer is surprising, because the final capsular polysaccharide product seems to contain no fatty acyl residue nor any other β-ketide (24). The sixth gene of complementation unit I, now called rkpF (15, 23), was proposed to code for an acyl carrier protein (ACP) (19), and here we describe the functional properties of the RkpF protein.
Cloning of the rkpF gene.
Total genomic DNA harboring genes involved in rhizobial capsular polysaccharide synthesis (rkp) was prepared from S. meliloti AK631 (21) grown in TY medium (3) at 29°C. By using PCR and specific oligonucleo- tides (GGAATACATATGATCGAAGGCAAATCGCCG CAG and AAAGGATCCTCACGGCTTGCTTACTTTTTCATGG), an open reading frame (ORF 6, now rkpF) was amplified from genomic DNA with Pfu polymerase. Suitable restriction sites (underlined) for cloning the rkpF gene in expression vector pET 9a (35) were introduced by PCR with the oligonucleotides. After restriction with NdeI and BamHI, the PCR-amplified DNA fragment was cloned into a pET 9a vector to obtain the expression plasmid pTB1003, in which the rkpF gene can be overexpressed under control of the T7 promoter. The correct in-frame cloning and the correct published sequence (19) (EMBL database, accession no. X64131) were demonstrated by DNA sequencing (data not shown). E. coli BL21(DE3) (35), which expresses the T7 polymerase under the control of the lac promoter, was transformed with pTB1003.
Expression and purification of the RkpF protein.
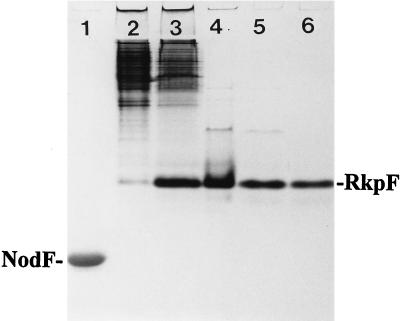
After E. coli BL21 (DE3) pTB1003 had been grown on complex Luria-Bertani (LB) medium in the presence of kanamycin (50 μg/ml) at 37°C on a gyratory shaker without or in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside [0.1 mM]), the protein profiles obtained after electrophoresis in nondenaturing (polyacrylamide gel electrophoresis [PAGE]) gels according the method of Jackowski and Rock (14) were very similar (Fig. 1). However, in the IPTG-induced situation, a major protein was visible which was present in much smaller amounts in the uninduced situation. The IPTG-inducible protein migrates much more slowly under such conditions than another rhizobial ACP, the NodF protein.
FIG. 1.
Overexpression and purification of RkpF protein. Purified ACP NodF (11) (lane 1) and cell extracts from E. coli BL21 (DE3) × pTB1003, grown without inducer (lane 2) or in the presence of IPTG (lane 3), dialyzed supernatant after a 60% isopropanol precipitation (lane 4), pooled fractions after chromatography on DE-52 (lane 5), and pooled fractions after chromatography on Sephadex G-50 (lane 6) were separated in a native 20% PAGE gel and stained with Coomassie blue.
