Summary
The liver possesses a unique regenerative ability to restore its original mass, in this regard, partial hepatectomy (PHx) and partial liver transplantation (PLTx) can be executed smoothly and safely, which has important implications for the treatment of liver disease. Liver regeneration (LR) can be the very complicated procedure that involves multiple cytokines and transcription factors that interact with each other to activate different signaling pathways. Activation of these pathways can drive the LR process, which can be divided into three stages, namely, the initiation, progression, and termination stages. Therefore, it is important to investigate the pathways involved in LR to elucidate the mechanism of LR. This study reviews the latest research on the key signaling pathways in the different stages of LR.
Subject areas: Health sciences, Biological sciences
Graphical abstract
Health sciences; Biological sciences
Introduction
The liver represents the biggest digestive organ, which has a critical effect on the synthesis, metabolism, storage, and redistribution of nutrients, carbohydrates, fats, and vitamins and is the core of the stable body metabolism.1 It is comprised by various cell types, including Kupffer cells (KCs), hepatocytes, hepatic stellate cells (HSCs), and hepatic sinusoidal endothelial cells (SECs).2 The aforementioned cell types are closely related to and interact with each other through various signaling pathways, thus exercising various functions of the liver.3,4 When the liver is exercising various functions, it is easy to be affected by external adverse factors and damaged. The damage of liver function will induce a variety of diseases, so the integrity of the liver function is very important for treating various liver diseases.
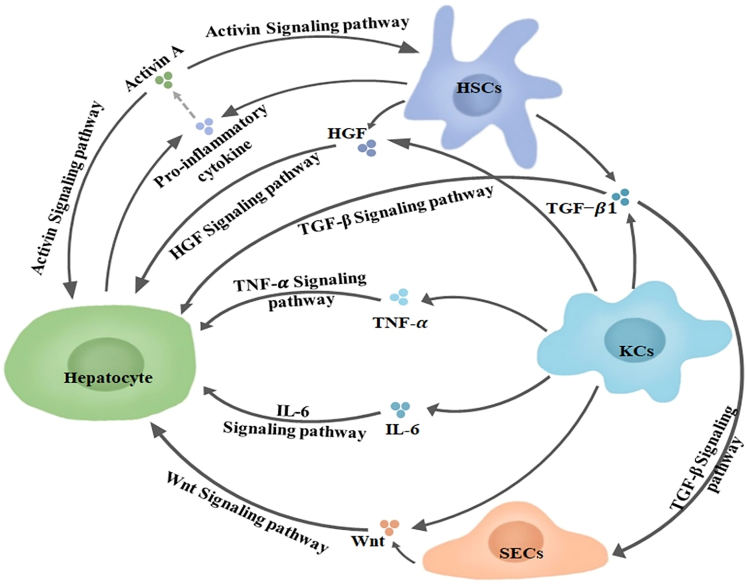
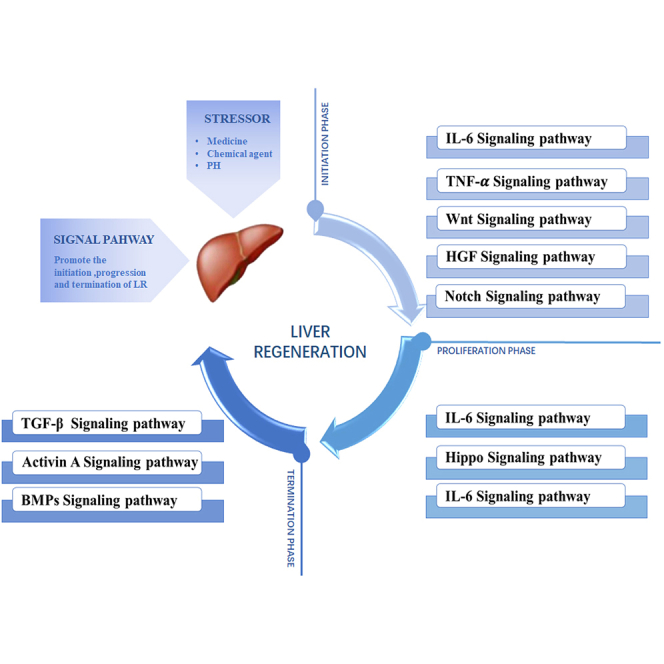
The liver is one of the few organs with the ability of regeneration, it can spontaneously return to its original mass when it is damaged.3,5 Hepatocytes are normally in the G0 phase.3,6 However, when they are subjected to stressors including drugs, chemicals, and PHx, hepatocytes will quickly re-enter the cell cycle, but once they recover to meet the minimum weight required for survival, the regeneration rate will decline.7,8,9 Hepatocyte proliferation plays an important role in restoring liver function and quality. The compensatory proliferation of remaining hepatocytes is the main mechanism of LR after acute and chronic liver injury.10 The differentiation of other cells in the liver into hepatocytes also plays an important role, such as biliary epithelial cells (BECs), liver progenitor cells (LPCs), etc.11,12,13 In addition, during the LR process, various cells in the liver transmit signals to hepatocytes in a paracrine way, prompting hepatocytes to re-enter the cell cycle to restore the original function of the liver.14 Crosstalk between this different cell types in the liver also plays an important role in LR.15 For instance, KCs and HSCs secrete transforming growth factor β(TGF-β1) and activate TGF-β signaling pathway to act on SECs and hepatocytes. KCs and HSCs secrete hepatocyte growth factor (HGF) and activate HGF Signaling pathway to act on hepatocytes. KCs secrete tumor necrosis factor α (TNF-α), interleukin-6 (IL-6) and activate TNF-α and IL-6 signaling pathway to act on hepatocytes. KCs and SECs secrete Wnt and activate Wnt signaling pathway to act on hepatocytes. HSCs and hepatocytes secrete pro-inflammatory cytokines, which act on hepatocytes or activate activin A secreted by BECs. Activin A acts on HSCs and hepatocytes through the activin signaling pathway. According to the physiological process, LR can be divided into three stages: initiation, progression, and termination.2 It has been widely believed that cytokines such as TNF-α, IL-6, epithelial growth factor (EGF), fibroblast growth factor (FGF), HGF, together with transcription factors like c-Jun, transcriptional activator 3 (STAT3), and activator protein-1 (AP-1), have critical effects on LR initiation and progression, which can promote the transformation of G0/G1 in hepatocytes and accelerate the cell cycle of hepatocytes and LR.16,17 TGF-β, along with the relevant family members, plays a role in the termination phase of LR, which can terminate LR by inhibiting hepatocyte proliferation.18
The unique regenerative capacity of the liver provides an idea for treating end-stage liver disease, and PHx or PLTx is an effective means for this purpose. LR can be the very complicated process, which is inseparable from the interactions between hepatocytes and between hepatocytes and other cells, while these interactions are completed by various cytokines and their mediated signaling pathways. Therefore, this study aims to review the recent research results on the signaling pathways related to LR, so as to providing certain basic theoretical support for the research on LR and the diagnosis and treatment of liver diseases. The main cells and their mediated signaling pathways during LR are summarized in Figure 1.
Figure 1.
The main activated signaling pathways in LR
KCs and HSCs secrete TGFβ1 and activate TGF-β signaling pathway to act on SECs and hepatocytes. KCs and HSCs secrete HGF and activate HGF Signaling pathway to act on hepatocytes. KCs secrete TNF-α, IL-6 and activate TNF-α and IL-6 signaling pathway to act on hepatocytes. KCs and SECs secrete Wnt and activate Wnt signaling pathway to act on hepatocytes. HSCs and hepatocytes secrete pro-inflammatory cytokine activating activin A and activin signaling pathway that acts on HSCs and hepatocytes. HSCs, Hepatic stellate cells; KCs, Kupffer cells; SECs, Hepatic sinusoidal endothelial cells; HGF, Hepatocyte growth factor; TG.F-β, Transforming growth factor β; TNF-α, Tumor necrosis factor-α; IL-6, Interleukin-6. Dotted line arrow represents activation between cytokines.
Signaling pathways in LR
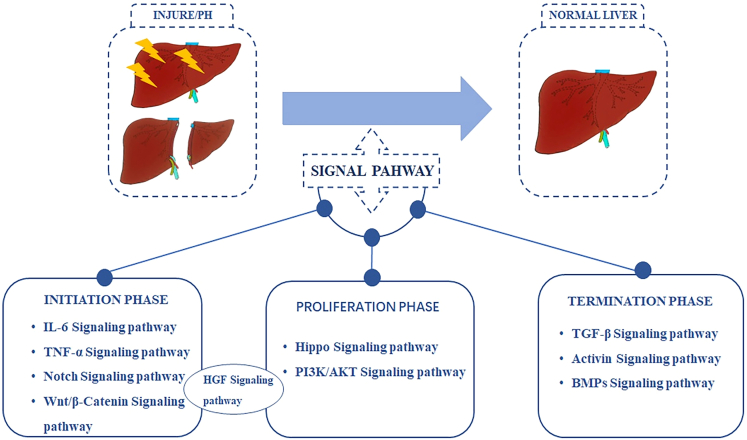
According to the three stages of LR, the relevant pathways in this process can be divided into three stages, including IL-6, TNF-α, Hippo, HGF, and TGF-β pathways, which play a critical part in the initiation, progression, and termination stages, respectively. Noteworthily, some pathways can participate in different stages of LR; for instance, the HGF pathway may be related to the initiation and progressive stages. These signaling pathways are not isolated, instead, they interact with each other to regulate the process of LR. The main signaling pathways in each stage of LR are summarized in Figure 2.
Figure 2.
The main signaling pathways at each phase of LR
According to the initiation, progression and termination stages of LR, the relevant signaling pathways in the process of LR can be divided into three stages. Some signaling pathways can be involved in different stages of LR. PHx, Partial hepatectomy; PI3K, Phosphoinositide 3-kinase; BMPs, bone morphogenetic proteins.
The initiation phase of liver regeneration
Under static or non-pathological situation, hepatocytes are at the G0 phase where mitosis is rare. In the case of liver damage or PHx, some cytokines are rapidly expressed, the status of receptors and transcription factors is rapidly changed, multiple signaling pathways are activated, and a large number of stationary hepatocytes and non-parenchymal hepatocytes are induced to re-enter the cell cycle, eventually restoring the liver mass.19,20 Cytokines such as IL-6, TNF-α, and HGF are necessary to initiate LR, and IL-6 and TNF-α have critical effects on the early signaling pathway of LR. IL-6 and TNF-α can promote hepatocyte transition from G0 into G1 and trigger early LR gene expression (like c-Fos, c-Myc, and c-Jun) by activating signal transduction and STAT3, as a result, hepatocytes enter the mitotic stage.21,22,23 HGF is another important factor participating in the initiation stage of LR, which promotes DNA synthesis and mitosis of hepatocytes in a paracrine manner.24 In addition to IL-6, TNF-α, and HGF-mediated signaling pathways, other mediated pathways like Notch and Wnt have a critical effect on the initiation stage of LR. Such main pathways related to the initiation stage of LR are summarized in Figure 3.
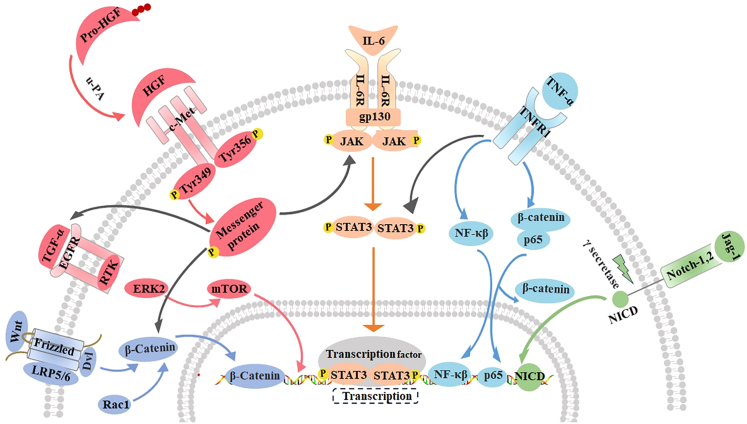
Figure 3.
The main signaling pathways in the initiation phase of LR
Wnt, HGF, IL-6, TNF-α, Notch mediated signaling pathway play an important role in the initiation stage of LR. Wnt binds to the receptor Frizzled to facilitate the transport of β-catenincin into the nucleus via factors such as Rac1. When pro-HGF is cut into active HGF by u-PA, it binds to c-Met and initiates a variety of downstream signaling pathways including IL-6 and TGF-α signaling. IL-6 binding to the receptor causes gp130 structure change which promotes the transfer of STAT3 dimer to the nucleus and upregulates its target gene. TNF-α binds to TNFR-1 to activate NF-κB, induce the dissociation of p65/β-catenin, promoting the nuclear translocation of p65.When the Notch ligand Jag-1 binds to the Notch receptor, the Notch receptor is activated and cleaved by the gamma secretase complex, and its subsequently the intracellular domain (NICD) is transferred to the nucleus and bound to DNA-binding proteins. TGF-α, Transforming growth factor α; EGFR, Epithelial growth factor receptor; RTK, protein receptor tyrosine kinase; mTOR, mechanistic target of rapamycin; u-PA, Urokinase-type plasminase activator; IL-6R, Interleukin-6 receptor; gp130, Glycoprotein 130; STAT3, Signal transducer and activator of transcription 3; TNFR-1, Tumor necrosis factor receptor-1; NF-Κβ, nuclear factor Κβ; NICD, Notch intracellular domain.
