Abstract
Epilepsy genetics is a rapidly developing field, in which novel disease-associated genes, novel mechanisms associated with epilepsy, and precision medicine approaches are continuously being identified. In the past decade, advances in genomic knowledge and analysis platforms have begun to make clinical genetic testing accessible for, in principle, people of all ages with epilepsy. For this reason, the Genetics Commission of the International League Against Epilepsy (ILAE) presents this update on clinical genetic testing practice, including current techniques, indications, yield of genetic testing, recommendations for pre- and post-test counseling, and follow-up after genetic testing is completed. We acknowledge that the resources vary across different settings but highlight that genetic diagnostic testing for epilepsy should be prioritized when the likelihood of an informative finding is high. Results of genetic testing, in particular the identification of causative genetic variants, are likely to improve individual care. We emphasize the importance of genetic testing for individuals with epilepsy as we enter the era of precision therapy.
Keywords: genetic epilepsy, next-generation sequencing, genetic counseling genetic testing, variant of uncertain significance, precision medicine
Summary
Genetic testing in epilepsies is a clinically useful tool through which a genetic diagnosis and improved prognostic counseling may be obtained, and in some cases, precision therapies may be employed. Genetic testing always requires informed consent and should utilize modern genomic strategies for identification and interpretation of genetic variants. The key points regarding clinical genetic diagnostics in individuals with epilepsy are:
• Epilepsies with a monogenic cause, especially severe epilepsies with early onset, are currently the primary target for diagnostic genetic testing.
• For most genetic epilepsy disorders, genetic heterogeneity has been described, i.e., variants in different genes can cause the same disorder.
• Genetic testing, as well as genetic counseling before and after testing, should be performed by appropriately qualified and trained professionals.
• In most cases, ES or GS (including CNV analysis) is currently recommended as first-line testing.
• Periodical genetic re-evaluation should be undertaken for individuals with suspected genetic epilepsy without a molecular genetic diagnosis. This includes re-analysis of previously acquired sequencing data and consideration of further testing based on new or evolving clinical information and availability of novel testing strategies.
We recommend genetic testing in the following conditions (provided no other clear cause has been identified):
• Severe childhood-onset epilepsies, particularly DEEs.
• Epilepsy with intellectual disability, autism, and/or other comorbidities.
• Progressive myoclonus epilepsies and progressive phenotypes generally.
• Non-acquired focal epilepsies in specific familial syndromes.
Genetic testing can be considered (rather than recommended) in the following conditions:
• Non-acquired focal, pharmacoresistant epilepsies in the setting of presurgical evaluation.
• Epilepsy in the setting of malformations of cortical development (which may require DNA from brain tissue to be tested in parallel with DNA from another tissue source, e.g., blood or saliva).
Epilepsy is one of the most common neurological diseases and represents a burden across the lifespan for 45.9 million people and their families worldwide [1].
Epilepsy classification currently incorporates age at onset, seizure types, electroencephalogram (EEG), and imaging results. Three main groups can be distinguished:
Focal epilepsies (FE) – ~60% of all epilepsies [2]
Generalized epilepsies (GE) – ~40 % of all epilepsies [2]
Developmental and epileptic encephalopathies (DEE) – rare; severe epilepsies that present early in life, accompanied by abnormal psychomotor development due to the underlying pathology as well as the epileptic activity, the relative contributions of which may be difficult to determine [3].
Genetic epilepsies are defined by a known or presumed underlying genetic etiology; the lack of an acquired cause, such as trauma or infection, is central to the conceptualization of genetic epilepsies. Familial aggregation and twin studies provided early evidence that epilepsy is highly heritable [4], and generalized epilepsies overall are more heritable than focal epilepsies, with 82% compared to 36% concordance rates in twin studies, respectively [5]. More recent work has highlighted the important role of non-inherited genetic contributions to epilepsy, in the form of de novo variants, especially in individuals with more severe epilepsy syndromes, or post-zygotic mosaic variants in many individuals with non-acquired focal lesions.
A precise epilepsy genetic diagnosis is important for individuals and their families as it has both clinical and personal utility. This is particularly true for the developmental and epileptic encephalopathies in which early genetic testing has also been shown to be cost-effective and end the invasive search for a cause [6, 7]. Identifying the causative gene can direct anti-seizure medication choice in up to 76% of young children with epilepsy [8]. Even in adults, treatment changes due to a genetic diagnosis after years of drug resistance has led to improved seizures, cognition and quality of life [9, 10]. In addition, precision therapies including both repurposed medication and genetic therapies are becoming available for some genetic epilepsies. The DEEs present when families are in their reproductive phase; therefore, knowing the genetic architecture of their child’s epilepsy informs reproductive choice and opens up options such as prenatal diagnostics and preimplantation genetic diagnosis and screening with IVF [7, 11]. Information on the natural progression of a genetic epilepsy enables families and clinicians to better prepare for potential comorbidities and to plan resources and support for the child’s future [12]. Finally, and not to be underestimated, a genetic answer can be a psychological turning point for a family as it alleviates parental guilt, facilitates grief processing, increases understanding and points them to family gene support networks which ultimately improves their quality of life [13, 14].
Since the last report from the International League Against Epilepsy (ILAE) [15], enormous progress has been made in gene discovery, genetic screening techniques, analysis strategies, and knowledge of different types of genetic variation, warranting this update. We recognize that the indications for genetic testing are evolving and that the interpretation of genetic test results may be challenging for the clinician who does not routinely request genetic testing or interpret genetic data. Complex cases may warrant referral to a clinical geneticist or genetic counselor. We summarize the latest developments in the field so that the growing body of knowledge of the genetics of epilepsy can be leveraged to select appropriate genetic tests for different clinical scenarios.
Basic genetic principles
Genetic traits in epilepsy and main modes of inheritance
We start this overview by briefly explaining basic principles of genetics including specific aspects for genetic epilepsies.
To understand genetic testing and its utility, it is important to understand the main modes of inheritance, to appreciate that many epilepsies have genetic underpinnings that do not follow a Mendelian pattern, and that some may be genetic even though they are not inherited (for more details see Helbig et al. [16]).
• Monogenic epilepsies
The so-called “monogenic” or “single gene” epilepsies are the main target of clinical genetic testing. These epilepsies are caused by a variation in a single gene and follow basic inheritance patterns (autosomal dominant [AD], autosomal recessive [AR], X-linked, mitochondrial; see table 1), even though additional genetic modifiers may still explain some of the phenotypic variation seen in these individuals [17]. Monogenic epilepsies are typically individually rare, but together comprise a significant proportion of the genetic epilepsies. Most familial self-limiting epilepsy syndromes have a monogenic cause, which are less common in isolated (non-familial) cases with GE or FE without developmental delay. Monogenic epilepsies also include epilepsies that arise from a de novo variant, such as many of the DEEs. A de novo variant occurs most often during gametogenesis and will be present in all cells of an individual, meaning standard clinical testing using DNA from blood lymphocytes or buccal samples should detect it. A de novo variant arising in the post-zygotic stage, however, results in a variant that is “mosaic” in an individual, meaning that it is present only in a fraction of cells and may potentially be restricted to only some tissues (e.g., brain) or cell populations (e.g., some neurons) and as such, may not be detectable with routine analysis of DNA extracted from blood lymphocytes.
Table 1.
Main modes of inheritance with recurrence risk and additional information.
| Mode of inheritance | Affected allele | Origin | Risk to offspring of inheriting the variant | Risk to siblings of inheriting the variant | Additional information |
|---|---|---|---|---|---|
| Autosomal dominant | One variant allele – heterozygous pathogenic variant | Inherited De novo |
50% 50% |
50% < 1 % (due to parental germline mosaicism that had been detected in <10% of cases [65]) |
Penetrance (some individuals with the variant allele may not be affected) Phenotypic expressivity (spectrum and severity of phenotype may differ between carriers of the variant) Note: phenotypes due to de novo variants are often severe and affected individuals may not reproduce |
| Autosomal recessive | Two variant alleles - homozygous (same variant) or -compound heterozygous (two different variants) | One from each parent (rarely one inherited and one de novo) | Zygosity in offspring depends on carrier status of partner | 25% | Heterozygous carriers of one of the variants are usually unaffected |
| X-linked | Hemizygous in males (one variant allele with no second allele) | Maternally inherited | All daughters are carriers, all sons are unaffected | 50% of sisters are carriers, 50% of brothers are affected | Heterozygous females are usually unaffected |
| De novo | < 1% for brothers (due to parental germline mosaicism that had been detected in <10% of cases [65]) | ||||
| Heterozygous in females | Inherited | 50% | Maternally transmitted: 50% Paternally transmitted: sisters 100%, brothers unaffected | Hemizygosity in males may be severe or even incompatible with life | |
| De novo | 50% | < 1 % (due to parental germline mosaicism that had been detected in <10% of cases [65]) | |||
| Homozygous/compound heterozygous in females | One from each parent (rarely one inherited and one de novo) | Daughters are carriers, all sons are unaffected | Heterozygous mother: 50% affected/50% carriers Homozygous mother: 100% of offspring is affected | Rarely in epilepsies | |
| Mitochondrial | Mitochondrial genome | Maternally inherited | Maternally transmitted: variable (depending on level of heteroplasmy) Paternally transmitted: 0% | Maternally transmitted: variable (depending on level of heteroplasmy) Paternally transmitted: 0% | Both females and males can have mitochondrial disease but only females transmit the disorder to their offspring [67] |
| Mosaic | Heterozygous in a fraction of cells/tissues | Post-zygotic de novo (after the 1-cell stage) | 0% if germline is unaffected Up to 50% if germline is affected by mosaicism | 0% | Detection depends on variant allele fraction and tissue being analyzed |
Causal genetic variations include single nucleotide variants (SNV) and copy number variants (CNV, e.g. deletions and duplications). Other causal genetic variants that are increasingly recognized include repeat expansions and complex structural rearrangements.
For more information on the proportions of FE, GE and DEE in which a monogenic cause is currently identified, see table 2.
Table 2.