For the purification of the IPTG-inducible protein, IPTG was added to a final concentration of 0.1 mM to a growing bacterial suspension of E. coli BL21 (DE3) × pTB1003 on LB medium at a density of 5 × 108 cells/ml. After 4 h of induction, cells were harvested and stored in a freezer at −20°C. Frozen cells (1.1 g) were suspended in 30 ml of buffer A (50 mM Tris-HCl [pH 6.8], 100 mM KCl) and passed three times through a French pressure cell at 20,000 lb/in2. The suspension was centrifuged at 6,800 × g for 30 min to obtain the cell extract. The cell extract was slowly stirred at 4°C, and isopropanol was added dropwise to a final concentration of 60% (vol/vol). After 60 min of incubation at 4°C, the precipitate obtained was removed by centrifugation at 6,800 × g for 30 min. The supernatant was dialyzed twice, each time for ca. 30 h against 1 liter of buffer B, which was a mixture of 50 mM Tris-HCl [pH 8.0] and 1 mM CHAPS {3-[(3-cholamidopropyl)-dimethyl-ammonio]-1-propanesulfonate}, and the dialysate was applied to a 30-ml column of DEAE-52–cellulose (Whatman), which had been equilibrated with buffer B. The column was washed with 90 ml of buffer B. Elution was performed with a linear gradient from 0 to 1 M NaCl in buffer B in a total volume of 90 ml. Fractions (1.5 ml) were collected, and aliquots were analyzed by PAGE. The RkpF protein eluted in a range from 0.15 to 0.3 M NaCl. Only those fractions in which the RkpF protein made up more than 90% of the total protein, as determined by densitometry, were combined. In order to obtain highly purified RkpF preparations, the concentrated RkpF-containing fractions from the DEAE-cellulose eluate were chromatographed on a Sephadex G-50 superfine (Pharmacia) column (2.6 cm by 80 cm). Elution was performed with 25 mM Tris–192 mM glycine at a flow rate of 36 ml/h. RkpF-containing fractions were pooled, and after this final purification step, the RkpF protein was found to be homogeneous (Fig. 1). From 300 ml of IPTG-induced cell suspension, 1.1 g of wet cells was obtained containing a total of 162 mg of protein. The combined fractions from the DEAE-cellulose chromatography contained 35 mg of protein, and the combined fractions after the molecular sieving step contained 5.4 mg of homogeneous RkpF protein, as determined by a micro-biuret method (12). Figure 1 shows the degree of purification obtained as a result of implementing the individual steps of purification.
The homogeneous RkpF protein migrates as a single band on sodium dodecyl sulfate (SDS) electrophoresis. In the system described by Schägger and von Jagow (29) with the separating gel containing 16.5% (6% C) and the stacking gel containing 4% (3% C) acrylamide, RkpF migrates with an apparent Mr of 5,000 (data not shown), much faster than expected from the predicted molecular weight for RkpF of 9,587, calculated from its sequence. Electrospray mass spectrometric analysis of purified RkpF protein that had been overexpressed in E. coli indeed gives a mass of 9,585. These results suggest that the RkpF protein isolated from E. coli consists of the plain protein product without any further substitution, and it was therefore used in subsequent experiments as an apo-RkpF preparation.
Interestingly, the ACP RkpF shows some unexpected properties. Following the algorithm of Bjellqvist et al. (5), the calculated pI of RkpF (4.84) is nearly 1 pH unit higher than the pI of classical ACPs (3.90 to 4.35) (6, 9, 20, 30, 32). Multifunctional peptide synthetases, involved in nonribosomal peptide synthesis, possess thiolation domains that can be cloned and expressed as individual peptidyl carrier proteins (PCPs), functional analogs of ACPs. It is interesting to note that in contrast with ACPs involved in fatty acid or β-ketide biosynthesis, PCPs have almost neutral pIs (pI = 6.07) (34). It remains to be seen whether RkpF is more similar to PCPs in its biochemical function than other, classical ACPs.
In vivo labeling of the RkpF protein with β-alanine.
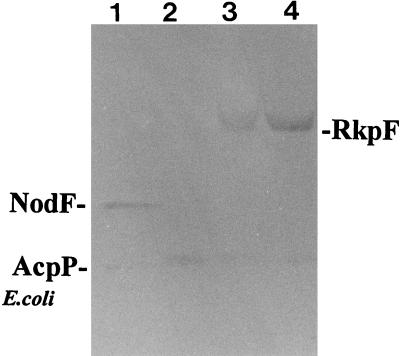
β-Alanine is a biosynthetic precursor of 4′-phosphopantetheine, the prosthetic group of ACPs (7, 10). If RkpF carries a 4′-phosphopantetheine, it should become labeled with radioactive β-alanine. By P1 cotransduction (18) of the panD mutation from E. coli SJ16 (13) to E. coli BL21 (DE3), the β-alanine auxotrophic expression strain OG7001 was constructed. OG7001 was transformed with pTB1003, and two cultures (1 ml each) of E. coli OG7001 × pTB1003 were grown on minimal medium M9 (18) in the presence of kanamycin (50 μg/ml). At a cell density of 2.5 × 108 cells/ml, 20 μCi of β-[3H (N)]alanine (specific activity, 60 Ci/mmol; American Radiolabeled Chemicals, St. Louis, Mo.) was added to each culture, and one culture was induced with IPTG (0.1 mM). After 4 h of incubation, cells were harvested and broken by successive treatments with sucrose, lysozyme, and EDTA. Cell extracts were obtained as supernatants after centrifugation at 7,000 × g for 5 min and were analyzed by PAGE and subsequent autoradiography, showing that an IPTG-inducible protein was labeled in vivo (Fig. 2). These results suggest that RkpF carries a β-alanine-derived prosthetic group, which is most likely 4′-phosphopantetheine, the usual prosthetic group of ACPs. The experiment also indicates that the posttranslational modification of adding the prosthetic group to the apoform of RkpF can be performed by an enzyme activity present in E. coli. However, the in vivo substitution of the overexpressed RkpF protein with 4′-phosphopantetheine is in no way complete, and under the conditions employed, more than 95% of the total RkpF proteins are still in their apoform (data not shown).
