Abstract
Objective:
Mast cells in the osteoarthritis (OA) synovium correlate with disease severity. This study aimed to further elucidate the role of mast cells in OA by RNA-Seq analysis and pharmacological blockade of the activity of histamine, a key mast cell mediator, in murine OA.
Methods:
We examined OA synovial tissues and fluids by flow cytometry, immunostaining, single-cell and bulk RNA-Seq, qPCR, and ELISA. Cetirizine, a histamine H1 receptor (H1R) antagonist, was used to treat the destabilization of the medial meniscus (DMM) mouse model of OA.
Results:
Flow cytometry and immunohistology analysis of OA synovial cells revealed KIT+ FcεRI+ and TPSAB1+ mast cells. Single-cell RNA-Seq of OA synovial cells identified the expression of prototypical mast cell markers KIT, TPSAB1, CPA3 and HDC, as well as distinctive markers HPGD, CAVIN2, IL1RL1, PRG2, and CKLF, confirmed by bulk RNA-Seq and qPCR. A mast cell prototypical marker expression score classified 40 OA patients into three synovial pathotypes: mast cell-high, -medium, and -low. Additionally, we detected mast cell mediators including histamine, tryptase AB1, CPA3, PRG2, CAVIN2, and CKLF in OA synovial fluids. Elevated H1R expression was detected in human OA synovium, and treatment of mice with the H1 receptor antagonist cetirizine reduced the severity and OA-related mediators in DMM.
Conclusion:
Based on differential expression of prototypical and distinct mast cell markers, human OA joints can be stratified into mast cell-high, -medium, and -low synovial tissue pathotypes. Pharmacologic blockade of histamine activity holds the potential to improve OA disease outcome.
Keywords: Osteoarthritis, RNA-Seq, mast cell, histamine, histamine H1 receptor (H1R)
INTRODUCTION
Osteoarthritis (OA) is the most prevalent arthritic disease, affects the aging population, and is a leading cause of disability1. However, its underlying pathogenic mechanisms remain elusive and disease-modifying therapy is lacking. OA is characterized by cartilage degeneration, synovitis, and formation of osteophytes1. Further understanding of the pathogenic mechanisms in OA could lead to the development of disease-modifying therapeutics.
Our previous work showed that in addition to complement factors2, integrins3, and CCL2/CCR24, mast cells in the OA synovium play an important role in disease pathogenesis. Mast cells contain granules rich in histamine, pro-inflammatory lipids, and enzymes5. Mast cells are sentinels of the innate immune system, poised to rapidly respond to exogenous pathogens and endogenous danger signals5. A wide variety of stimuli can influence degranulation of mast cells and release of pre-formed mediators including histamine, tryptases, pro-inflammatory lipids, cytokines, and chemokines6, 7.
As unique cells of the hematopoietic lineage, mast cells express the myeloid lineage marker c-Kit, encoded by the KIT gene, which is a transmembrane growth factor receptor with intrinsic tyrosine kinase activity8. KIT is highly expressed in hematopoietic stem cells from the bone marrow and its activity is critical for hematopoiesis as well as for the proliferation, survival, differentiation and homing of these cells9. Expression of KIT is generally lost during the differentiation process of most hematopoietic cells, except mast cells, which retain KIT throughout their lifespan8. The specific ligand for c-Kit, Kit ligand, also known as stem cell factor (SCF), is encoded by the KITLG gene8. SCF is an important growth factor for human mast cells. SCF induces chemotaxis and survival of mast cells as well as proliferation and differentiation of immature mast cells from CD34+ progenitors10.
In mast cells, tryptases (α-tryptase and β-tryptase) are the most abundant secretory granule-derived serine proteinases and used as a marker for the activation of mast cells. Tryptases from synovial mast cells may degrade different joint components and are implicated in OA pathogenesis11. Another protease found in mast cell secretory granules is carboxypeptidase A3 (CPA3)12, also known as mast cell carboxypeptidase A, which is encoded by CPA3 gene. The CPA3 expression has only been detected in mast cells and MC-like cell lines13. CPA3 degrades proteins and peptides and is also involved in OA pathogenesis11. Both CPA3 and TPSAB1 are only detected in mast cells14.
Histamine is a critical mediator of mast cell-mediated anaphylaxis. It is synthesized primarily by mast cells, basophils, and histaminergic neurons in the basal ganglia of the brain and enterochromaffin-like cells (ECL) in the stomach. Mast cells, histamine, and the histamine H1 receptor (H1R) are involved in different inflammatory disease conditions. A H1R antagonist, cetirizine, exerts its beneficial effects on viral myocarditis by suppressing expression of pro-inflammatory cytokines15. In the cross-sectional analyses of Osteoarthritis Initiative (OAI) cohort data, Shirinsky reported H1-antihistamines were associated with decreased prevalence of radiographic knee OA16.
In this study, we sought to identify markers that characterize synovial mast cells in OA. Using single-cell Smart-Seq2 and bulk RNA-Seq analysis of OA synovial cells, we identified common and distinctive markers of OA synovial mast cells, which were used to classify OA patients in bulk RNA-Seq analysis. Our results were confirmed by quantitative PCR (qPCR). We used ELISA to demonstrate significant pathogenic levels of mast cell mediators in human OA synovial fluids. Since mast cell histamine and H1R are both involved in the release of pro-inflammatory mediators and matrix metallopeptidases (MMPs), we examined the effects of the H1R antagonist cetirizine on the DMM mouse model of OA and demonstrated that it protects against cartilage degeneration.
MATERIALS AND METHODS
Patient samples
Synovium samples were obtained from patients with knee OA and RA who underwent total knee arthroplasty after written informed consent under human subject’s protocols approved by the Institutional Review Board at Stanford University. OA synovial tissues were used for immunostaining (n=11), single cell Smart-Seq2 RNA-Seq (n=2), bulk RNA-Seq (OA, n=40; RA, n=9), and qPCR (n=11). OA synovial fluids were used for ELISA analyses (n=21).
Human synovial cell isolation, flow cytometry, and cell sorting
Synovial tissue was isolated from OA patients who had total knee arthroplasty. The tissue was digested with 1.5mg/mL type 2 Collagenase, 0.7mg/mL hyaluronidase, and 0.3mg/mL DNase I (1–2g tissue/10mL digestion buffer) on a rotating shaker at 270rpm and 37°C for 4 hours. The dispersed cells were either used to seed the wells in 96-well PCR plates (Eppendorf) for single-cell Smart-Seq2 RNA-Seq or to perform flow cytometry analysis. Cells were stained with lineage, c-Kit, FcεRI, CD34, and Integrinβ7 antibodies (BD).
Immunostaining of synovial tissue
Frozen sections of human OA synovial tissue were fixed in 4% paraformaldehyde (PFA) for 1 hour, blocked, and sections were incubated with primary antibody specific to tryptase (1:100, Abcam #ab2378) or mouse isotype control antibody (1μg/mL, Abcam #ab170190). After washing, samples were incubated with goat anti-mouse IgG (Invitrogen), washed again, and mounted with anti-fade mounting medium that contained DAPI (Invitrogen). Stained sections were examined under either a Keyence microscope (Keyence) to obtain stitched images, or a confocal microscope (Zeiss) to obtain magnified images, and positive cells were quantified by Image J. Percentage of mast cells in total synovial cells was calculated by tryptase+ signal / DAPI+ signal. The quantification was repeated twice, and the average numbers were used for data analysis.
Single-cell Smart-Seq2 RNA-Seq
Synovial cells isolated from OA synovial tissue were seeded into 96-well PCR plates and sequenced by the Smart-Seq2 method (Broad Institute, MA). The sequencing data were aligned to human genome reference GRCh38 using STAR aligner17, and QC was performed with FastQC and multiQC packages. Data were analyzed with the Seurat platform (R package). During data QC, any coverage for less than 10,000 genes, and genes that have coverage for less than 1% of cells were removed. After QC, two synovial cell sequencing datasets were removed. For each dataset, we followed the default processes proposed by Seurat to perform clustering of cells. To this end, we first normalized the gene counts by the total library size of each cell and applied log-transformation, centered data by using Seurat’s ScaleData function, identified 5,000 most variable genes in the dataset, and applied principal component analysis (PCA) to obtain PCA-ranked dimensions. We used 20 principal components of the PCA data to perform UMAP clustering with default parameters, as described by Seurat18. Differentially expressed genes within each cluster were analyzed using FindMarkers function with Wilcoxon Rank Sum test and the Benjamini–Hochberg method to adjust the p-values for multiple testing. Heatmaps of the top markers for each cluster were generated using ComplexHeatmap (R packages).
