Abstract
目的
发现并鉴定肺结节患者与健康人群唾液微生物菌群特征差异及其潜在作用,以期为肺结节早期预警提供新的候选生物标志物。
方法
对肺结节(pulmonary nodule, PN)组(n=173)和健康对照(healthy control, HC)组(n=40)的唾液样本进行16S rRNA测序,比较两组人群唾液微生物群的多样性、组成和差异物种等特征,及其功能改变情况。使用随机森林算法识别唾液微生物标志物,并用曲线下面积(area under the curve, AUC)评估其对PN的预测效能。最后,基于PICRUSt2菌群功能预测分析,对唾液样本中差异基因的生物学功能及潜在作用机制进行初步探索。
结果
与HC相比,PN组唾液样本的微生物α、β多样性较高( P<0.05),且PN组唾液微生物的群落组成和丰度与HC组相比差异有统计学意义(P<0.05)。随机森林算法对差异微生物种进行筛选,Porphyromonas、Haemophilus和Fusobacterium构成了最优标志物集(AUC=0.79,95%置信区间:0.71~0.86),可有效区分PN。差异菌群的生物信息学功能显示,PN患者唾液微生物在免疫缺陷和氧化还原稳态相关的蛋白/分子功能表现出显著富集。群的生物信息学功能显示,PN患者唾液微生物在免疫缺陷和氧化还原稳态相关的蛋白/分子功能表现出显著富集。
结论
唾液微生物群的变化与PN密切相关,其可能驱动了肺结节或肺“结癌转化”的发生,提示唾液微生物具有作为PN早期诊断新型无创体液标志物的潜力。
Keywords: 唾液微生物组, 肺结节, 液体活检, 生物标志物, 前瞻性试验
Abstract
Objective
To uncover and identify the differences in salivary microbiota profiles and their potential roles between patients with pulmonary nodules (PN) and healthy controls, and to propose new candidate biomarkers for the early warning of PN.
Methods
16S rRNA amplicon sequencing was performed with the saliva samples of 173 PN patients, or the PN group, and 40 health controls, or the HC group, to compare the characteristics, including diversity, community composition, differential species, and functional changes of salivary microbiota in the two groups. Random forest algorithm was used to identify salivary microbial markers of PN and their predictive value for PN was assessed by area under the curve (AUC). Finally, the biological functions and potential mechanisms of differentially-expressed genes in saliva samples were preliminarily investigated on the basis of predictive functional profiling of Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt2).
Results
The α diversity and β diversity of salivary microbiota in the PN group were higher than those in the HC group (P<0.05). Furthermore, there were significant differences in the community composition and the abundance of oral microorganisms between the PN and the HC groups (P<0.05). Random forest algorithm was applied to identify differential microbial species. Porphyromonas, Haemophilus, and Fusobacterium constituted the optimal marker sets (AUC=0.79, 95% confidence interval: 0.71-0.86), which can be used to effectively identify patients with PN. Bioinformatics analysis of the differentially-expressed genes revealed that patients with PN showed significant enrichment in protein/molecular functions involved in immune deficiency and redox homeostasis.
Conclusion
Changes in salivary microbiota are closely associated with PN and may induce the development of PN or malignant transformation of PN, which indicates the potential of salivary microbiota to be used as a new non-invasive humoral marker for the early diagnosis of PN.