Yield of genetic testing and recommended testing strategy.
| Diagnosis | Proportion of individuals with detectable pathogenic variant (s) | Genetic test(s) indicated | |
|---|---|---|---|
| Focal epilepsies | |||
| Familial self-limited neonatal / neonatal-infantile epilepsy (SeLNE, SeLNIE) Previously benign familial neonatal/neonatal-infantile epilepsy BFNE/BFNIE |
> 90% of familial cases | ES/GS* | |
| Self limited familial infantile epilepsy (SeLIE) Previously benign familial infantile epilepsy [BFIE] |
> 90% of familial cases | ES/GS* | |
| Autosomal dominant sleep related hypermotor epilepsy (ADSHE) Previously autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE) | ~ 30% of familial cases | ES/GS* | |
| Autosomal dominant epilepsy with auditory features (ADEAF) | ~ 50% of familial cases | ES/GS* | |
| Familial focal epilepsy with variable foci | ~ 80% of familial cases | ES/GS* | |
| Isolated FE | Rarely due to identified monogenic causes | ES/GS* only to be considered in case of: - positive family history - additional symptoms (e.g. ID, autism, dysmorphology, etc.) - pharmacoresistance |
|
| Generalized epilepsies | |||
| Genetic epilepsy with febrile seizures plus (GEFS+) | ~ 30% of familial cases | ES/GS* | |
| Familial adult myoclonic epilepsy (FAME) | Up to 90%: repeat expansion | RP-PCR | |
| Idiopathic generalized epilepsy (IGE) Childhood absence epilepsy (CAE) Juvenile absence epilepsy (JAE) Juvenile myoclonic epilepsy (JME) Generalised tonic-clonic seizures alone (GTCA) |
Rarely due to identified monogenic causes | ES/GS* only to be considered in case of: - positive family history - additional symptoms (e.g. ID, autism, dysmorphology, etc.) - pharmacoresistance |
|
| Isolated GE | Rarely due to identified monogenic causes | ES/GS* only to be considered in case of: - positive family history - additional symptoms (e.g. ID, autism, dysmorphology, etc.) - pharmacoresistance |
|
| Developmental and epileptic encephalopathies (DEE) | |||
| Unspecified DEE and related phenotypes (e.g. PME, complex MCD, Fragile X, etc.) | 50% | ES/GS* and/or fragment length analysis | |
| Neonatal | Early-infantile DEE | ~ 60% | ES/GS* |
| Infantile | Dravet syndrome | ~ 90% of cases SCN1A de novo, 5% inherited | ES/GS* |
| Epilepsy of infancy with migrating focal seizures (EIMFS) | ~ 70% | ES/GS* | |
| Infantile epileptic spasms syndrome (previously West syndrome) | ~ 30% | ES/GS* | |
| Epilepsy in females with mental retardation (EFMR) (e.g., PCDH19-clustering epilepsy) | Nearly all PCDH19 (de novo or inherited) | ES/GS* | |
| Early-onset absence epilepsy (EOAE) | Up to 10% | ES/GS* | |
| Childhood | Epilepsy with myoclonic-atonic seizures (EMAtS; Doose syndrome) | Genetically heterogeneous | ES/GS* |
| Lennox-Gastaut syndrome | ~ 30% | ES/GS* | |
| Developmental and/or epileptic encephalopathy with spike-wave activation in sleep (D/EE-SWAS) | ~ 20% | ES | |
| Syndromes suggesting chromosomal rearrangements | CMA (karyotyping can be considered, e.g. when ring chromosomes are suspected) | ||
CMA: chromosomal micro array; ES: exome sequencing; RP-PCR: repeat primed PCR; PME: progressive myoclonus epilepsy, MCD: malformation of cortical development.
Exome or Genome sequencing is indicated, depending on availability and local standards. If neither is available, panel sequencing (including all relevant disease genes – see supplementary table 1) should be considered as an alternative option.
• Epilepsies with complex genetic patterns
Genetics also plays an important role in many common forms of epilepsy, including genetic generalized epilepsy (GGE) and non-acquired focal epilepsies (NAFE). Although some large GGE pedigrees have been described, the risk of developing epilepsy for first-degree family members of a person with epilepsy is only 3–8% [18], which is considerably lower than would be expected for disorders thought to be caused by autosomal dominantly inherited variants. The majority of these common epilepsies are thought to have a multifactorial etiology, likely involving multiple genes (oligogenic or polygenic) and possibly contributions from environmental or epigenetic factors (e.g. changes that affect gene activity and expression). To date, several genetic risk factors, or susceptibility alleles, have been identified for common epilepsies (ILAE Consortium on Complex Epilepsies 2018), but translation of these findings into clinical care is still in its infancy. Nevertheless, clinical implementation of polygenic risk scores may be expected in the medium term, which, for example, might aid in diagnostic issues and risk stratification.
It is also important to note that genetic testing and results reflect knowledge at the time of testing. Unremarkable results of genetic testing should be regularly re-evaluated by the referring clinician or appropriate specialists. After an appropriate time frame (e.g., two years), consideration should be given to: (1) re-analysis of previous genetic sequencing data; (2) performing additional investigations using new forms of genetic testing due to the rapid pace of technological progress, genetic discoveries and increase of knowledge (see Section Outlook); and (3) in some selected cases, evaluation of whether the appropriate tissue had been examined [2].
Genetic testing methods
The range of tests available has evolved considerably since the previous ILAE report on genetic testing [15], and their yields are reviewed in a recent systematic evidence review [19] (see table 3). Each testing method has advantages and limitations. The tests most commonly used in genetic diagnostics aim to detect causative SNV or CNV.
Table 3.
Current diagnostic yield of genetic tests in epilepsy.
• Next-generation sequencing (NGS)
NGS modalities include exome sequencing (ES) and genome sequencing (GS), as well as epilepsy-focused gene panels. Due to the high genetic heterogeneity of most epilepsies, NGS is generally considered the methodology of choice for diagnostic testing and should be adopted as a first-line investigation [19, 20]. In addition, NGS has the benefit of enabling comprehensive detection of both SNV and CNV and is usually more cost-efficient compared to other methods [6, 21].
ES / Trio ES
ES comprises simultaneous sequencing of the entire coding sequence and surrounding intronic regions of the human genome, enabling identification of SNV as well as CNV. Intra- and inter-genic non-coding regions are usually not targeted, except for splice sites near the exons. Due to the high pace of ongoing gene discovery and the expansion of phenotypes associated with known disease genes, the analysis of “in silico” panels has become standard in many laboratories; this entails generating exome data and performing a dedicated analysis targeting all genes associated with a given disorder. This approach enables the use of state of the art and, in principle, contemporary in silico panels of epilepsy genes [22]. Recent efforts to curate gene-disease associations have incorporated data derived from OMIM, ClinVar, HPO, and manual curation of recently-published genes. Additional resources to inform variant interpretation are publicly available to the wider community (including genetic testing laboratories) and include PanelApp [23], PanelApp Australia [24], GenCC [25], Gene2Phenotype [26], SysID [27], and ClinGen [28].
GS / Trio GS
GS comprises sequencing of the entire human genome, enabling identification of SNV and CNV in coding regions as well as in intronic, intra- and inter-genic non-coding regions, thus improving the yield of genetic testing [29]. All advantages of ES are also applicable to GS. Further, GS will be increasingly useful as particular types of variants (e.g. repeat expansions [30] or structural variants) can potentially be detected more easily (see below). GS still poses challenges related to interpretation of non-coding variants and data storage. GS analysis typically yields even more variants of uncertain significance than ES, and sequencing both parents and offspring (trio approach) can greatly facilitate variant interpretation.
• Epilepsy panel sequencing
Panel sequencing, based on targeted enrichment of epilepsy genes, comprises simultaneous sequencing of the coding and surrounding intronic regions of selected genes. Only genes included in the panel design can be evaluated, and thus the panel composition is often outdated quickly after its implementation due to the rapid pace of ongoing gene discovery [31]. With recent demonstration of the higher yield of ES and GS compared to panels, and with the continuing reduction in costs for ES/GS, we recommend considering epilepsy panels only if ES/GS is not available, or if deeper sequencing of certain genes is indicated, e.g., if mosaicism is suspected.
GS also allows for the analysis of mitochondrial DNA providing another increase in diagnostic yield for a group of disorders that was previously challenging to diagnose. In essence, ES or GS should be the first diagnostic test in the epilepsies, barring any specific clinical findings warranting a different approach. We acknowledge that resources may not always be available to conduct these studies and advocate for the most comprehensive degree of testing available to be undertaken in any given setting.
• Chromosomal microarray (CMA)
In total, 1.5–3% of all common epilepsies are associated with CNV [32]. In DEE, the diagnostic yield increases up to 16% [33, 34]. NGS enables CNV and SNV to be detected in a single test, making microarray redundant in certain settings.
• Sanger sequencing
Single gene sequencing by polymerase chain reaction (PCR) and Sanger sequencing has almost become obsolete within a routine diagnostic work-up. Even in scenarios for which a variant in a particular gene can be predicted with relatively high confidence (e.g., SCN1A in Dravet syndrome, or MECP2 in Rett syndrome), tests such as panel sequencing that employ NGS are preferred over Sanger sequencing of a single gene due to three main reasons: 1) PCR appears to be vulnerable to false negatives (e.g., allelic drop-out due to primer drop-out [35]; (2) CNV, such as intragenic deletions, are not detected by PCR; and (3) there may still be genetic heterogeneity among the potentially small proportion of differential diagnoses that require more comprehensive screening. However, Sanger sequencing is still valuable as a confirmatory diagnostic procedure to validate previously identified SNV or for familial segregation analysis.
• Karyotyping
Classic karyotyping has been surpassed by CMA for CNV detection, but may still be used to resolve gross structural rearrangements (e.g., translocations, inversions, ring chromosomes) or numerical chromosomal abnormalities (aneuploidy), though this is rarely requested for people with epilepsy. Individuals with syndromes such as Down syndrome (trisomy 21) will typically have been diagnosed clinically prior to the onset of epilepsy. Karyotyping should, however, still be requested if a ring chromosomal disorder is clinically suspected, as this is often missed by CMA. As the majority of ring chromosome 20 individuals are mosaic, analysis of at least 100 metaphases is necessary.