Bulk RNA-Seq
Total RNA was extracted from human OA and RA synovial tissues with a RNeasy kit (Qiagen) and quantified by Tapestation (Agilent). RNA samples were sequenced at New York Genome Center.
Sequence reads were mapped to the human genome (GRCh38) reference using STAR17 with default parameters. Rsubread featureCounts (R package) was used to generate read counts and edgeR (R package) to analyze differentially expressed (DE) genes using gene models for hg38 downloaded from UCSC (www.genome.ucsc.edu).
The abundance of gene expression was calculated as count-per-million (CPM) reads. Genes with less than three CPM in at least three samples were filtered out. The filtered libraries were normalized using the trimmed mean of M-values (TMM) normalization method19. Adjusted p-values (padj) for multiple testing, using Benjamini-Hochberg to estimate the false discovery rate (FDR), were calculated for final estimation of DE significance. The mast cell score was calculated by measuring the average normalized expression of the KIT, TPSAB1, CPA3, and HDC genes from the bulk-RNA-seq data. The average values were log-transformed and scaled using scales R package. Based on the calculated mast score, we performed hierarchical clustering with Euclidean distance across the OA and RA samples. This resulted in clustering of the OA and RA samples into three groups: mast cell gene expression scores High, Medium, and Low. Data visualization and heatmaps were generated using ggplot2 and ComplexHeatmap (R packages). Quantiles were calculated in R with the ‘quantile’ function for continuous data20.
Quantitative PCR (qPCR)
RNA was isolated from human or murine OA synovial tissues using RNeasy Plus Mini Kit (Qiagen), reverse transcription was performed using oligo(dT)18 (Applied Biosystem), and analyzed by qPCR (Applied Biosystems). Results were normalized to 18s or β-actin RNA (2−Δ(ΔCt)).
ELISA of human OA synovial fluids
OA synovial fluids were analyzed by ELISA for histamine, tryptase AB1, mast cell carboxypeptidase A (CPA3), major basic protein (PRG2), caveolae associated protein 2 (CAVIN2), and chemokine-like factor (CKLF) (LSBio, ENZO, Biomatik) according to the manufacturers’ instructions.
Surgical induction of osteoarthritis in mice
Mouse studies were performed in accordance with recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Animal protocols were approved by the Stanford University Administrative Panel on Laboratory Animal Care and by the VA Palo Alto Health Care System Institutional Animal Care and Use Committee. C57BL/6J male mice (Stock # 000664) were obtained from The Jackson Laboratory. Destabilization of the medial meniscus (DMM) was performed as described previously21. Seven to fifteen mice were used per experiment arm. Drug treatment experiments were repeated at least three times.
Pharmacologic treatment of murine osteoarthritis
Twenty-week-old male C57BL/6J mice were randomized by cage to receive DMM surgery and treated by vehicle (PBS) or 100μg/day cetirizine (AK Scientific) once daily by oral gavage for 12 weeks (beginning 24h after DMM surgery). Mice were sacrificed 12 weeks after DMM. The impacted knees or synovial tissues from the impacted knee were isolated for histologic assessment of osteoarthritic development or qPCR to evaluate the expression of OA-related mediators.
Histologic assessment of osteoarthritic development in mice
Stifle joints were harvested 12 weeks after DMM and fixed in 10% neutral buffered formalin followed by decalcification in formic acid for 48 hours. Joints were then embedded in paraffin, and 6μm sections cut from three separate levels of the joint and stained with Safranin-O for assessment of cartilage damage. Cartilage degeneration was evaluated by two blinded observers using a modified version of a described scoring system22 as previously described2. In brief, cartilage degeneration was calculated by depth of cartilage degeneration (score of 0–4) × width of cartilage degeneration (with a score of one denoting one-third of the surface area, a score of two denoting two-thirds of the surface area, and a score of two denoting the whole surface area) in each third of the femoral-medial and tibial-medial condyles. Scores for the six regions were then added.
Statistical Analyses
For analyses involving a single comparison of RNA-Seq genes presented in boxplots, one-way Anova with post-hoc Tukey test was used. For the murine cartilage degeneration or qPCR statistical comparisons were a Mann-Whitney U test.
RESULTS
Mast cells with c-Kit, FcεRI, and tryptase in sublining of human OA synovium
Single-cell suspensions were generated from OA synovium, and stained with a Lineage cocktail (CD3, CD14, CD16, CD19, CD20, and CD56; markers for T cells, B cells, monocytes, and NK cells) along with antibodies specific for the mast cell and mast cell subpopulation markers c-Kit, FcεRI, Integrin β7, and CD34. Flow cytometry analysis demonstrated that approximately 1.67% of the Lineage− cells expressed both c-Kit and FcεRI (Fig 1A, middle panel), and that approximately 1.36% of the Lineage− cells were c-Kit+ FcεRI+ integrin β7− CD34− (Fig. 1A, upper right panel, as shown in red circles). We considered the Lineage− c-Kit+ FcεRI+ population as total mast cells, and the Lineage− c-Kit+ FcεRI+ integrin β7− CD34− population as mature mast cells in synovium, because mast cells lose expression of integrin β7 and CD34 during maturation23, 24.
Figure 1. Analysis of OA synovial mast cells using flow cytometry and immunofluorescence.
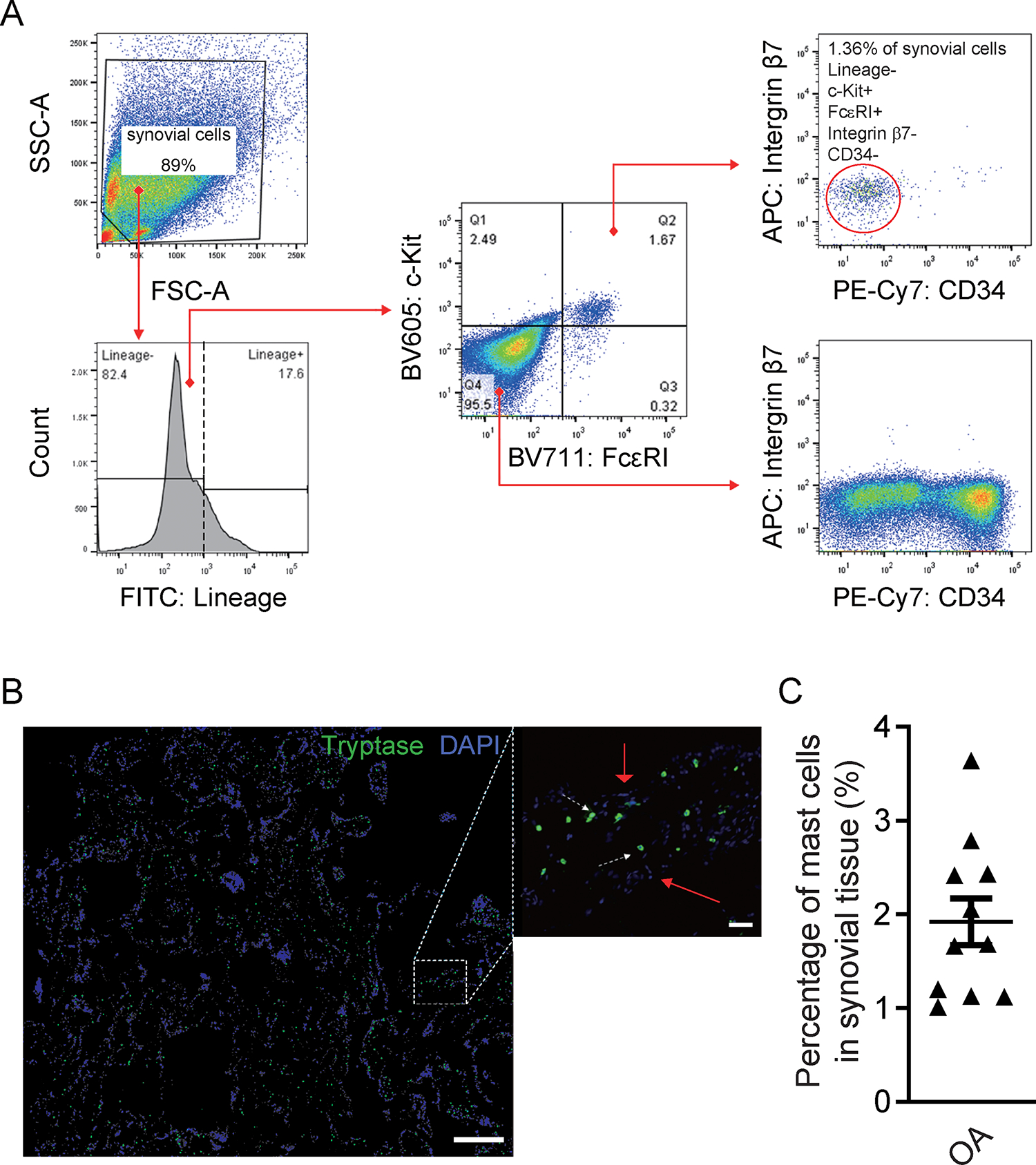
(A) Flow cytometry analysis of dissociated OA synovial cells shows Lineage− (negative for CD3, CD14, CD16, CD19, CD20, and CD56), c-Kit+, FcεRI+, CD34−, Integrinβ7− mast cells. Gating analysis shows expression of lineage markers (lower left) in the pregated synovial cell population based on their forward scatter/side scatter (FSC/SSC) profile (upper left). Expression of c-Kit and FcεRI (middle panel) within the Lineage− population, and analysis of Integrin β7 and CD34 expression gated either on c-Kit+ FcεRI+ cells (upper right), or double-negative (c-Kit− FcεRI−) cells (lower right), are shown. (B) Stitched immunofluorescence 10. images (left panel) show the distribution of tryptase+ mast cells (green) in OA synovial tissue; the 20. image (middle panel) shows tryptase+ mast cells (green) within the sublining area of the OA synovium. White arrowheads indicate tryptase AB1+ mast cells in the sublining synovial area; red arrowheads indicate DAPI+ synovial cells. (C) Quantification of mast cell numbers in the OA synovium (n=11). Mast cells in the OA synovium range from 1.01% to 3.64%. Scale bar represents 100μm, right 10μm. The data presented are representative of three independent experiments.