Keywords: Oral microbiome, Pulmonary nodule, Liquid biopsy, Biomarkers, Prospective trial
肺结节(pulmonary nodule, PN)存在癌变风险,且人群激增[1-2]。仅仅依靠以CT影像为主的随访监测手段难以实现PN临床处置与管理决策的精准化制定[3],更重要的是,CT筛查不仅存在辐射,还会导致患者进行不必要的检查/有创手术和延误最佳诊疗时机,其过度诊疗带来的身心负担已成为社会性问题[4]。因此,在体液层面上不断拓展无创、无辐射检测的PN稳健生物标志物和实现PN精准诊断,是聚焦肺癌这一重大疾病前期状态以实现其超早期、精准诊疗的有效路径和关键策略,也是近年来许多团队持续性探索的热点方向[5]。
微生物组在健康和疾病中到底扮演了什么样的角色?这一话题作为全世界最前沿的125个科学问题之一,近年来受到广泛关注[6]。更重要的是,大量证据不断提示相较于遗传因素,微生物作为一种环境刺激因子,诱导肺癌发生更为常见,对肺癌易感性的贡献更大[7-9]。纵观肺癌与微生物,不难发现,既往许多研究更多的是从肠道菌群视角,为溯源肺癌发生发展提供新的见解[10-12]。众所周知,肺主呼吸,与外界环境直接相通,存在动态平衡的微生物群落——口腔微生物也是肺部微生物最主要的来源,新近研究更进一步提出了唾液生物样本应作为疾病诊断首选目标样本的全新观点[13-14]。然而,包括唾液在内的口腔生物样本能否作为PN精准诊断的潜在液体活检标志物,目前的证据依然十分有限,其潜力尚未可知。
因此,本研究基于16S rRNA测序技术,开展一项前瞻性、非随机、同期对照试验,分析PN患者与健康人群唾液样本微生物群的特征和功能,发现可能对PN有预测作用的候选唾液微生物标志物,并初步探讨唾液微生物组如何影响PN的发生发展。
1. 材料和方法
1.1. 样本来源
2022年7月–2023年3月在成都中医药大学附属医院、四川省肿瘤医院、成都市中西医结合医院进行受试者招募。本研究遵循《赫尔辛基宣言》,获成都中医药大学附属医院伦理委员会审查批准(伦理批件号:2022KL-051),并在中国临床试验注册中心进行登记注册(注册号:ChiCTR2200062140),所有受试者均已签署书面知情同意书。
纳入标准:①肺结节患者的诊断标准参照《肺结节诊治中国专家共识(2018年版)》[15],肺结节直径≤3 cm,CT影像上不透明,无肺不张、肺门增大和胸腔积液;②健康人群胸部CT影像学无肺结节及其他肺部异常现象;③无其他肿瘤病史;④无呼吸系统相关手术史;⑤年龄18~80岁,性别不限。
排除标准:①有未经治疗的传染性疾病病史;②有自身免疫性疾病病史;③患有呼吸道感染疾病、口腔疾病等合并疾病。
1.2. 样本信息采集
一般信息采集:性别、年龄、吸烟史、个人肿瘤史等资料;肺CT影像学资料,并参照美国胸科医师学会(American College of Chest Physicians, ACCP)推荐的Mayo恶性概率预测模型确定肺结节风险分层:<5%(低危)、5%~65%(中危)、>65%(高危)。纯磨玻璃肺结节≤5 mm(低危),>5 mm(中危);部分实性肺结节≤8 mm(中危),>8 mm(高危)。口腔生物样本采集:受试者采样前饮用水漱口,采集非刺激性唾液2~3 mL于无菌EP管(与刺激性唾液相比,非刺激性唾液的分泌率低,在口腔中停留时间长,更容易捕获来自口腔各个部位的微生物样本)[16]。随后,干冰保存,4 h内转运至实验室−80 ℃冰箱等待后续实验。
1.3. 微生物DNA提取和测序
按照制造商的说明书(Omega Bio-tek, Norcross, GA, U.S.)进行微生物群落总基因组DNA提取,并使用NanoDrop2000(美国Thermo Scientific公司)对DNA浓度和纯度进行质检。随后以提取出的DNA为模板进行PCR扩增。使用NEXTFLEX® Rapid DNA-Seq Kit对纯化后的PCR产物进行建库。测序通过Illumina NovaSeq PE250平台进行。具体操作委托上海美吉生物医药科技有限公司完成。
1.4. 样本量计算
本研究使用PASS软件(PASS 15. NCSS, USA)的平行组间比较进行样本量估算。基于《人体微生物组临床试验指南》[17]和课题组前期预试验的α多样性Sobs指数结果:健康人群组Sobs指数均值为356,肺结节组Sobs指数均值为477,两组的Sobs指数标准差为215.3。设定两组分配比例为0.25,双侧检验水准为0.05,检验效能1−β为90%,并考虑到约10%的脱落率,最终共需纳入234例受试者。
1.5. 生物信息学分析
通过fastp在线平台(https://github.com/OpenGene/fastp, version 0.19.6)完成原始序列质控。使用FLASH(http://www.cbcb.umd.edu/software/flash, version 1.2.11)软件进行拼接。DADA2对序列进行降噪,DADA2降噪处理之后的序列则为ASVs(即扩增子序列变体)。物种注释方法采用classify-sklearn(Naive Bayes),分类置信度为0.7。为了尽量减少测序深度对后续α多样性和β多样性数据分析的影响,将所有样本序列数抽平至10000。基于Sliva 16S rRNA基因数据库(version 138, http://www.arb-silva.de),使用QIIME2(version 2022.2)中的Naive bayes分类器进行物种分类学分析。Mothur软件计算α多样性指数。采用基于ANOSIM/Bray-Curtis算法的主成分分析(principal co-ordinates analysis, PCoA)方法计算β多样性指数。Wilcoxon秩和检验的线性判别分析(linear discriminant analysis effect size, LEfSe)估计组间物种丰度差异(LDA>3, P<0.05)。随机森林算法进行疾病预测效能判别。基于Bray-Curtis距离的db-RDA分析评估临床特征与唾液微生物群落之间的关系。采用PICRUSt2功能谱预测分析差异微生物群落的功能。
