Abstract
Spo0A activates transcription in Bacillus subtilis from promoters that are used by two types of RNA polymerase, RNA polymerase containing the primary sigma factor, ςA, and RNA polymerase containing a secondary sigma factor, known as ςH. The region of ςA near positions 356 to 359 is required for Spo0A-dependent promoter activation, possibly because Spo0A interacts with this region of ςA at these promoters. To determine if the amino acids in the corresponding region of ςH are also important in Spo0A-dependent promoter activation, we examined the effects of single alanine substitutions at 10 positions in ςH (201 to 210). Two alanine substitutions in ςH, at glutamine 201 (Q201A) and at arginine 205 (R205A), significantly decreased activity from the Spo0A-dependent, ςH-dependent promoter spoIIA but did not affect expression from the ςH-dependent, Spo0A-independent promoters citGp2 and spoVG. Therefore, promoter activation by Spo0A requires homologous regions in ςA and ςH. A mutant form of Spo0A, S231F, that suppresses the sporulation defect caused by several amino acid substitutions in ςA did not suppress the sporulation defects caused by the Q201A and R205A substitutions in ςH. This result and others indicate that different surfaces of Spo0A probably interact with ςA and ςH RNA polymerases.
Spo0A is a DNA binding protein in Bacillus subtilis that is essential for the initiation of endospore formation (reviewed in reference 6). Spo0A activates transcription from promoters that are used by two types of RNA polymerase, RNA polymerase containing the primary sigma factor, ςA, and RNA polymerase containing a secondary sigma factor, known as ςH (12, 15, 17). Three sporulation-specific promoters that are activated by Spo0A have been characterized extensively: spoIIG and spoIIE, which are ςA dependent, and spoIIA, which is ςH dependent (7, 18, 20). Spo0A binds to these promoters at sites overlapping the −35 region and may interact with the sigma subunit of RNA polymerase. Baldus et al. (1) found that transcription from the ςA-dependent, Spo0A-dependent promoters spoIIG and spoIIE was reduced in mutants of B. subtilis in which ςA contained either of two single-amino-acid substitutions, lysine at position 356 replaced by glutamate (K356E) or histidine at position 359 replaced by arginine (H359R). However, these substitutions had no effect on the use of a Spo0A-independent promoter, tms. Moreover, alanine substitutions at positions 356 and 359 had similar Spo0A-specific effects (14). A single-amino-acid substitution in Spo0A, serine at position 231 replaced by a phenylalanine (S231F), was recently shown to partially suppress the effect of the H359R mutation in ςA (2). These results support the hypothesis that spoIIG and spoIIE promoter activation by Spo0A requires the region of ςA near positions 356 to 359, possibly because Spo0A interacts with this region of ςA at these promoters.
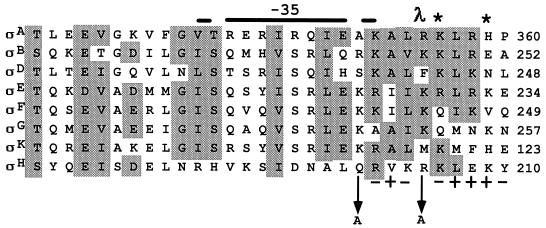
To determine if the amino acids in the corresponding region of ςH are also important in Spo0A-dependent promoter activation, we examined the effects of single alanine substitutions at 10 positions in ςH, from 201 to 210 (Fig. 1). We expected that strains in which an alanine substitution in ςH resulted in loss of interaction with Spo0A would exhibit reduced sporulation efficiency because utilization of ςH-dependent, Spo0A-dependent promoters such as spoIIA is required for endospore formation. Moreover, if the alanine substitution in ςH specifically prevented its interaction with Spo0A, then the ςH mutant would retain the ability to direct transcription from promoters such as citG and spoVG, since use of these promoters does not require direct interaction with Spo0A (4). The sigH allele on plasmid pJB3 was mutagenized in vitro by a multiple-step PCR procedure (3) and used to replace the wild-type allele in the B. subtilis chromosome by transformation of strain JH642 (Table 1) to spectinomycin resistance as described previously (1). Spectinomycin-resistant transformants resulted from a single crossover event between the plasmid-encoded sigH allele and the chromosomal sigH allele. Four sigH mutant alleles, which resulted in the substitution of alanine for amino acids at positions 203, 207, 208, and 209 in ςH, produced transformants that appeared to sporulate as efficiently as the wild-type strain on DSM agar (13). Six alleles of sigH produced strains that appeared sporulation deficient (spo) on DSM agar (Fig. 1). Of these, four alleles (producing alanine substitutions at positions 202, 204, 206, and 210 in ςH [Fig. 1]) appeared to inactivate all ςH-dependent promoter activity (measured as described in reference 1; data not shown) and were not further studied. The remaining two alleles, which resulted in substitutions at 201 and 205 in ςH, each caused a specific decrease in the Spo0A-dependent, ςH-dependent spoIIA promoter activity, while the Spo0A-independent, ςH-dependent promoters spoVG and citGp2 appeared to be active (data not shown).
