Abstract
Proteins harboring a C-terminal cell wall sorting signal are covalently linked to pentaglycine acceptors within the staphylococcal peptidoglycan. This pentaglycine was modified when the lysostaphin immunity factor (Lif) of Staphylococcus simulans was expressed in Staphylococcus carnosus, likely by the exchange of two glycine residues for serine residues. A reporter protein was efficiently linked to the modified acceptor, indicating that the sorting reaction is not strictly dependent on the wild-type structures of the acceptors.
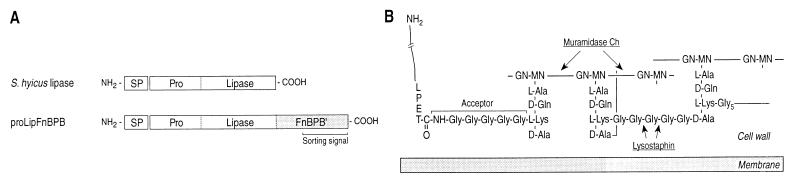
Proteins covalently anchored to the cell wall of gram-positive bacteria contain, in addition to a cleavable N-terminal signal peptide, a C-terminal cell wall sorting signal, starting with the conserved LPXTG sequence motif, followed by a hydrophobic region and a charged tail (13). A postulated sorting machinery cleaves the surface protein between the threonine and the glycine residues of the LPXTG sequence motif (8). In Staphylococcus aureus, the carboxyl group of the threonine is subsequently amide linked to the acceptor, the pentaglycine of a branched anchor peptide [NH2-Ala-γ-Gln-Lys-(NH2-Gly5)-Ala-COOH], which in turn is linked to the glycan backbone of the peptidoglycan (18). The branched anchor peptides appear not to be substituted at their C-terminal d-alanine, and this seems to be their only structural difference from the branched wall peptides that cross-link the peptidoglycan via their interpeptide chains (18).
A number of point mutations within the cell wall sorting signal have been described as affecting cell wall sorting of surface proteins (13, 14). Previous work left unanswered whether the sorting machinery displays a similar specificity to the acceptors of surface proteins. To address this question, we sought a way to modify the pentaglycine of branched anchor peptides. In S. aureus, Staphylococcus simulans, Staphylococcus carnosus, and other staphylococci, the interpeptide chains are composed of five glycine residues (10, 11). This pentaglycine is the target of lysostaphin (1), which cleaves between the third and fourth glycine residues (12). Lif, the lysostaphin immunity factor of S. simulans bv. staphylolyticus, causes the incorporation of two serine residues into the interpeptide chains, thereby conferring lysostaphin resistance (17). The branched wall peptides and the branched anchor peptides are thought to be synthesized in the same way (18). We therefore assumed that Lif would not only cause selective serine incorporation into the interpeptide chains but would also alter the acceptors of surface proteins. In this work, the effect of Lif on the pentaglycine of branched anchor peptides, on secretion, and on cell wall anchoring of surface proteins was studied in S. carnosus.
Bacterial strains, culture conditions, and cell wall lytic enzymes.
The wild-type strain S. carnosus TM300 (4) was transformed (5) and cultivated at 30°C in basic broth (BB) (15). When appropriate, BB was supplemented with chloramphenicol (10 mg liter−1) or tetracycline (25 mg liter−1). Genes of interest were expressed under the control of the xylose promoter-repressor system of Staphylococcus xylosus (19) and were induced as described previously (15). Muramidase Ch (6) was a generous gift of J. Hash, Nashville, Tenn. The lysostaphin preparation used in this study was purified to homogeneity and showed no contaminating proteins in Coomassie blue-stained polyacrylamide gels.
Lif does not interfere with secretion or with cell wall anchoring of proteins in S. carnosus.
Expression of lif from plasmid pCXlif renders the interpeptide chains of S. carnosus resistant to lysostaphin (17). To study whether these changes influence secretion or cell wall anchoring of proteins, two different reporter enzymes were used (Fig. 1A): (i) authentic Staphylococcus hyicus lipase, a secreted enzyme encoded on plasmid pTX15 (9), and (ii) ProLipFnBPB, a hybrid protein consisting of S. hyicus lipase fused to the C-terminal region of S. aureus fibronectin binding protein B (7), which is covalently anchored to the cell wall in an enzymatically active conformation (15). ProLipFnBPB is encoded on plasmid pTX30, which was constructed by inserting a BamHI-NarI fragment from pCX30 (15) into the respective restriction sites of pTX15. To first test the influence of the reporter enzymes on the lysostaphin-resistant phenotype of cells expressing lif, S. carnosus TM300 and cells producing S. hyicus lipase or proLipFnBPB in the presence or absence of Lif (pCXlif) were washed and resuspended in lysostaphin buffer (0.15 M NaCl, 50 mM Tris-HCl, pH 7.9) to an optical density of 0.50 (at 590 nm). Cells that did not harbor pCXlif were lysed by lysostaphin (10 μg ml−1), and the optical density at 590 nm decreased to about 0.1 in 10 min at 30°C. In contrast, the optical density of cells that carried pCXlif did not decrease, indicating that coexpression of the lipase or proLipFnBPB did not compromise the Lif-mediated lysostaphin resistance of S. carnosus.