1.6. 统计学方法
所有实验数据均采用SPSS 26.0软件进行实验数据统计分析。连续变量若呈正态分布,则以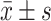 表示,并使用独立样本Student’s t检验进行组间比较;若为非正态分布,则以中位数和四分位数表示,并使用Wilcoxon符号秩和检验进行组间比较。分类变量以例数(%)表示,Wilcoxon符号秩和检验被用于等级分类变量的组间检验;当分类变量之间没有等级关联时,使用卡方检验。P<0.05为差异有统计学意义。本研究采用错误发现率(false discovery rate, FDR)法对P值进行校正,通过比较调整P值与预设的显著性水平,确定差异显著的微生物群。
表示,并使用独立样本Student’s t检验进行组间比较;若为非正态分布,则以中位数和四分位数表示,并使用Wilcoxon符号秩和检验进行组间比较。分类变量以例数(%)表示,Wilcoxon符号秩和检验被用于等级分类变量的组间检验;当分类变量之间没有等级关联时,使用卡方检验。P<0.05为差异有统计学意义。本研究采用错误发现率(false discovery rate, FDR)法对P值进行校正,通过比较调整P值与预设的显著性水平,确定差异显著的微生物群。
2. 结果
2.1. 受试者的临床特征
本研究共招募受试者234例,在排除不合格样本后,最终173例PN患者和40例健康人的唾液样本被纳入最终分析(图1)。受试者人口统计学和临床特征组间比较结果显示:除外吸烟史,在年龄、性别等基线特征方面,两组间差异均无统计学意义(表1)。
图 1.
Flowchart showing the subject recruitment process
项目招募流程图
表 1. Patient characteristics.
患者临床特征
| Characteristic | PN group (n=173) |
HC group (n=40) |
P |
| IQR: interquartile range; PN: pulmonary nodule; HC: healthy control. a Wilcoxon rank sum test; b Chi-square test; c Yates' correction. | |||
| Age/yr., median (IQR)a | 45 (32, 55) | 42 (33, 48) | 0.39 |
| Sex/case (%)b | 0.25 | ||
| Male | 69 (32.4) | 12 (5.6) | |
| Female | 104 (48.8) | 28 (13.1) | |
| Smoking history/case (%)b | <0.001 | ||
| No | 134 (62.9) | 40 (18.8) | |
| Yes | 39 (18.3) | 0 (0.0) | |
| Personal tumor history/case (%)c | 0.99 | ||
| No | 167 (78.4) | 39 (18.3) | |
| Yes | 6 (2.8) | 1 (0.5) | |
| Risk stratification of PN/case (%) | |||
| Unstratified | 9 (5.2) | / | |
| Low risk | 100 (57.8) | / | |
| Moderate risk | 58 (33.5) | / | |
| High risk | 6 (3.5) | / | |
2.2. 微生物组学数据质控与ASV分析
213份样本原始数据质控后,获得优化序列数19236964,碱基数8117947662,平均序列长度422 bp。Pan/Core物种分析曲线均相对趋于平缓,表明本实验的样本量充足(图2A、2B),Rank-Abundance曲线在水平方向横轴上的范围较大,表明本实验样本具有较高的微生物物种丰富度和均匀度(图2C)。
图 2.

Overview of the raw data of the sample
样本原始数据概况
PN: pulmonary nodule; HC: healthy control. A, Pan analysis to observe the changes in the total number of species in the HC and the PN groups as the number of samples increases. B, Core analysis to observe the changes in the numbers of core species in the HC and the PN groups as the sample size increases. C, Rank abundance curves of the HC and the PN groups.