• Other variant types and detection methods
Although desirable, a “one-for-all” genetic test is not yet established. In specific cases, it might be necessary to consider additional specific genetic testing such as:
Detection of repeat expansion disorders (e.g., FraX, FAME)
In case of suspicion of Fragile X syndrome, a test for the expansion of the CGG triplet repeat within the X-linked FMR1 (fragile X mental retardation 1) gene should be considered. Based on recent findings, the historical first-tier status of FraX testing in neurodevelopmental disorders has been questioned and, in the absence of suggestive clinical features, FraX should usually be relegated to second-tier testing [36]. Another form of epilepsy due to repeat expansion is familial adult myoclonic epilepsy (FAME) [37]. The genetic variant underlying FAME is an intronic repeat expansion (pentamers; an expanded TTTTA or insertion of TTTCA) in one of six genes reported so far (STARD7, YEATS2, RAPGEF2, MARCHF6, SAMD12 and TNRC6A) located on different chromosomes [37].
Methylation analysis (e.g., Angelman syndrome)
In cases with a high level of clinical suspicion of Angelman syndrome, analysis of the parent-specific DNA methylation imprints at chromosome 15q11.2-q13 by MS-MLPA (methylation-specific multiplex ligation-dependent probe amplification analysis), with a focus on deletions, UPD (uniparental disomy) and imprinting defects of the region 15q11.2-q13, could be considered prior to ES/GS [38].
Pre-test considerations for the referring clinician
A thorough delineation of the individual’s phenotype is valuable for test selection and interpretation of test results. Based on the phenotype, the clinician can form a hypothesis about which gene or genes might be responsible for an individual’s epilepsy and prioritize which type of testing is most appropriate. Genetic counseling (table 4) should be provided to individuals and families before genetic tests are ordered, and should delineate the reasons for testing, anticipated results and their interpretation, limitations of the testing modalities to be implemented, and possible next steps if the initial evaluation is unrevealing. Interpretation of genetic testing results requires phenotyping prior to genetic testing, a principle that is incorporated into current guidelines for variant interpretation, including those of the American College of Medical Genetics and Genomics [39]. For that reason, the phenotypic features (including epilepsy and other relevant features) need to be provided to the diagnostic laboratory prior to analysis (figure 1). Human Phenotype Ontology (HPO)-based phenotypic descriptions [40] represents a suitable option to provide a standard terminology that facilitates communication across laboratories.
Table 4.
Counselling aspects and general considerations for genetic testing in individuals with epilepsy.
|
Counselling aspects to be considered with affected individual/legal guardian) before genetic testing: • Explanation of the indication for genetic testing in the individual case • Explanation of test choice • Discussion of possible outcomes, e.g., definitive result vs. variant of uncertain significance (VUS) vs. ‘negative’ result • Explanation of potential positive results • Discussion of potential effects of results on non-medical issues (e.g., health insurance, social stigma, family dynamics) • Discussion of the limitations of interpretation • Outline of expected possibilities for precision medicine • Discussion of coverage of costs, if relevant • Discussion of potential next steps if initial results are unrevealing (e.g., for re-analysis or additional testing) |
|
General aspects for the clinician to consider before genetic testing: • Test selection based on individual phenotype • Listing of clinical features to the laboratory (e.g., HPO-based list of features) • Informed consent for genetic testing method(s) • Consideration of alternatives to clinical testing (e.g., research) if costs are prohibitive. |
|
Counselling aspects to be considered after genetic testing: • Explanation of results and their impact on diagnosis, surveillance, and prognosis • Discussion of next steps if results do not provide a genetic diagnosis • Impact on comorbidities • Discussion of therapeutic implications • Impact on psychological wellbeing • Impact on further family planning and potentially other family members • Impact on social circumstances • Discussion of interpretation limits – inclusive positive or negative results and VUS. |
Figure 1.
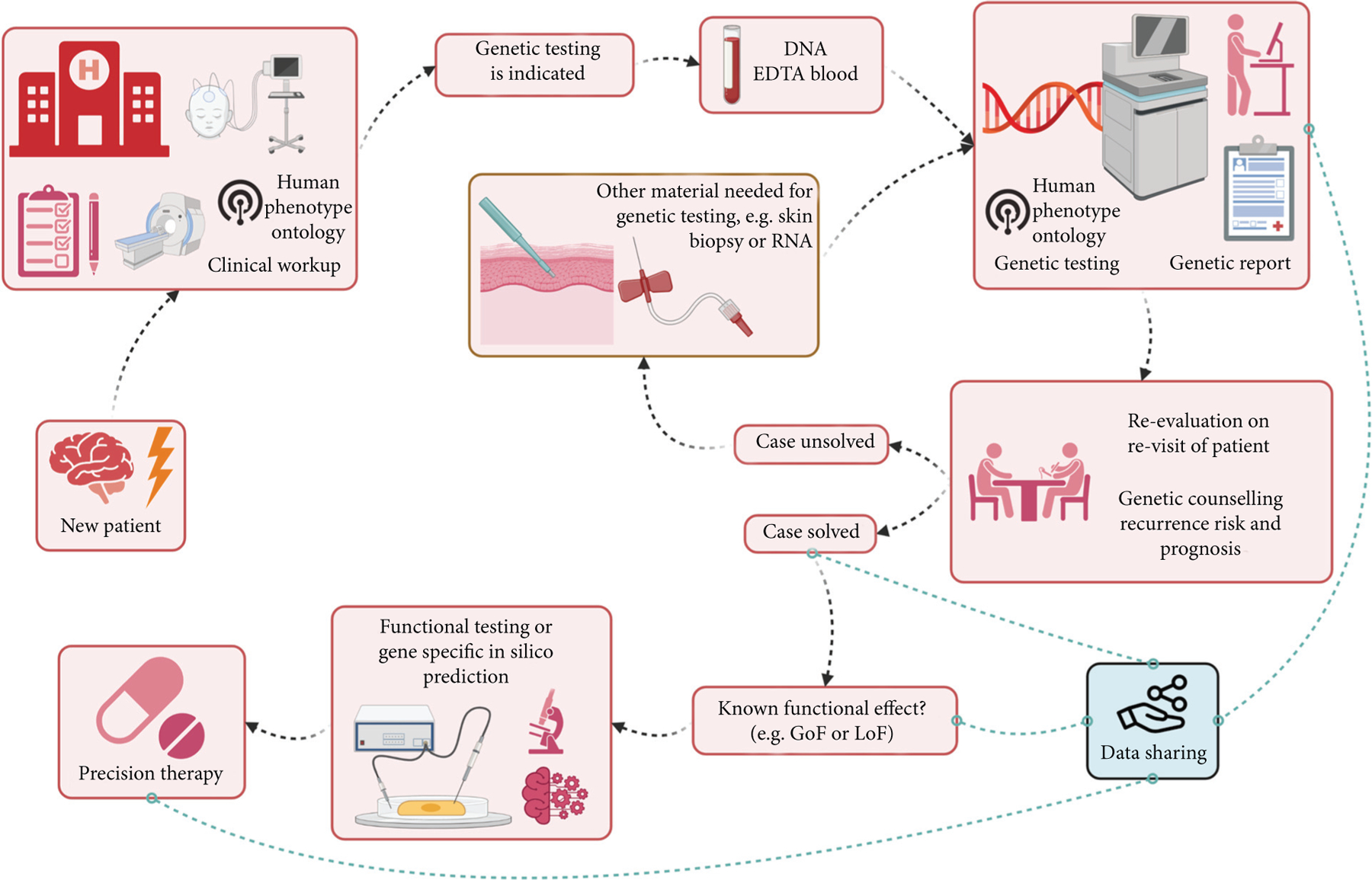
Workflow of genetic testing. The workflow of genetic testing is indicated by the black dotted arrow lines. Blue dotted lines indicate possible scenarios that will depend on individual circumstances (created with BioRender.com).
Which genetic test is indicated first in which epilepsies?
Testing should be considered in epilepsy types with a reasonably high pre-test probability of a genetic cause being identified and, especially, if the results may lead to improved care for the individual (see also tables 2 and 5). Overall, the likelihood of identifying a genetic cause decreases with increasing age at onset of the epilepsy; the greatest proportion of genetic epilepsies manifests in the neonatal period, followed by infancy. In this age period, the diagnostic yield of genetic testing may reach up to 60% [41]. However, age at testing (as opposed to the age at onset of epilepsy) should not influence the decision to test or the type of test chosen [33, 42, 43]. Individuals who are now adults who had early-onset epilepsy likely presented in the era before genetic testing was widely available, and should be considered candidates for testing. Clinical utility of genetic testing is highest in the more severe, drug-resistant epilepsies [44, 45]. Overall, the most obvious indication, in terms of clinical utility and diagnostic yield, is for people with early-onset DEE or neurodevelopmental disorders with epilepsy. The presence of comorbid conditions, such as intellectual disability, autism, dysmorphic features or multi-system symptoms increases the likelihood of a genetic finding [46]. Testing of individuals with drug-resistant non-acquired epilepsy without such comorbidities could be useful as identification of an underlying genetic cause might lead to a more targeted treatment [47].
Table 5.