FIG. 2.
RkpF protein is labeled in vivo with β-[3H]alanine. Cell extracts from cells grown in media containing radiolabeled β-alanine were analyzed by 20% PAGE under native conditions and subsequent autoradiography. The various lanes show E. coli OG7001 containing the NodF-overexpressing plasmid pMP2301 induced with IPTG (lane 1), E. coli OG7001 × pET9a (lane 2), E. coli OG7001 × pTB1003 without inducer (lane 3), and E. coli OG7001 × pTB1003 induced with IPTG (lane 4).
In vitro labeling of the RkpF protein with CoA.
Holo-ACP synthase (AcpS) from E. coli is the enzyme responsible for the posttranslational modification of the AcpP protein in order to obtain the 4′-phosphopantetheine-bearing holoform of the constitutive ACP. The gene coding for AcpS has recently been cloned and characterized (17). Holo-ACP synthase (AcpS) was purified from an AcpS-overexpressing E. coli strain, BL21 (DE3) × pDPJ (kindly supplied by Dr. Ralph Lambalot, Harvard Medical School, Boston, Mass.), similarly to the purification described by Lambalot and Walsh (17). Cell extracts of IPTG-induced cells were subjected to chromatography on a DE52 column under conditions described previously (17), and nonadsorbed fractions were chromatographed on an SP-Sephadex C-50 column under conditions described previously (17). Some of the fractions eluted contained a homogeneous protein with a subunit molecular weight of 14,000 as judged from electrophoresis in 15% polyacrylamide gels containing 0.1% (wt/vol) SDS by the method of Laemmli (16) (data not shown), the molecular weight expected for an AcpS protein subunit (17). Such homogeneous AcpS preparations were used routinely for in vitro conversion of apo-RkpF to holo-RkpF.
In order to determine whether AcpS was the E. coli enzyme responsible for the attachment of the 4′-phosphopantetheine to the RkpF protein, we incubated apo-RkpF with radiolabeled coenzyme A (CoA) in the presence of homogeneous AcpS protein. In a total volume of 150 μl, different amounts of apo-RkpF protein were incubated with [3H]CoA (3.8 μM; specific radioactivity, 4.7 Ci/mmol; kind gift of Hans von Döhren, Technical University of Berlin), MgCl2 (5 mM), dithiothreitol (DTT) (0.33 μM), and 100 μg of homogeneous holo-ACP synthase (AcpS) at 37°C for 45 min in 10 mM Tris-HCl (pH 8.0), respectively. The reaction was stopped by the addition of 500 μl of 20% trichloroacetic acid, and the precipitated proteins were adsorbed to a filter (0.45-μm pore diameter). Non-protein-bound radioactivity was washed away from the filters with acetic acid, and the amount of 3H-labeled protein was quantified by liquid scintillation counting. The results given in Table 1 show that AcpS is able to transfer the radiolabeled part of CoA to apo-RkpF and that the amount of radiolabel transferred depends on the amount of apo-RkpF present. When AcpS was absent, no transfer occurred. Analysis of the radiolabeled RkpF sample on a native PAGE gel indicated that it migrates slightly faster than apo-RkpF (data not shown).
TABLE 1.