2.3. 微生物多样性及差异分析
α多样性通过计算Sobs指数和Shannon指数描述微生物群落内的物种丰富度。与HC组相比,PN组的Sobs指数(496.11 vs. 378.95,P=0.000167)(图3A)和Shannon指数(6.05 vs. 3.89,P=0.005309)(图3B)均较高,差异有统计学意义(P<0.05),表明PN患者的唾液微生物群在物种水平上的α多样性高于健康人群。
图 3.

Analysis of the α diversity and the β diversity indices of the PN group and the HC group
PN组和HC组间α和β多样性指数的分析
PN: pulmonary nodule; HC: healthy control. A, Comparison of Sobs index between the PN group and the HC group. B, Comparison of Shannon index between the PN group and the HC group. C, Scatter plot of the of β diversity comparative analysis conducted with PCoA of the PN group and the HC group. D, Scatter plot of the β diversity comparative analysis conducted with NMDS of the PN group and the HC group.
β多样性能评估两个群落之间整体微生物群的物种多样性。通过PCoA分析和NMDS分析方法对唾液样本的β多样性进行组间差异的双重检验(图3C、3D)。结果显示,与组内差异相比,PN和HC组的唾液样本菌群构成的组间差异更显著(R=0.1175,P=0.01;R2=0.0379,P=0.001),表明PN患者与健康人群之间的唾液菌群相似性不同。
2.4. 微生物群落组成分析
属水平Venn图结果显示(图4A),PN组独有物种数为188,HC组独有物种数为31,两组共有物种数244个。为了进一步分析PN组与HC组的微生物群落差异,使用群落结构Bar图、Heatmap图和Circos图(图4B~4D)比较PN组与HC组微生物群组成的相对丰度,PN组物种相对丰度排名前5位的菌属分别是:Streptococcus(23.86%)、Rothia(12.83%)、Prevotella(8.40%)、Actinomyces(6.65%)、Veillonella(5.59%);而在HC组中前5位的菌属则是:Streptococcus(24.94%)、Rothia(10.82%)、Haemophilus(9.06%)、Neisseria(8.72%)、Veillonella(6.33%)。
图 4.

Analysis of the microbial community composition in the PN group and the HC group
PN组与HC组的微生物群落组成分析
PN: pulmonary nodule; HC: healthy control. A, Venn diagram of the genera of the PN group and the HC group. B, Relative abundance (%) of the genera in the the PN and the HC groups. C, Heatmap of the salivary sample similarity and difference matrix. D, The co-occurrence relationships of core bacteria between the PN and the HC groups.
2.5. 微生物物种差异分析
用Wilcoxon秩和检验(图5)和LEfSe分析(图6)对物种差异进行了双重验证,以鉴定潜在的唾液微生物标志物。与HC组相比,具有组间显著差异的物种有9种(LDA>2, P<0.05, FDR校正),包括Neisseria、Prevotella、Haemophilus、Actinomyces、Porphyromonas、Fusobacterium、7M7x、Granulicatella和Selenomonas。同时,基于属水平的ANCOM差异检验火山图结果显示,唾液样本具有丰富的差异性物种(图7)。
图 5.
Analysis of the microbiota species differences between patients with PN and healthy controls based on Wilcoxon rank-sum test
基于Wilcoxon秩和检验的PN患者与HC人群微生物物种差异分析
** 0.001<P≤0.01, *** P≤0.001. PN: pulmonary nodule; HC: healthy control.
图 6.

Microbiota species differences between patients with PN and healthy controls based on LEfSe analysis
基于LEfSe的PN患者与HC人群微生物物种差异分析
PN: pulmonary nodule; HC: healthy control. A, Cladogram plot of LEfSe analysis indicating the enriched taxa of saliva microbiome in the PN and the HC groups. The central point represents the root of the tree (bacteria), and each ring represents the next lower taxonomic level. The diameter of each circle represents the relative abundance of the taxon. B, Histogram of linear discriminant analysis (LDA) scores (>3) for differentially abundant genera between the two groups.
图 7.
The volcano plot of the comparison of differences in abundance of common species by ANCOM analysis
基于ANCOM分析的物种丰度差异火山图
Each dot on the plot represents a genus, with the ordinate representing the W value and the abscissa representing the Center Log Transform (clr). The clr represents the degree of difference in sample abundance between groups, with larger absolute values indicating greater relative abundance differences.