FIG. 1.
Structures of reporter proteins and the staphylococcal peptidoglycan. (A) Schematic diagrams showing the domains of the S. hyicus lipase and proLipFnBPB, a reporter enzyme for cell wall anchoring, consisting of the cleavable signal peptide (SP), the propeptide (Pro), and the catalytic domain (Lipase) of S. hyicus lipase fused to the C-terminal region of the S. aureus fibronectin binding protein B (FnBPB′). FnBPB′ comprises the complete cell wall-spanning region and the cell wall sorting signal of the binding protein. (B) Structure of staphylococcal peptidoglycan with a C-terminally processed surface protein attached to the lysostaphin-sensitive wild-type pentaglycine acceptor of a branched anchor peptide. Cleavage sites of cell wall lytic enzymes used in this study are indicated (modified after references 16 and 18).
The influence of Lif on secretion and cell wall anchoring of proteins was studied by determining the distribution of the lipase activity between the cell surface and the culture supernatants in clones producing S. hyicus lipase or proLipFnBPB. Cultures were separated into cell pellets and culture supernatants by centrifugation. The pellets were washed three times and resuspended in BB. Dilutions of the culture supernatants, the resuspended cells, or the proteins released by lysostaphin treatment (80 μg ml−1 in BB; 30 min at 37°C) from the walls of cells harboring only pTX30 were mixed with reaction buffer (10 mM CaCl2, 0.1% Triton X-100, and 20 mM Tris-HCl, pH 8.5, containing 5 mM of the chromogenic lipase substrate p-nitrophenyl caprylate [Sigma]). The hydrolysis of the substrate was monitored in a reaction volume of 100 μl at 405 nm for 10 min at 30°C with a SpectraMax 340 microplate reader (Molecular Devices) against the respective samples derived from wild-type S. carnosus TM300. All lipase activities were determined in quintuplicate in four independent experiments. Due to steric hinderance the specific activity of cell wall-immobilized proLipFnBPB is lower than that of proLipFnBPB released from the cells (15). A correction factor (1.25), determined by comparing the specific lipase activity of cell wall-immobilized and lysostaphin-solubilized proLipFnBPB in cells that did not express lif, was also used to correct the activity of proLipFnBPB anchored to the walls of lif-expressing cells.
Cells harboring pTX15 secreted 99.2% of the total lipase activity into the culture supernatant, compared to 99.1% secreted by cells containing pTX15 and pCXlif. In contrast, cells carrying pTX30 displayed 85.1% of the total lipase activity at their surfaces, compared to 84.5% in cells harboring pTX30 and pCXlif. Thus, lif expression does not interfere with the secretion of the lipase or the covalent anchoring of prolipFnBPB.
Effect of Lif on the branched anchor peptides.
Branched anchor peptides that tether surface proteins to the staphylococcal cell wall do not contribute to the cross-linking of the peptidoglycan (18). Therefore, a possibility remained that the branched anchor peptides of lif-expressing cells were still equipped with wild-type, i.e., lysostaphin-sensitive, pentaglycine acceptors. Incubation of washed cells in the presence of lysostaphin (80 μg ml−1 in BB; 30 min at 37°C) released 8% of the lipase activity from cells producing both proLipFnBPB and Lif, whereas the same procedure released 100% of lipase activity from cells producing only proLipFnBPB. These results indicated that in lif-expressing cells at least some of the surface proteins were attached to acceptors that were sensitive to lysostaphin, probably because they had retained the wild-type pentaglycine structure.