Holo-ACP synthase (AcpS) from E. coli uses RkpF as a substratea
| Amt (μg) of:
|
Protein-linked radio- activity (pmol) | |
|---|---|---|
| AcpS | Apo-RkpF | |
| 100 | 0 | 0.54 |
| 100 | 10 | 18.1 ± 1.08 |
| 0 | 10 | 0.78 |
| 100 | 255 | 69.6 ± 5.1 |
| 0 | 255 | 1.06 |
Apo-RkpF was incubated with [3H]CoA in the presence of holo-ACP synthase (AcpS) in order to obtain protein-linked radioactivity. The total radioactivity used in each assay was 190,687 cpm (570 pmol). All determinations were done in duplicate.
Apo-RkpF and holo-RkpF show different mobilities on native PAGE.
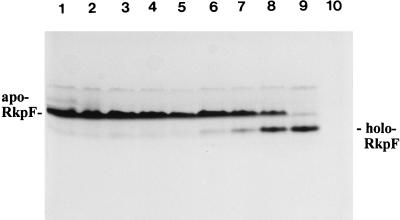
In order to test whether we could quantitatively convert apo-RkpF to holo-RkpF, we repeated the incubation of apo-RkpF with higher concentrations of CoA. For in vitro conversion of apo-RkpF to holo-RkpF, a sample of apo-RkpF protein (38 μg) was incubated with CoA (300 μM), MgCl2 (5 mM), DTT (10 mM), and homogeneous holo-ACP synthase (8.5 μg) at 37°C for 2 h in 10 mM Tris-HCl (pH 8.0) in a total volume of 200 μl. Over a time course, such a reaction was stopped by adding EGTA (pH 8.0) to a final concentration of 80 mM. The analysis of the different forms of RkpF was performed with a native PAGE gel, and the results are shown in Fig. 3. The time course of a quantitative conversion of apo-RkpF into holo-RkpF can readily be followed. If the AcpS activity is inhibited by the presence of EGTA, apo-RkpF is not converted to holo-RkpF.
FIG. 3.
Quantitative conversion of apo-RkpF into holo-RkpF protein. Apo-RkpF was incubated with CoA in the presence of holo-ACP synthase (AcpS), and the reaction was stopped after different incubation times with EGTA. A Coomassie-stained gel of native 20% PAGE is shown. The various lanes show reaction mixture lacking CoA (lane 1), reaction mixture lacking AcpS (lane 2), reaction mixture in which EGTA was added before the incubation was started (incubation time, 128 min) (lane 3), 0-min incubation (lane 4), 8-min incubation (lane 5), 16-min incubation (lane 6), 32-min incubation (lane 7), 64-min incubation (lane 8), 128-min incubation (lane 9), and reaction mixture lacking RkpF (lane 10).
By analysis with native PAGE, even apo-RkpF protein and holo-RkpF protein migrate differently. Holo-RkpF migrates faster (Rf, 0.69) on native 20% PAGE than apo-RkpF (Rf, 0.54), suggesting a more compact structure for holo-RkpF than for apo-RkpF. Jackowski and Rock showed for the constitutive ACP (AcpP) from E. coli that the apoform and the holoform could be separated by conformationally sensitive gel electrophoresis (14). The separation of apo-AcpP from holo-AcpP was possible only when electrophoretic separation was performed at 37°C. We have tested the separation of apo-RkpF and holo-RkpF over a temperature range from 15 to 55°C, and we see equally good separation of the two forms over the whole temperature range (data not shown).
Masses of apo-RkpF and holo-RkpF.