FIG. 1.
Amino acid alignment of the −35 recognition regions in the carboxyl terminus of sigma factors in B. subtilis. Conserved regions are shaded according to Sun et al. (16). The position number for the last amino acid shown in each sigma factor is indicated. ς70 of E. coli is not shown, but ς70 and ςA are almost identical in this region. Amino acid substitutions are indicated by the arrow from the wild-type amino acid to the altered amino acid. The + and − signs below the ςH sequence indicate the phenotypes of alanine substitutions at that amino acid in ςH. A + sign indicates an allele which produced a functionally wild-type ςH protein, and a − sign designates a completely inactive ςH protein. The λ indicates an amino acid substitution at 596 in ς70, R596H, which specifically suppresses the D38N mutation in λ cI (8, 9). The asterisk indicates amino acid substitutions in ςA (K356E and H359R) which specifically prevent transcription from Spo0A-dependent promoters (1).
TABLE 1.
Bacterial strains and bacteriophages
| Strain or phage | Derivation or genotype | Reference or source |
|---|---|---|
| Strain | ||
| JH642 | trpC2 phe-1 | J. A. Hoch |
| AM878 | amyE::φ(citGp2-lacZ cat) | 4 |
| ZB307A | Spβc2del2::Tn917::pSK10δ6 | 21 |
| EUC97Q1 | JH642 transformed with pJB3sigHQ201A | This work |
| EUC97R1 | JH642 transformed with pJB3sigHR205A | This work |
| EUC9729 | JH642 transformed with pCB2spo0Awta | This work |
| EUC9733 | JH642 transformed with pCB2spo0AS231F | This work |
| EUC9791 | EUC97Q1 transformed with pCB2spo0Awt | This work |
| EUC9792 | EUC97Q1 transformed with pCB2spo0AS231F | This work |
| EUC9793 | EUC97R1 transformed with pCB2spo0Awt | This work |
| EUC9794 | EUC97R1 transformed with pCB2spo0AS231F | This work |
| EUC97Q2 | EUC97Q1 transduced with SPβ spoIIA-lacZ | This work |
| EUC97Q3 | EUC97Q1 transformed with AM878 chromosomal DNA | This work |
| EUC97Q4 | EUC97Q1 transduced with SPβ spoVG-lacZ | This work |
| EUC97R2 | EUC97Q1 transduced with SPβ spoIIA-lacZ | This work |
| EUC97R3 | EUC97R1 transformed with AM878 chromosomal DNA | This work |
| EUC97R4 | EUC97R1 transduced with SPβ spoVG-lacZ | This work |
| Bacteriophage | ||
| SPβ spoIIA-lacZ | 19 | |
| SPβ spoVG-lacZ | 21 | |
| SPβ spoIIG-lacZ | 11 |
wt, wild-type allele.
In order to examine the quantitative effects of these amino acid substitutions in ςH, we isolated derivatives of our sigH mutants that had lost the integrated plasmid but retained the mutant sigH allele. To isolate these strains, we screened for sporulation-deficient (spo; determined as described in reference 1), spectinomycin-sensitive mutants after transformation of B. subtilis JH642 (trpC2 phe-1) to tryptophan prototrophy with a mixture of 40 ng of B. subtilis ZB307A chromosomal DNA (Table 1) and 2 μg of the pJB3sigHQ201A and pJB3sigHR205A plasmids. Transformants were selected on minimal plates for sporulation containing 50 μg of phenylalanine and screened for spectinomycin sensitivity and a spo phenotype. DNA sequence analysis demonstrated that the spo mutants, but none of the Spo+ isolates, contained the Q201A or R205A mutations in sigH. Two of these spo strains, EUC97Q1 and EUC97R1, which express ςH Q201A and R205A, respectively, produced only about 1% of the spores made by the strain containing wild-type sigH (Table 2).