To map the distribution of mast cells in the human OA synovium, we performed immunostaining for tryptase AB1, which is a highly specific marker for mast cells (6). Stitched immunostaining images of OA synovial tissue showed that tryptase AB1+ mast cells were distributed throughout the synovium (Fig. 1B, left panel), and located within the sublining area (Fig. 1B, middle panel). We quantified the numbers of mast cells (green, tryptase AB1+) and total synovial cells (blue, DAPI+), and found that the OA synovium exhibited a range of mast cell levels, with mast cells constituting 1.01% – 3.64% of synovial cells (Fig. 1C).
Characterization of OA synovial mast cells by single cell RNA-Seq
To further characterize OA synovial mast cells, we performed single cell RNA-Seq of dissociated OA synovial cells. Sequenced OA synovial cells fulfilled quality control criteria (sequenced genes including <5% mitochondrial genes and >600 cellular genes)25. Using unsupervised methods to analyze differentially expressed genes, the OA synovial cells formed six distinct major clusters (Fig. 2A). This includes macrophages, two clusters of synovial lining fibroblasts (SLF), two clusters of synovial sublining fibroblasts (SSF), and mast cells. Genes with significant differential expression between clusters (adjusted p <0.01 & log fold-change >1) were used as unique markers for each cluster and the expression of the top genes are presented in a heatmap (Fig. 2B). We used the macrophage receptor with collagenous structure (MARCO) as a marker for macrophages26; Lubricin/Proteoglycan 4 (PRG4) and matrix metallopeptidase 3 (MMP3) as markers for the synovial lining fibroblasts27; and matrix metallopeptidase 2 (MMP2) as a marker for the synovial sublining fibroblasts that is known to be decreased in synovial lining fibroblast compared to the sublining cells28.
Figure 2. Identification of prototypical and distinctive mast cell marker genes in the OA synovium by single-cell RNA-Seq.
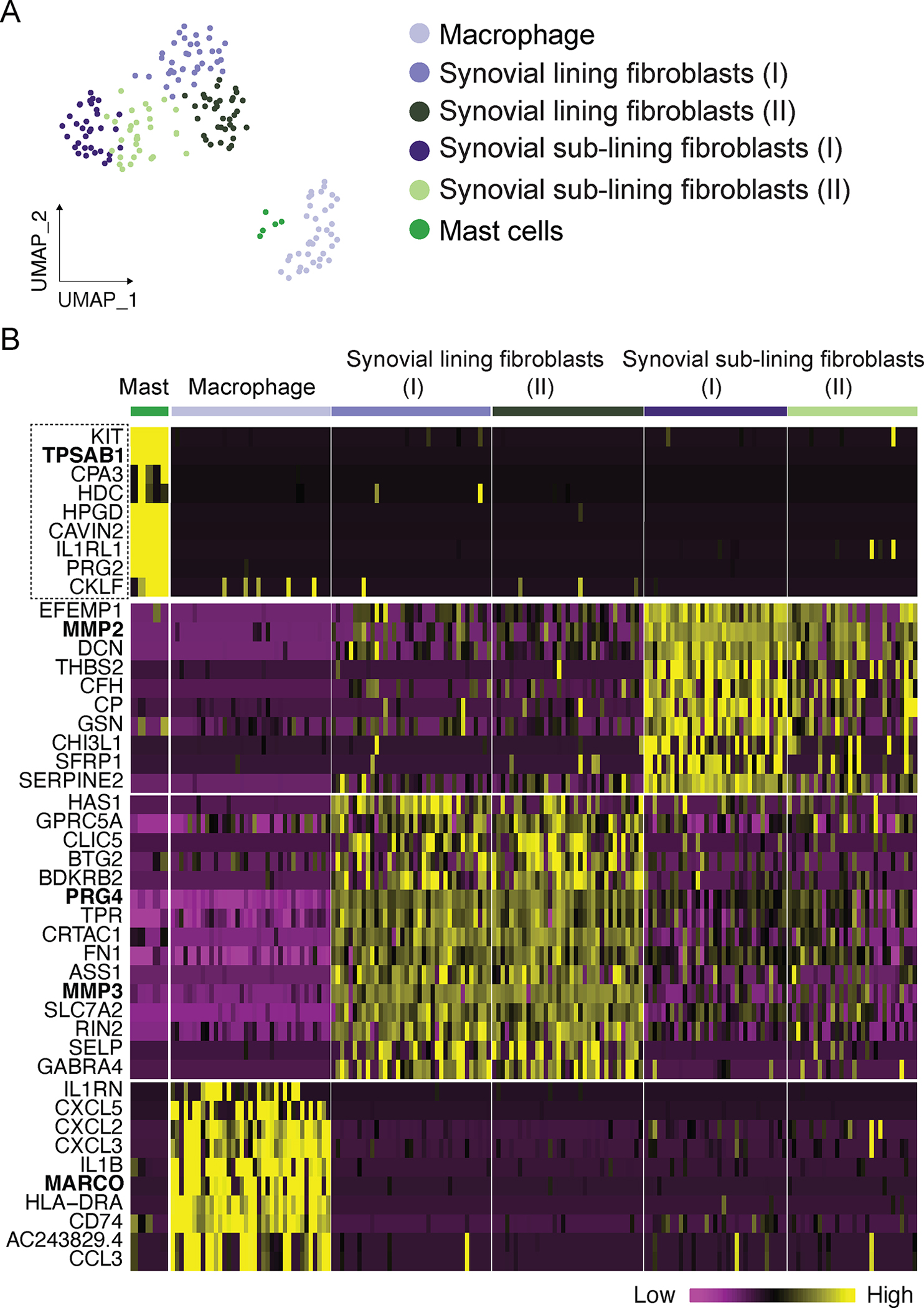
OA synovial tissue was digested to single-cell suspensions and Smart-Seq2 RNA-Seq was performed. (A) OA synovial cell single-cell Smart-Seq2 RNA-Seq analysis. Sequenced cells from human OA synovial tissues were grouped into six clusters using unsupervised approaches. Mast cells were clustered together. (B) The expression of top significant markers for each cluster are presented as heatmap. The dashed line box highlights the mast cell markers, including the characteristic mast cell marker genes KIT, TPSAB1, CPA3, HDC, and the distinctive mast cell markers HPGD, CAVIN2, IL1RL1, PRG2, and CKLF, most of which were only expressed in the synovial mast cell cluster, but not expressed or expressed at a lower level in the other cell clusters of the sequenced OA synovial cells. The unique markers for each cluster are highlighted in bold, including TPSAB1, MARCO, PRG4, MMP3 and MMP2 gene names.