Examples of precision therapies based on genetic findings in epilepsy.
| Genes | Proteins | Main pathophysiology | Potential precision treatment approaches | Evidence | Reference |
|---|---|---|---|---|---|
|
ALDH7A1
PNPO PROSC |
Aldehydedehydrogenase Pyridoxine phosphate oxidase Pyridoxine phosphate binding protein |
Vitamin B6 deficiency | Supplementation with pyridoxine Supplementation with pyridoxal-5-phosphate |
+ | Mills et al., 2014 [69] Darin et al., 2016 [70] |
| CAD | Trifunctional protein (CPSase, ATCase, DHOase) in pyrimidine biosynthesis | Deficiency in pyrimidine biosynthesis | Supplementation with uridine | + | Koch et al., 2017 [71] |
|
CHRNA4
CHRNB2 CHRNA2 |
Nicotinic acetylcholine receptor (AChR) | Desensitization of the nicotinic AChR | Nicotine | + | Fox et al., 2021 [72] Lossius et al., 2020 [73] |
|
GRIN1
GRIN2A GRIN2B GRIN2D |
Glutamate receptor (NMDAR) | GoF/LoF | Memantine, dextrometorphane, ketamine for GOF, Serine for LOF | + | Pierson et al., 2014 [74] Gale et al., 2021 [75] Amador et al., 2020 [76] Soto et al., 2019 [63] Krey et al., 2022 [64] |
| KCNA2 | Voltage-gated K+ channel KV1.2 (A-type) | Loss or gain of function (or a mixture of both) | 4-aminopyridine for GOF or some GOF+LOF variants to reduce channel overactivity | + | Syrbe et al., 2015 [77] Hedrich et al., 2021 [78] |
|
KCNQ2
KCNQ3 |
Voltage-gated K+ channels KV7.2, KV7.3 (M-type) | Loss or gain of function, depending on variant | Na+ channel blockers for LOF variants (indirect effect blocking increased neuronal firing induced by reduced activity of K+ channels); KV7.2/KV7.3 channel activators, such as ezogabine/retigabine | + | Pisano et al., 2015 [79] Sands et al., 2016 [80] Nissenkorn et al., 2021 [81] Orhan et al., 2014 [82] Millichap et al., 2016 [83] Vanoye et al., 2022 [84] |
|
SCN1A
SCN2A SCN8A |
Voltage-gated Na+ channels NaV1.1, NaV1.2, NaV1.6 | LOF or GOF of Na+ channel function depending on individual variants | Na+ channel blockers for GOF (to reduce channel overactivity), avoid such drugs for LOF variants (which may enhance reduced channel activity) | + | Guerrini et al., 1998 [85] Wolff et al., 2017 [86] Johannesen et al., 2021 [43] |
| SLC2A1 | Glucose transporter type 1 (GLUT1) | Reduced glucose transport across the blood-brain barrier | Ketogenic diet, providing ketone bodies as alternative fuel instead of glucose | + | Klepper et al., 2020 [87] |
|
TSC1
TSC2 |
Hamartin, Tuberin | mTOR disinhibition | Everolimus, Sirolimus (mTOR inhibitor) | ++ | French et al., 2016 [88] |
: evidence from retrospective case series or clinical experiences from study groups
: evidence from a controlled clinical trial; GoF: gain of function; LoF: loss of function; AChR: acetylcholine receptor; AMPAR: a-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor; mTOR : mechanistic target of rapamycin.
Diagnostic genetic testing has not been as widely pursued in drug-responsive epilepsy. A notable exception would be self-limiting neonatal or infantile-onset familial epilepsy syndromes (e.g., BFNE, BFIE, BFNIE), as early genetic diagnosis would reduce further investigation in a neonate or infant, underpin prognostic counseling, and promote earlier modification of treatment. Individuals from larger families with self-limiting epilepsy syndromes of childhood or adolescent onset might benefit from genetic testing if there is an active question about genetic diagnosis, prognosis and recurrence risk. Special attention should, however, be paid during pre-test counseling to aspects such as diagnostic yield, reduced penetrance, and variable expressivity of disease-causing genes (supplementary table 1). The diagnostic yield of any genetic test remains low in sporadic/isolated GE or FE. In GE and FE, genetic testing can, however, also be applied in specific clinical scenarios (see table 2).
The biggest advantages of ES and GS over a targeted panel analysis are: (1) to not be restricted to the analysis of a limited number of genes (however, the possibility remains to perform in silico panels based on ES/GS) and have the possibility of later re-analysis when new disease genes have been detected; and (2) the possibility to perform a broad CNV analysis.
Cost-effectiveness of various genetic testing strategies for individuals with epilepsy is dynamic and depends on the clinical scenario as well as overall yields and costs that continue to change over time. The decision about which test method to use is often made by the clinician with, in experienced laboratories, the laboratory geneticist, who typically re-evaluates the request; further discussion with the requesting clinician may be valuable.
Post-test considerations by the referring clinician after receiving the genetic test result
Interpretation of genetic testing results
In 2015, the American College of Medical Genetics and Genomics (ACMG) developed guidance for the interpretation of sequence variants. This report recommends the use of specific standard terminology - “pathogenic,” “likely pathogenic,” “uncertain significance”, “likely benign”, and “benign” - to describe variants identified in genes that cause Mendelian disorders [39]. Based on these guidelines, the ACMG and the Clinical Genome Resource (ClinGen) also published a joint consensus recommendation for the interpretation of constitutional CNV [48].
Post-test considerations depend on the results of genetic testing. Three possible scenarios exist (figure 1).
• The clinical features are fully explained by the detected genetic cause – the case is “solved”
In most cases, a confirmed genetic diagnosis means the end of a diagnostic odyssey. Genetic counseling should be offered, taking into account what is known about prognosis and disease presentation of other individuals with the same genetic disorder. In light of the ever-growing possibilities of precision medicine, it is important to determine whether there are any therapeutic implications for the individual. For some genes, therapeutic decisions depend on the functional consequence of the specific variant (e.g. loss or gain of function). If there is no functional data on the specific genetic variant available, it is worth contacting a group working on this gene asking for the possibility to initiate such testing (on a research basis; see Section Post-test considerations by the genetic testing laboratory and further clinical involvement). Finally, it is useful to determine whether there is a registry/natural history study or an ongoing drug trial in which the individual can be enrolled.
• A variant of uncertain significance (VUS) is detected – it currently remains unclear whether the case is solved or not
The detected variant cannot be confirmed to be the cause of the individual’s condition at this time, and its relevance to the phenotype thus remains unclear. Typically, variant interpretation is greatly facilitated by the availability of detailed phenotypic data, including both epilepsy-related and non-epilepsy related features. So, retrospective deep phenotyping by the clinician to evaluate if the gene and the individual’s phenotype match is mandatory. Additional diagnostic testing, such as MRI or enzymatic assays, may be appropriate. In addition, segregation analysis through testing of parents or other relatives with known disease status can help to re-classify the variant as (likely) benign or (likely) pathogenic. For some variants, though not yet widely clinically available, RNA sequencing may be informative, for example to evaluate expression of a gene if a variant is predicted to affect splicing and reduce the gene’s expression. In addition, functional testing of the identified variant by a research group with functional expertise may be helpful to evaluate the potential impact of the variant on the gene’s function. All options should be addressed during genetic counseling.
• No clinically relevant genetic cause is identified
An uninformative test result does not mean that a genetic cause is excluded, but rather that a genetic cause can not be determined with the methodology employed or available at the time of testing. A potentially causative genetic variant may have escaped detection due to technical issues, or may not be classified as (likely) pathogenic due to insufficient scientific knowledge about the impact of the variant. In addition, oligogenic or polygenic causes are typically not yet diagnostically identified. Thus, an inconclusive genetic test result should lead either to a re-evaluation of the generated genetic data after an appropriate time interval or to further genetic testing with a different complementary method (see Section Further clinical follow-up after genetic testing).
Post-test considerations by the genetic testing laboratory and further clinical involvement
The laboratory should provide the referring clinician with an easily understandable interpretation of the test results and clear recommendations for further practice. If available, results of functional data on the identified variant should be mentioned in the report. If no functional data is available, it may be possible to predict the variant effect with the growing number of gene-family specific in silico prediction tools [49, 50]. This is especially relevant if both loss- and gain-of-function variants are described for the gene of interest, as they often require different precision medicine approaches. Additional information that might be useful to the individual comprises online resources, contact information of family-led organizations as well as information on ongoing research efforts, especially about ongoing clinical trials (see Section Implications of genetic diagnosis for precision medicine). Efforts to harmonize genetic test reports with recommendations that are comprehensible to non-specialists and also affected individuals and their families are ongoing [51]. All these aspects may be addressed during genetic counseling.
The genetic testing laboratory is responsible for the regular upload of identified variants to public resources such as ClinVar (www.ncbi.nlm.nih.gov/clinvar) to facilitate global variant interpretation and also enable feedback. Ideally, the laboratory should also establish continuous re-evaluation procedures of genetic test results and should report updated results to the clinician (e.g. a VUS has since been reported to be de novo in another affected individual in ClinVar and therefore is now more likely to be considered causative).
Implications of genetic diagnosis for precision medicine
Identifying the precise cause of an individual’s epilepsy is presently still the main reason for performing clinical genetic testing. In addition to providing diagnostic certainty, a genetic diagnosis can inform on prognosis and recurrence risk. A genetic diagnosis can ultimately lead to a more precise treatment and better individualized care (figure 1). While genetic diagnoses influence treatment for a growing number of genetic epilepsies (table 5), precision treatment remains an area of promise that has yet to be achieved for the majority of individuals with genetic epilepsies. The goals of precision treatment for epilepsy include improved seizure control, improved cognitive function, relief from other (neurological or non-neurological) comorbidities, and improved survival (e.g. reduced risk of SUDEP). A longstanding example of precision medicine for epilepsy is supplementation of metabolites in the setting of a genetic metabolic defect (e.g., pyridoxine for ALDH7A1 or PNPO, uridine for CAD variants) (table 5). Precision therapy for genetic epilepsies may broadly include changes to a treatment regimen on the basis of a variant in a given gene, such as addition of a specific anti-seizure medication (ASM) that has been reported to be useful in that setting (e.g., sodium channel blockers for loss-of-function KCNQ2 variants or for gain-of-function SCN2A variants). In contrast, some ASMs should be avoided in the setting of a given genetic diagnosis (e.g., sodium channel blockers in individuals with loss-of-function SCN1A variants) (see table 5 for more examples). It is important to note that most of these examples are based on collective anecdotes rather than controlled clinical trials, and long-term outcomes from such treatment changes are still to be documented.