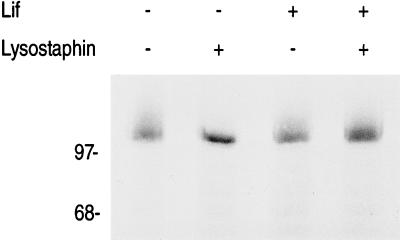
To exclude the possibility that proLipFnBPB was noncovalently anchored to the cell wall of lif-expressing cells, we employed a strategy based on the sequential use of muramidase Ch and lysostaphin. Muramidase Ch hydrolyzes the β-1,4 linkage of N-acetylmuramic acid and N-acetylglucosamine (Fig. 1B) (3). It cleaves at some distance from the anchoring points of surface proteins, thereby solubilizing those proteins attached to cell wall fragments of variable length (13). In contrast, surface proteins that are noncovalently anchored to the cell wall are solubilized by muramidase Ch with a uniform molecular mass (13). Pellets derived from 500 μl of a culture were washed three times in water and precipitated with trichloroacetic acid (7%, wt/vol) for 20 min on ice. After centrifugation, the precipitates were washed twice in acetone and dried under vacuum. The precipitates were resuspended in 170 μl of BB containing muramidase Ch (100 μg ml−1) and incubated for 3 h at 37°C. The samples were centrifuged, and each supernatant was divided into two aliquots of 80 μl. Prior to incubation for 30 min at 37°C, either 20 μl of water or 20 μl of lysostaphin stock solution (400 μg ml−1) was added to each aliquot. Subsequently, the samples were concentrated, and the aliquots were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting. Muramidase Ch released prolipFnBPB efficiently from the cell wall of S. carnosus, regardless of whether lif was expressed or not. In both cases, a spectrum of lipase-specific signals was visualized as a smear in immunoblots (Fig. 2). When prolipFnBPB was released with muramidase Ch and digested with lysostaphin prior to immunoblotting, the attached cell wall fragments of variable lengths were quantitatively removed only in samples derived from cells that did not express lif, transforming the smear into a signal of a uniform molecular mass. In contrast, surface proteins released from lif-expressing cells were not sensitive to lysostaphin (Fig. 2).
FIG. 2.
Lysostaphin sensitivity of branched anchor peptides solubilized with muramidase Ch from the cell wall of S. carnosus. Cells synthesizing proLipFnBPB encoded on plasmid pTX30 in the presence (+) or absence (−) of Lif (pCXlif) were washed, trichloroacetic acid-precipitated, and digested with muramidase Ch. Subsequently, the solubilized hybrid proteins were incubated with (+) or without (−) lysostaphin prior to Tricine–SDS-PAGE (10% acrylamide) and immunoblotting with prolipase-specific antiserum as described previously (15). The molecular masses of standard proteins (in kDa) are indicated on the left.
S. carnosus releases surface proteins linked to cell wall fragments into the culture supernatant.
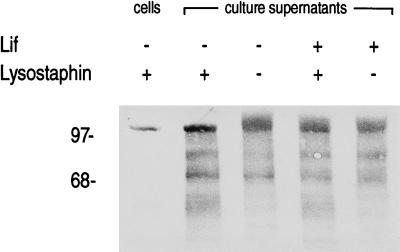
When proLipFnBPB was expressed from low-copy-number plasmid pCX30, about 5% of total lipase activity was found in the culture supernatant (15). In contrast, 15% was released from cells expressing proLipFnBPB from medium-copy-number plasmid pTX30 (see above). Coexpression of unrelated surface proteins did not increase this natural release of lipase activity (data not shown), excluding the possibility that a gene dosage effect, i.e., saturation of the sorting machinery, was responsible for the phenomenon. Concentrated culture supernatants of cells harboring plasmid pCXlif and/or pTX30 were collected, concentrated, and analyzed by immunoblotting to study further the molecular basis of the release of lipase activity by cells producing proLipFnBPB. ProLipFnBPB released with lysostaphin from the peptidoglycan of cells harboring only plasmid pTX30 was included as a reference in this analysis. In addition to degradation products which had electrophoretic mobilities higher than the reference, smears of lipase-specific signals were observed in the culture supernatants (Fig. 3). Incubation with lysostaphin (80 μg ml−1 in BB; 30 min at 37°C) prior to SDS-PAGE and immunoblotting had an effect only on the patterns found in the culture supernatants of cells that did not express lif. In this case, the smear of lipase-specific signals was transformed into a distinct band that migrated at the same electrophoretic mobility as the reference. The lipase-specific signals in the culture supernatant of cells synthesizing Lif and proLipFnBPB were not influenced by lysostaphin (Fig. 3). These findings indicate that release of proLipFnBPB occurred after its covalent linkage to the (modified) acceptors, suggesting that the release was caused—predominantly involving a muramidase activity—by natural cell wall turnover, a process widespread among gram-positive bacteria (2).
FIG. 3.
Lysostaphin sensitivity of proLipFnBPB naturally released by S. carnosus into the culture supernatant during growth. Culture supernatants derived from clones synthesizing proLipFnBPB (pTX30) in the presence (+) or absence (−) of Lif (pCXlif) were collected and incubated with (+) or without (−) lysostaphin. As a reference, proLipFnBPB released with lysostaphin from the cell wall of S. carnosus harboring only pTX30 was included. Proteins were separated by Tricine–SDS-PAGE (10% acrylamide) and immunoblotted with prolipase-specific antiserum. The molecular masses of standard proteins (in kDa) are indicated on the left.
Conclusion.