2.6. 肺结节的潜在唾液微生物标志物
为了进一步确定与PN相关的特征唾液微生物种类,通过随机森林模型判别唾液微生物区系的疾病预测效率。两组唾液样本中微生物群落的分布均匀,选取具有前11个重要特征的物种,曲线下面积(area under the curve, AUC)验证曲线的最高准确率为0.8749(图8A、8B)。在从文献中排除8个不可分类且临床相关性较小的属后,Porphyromonas、Haemophilu和Fusobacterium形成了最佳的生物标志物集合。受试者工作特征(receiver operating characteristic, ROC)曲线预测效率的AUC值达到0.79(95%CI:0.71~0.86)(图8C)。
图 8.

Potential microbial biomarkers for PN
PN潜在唾液微生物标志物
PN: pulmonary nodule; HC: healthy control. A, Two-dimensional scatter plot of saliva samples based on random forest algorithm in two groups. B, Bar plot of species importance at the genus level performed by random forest algorithm. C, Trend graph of accuracy rate increasing by using top important features. D, ROC curve of saliva samples for predicting PNs at the genus level.
2.7. 唾液微生物与受试者临床特征的相关性分析
方差膨胀因子(variance inflation factor, VIF)<5可确定影响微生物群落的环境因素。如表2所示,受试者临床特征环境因素的VIF值在鉴定前后没有变化。db-RDA分析评估了受试者临床特征与唾液微生物群落组成的相关性。结果显示,所有样本点聚类形成了两个cluster,表明临床特征对PN患者的唾液微生物群落组成具有潜在影响(图9A,表3)。Spearman相关系数结果显示(图9B),PN患者年龄、性别、吸烟史与唾液微生物群呈正或负相关(P<0.001,r>0.6或r<−0.6),而个人肿瘤史与唾液微生物群无显著关系。
表 2. Variance inflation factor collinearity analysis.
方差膨胀因子共线性分析
| Environmental factor | VIF value | |
| Before screening | After screening | |
| VIF: variance inflation factor. | ||
| Age | 1.08610272033186 | 1.08610272033186 |
| Smoking history | 1.37991739233775 | 1.37991739233775 |
| Sex | 1.36666770970373 | 1.36666770970373 |
| Personal history of cancer | 1.01563553235591 | 1.01563553235591 |
图 9.
Correlation analysis of salivary microbiota and clinical features of PN
唾液微生物群与PN临床特征相关性分析
* 0.01<P≤ 0.05, ** 0.001<P≤0.01, *** P≤0.001. PN: pulmonary nodule; HC: healthy control. A, db-RDA analysis based on Bray-Curtis distance between the environmental factors and bacterial groups (at the genus level). B, Heatmap diagram based on Spearman correlation analysis between the environmental factors and bacterial groups (at the genus level). a, Age; b, gender; c, smoking status; d, personal tumor history.
表 3. db-RDA environmental factor analysis.
db-RDA环境因子分析
| Environmental factor | CAP1 | CAP2 | R 2 | P |
| CAP: constrained analysis of principal coordinates; db-RDA: distance-based redundancy analysis. | ||||
| Age | 0.9998 | 0.0177 | 0.1876 | 0.001 |
| Sex | 0.66901 | −0.7238 | 0.1150 | 0.001 |
| Smoking status | 0.9998 | −0.0200 | 0.1430 | 0.001 |
| Personal history of cancer | −0.9356 | 0.9877 | 0.0210 | 0.124 |
2.8. 口腔唾液微生物及其差异基因的生物信息学分析
KEGG orthology(KO)功能丰度统计结果表明,PN组唾液微生物富集在细胞骨架蛋白RodZ、假尿苷2457合酶、通用应激蛋白E、溶血素激活/分泌蛋白、NADPH脱氢酶、γ-谷氨酰腐胺氧化等相关功能基因(图10)。COG功能分类统计结果显示,PN组在RNA加工和修饰、能量产生和转换、细胞周期控制、氨基酸运输和代谢等相关功能基因的预测丰度则功能方面丰度显著降低(图11)。这些数据提示唾液微生物组可能通过氧化还原、免疫逃逸及能量代谢等通路参与PN疾病进程。
图 10.

Statistical validation of KEGG functional abundance of microbial species in PN patients and HC subjects
PN患者与HC人群微生物物种KEGG功能丰度统计
PN: pulmonary nodule; HC: healthy control. A, Q-value heatmap of KEGG function enrichment in two groups. B, Histogram of KEGG pathway enrichment analysis of the PN and the HC groups.
图 11.