If RkpF samples were further analyzed by electrospray mass spectrometry, they were first desalted by repeated washes with water by using Centricon TM3 devices from Amicon. The two RkpF proteins were dissolved in acetonitrile-water (50:50 [vol/vol], 5% HCOOH) yielding final concentrations of about 1.5 μg/μl for the apoprotein and 3.3 μg/μl for the holoprotein. Positive-mode electrospray mass spectra were obtained on a VG Platform II single quadrupole mass spectrometer. Aliquots of 10 μl of the samples were infused in a mobile phase of acetonitrile-water (50:50 [vol/vol]) and introduced into the electrospray source at a flow rate of 5 μl/min. Spectra were scanned at a speed of 10 s for m/z 700 to 1,800, with a cone voltage of 60 V, and recorded and processed with MassLynx software, version 2.0. Mass calibration was performed by multiple-ion monitoring of horse heart myoglobin signals. Electrospray ionization mass spectrometry allowed the average masses of the apo-RkpF and holo-RkpF proteins to be determined. The experimental (9,585.0 ± 0.5) and calculated (9,587.1) masses of the apo-RkpF protein and the experimental (9,926.6 ± 1.9) and calculated (9,927.4) masses of the holo-RkpF protein demonstrate that both proteins have average masses that match the calculated masses very well. The mass difference of 342 atomic mass units (amu) between the values obtained for the apo- and holoforms is in agreement with that expected (340 amu) for 4′-phosphopantetheine substitution of the holoform. RkpF therefore carries the 4′-phosphopantetheine prosthetic group characteristic for ACPs involved in fatty acyl and β-ketide biosynthesis. The 4′-phosphopantetheine residue is quite different from the prosthetic group of ACPs associated with citrate lyase (28), citramalate lyase, and malonate decarboxylase (2), the 2′-(5"-phosphoribosyl)-3′-dephospho-CoA, which has a mass of 959 and for which a mass difference of 941 amu would be expected between the respective apo- and holoforms of ACPs. The latter three enzymatic systems are all involved in C-C bond cleavage reactions. These data show that the overexpressed protein has the molecular mass predicted from the gene sequence and that the holo-ACP synthase does indeed transfer a 4′-phosphopantetheine group to form holo-RkpF protein.
In summary, our data show that RkpF is indeed a novel ACP which presumably functions in combination with the other gene products of the complementation unit I (RkpA-RkpE) in the synthesis of an unusual fatty acid or β-ketide. Studies designed to identify the product formed by RkpA-RkpF are presently under way. A model for rhizobial capsular polysaccharide biosynthesis proposed by Reuhs et al. (24) suggests that the fatty acyl or β-ketide product formed by RkpA-RkpF might function as a membrane anchor during the polymerization of the polysaccharide subunits and during targeting of the newly formed RKP to the bacterial surface. At the bacterial surface, the mature RKP would be released from the fatty acyl or β-ketide residue.
Until very recently, only ACPs expressed constitutively and involved in the biosynthesis of essential fatty acids were known. The discovery of specialized ACPs for the biosynthesis of polyunsaturated fatty acids in S. meliloti (9) and in Rhizobium leguminosarum (10, 32) and the discovery of β-ketide antibiotics in Streptomyces (4, 33) were thought to represent unusual cases involved in complex secondary metabolism. Recently another ACP (AcpXL) was isolated from R. leguminosarum and characterized (6). AcpXL is able to transfer 27-hydroxyoctacosanoic acid to lipid IVA during lipid A biosynthesis. Thus, to date, from the genera Rhizobium and Sinorhizobium, four different ACPs have been characterized: the constitutive AcpP (20), the flavonoid-inducible NodF (10, 27), AcpXL (6), and RkpF (19). The overall amino acid sequence similarities of these four ACPs vary between 26 and 32% (6), and a well-conserved amino acid region can be observed only around the phosphopantetheine-binding site. A three-dimensional structure of the constitutive ACP (AcpP) has been obtained. Recently, initial characterizations of the secondary structure and general tertiary fold of the NodF protein (11) and of the structure of the Act protein (8) have shown that the overall structure of ACPs is surprisingly well conserved.
Acknowledgments
This research was supported by a grant (Ge 556/4-1) from the Deutsche Forschungsgemeinschaft.
Plasmid pDPJ was kindly supplied by Ralph Lambalot (Harvard Medical School, Boston, Mass.). We thank Hans von Döhren and Eva Pfeiffer for the kind gift of radiolabeled CoA and for stimulating discussions, as well as H. Görisch for critically reading the manuscript.