IL-6 pathway
IL-6, the cytokine in the chemokine family, is synthesized and secreted by KCs, monocytes and T cells,25 and it initiates its signaling pathway via the hexamer complex composed of the receptor IL-6R and glycoprotein 130(gp130).26 IL-6 pathway mediates various physiological activities, including the regulation of lymphocyte survival and activation, the induction of acute phase protein (APP) synthesis in the liver, involvement in inflammation, and promotion of blood cell development.27,28,29 In addition, it also has a crucial effect on LR.30,31,32
IL-6 is mainly responsible for sending mitotic signals into hepatocytes via Jak/STAT3 pathway, besides, it exerts an essential effect on the initiation stage of LR and controls 40% of early gene expression in LR.33,34,35 As discovered in the latest study, regional damage of liver tissue (the area around the portal vein) stimulates KCs to aggregate and activate at the injuried site, regionally activated KCs promote the formation of liver progenitor cell-like cells (LPLCs), then IL-6 produced by LPLCs triggers progenitor cell-related genes (RRGs) expression and promote LR by activating STAT3.36 LPCs, also known as oval cells in rodents, which are bipotent cells capable of producing hepatocytes and BECs.37 LPCs can also be activated under severe chronic liver injury, differentiating into hepatocytes that promote LR.38 In zebrafish, several research groups have found that LPCs can differentiate into functional hepatocytes following severe hepatocyte depletion.39,40,41 Subsequently, LPC to hepatocytes conversion was also verified in the mouse system by positive lineage tracing.42 In addition to stimulating KCs to produce IL-6, HSCs, macrophages, and KCs can also be stimulated to produce IL-6, thus activating the IL-6/Jak/STAT3 signaling pathway to promote LR. HSCs can secrete IL-6 and induce YAP to promote LR by the activation of STAT3.43 Transcriptional co-activators containing the PDZ-binding motif (TAZ) can activate IL-6/STAT3 signaling pathway by stimulating macrophages to produce IL-6.44 Vagus nerve signals can stimulate KCs to produce IL-6, which then activates the transcription factor protein forkhead box protein M1(FoxM1) in hepatocytes via IL-6/STAT3 pathway, thereby regulating the cell cycle.45 In addition, exogenous melatonin (MLT) promotes LR by inducing the monocyte-promoted IL-6 and activating the IL-6/STAT3 signaling pathway.46 Osteopontin (OPN) induces IL-6 production in KCs, which in turn activates STAT3 to promote LR.47 According to Bahn and Cheng et al., IL-6/STAT3 pathway was activated by prominin-1 and heme oxygenase-1 (HO-1). Prominin-1, a lipid raft protein, promoted LR by recruiting gp130 into the lipid raft to activate the IL-6/STAT3 pathway.48 Heme oxygenase-1 (HO-1) also induces LR through triggering IL-6/STAT3 pathway.49 Based on Xie and Kremer et al., initiating IL-6/STAT3 pathway restricted the PHx-induced LR, when IL-6/STAT3 pathway was inhibited via myeloid peroxisome proliferator-activated receptor α (PPARα), PHx-induced LR was limited.50 Moreover, Kremer et al. reported that when Smad3 was absent, IL-6/STAT3 activation was hindered during mouse LR.51 As reported by Piobbico et al., IL-6/STAT3 pathway was regulated by IL-17, and the lack of IL-17 resulted in the low serum concentration of IL-6 and failure to activate STAT3 in the first few hours after PHx, resulting in the decreased expression of early LR genes.20 In addition, IL-6 pathway also exerts a critical effect on human living donor liver transplantation (LDLT). According to Chae et al., a high serum IL-6 level before surgery was more conducive to postoperative LR in LDLT patients.52
In line with the aforementioned data, IL-6 pathway shows a crucial effect on the initiation stage of LR. Although there are numerous studies on IL-6 pathway in recent years, the specific underlying mechanisms and the upstream and downstream relationships remain to be further investigated in future studies.
TNF-α signaling pathway
Discovered by Carswell in 1975, TNF-α is a multifunctional cytokine that has a key impact on different immune and inflammatory responses.53,54 It exists in two forms, namely, transmembrane precursor protein (mTNF) and soluble protein (sTNF). Of them, mTNF produces sTNF via the action of TNF-converting enzyme. Both sTNF and mTNF are biologically active, which transmit signals by binding to TNFR1 and TNFR2, the two membrane receptors with different structures.55 TNFR2 is almost exclusively expressed in hematopoietic cells and endothelial cells during homeostasis,54 while TNFR1 is universally distributed in every cell type, important for LR, and has a necessary effect on activating NF-kβ pathway.56
TNF-α participates in numerous liver biological responses and exerts a critical effect on hepatitis, liver fibrosis, liver failure, and LR.57,58,59,60 After PHx, TNF-α activates NF-κB by binding to TNFR-1, which translocates NF-κB into the nucleus and induces IL-6 expression, subsequently, IL-6 activates STAT3 to promote LR.61 When TNF-α is blocked, the production of IL-6 and LR will be inhibited.62 Recently, indoleamine 2, 3-dioxygenase (Ido1/2) deficiency is found to activate the nuclear factor kB (NF-kB), leading to the production of TNF-α, which in turn promotes LR.63,64 According to Nejak-Bowen et al., TNF-α pathway induced decomposition of the p65/β-catenin complex, promoted the nuclear translocation of p65, and thus upregulated cyclin D1 and β-catenin expression as well as hepatocyte proliferation in vivo and in vitro.65 The TNF-α pathway can be regulated by proteases ADAM17 and F13A in the mouse models, which promotes early LR after PHx by regulating the serum levels of TNFR1 and TNF-α.66,67 Wang et al. showed that the loss of Golgi protein 73 (GP73) led to the downregulation of TNF-α and cell cycle-related proteins, thus delaying the initiation of LR in mice.68 Studies have exhibited that increasing TNF-α expression enhanced hepatocyte proliferation and accelerated LR in PHx rats.69 Zhang et al. reported that MiR-199a was an important regulator of TNF-α signaling pathway, which regulated LR and hepatocyte proliferation in rats by targeting TNF-α/TNFR1 pathway.70 As demonstrated by Lee et al., human placental hydrolyzate (hPH) activated TNF-α pathway through activating TNF-α secretion in KCs, thereby promoting LR in rats.71
In addition to mouse and rat models, researchers also suggest that TNF-α pathway exerts an important effect on LR in rabbits and humans. For example, Fathi et al. suggested that TNF-α expression significantly increased at 90 min after hepatectomy among hepatocellular carcinoma (HCC) cases, which was consistent with animal studies showing the critical effect of TNF-α pathway on the initiation stage of LR.72 By constructing a rabbit HCC model, Guo et al. indicated that TNF-α might be used as a combination target for transcatheter hepatic arterial chemoembolization (TACE) as well as portal vein embolization (PVE), and TACE+PVE enhanced LR by inducing the high TNF-α expression level.73
The aforementioned studies provide strong evidence supporting the importance of TNF-α/TNFR1 pathway for LR. However, the function of TNF-α pathway is not a single in LR. Apart from activating the initiation stage of LR, its mediated apoptosis pathway can also regulate the termination stage of LR.74 Therefore, the different roles of TNF-α pathway in various stages of LR deserve further investigations.
Notch pathway
Notch pathway shows a high conservation degree, which is universally expressed within tissues and can regulate cell growth, differentiation, and proliferation.75,76,77,78 In mammals, the Notch system includes four transmembrane Notch receptors (Notch1, -2, -3, -4), two ligands Jagged (Jag-1, -2), together with delta-like proteins (Delta-1, -3, and -4).79 In recent years, this pathway is demonstrated in a number of clinical observations and basic studies to exert an important effect on metabolism, vascular physiology, bile duct differentiation, and LR.80,81,82,83,84,85 Moreover, great attention has been paid to LR. Notch pathway represents an earliest pathway that is activated within 15–30 min after PHx. Proteins involved in Notch signaling pathway are significantly upregulated at 1–5 days after PHx,86,87 and pre-PHx silencing of Notch1 and Jag-1 impairs hepatocyte proliferation and regeneration, and disrupts the cell cycle progression of hepatocytes.88,89 These results indicate the critical effect of this pathway on LR after PHx.
Hypoxia inducible factor-1 alpha (HIF-1α) is another downstream pathway of the Notch signaling pathway, which is related to regulating downstream molecules, like vascular endothelial growth factor (VEGF), and shows an active regulatory effect on LR.90 As reported by Li et al., Notch- HIF-1α pathway played an essential part in promoting LR by enhancing hepatocytes proliferation.91 Moreover, they also found that the LR rate decreased after Notch signaling was destroyed by inhibitors, suggesting that liver/body weight ratio together with Ki-67 positive and PCNA positive cell numbers of experimental group significantly decreased compared with control group. The hairy and enhancer of split homolog-1(Hes1) and VEGF mRNA expression, along with the Notch intracellular domain (NICD) and HIF-1α protein levels, was downregulated.91 In addition, Notch signaling exerts a necessary effect on keeping liver homeostasis and hepatocyte transformation into BECs when the liver is damaged. In PHx and severe liver injury, hepatocytes and BECs can repair the lost liver function and mass via their own proliferation and division.92 Therefore, when hepatocyte proliferation is impaired, and BECs, as facultative liver stem cells, lead to the emergence of a large number of BECs-derived hepatocytes.42 Chronically injured mature hepatocytes can also be dedifferentiated into LPCs, once the liver injury was gone, these hepatocyte-derived LPCs reverted to hepatocytes.13 Overexpression of constitutive active YAP1 or Notch in mature hepatocytes can transform hepatocytes into LPCs to support this hepatocyte origin.93,94 In addition, BECs can be dedifferentiated into LPCs and then differentiated into hepatocytes to promote LR.11,12 Recent studies have shown that BECs clonally expand as self-renewing liver organoids that retain their differentiation capacity into both hepatocytes and BECs.95,96,97 Activation of Notch signaling pathway can also induce hepatocytes reprogramming into BECs, which can also promote LR.10,93 Furthermore, Notch participates in biliary regeneration and basically promotes cholangiocyte characteristics.98 The activation of Notch signaling suppresses BECs-to-TLPCs induction and promotes hepatocytes to BECs conversion in injured livers.84 Notch pathway is related to various additional pathways, while the mechanisms of the aforementioned signaling pathways in promoting LR still need to be explored.
Wnt/β-catenin signaling pathway
Wnt pathway can be extensively distributed within vertebrates and invertebrates, and it shows a high conservation degree in the evolution of species.99 Wnt ligands are mainly secreted by endothelial cells and non-parenchymal cells of KCs, which function in the autocrine or paracrine manner.100 After being secreted, Wnt can interact with specific receptors on the cell surface, later activate downstream β-catenin through a series of downstream protein phosphorylation and dephosphorylation processes to initiate the classical Wnt/β-catenin signaling cascade, and ultimately induce target gene expression like c-Myc and cyclinD1.101 The Wnt signaling pathway, along with the components, exhibits a critical effect on embryogenesis and is essential for normal cell growth and proliferation.102,103
Wnt/β-catenin pathway exerts an important effect on organ development and is also important for liver development, such as hepatocyte differentiation, hepatoblast growth, and differentiation.104 Monga et al. first discovered that β-catenin played a role in rat LR after PHx and that β-catenin activated numerous target genes related to LR initiation.105 A recent study finds that Wnt/β-catenin signaling promotes LR in mice by promoting the transformation of transitional liver progenitor cells (TLPCs) into hepatocytes,84 consistent with ectopic activation of Wnt pathway inducing LPCs to hepatocytes conversion in mice with biliary injury.98 The transformation and interaction between LPCs and hepatocytes in LR have been summarized in the IL-6 pathway. In addition, Wnt/β-catenin signaling can also promote the transdifferentiation of damaged hepatocytes into BECs and repair the injured liver in chronic liver injury models.106 The transformation and interaction between BECs and hepatocytes in LR have been summarized in the Notch pathway. According to Li et al., activating Wnt/β-catenin pathway promoted LR during non-alcoholic fatty liver disease (NAFLD).107 Jung, Seo et al. also proved that activating or enhancing Wnt/β-catenin pathway promoted LR.108,109,110 Other studies have also demonstrated that certain genes act as the regulators of Wnt/β-catenin pathway, which accelerates hepatocyte growth and LR by regulating Wnt/β-catenin pathway.111,112 Yin and Clemens et al. proved that Wnt/β-catenin pathway had a crucial effect on the initiation stage of LR from the opposite side, and reducing glycogen synthase kinase-3β (GSK-3β) phosphorylation affected the Wnt/β-catenin pathway, thereby inhibiting LR.113 After the excessive expression of acetaminophen (APAP), phosphatidic acid (PA) can inhibit glycogen synthetase kinase-3β (GSK-3β), a component in Wnt/β-catenin pathway, thereby suppressing Wnt/β-catenin pathway and hindering LR.114
Wnt/β-catenin signaling also shows a critical effect on LR after transplantation. As revealed by Ma et al., activating Wnt/β-catenin pathway accelerated the regeneration in liver transplantation models via pharmacological preconditioning.115 Oliva-Vilarnau et al. demonstrated that Wnt/β-catenin pathway had an important effect on human hepatocyte regeneration through constructing a three-dimensional spherical model of human hepatocytes.116 Collectively, Wnt/β-catenin pathway shows an essential impact on the initiation stage of LR.
HGF signaling pathway
As the multifunctional cytokine, HGF can be detected within diverse cell types, like HSCs, vascular endothelial cells (ECs) or KCs, and its receptor is c-Met.117 The biological functions of HGF are broad and varied: targeted destruction of HGF or c-Met during embryonic development disrupts liver and placenta development.118 Recombinant human HGF (rh-HGF) inhibits mouse hepatocyte death and stabilizes structural and vascular integrity.119 HGF attenuates APAP-induced hepatocyte necrosis and improves mouse liver failure.120
The level of HGF in the plasma has increased 17 times at 2h after PHx, well before the onset of DNA synthesis in the liver, while HGF is absent in liver residues.121 This suggests that the increase in plasma HGF is due to the production of the hepatic extranet, which is transported to the damaged liver by paracrine for initiating hepatocyte proliferation and LR. According to endogenous HGF expression, LR can be divided into two stages. In phase I (0–3 h after PHx), also known as depletion phase, HGF is derived from the transcription of HGF gene in KCs and ECs of normal liver, with a decrease in the level of both inactive form (pro-HGF) and active form.122 In phase II (from 3 to 48 h or more after PHx), named as the production phase, HGF is newly synthesized by ECs and HSCs and characterized by a significant increase in both forms.121,123 After PHx, the tyrosine phosphorylation of c-Met occurs within 5 min and increases gradually, reaching a peak at 60 min.124 In animal experiments, HGF activates LR at the onset of injury and improves NASH liver function, and is associated with JAK2-STAT3 phosphorylation and inhibition of inflammation.24 It is a highly active stimulator of DNA synthesis in hepatocytes and is active at concentrations as low as 1 ng/ml.125 HGF is elevated in the liver of rats with intestinal ischemia reperfusion (IIR) at 6 h and 72 h after reperfusion.126 These results suggest that the activation of LR by HGF is related to the promotion of hepatocyte DNA synthesis, improvement of inflammation, improvement of hepatocyte function, and activation of downstream signaling pathways. It also implies that HGF is involved in progression of LR.
The HGF secreted is usually stored in the pro-HGF form within extracellular matrix (ECM). When the liver is damaged, pro-HGF is triggered via the urokinase-type plasminase activator (u-PA). After activation, HGF promotes hepatocyte proliferation by binding to the receptor c-Met onto hepatocyte surface.127 HGF/c-Met pathway is important for protecting and regenerating tissues. Under diverse injury and disease models, HGF can promote cell growth as well as tissue regeneration, in the meantime of improving fibrosis and chronic inflammation.128 At the same time, HGF also has an important role in the initiation stage of LR, which promotes DNA synthesis and hepatocyte mitosis in a paracrine manner.24 In the early stage of LR, HGF pathway can affect cell proliferation, growth, and survival by activating various downstream pathways such as PI3K/AKT, JAK/STAT3, and Ras/Raf pathways.24,129,130 In addition, HGF pathway also interacts with TGF-α pathway to directly stimulate DNA synthesis in hepatocytes and promote hepatocytes to enter G1 phase from G0 phase.131 In cell models, HGF/c-Met signaling is found to block the bone morphogenetic protein (BMP-9)-mediated apoptosis, thereby regulating the survival of liver progenitor cells.132 In mouse models, the HGF signaling pathway can be activated by ursodeoxycholic acid (UDCA), platelet-derived growth factor (PDGF), and endocan (a pro-angiogenic protein), thereby accelerating hepatocyte proliferation and promoting LR.133,134,135 Furthermore, Huang et al. found that inhibition of C-C motif chemokine ligand (CCL) 5 enhanced the secretion of HGF from macrophages via forkhead box O3(FoxO3a) pathway, thus accelerating hepatocyte proliferation and promoting LR.136
Therefore, HGF/c-Met pathway exerts a crucial effect on initiating LR, which is related to DNA synthesis, inflammation, hepatocyte function, and mitosis. Currently, there are many clinical trials using the HGF gene in clinical management,137,138,139 suggesting that it is the candidate therapeutic target of human liver disease, so further studies are needed to investigate the impact of HGF pathway on LR. The mechanism of signaling pathway in the initiation stage of LR is summarized in Table 1.