The mast cell cluster was classified based on expression of the prototypical mast cell markers KIT, TPSAB1, CPA3 and HDC (Fig. 2B). Additionally, the OA synovial mast cells also expressed distinct genes including HPGD, CAVIN2, and IL1RL1, as well as genes typically expressed by other cell types, such as PRG2, which is predominantly expressed in eosinophils (Fig. 2B). The mast cells also expressed a unique chemokine gene CKLF (Fig. 2B), known as chemokine-like factor or chemokine-like factor 1.
Classification of OA patients based on a synovial mast cell gene expression profile
To transcriptionally characterize mast cells and their levels in the OA and RA synovium, bulk RNA-Seq was performed on synovial tissues from 40 OA patients and 9 RA patients. The normalized RNA-Seq gene expression levels were used to calculate a mast cell score based on the expression levels of four prototypical mast cell genes: KIT, TPSAB1, CPA3, and HDC. Based on the mast cell score, we performed hierarchical clustering with Euclidean distance across the OA and RA samples. This resulted in clustering of the OA and RA samples into three groups: mast cell gene expression scores High, Medium, and Low. The expression of KIT, TPSAB1, CPA3 and HDC revealed significant differences between the three groups (Fig. 3A–D), and their expression patterns were visualized using a heatmap (Fig. 3F, Sup. Fig. 1, and Sup. Fig. 3). As a reference, the expression level of housekeeping genes NEAT1 did not show significant differences in its expression level between the mast cell-high, mast cell-medium, and mast cell-low groups of OA patients (Fig. 3E).
Figure 3. A mast cell gene expression module classified the OA synovium into three synovial pathotypes.
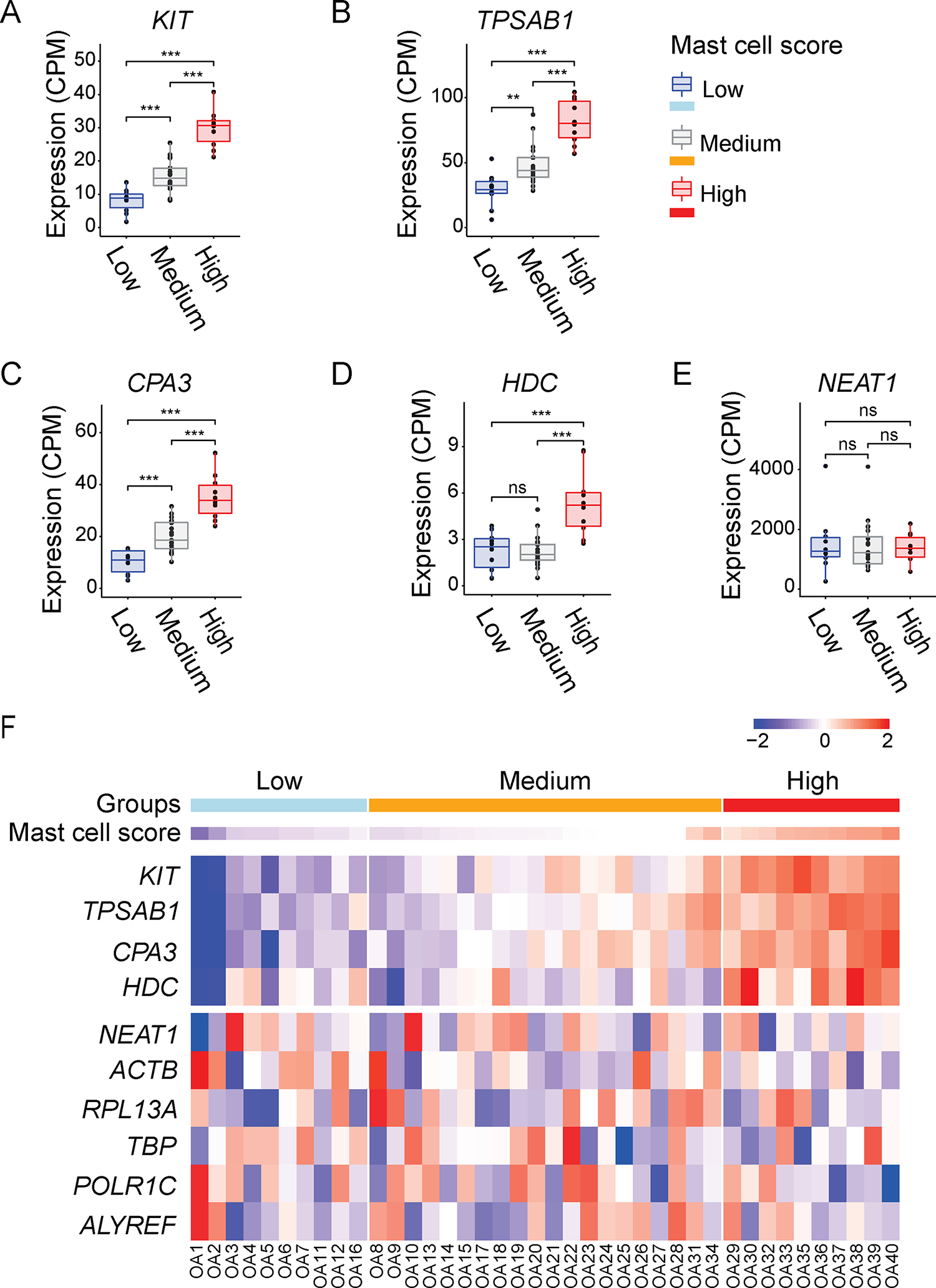
Total RNA was isolated from synovial tissues of OA patients who underwent total knee arthroplasty (n=40) and performed bulk RNA-Seq. (A-D) Boxplots of KIT, TPSAB1, CPA3, and HDC expression levels in the synovium of three groups of OA patients (n=40) with High, Medium or Low mast cell scores. (E) Boxplot of NEAT1 expression level as housekeeping gene in the synovium of three groups of OA patients (n=40) with High, Medium, or Low mast cell scores. The dots represent the normalized count-per-million (CPM) for each individual. NS, p>0.05; ★p≤0.05; ★★p≤0.01; ★★★p≤0.001 of one-way ANOVA with post-hoc Tukey test. (F) Heatmap of KIT, TPSAB1, CPA3, and HDC, and housekeeping genes NEAT1 expression levels across the OA patients (n=40) with High, Medium, or Low mast cell scores. The colors represent the scaled gene expression levels ranging from low (blue) to high (red) mRNA expression. Scale bar represents the scaled gene expression levels from low to high.
OA synovial mast cells express distinctive mast cell markers
To validate the synovial mast cell gene expression from single-cell Smart-Seq2, we performed differential expression (DE) analysis among the mast cell High, Medium, and Low OA groups. Among the DE genes, we detected two mast cell prototypical markers with different expression levels, for which no statistically significant expressions were detected by Smart-Seq2. These two genes are FCER1A and MS4A2, which encode IgE receptor FcεRI subunits FCER1A and FCER1B respectively. We also found FCER1A and MS4A2 expression levels were positively correlated with other mast cell prototypical genes in 40 OA synovial samples and exhibited significant up-regulation in the H OA group (Fig. 4A–B). Consistent with the single-cell Smart-Seq2 data, the distinctive synovial mast cell markers including HPGD, CAVIN2, and IL1RL1 were expressed in the synovial tissues of 40 OA patients, and at higher levels in the High OA group (Fig. 4C–E). KITLG, a mast cell growth factor or c-KIT ligand, and IL33, an IL1RL1 ligand, were also expressed in the OA synovial tissues and enriched in the High OA group (Fig. F–G). In comparison, no significant change in NEAT1 expression was detected in any synovial tissues of the three groups of OA patients (Fig. 4H). The heatmap shows the expression patterns of FCER1A, MS4A2, HPGD, CAVIN2, IL1RL1, KITLG, IL33, and housekeeping gene NEAT1 in individual samples with their calculated mast cell scores (Fig. 4I). Although single-cell Smart-Seq2 analysis identified the expression of PRG2 among the mast cells, the expression level of PRG2 measured by bulk RNA-Seq was below the threshold (> 3CPM in at least three cells). The bulk RNA-Seq data from the OA synovium suggest that the prototypical and distinctive mast cell markers such as KIT, TPSAB1, CPA3, HDC, FCER1A, MS4A2, HPGD, CAVIN2, IL1RL1, KITLG, and IL33 may be considered as biomarkers of OA.
Figure 4. Expression of prototypical and distinctive mast cell markers in the OA synovium.