REFERENCES
- 1.Becquart-de Kozak I, Reuhs B L, Buffard D, Breda C, Kim J S, Esnault R, Kondorosi A. Role of the K-antigen subgroup of capsular polysaccharides in the early recognition process between Rhizobium meliloti and alfalfa leaves. Mol Plant-Microbe Interact. 1997;10:114–123. [Google Scholar]
- 2.Berg M, Hilbi H, Dimroth P. The acyl carrier protein of malonate decarboxylase of Malonomonas rubra contains 2′-(5′-phosphoribosyl)-3′-dephosphocoenzyme A as a prosthetic group. Biochemistry. 1996;35:4689–4696. doi: 10.1021/bi952873p. [DOI] [PubMed] [Google Scholar]
- 3.Beringer J E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 4.Bibb M J, Biro S, Motamedi H, Collins J F, Hutchinson C R. Analysis of the nucleotide sequence of the Streptomyces glaucescens tcmI genes provides key information about the enzymology of polyketide antibiotic biosynthesis. EMBO J. 1989;8:2727–2736. doi: 10.1002/j.1460-2075.1989.tb08414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjellqvist B, Hughes G J, Pasquali C, Paquet N, Ravier F, Sanchez J-C, Frutiger S, Hochstrasser D F. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14:1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- 6.Brozek K A, Carlson R W, Raetz C H R. A special acyl carrier protein for transferring long hydroxylated fatty acids to lipid A in Rhizobium. J Biol Chem. 1996;271:32126–32136. doi: 10.1074/jbc.271.50.32126. [DOI] [PubMed] [Google Scholar]
- 7.Cronan J E., Jr β-Alanine synthesis in Escherichia coli. J Bacteriol. 1980;141:1291–1297. doi: 10.1128/jb.141.3.1291-1297.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crump M P, Crosby J, Dempsey C E, Parkinson J A, Murray M, Hopwood D A, Simpson T J. Solution structure of the actinorhodin polyketide synthase acyl carrier protein from Streptomyces coelicolor A3(2) Biochemistry. 1997;36:6000–6008. doi: 10.1021/bi970006+. [DOI] [PubMed] [Google Scholar]
- 9.Debellé F, Sharma S B. Nucleotide sequence of Rhizobium meliloti RCR2011 genes involved in host-specificity of nodulation. Nucleic Acids Res. 1986;14:7453–7471. doi: 10.1093/nar/14.18.7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geiger O, Spaink H P, Kennedy E P. Isolation of the Rhizobium leguminosarum NodF nodulation protein: NodF carries a 4′-phosphopantetheine prosthetic group. J Bacteriol. 1991;173:2872–2878. doi: 10.1128/jb.173.9.2872-2878.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghose R, Geiger O, Prestegard J H. NMR investigations of the structural properties of the nodulation protein, NodF, from Rhizobium leguminosarum and its homology with Escherichia coli acyl carrier protein. FEBS Lett. 1996;388:66–72. doi: 10.1016/0014-5793(96)00512-1. [DOI] [PubMed] [Google Scholar]
- 12.Itzhaki R F, Gill D M. A micro-biuret method for estimating protein. Anal Biochem. 1964;9:401–410. doi: 10.1016/0003-2697(64)90200-3. [DOI] [PubMed] [Google Scholar]
- 13.Jackowski S, Rock C O. Regulation of coenzyme A biosynthesis. J Bacteriol. 1981;148:926–932. doi: 10.1128/jb.148.3.926-932.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackowski S, Rock C O. Ratio of active to inactive forms of acyl carrier protein in Escherichia coli. J Biol Chem. 1983;258:15186–15191. [PubMed] [Google Scholar]
- 15.Kiss E, Reuhs B L, Kim J S, Kereszt A, Petrovics G, Putnoky P, Dusha I, Carlson R W, Kondorosi A. The rkpGHI and -J genes are involved in capsular polysaccharide production by Rhizobium meliloti. J Bacteriol. 1997;179:2132–2140. doi: 10.1128/jb.179.7.2132-2140.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Lambalot R H, Walsh C T. Cloning, overproduction, and characterization of the Escherichia coli holo-acyl carrier protein synthase. J Biol Chem. 1995;270:24658–24661. doi: 10.1074/jbc.270.42.24658. [DOI] [PubMed] [Google Scholar]
- 18.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 19.Petrovics G, Putnoky P, Reuhs B, Kim J, Thorp T A, Noel K D, Carlson R W, Kondorosi A. The presence of a novel type surface polysaccharide in Rhizobium meliloti requires a new fatty acid synthase-like gene cluster involved in symbiotic nodule development. Mol Microbiol. 1993;8:1083–1094. doi: 10.1111/j.1365-2958.1993.tb01653.x. [DOI] [PubMed] [Google Scholar]