Table 1.
The mechanism of signaling pathway in the initiation stage of LR
| Signal pathway | Mechanism | Reference |
|---|---|---|
| IL-6 signaling pathway | Stimulating LPLCs, HSC, macrophages, monocytes and KCs to produce IL-6, thereby activating the IL-6/Jak/STAT3 signaling pathway | Li et al.36 Cheng et al.43 Kim et al.44 Izumi et al.45 Song et al.46 Wen et al.47 |
| Prominin-1 promoted LR by recruiting gp130 into the lipid raft to activate the IL-6/STAT3 pathway, HO-1 promoted LR activation of IL-6/STAT3 signaling pathway by regulating IL-6, STAT3, c-Myc and c-Jun. | Bahn et al.48 Cheng et al.49 |
|
| PPARα inhibited PHx-induced LR via the IL-6/STAT3 signaling pathway. | Xie et al.50 | |
| Smad3 deletion inhibited LR By inhibiting IL-6/STAT3 pathway in early LR in mice | Kremer et al.51 | |
| Lack of IL-7 resulted in the low serum concentration of IL-6 and failure to activate STAT3in the first few hours after PHx, resulting in the decreased expression of early LR genes. | Piobbico et al.20 | |
| A high serum IL-6 level before surgery was more conducive to postoperative LR in LDLT patients | Chae et al.52 | |
| TNF-α signaling pathway | Indoleamine 2, 3-dioxygenase (Ido1/2) deficiency activated NF-kβ, resulting in the production of TNF-α, which in turn promotes LR | Ando et al.63 Ogiso et al.64 |
| TNF-α pathway induced decomposition of the p65/β-catenin complex, promoted the nuclear translocation of p65, and thus upregulated cyclin D1 and β-catenin expression as well as hepatocyte proliferation in vivo and in vitro. | Nejak-Bowen et al.65 | |
| Proteases ADAM17 and F13A promoted early LR after PHx by regulating serum levels of TNFR1 and TNF-α. | Yoshiya et al.66 Zbodakova et al.67 | |
| Loss of GP73 led to the downregulation of TNF-α and cell cycle related proteins, thus delaying the initiation of LR in mice. | Wang et al.68 | |
| TNF-α signaling pathway | MiR-199a regulated LR and hepatocyte proliferation by targeting the TNF-α/TNFR1 signaling pathway. | Zhang et al.70 |
| hPH activated Kupffer cells to secrete TNF-α to activate the TNF-α signaling pathway and promote LR. | Lee et al.71 | |
| TACE+PVE enhanced LR by inducing high levels of TNF-α expression. | Guo et al.73 | |
| Notch signaling pathway | HIF-1α is related to regulating downstream molecules, like vascular endothelial growth factor (VEGF), and shows an active regulatory effect on LR. | Ahluwalia et al.90 |
| Notch-HIF-1α signaling pathway plays an important role in hepatocyte proliferation and LR by regulating Hes1, VEGF, NICD and HIF-1α level. | Li et al.91 | |
| Notch signaling plays an important role in maintaining liver homeostasis and hepatocyte transformation into bile duct cells after liver injury. | Huang et al.10 | |
| Wnt/β-Catenin signaling pathway | Wnt/β-catenin signaling promoted the transformation of TLPCs into hepatocytes to promote LR in mice. | Pu et al.84 |
| The Wnt/β-catenin signaling pathway is an activator or enhancer of LR. | Li et al.107 Jung et al.108 Li et al.109 Seo et al.110 |
|
| T3, TRβ and SNHG12 accelerated hepatocyte proliferation and LR by regulating Wnt/β-catenin signaling. | Hönes et al.111 Zhu et al.112 |
|
| Reducing the phosphorylation level of GSK-3β can inhibit the wnt/β-catenin signaling pathway, thereby inhibiting LR. | Yin et al.112 Clemens et al.114 | |
| Activating Wnt/β-catenin pathway accelerated the regeneration in liver transplantation models via pharmacological preconditioning. | Ma et al.115 | |
| HGF signaling pathway | HGF pathway can affect cell proliferation, growth and survival by activating various downstream pathways such as PI3K/AKT, JAK/STAT3 and Ras/Raf pathways in the early stage of LR. | Zhao et al.24 Li et al.129 Okano et al.130 |
| HGF/c-Met signaling can regulate the survival of liver progenitor cells by blocking BMP9 -mediated apoptosis. | Addante et al.132 | |
| HGF signaling pathway can be activated by UDCA, PDGF, Endoca, etc. | Dong et al.133 Qian et al.134 Zhao et al.135 |
|
| Inhibition of CCL5 enhanced the secretion of HGF from repair macrophages through FoxO3a pathway. | Huang et al.136 |
The progression stage of LR
With the completion of the initiation stage, hepatocytes begin the DNA replication procedure, and nearly 95% of hepatocytes are enrolled in cell cycle to initiate proliferation.4 Hepatocyte proliferation is a mitotic process in which hepatocytes enter the cell cycle under the combined stimulation of diverse cytokines, growth factors, and cyclin CDKs. The progression of hepatocyte cycle includes the activation of several metabolic pathways, such as Hippo, HGF/c-Met and PI3K-AKT pathways, which jointly regulate the proliferation of hepatocytes. The main signaling pathways in the progression stage of LR are summarized in Figure 4.
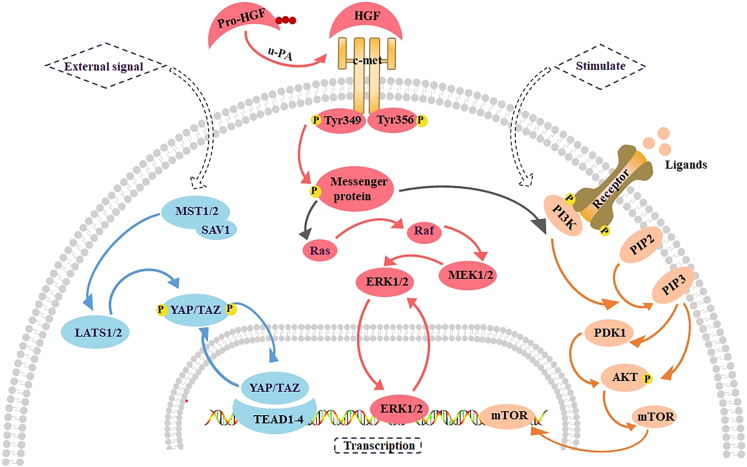
Figure 4.
The main signaling pathways in the progression phase of LR
Extracellular signal activates LATS1/2 by a complex Mst1/2 kinase and SAV1, which enables YAP/TAZ to be transported to the nucleus and interact with TEAD1-4 and other transcription factors. When pro-HGF is activated by u-PA, it can bind to c-Met to initiate a variety of downstream signaling pathways, including ERK1/2 signaling pathway and PI3K/AKT signaling pathway. When ligand-receptor binding or the PI3K/AKT signaling pathway is stimulated, PI3K promotes the conversion of from PIP2 to PIP3. PIP3 can act on AKT directly or through PDK1 on AKT, thereby activating mTOR. TEAD, Transcriptional enhanced associate domain; PIP2, Phosphatidylinositol 4, 5-diphosphate; PIP3, Phosphatidylinositol 3,4, 5-triphosphate; PDK1, 3-phosphate dependent protein kinase 1.
Hippo pathway
Hippo signaling pathway was initially discovered from Drosophila melanogaster, and it represents a conserved organ size regulator whose abnormal regulation leads to tumorigenesis.140 The kinase cascade from tumor suppressor Hippo (Mst1 as well as Mst2 within mammals) to oncoproteins Yki (YAP together with TAZ within mammals) is the heart of the pathway, which are the transcriptional coactivators for target genes related to cell growth and survival.141 It provides directions for investigating genes responsible for controlling hepatic proliferation and growth. Hippo pathway within mammalian livers sheds light on how it drives disease progression, such as from fatty liver to liver cancer.141 Therefore, numerous studies target Hippo pathway to promote LR while preventing and treating liver disease as the candidate therapeutic target.
In recent studies, Hippo pathway plays a critical role in regulating liver size, development, regeneration, homeostasis and metabolism, and its perturbations can contribute to frequent liver diseases, like fatty liver disease or even liver cancer. As a result, pharmacologically targeting Hippo pathway is adopted for promoting LR and preventing liver disease progression.142 Dong and Camargo et al. found that overexpression of YAP led to liver enlargement, thereby producing a liver five times larger than normal.140,143 The overgrowth was regulated via the enhanced hepatocyte growth and the decreased apoptosis. Stopping YAP up-regulation reversed the phenotype, and the liver then returned to the normal size rapidly.140,143 YAP up-regulation led to HCC occurrence.143 The aforementioned studies indicate that Hippo pathway has a critical effect on LR and liver disease, suggesting that it may be a therapeutic target for liver diseases in the future.
PI3K/AKT signaling pathway
As the intracellular phosphatidylinositol kinase, PI3K possesses the serine/threonine (Ser/Thr) kinase activities. The activated PI3K can phosphorylate phosphatidylinositol 4, 5-diphosphate (PIP2) into phosphatidylinositol 3,4, 5-triphosphate (PIP3) and enrolls AKT and inosine 3-phosphate dependent protein kinase 1 (PDK1) onto plasma membrane, where PDK1 phosphorylation activates AKT.144,145 As the serine/threonine kinase, AKT is involved in numerous cell activities and functions as a downstream effector for cell proliferation and survival regulated through growth factors.146,147 PI3K/AKT pathway accounts for a major intracellular signaling pathway, which participates in the regulation of cell cycle, cell growth, apoptosis, metabolism, and angiogenesis through the communication of its associated upstream together with downstream factors, and it can be activated in a variety of physiological activities.148,149,150
PI3K/AKT plays a key role in LR.151 Recent studies have suggested that dietary salidroside relieves lipid metabolic disorder and inflammatory response in the liver of laying hens via PI3K/AKT/Gsk3-β pathway, and promotes hepatocyte regeneration.152 In zebrafish, activation of the Farnesoid X receptor damaged the liver progenitor cells-mediated LR via the PTEN-PI3K-AKT-mTOR axis.153 Zhang et al. found that when PI3K-AKT and insulin signaling pathways were suppressed, autophagy was inhibited, apoptosis was enhanced, proliferation was impaired, and cell death was promoted in hepatocytes of aquaporin-9 knockout mice.154 Lin et al. revealed that translation control tumor protein (TCTP) promoted LR in mice by activating PI3K/AKT signaling pathway through interaction with mTORC2.155 Zhong et al. found that Panax notoginseng saponins (PNSs) promoted LR in mice by activating the PI3K/AKT/mTOR pathway.156 Collectively, activation of PI3K/AKT pathway promotes hepatocyte proliferation and LR.
HGF signaling pathway
HGF is a cytokine that can stimulate hepatocyte proliferation. It also acts on diverse cell types like hematopoietic cells, epithelial cells, and vascular endothelial cells and is a multifunctional factor that regulates the growth, movement, and morphogenesis of a variety of cells. It exerts a critical effect on embryogenesis, angiogenesis, organ/tissue regeneration, wound healing, carcinogenesis, and morphogenesis through paracrine or autocrine mechanisms, under the assistance of epithelial interstitial interactions.127
HGF has a critical effect on the initiation stage of LR, and is mainly responsible for initiating LR after injury.24 In addition, it is crucial for hepatocyte proliferation in the progressive stage of LR. Unlike the initial phase, it has significant functions in promoting cell proliferation, cell division, angiogenesis and cell motility.157 After PHx, HGF activity in the remnant liver begins to increase within 24 h, involving the first peak of DNA synthesis in the progressive stage of LR.158,159 However, the first increase in HGF activity in the plasma is noted 3 h after PH, which is much earlier than the initial DNA synthesis in the remnant liver. At this point, HGF mRNA levels were significantly elevated in intact distal organs such as the lungs, kidneys, and spleen, but not in the residual liver.158,160 uggesting that HGF is initially produced by the extrahepatic organs and transported to the residual liver by the “endocrine” system. Ishiki et al. demonstrated that HGF in the blood promotes LR: when recombinant human HGF (rh-HGF) was injected intravenously into hepatectomy mice, hepatocyte replication was enhanced.161 In contrast, anti-HGF IgG reduces the regenerative response of hepatocytes after liver injury (G1/S progression).162,163 HGF is an angiogenic factor in vitro,164,165,166,167 which plays an important role in angiogenesis of vascular cells, addition of HGF can induce angiogenesis and improve regional hypoxia in rabbit hindlimb ischemia model as cytokine supplement therapy.168 In addition, HGF stimulates DNA synthesis of serum-free primary hepatocytes and has a critical effect on physiological regulation of tissue and organ development.169 HGF promotes cell development, cell protection, DNA synthesis, and cell cycle progression through the direct binding to a certain tyrosine kinase receptor c-Met.24
During LR, HGF/c-Met is a key promoter that activates downstream pathways including JAK/STAT3, PI3K/AKT/NF-κB, and Ras/Raf pathways and affects cell proliferation, growth, and survival. It has important clinical significance for liver fibrosis, hepatocyte regeneration after inflammation, and LR after transplantation.24 Cheng et al. suggested that peroxisome proliferator-activated receptor γ (PPARγ) inhibited LR after PHx by negatively regulating HGF/c-Met/ERK1/2 pathway.170 Besides, Gui et al. discovered that suppressor of cytokine signaling1(SOCS1) controlled LR through modulating HGF pathway within hepatocytes. They found that the LR rate was accelerated, DNA synthesis was enhanced and the early phosphorylation level of Gab1, the downstream signal of c-Met, was upregulated in SOCS1 (−/−) mice.171 HGF/c-Met is related not only to G1/S phase but also to G2/M phase. According to Factor et al., c-Met loss within hepatocytes not only reduced G1/S progression and postponed post-PHx liver recovery but also blocked the early/middle G2 phase. Mechanism studies have indicated that these phenomena are related to the lack of ERK1/2 activation as well as the deficiency of histone H3 phosphorylation and chromatin condensation.172 HGF/c-Met signaling pathway is also positively correlated with the cell cycle progression and cell proliferation of hepatocytes in the zebrafish LR model.173 In addition, HGF also induced the expression of cyclin D1 and various cell cycle core proteins through the mitogen-activated protein kinase (MAPK) family member P38 and the STAT3/AKT/MAPK signaling pathway.174,175 Consequently, HGF pathway is important for regulating hepatocyte cycle, cell growth, and LR, and can be used as a key regulatory factor for tissue regeneration. The mechanism of signaling pathway in the progression stage of LR is summarized in Table 2.