TABLE 2.
Effects of mutations on sporulation
| Strain | Allele
|
No. of heat-resistant spores/ml | |
|---|---|---|---|
| sigH | spo0A | ||
| JH642 | wta | wt | 6.0 × 108 |
| EUC97Q1 | Q201A | wt | 1.1 × 106 |
| EUC97R1 | R205A | wt | 7.7 × 105 |
| EUC9729 | wt | wtb | 4.0 × 108 |
| EUC9733 | wt | S231Fb | 1.1 × 109 |
| EUC9791 | Q201A | wtb | 1.4 × 107 |
| EUC9792 | Q201A | S231Fb | 4.9 × 107 |
| EUC9793 | R205A | wtb | 5.2 × 106 |
| EUC9794 | R205A | S231Fb | 6.2 × 107 |
wt, wild-type allele.
Strain was transformed with pCB2spo0A.
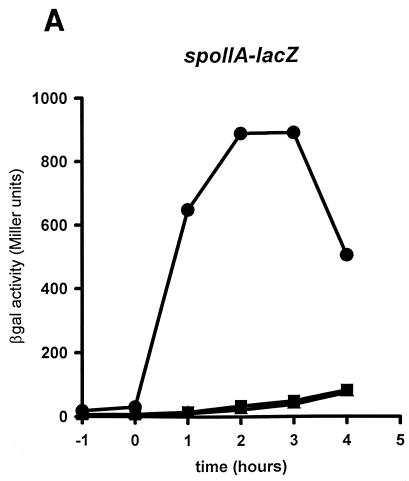
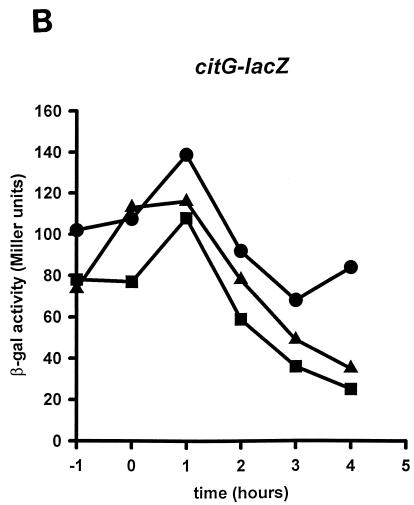
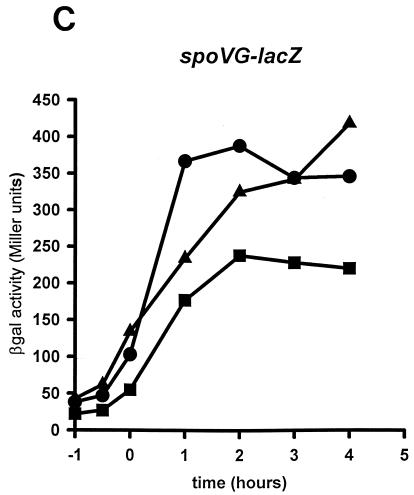
To monitor the effects of each amino acid substitution on the activation of specific promoters, we isolated an isogenic set of strains containing operon fusions of lacZ to spoIIG, spoIIA, or spoVG or to citG as described previously (1) (Table 1). The ςH Q201A and R205A mutations decreased the expression from the ςH-dependent, Spo0A-dependent promoter, spoIIA, more than 10-fold (Fig. 2A). The amount of activity in these strains is similar to that measured in strains expressing a Spo0A nonsense allele, which retains only 5% of wild-type spoIIA activity (2). However, the strains expressing the sigHQ201A and sigHR205A alleles displayed wild-type levels of the ςH-dependent, Spo0A-independent promoters citGp2 and spoVG (Fig. 2B and C). To determine if spoIIA promoter activity was indirectly reduced in the sigH mutant strains because of a decreased level of Spo0A, we measured the activity of the ςA-dependent, Spo0A-dependent promoter spoIIG and found it to be about 25% of the activity measured in the isogenic wild-type strain (data not shown). These levels are significantly higher than those in an otherwise isogenic strain expressing a spo0A nonsense allele (7.4% of wild-type spoIIG activity [2]). These results suggest that the side chains of Q201 and R205 of ςH are important for wild-type levels of Spo0A-dependent, ςH-dependent spoIIA promoter activity. On the other hand, these amino acid residues are not required for activity of the Spo0A-independent, ςH-dependent promoters citGp2 or spoVG.