- 20.Platt M W, Miller K J, Lane W S, Kennedy E P. Isolation and characterization of the constitutive acyl carrier protein from Rhizobium meliloti. J Bacteriol. 1990;172:5440–5444. doi: 10.1128/jb.172.9.5440-5444.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Putnoky P, Kondorosi A. Two gene clusters of Rhizobium meliloti code for early essential nodulation functions and a third influences nodulation efficiency. J Bacteriol. 1986;167:881–887. doi: 10.1128/jb.167.3.881-887.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Putnoky P, Petrovics G, Kereszt A, Grosskopf E, Ha D T C, Banfalvi Z, Kondorosi A. Rhizobium meliloti lipopolysaccharide and exopolysaccharide can have the same function in the plant-bacterium interaction. J Bacteriol. 1990;172:5450–5458. doi: 10.1128/jb.172.9.5450-5458.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reuhs B L. Acidic capsular polysaccharides (K antigens) of Rhizobium. In: Stacey G, Mullin B, Gresshoff P M, editors. Biology of plant-microbe interactions. St. Paul, Minn: International Society for Molecular Plant-Microbe Interactions; 1996. pp. 331–336. [Google Scholar]
- 24.Reuhs B L, Carlson R W, Kim J S. Rhizobium fredii and Rhizobium meliloti produce 3-deoxy-d-manno-2-octulosonic acid-containing polysaccharides that are structurally analogous to group II K antigens (capsular polysaccharides) found in Escherichia coli. J Bacteriol. 1993;175:3570–3580. doi: 10.1128/jb.175.11.3570-3580.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reuhs B L, Kim J S, Badgett A, Carlson R W. Production of cell-associated polysaccharides of Rhizobium fredii USDA205 is modulated by apigenin and host root extract. Mol Plant-Microbe Interact. 1994;7:240–247. doi: 10.1094/mpmi-7-0240. [DOI] [PubMed] [Google Scholar]
- 26.Reuhs B L, Williams M N V, Kim J S, Carlson R W, Côté F. Suppression of the Fix− phenotype of Rhizobium meliloti exoB mutants by lpsZ is correlated to a modified expression of the K polysaccharide. J Bacteriol. 1995;177:4289–4296. doi: 10.1128/jb.177.15.4289-4296.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritsema T, Geiger O, van Dillewijn P, Lugtenberg B J J, Spaink H P. Serine residue 45 of nodulation protein NodF from Rhizobium leguminosarum bv. viciae is essential for its biological function. J Bacteriol. 1994;176:7740–7743. doi: 10.1128/jb.176.24.7740-7743.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson J B, Singh M, Srere P A. Structure of the prosthetic group of Klebsiella aerogenes citrate (pro-3S)-lyase. Proc Natl Acad Sci USA. 1976;73:1872–1876. doi: 10.1073/pnas.73.6.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 30.Schofield P R, Watson J M. DNA sequence of Rhizobium trifolii nodulation genes reveals a reiterated and potentially regulatory sequence preceding nodABC and nodFE. Nucleic Acids Res. 1986;14:2891–2903. doi: 10.1093/nar/14.7.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schultze M, Kondorosi E, Ratet P, Buire M, Kondorosi A. Cell and molecular biology of Rhizobium-plant interactions. Int Rev Cytol. 1994;156:1–75. [Google Scholar]
- 32.Shearman C A, Rossen L, Johnston A W B, Downie J A. The Rhizobium leguminosarum nodulation gene nodF encodes a polypeptide similar to acyl carrier protein and is regulated by nodD plus a factor in pea root exudate. EMBO J. 1986;5:647–652. doi: 10.1002/j.1460-2075.1986.tb04262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman D H, Malpartida F, Bibb M J, Kieser H M, Bibb M J, Hopwood D A. Structure and deduced function of the granaticin-producing polyketide synthase gene cluster of Streptomyces violaceoruber Tu22. EMBO J. 1989;8:2717–2725. doi: 10.1002/j.1460-2075.1989.tb08413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stachelhaus T, Hüser A, Marahiel M A. Biochemical characterization of peptidyl carrier protein (PCP), the thiolation domain of multifunctional peptide synthetases. Chem Biol. 1996;3:913–921. doi: 10.1016/s1074-5521(96)90180-5. [DOI] [PubMed] [Google Scholar]
- 35.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]