Table 2.
The mechanism of signaling pathway in the progression stage of LR
| Signal pathway | Mechanism | Reference |
|---|---|---|
| Hippo Pathway | Overexpression of YAP led to liver enlargement, thereby producing a liver five times larger than normal. Stopping YAP up-regulation reversed the phenotype, and the liver then returned to the normal size rapidly. | Dong et al.140 Camargo et al.143 |
| YAP up-regulation led to HCC occurrence. YAP1 expression levels were strikingly correlated with the expression levels of both cyclinD1 and BclXL. | Camargo et al.143 | |
| PI3K/AKT signaling pathway | Salidroside promoted hepatocyte regeneration through PI3K/AKT/Gsk3-β pathway. | Cui et al.152 |
| Farnesoid X receptors damaged liver progenitor cell-mediated LR via the PTEN-PI3K-AKT-mTOR axis. | Jung et al.153 | |
| In hepatocytes of Aquaporin-9 knockout mice, the PI3K-AKT and insulin signaling pathways were suppressed and hepatocyte proliferation was impaired. | Zhang et al.154 | |
| TCTP promoted LR in mice by activating PI3K/AKT signaling pathway through interaction with mTORC2. | Lin et al.155 | |
| PNS promoted LR through activation of PI3K/AKT/mTOR and upregulated the AKT/Bad cell pathways in mice. | Zhong et al.156 | |
| HGF signaling pathway | PPARγ inhibited LR after PHx by negatively regulating HGF/c-Met/ERK1/2 pathway. | Cheng et al.170 |
| SOCS1 controlled LR through modulating HGF pathway within hepatocytes. LR rate was accelerated, DNA synthesis was enhanced and the early phosphorylation level of Gab1, the downstream signal of c-Met, was upregulated in SOCS1 (−/−) mice. | Gui et al.171 | |
| HGF signaling pathway | HGF/c-Met signaling pathway was positively correlated with hepatocyte cycle progression and cell proliferation in the zebrafish LR model. | Chiang et al.173 |
| HGF induced cyclin D1 expression via MAPK family member P38. | Mohammed et al.174 | |
| HGF activated the STAT3/AKT/MAPK signaling pathway and induced the expression of various cell cycle core proteins. | Li et al.175 |
The termination stage of LR
After the liver returns to the original liver weight during LR, it will cause cancer if the hepatocytes continue to proliferate, so there must be a termination stage in the process of LR to inhibit the abnormal proliferation of hepatocytes.176 Studies have reported that when the liver mass/body weight ratio returns to 2.5%, the LR rate will slow down.177 When the liver is restored to the appropriate size and basic function, various signaling pathways associated with the termination stage of LR are activated to slow down LR, among which the most extensively studied signaling pathways are mediated by the TGF-β family including three TGF-β isomers (TGF-β1/-β2/-β3), bone morphogenetic proteins (BMPs), and activins.178 It is generally believed that TGF-β, activin, and BMPs-mediated signaling pathways have a critical effect on the termination stage of LR.18 Those main pathways related to the termination stage of LR are summarized in Figure 5.
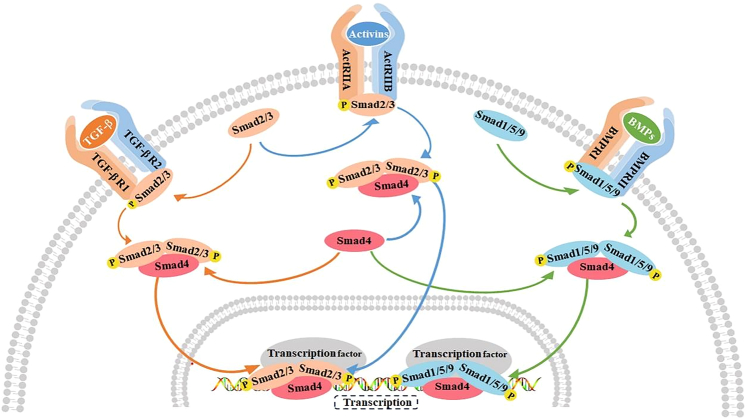
Figure 5.
The main signaling pathways in the termination phase of LR
TGF-β, activin, and BMPs-mediated signaling pathways play an important role in the termination phase of LR. TGF-β binds to the receptor TGF-βR1/TGF-βR2 to recruit and phosphorylate Smad2/3. The activin binds to the receptor ActRⅡA/ActRⅡB and also recruits and phosphorylates Smad2/3. BMP binds to the receptor BMPRⅠ/BMPRⅡ and recruits and phosphorylates Smad1/5/9. Smad4 is a common transduction factor of these three signaling pathways, and Smad2/3 and Smad1/5/9 bind to Smad4 after phosphorylation, and are eventually transported to the nucleus to bind to transcription factors. TGF-βR1, TGF-β receptors1; TGF-βR2, TGF-β receptors2; Smad, Small mother against decapentaplegic; BMPRⅠ, Bone morphogenetic proteins receptors Ⅰ; BMPRⅡ, Bone morphogenetic proteins receptors Ⅱ.
TGF-β signaling pathway
TGF-β was first discovered and named in 1978 by De Larco et al., who purified a secreted polypeptide from fibroblasts infected with MSV oncovirus, which allowed the wild proliferation of normal fibroblasts in soft AGAR and was thus named “transforming growth factor”.179 The TGF-β family of pathways are critical for different physiological events, such as cell growth, differentiation, invasion, and death, and are essential for the homeostasis of tissues and organs.180 After the binding of TGF-β to type I/type II TGF-β receptors (TGF-βR1 as well as TGF-βR2), TGF-β activates the TGF-β signaling pathway, which in turn regulates cell growth and death. After activation, type I receptor phosphorylates Smad2 and Smad3 while binding to Smad4 to form the Smad complex. Smad complexes enter the nucleus to interact with various transcription factors for regulating target gene transcription.181
TGF-β has a critical effect on the termination stage of LR, which accounts for a powerful inhibitor of hepatocyte proliferation. Recent studies have indicated that TGF-β can be used as a target for Ahnak (a large 700 kDa protein), G protein-coupled receptor 50 (GRP50), and growth differentiation factor (GDF) 11 to inhibit hepatocyte proliferation and LR by blocking the cell cycle.182,183,184 TGF-β also acts as a target for integrin αvβ8, LSKL peptide (an inhibitor of platelet reactive protein TSP-1), simvastatin (Sim, a drug used to treat hypercholesterolemia), which can block the TGF-β signaling pathway, thereby delaying the termination of LR.185,186,187 These studies also demonstrate that TGF-β signaling is tightly associated with the termination stage of LR. Masuda, Karkampouna et al. showed that TGF-βR-I inhibitors LY2157299 and LY364947 blocked the inhibition of TGF-β pathway on hepatocytes by blocking the downstream signaling of TGF-βI receptor kinase.188,189 Chen et al. reported that down-regulating TGF-βR1 weakened the inhibitory effect of TGF-β signaling pathway on LR in mice.190 According to the studies of Bird, Heo et al., regulating signaling factors like TGF-βR1 and Smad within TGF-β pathway blocked TGF-β signaling pathway, thus inhibiting the termination of LR and promoting hepatocyte proliferation.191,192 Inhibition of TGF-β pathway is an effective therapeutic strategy to promote hepatocyte proliferation and LR. Therefore, the TGF-β signaling pathway deserves further investigations.
Activin signaling pathway and BMPs signaling pathway
Activins, belonging to TGF-β superfamily, are first extracted due to their abilities to induce the release of follicle-stimulating hormone (FSH) from the pituitary.193 Activins exist in three basic molecular forms, including activin A (βAβA), activin B (βBβB), and together with activin AB (βAβB).194 Activin A is structurally similar to other family members, yet maintains its own unique function. It is related to numerous cellular activities like growth, differentiation, and apoptosis, and is also a key regulator of inflammation.194 Activin A can combine with activin type II receptors (ActRIIA and ActRIIB) to recruit the trans-phosphorylated companion activin type I receptor (ALK4). The activated ALK4 induces the activation and nuclear translocation of Smad protein and regulates the expression of target genes.195 Activin A can inhibit DNA synthesis in hepatocytes in an autocrine manner during LR to exert its role as a negative regulator.196 Recent studies have shown that activin A released by endothelial cells as a transcription target of Kruppel-like transcription factor 2 (KLF2) can negatively regulate hepatocyte proliferation and LR.197 Lv, Chen et al. found that activin A signaling pathway induced the growth arrest and apoptosis of hepatocytes both in vitro and in vivo.198,199 Chen et al. suggested that activin A inhibited the process of LR by activating the Smad signaling pathway to up-regulate p15INK4B and p21WAF1/Cip1 and down-regulate cyclin D1 and cyclin E.198 Other studies have indicated that the expression of ActRIIB promotes the phosphorylation of Smad2/3 and eliminates the role of miR-194 in hepatocyte growth, thus inhibiting hepatocyte proliferation and apoptosis.200
BMPs, the greatest TGF-β subfamily, consist of over 15 ligands and is divided into about 20 different BMPs including BMP-2/-4/-7/-9, according to the similarity of their amino acid sequences.201 In the earliest studies, BMP can promote cartilage and bone generation as well as fracture repair, while regulating the differentiation and proliferation of chondrocytes and osteoblasts in vitro.202 In subsequent studies, BMPs are also found to be the key regulators of adult tissue homeostasis, regulating diverse biological events like growth, survival, and differentiation. Therefore, it exerts a critical effect on embryology, tissue regeneration, or even cancer.203,204 The liver is a target organ of BMP action, and it has a critical effect on the termination stage of LR.205 BMP ligands in the typical BMP pathway combine with types I/II BMP receptors for the formation of complexes related to the activation and phosphorylation of receptors, which thereby regulate SAMAD-1, SAMAD-5, or SAMAD-9. Phosphorylated Smad −1, Smad −5, or Smad −9 (p Smad 1/5/9) binds to Smad 4 and translocates heterocomplex in the nucleus for regulating the expression of target genes.206 BMPs-mediated signaling pathway is relatively complex. Firstly, BMPs mainly bind to phosphorylated Smad1/5/8 receptor to inhibit LR.207 Secondly, different subunits of BMPs may have different or even opposite effects, for example, BMP-7 promotes hepatocyte proliferation.208 However, BMP-2, BMP-4, and BMP-9 inhibit the proliferation of hepatocytes.132,209,210,211
The aforementioned studies indicate that activins and BMPs are critical for the termination stage of LR. Nonetheless, further research is warranted to verify and illustrate the specific mechanisms of activins and BMPs in the termination stage of LR. The mechanism of signaling pathway in in the termination stage of LR is summarized in Table 3.
Table 3.
The mechanism of signaling pathway in the termination stage of LR
| Signal pathway | Mechanism | Reference |
|---|---|---|
| TGF-β signaling pathway | Ahnak depletion accelerated LR by modulating the TGF-β/Smad signaling pathway | Yang et al.184 |
| The orphan GPR50 receptor interacting with TβRI induces G1/S-phase cell-cycle arrest via Smad3-p27/p21 in BRL-3A cells. | Chang et al.182 | |
| GDF11 inhibited cell cycle progression and LR through TGF-β- Smad 2/3 signaling pathway. | Wang et al.183 | |
| Loss of Integrin αvβ8 in Murine Hepatocytes Accelerates LR. | Greenhalgh et al.186 | |
| TGF-β signaling pathway | Effect of LSKL peptide on thrombospondin 1-mediated TGF-β signal activation and LR after hepatectomy in an experimental model. | Kuroki et al.187 |
| LY2157299 and LY364947 block downstream signaling of TGF-βI receptor kinases, thereby blocking the TGF-β pathway. | Karkampouna et al.188 Masuda et al.189 |
|
| Regulation of TGF-βR1, Smad, and other signaling factors in the TGF-β signaling pathway impeded TGF-β signaling. | Chen et al.190 Bird et al.191 Heo et al.192 |
|
| Activin signaling pathway | Activin A, as a transcriptional target of KLF2, negatively regulated hepatocyte proliferation and LR. | Manavski et al.197 |
| ActRIIB eliminated the effects of miR-194 on the proliferation of hepatocytes by promoting the phosphorylation of Smad2/3. | Gao et al.200 | |
| Activin A activated the Smad signaling pathway by up-regulating p15INK4B and p21WAF1/Cip1, and down-regulating cyclin D1 and cyclin E. | Chen et al.198 | |
| Activin A induced growth arrest through a Smad - dependent pathway in hepatic progenitor cells | Chen et al.198 | |
| BMPs signaling pathway | BMPs inhibited LR by binding to the phosphate Smad1/5/8 receptor. | Katagiri et al.206 Tsugawa et al.207 |
| BMP-2, BMP-4 and BMP-9 mediated signaling pathways inhibited the proliferation of hepatocytes. | Addante et al.132 Breitkopf-Heinlein et al.209 Do et al.210 Xu et al.211 |
Conclusions and prospects
In the last few decades, great progresses are achieved in the biological understanding and clinical application of LR. Clinical studies have found that preoperative high levels of serum IL-6 and TNF-α contribute to early regeneration after LDLT.52 The expressions of IL-6 and TNF-α are significantly increased in HCC patients after hepatectomy.72 During liver cell transplantation, localized direct delivery of Wnt3a and Wnt5a to the cells might be beneficial to the successful LR.212 After PHx, the C-C motif chemokine ligand (CCL5) is increased in healthy donors of living donor liver transplantation (LT)and inhibition of CCL5 enhances HGF secretion in repair macrophages, thereby activating the HGF signaling pathway to improve LR. Reduce the mortality rate of liver failure after hepatectomy.136 Due to lack of nephrotoxicity and anti-cancer effect, mTOR inhibitors can be used in patients after 30 days of PLTx to promote PLTx success.213,214 Although there have been some advances in clinical application, but the actual mechanism of LR remains unclear, and its current research results are far from meeting the needs of clinical treatment of liver diseases. The clinical application of LR still requires further research to provide theoretical basis for treating liver transplantation, liver failure, or other end-stage liver disease.
The initiation, maintenance, and termination of LR are strictly regulated by many cytokines and their mediated signaling pathways. The present study pays special attention to main pathways in each stage of LR. LR is a complex process, and a single factor or a single signaling pathway cannot complete the whole process independently. Instead, the combined effect of multiple signaling pathways is needed. During LR, if one pathway is blocked, the others will compensate for it and return to almost the normal liver quality. In the process of LR, inflammation-related signaling pathways run through the whole process and have the role of initiating tissue repair and regeneration.215 IL-6 and TNF-α are the most widely studied pro-inflammatory factors, and their mediated signaling pathways play a major role in the process of LR. In addition to inflammatory pathways, metabolic pathways also play an important role in the process of LR.