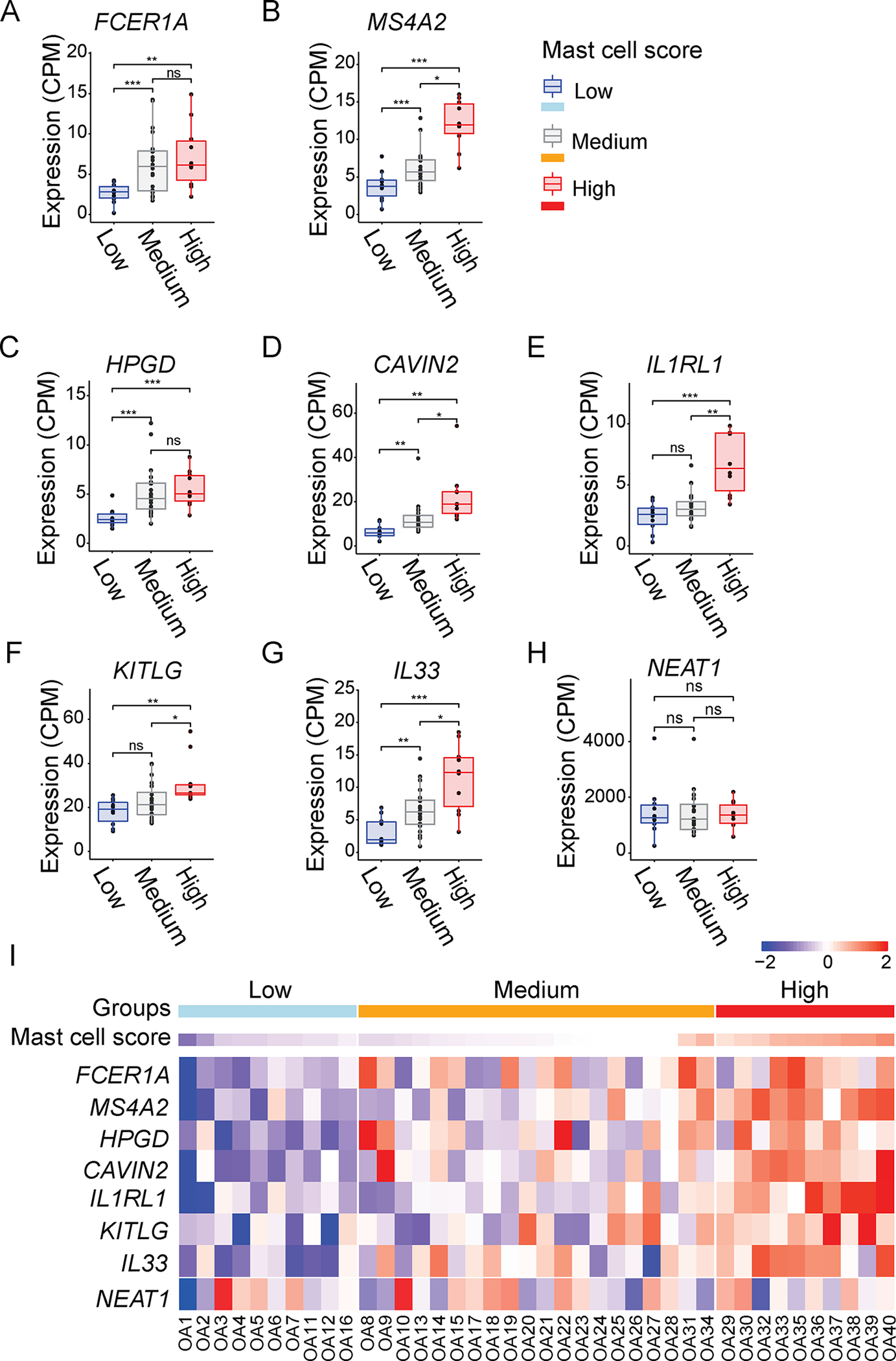
(A-G) Boxplots of FCER1A, MS4A2, HPGD, CAVIN2, IL1RL1, KITLG, IL33, expression levels in the synovium of three groups of OA patients (n=40) with High, Medium and Low mast cell scores. (H) Boxplot of NEAT1 expression level in the synovium of three groups of OA patients (n=40) with High, Medium and Low mast cell scores. The dots represent the normalized CPM for each individual. NS, p>0.05; ★p≤0.05; ★★p≤0.01; ★★★p≤0.001 of one-way ANOVA with post-hoc Tukey test. (I) Heatmap of FCER1A, MS4A2, HPGD, CAVIN2, IL1RL1, KITLG, IL33 and NEAT1 expression levels across the OA patients with total knee arthroplasty (n=40), and High, Medium or Low mast cell scores. The colors represent the scaled gene expression ranging from low (blue) to high (red) mRNA expression. Scale bar represents the scaled gene expression levels from low to high.
qPCR and ELISA validation of OA synovial mast cell biomarkers
We performed qPCR on OA synovial tissue samples to validate and further quantify the expression levels of mast cell markers identified by single-cell Smart-Seq2 and bulk RNA-Seq analyses. These markers included those common among mast cells (e.g., KIT, TPSAB1, CPA3, HDC, FCER1A, and MS4A2), and those distinctly expressed in synovial mast cells (e.g., HPGD, PRG2, CAVIN2, IL1RL1, and CKLF) and mast cell proliferation and differentiation (e.g., KITLG and IL33). Our qPCR data confirmed that these markers are expressed in human OA synovial tissues at relatively abundant levels (Fig. 5A–B).
Figure 5. Expression of prototypical and distinctive mast cell markers, and high levels of mast cell mediators, in OA synovium and synovial fluids.
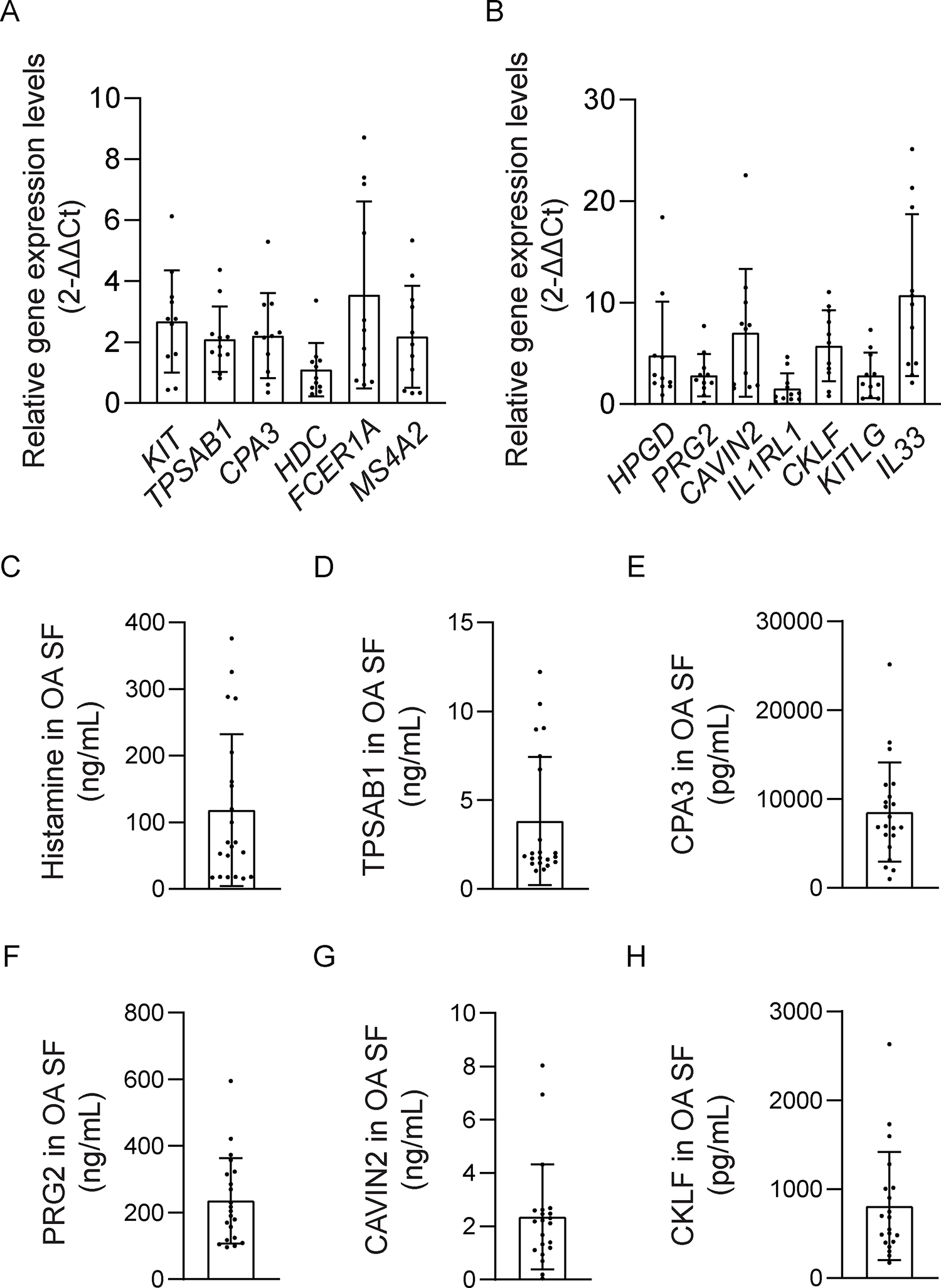
(A-B) Total RNA was isolated from synovial tissues of OA patients who underwent total knee arthroplasty (n=11) to perform qPCR. qPCR data show prototypical and distinctive mast cell marker genes including KIT, TPSAB1, CPA3, HDC, FCER1A, MS4A2, HPGD, CAVIN2, PRG2, IL1RL1, CKLF, mast cell growth factors KITLG, and IL1RL1 ligand IL33 were expressed at high levels in synovial tissues of OA patients. Data are shown as results of one representative experiment with at least three repeats. The qPCR experiments included technical triplicates. (C-H) ELISA analysis of OA-related mast cell mediators, including histamine, tryptaseAB1, CPA3, PRG2, CAVIN2, and CKLF, in OA synovial fluid from patients underwent total knee arthroplasty (n=21). Data shown are from one representative experiment with technical duplicates, for which results were confirmed in three independent experiments.