Statistical validation of COG functional abundance of microbial species in PN patients and HC subjects
PN患者与HC人群微生物物种COG功能丰度统计
PN: pulmonary nodule; HC: healthy control. Box-plot of COG functional classification (top) and histogram of COG pathway enrichment analysis of the PN and the HC groups (bottom).
3. 讨论
人类口腔生物组织标本因其收集方便、无创性等特点,越来越多地被用于肺癌早期诊疗方面[18]。同时,随着高通量测序技术的进步,来自包括微生物组学在内的多组学检测手段已证实唾液生物样本作为早期肺癌临床筛选和检测生物标志物的潜力[19]。然而,这些研究大多数定位于发现肺癌与健康人群唾液微生物之间的差异[20-21],并未聚焦于肺癌早期状态——PN深入探索唾液微生物组作为早期无创诊断体液标志物的潜力。
本研究首次通过16S rRNA基因测序技术鉴定和证实了PN患者和HC人群唾液微生物特征与差异。Pan/Core物种与稀释曲线分析结果均显示本实验的样本量及测序深度充足,在此基础上开展后续分析的可靠性和稳定性更好。越来越多的证据表明,微生物组的多样性改变与疾病风险有关[22]。本研究发现PN患者唾液微生物群α和β多样性指数高于健康人群,表明PN患者的唾液微生物群中存在生态失调。值得注意的是,ZENG等[23]使用16S rRNA测序技术,报告了与健康受试者相比,肺癌患者唾液微生物群的丰富性和多样性更高,这一结果与本研究的α和β多样性组间差异检验结果一致。然而,SHI等[24]和TSAY等[25]发现肺癌患者和健康受试者之间的多样性没有差异。上述结果的差异可能是由于生活环境、收集的样本数量、对照组或测序数据分析方式不同造成。随后,本研究发现在PN患者唾液样本中Prevotella、Porphyromonas、Actinomyces、Selenomonas、Granulicatella和7M7x等菌属显著富集,在Wilcoxon秩和检验和LfFSe分析中也获得了相同的结果,提示了表征PN患者唾液微生物特性和差异的结果具有稳定性和可靠性。此外,几项研究将唾液微生物作为新型体液标志物在鉴别肺癌与健康人、肺癌与肺部良性病变等方面的潜力[26-28],在一定程度上提示了唾液微生物标志物在“健康状态-PN-肺癌”的演进过程中可能具有潜在的诊断价值。
为了更准确地评价唾液作为新型无创体液标志物的潜能,基于AUC验证的随机森林模型对唾液样本进行疾病预测效能判别分析,确定了3种微生物属Porphyromonas、Haemophilus和Fusobacterium为最佳预测因子,其可能有助于PN的早期诊断,但确切机制需要进一步探索。因此,基于PICRUSt2分析,初步探索了PN患者和健康人群在唾液样本中差异基因的生物信息学功能。结果发现PN患者唾液微生物与细胞氧化还原稳态、免疫调节等直接相关蛋白/分子功能的富集最为明显。细胞氧化还原稳态是微生物群维持与宿主共生关系的关键指标,该功能的下调可能导致宿主的菌群失调和炎症[29],慢性炎症则是导致包括肺癌、PN等在内疾病发展的关键一环[30]。此外,有证据表明微生物组可以塑造适应性免疫,以逃避免疫监视[31]。在肺癌最新的研究中发现,与能量代谢相关的鞭毛装配途径出现明显富集,细菌迁移能力和细胞发育水平增加,但免疫相关功能却出现下降[32]。因此,免疫缺陷和能量代谢异常可能导致微生物易位驱动下的PN或肺“结-癌转化”的发生。
本研究存在以下局限性:首先是基于前瞻性非随机对照试验的真实世界研究,虽然具有更接近临床实际情况的优点,但和随机对照试验相比,数据本质上混杂因素较多,尽管研究使用了严格的统计学方法,仍可能存在潜在偏倚,因此本研究的结果需在后续独立研究中加以证实;其次,由于患者为前瞻性纳入,且属于真实世界研究,总样本量为213例,后续仍需要更大样本量的真实数据进行验证;最后,尽管本研究基于PICRUSt2对差异功能基因进行了生物信息学预测分析,但并未对其作用机制和潜在的分子调控网络进行深入挖掘探索与实验验证,这也是今后研究值得深入探索和挖掘的前沿方向之一。
本研究提示Porphyromonas、Haemophilus和Fusobacterium等唾液微生物可作为PN精准诊断的潜在生物标志物。此外,唾液微生物参与PN发生发展可能与其诱导宿主免疫缺陷和细胞氧化还原稳态异常相关。
* * *
作者贡献声明 任益锋负责论文构思、数据审编、正式分析、软件、验证、可视化和初稿写作,马琼负责正式分析、研究方法、可视化和初稿写作,李芳负责调查研究、研究方法和软件,曾潇和谭施言负责调查研究、软件和可视化,付西负责研究项目管理、软件和可视化,郑川负责研究项目管理、提供资源和监督指导,由凤鸣和李雪珂负责论文构思、经费获取、验证和审读与编辑写作。所有作者已经同意将文章提交给本刊,且对将要发表的版本进行最终定稿,并同意对工作的所有方面负责。
利益冲突 所有作者均声明不存在利益冲突
Funding Statement
四川省科技厅重大科技专项(No. 2022ZDZX0022)和四川省自然科学基金青年项目(No. 2023NSFSC1815)资助
Contributor Information
益锋 任 (Yifeng REN), Email: ryftcm.dr@yahoo.com.