Following PHx, the metabolic of the LR is enormously required to meet DNA replication and cell division, with lipids being the main source of energy.216 mTORC2 promotes fatty acid (FA) metabolism through the glucose-peroxisom- activated receptor α(PPAR-α) pathway and promotes LR.217 Bile acid signaling is also required for LR, bile acids are important liver products, and their levels are tightly regulated. Huang et al. find that nuclear receptor-dependent bile acid signaling is required for normal LR, elevated bile acid levels accelerate regeneration, and decreased levels inhibit liver regrowth, as does the absence of the primary nuclear bile acid receptor FXR.218 Belenguer et al. have found that RNF43/ZNRF3 deficiency predisposes to HCC by impairing LR and altering lipid metabolic ground-state.219 Glucose metabolism is also involved in LR. Aquaporin-9 (AQP9) promotes LR after PHx by regulating glucose metabolism in hepatocytes.154 Insulin exacerbates hepatic ischemia/reperfusion(I/R) injury by energy depletion through the IRS-2/SREBP-1c pathway.220 MSC transplantation improves the glucose metabolism and survival rate in the post-hepatectomy liver failure (PHLF) model, which may be related to the AKT/GSK-3β/β-catenin pathway.221 Amino acid metabolism is also involved in LR. Studies have found that abnormal s-adenosylmethionine (SAM) is prone to liver disease.222 Nelsen et al. have found that amino acids can also regulate hepatocytes proliferation by regulating the expression of cyclin D1.223 Feeding amino acids led to liver DNA synthesis and mitotic explosion in rats that were not fed protein for 3 days,224 suggesting an important role for amino acid metabolism in LR. The effect of metabolism on LR cannot be ignored because the replication of hepatocytes requires a large number of proteins, lipids, and nucleic acids. Therefore, this indicates that it is not feasible to regulate a certain factor or a certain signaling pathway to solve the problems about LR. In the future, with the continuous understanding of LR signaling pathways, it is possible to simultaneously regulate multiple signaling pathways to enhance LR, thereby improving the success rate of LDLT, PHx treatment of liver, and other operations.
In addition, numerous studies have verified that the signaling pathways play important roles in LR under different physiological and pathological conditions. For instance, studies have shown that activating signaling pathways such as HGF, EGFR, and TNF-α prevents acute liver failure and promotes LR.225 In PHx after cirrhosis, the IL-6 signaling pathway can maintain compensatory LR by enhancing STAT3 activation,226 the continuous activation of IL-6 signaling pathway can lead to the development of HCC. In mice with carbon tetrachloride (CCl4),-induced cirrhosis, the TGF-β receptor type I(TGFβR1) kinase inhibitor galunisertib (LY2157299) can block the liver fibrosis process and promote LR by inhibiting the TGF-β signaling pathway.189 In alcoholic hepatitis (AH) samples, overactivation of YAP leads to loss of key biological functions such as regenerative capacity, and targeting YAP is an effective strategy for treating patients with AH or alcohol-related cirrhosis.227 In liver ischemia-reperfusion injury (IRI), Hippo signaling pathway plays a role in promoting liver repair and regeneration.228 In addition, the promotion role of Hippo signaling pathway in LR after IRI is related to the transformation of LPCs, and the activation of YAP promotes the dedifferentiation of hepatocytes into LPCs, which can differentiate into BECs and further promote LR.229 When hepatocyte proliferation is impaired, LPCs are activated and differentiate into BECs and hepatocytes to promote LR and maintain liver stability. The crosstalk between HGF/c-Met signaling pathway and BMP9 can also promote the mutual transformation between hepatocytes, BECs, and LPCs.132 The relationship between signal pathway and cell transformation provides theoretical basis and direction for the treatment of patients with liver disease. Therefore, more research is needed to understand the signaling pathways of LR under different physiological and pathological conditions for clinical medicine.
At present, LR is mainly studied by establishing animal models (rats, mice, and zebrafish) in vivo and by culturing hepatocytes in vitro. However, both animal model experiments and cell model experiments are inadequate. Due to species differences, conclusions inferred from animal models cannot be directly applied in human beings. The liver is comprised by various cell types, while cells can secrete different molecules to form different signaling pathways and interact with each other to jointly regulate LR. Consequently, during in vitro hepatocyte culture, it is impossible to accurately obtain a complete signal map of the upstream and downstream cell molecules and between signaling pathways. Therefore, in the future study on LR-related pathways, the organoid model of the liver can be considered, so that the relationship between cells, molecules, and signaling pathways in LR process can be elaborated, and the theoretical results of LR can thus be truly applied in clinical practice.
Acknowledgments
We thank Dr. Jianlin Guo for his excellent language editing work for this manuscript.
Funding: Our research has been funded by the Startup package (GY) of Henan Normal University, Overseas Expertise Introduction Center for Discipline Innovation of Pulmonary Fibrosis (111 Project), National Key R&D Program of China (2019YFE0119500), National Research Project Cultivation Fund of Henan Normal University (20210390), Doctoral Research Foundation of Henan Normal University (5101049170192) as well as Scientific and technological breakthroughs Project of Henan (222102310291, 222102310589).
Author contributions
C.F.S. and Y.B.Z. searched and sorted literatures. B.Y.Y. and C.F.S. refined the drafted manuscript. C.Y.Z. and G.Y.Y. conceived and coordinated the project. C.F.S. and C.Y.Z. wrote the manuscript. All of the authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Chunyan Zhang, Email: zcy1119sdc@163.com.
GuoYing Yu, Email: guoyingyu@htu.edu.cn.
References
- 1.Taki-Eldin A., Zhou L., Xie H.Y., Zheng S.S. Liver regeneration after liver transplantation. Eur. Surg. Res. 2012;48:139–153. doi: 10.1159/000337865. [DOI] [PubMed] [Google Scholar]
- 2.Pahlavan P.S., Feldmann R.E., Jr., Zavos C., Kountouras J. Prometheus' challenge: molecular, cellular and systemic aspects of liver regeneration. J. Surg. Res. 2006;134:238–251. doi: 10.1016/j.jss.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Fujiyoshi M., Ozaki M. Molecular mechanisms of liver regeneration and protection for treatment of liver dysfunction and diseases. J. Hepatobiliary. Pancreat. Sci. 2011;18:13–22. doi: 10.1007/s00534-010-0304-2. [DOI] [PubMed] [Google Scholar]
- 4.Taub R. Liver regeneration: from myth to mechanism. Nat. Rev. Mol. Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 5.Minuk G.Y. Hepatic regeneration: If it ain't broke, don't fix it. Can. J. Gastroenterol. 2003;17:418–424. doi: 10.1155/2003/615403. [DOI] [PubMed] [Google Scholar]
- 6.Terui K., Ozaki M. The role of STAT3 in liver regeneration. Drugs Today. 2005;41:461–469. doi: 10.1358/dot.2005.41.7.893622. [DOI] [PubMed] [Google Scholar]
- 7.Haga S., Ogawa W., Inoue H., Terui K., Ogino T., Igarashi R., Takeda K., Akira S., Enosawa S., Furukawa H., et al. Compensatory recovery of liver mass by Akt-mediated hepatocellular hypertrophy in liver-specific STAT3-deficient mice. J. Hepatol. 2005;43:799–807. doi: 10.1016/j.jhep.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 8.Michalopoulos G.K., DeFrances M.C. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 9.Ozaki M. Cellular and molecular mechanisms of liver regeneration: Proliferation, growth, death and protection of hepatocytes. Semin. Cell Dev. Biol. 2020;100:62–73. doi: 10.1016/j.semcdb.2019.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Huang R., Zhang X., Gracia-Sancho J., Xie W.F. Liver regeneration: Cellular origin and molecular mechanisms. Liver Int. 2022;42:1486–1495. doi: 10.1111/liv.15174. [DOI] [PubMed] [Google Scholar]
- 11.Deng X., Zhang X., Li W., Feng R.X., Li L., Yi G.R., Zhang X.N., Yin C., Yu H.Y., Zhang J.P., et al. Chronic Liver Injury Induces Conversion of Biliary Epithelial Cells into Hepatocytes. Cell Stem Cell. 2018;23:114–122.e3. doi: 10.1016/j.stem.2018.05.022. [DOI] [PubMed] [Google Scholar]
- 12.Manco R., Clerbaux L.A., Verhulst S., Bou Nader M., Sempoux C., Ambroise J., Bearzatto B., Gala J.L., Horsmans Y., van Grunsven L., et al. Reactive cholangiocytes differentiate into proliferative hepatocytes with efficient DNA repair in mice with chronic liver injury. J. Hepatol. 2019;70:1180–1191. doi: 10.1016/j.jhep.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Tarlow B.D., Pelz C., Naugler W.E., Wakefield L., Wilson E.M., Finegold M.J., Grompe M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell. 2014;15:605–618. doi: 10.1016/j.stem.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Preziosi M.E., Monga S.P. Update on the Mechanisms of Liver Regeneration. Semin. Liver Dis. 2017;37:141–151. doi: 10.1055/s-0037-1601351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shu W., Yang M., Yang J., Lin S., Wei X., Xu X. Cellular crosstalk during liver regeneration: unity in diversity. Cell Commun. Signal. 2022;20:117. doi: 10.1186/s12964-022-00918-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webber E.M., Bruix J., Pierce R.H., Fausto N. Tumor necrosis factor primes hepatocytes for DNA replication in the rat. Hepatology. 1998;28:1226–1234. doi: 10.1002/hep.510280509. [DOI] [PubMed] [Google Scholar]
- 17.Yagi S., Hirata M., Miyachi Y., Uemoto S. Liver Regeneration after Hepatectomy and Partial Liver Transplantation. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21218414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilgenkrantz H., Collin de l'Hortet A. New insights into liver regeneration. Clin. Res. Hepatol. Gastroenterol. 2011;35:623–629. doi: 10.1016/j.clinre.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Michalopoulos G.K., Bhushan B. Liver regeneration: biological and pathological mechanisms and implications. Nat. Rev. Gastroenterol. Hepatol. 2021;18:40–55. doi: 10.1038/s41575-020-0342-4. [DOI] [PubMed] [Google Scholar]
- 20.Piobbico D., Bartoli D., Pieroni S., De Luca A., Castelli M., Romani L., Servillo G., Della-Fazia M.A. Role of IL-17RA in the proliferative priming of hepatocytes in liver regeneration. Cell Cycle. 2018;17:2423–2435. doi: 10.1080/15384101.2018.1542893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters M., Blinn G., Jostock T., Schirmacher P., Meyer zum Büschenfelde K.H., Galle P.R., Rose-John S. Combined interleukin 6 and soluble interleukin 6 receptor accelerates murine liver regeneration. Gastroenterology. 2000;119:1663–1671. doi: 10.1053/gast.2000.20236. [DOI] [PubMed] [Google Scholar]
- 22.Su A.I., Guidotti L.G., Pezacki J.P., Chisari F.V., Schultz P.G. Gene expression during the priming phase of liver regeneration after partial hepatectomy in mice. Proc. Natl. Acad. Sci. USA. 2002;99:11181–11186. doi: 10.1073/pnas.122359899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmers T.A., McKillop I.H., Pierce R.H., Yoo J.Y., Koniaris L.G. Massive liver growth in mice induced by systemic interleukin 6 administration. Hepatology. 2003;38:326–334. doi: 10.1053/jhep.2003.50318. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y., Ye W., Wang Y.D., Chen W.D. HGF/c-Met: A Key Promoter in Liver Regeneration. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.808855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirano T. IL-6 in inflammation, autoimmunity and cancer. Int. Immunol. 2021;33:127–148. doi: 10.1093/intimm/dxaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaur S., Bansal Y., Kumar R., Bansal G. A panoramic review of IL-6: Structure, pathophysiological roles and inhibitors. Bioorg. Med. Chem. 2020;28 doi: 10.1016/j.bmc.2020.115327. [DOI] [PubMed] [Google Scholar]
- 27.Choy E.H., De Benedetti F., Takeuchi T., Hashizume M., John M.R., Kishimoto T. Translating IL-6 biology into effective treatments. Nat. Rev. Rheumatol. 2020;16:335–345. doi: 10.1038/s41584-020-0419-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mesquida M., Molins B., Llorenç V., de la Maza M.S., Adán A. Targeting interleukin-6 in autoimmune uveitis. Autoimmun. Rev. 2017;16:1079–1089. doi: 10.1016/j.autrev.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Rose-John S. Interleukin-6 Family Cytokines. Cold Spring Harb. Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a028415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al-Ghamdi T.H., Atta I.S., El-Refaei M. Role of interleukin 6 in liver cell regeneration after hemi-hepatectomy, correlation with liver enzymes and flow cytometric study. Clin. Exp. Hepatol. 2020;6:42–48. doi: 10.5114/ceh.2020.93055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naseem S., Hussain T., Manzoor S. Interleukin-6: A promising cytokine to support liver regeneration and adaptive immunity in liver pathologies. Cytokine Growth Factor Rev. 2018;39:36–45. doi: 10.1016/j.cytogfr.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt-Arras D., Rose-John S. IL-6 pathway in the liver: From physiopathology to therapy. J. Hepatol. 2016;64:1403–1415. doi: 10.1016/j.jhep.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Cressman D.E., Greenbaum L.E., DeAngelis R.A., Ciliberto G., Furth E.E., Poli V., Taub R. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 34.Hunter C.A., Jones S.A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 35.White P., Brestelli J.E., Kaestner K.H., Greenbaum L.E. Identification of transcriptional networks during liver regeneration. J. Biol. Chem. 2005;280:3715–3722. doi: 10.1074/jbc.M410844200. [DOI] [PubMed] [Google Scholar]