We then examined the levels of mast cell mediators in synovial fluids from OA patients who underwent total knee arthroplasty. Synovial fluids from OA patients were analyzed for the levels of histamine, TPSAB1, CPA3, PRG2, CAVIN2, and CKLF by ELISA. We detected an average level of 118.19 ng/mL for histamine, 3.83 ng/mL for TPSAB1, 8529.45 pg/mL for CPA3, 235.48 ng/mL for PRG2, 2.36 ng/mL for CAVIN2, and 809.7 pg/mL for CKLF (Fig. 5C–H). The qPCR and ELISA results indicated increased levels of mast cell mediators in the OA synovium and synovial fluids, and the usage of mast cell mediators as biomarkers in OA.
Blockade of H1 receptor reduces the severity of OA in the DMM mouse model
Histamine is a mast cell inflammation mediator. The bulk RNA-Seq data reveals 40 OA synovial tissues express high expression levels of histamine H1R (Sup. Fig. 2). A clinical study showed that histamine H1R blocker attenuates human OA16. As a result, we investigated cetirizine, a histamine H1 receptor antagonist, in the treatment of destabilization of the medial meniscus (DMM) model of OA. Cetirizine markedly attenuated cartilage degeneration in C57B/6J mice subjected to DMM, as shown in representative safranin-O-stained section images of the medial region of stifle joints (Fig. 6A; arrowheads pointing to the cartilage degeneration area) and quantification of cartilage degeneration (Fig. 6B). Treatment with cetirizine, as compared to vehicle, significantly reduced the severity of cartilage degeneration in the DMM model of OA (p ≤0.001).
Figure 6. A H1R antagonist cetirizine attenuated surgical induced OA in mice.
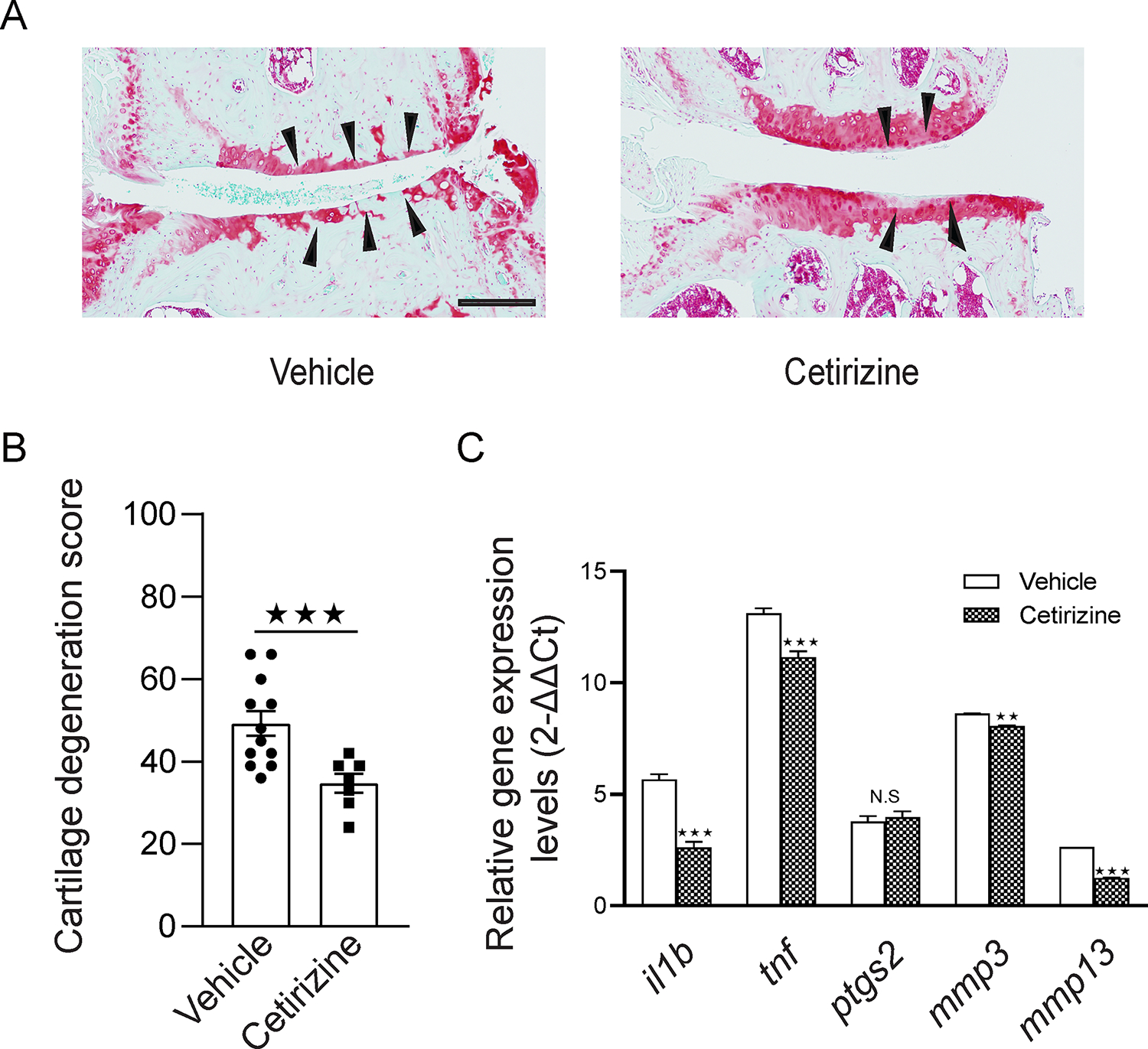
(A) Representative cartilage degeneration in safranin-O-stained sections of the medial region of stifle joints from mice subjected to DMM and immediately treated orally for 12 weeks with H1R antagonist cetirizine (n=7) or Vehicle (n=12). Black arrowheads indicate areas of cartilage degeneration. Scale bar 200μm. (B) Quantification of cartilage degeneration. Mouse DMM data are the mean ± SEM of one experiment and are representative of ≥3 independent experiments. NS, p>0.05; ★p≤0.05; ★★p≤0.01, ★★★p≤0.001 by Mann-Whitney U test. (C) RNA isolated from stifle knee joints of mice subjected to DMM and treated with vehicle or cetirizine, and transcribed to cDNA to perform qPCR. The data show that cetirizine significantly inhibited the expression of OA-related genes including IL1B, TNF, MMP3, and MMP13 (n=5) in the synovium of DMM mice compared with vehicle control. The qPCR experiments included technical triplicates. NS, p>0.05; ★p≤0.05; ★★p≤0.01, ★★★p≤0.001 by Mann-Whitney U test.
We also analyzed expression levels of OA-related mediator genes IL1B, TNF, PTGS2, MMP3, and MMP13 in OA synovial tissues from vehicle and cetirizine-treated mice. RNAs from vehicle and cetirizine-treated groups (n=5/group) were used to quantify the expression levels of OA-related mediator genes by qPCR. The results showed cetirizine significantly inhibited the expression of OA-related genes, such as IL1B, TNF, MMP3, and MMP13 (p <0.001; Fig. 6C).