雪珂 李 (Xueke LI), Email: 2017202040046@whu.edu.cn.
References
- 1.PINSKY P, MILLER E, FARIS N, et al Pulmonary nodules, lung cancer screening, and lung cancer in the medicare population. Chest. 2023;163(5):1304–1313. doi: 10.1016/j.chest.2022.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.TOUMAZIS I, BASTANI M, HAN S S, et al Risk-based lung cancer screening: a systematic review. Lung Cancer. 2020;147:154–186. doi: 10.1016/j.lungcan.2020.07.007. [DOI] [PubMed] [Google Scholar]
- 3.何新, 陈楠, 林锋 肺磨玻璃结节的诊断与临床对策. 中国肿瘤外科杂志. 2018;10(4):212–216. doi: 10.3969/j.issn.1674-4136.2018.04.002. [DOI] [Google Scholar]
- 4.郭献平, 朱晓莉 肺结节患者临床特征及情绪研究进展. 东南大学学报(医学版) 2020;39(5):688–692. doi: 10.3969/j.issn.1671-6264.2020.05.027. [DOI] [Google Scholar]
- 5.KAMMER M N, LAKHANI D A, BALAR A B, et al Integrated biomarkers for the management of indeterminate pulmonary nodules. Am J Respir Crit Care Med. 2021;204(11):1306–1316. doi: 10.1164/rccm.202012-4438OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.SANDERS S J S, AAAS CUSTOM PUBLISHING OFFICE: WASHINGTON, DC, USA. 125 questions: exploration and discovery. Science, 2021. https://www.science.org/content/resource/125-questions-exploration-and-discovery.
- 7.PIZZO F, MAROCCIA Z, HAMMARBERG FERRI I, et al Role of the microbiota in lung cancer: insights on prevention and treatment. Int J Mol Sci. 2022;23(11):6138. doi: 10.3390/ijms23116138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.FU A, YAO B, DONG T, et al Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. 2022;185(8):1356–1372.e26. doi: 10.1016/j.cell.2022.02.027. [DOI] [PubMed] [Google Scholar]
- 9.BAEK A E Bacteria benefit tumor cells. Sci Signal. 2022;15(729):eabq4492. doi: 10.1126/scisignal.abq4492. [DOI] [PubMed] [Google Scholar]
- 10.ZHENG Y, FANG Z, XUE Y, et al Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes. 2020;1(4):1030–1042. doi: 10.1080/19490976.2020.1737487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ROVIELLO G, IANNONE L F, BERSANELLI M, et al The gut microbiome and efficacy of cancer immunotherapy. Pharmacol Ther. 2022;231:107973. doi: 10.1016/j.pharmthera.2021.107973. [DOI] [PubMed] [Google Scholar]
- 12.MANOS J The human microbiome in disease and pathology. APMIS. 2022;130(12):690–705. doi: 10.1111/apm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MAMMEN M J, SCANNAPIECO F A, SETHI S Oral-lung microbiome interactions in lung diseases. Periodontology. 2000;83(1):234–241. doi: 10.1111/prd.12301. [DOI] [PubMed] [Google Scholar]
- 14.LIU B, LI Y, SUO L, et al Characterizing microbiota and metabolomics analysis to identify candidate biomarkers in lung cancer. Front Oncol. 2022;12:1058436. doi: 10.3389/fonc.2022.1058436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.张晓菊, 白莉, 金发光, 等 肺结节诊治中国专家共识(2018年版) 中华结核和呼吸杂志. 2018;41(10):763–771. doi: 10.3760/cma.j.issn.1001-0939.2018.10.004. [DOI] [Google Scholar]
- 16.KAWASAKI M, IKEDA Y, IKEDA E, et al Oral infectious bacteria in dental plaque and saliva as risk factors in patients with esophageal cancer. Cancer. 2021;127(4):512–519. doi: 10.1002/cncr.33316. [DOI] [PubMed] [Google Scholar]