Further clinical follow-up after genetic testing
When an individual has previously undergone genetic testing without conclusive findings, periodic re-analysis of existing NGS data or initiation genetic re-testing with newer, more sensitive technologies is warranted. Re-testing or re-analysis of data has been proven to lead to positive results in individuals who previously tested negative [52]. The timing of this evaluation should be governed by clinical need and technological advances, as well as the availability of new knowledge. At that moment, an update of the phenotype (e.g. changes in features or novel aspects) is invaluable. Re-analysis of existing data includes reviewing of VUS in light of growing knowledge, and use of improved methods to detect both SNV and CNV. How such a re-evaluation of existing genomic data takes place will vary from setting to setting; in most cases, actively contacting the clinical laboratory is the first step, and in some cases, research re-analysis may be required (figure 1). If the referring clinician is unsure whether to initiate re-testing, guidance and advice from a genetic testing laboratory should be sought.
If a genetic cause has previously been identified, regular re-evaluation may be necessary to determine whether novel possibilities in precision therapy have emerged since the last consultation.
Benefits and limitations of genetic testing
The benefits, risks, and limitations of genetic testing were discussed comprehensively by Ottman et al. in 2010 (see their table 6 and section on “potential benefits and harms” [15]). Since then, there has been significant progress in the handling of secondary findings [53–55]. Secondary findings are pathogenic SNV or CNV unrelated to the primary indication for the testing. The broader the scope of the applied diagnostic method, the more likely secondary findings will emerge. Secondary findings without treatment consequences are considered “non-actionable” and are generally not reported in the results of genetic testing. By contrast, “actionable” secondary findings with treatment or prevention consequences can be reported back to the individual if this was agreed in the original written informed consent. The goal of reporting these secondary findings is to provide healthcare benefits by preventing primary or secondary complications. The yield of actionable secondary findings in individuals with epilepsy ranges from 2 to 4% [42].
Table 6.
Frequently asked questions about the legal implications of genetic testing.
| Topic | Examples from countries with legislation on genetic testing | Suggestion on how to address this issue in countries where no legislation is in place |
|---|---|---|
| Who is qualified to order genetic testing? |
Diagnostic: any clinician (e.g. US, many EU countries) |
Any medical practitioner |
|
Predictive: medical geneticist or medical specialist (e.g. most countries in EU) |
Specialist in the condition being tested | |
| Who should communicate the genetic testing result to the individual? | Requesting clinician or – optionally – a medical geneticist (e.g. most countries in EU) |
Practitioner who ordered genetic testing or a genetic counsellor / medical geneticist |
| Who in addition to the individual should be made aware of the genetic testing result? |
Doctors: Practitioners who ordered genetic testing receive report. Other practitioners involved in treatment of an individual can or should be informed according to individual consent. (e.g. most countries in EU) |
Practitioner who ordered genetic testing receives report. Other practitioners involved in treatment of an individual should be informed according to individual consent |
| Relatives: Delivery of information to relevant family members is usually not regulated and is in responsibility of the index individual or his/her legally appointed guardian. Genetic counselling is then offered upon request. | Delivery of information to relevant family members is the responsibility of the index individual. Genetic counselling should then be offered upon request. | |
| Insurances/employer: National regulations on delivery of information to health care providers or employers differ. Some countries have legislation that prohibits health care providers and insurers from requesting or utilizing genetic testing results. (e.g. UK, USA, Switzerland) | Health care providers should have access and options restricted to utilize genetic testing results. | |
| What sort of secondary findings can be detected and to what extent should they be reported? | Handling of secondary findings usually depends on individual consent. Extent, definition, integrity and screening quality of relevant additional findings are usually not regulated by legislation and varies greatly between testing laboratories. | Depending on the individual’s consent, actionable secondary findings (i.e. according to ACMG guidelines; Richards et al., 2015 [39]) should actively be screened for by the laboratory with same comprehensive screening quality as the primary diagnostic genetic testing approach. Potential results should be communicated to the individual by a genetic counsellor or medical geneticist. Non-actionable secondary findings should not be looked for and not be reported back. |
| What happens if a genetic testing result questions assumed family relations? | Questioning or confirmation of genetic relations is not the subject of medical genetic but rather of forensic testing. There may be various reasons for discordant findings, e.g. sample mix-up, semen or egg donation, adoption, bone marrow transplantation. Handling of discordant findings from genetic testing is usually not regulated by legislation. | It is advisable to communicate that a genetic testing approach was inconclusive than to actively question family relations. |
| Should a negative genetic test result be re-evaluated at a later time and how should a potentially novel result be managed? | Re-evaluation of negative genetic testing is usually not regulated by legislation and is offered by some testing laboratories upon request. | Re-evaluation of negative genetic testing should be possible upon request after, e.g. after a few years. Novel results (diagnostic or incidental findings) should be managed as in normal diagnostic settings. It is advisable to address this issue in the individual consent form. |
| Will genetic material be stored after genetic testing? | Accredited diagnostic laboratories are obligated to store DNA (or any patient specimen) for a specified period of time, as part of their laboratory accreditation and quality management systems. Storage of DNA obtained for diagnostic testing may not be regulated by the country’s legislation. However, the patient’s consent is usually required for future use of the DNA or the generated data (re-analysis, re-testing, research etc.). | The individual should be offered the option to consent for unlimited storage of tissue / DNA sample. |
| Will individual records be stored after genetic testing? | Diverse spectrum of legislation with respect to time scale until individual data will be deleted. | The individual should be offered the option to consent for unlimited storage of records. |
| Can a laboratory perform an upload of anonymized genetic findings to e.g. population databases? | Some countries have regulations that allow or even require an upload of anonymized variants to variant databases without particular individual consent. (e.g. Germany) | An upload of anonymized variants to variant databases without individual consent is desirable, however, countries will have variable ethical requirements often based on cultural beliefs that need to be considered. |
A list of genes that are associated with actionable secondary findings is maintained by ACMG and currently comprises 73 genes (ACMG SF v3.0 [56]), mostly corresponding to cancer predisposition, cardiac conduction disease and metabolic disorders. Note that this list is periodically updated, and the number of genes included is likely to increase over time. Since the genes and variants and their associated conditions are typically beyond the scope of expertise of the epileptologist or genetic counsel- or experienced in epilepsy genetics, it is advised that clinicians seek expertise from the appropriate colleagues before reporting these findings and their associated recommendations to individuals and families.
A field of active discussion is whether genetic findings may also influence decision-making related to epilepsy surgery. To date there has not been a large-scale systematic evaluation of the relationship between the presence of a genetic diagnosis, its type and surgical outcome. In general, detection of a pathogenic variant is not an absolute contraindication for epilepsy surgery [57, 58], but each case must be evaluated taking into account current knowledge on the specific genetic disorder, its natural disease course, and the individual case characteristics; in such cases, it would be prudent to include a clinician with genetic epilepsy expertise in the multidisciplinary epilepsy surgery consensus meeting.
Despite these benefits, one of the most relevant limitations is the restricted implementation of genetic testing in routine clinical practice. In many health systems globally, genetic testing is not included as part of routine health care or analysis techniques may be outdated, which results in limited or no access to testing or substantial costs to the individual and family.
Legal implications of genetic testing
Many countries have their own legislation regulating various aspects of genetic testing, but the details differ substantially, and some jurisdictions do not have specific regulations at all [59]. The differences in regulations generally revolve around the reasons for testing (i.e., diagnostic, carrier, predictive, prenatal). Table 6 lists various questions regarding genetic testing, examples of how these questions are being addressed in some countries, as well as suggestions on how questions may be handled in countries where no relevant legislation is yet in place. In addition to legal requirements, there may be local or regional requirements (e.g., insurance companies in some states in the US requiring a medical doctor with genetics training to order a genetic test).
Outlook
The pace of new discoveries in genomic medicine is rapid [31], making it a challenge for all parties to stay informed with state-of-the-art information at all times. Several future directions are briefly outlined here with more detail available in other publications [60]. As sequencing costs decrease, it is anticipated that GS will eventually replace ES in the coming years as a first-line genetic test for the epilepsies, as is already the case in some countries. The interpretation of non-coding genetic variation is still in its infancy and there will likely be a transition period with increased uncertainty with respect to results of GS due to an even larger number of VUS emerging per test. With time and increased experience, other opportunities derived from GS will unfold, such as calculation of polygenic risk scores for epilepsy, and more accurate and comprehensive detection of repeat expansions and structural variants. Additional methodologies will likely find their way into the standard portfolio of genetic testing, such as RNA sequencing, methylome analysis and long-read sequencing. For these analyses, DNA from lymphocytes is not always the best representative source, and skin biopsies or liquid biopsies [61] will likely complement current source materials for genetic testing. Furthermore, we expect that precision medicine approaches, including both the rational use of (repurposed) drugs and more advanced antisense oligonucleotide or gene therapy approaches, will be established for an increasing number of genetic epilepsies. To reach this goal, mechanistic insights in the molecular biology of individual genetic epilepsies, pre-clinical data, knowledge on the natural history of each disorder, and appropriately designed clinical trials will all be needed to support their use. We therefore encourage clinicians and genetic testing laboratories to include individuals in ongoing research efforts to advance knowledge on treatment and management of rare genetic epilepsies.
To facilitate the increased possibilities and outcomes, new forms of communication between referring clinicians and genetic testing may help influence the standard of care [62]. Genetic testing has already become routine practice in some countries for selected groups of individuals, such as those with DEE. We anticipate that with increasing demonstration of the impact of genetic testing on the care of individual patients, it will take a more prominent role in clinical practice, to the point that it will become as much a part of diagnostic evaluation as EEG and MRI in the evaluation of individuals with epilepsy.
Example cases
Case 1: An individual with a focal epilepsy
The individual was sent to an epilepsy center at 46 years of age. He had suffered from a drug-resistant form of epilepsy since six years of age with frequent focal seizures with loss of awareness, and bilateral tonic-clonic seizures which occurred up to four times a week. More than 10 antiseizure medications had been tried alone or in combination. Non-progressive tubers were identified on neuroimaging in the right frontal, pre- and postcentral regions in the right hemisphere and left temporal and bilateral occipital regions. A diagnosis of tuberous sclerosis was suspected and a pathogenic variant in the TSC1 gene was detected subsequently. Everolimus was started without changing the antiseizure medications at the individual’s request. The individual attained full control of seizures with this therapeutic regime for several months and the medication was well tolerated.