FIG. 2.
Effects of amino acid substitutions in sigH on spoIIA, citGp2, and spoVG promoter activities. B. subtilis JH642 (wild-type sigH) (circles), EUC97Q1 (sigHQ201A) (squares), and EUC97R1 (sigHR205A) (triangles) containing the spoIIA (A), citGp2 (B), or spoVG (C) promoter-lacZ fusions were grown in DSM liquid medium. Samples were taken from cultures growing at mid-log phase (T1), at the end of exponential growth (T0), and at 1-h intervals after the onset of stationary phase (T1 to T4) and were then assayed for β-galactosidase (βgal) activity (11). An independent transductant from each strain was assayed for β-galactosidase activity and found to express essentially the same levels of activity as those shown (data not shown).
The sporulation-defective phenotype caused by an amino acid substitution in ςA (H359R) is suppressed by a single-amino-acid substitution (S231F) in Spo0A (2). We wished to determine whether the S231F form of Spo0A suppressed the spo phenotype of the ςH mutant strains. We transformed strains EUC97Q1, EUC97R1, and the wild-type parent, JH642, with plasmids pCB2spo0Awt as a control and pCB2spo0AS231F as described previously (2). Analysis of the sporulation efficiency of the resultant strains (Table 2) indicated that the spo0AS231F allele failed to suppress the spo phenotype caused by the amino acid substitutions in ςH. However, the spo0AS231F allele slightly increased the sporulation efficiency of an otherwise isogenic strain containing a wild-type sigH allele (Table 2).
Our results support the model in which spoIIA promoter activation requires an interaction between Spo0A and ςH. Two single alanine substitutions in ςH, at glutamine 201 (Q201A) and at arginine 205 (R205A), which significantly decrease activity from the Spo0A-dependent, ςH-dependent promoter spoIIA, do not affect expression from the ςH-dependent, Spo0A-independent promoters citGp2 and spoVG. These two amino acids lie in the region of ςH corresponding to the region in ςA that has been implicated in an interaction with Spo0A at the spoIIG and spoIIE promoters (1, 2, 14) (Fig. 1). Therefore, Spo0A appears to activate transcription by interacting with homologous regions in ςA and ςH. A homologous region in ς70 of Escherichia coli is also suspected to be involved in interaction with cI from phage lambda (8, 9) (Fig. 1).
It is not known if the same surface of Spo0A or different surfaces of Spo0A are important in its activation of ςA- and ςH-dependent promoters. An amino acid substitution in Spo0A (S231F) that was previously identified as a suppressor mutation of the sporulation defect caused by an amino acid substitution in ςA, H359R (2), displayed little or no suppression of the sporulation defects of the ςH mutants Q201A and R205A. Alanine substitutions in the 229 to 233 region of Spo0A did not reduce spoIIA transcription (2), and Hatt and Youngman (5) have isolated five additional mutant forms of Spo0A in the region of 227 to 240 that prevented spoIIG and spoIIE transcription, without affecting transcription from spoIIA. Therefore, another region of Spo0A may interact with ςH RNA polymerase to stimulate its activity. An amino acid substitution in Spo0A, A257T, has been found to prevent spoIIA transcription but not abrB repression (10). These results are consistent with the model that different amino acids in Spo0A are involved in the activation of ςH and ςA RNA polymerases. The ability of response regulators to interact with multiple sigma factors may increase the variety of responses made by bacteria with a limited number of transcription factors.
Acknowledgments
This work was supported by PHS grant GM54395 to C.P.M. from the National Institutes of Health.