DISCUSSION
A major challenge and opportunity in OA research are development of therapeutics that can prevent and treat OA. Herein, we used RNA-Seq to characterize mast cells in OA synovium. We demonstrated that human OA synovial mast cells expressed high levels of HDC, which plays an integral role in the synthesis of histamine in synovial mast cells29. Furthermore, the elevated expression of HDC in OA samples with high mast cell scores was highly significant as compared to samples with low cell mast scores. Our bulk RNA-Seq data showed H1R is highly expressed in OA synovium, including in mast cell-high, -medium and -low OA synovial tissues. Based on our RNA-Seq findings of increased HDC and H1R expression in OA synovium, and pathogenic level of histamine in OA synovial fluid, and previously reported clinical data16, we used the H1R antagonist cetirizine to treat the DMM mouse model of OA. Our results demonstrate that cetirizine reduced the development of OA in mice. Together, our findings suggest that antihistamine agents hold potential as a new therapeutic approach to prevent the development of OA. We anticipate that cetirizine exerts its effect by blocking mast cell-released histamine on H1-receptor-expressing cells, including synovial lining and sublining fibroblasts, macrophages, mast cells, and other cell types such as sensory neurons which express HRH1 etc. Cetirizine may not influence the mast cell proliferation in OA synovium, the development of some agents which can block mast cell proliferation and degranulation in the OA joints may open a new door for OA prevention and treatment.
Prior studies demonstrated that mast cell density (cells/mm2) is significantly increased in OA and RA synovium when compared to healthy synovium32, 33, 34, 35, 36. Mast cell numbers were previously described to correlate with the degree of synovitis. A major challenge in the field of OA is elucidation of the underlying pathotypes of OA joints. Based on previous research, we examined various cell types, growth factors, and inflammation mediators in human OA synovium samples from total knee replacement to identify markers that can classify OA patients. Bulk and single cell transcriptomic analyses demonstrated that human OA synovial tissues express genes encoding common and distinctive mast cell markers. We created a mast cell gene expression score based on the expression levels of prototypical mast cell genes. In this study, we classified 40 OA patients into High, Medium, and Low mast cell score groups by the expression levels of common mast cell markers including KIT, TPSAB1, CPA3, and HDC. Due to the limitations of the single-cell RNA-Seq technique, we did not observe significant expression of FCER1A or MS4A2 in our analyses. Nonetheless, we confirmed that the expression pattern of FCER1A and MS4A2 within three OA groups is consistent with the other common markers. Overall, our data suggests the use of common mast cell hallmarks including KIT, TPSAB1, CPA3, and HDC to classify OA patients. It is possible that the mast cell -high vs. mast cell-low pathotypes, which are mediated by different underlying pathogenic mechanisms, exhibit differential inflammation mediators and disease progression that shows differential response to treatments.
In this study, we reported several unique synovial mast cell markers. The gene HPGD encodes a prostaglandin-catabolizing enzyme NAD+-dependent 15-hydroxyprostaglandin dehydrogenase (15-PGDH). 15-PGDHs are considered as the key enzymes responsible for the biological inactivation of prostaglandins and related eicosanoids30. We found that HPGD is expressed specifically in mast cells in the OA synovial tissue, and the expression of HPGD in OA samples with high mast score was significantly elevated (p = 0.0005) compared to samples with low mast cell scores.
Caveolae are small, specialized trafficking vesicles involved in organization of signal transduction. Caveolae-associated protein 2 (CAVIN2), also known as serum deprivation-response protein (SDPR), is a calcium-independent phospholipid-binding protein whose expression increases in serum-starved cells. This protein is a substrate for protein kinase C (PKC) phosphorylation, and recruits both polymerase I and caveolae-associated protein 1 (CAVIN1), also known as transcript release factor (PTRF), to caveolae. Deletion of this protein increases the apparent size of cavin complexes31 and its overexpression changes caveolae to tube-like morphology. Expression of Cavin2 in the high mast cell OA group and its involvement in the size of caveolae, suggests that CAVIN2 may be involved in mast cell granule membrane assembly and the formation of suitable sized granules.
The IL1RL1 gene encodes interleukin 1 receptor-like 1 (IL1RL1 or ST2), with IL33 as its ligand. The synovial mast cells in our analysis expressed high levels of IL1RL1, enriched in the High OA group. Further investigation is needed to determine whether IL1RL1 is involved in the development of OA mast cells or synovial inflammation by recruiting immune cells through ligand-receptor binding.
Proteoglycan 2 (PRG2), also called eosinophil granule major basic protein (MBP), is the predominant constituent of the crystalline core of the eosinophil granule. PRG2 is directly implicated in epithelial cell damage, exfoliation, and bronchospasm in allergic diseases. In our study, the eosinophil marker PRG2 exclusively expressed in the synovial OA mast cells indicates that mast cells may be the source of this protein, which may contribute to OA pathobiology.
The mast cell mediators in human OA synovial fluid may contribute to deterioration in the development of OA. We previously demonstrated that tryptase plays a pathogenic role in accelerating cartilage and joint breakdown in a mouse model of OA. Some mediators (e.g., histamine and tryptase) have been well studied, while others remain poorly understood. Our results strengthen the possibility to use mast cell mediators as OA biomarkers.
Here, we used single-cell and bulk RNA-Seq to identify mast cell biomarkers and mast cell-high, medium, and low OA synovial tissue pathotypes in human OA. Our results demonstrate that the human knee OA is characterized by a range of mast cell levels, likely representing distinct synovial pathotypes arising from different underlying disease mechanisms. We further show that pharmacological blockade of H1R protects against OA development and progression in DMM mice. Future investigation is needed to fully define the role of histamine and other mast cell mediators in OA pathogenesis, and to determine if blockade of H1R and/or inhibition of mast cells will provide therapeutic benefit in OA.
Supplementary Material
Supplemental Figure 1. Heatmap of the differential expressed genes in OA synovial bulk RNA-Seq.
The gene expression levels across the OA patients with total knee arthroplasty (n=40) were grouped into mast cell scores High, Medium, and Low. (A) The top 50 significant (FDR <0.05) upregulated genes in patients with High mast cell scores. (B) The top 50 significant (FDR <0.05) upregulated genes in patients with Low mast cell scores. The colors represent the scaled gene expression ranging from low (blue) to high (red) mRNA expression. Scale bar represents the scaled gene expression levels from low to high.
Supplemental Figure 2. Expression of HRH1 in OA synovial bulk RNA-Seq.
(A) Genes ranked by the normalized expression logCPM values, from low to high. The line indicates the 75% quantile of the expressed genes. HRH1 is highlighted in blue, and displayed with Fibronectin-1 (FN1) and Clusterin (CLU), two of the most highly expressed genes. (B) Boxplots of HRH1 expression levels in the synovium of three groups of OA patients (n=40) with High, Medium, or Low mast cell scores.
Supplemental Figure 3: Expression of the mast cell-related marker genes in OA and RA synovium by bulk RNA-seq.
Heatmap of KIT, TPSAB1, CPA3, HDC, FCER1A, MS4A2, HPGD, CAVIN2, IL1RL1, KITLG, IL33 and housekeeping genes NEAT1 and ACTB expression levels across the OA patients (n=40) and RA patients (n=9) with High, Medium, or Low mast cell scores. The colors represent the scaled gene expression levels ranging from low (blue) to high (red) mRNA expression. Scale bar represents the scaled gene expression levels from low to high.
Supplemental Table 1: Demographic and clinical characters of the 40 OA patients
Acknowledgements
The authors thank Jai Sharma, Jocelyn Li, Lauren Soh, and Annie Zhou for their bio-statistical support.
Funding sources
These studies were supported by VA I01RX002689, VA BX004713, and DOD W81XWH-18-1-0590 to W.H.R. The funding sources played no role other than providing funding.
Footnotes
Conflict of interest
The authors declare no conflicts of interest.
Competing interests
The authors declare no competing interests.