- 36.Li L., Cui L., Lin P., Liu Z., Bao S., Ma X., Nan H., Zhu W., Cen J., Mao Y., et al. Kupffer-cell-derived IL-6 is repurposed for hepatocyte dedifferentiation via activating progenitor genes from injury-specific enhancers. Cell Stem Cell. 2023;30:283–299.e9. doi: 10.1016/j.stem.2023.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Gennero L., Roos M.A., Sperber K., Denysenko T., Bernabei P., Calisti G.F., Papotti M., Cappia S., Pagni R., Aimo G., et al. Pluripotent plasticity of stem cells and liver repopulation. Cell Biochem. Funct. 2010;28:178–189. doi: 10.1002/cbf.1630. [DOI] [PubMed] [Google Scholar]
- 38.Ko S., Russell J.O., Molina L.M., Monga S.P. Liver Progenitors and Adult Cell Plasticity in Hepatic Injury and Repair: Knowns and Unknowns. Annu. Rev. Pathol. 2020;15:23–50. doi: 10.1146/annurev-pathmechdis-012419-032824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi T.Y., Ninov N., Stainier D.Y.R., Shin D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology. 2014;146:776–788. doi: 10.1053/j.gastro.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He J., Lu H., Zou Q., Luo L. Regeneration of liver after extreme hepatocyte loss occurs mainly via biliary transdifferentiation in zebrafish. Gastroenterology. 2014;146:789–800.e8. doi: 10.1053/j.gastro.2013.11.045. [DOI] [PubMed] [Google Scholar]
- 41.Huang M., Chang A., Choi M., Zhou D., Anania F.A., Shin C.H. Antagonistic interaction between Wnt and Notch activity modulates the regenerative capacity of a zebrafish fibrotic liver model. Hepatology. 2014;60:1753–1766. doi: 10.1002/hep.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raven A., Lu W.Y., Man T.Y., Ferreira-Gonzalez S., O'Duibhir E., Dwyer B.J., Thomson J.P., Meehan R.R., Bogorad R., Koteliansky V., et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature. 2017;547:350–354. doi: 10.1038/nature23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng N., Kim K.H., Lau L.F. Senescent hepatic stellate cells promote liver regeneration through IL-6 and ligands of CXCR2. JCI Insight. 2022;7 doi: 10.1172/jci.insight.158207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim A.R., Park J.I., Oh H.T., Kim K.M., Hwang J.H., Jeong M.G., Kim E.H., Hwang E.S., Hong J.H. TAZ stimulates liver regeneration through interleukin-6-induced hepatocyte proliferation and inhibition of cell death after liver injury. FASEB J. 2019;33:5914–5923. doi: 10.1096/fj.201801256RR. [DOI] [PubMed] [Google Scholar]
- 45.Izumi T., Imai J., Yamamoto J., Kawana Y., Endo A., Sugawara H., Kohata M., Asai Y., Takahashi K., Kodama S., et al. Vagus-macrophage-hepatocyte link promotes post-injury liver regeneration and whole-body survival through hepatic FoxM1 activation. Nat. Commun. 2018;9:5300. doi: 10.1038/s41467-018-07747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song Z., Humar B., Gupta A., Maurizio E., Borgeaud N., Graf R., Clavien P.A., Tian Y. Exogenous melatonin protects small-for-size liver grafts by promoting monocyte infiltration and releases interleukin-6. J. Pineal Res. 2018;65 doi: 10.1111/jpi.12486. [DOI] [PubMed] [Google Scholar]
- 47.Wen Y., Feng D., Wu H., Liu W., Li H., Wang F., Xia Q., Gao W.Q., Kong X. Defective Initiation of Liver Regeneration in Osteopontin-Deficient Mice after Partial Hepatectomy due to Insufficient Activation of IL-6/Stat3 Pathway. Int. J. Biol. Sci. 2015;11:1236–1247. doi: 10.7150/ijbs.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bahn M.S., Yu D.M., Lee M., Jo S.J., Lee J.W., Kim H.C., Lee H., Kim H.L., Kim A., Hong J.H., et al. Central role of Prominin-1 in lipid rafts during liver regeneration. Nat. Commun. 2022;13:6219. doi: 10.1038/s41467-022-33969-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng B., Xie H., Jia J., Wu M., Guo J., Zhang Y., Liu Y., Zhou J., He N. Deceleration of Liver Regeneration by Knockdown of Heme Oxygenase-1 is Associated With Impairment of Liver Injury Recovery After Reduced-Size Liver Transplantation in Rats. Transplant. Proc. 2020;52:1001–1006. doi: 10.1016/j.transproceed.2019.11.051. [DOI] [PubMed] [Google Scholar]
- 50.Xie G., Song Y., Li N., Zhang Z., Wang X., Liu Y., Jiao S., Wei M., Yu B., Wang Y., et al. Myeloid peroxisome proliferator-activated receptor α deficiency accelerates liver regeneration via IL-6/STAT3 pathway after 2/3 partial hepatectomy in mice. Hepatobiliary Surg. Nutr. 2022;11:199–211. doi: 10.21037/hbsn-20-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kremer M., Son G., Zhang K., Moore S.M., Norris A., Manzini G., Wheeler M.D., Hines I.N. Smad3 signaling in the regenerating liver: implications for the regulation of IL-6 expression. Transpl. Int. 2014;27:748–758. doi: 10.1111/tri.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chae M.S., Moon K.U., Chung H.S., Park C.S., Lee J., Choi J.H., Hong S.H. Serum interleukin-6 and tumor necrosis factor-α are associated with early graft regeneration after living donor liver transplantation. PLoS One. 2018;13 doi: 10.1371/journal.pone.0195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carswell E.A., Old L.J., Kassel R.L., Green S., Fiore N., Williamson B. An endotoxin-induced serum factor that causes necrosis of tumors. Proc. Natl. Acad. Sci. USA. 1975;72:3666–3670. doi: 10.1073/pnas.72.9.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tiegs G., Horst A.K. TNF in the liver: targeting a central player in inflammation. Semin. Immunopathol. 2022;44:445–459. doi: 10.1007/s00281-022-00910-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dostert C., Grusdat M., Letellier E., Brenner D. The TNF Family of Ligands and Receptors: Communication Modules in the Immune System and Beyond. Physiol. Rev. 2019;99:115–160. doi: 10.1152/physrev.00045.2017. [DOI] [PubMed] [Google Scholar]
- 56.Liedtke C., Trautwein C. The role of TNF and Fas dependent signaling in animal models of inflammatory liver injury and liver cancer. Eur. J. Cell Biol. 2012;91:582–589. doi: 10.1016/j.ejcb.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 57.Alanazi A., Nagi M.N., Alhareth D.Y., Al-Hamamah M.A., Mahmoud M.A., Ahmad S.F., Ansari M.A., Nadeem A., Bakheet S.A., Harisa G.I., Attia S.M. Crosstalk of TNF-α, IFN-γ, NF-kB, STAT1 and redox signaling in lipopolysaccharide/d-galactosamine/dimethylsulfoxide-induced fulminant hepatic failure in mice. Saudi Pharm. J. 2023;31:370–381. doi: 10.1016/j.jsps.2023.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Elshal M., Hazem S.H. Escin suppresses immune cell infiltration and selectively modulates Nrf2/HO-1, TNF-α/JNK, and IL-22/STAT3 signaling pathways in concanavalin A-induced autoimmune hepatitis in mice. Inflammopharmacology. 2022;30:2317–2329. doi: 10.1007/s10787-022-01058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kimura M., Moteki H., Ogihara M. Role of Hepatocyte Growth Regulators in Liver Regeneration. Cells. 2023;12:208. doi: 10.3390/cells12020208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Luan X., Chen P., Li Y., Yuan X., Miao L., Zhang P., Cao Q., Song X., Di G. TNF-α/IL-1β-licensed hADSCs alleviate cholestatic liver injury and fibrosis in mice via COX-2/PGE2 pathway. Stem Cell Res. Ther. 2023;14:100. doi: 10.1186/s13287-023-03342-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Libermann T.A., Baltimore D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol. Cell Biol. 1990;10:2327–2334. doi: 10.1128/mcb.10.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kimura T., Sakaida I., Terai S., Matsumura Y., Uchida K., Okita K. Inhibition of tumor necrosis factor-alpha production retards liver regeneration after partial hepatectomy in rats. Biochem. Biophys. Res. Commun. 1997;231:557–560. doi: 10.1006/bbrc.1997.6135. [DOI] [PubMed] [Google Scholar]
- 63.Ando T., Hoshi M., Tezuka H., Ito H., Nakamoto K., Yamamoto Y., Saito K. Absence of indoleamine 2,3-dioxygenase 2 promotes liver regeneration after partial hepatectomy in mice. Mol. Med. Rep. 2023;27 doi: 10.3892/mmr.2022.12911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ogiso H., Ito H., Kanbe A., Ando T., Hara A., Shimizu M., Moriwaki H., Seishima M. The Inhibition of Indoleamine 2,3-Dioxygenase Accelerates Early Liver Regeneration in Mice After Partial Hepatectomy. Dig. Dis. Sci. 2017;62:2386–2396. doi: 10.1007/s10620-017-4651-6. [DOI] [PubMed] [Google Scholar]
- 65.Nejak-Bowen K., Moghe A., Cornuet P., Preziosi M., Nagarajan S., Monga S.P. Role and Regulation of p65/β-Catenin Association During Liver Injury and Regeneration: A "Complex" Relationship. Gene Expr. 2017;17:219–235. doi: 10.3727/105221617X695762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoshiya S., Shirabe K., Imai D., Toshima T., Yamashita Y.i., Ikegami T., Okano S., Yoshizumi T., Kawanaka H., Maehara Y. Blockade of the apelin-APJ system promotes mouse liver regeneration by activating Kupffer cells after partial hepatectomy. J. Gastroenterol. 2015;50:573–582. doi: 10.1007/s00535-014-0992-5. [DOI] [PubMed] [Google Scholar]
- 67.Zbodakova O., Chalupsky K., Sarnova L., Kasparek P., Jirouskova M., Gregor M., Sedlacek R. ADAM10 and ADAM17 regulate EGFR, c-Met and TNF RI signalling in liver regeneration and fibrosis. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-90716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J., Ning J., Qian X., Zhang T., Yao M., Wang J., Chen X., Lu F. Deletion of Golgi protein 73 delayed hepatocyte proliferation of mouse in the early stages of liver regeneration. J. Gastroenterol. Hepatol. 2021;36:1346–1356. doi: 10.1111/jgh.15315. [DOI] [PubMed] [Google Scholar]
- 69.Xie C., Zhang Z., Yang M., Cao C., Zhou Y., Zhu Z., Gong W., Xu C., Yan L., Hu Z., et al. Lactiplantibacillus plantarum AR113 Exhibit Accelerated Liver Regeneration by Regulating Gut Microbiota and Plasma Glycerophospholipid. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.800470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang C., Ye B., Wei J., Wang Q., Xu C., Yu G. MiR-199a-5p regulates rat liver regeneration and hepatocyte proliferation by targeting TNF-α TNFR1/TRADD/CASPASE8/CASPASE3 signalling pathway. Artif. Cells, Nanomed. Biotechnol. 2019;47:4110–4118. doi: 10.1080/21691401.2019.1683566. [DOI] [PubMed] [Google Scholar]
- 71.Lee T.H., Park D.S., Jang J.Y., Lee I., Kim J.M., Choi G.S., Oh C.T., Kim J.Y., Han H.J., Han B.S., Joh J.W. Human Placenta Hydrolysate Promotes Liver Regeneration via Activation of the Cytokine/Growth Factor-Mediated Pathway and Anti-oxidative Effect. Biol. Pharm. Bull. 2019;42:607–616. doi: 10.1248/bpb.b18-00712. [DOI] [PubMed] [Google Scholar]
- 72.Fathi F., Sanei B., Ganjalikhani Hakemi M., Saidi R.F., Rezaei A. Liver Resection Promotes (Regulates) Proinflammatory Cytokines in Patients with Hepatocellular Carcinoma. Can. J. Gastroenterol. Hepatol. 2021;2021 doi: 10.1155/2021/5593655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo W.C., He X.F., Li Y.H., Li Z.H., Mei Q.L., Chen Y. The effect of sequential transcatheter arterial chemoembolization (TACE) and portal venous embolizations (PVE) vs. TACE or PVE alone on rabbit VX2 liver carcinoma and on liver regeneration. Eur. Rev. Med. Pharmacol. Sci. 2016;20:3186–3193. [PubMed] [Google Scholar]
- 74.Chen X., Xu C. High-throughput analysis of tumor necrosis factor signaling pathways in eight cell types during rat hepatic regeneration. Inflammation. 2012;35:1538–1548. doi: 10.1007/s10753-012-9469-y. [DOI] [PubMed] [Google Scholar]
- 75.Geisler F., Strazzabosco M. Emerging roles of Notch signaling in liver disease. Hepatology. 2015;61:382–392. doi: 10.1002/hep.27268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kessler M., Hoffmann K., Brinkmann V., Thieck O., Jackisch S., Toelle B., Berger H., Mollenkopf H.J., Mangler M., Sehouli J., et al. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat. Commun. 2015;6:8989. doi: 10.1038/ncomms9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kozlovskaja-Gumbrienė A., Yi R., Alexander R., Aman A., Jiskra R., Nagelberg D., Knaut H., McClain M., Piotrowski T. Proliferation-independent regulation of organ size by Fgf/Notch signaling. Elife. 2017;6 doi: 10.7554/eLife.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou W., He Q., Zhang C., He X., Cui Z., Liu F., Li W. BLOS2 Negatively Regulates Notch Signaling during Neural and Hematopoietic Stem and Progenitor Cell Development. Elife. 2016;5:e18108. doi: 10.7554/eLife.18108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Morell C.M., Strazzabosco M. Notch signaling and new therapeutic options in liver disease. J. Hepatol. 2014;60:885–890. doi: 10.1016/j.jhep.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 80.Duan J.L., Zhou Z.Y., Ruan B., Fang Z.Q., Ding J., Liu J.J., Song P., Xu H., Xu C., Yue Z.S., et al. Notch-Regulated c-Kit-Positive Liver Sinusoidal Endothelial Cells Contribute to Liver Zonation and Regeneration. Cell. Mol. Gastroenterol. Hepatol. 2022;13:1741–1756. doi: 10.1016/j.jcmgh.2022.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hasan S.S., Fischer A. Notch Signaling in the Vasculature: Angiogenesis and Angiocrine Functions. Cold Spring Harb. Perspect. Med. 2023;13 doi: 10.1101/cshperspect.a041166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jia W., Zhang Y., Wang X., Luo L., Sun H., Jiang Y., Wang J., Mao Q., Guo Y., Kong L., et al. KRT5 mutation regulate melanin metabolism through notch signalling pathway between keratinocytes and melanocytes. Exp. Dermatol. 2023;32:752–765. doi: 10.1111/exd.14761. [DOI] [PubMed] [Google Scholar]
- 83.Kim H.J., Kim G., Chi K.Y., Kim H., Jang Y.J., Jo S., Lee J., Lee Y., Woo D.H., Han C., et al. Generation of multilineage liver organoids with luminal vasculature and bile ducts from human pluripotent stem cells via modulation of Notch signaling. Stem Cell Res. Ther. 2023;14:19. doi: 10.1186/s13287-023-03235-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pu W., Zhu H., Zhang M., Pikiolek M., Ercan C., Li J., Huang X., Han X., Zhang Z., Lv Z., et al. Bipotent transitional liver progenitor cells contribute to liver regeneration. Nat. Genet. 2023;55:651–664. doi: 10.1038/s41588-023-01335-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao T., Li X., Qian H., Miao X., Zhu Y., Wang J., Hui J., Zhou L., Ye L. PM(2.5) induces the abnormal lipid metabolism and leads to atherosclerosis via Notch signaling pathway in rats. Toxicology. 2023;485 doi: 10.1016/j.tox.2022.153415. [DOI] [PubMed] [Google Scholar]