REFERENCES
- 1.Baldus J M, Buckner C M, Moran C P., Jr Evidence that the transcriptional activator Spo0A interacts with two sigma factors in Bacillus subtilis. Mol Microbiol. 1995;17:281–290. doi: 10.1111/j.1365-2958.1995.mmi_17020281.x. [DOI] [PubMed] [Google Scholar]
- 2.Buckner C M, Schyns G, Moran C P., Jr A region in the Bacillus subtilis transcription factor Spo0A that is important for spoIIG promoter activation. J Bacteriol. 1998;180:3578–3583. doi: 10.1128/jb.180.14.3578-3583.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cormack B. Mutagenesis of cloned DNA. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. pp. 857–859. [Google Scholar]
- 4.Feavers I M, Price V, Moir A. The regulation of the fumarase (citG) gene of Bacillus subtilis 168. Mol Gen Genet. 1988;211:465–471. doi: 10.1007/BF00425702. [DOI] [PubMed] [Google Scholar]
- 5.Hatt J K, Youngman P. Spo0A mutants of Bacillus subtilis with sigma factor-specific defects in transcription activation. J Bacteriol. 1998;180:3584–3591. doi: 10.1128/jb.180.14.3584-3591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoch J A. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu Rev Microbiol. 1993;47:441–465. doi: 10.1146/annurev.mi.47.100193.002301. [DOI] [PubMed] [Google Scholar]
- 7.Kenney T J, York K, Youngman P, Moran C P., Jr Genetic evidence that RNA polymerase associated with ςA uses a sporulation-specific promoter in Bacillus subtilis. Proc Natl Acad Sci USA. 1989;86:9109–9113. doi: 10.1073/pnas.86.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M, McClure W R, Susskind M M. Changing the mechanism of transcriptional activation by phage lambda repressor. Proc Natl Acad Sci USA. 1997;94:3691–3696. doi: 10.1073/pnas.94.8.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, Moyle H, Susskind M M. Target of the transcriptional activation function of phage lambda repressor protein. Science. 1994;263:75–77. doi: 10.1126/science.8272867. [DOI] [PubMed] [Google Scholar]
- 10.Perego M, Wu J-J, Spiegelman G B, Hoch J A. Mutational dissociation of the positive and negative regulatory properties of the Spo0A sporulation transcription factor of Bacillus subtilis. Gene. 1991;100:207–212. doi: 10.1016/0378-1119(91)90368-l. [DOI] [PubMed] [Google Scholar]
- 11.Satola S, Kirchman P, Moran C P., Jr Spo0A binds to a promoter used by ςA RNA polymerase during sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1991;88:4533–4537. doi: 10.1073/pnas.88.10.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satola S W, Baldus J M, Moran C P., Jr Binding of Spo0A stimulates spoIIG promoter activity in Bacillus subtilis. J Bacteriol. 1992;174:1448–1453. doi: 10.1128/jb.174.5.1448-1453.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaeffer P, Millet J, Aubert J P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schyns G, Buckner C M, Moran C P., Jr Activation of the Bacillus subtilis spoIIG promoter requires interaction of Spo0A and the sigma subunit of RNA polymerase. J Bacteriol. 1997;179:5605–5608. doi: 10.1128/jb.179.17.5605-5608.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strauch M A, Trach K A, Hoch J A. Spo0A activates and represses its own synthesis by binding at its dual promoters. Biochimie. 1992;74:619–626. doi: 10.1016/0300-9084(92)90133-y. [DOI] [PubMed] [Google Scholar]
- 16.Sun D, Straiger P, Setlow P. Identification of a new ς-factor involved in compartmentalized gene expression in Bacillus subtilis. Genes Dev. 1989;3:141–149. doi: 10.1101/gad.3.2.141. [DOI] [PubMed] [Google Scholar]
- 17.Trach K, Burbulys D, Strauch M, Wu J, Dhillon N, Jonas R, Hanstein C, Kallio P, Perego M, Bird T, Spiegelman G, Fogher C, Hoch J A. Control of the initiation of sporulation in Bacillus subtilis by a phosphorelay. Res Microbiol. 1991;142:815–823. doi: 10.1016/0923-2508(91)90060-n. [DOI] [PubMed] [Google Scholar]
- 18.Wu J, Piggot P J, Tatti K M, Moran C P., Jr Transcription of the Bacillus subtilis spoIIA locus. Gene. 1991;101:113–116. doi: 10.1016/0378-1119(91)90231-y. [DOI] [PubMed] [Google Scholar]
- 19.Wu J J, Howard M G, Piggot P J. Regulation of transcription of the Bacillus subtilis spoIIA locus. J Bacteriol. 1989;171:692–698. doi: 10.1128/jb.171.2.692-698.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.York K, Kenney T J, Satola S, Moran C P, Jr, Poth H, Youngman P. Spo0A controls the ςA-dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE. J Bacteriol. 1992;174:2648–2658. doi: 10.1128/jb.174.8.2648-2658.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuber P, Losick R. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J Bacteriol. 1987;169:2223–2230. doi: 10.1128/jb.169.5.2223-2230.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]