Case 2: An individual with a idiopathic generalized epilepsy
An individual developed bilateral myoclonic seizures of the arms at 13 years of age. These appeared in the first hour after awakening, and interfered with routine activities, such as having breakfast and personal hygiene. During the rest of the day, myoclonic seizures rarely occurred. There was no family history of epilepsy. At 15 years of age, he had his first generalized tonic-clonic seizure after sleep deprivation. The neurological examination and MRI of the brain was normal. The EEG showed frequent generalized epileptic discharges; a diagnosis of juvenile myoclonic epilepsy (JME) was made. He started valproic acid which was well tolerated, and this resulted in seizure freedom. At 30 years of age, he married and had his first child. He was concerned that he might pass the disease on to his child and went to his epileptologist. Polygenic inheritance was assumed, and recurrence risk for offspring was estimated to be 3–8% [18]. Genetic diagnostics were not performed as the diagnostic yield and clinical utility were considered to be low.
Case 3: An individual with DEE
A three-year-old female, born at term, had first seizures at the age of one year. The clinician ordered genetic testing and provided “epilepsy” as the sole phenotypic information. The laboratory initiated exome sequencing and simultaneously requested additional phenotypic details from the ordering clinician. With more time on hand, the referring clinicians informed the laboratory about daily refractory generalized seizures, severe developmental delay, behavior abnormalities, muscular hypotonia and cortical visual impairment. A fast-track trio-ES identified a pathogenic de novo variant in GRIN2B, encoding a subunit of the N-methyl-D-aspartate receptor (NMDAR). The identified missense variant is previously described and associated with ID and epilepsy in multiple individuals with a consistent phenotype. Published functional data suggests a loss-of-function effect. Thus, the laboratory recommended to consider treatment with L-serine [63, 64]. Using this precision medicine approach, parents and clinicians noted behavioral improvements and a reduced seizure frequency within the next few weeks. ■
Competencies and learning objectives.
To gain awareness and understanding of genetic causes of epilepsy
To learn about important aspects of genetic counseling before and after genetic testing
To learn about the different types of tests available
To be able to decide which genetic test should be performed in which type of epilepsy
To consider precision medicine implications of genetic test results
Acknowledgements and disclosures.
SW received funding from FWO (1861419N) and the Queen Elisabeth Medical Foundation. IK received funding from the German Society of Epileptology through an Otfrid-Foerster-Stipendium.
SMS is supported by the Epilepsy Society (UK). This work was partly carried out at NIHR University College London Hospitals Biomedical Research Centre, which receives a proportion of funding from the UK Department of Health’s NIHR Biomedical Research Centres funding scheme.
IH, HL, and YW are supported by Research Unit FOR-2715 of the German Research Foundation (DFG, grants He5415/7–2, Le1030/16–2 and/23–1, We4896/4–2) and HL and YW by Treat-ION network of the Federal Ministry for Education and Research (BMBF, grants 01GM1907A/B).
Members of the ILAE Genetics Commission or the Task Force on Clinical Genetic Testing in the Epilepsies:
Samuel F. Berkovic4, J. Helen Cross23, Ingo Helbig5−10, Holger Lerche12, Daniel Lowenstein24, Heather C. Mefford25, Piero Perucca26, Annapurna Poduri16, Nigel C.K. Tan27, Hande Caglayan28, Katherine Helbig29, Michael S. Hildebrand11, Johannes R. Lemke1,22, Lynette Sadleir17, Gagandeep Singh20, Sanjay Sisodiya18, Yvonne Weber12,21, Sarah Weckhuysen19
23University College London National Institute for Health Research Biomedical Research Centre, Great Ormond Street Institute of Child Health, London, UK
24Department of Neurology, University of California, San Francisco, USA
25Division of Genetic Medicine, Department of Pediatrics, University of Washington, Seattle, Washington, USA
26The Royal Melbourne Hospital, The University of Melbourne, Australia
27Department of Neurology, National Neuroscience Institute, Singapore
28Departmentof Molecular Biology and Genetics, Bogaziçi University, Istanbul, Turkey
29Division of Neurology, Children’s Hospital of Philadelphia, Philadelphia, PA, USA
30Dayanand Medical College, Ludhiana, Punjab, India
TEST YOURSELF
- Which statement is correct?
- Genetic epilepsies are defined by a known or presumed underlying genetic etiology and by the lack of an acquired cause.
- Most genetic epilepsies follow Mendelian inheritance.
- Twin studies were uninformative concerning genetic risk in epilepsy.
- Autosomal recessive inheritance is only seen in consanguineous families.
- Once a genetic test is negative, subsequent testing is not necessary.
- What are the principles of autosomal dominant inheritance?
- The risk of transmitting the pathogenic variant from an affected individual to his/her offspring is 50% with each pregnancy.
- Only females are affected.
- Affected individuals occur in every second generation.
- Genetic testing is not necessary in families with an autosomal dominant mode of inheritance.
- A variant in a gene associated with autosomal dominant inheritance cannot be found in blood samples.
- Which aspects should be considered in genetic counseling after genetic testing?
- Variants of uncertain significance can be ignored.
- Implications of a positive result should be discussed in detail.
- Family planning is not influenced by the result.
- All family members should be contacted by the treating physician to organize genetic testing.
- In the case of a negative result, the individual does not have a genetic epilepsy.
- Which aspects should be considered if individuals ask for the benefits of genetic testing in epilepsy?
- There are no clinical benefits yet.
- There is no need for further neurological follow-up.
- A definite diagnosis can be an important benefit for the individual.
- Establishing a genetic diagnosis always leads to a more precise therapy.
- All individuals have improved outcomes as a result of genetic testing.
- Which is correct about genetic testing methods for individuals with epilepsy?
- Analysis of copy number variations is irrelevant in the genetics of epilepsy.
- Single gene sequencing is the most cost-effective method.
- Most epilepsy syndromes are associated with changes in one gene.
- Exome sequencing gives information about variants in the coding regions of genes.
- Karyotyping should be performed as the first step in all cases.
- How commonly are pathogenic or likely pathogenic copy number variations identified in individuals with developmental and epileptic encephalopathy?
- < 20 %
- 20–30 %
- 30–40 %
- 50–80 %
- > 80%
- Which of the following statements is correct regarding genetic re-testing?
- Genetic re-testing can be informative when new knowledge becomes available.
- Genetic re-testing should be performed only if the diagnosis of the individual has changed.
- Genetic re-testing in epilepsy is unnecessary.
- Re-testing should be performed not earlier than 10 years after the last testing was performed.
- Genetic re-testing almost always produces a conclusive result.
- In which group of people does routine clinical genetic testing currently have the highest yield?
- People with genetic generalized epilepsies, such as absence epilepsies
- People with developmental and epileptic encephalopathy
- People with lesional epilepsies, such as those with hippocampal sclerosis or focal cortical dysplasia
- In all people with epilepsy who ask about the risk of epilepsy in their children
- All cases of childhood-onset epilepsy
- Who should communicate a genetic test result to clinically affected individuals (valid in most countries)?
- Any clinician (physician, genetic counselor, nurse)
- Family members
- A geneticist or a clinician familiar with the situation of the individual, genetic epilepsies, and the test that was performed.
- Only a clinical geneticist
- Only a neuropediatrician/pediatric neurologist
- Regarding precision medicine, which statement is correct?
- Individuals with pathogenic variants in SLC2A1 and neurological symptoms should consider treatment with the ketogenic diet.
- Sodium channel blockers should generally be avoided in individuals with loss-of-function SCN1A variants.
- Sodium channel blockers should be considered in individuals with gain-of-function sodium channel variants.
- Administration of vitamin B6 is essential in individuals with pathogenic variants in the PNPO gene.
- All of the above.
Note: Reading the manuscript provides an answer to all questions. Correct answers may be accessed on the website, www.epilepticdisorders.com.
Footnotes
Supplementary material.
Supplementary data accompanying the manuscript are available at www.epilepticdisorders.com.
None of the authors have any conflicts of interest to declare.