- 86.Morell C.M., Fiorotto R., Fabris L., Strazzabosco M. Notch signalling beyond liver development: emerging concepts in liver repair and oncogenesis. Clin. Res. Hepatol. Gastroenterol. 2013;37:447–454. doi: 10.1016/j.clinre.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 87.Stanger B.Z., Greenbaum L. The role of paracrine signals during liver regeneration. Hepatology. 2012;56:1577–1579. doi: 10.1002/hep.25911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Köhler C., Bell A.W., Bowen W.C., Monga S.P., Fleig W., Michalopoulos G.K. Expression of Notch-1 and its ligand Jagged-1 in rat liver during liver regeneration. Hepatology. 2004;39:1056–1065. doi: 10.1002/hep.20156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang F., Zhang J., Li X., Li B., Tao K., Yue S. Notch signaling pathway regulates cell cycle in proliferating hepatocytes involved in liver regeneration. J. Gastroenterol. Hepatol. 2018;33:1538–1547. doi: 10.1111/jgh.14110. [DOI] [PubMed] [Google Scholar]
- 90.Ahluwalia A., Tarnawski A.S. Critical role of hypoxia sensor--HIF-1α in VEGF gene activation. Implications for angiogenesis and tissue injury healing. Curr. Med. Chem. 2012;19:90–97. doi: 10.2174/092986712803413944. [DOI] [PubMed] [Google Scholar]
- 91.Li Y., Xu Y., Wang R., Li W., He W., Luo X., Ye Y. Expression of Notch-Hif-1α signaling pathway in liver regeneration of rats. J. Int. Med. Res. 2020;48 doi: 10.1177/0300060520943790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Michalopoulos G.K. The Regenerative Altruism of Hepatocytes and Cholangiocytes. Cell Stem Cell. 2018;23:11–12. doi: 10.1016/j.stem.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 93.Yanger K., Zong Y., Maggs L.R., Shapira S.N., Maddipati R., Aiello N.M., Thung S.N., Wells R.G., Greenbaum L.E., Stanger B.Z. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yimlamai D., Christodoulou C., Galli G.G., Yanger K., Pepe-Mooney B., Gurung B., Shrestha K., Cahan P., Stanger B.Z., Camargo F.D. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aloia L., McKie M.A., Vernaz G., Cordero-Espinoza L., Aleksieva N., van den Ameele J., Antonica F., Font-Cunill B., Raven A., Aiese Cigliano R., et al. Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration. Nat. Cell Biol. 2019;21:1321–1333. doi: 10.1038/s41556-019-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fausto N., Campbell J.S. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mech. Dev. 2003;120:117–130. doi: 10.1016/s0925-4773(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 97.Michalopoulos G.K., Khan Z. Liver Stem Cells: Experimental Findings and Implications for Human Liver Disease. Gastroenterology. 2015;149:876–882. doi: 10.1053/j.gastro.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boulter L., Govaere O., Bird T.G., Radulescu S., Ramachandran P., Pellicoro A., Ridgway R.A., Seo S.S., Spee B., Van Rooijen N., et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat. Med. 2012;18:572–579. doi: 10.1038/nm.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zou G., Park J.I. Wnt signaling in liver regeneration, disease, and cancer. Clin. Mol. Hepatol. 2023;29:33–50. doi: 10.3350/cmh.2022.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ding B.S., Nolan D.J., Butler J.M., James D., Babazadeh A.O., Rosenwaks Z., Mittal V., Kobayashi H., Shido K., Lyden D., et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Monga S.P.S. Role of Wnt/β-catenin signaling in liver metabolism and cancer. Int. J. Biochem. Cell Biol. 2011;43:1021–1029. doi: 10.1016/j.biocel.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cadigan K.M., Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 103.Young C.S., Kitamura M., Hardy S., Kitajewski J. Wnt-1 induces growth, cytosolic beta-catenin, and Tcf/Lef transcriptional activation in Rat-1 fibroblasts. Mol. Cell Biol. 1998;18:2474–2485. doi: 10.1128/mcb.18.5.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.So J., Martin B.L., Kimelman D., Shin D. Wnt/β-catenin signaling cell-autonomously converts non-hepatic endodermal cells to a liver fate. Biol. Open. 2013;2:30–36. doi: 10.1242/bio.20122857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Monga S.P., Pediaditakis P., Mule K., Stolz D.B., Michalopoulos G.K. Changes in WNT/beta-catenin pathway during regulated growth in rat liver regeneration. Hepatology. 2001;33:1098–1109. doi: 10.1053/jhep.2001.23786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Okabe H., Yang J., Sylakowski K., Yovchev M., Miyagawa Y., Nagarajan S., Chikina M., Thompson M., Oertel M., Baba H., et al. Wnt signaling regulates hepatobiliary repair following cholestatic liver injury in mice. Hepatology. 2016;64:1652–1666. doi: 10.1002/hep.28774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li Q., Gong Y., Wang Y., Liu B., Chu Y., Gui S., Zheng Y., Chen X. Sirt1 Promotes the Restoration of Hepatic Progenitor Cell (HPC)-Mediated Liver Fatty Injury in NAFLD Through Activating the Wnt/β-Catenin Signal Pathway. Front. Nutr. 2021;8 doi: 10.3389/fnut.2021.791861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jung Y.S., Stratton S.A., Lee S.H., Kim M.J., Jun S., Zhang J., Zheng B., Cervantes C.L., Cha J.H., Barton M.C., Park J.I. TMEM9-v-ATPase Activates Wnt/β-Catenin Signaling Via APC Lysosomal Degradation for Liver Regeneration and Tumorigenesis. Hepatology. 2021;73:776–794. doi: 10.1002/hep.31305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li N., Kong M., Zeng S., Hao C., Li M., Li L., Xu Z., Zhu M., Xu Y. Brahma related gene 1 (Brg1) contributes to liver regeneration by epigenetically activating the Wnt/β-catenin pathway in mice. FASEB J. 2019;33:327–338. doi: 10.1096/fj.201800197R. [DOI] [PubMed] [Google Scholar]
- 110.Seo S.H., Kim E., Yoon M., Lee S.H., Park B.H., Choi K.Y. Metabolic improvement and liver regeneration by inhibiting CXXC5 function for non-alcoholic steatohepatitis treatment. Exp. Mol. Med. 2022;54:1511–1523. doi: 10.1038/s12276-022-00851-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hönes G.S., Kerp H., Hoppe C., Kowalczyk M., Zwanziger D., Baba H.A., Führer D., Moeller L.C. Canonical Thyroid Hormone Receptor β Action Stimulates Hepatocyte Proliferation in Male Mice. Endocrinology. 2022;163 doi: 10.1210/endocr/bqac003. [DOI] [PubMed] [Google Scholar]
- 112.Zhu Y., Qiu Z., Zhang Y., Li B., Jiang X. Partial hepatectomy-induced upregulation of SNHG12 promotes hepatocyte proliferation and liver regeneration. Mol. Med. Rep. 2020;21:1089–1096. doi: 10.3892/mmr.2019.10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yin Y., Kong D., He K., Xia Q. Aurora kinase A regulates liver regeneration through macrophages polarization and Wnt/β-catenin signalling. Liver Int. 2022;42:468–478. doi: 10.1111/liv.15094. [DOI] [PubMed] [Google Scholar]
- 114.Clemens M.M., Kennon-McGill S., Apte U., James L.P., Finck B.N., McGill M.R. The inhibitor of glycerol 3-phosphate acyltransferase FSG67 blunts liver regeneration after acetaminophen overdose by altering GSK3β and Wnt/β-catenin signaling. Food Chem. Toxicol. 2019;125:279–288. doi: 10.1016/j.fct.2019.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ma Y., Lv X., He J., Liu T., Wen S., Wang L. Wnt agonist stimulates liver regeneration after small-for-size liver transplantation in rats. Hepatol. Res. 2016;46:E154–E164. doi: 10.1111/hepr.12553. [DOI] [PubMed] [Google Scholar]
- 116.Oliva-Vilarnau N., Vorrink S.U., Ingelman-Sundberg M., Lauschke V.M. A 3D Cell Culture Model Identifies Wnt/β-Catenin Mediated Inhibition of p53 as a Critical Step during Human Hepatocyte Regeneration. Adv. Sci. 2020;7 doi: 10.1002/advs.202000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Trusolino L., Bertotti A., Comoglio P.M. MET signalling: principles and functions in development, organ regeneration and cancer. Nat. Rev. Mol. Cell Biol. 2010;11:834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 118.Uehara Y., Minowa O., Mori C., Shiota K., Kuno J., Noda T., Kitamura N. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature. 1995;373:702–705. doi: 10.1038/373702a0. [DOI] [PubMed] [Google Scholar]
- 119.Motoi S., Toyoda H., Obara T., Ohta E., Arita Y., Negishi K., Moriya K., Kuboi Y., Soejima M., Imai T., et al. Anti-Apoptotic Effects of Recombinant Human Hepatocyte Growth Factor on Hepatocytes Were Associated with Intrahepatic Hemorrhage Suppression Indicated by the Preservation of Prothrombin Time. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20081821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang P., Cui Y., Wang J., Liu D., Tian Y., Liu K., Wang X., Liu L., He Y., Pei Y., et al. Mesenchymal stem cells protect against acetaminophen hepatotoxicity by secreting regenerative cytokine hepatocyte growth factor. Stem Cell Res. Ther. 2022;13:94. doi: 10.1186/s13287-022-02754-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lindroos P.M., Zarnegar R., Michalopoulos G.K. Hepatocyte growth factor (hepatopoietin A) rapidly increases in plasma before DNA synthesis and liver regeneration stimulated by partial hepatectomy and carbon tetrachloride administration. Hepatology. 1991;13:743–750. [PubMed] [Google Scholar]
- 122.Maher J.J. Cell-specific expression of hepatocyte growth factor in liver. Upregulation in sinusoidal endothelial cells after carbon tetrachloride. J. Clin. Invest. 1993;91:2244–2252. doi: 10.1172/JCI116451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pediaditakis P., Lopez-Talavera J.C., Petersen B., Monga S.P., Michalopoulos G.K. The processing and utilization of hepatocyte growth factor/scatter factor following partial hepatectomy in the rat. Hepatology. 2001;34:688–693. doi: 10.1053/jhep.2001.27811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Stolz D.B., Mars W.M., Petersen B.E., Kim T.H., Michalopoulos G.K. Growth factor signal transduction immediately after two-thirds partial hepatectomy in the rat. Cancer Res. 1999;59:3954–3960. [PubMed] [Google Scholar]
- 125.Selden A.C., Hodgson H.J. Growth factors and the liver. Gut. 1991;32:601–603. doi: 10.1136/gut.32.6.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liu J., Pan G., Liang T., Huang P. HGF/c-Met signaling mediated mesenchymal stem cell-induced liver recovery in intestinal ischemia reperfusion model. Int. J. Med. Sci. 2014;11:626–633. doi: 10.7150/ijms.8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huh C.G., Factor V.M., Sánchez A., Uchida K., Conner E.A., Thorgeirsson S.S. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc. Natl. Acad. Sci. USA. 2004;101:4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jangphattananont N., Sato H., Imamura R., Sakai K., Terakado Y., Murakami K., Barker N., Oshima H., Oshima M., Takagi J., et al. Distinct Localization of Mature HGF from its Precursor Form in Developing and Repairing the Stomach. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20122955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li N., Dou Z., Liu J., Chai B., Li Y., An X., Chu P., Zhang X. Therapeutic Effect of HGF on NASH Mice Through HGF/c-Met and JAK2-STAT3 Signalling Pathway. Ann. Hepatol. 2018;17:501–510. doi: 10.5604/01.3001.0011.7395. [DOI] [PubMed] [Google Scholar]
- 130.Okano J.i., Shiota G., Matsumoto K., Yasui S., Kurimasa A., Hisatome I., Steinberg P., Murawaki Y. Hepatocyte growth factor exerts a proliferative effect on oval cells through the PI3K/AKT signaling pathway. Biochem. Biophys. Res. Commun. 2003;309:298–304. doi: 10.1016/j.bbrc.2003.04.002. [DOI] [PubMed] [Google Scholar]
- 131.Scheving L.A., Stevenson M.C., Taylormoore J.M., Traxler P., Russell W.E. Integral role of the EGF receptor in HGF-mediated hepatocyte proliferation. Biochem. Biophys. Res. Commun. 2002;290:197–203. doi: 10.1006/bbrc.2001.6157. [DOI] [PubMed] [Google Scholar]
- 132.Addante A., Roncero C., Lazcanoiturburu N., Méndez R., Almalé L., García-Álvaro M., Ten Dijke P., Fabregat I., Herrera B., Sánchez A. A Signaling Crosstalk between BMP9 and HGF/c-Met Regulates Mouse Adult Liver Progenitor Cell Survival. Cells. 2020;9:752. doi: 10.3390/cells9030752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dong X., Luo Y., Lu S., Ma H., Zhang W., Zhu Y., Sun G., Sun X. Ursodesoxycholic acid alleviates liver fibrosis via proregeneration by activation of the ID1-WNT2/HGF signaling pathway. Clin. Transl. Med. 2021;11:e296. doi: 10.1002/ctm2.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Qian J., Liu J., Hong L., Lu H., Guo D., Liu Z., Zhou L., Yin S., Zheng S. Upregulation of PDGF Mediates Robust Liver Regeneration after Nanosecond Pulsed Electric Field Ablation by Promoting the HGF/c-Met Pathway. BioMed Res. Int. 2020;2020 doi: 10.1155/2020/3635787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhao L., Dai C., Gong Q. Changes of Endocan and its effect on hepatic stem cells during the rapid proliferation process of residual liver after ALPPS procedure. Cell Biochem. Funct. 2020;38:817–825. doi: 10.1002/cbf.3553. [DOI] [PubMed] [Google Scholar]
- 136.Huang M., Jiao J., Cai H., Zhang Y., Xia Y., Lin J., Shang Z., Qian Y., Wang F., Wu H., et al. C-C motif chemokine ligand 5 confines liver regeneration by down-regulating reparative macrophage-derived hepatocyte growth factor in a forkhead box O 3a-dependent manner. Hepatology. 2022;76:1706–1722. doi: 10.1002/hep.32458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hoffmann K., Nagel A.J., Tanabe K., Fuchs J., Dehlke K., Ghamarnejad O., Lemekhova A., Mehrabi A. Markers of liver regeneration-the role of growth factors and cytokines: a systematic review. BMC Surg. 2020;20:31. doi: 10.1186/s12893-019-0664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lehwald N., Duhme C., Wildner M., Kuhn S., Fürst G., Forbes S.J., Jonas S., Robson S.C., Knoefel W.T., Schmelzle M., Schulte Am Esch J. HGF and SDF-1-mediated mobilization of CD133+ BMSC for hepatic regeneration following extensive liver resection. Liver Int. 2014;34:89–101. doi: 10.1111/liv.12195. [DOI] [PubMed] [Google Scholar]