- 17.QIAN X B, CHEN T, XU Y P, et al A guide to human microbiome research: study design, sample collection, and bioinformatics analysis. Chin Med J. 2020;133(15):1844–1855. doi: 10.1097/CM9.0000000000000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.SONG M, BAI H, ZHANG P, et al Promising applications of human-derived saliva biomarker testing in clinical diagnostics. Int J Oral Sci. 2023;15(1):2. doi: 10.1038/s41368-022-00209-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WANG X, KACZOR-URBANOWICZ K E, WONG D T Salivary biomarkers in cancer detection. Med Oncol. 2017;34(1):7. doi: 10.1007/s12032-016-0863-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.VOGTMANN E, HUA X, YU G, et al The oral microbiome and lung cancer risk: an analysis of 3 prospective cohort studies. J Natl Cancer Inst. 2022;114(11):1501–1510. doi: 10.1093/jnci/djac149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CHRISTIANI D C The oral microbiome and lung cancer risk. Thorax. 2021;76(3):216–217. doi: 10.1136/thoraxjnl-2020-216385. [DOI] [PubMed] [Google Scholar]
- 22.RIQUELME E, ZHANG Y, ZHANG L, et al Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178(4):795–806.e712. doi: 10.1016/j.cell.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ZENG W, ZHAO C, YU M, et al Alterations of lung microbiota in patients with non-small cell lung cancer. Bioengineered. 2022;13(3):6665–6677. doi: 10.1080/21655979.2022.2045843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.SHI J, YANG Y, XIE H, et al Association of oral microbiota with lung cancer risk in a low-income population in the Southeastern USA. Cancer Causes Control. 2021;32(12):1423–1432. doi: 10.1007/s10552-021-01490-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.TSAY J J, WU B G, BADRI M H, et al Airway microbiota is associated with upregulation of the PI3K pathway in lung cancer. Am J Respir Crit Care Med. 2018;198(9):1188–1198. doi: 10.1164/rccm.201710-2118OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.BINGULA R, FILAIRE E, MOLNAR I, et al Characterisation of microbiota in saliva, bronchoalveolar lavage fluid, non-malignant, peritumoural and tumour tissue in non-small cell lung cancer patients: a cross-sectional clinical trial. Respir Res. 2020;21(1):129. doi: 10.1186/s12931-020-01392-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.SUN Y, LIU Y, LI J, et al Characterization of lung and oral microbiomes in lung cancer patients using culturomics and 16S rRNA gene sequencing. Microbiol Spect. 2023;11(3):e0031423. doi: 10.1128/spectrum.00314-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.YING K L, BRASKY T M, FREUDENHEIM J L, et al Saliva and lung microbiome associations with electronic cigarette use and smoking. Cancer Prev Res (Phila) 2022;15(7):435–446. doi: 10.1158/1940-6207.Capr-21-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LIAN W S, WANG F S, CHEN Y S, et al Gut microbiota ecosystem governance of host inflammation, mitochondrial respiration and skeletal homeostasis. Biomedicines. 2022;10(4):860. doi: 10.3390/biomedicines10040860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LIU C H, CHEN Z, CHEN K, et al Lipopolysaccharide-mediated chronic inflammation promotes tobacco carcinogen-induced lung cancer and determines the efficacy of immunotherapy. Cancer Res. 2021;81(1):144–157. doi: 10.1158/0008-5472.Can-20-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.FRAUENFELDER T, LANDSMANN A Pulmonary nodules and pneumonia: a diagnostic guideline. Der Radiologe. 2022;62(2):109–119. doi: 10.1007/s00117-021-00953-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.INVERNIZZI R, LLOYD C M, MOLYNEAUX P L Respiratory microbiome and epithelial interactions shape immunity in the lungs. Immunology. 2020;160(2):171–182. doi: 10.1111/imm.13195. [DOI] [PMC free article] [PubMed] [Google Scholar]