References
- 1.Beghi E, Giussani G, Nichols E, Abd-Allah F, Abdela J, Abdelalim A, et al. Global, regional, and national burden of epilepsy, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol 2019; 18(4): 357–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hauser WA, Kurland LT. The epidemiology of epilepsy in Rochester, Minnesota, 1935 through 1967. Epilepsia 1975; 16 (1): 1–66. [DOI] [PubMed] [Google Scholar]
- 3.Scheffer IE, Berkovic S, Capovilla G, Connolly MB, French J, Guilhoto L, et al. ILAE classification of the epilepsies: position paper of the ILAE commission for classification and terminology. Epilepsia 2017; 58(4): 512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helbig I, Scheffer IE, Mulley JC, Berkovic SF. Navigating the channels and beyond: unravelling the genetics of the epilepsies. Lancet Neurol 2008; 7(3): 231–45. [DOI] [PubMed] [Google Scholar]
- 5.Berkovic SF, Howell RA, Hay DA, Hopper JL. Epilepsies in twins: genetics of the major epilepsy syndromes. Ann Neurol 1998; 43(4): 435–45. [DOI] [PubMed] [Google Scholar]
- 6.Howell KB, Eggers S, Dalziel K, Riseley J, Mandelstam S, Myers CT, et al. A population-based cost-effectiveness study of early genetic testing in severe epilepsies of infancy. Epilepsia 2018; 59(6): 1177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papuc SM, Abela L, Steindl K, Begemann A, Simmons TL, Schmitt B, et al. The role of recessive inheritance in early-onset epileptic encephalopathies: a combined whole-exome sequencing and copy number study. Eur J Hum Genet 2019; 27(3): 408–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Symonds JD, Zuberi SM, Stewart K, McLellan A, O’Regan M, MacLeod S, et al. Incidence and phenotypes of childhood-onset genetic epilepsies: a prospective population-based national cohort. Brain 2019; 142(8): 2303–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catarino CB, Liu JYW, Liagkouras I, Gibbons VS, Labrum RW, Ellis R, et al. Dravet syndrome as epileptic encephalopathy: evidence from long-term course and neuropathology. Brain 2011; 134(Pt 10): 2982–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balestrini S, Sisodiya SM. Audit of use of stiripentol in adults with Dravet syndrome. Acta Neurol Scand 2017; 135(1): 73–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer EE, Schofield D, Shrestha R, Kandula T, Macintosh R, Lawson JA, et al. Integrating exome sequencing into a diagnostic pathway for epileptic encephalopathy: evidence of clinical utility and cost effectiveness. Mol Genet Genomic Med 2018; 6(2): 186–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer EE, Howell K, Scheffer IE. Natural History Studies and Clinical Trial Readiness for Genetic Developmental and Epileptic Encephalopathies. Neurotherapeutics 2021; 18(3): 1432–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeffrey JS, Leathem J, King C, Mefford HC, Ross K, Sadleir LG. Developmental and epileptic encephalopathy: Personal utility of a genetic diagnosis for families. Epilepsia Open 2021; 6(1): 149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vears DF, Dunn KL, Wake SA, Scheffer IE. It’s good to know”: experiences of gene identification and result disclosure in familial epilepsies. Epilepsy Res 2015; 112: 64–71. [DOI] [PubMed] [Google Scholar]
- 15.Ottman R, Hirose S, Jain S, Lerche H, Lopes-Cendes I, Noebels JL, et al. Genetic testing in the epilepsies-Report of the ILAE Genetics Commission. Epilepsia 2010; 51(4): 655–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helbig I, Heinzen EL, Mefford HC, the ILAE Genetics Commission. Primer Part 1-The building blocks of epilepsy genetics. Epilepsia 2016; 57(6): 861–8. [DOI] [PubMed] [Google Scholar]
- 17.Niemi MEK, Martin HC, Rice DL, Gallone G, Gordon S, Kelemen M, et al. Common genetic variants contribute to risk of rare severe neurodevelopmental disorders. Nature 2018; 562(7726): 268–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peljto AL, Barker-Cummings C, Vasoli VM, Leibson CL, Hauser WA, Buchhalter JR, et al. Familial risk of epilepsy: a population-based study. Brain 2014; 137(3): 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheidley BR, Malinowski J, Bergner AL, Bier L, Gloss DS, Mu W, et al. Genetic testing for the epilepsies: a systematic review. Epilepsia 2022; 63(2): 375–87. [DOI] [PubMed] [Google Scholar]
- 20.Lemke JR, High-Throughput. Sequencing as first-tier diagnostics in congenital and early-onset disorders. JAMA Pediatr 2017; 171(9): 833. [DOI] [PubMed] [Google Scholar]
- 21.Klau J, Abou Jamra R, Radtke M, Oppermann H, Lemke JR, Beblo S, et al. Exome first approach to reduce diagnostic costs and time - retrospective analysis of 111 individuals with rare neurodevelopmental disorders. Eur J Hum Genet 2022; 30(1): 117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jauss RT, Popp B, Platzer K, Jamra R. MorbidGenes-Panel-v2022–02.1 [Internet]. Zenodo, 2022. https://zenodo.org/record/6136995 [Google Scholar]
- 23.Martin AR, Williams E, Foulger RE, Leigh S, Daugherty LC, Niblock O, et al. PanelApp crowdsources expert knowledge to establish consensus diagnostic gene panels. Nat Genet 2019; 51(11): 1560–5. [DOI] [PubMed] [Google Scholar]
- 24.Stark Z, Foulger RE, Williams E, Thompson BA, Patel C, Lunke S, et al. Scaling national and international improvement in virtual gene panel curation via a collaborative approach to discordance resolution. Am J Hum Genet 2021; 108(9): 1551–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiStefano MT, Goehringer S, Babb L, Alkuraya FS, Amberger J, Amin M, et al. The gene curation coalition: a global effort to harmonize gene-disease evidence resources [Internet]. Medrxiv, 2022. 10.1101/2022.01.03.21268593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thormann A, Halachev M, McLaren W, Moore DJ, Svinti V, Campbell A, et al. Flexible and scalable diagnostic filtering of genomic variants using G2P with Ensembl VEP. Nat Commun 2019; 10(1): 2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kochinke K, Zweier C, Nijhof B, Fenckova M, Cizek P, Honti F, et al. Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am J Hum Genet 2016; 98(1): 149–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helbig I, Riggs ER, Barry C-A, Klein KM, Dyment D, Thaxton C, et al. The clingen epilepsy gene curation expert panel-bridging the divide between clinical domain knowledge and formal gene curation criteria. Hum Mutat 2018; 39(11): 1476–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmer EE, Sachdev R, Macintosh R, Melo US, Mundlos S, Righetti S, et al. Diagnostic yield of whole genome sequencing after nondiagnostic exome sequencing or gene panel in developmental and epileptic encephalopathies. Neurology 2021; 96(13): e1770–82. [DOI] [PubMed] [Google Scholar]
- 30.Ibañez K, Polke J, Hagelstrom RT, Dolzhenko E, Pasko D, Thomas ERA, et al. Whole genome sequencing for the diagnosis of neurological repeat expansion disorders in the UK: a retrospective diagnostic accuracy and prospective clinical validation study. Lancet Neurol 2022; 21(3): 234–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bamshad MJ, Nickerson DA, Chong JX. Mendelian gene discovery: fast and furious with no end in sight. Am J Hum Genet 2019; 105(3): 448–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niestroj L-M, Perez-Palma E, Howrigan DP, Zhou Y, Cheng F, Saarentaus E, et al. Epilepsy subtype-specific copy number burden observed in a genome-wide study of 17 458 subjects. Brain 2020; 143(7): 2106–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borlot F, Regan BM, Bassett AS, Stavropoulos DJ, Andrade DM. Prevalence of pathogenic copy number variation in adults with pediatric-onset epilepsy and intellectual disability. JAMA Neurol 2017; 74(11): 1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson H, Shen Y, Avallone J, Sheidley BR, Pinsky R, Bergin AM, et al. Copy number variation plays an important role in clinical epilepsy. Ann Neurol 2014; 75(6): 943–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Djémié T, Weckhuysen S, von Spiczak S, Carvill GL, Jaehn J, Anttonen A-K, et al. Pitfalls in genetic testing: the story of missed SCN1A mutations. Mol Genet Genomic Med 2016; 4 (4): 457–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borch LA, Parboosingh J, Thomas MA, Veale P. Reevaluating the first-tier status of fragile X testing in neurodevelopmental disorders. Genet Med 2020; 22(6): 1036–9. [DOI] [PubMed] [Google Scholar]
- 37.Peters L, Depienne C, Klebe S. Familial adult myoclonic epilepsy (FAME): clinical features, molecular characteristics, pathophysiological aspects and diagnostic work-up. Medizinische Genetik 2022; 33(4): 311–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dagli AI, Mathews J, Williams CA. Angelman Syndrome. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJ, Gripp KW, et al. , editors. GeneReviews® [Internet]. University of Washington, Seattle: Seattle (WA), 1993. http://www.ncbi.nlm.nih.gov/books/NBK1144/ [Google Scholar]
- 39.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17(5): 405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis-Smith D, Galer PD, Balagura G, Kearney H, Ganesan S, Cosico M, et al. Modeling seizures in the Human Phenotype Ontology according to contemporary ILAE concepts makes big phenotypic data tractable. Epilepsia 2021; 62(6): 1293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stödberg T, Tomson T, Barbaro M, Stranneheim H, Anderlid B-M, Carlsson S, et al. Epilepsy syndromes, etiologies, and the use of next-generation sequencing in epilepsy presenting in the first 2 years of life: a population-based study. Epilepsia 2020; 61(11): 2486–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benson KA, White M, Allen NM, Byrne S, Carton R, Comerford E, et al. A comparison of genomic diagnostics in adults and children with epilepsy and comorbid intellectual disability. Eur J Hum Genet 2020; 28(8): 1066–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johannesen KM, Nikanorova N, Marjanovic D, Pavbro A, Larsen LHG, Rubboli G, et al. Utility of genetic testing for therapeutic decision-making in adults with epilepsy. Epilepsia 2020; 61(6): 1234–9. [DOI] [PubMed] [Google Scholar]