REFERENCES
- 1.Robinson WH, Lepus CM, Wang Q, Raghu H, Mao R, Lindstrom TM, et al. Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat Rev Rheumatol 2016; 12: 580–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Q, Rozelle AL, Lepus CM, Scanzello CR, Song JJ, Larsen DM, et al. Identification of a central role for complement in osteoarthritis. Nat Med 2011; 17: 1674–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Q, Onuma K, Liu C, Wong H, Bloom MS, Elliott EE, et al. Dysregulated integrin alphaVbeta3 and CD47 signaling promotes joint inflammation, cartilage breakdown, and progression of osteoarthritis. JCI Insight 2019; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raghu H, Lepus CM, Wang Q, Wong HH, Lingampalli N, Oliviero F, et al. CCL2/CCR2, but not CCL5/CCR5, mediates monocyte recruitment, inflammation and cartilage destruction in osteoarthritis. Ann Rheum Dis 2017; 76: 914–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischoff SC. Role of mast cells in allergic and non-allergic immune responses: comparison of human and murine data. Nat Rev Immunol 2007; 7: 93–104. [DOI] [PubMed] [Google Scholar]
- 6.Yu Y, Blokhuis BR, Garssen J, Redegeld FA. Non-IgE mediated mast cell activation. Eur J Pharmacol 2016; 778: 33–43. [DOI] [PubMed] [Google Scholar]
- 7.Theoharides TC, Alysandratos KD, Angelidou A, Delivanis DA, Sismanopoulos N, Zhang B, et al. Mast cells and inflammation. Biochim Biophys Acta 2012; 1822: 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruse G, Metcalfe DD, Olivera A. Functional deregulation of KIT: link to mast cell proliferative diseases and other neoplasms. Immunol Allergy Clin North Am 2014; 34: 219–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ashman LK. The biology of stem cell factor and its receptor C-kit. Int J Biochem Cell Biol 1999; 31: 1037–1051. [DOI] [PubMed] [Google Scholar]
- 10.Da Silva CA, Reber L, Frossard N. Stem cell factor expression, mast cells and inflammation in asthma. Fundam Clin Pharmacol 2006; 20: 21–39. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Lepus CM, Raghu H, Reber LL, Tsai MM, Wong HH, et al. IgE-mediated mast cell activation promotes inflammation and cartilage destruction in osteoarthritis. Elife 2019; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douaiher J, Succar J, Lancerotto L, Gurish MF, Orgill DP, Hamilton MJ, et al. Development of mast cells and importance of their tryptase and chymase serine proteases in inflammation and wound healing. Adv Immunol 2014; 122: 211–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reynolds DS, Gurley DS, Austen KF. Cloning and characterization of the novel gene for mast cell carboxypeptidase A. J Clin Invest 1992; 89: 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Payne V, Kam PC. Mast cell tryptase: a review of its physiology and clinical significance. Anaesthesia 2004; 59: 695–703. [DOI] [PubMed] [Google Scholar]
- 15.Matsumori A, Yamamoto K, Shimada M. Cetirizine a histamine H1 receptor antagonist improves viral myocarditis. J Inflamm (Lond) 2010; 7: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shirinsky I, Shirinsky V. H1-antihistamines are associated with lower prevalence of radiographic knee osteoarthritis: a cross-sectional analysis of the Osteoarthritis Initiative data. Arthritis Res Ther 2018; 20: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018; 36: 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010; 11: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyndman RJF Y Sample quantiles in statistical packages. American Statistician 1996; 50: 361–365. [Google Scholar]
- 21.Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage 2007; 15: 1061–1069. [DOI] [PubMed] [Google Scholar]
- 22.Kamekura S, Hoshi K, Shimoaka T, Chung U, Chikuda H, Yamada T, et al. Osteoarthritis development in novel experimental mouse models induced by knee joint instability. Osteoarthritis Cartilage 2005; 13: 632–641. [DOI] [PubMed] [Google Scholar]
- 23.Kirshenbaum AS, Goff JP, Semere T, Foster B, Scott LM, Metcalfe DD. Demonstration that human mast cells arise from a progenitor cell population that is CD34(+), c-kit(+), and expresses aminopeptidase N (CD13). Blood 1999; 94: 2333–2342. [PubMed] [Google Scholar]
- 24.Yamada D, Kadono T, Masui Y, Yanaba K, Sato S. beta7 Integrin controls mast cell recruitment, whereas alphaE integrin modulates the number and function of CD8+ T cells in immune complex-mediated tissue injury. J Immunol 2014; 192: 4112–4121. [DOI] [PubMed] [Google Scholar]
- 25.Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol 2019; 15: e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraal G, van der Laan LJ, Elomaa O, Tryggvason K. The macrophage receptor MARCO. Microbes Infect 2000; 2: 313–316. [DOI] [PubMed] [Google Scholar]
- 27.Mizoguchi F, Slowikowski K, Wei K, Marshall JL, Rao DA, Chang SK, et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat Commun 2018; 9: 789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pakozdi A, Amin MA, Haas CS, Martinez RJ, Haines GK 3rd, Santos LL, et al. Macrophage migration inhibitory factor: a mediator of matrix metalloproteinase-2 production in rheumatoid arthritis. Arthritis Res Ther 2006; 8: R132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang H, Li Y, Liang J, Finkelman FD. Molecular Regulation of Histamine Synthesis. Front Immunol 2018; 9: 1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tai HH, Ensor CM, Tong M, Zhou H, Yan F. Prostaglandin catabolizing enzymes. Prostaglandins Other Lipid Mediat 2002; 68–69: 483–493. [DOI] [PubMed] [Google Scholar]
- 31.Hansen CG, Shvets E, Howard G, Riento K, Nichols BJ. Deletion of cavin genes reveals tissue-specific mechanisms for morphogenesis of endothelial caveolae. Nat Commun 2013; 4: 1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dean G, Hoyland JA, Denton J, Donn RP, Freemont AJ. Mast cells in the synovium and synovial fluid in osteoarthritis. Br J Rheumatol. 1993. Aug;32(8):671–5. [DOI] [PubMed] [Google Scholar]
- 33.Gotis-Graham I, McNeil HP. Mast cell responses in rheumatoid synovium. Association of the MCTC subset with matrix turnover and clinical progression. Arthritis Rheum. 1997. Mar;40(3):479–89. [DOI] [PubMed] [Google Scholar]
- 34.Buckley MG, Gallagher PJ, Walls AF. Mast cell subpopulations in the synovial tissue of patients with osteoarthritis: selective increase in numbers of tryptase-positive, chymase-negative mast cells. J Pathol. 1998. Sep;186(1):67–74. [DOI] [PubMed] [Google Scholar]
- 35.Pu J, Nishida K, Inoue H, Asahara H, Ohtsuka A, Murakami T. Mast cells in osteoarthritic and rheumatoid arthritic synovial tissues of the human knee. Acta Med Okayama. 1998. Feb;52(1):35–9. [DOI] [PubMed] [Google Scholar]
- 36.de Lange-Brokaar BJ, Kloppenburg M, Andersen SN, Dorjée AL, Yusuf E, Herb-van Toorn L, Kroon HM, Zuurmond AM, Stojanovic-Susulic V, Bloem JL, Nelissen RG, Toes RE, Ioan-Facsinay A. Characterization of synovial mast cells in knee osteoarthritis : association with clinical parameters. Osteoarthritis Cartilage. 2016. Apr;24(4):664–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Heatmap of the differential expressed genes in OA synovial bulk RNA-Seq.
The gene expression levels across the OA patients with total knee arthroplasty (n=40) were grouped into mast cell scores High, Medium, and Low. (A) The top 50 significant (FDR <0.05) upregulated genes in patients with High mast cell scores. (B) The top 50 significant (FDR <0.05) upregulated genes in patients with Low mast cell scores. The colors represent the scaled gene expression ranging from low (blue) to high (red) mRNA expression. Scale bar represents the scaled gene expression levels from low to high.
Supplemental Figure 2. Expression of HRH1 in OA synovial bulk RNA-Seq.
(A) Genes ranked by the normalized expression logCPM values, from low to high. The line indicates the 75% quantile of the expressed genes. HRH1 is highlighted in blue, and displayed with Fibronectin-1 (FN1) and Clusterin (CLU), two of the most highly expressed genes. (B) Boxplots of HRH1 expression levels in the synovium of three groups of OA patients (n=40) with High, Medium, or Low mast cell scores.
Supplemental Figure 3: Expression of the mast cell-related marker genes in OA and RA synovium by bulk RNA-seq.
Heatmap of KIT, TPSAB1, CPA3, HDC, FCER1A, MS4A2, HPGD, CAVIN2, IL1RL1, KITLG, IL33 and housekeeping genes NEAT1 and ACTB expression levels across the OA patients (n=40) and RA patients (n=9) with High, Medium, or Low mast cell scores. The colors represent the scaled gene expression levels ranging from low (blue) to high (red) mRNA expression. Scale bar represents the scaled gene expression levels from low to high.
Supplemental Table 1: Demographic and clinical characters of the 40 OA patients