This work demonstrates that in S. carnosus the acceptor of surface proteins, the pentaglycine of branched anchor peptides, has a modified amino acid composition when Lif is expressed. Branched anchor peptides are thought to originate from the same pool of peptidoglycan precursors as the branched wall peptides that cross-link the peptidoglycan via pentaglycine interpeptide chains (18). The fact that Lif confers lysostaphin resistance on both structures strongly suggests that the acceptors acquire the same modification as the interpeptide chains, which has previously been shown to be the exchange of two glycine residues for two serine residues (17). Since surface proteins were linked to modified acceptors as efficiently as to wild-type acceptors, the cell wall sorting reaction seems not to be strictly dependent on their wild-type pentaglycine structures.
Acknowledgments
We are indebted to Silke Egner for excellent technical assistance. We thank Vera Augsburger and Regine Stemmler for technical assistance and Karen A. Brune for critically reading the manuscript. We are grateful to John Hash for the generous gift of muramidase Ch.
This work was supported by grants from the BMFT-BEO and by Evotec BioSystems Ltd., Hamburg, Germany.
REFERENCES
- 1.Browder H P, Zygmunt J R, Young J R, Tavormina P A. Lysostaphin: enzymatic mode of action. Biochem Biophys Res Commun. 1965;19:383–389. doi: 10.1016/0006-291x(65)90473-0. [DOI] [PubMed] [Google Scholar]
- 2.Doyle R J, Chaloupka J, Vinter V. Turnover of cell walls in microorganisms. Microbiol Rev. 1988;52:554–567. doi: 10.1128/mr.52.4.554-567.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghuysen J-M. Use of bacteriolytic enzymes in determination of wall structure and their role in cell metabolism. Bacteriol Rev. 1968;32:425–464. [PMC free article] [PubMed] [Google Scholar]
- 4.Götz F. Staphylococcus carnosus: a new host organism for gene cloning and protein production. Soc Appl Bacteriol Symp Ser. 1990;19:495–535. doi: 10.1111/j.1365-2672.1990.tb01797.x. [DOI] [PubMed] [Google Scholar]
- 5.Götz F, Schumacher B. Improvements of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol Lett. 1987;40:285–288. [Google Scholar]
- 6.Hash J H, Rothlauf M V. The N,O-diacetylmuramidase of Chalaropsis species. J Biol Chem. 1967;242:5586–5590. [PubMed] [Google Scholar]
- 7.Jönsson K, Signäs C, Müller H-P, Lindberg M. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur J Biochem. 1991;202:1041–1048. doi: 10.1111/j.1432-1033.1991.tb16468.x. [DOI] [PubMed] [Google Scholar]
- 8.Navarre W W, Schneewind O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in Gram-positive bacteria. Mol Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- 9.Peschel A, Ottenwälder B, Götz F. Inducible production and cellular location of epidermin biosynthetic enzyme EpiB using an improved staphylococcal expression system. FEMS Microbiol Lett. 1996;137:279–284. doi: 10.1111/j.1574-6968.1996.tb08119.x. [DOI] [PubMed] [Google Scholar]
- 10.Schleifer K H, Fischer U. Description of a new species of the genus Staphylococcus: Staphylococcus carnosus. Int J Syst Bacteriol. 1982;32:153–156. [Google Scholar]
- 11.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schneewind O, Fowler A, Faull K F. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 13.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneewind O, Model P, Fischetti V A. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 15.Strauss A, Götz F. In vivo immobilization of enzymatically active polypeptides on the cell surface of Staphylococcus carnosus. Mol Microbiol. 1996;21:491–500. doi: 10.1111/j.1365-2958.1996.tb02558.x. [DOI] [PubMed] [Google Scholar]
- 16.Strominger J L, Ghuysen J-M. Mechanisms of enzymatic bacteriolysis. Science. 1967;156:213–221. doi: 10.1126/science.156.3772.213. [DOI] [PubMed] [Google Scholar]
- 17.Thumm G, Götz F. Studies on prolysostaphin processing and characterization of the lysostaphin immunity factor (Lif) of Staphylococcus simulans biovar staphylolyticus. Mol Microbiol. 1997;23:1251–1265. doi: 10.1046/j.1365-2958.1997.2911657.x. [DOI] [PubMed] [Google Scholar]
- 18.Ton-That H, Faull K F, Schneewind O. Anchor structure of staphylococcal surface proteins. J Biol Chem. 1997;272:22285–22292. doi: 10.1074/jbc.272.35.22285. [DOI] [PubMed] [Google Scholar]
- 19.Wieland K P, Wieland B, Götz F. A promoter-screening plasmid and xylose-inducible, glucose-repressible expression vectors for Staphylococcus carnosus. Gene. 1995;158:91–96. doi: 10.1016/0378-1119(95)00137-u. [DOI] [PubMed] [Google Scholar]