- 44.Møller RS, Larsen LHG, Johannesen KM, Talvik I, Talvik T, Vaher U, et al. Gene panel testing in epileptic encephalopathies and familial epilepsies. Mol Syndromol 2016; 7(4): 210–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Demarest ST, Brooks-Kayal A. From molecules to medicines: the dawn of targeted therapies for genetic epilepsies. Nat Rev Neurol 2018; 14(12): 735–45. [DOI] [PubMed] [Google Scholar]
- 46.Truty R, Patil N, Sankar R, Sullivan J, Millichap J, Carvill G, et al. Possible precision medicine implications from genetic testing using combined detection of sequence and intragenic copy number variants in a large cohort with childhood epilepsy. Epilepsia Open 2019; 4(3): 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCormack M, McGinty RN, Zhu X, Slattery L, Heinzen EL, Costello DJ, et al. De novo mutations in patients with chronic ultra-refractory epilepsy with onset after age five years. Eur J Med Genet 2020; 63(1): 103625. [DOI] [PubMed] [Google Scholar]
- 48.Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med 2020; 22(2): 245–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heyne HO, Baez-Nieto D, Iqbal S, Palmer DS, Brunklaus A, May P, et al. Predicting functional effects of missense variants in voltage-gated sodium and calcium channels. Sci Transl Med 2020; 12(556): eaay6848. [DOI] [PubMed] [Google Scholar]
- 50.Boßelmann CM, Hedrich UBS, Müller P, Sonnenberg L, Parthasarathy S, Helbig I, et al. Predicting the functional effects of voltage-gated potassium channel missense variants with multi-task learning [Internet]. Bioinformatics, 2021. 10.1101/2021.12.02.470894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farmer GD, Gray H, Chandratillake G, Raymond FL, Freeman ALJ. Recommendations for designing genetic test reports to be understood by patients and non-specialists. Eur J Hum Genet 2020; 28(7): 885–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Lange IM, Koudijs MJ, van ‘t Slot R, Sonsma ACM, Mulder F, Carbo EC, et al. Assessment of parental mosaicism in SCN1A -related epilepsy by single-molecule molecular inversion probes and next-generation sequencing. J Med Genet 2019; 56(2): 75–80. [DOI] [PubMed] [Google Scholar]
- 53.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013; 15(7): 565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med 2017; 19(2): 249–55. [DOI] [PubMed] [Google Scholar]
- 55.Miller DT, Lee K, Gordon AS, Amendola LM, Adelman K, Bale SJ, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2021 update: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2021; 23(8): 1391–8. [DOI] [PubMed] [Google Scholar]
- 56.Miller DT, Lee K, Chung WK, Gordon AS, Herman GE, Klein TE, et al. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: a policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet Med 2021; 23(8): 1381–90. [DOI] [PubMed] [Google Scholar]
- 57.Sanders MWCB, Lemmens CMC, Jansen FE, Brilstra EH, Koeleman BPC, Braun KPJ, et al. Implications of genetic diagnostics in epilepsy surgery candidates: A single-center cohort study. Epilepsia Open 2019; 4(4): 609–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stevelink R, Sanders MWCB, Tuinman MP, Brilstra EH, Koeleman BPC, Jansen FE, et al. Epilepsy surgery for patients with genetic refractory epilepsy: a systematic review. Epileptic Disorders 2018; 20(2): 99–115. [DOI] [PubMed] [Google Scholar]
- 59.Kiess W, Bornehag C-G, Gennings C, editors. Preliminaries [Internet]. Pediatr Adolesc Med 2018; 21: 1–8. https://www.karger.com/Article/FullText/481317 [Google Scholar]
- 60.Hartley T, Lemire G, Kernohan KD, Howley HE, Adams DR, Boycott KM. New diagnostic approaches for undiagnosed rare genetic diseases. Annu Rev Genomics Hum Genet 2020; 21: 351–72. [DOI] [PubMed] [Google Scholar]
- 61.Ye Z, Chatterton Z, Pflueger J, Damiano JA, McQuillan L, Harvey AS, et al. Cerebrospinal fluid liquid biopsy for detecting somatic mosaicism in brain. Brain Commun 2021; 3(1): fcaa235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pearson NM, Stolte C, Shi K, Beren F, Abul-Husn NS, Bertier G, et al. GenomeDiver: a platform for phenotype-guided medical genomic diagnosis. Genet Med 2021; 23(10): 1998–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soto D, Olivella M, Grau C, Armstrong J, Alcon C, Gasull X, et al. L-Serine dietary supplementation is associated with clinical improvement of loss-of-function GRIN2B-related pediatric encephalopathy. Sci Signal 2019; 12(586): eaaw0936. [DOI] [PubMed] [Google Scholar]
- 64.Krey I, von Spiczak S, Johannesen KM, Hikel C, Kurlemann G, Muhle H, et al. L-serine treatment is associated with improvements in behavior, EEG, and seizure frequency in individuals with GRIN-related disorders due to null variants. Neurotherapeutics 2022; 19(1): 334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Møller RS, Liebmann N, Larsen LHG, Stiller M, Hentschel J, Kako N, et al. Parental mosaicism in epilepsies due to alleged de novo variants. Epilepsia 2019; 60(6): e63–6. [DOI] [PubMed] [Google Scholar]
- 66.Myers CT, Hollingsworth G, Muir AM, Schneider AL, Thuesmunn Z, Knupp A, et al. Parental Mosaicismin “denovo” epileptic encephalopathies. N Engl J Med 2018; 378(17): 1646–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zsurka G, Kunz WS. Mitochondrial dysfunction and seizures: the neuronal energy crisis. Lancet Neurol 2015; 14 (9): 956–66. [DOI] [PubMed] [Google Scholar]
- 68.Sánchez Fernández I, Gaínza-Lein M, Lamb N, Loddenkemper T. Meta-analysis and cost-effectiveness of second-line antiepileptic drugs for status epilepticus. Neurology 2019; 92(20): e2339–48. [DOI] [PubMed] [Google Scholar]
- 69.Mills PB, Camuzeaux SSM, Footitt EJ, Mills KA, Gissen P, Fisher L, et al. Epilepsy due to PNPO mutations: genotype, environment and treatment affect presentation and outcome. Brain 2014; 137(Pt 5): 1350–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darin N, Reid E, Prunetti L, Samuelsson L, Husain RA, Wilson M, et al. Mutations in PROSC disrupt cellular pyridoxal phosphate homeostasis and cause vitamin-B6-dependent epilepsy. Am J Hum Genet 2016; 99(6): 1325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koch J, Mayr JA, Alhaddad B, Rauscher C, Bierau J, Kovacs-Nagy R, et al. CAD mutations and uridine-responsive epileptic encephalopathy. Brain 2017; 140(2): 279–86. [DOI] [PubMed] [Google Scholar]
- 72.Fox J, Thodeson DM, Dolce AM. Nicotine: a targeted therapy for epilepsy due to nAChR gene variants. J Child Neurol 2021; 36(5): 371–7. [DOI] [PubMed] [Google Scholar]
- 73.Lossius K, de Saint Martin A, Myren-Svelstad S, Bjørnvold M, Minken G, Seegmuller C, et al. Remarkable effect of transdermal nicotine in children with CHRNA4-related autosomal dominant sleep-related hypermotor epilepsy. Epilepsy Behav 2020; 105: 106944. [DOI] [PubMed] [Google Scholar]
- 74.Pierson TM, Yuan H, Marsh ED, Fuentes-Fajardo K, Adams DR, Markello T, et al. GRIN2A mutation and early-onset epileptic encephalopathy: personalized therapy with memantine. Ann Clin Transl Neurol 2014; 1(3): 190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gale JR, Kosobucki GJ, Hartnett-Scott KA, Aizenman E. Imprecision in precision medicine: differential response of a disease-linked GluN2A mutant to NMDA channel blockers. Front Pharmacol 2021; 12: 773455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amador A, Bostick CD, Olson H, Peters J, Camp CR, Krizay D, et al. Modelling and treating GRIN2A developmental and epileptic encephalopathy in mice. Brain 2020; 143(7): 2039–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Syrbe S, Hedrich UBS, Riesch E, Djémié T, Müller S, Møller RS, et al. De novo loss- or gain-of-function mutations in KCNA2 cause epileptic encephalopathy. Nat Genet 2015; 47(4): 393–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hedrich UBS, Lauxmann S, Wolff M, Synofzik M, Bast T, Binelli A, et al. 4-Aminopyridine is a promising treatment option for patients with gain-of-function KCNA2-encephalopathy. Sci Transl Med 2021; 13(609): eaaz4957. [DOI] [PubMed] [Google Scholar]
- 79.Pisano T, Numis AL, Heavin SB, Weckhuysen S, Angriman M, Suls A, et al. Early and effective treatment of KCNQ2 encephalopathy. Epilepsia 2015; 56(5): 685–91. [DOI] [PubMed] [Google Scholar]
- 80.Sands TT, Balestri M, Bellini G, Mulkey SB, Danhaive O, Bakken EH, et al. Rapid and safe response to low-dose carbamazepine in neonatal epilepsy. Epilepsia 2016; 57(12): 2019–30. [DOI] [PubMed] [Google Scholar]
- 81.Nissenkorn A, Kornilov P, Peretz A, Blumkin L, Heimer G, Ben-Zeev B, et al. Personalized treatment with retigabine for pharmacoresistant epilepsy arising from a pathogenic variant in the KCNQ2 selectivity filter. Epileptic Disord 2021; 23(5): 695–705. [DOI] [PubMed] [Google Scholar]
- 82.Orhan G, Bock M, Schepers D, Ilina EI, Reichel SN, Löffler H, et al. Dominant-negative effects of KCNQ2 mutations are associated with epileptic encephalopathy: KCNQ2 defects in EE. Ann Neurol 2014; 75(3): 382–94. [DOI] [PubMed] [Google Scholar]
- 83.Millichap JJ, Park KL, Tsuchida T, Ben-Zeev B, Carmant L, Flamini R, et al. KCNQ2 encephalopathy: features, mutational hot spots, and ezogabine treatment of 11 patients. Neurol Genet 2016; 2(5): e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vanoye CG, Desai RR, Ji Z, Adusumilli S, Jairam N, Ghabra N, et al. High-throughput evaluation of epilepsy-associated KCNQ2 variants reveals functional and pharmacological heterogeneity. JCI Insight 2022; 7(5): e156314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guerrini R, Dravet C, Genton P, Belmonte A, Kaminska A, Dulac O. Lamotrigine and seizure aggravation in severe myoclonic epilepsy. Epilepsia 1998; 39(5): 508–12. [DOI] [PubMed] [Google Scholar]
- 86.Wolff M, Johannesen KM, Hedrich UBS, Masnada S, Rubboli G, Gardella E, et al. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain 2017; 140(5): 1316–36. [DOI] [PubMed] [Google Scholar]
- 87.Klepper J, Akman C, Armeno M, Auvin S, Cervenka M, Cross HJ, et al. Glut1 deficiency syndrome (Glut1DS): state of the art in 2020 and recommendations of the international Glut1DS study group. Epilepsia Open 2020; 5 (3): 354–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.French JA, Lawson JA, Yapici Z, Ikeda H, Polster T, Nabbout R, et al. Adjunctive everolimus therapy for treatment-resistant focal-onset seizures associated with tuberous sclerosis (EXIST-3): a phase 3, randomised, double-blind, placebo-controlled study. The Lancet 2016; 388(10056): 2153–63. [DOI] [PubMed] [Google Scholar]