- 139.Takeuchi E., Nimura Y., Nagino M., Kurumiya Y., Maeda A., Kamiya J., Kondo S., Kanai M., Miyachi M., Uesaka K., Yoshida S. Human hepatocyte growth factor in bile: an indicator of posthepatectomy liver function in patients with biliary tract carcinoma. Hepatology. 1997;26:1092–1099. doi: 10.1053/jhep.1997.v26.pm0009362347. [DOI] [PubMed] [Google Scholar]
- 140.Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S.A., Gayyed M.F., Anders R.A., Maitra A., Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pan D. The hippo signaling pathway in development and cancer. Dev. Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Driskill J.H., Pan D. The Hippo Pathway in Liver Homeostasis and Pathophysiology. Annu. Rev. Pathol. 2021;16:299–322. doi: 10.1146/annurev-pathol-030420-105050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Camargo F.D., Gokhale S., Johnnidis J.B., Fu D., Bell G.W., Jaenisch R., Brummelkamp T.R. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 144.Dieterle A.M., Böhler P., Keppeler H., Alers S., Berleth N., Drießen S., Hieke N., Pietkiewicz S., Löffler A.S., Peter C., et al. PDK1 controls upstream PI3K expression and PIP3 generation. Oncogene. 2014;33:3043–3053. doi: 10.1038/onc.2013.266. [DOI] [PubMed] [Google Scholar]
- 145.Manning B.D., Toker A. AKT/PKB Signaling: Navigating the Network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ha J., Lee S., Park J., Seo J., Kang E., Yoon H., Kim B.R., Lee H.K., Ryu S.E., Cho S. Identification of a novel inhibitor of liver cancer cell invasion and proliferation through regulation of Akt and Twist1. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-95933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sun N., Wang J., Dou X., Wang Y., Yang Y., Xiao D., Zhao P., Li J., Wang S., Gu P., Ji J. A chiral microenvironment promotes retinal progenitor cell proliferation by activating the Akt and ERK pathways. Biomater. Sci. 2022;10:5938–5946. doi: 10.1039/d2bm00886f. [DOI] [PubMed] [Google Scholar]
- 148.DeBerardinis R.J., Lum J.J., Hatzivassiliou G., Thompson C.B. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 149.Miricescu D., Totan A., Stanescu-Spinu I.I., Badoiu S.C., Stefani C., Greabu M. PI3K/AKT/mTOR Signaling Pathway in Breast Cancer: From Molecular Landscape to Clinical Aspects. Int. J. Mol. Sci. 2020;22 doi: 10.3390/ijms22010173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wei D., Rui B., Qingquan F., Chen C., Ping H.Y., Xiaoling S., Hao W., Jun G. KIF11 promotes cell proliferation via ERBB2/PI3K/AKT signaling pathway in gallbladder cancer. Int. J. Biol. Sci. 2021;17:514–526. doi: 10.7150/ijbs.54074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Valizadeh A., Majidinia M., Samadi-Kafil H., Yousefi M., Yousefi B. The roles of signaling pathways in liver repair and regeneration. J. Cell. Physiol. 2019;234:14966–14974. doi: 10.1002/jcp.28336. [DOI] [PubMed] [Google Scholar]
- 152.Cui Z., Jin N., Amevor F.K., Shu G., Du X., Kang X., Ning Z., Deng X., Tian Y., Zhu Q., et al. Dietary supplementation of salidroside alleviates liver lipid metabolism disorder and inflammatory response to promote hepatocyte regeneration via PI3K/AKT/Gsk3-β pathway. Poult. Sci. 2022;101 doi: 10.1016/j.psj.2022.102034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jung K., Kim M., So J., Lee S.H., Ko S., Shin D. Farnesoid X Receptor Activation Impairs Liver Progenitor Cell-Mediated Liver Regeneration via the PTEN-PI3K-AKT-mTOR Axis in Zebrafish. Hepatology. 2021;74:397–410. doi: 10.1002/hep.31679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang B., Lv D., Chen Y., Nie W., Jiao Y., Zhang J., Zhou X., Wu X., Chen S., Ma T. Aquaporin-9 facilitates liver regeneration following hepatectomy. Redox Biol. 2022;50 doi: 10.1016/j.redox.2022.102246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lin Z., Zhang X., Wang J., Liu W., Liu Q., Ye Y., Dai B., Guo D., Zhang P., Yang P., et al. Correction: Translationally controlled tumor protein promotes liver regeneration by activating mTORC2/AKT signaling. Cell Death Dis. 2020;11:114. doi: 10.1038/s41419-020-2311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhong H., Wu H., Bai H., Wang M., Wen J., Gong J., Miao M., Yuan F. Panax notoginseng saponins promote liver regeneration through activation of the PI3K/AKT/mTOR cell proliferation pathway and upregulation of the AKT/Bad cell survival pathway in mice. BMC Complement. Altern. Med. 2019;19:122. doi: 10.1186/s12906-019-2536-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nakamura T., Mizuno S. The discovery of hepatocyte growth factor (HGF) and its significance for cell biology, life sciences and clinical medicine. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010;86:588–610. doi: 10.2183/pjab.86.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kinoshita T., Hirao S., Matsumoto K., Nakamura T. Possible endocrine control by hepatocyte growth factor of liver regeneration after partial hepatectomy. Biochem. Biophys. Res. Commun. 1991;177:330–335. doi: 10.1016/0006-291x(91)91987-n. [DOI] [PubMed] [Google Scholar]
- 159.Raschzok N., Werner W., Sallmon H., Billecke N., Dame C., Neuhaus P., Sauer I.M. Temporal expression profiles indicate a primary function for microRNA during the peak of DNA replication after rat partial hepatectomy. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011;300:R1363–R1372. doi: 10.1152/ajpregu.00632.2010. [DOI] [PubMed] [Google Scholar]
- 160.Yanagita K., Nagaike M., Ishibashi H., Niho Y., Matsumoto K., Nakamura T. Lung may have an endocrine function producing hepatocyte growth factor in response to injury of distal organs. Biochem. Biophys. Res. Commun. 1992;182:802–809. doi: 10.1016/0006-291x(92)91803-x. [DOI] [PubMed] [Google Scholar]
- 161.Ishiki Y., Ohnishi H., Muto Y., Matsumoto K., Nakamura T. Direct evidence that hepatocyte growth factor is a hepatotrophic factor for liver regeneration and has a potent antihepatitis effect in vivo. Hepatology. 1992;16:1227–1235. [PubMed] [Google Scholar]
- 162.Burr A.W., Toole K., Chapman C., Hines J.E., Burt A.D. Anti-hepatocyte growth factor antibody inhibits hepatocyte proliferation during liver regeneration. J. Pathol. 1998;185:298–302. doi: 10.1002/(SICI)1096-9896(199807)185:3<298::AID-PATH88>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 163.Li Z., Mizuno S., Nakamura T. Antinecrotic and antiapoptotic effects of hepatocyte growth factor on cholestatic hepatitis in a mouse model of bile-obstructive diseases. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;292:G639–G646. doi: 10.1152/ajpgi.00292.2006. [DOI] [PubMed] [Google Scholar]
- 164.Hilkens P., Fanton Y., Martens W., Gervois P., Struys T., Politis C., Lambrichts I., Bronckaers A. Pro-angiogenic impact of dental stem cells in vitro and in vivo. Stem Cell Res. 2014;12:778–790. doi: 10.1016/j.scr.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 165.Nakamura Y., Morishita R., Higaki J., Kida I., Aoki M., Moriguchi A., Yamada K., Hayashi S., Yo Y., Nakano H., et al. Hepatocyte growth factor is a novel member of the endothelium-specific growth factors: additive stimulatory effect of hepatocyte growth factor with basic fibroblast growth factor but not with vascular endothelial growth factor. J. Hypertens. 1996;14:1067–1072. doi: 10.1097/00004872-199609000-00004. [DOI] [PubMed] [Google Scholar]
- 166.Uda Y., Hirano T., Son G., Iimuro Y., Uyama N., Yamanaka J., Mori A., Arii S., Fujimoto J. Angiogenesis is crucial for liver regeneration after partial hepatectomy. Surgery. 2013;153:70–77. doi: 10.1016/j.surg.2012.06.021. [DOI] [PubMed] [Google Scholar]
- 167.Vimalraj S. A concise review of VEGF, PDGF, FGF, Notch, angiopoietin, and HGF signalling in tumor angiogenesis with a focus on alternative approaches and future directions. Int. J. Biol. Macromol. 2022;221:1428–1438. doi: 10.1016/j.ijbiomac.2022.09.129. [DOI] [PubMed] [Google Scholar]
- 168.Morishita R., Nakamura S., Hayashi S., Taniyama Y., Moriguchi A., Nagano T., Taiji M., Noguchi H., Takeshita S., Matsumoto K., et al. Therapeutic angiogenesis induced by human recombinant hepatocyte growth factor in rabbit hind limb ischemia model as cytokine supplement therapy. Hypertension. 1999;33:1379–1384. doi: 10.1161/01.hyp.33.6.1379. [DOI] [PubMed] [Google Scholar]
- 169.Kimura M., Moteki H., Ogihara M. Inhibitory effects of dexamethasone on hepatocyte growth factor-induced DNA synthesis and proliferation in primary cultures of adult rat hepatocytes. J. Pharmacol. Sci. 2011;115:390–398. doi: 10.1254/jphs.10302fp. [DOI] [PubMed] [Google Scholar]
- 170.Cheng Z., Liu L., Zhang X.J., Lu M., Wang Y., Assfalg V., Laschinger M., von Figura G., Sunami Y., Michalski C.W., et al. Peroxisome Proliferator-Activated Receptor gamma negatively regulates liver regeneration after partial hepatectomy via the HGF/c-Met/ERK1/2 pathways. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-30426-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Gui Y., Yeganeh M., Ramanathan S., Leblanc C., Pomerleau V., Ferbeyre G., Saucier C., Ilangumaran S. SOCS1 controls liver regeneration by regulating HGF signaling in hepatocytes. J. Hepatol. 2011;55:1300–1308. doi: 10.1016/j.jhep.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 172.Factor V.M., Seo D., Ishikawa T., Kaposi-Novak P., Marquardt J.U., Andersen J.B., Conner E.A., Thorgeirsson S.S. Loss of c-Met disrupts gene expression program required for G2/M progression during liver regeneration in mice. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Chiang K.Y., Li Y.W., Li Y.H., Huang S.J., Wu C.L., Gong H.Y., Wu J.L. Progranulin A Promotes Compensatory Hepatocyte Proliferation via HGF/c-Met Signaling after Partial Hepatectomy in Zebrafish. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms222011217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mohammed F.F., Pennington C.J., Kassiri Z., Rubin J.S., Soloway P.D., Ruther U., Edwards D.R., Khokha R. Metalloproteinase inhibitor TIMP-1 affects hepatocyte cell cycle via HGF activation in murine liver regeneration. Hepatology. 2005;41:857–867. doi: 10.1002/hep.20618. [DOI] [PubMed] [Google Scholar]
- 175.Li X., Sun J., Fan X., Guan L., Li D., Zhou Y., Zeng X., Chen Y., Zhang H., Xu L., et al. Schisandrol B promotes liver regeneration after partial hepatectomy in mice. Eur. J. Pharmacol. 2018;818:96–102. doi: 10.1016/j.ejphar.2017.10.044. [DOI] [PubMed] [Google Scholar]
- 176.Liu M., Chen P. Proliferation-inhibiting pathways in liver regeneration (Review) Mol. Med. Rep. 2017;16:23–35. doi: 10.3892/mmr.2017.6613. [DOI] [PubMed] [Google Scholar]
- 177.Tao Y., Wang M., Chen E., Tang H. Liver Regeneration: Analysis of the Main Relevant Signaling Molecules. Mediators Inflamm. 2017;2017 doi: 10.1155/2017/4256352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Derynck R., Budi E.H. Specificity, versatility, and control of TGF-β family signaling. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aav5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.de Larco J.E., Todaro G.J. Growth factors from murine sarcoma virus-transformed cells. Proc. Natl. Acad. Sci. USA. 1978;75:4001–4005. doi: 10.1073/pnas.75.8.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Fabregat I., Moreno-Càceres J., Sánchez A., Dooley S., Dewidar B., Giannelli G., Ten Dijke P., IT-LIVER Consortium TGF-β signalling and liver disease. FEBS J. 2016;283:2219–2232. doi: 10.1111/febs.13665. [DOI] [PubMed] [Google Scholar]
- 181.Ren L.L., Li X.J., Duan T.T., Li Z.H., Yang J.Z., Zhang Y.M., Zou L., Miao H., Zhao Y.Y. Transforming growth factor-β signaling: From tissue fibrosis to therapeutic opportunities. Chem. Biol. Interact. 2023;369 doi: 10.1016/j.cbi.2022.110289. [DOI] [PubMed] [Google Scholar]
- 182.Chang C., Wang D., Xi L., Guo X., Wang G., Yu G. The orphan GPR50 receptor interacting with TβRI induces G1/S-phase cell cycle arrest via Smad3-p27/p21 in BRL-3A cells. Biochem. Pharmacol. 2022;202 doi: 10.1016/j.bcp.2022.115117. [DOI] [PubMed] [Google Scholar]
- 183.Wang W., Yang X., Yang J., Liu S., Lv Y., Zhang C., Dong W., Liu A. GDF11 impairs liver regeneration in mice after partial hepatectomy. Clin. Sci. 2019;133:2069–2084. doi: 10.1042/CS20190441. [DOI] [PubMed] [Google Scholar]
- 184.Yang I., Son Y., Shin J.H., Kim I.Y., Seong J.K. Ahnak depletion accelerates liver regeneration by modulating the TGF-β/Smad signaling pathway. BMB Rep. 2022;55:401–406. doi: 10.5483/BMBRep.2022.55.8.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fang D., He Y., Luan Z. Simvastatin augments activation of liver regeneration through attenuating transforming growth factor-β1 induced-apoptosis in obstructive jaundice rats. Exp. Ther. Med. 2017;14:4839–4845. doi: 10.3892/etm.2017.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Greenhalgh S.N., Matchett K.P., Taylor R.S., Huang K., Li J.T., Saeteurn K., Donnelly M.C., Simpson E.E.M., Pollack J.L., Atakilit A., et al. Loss of Integrin αvβ8 in Murine Hepatocytes Accelerates Liver Regeneration. Am. J. Pathol. 2019;189:258–271. doi: 10.1016/j.ajpath.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kuroki H., Hayashi H., Nakagawa S., Sakamoto K., Higashi T., Nitta H., Hashimoto D., Chikamoto A., Beppu T., Baba H. Effect of LSKL peptide on thrombospondin 1-mediated transforming growth factor β signal activation and liver regeneration after hepatectomy in an experimental model. Br. J. Surg. 2015;102:813–825. doi: 10.1002/bjs.9765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Karkampouna S., Goumans M.J., Ten Dijke P., Dooley S., Kruithof-de Julio M. Inhibition of TGFβ type I receptor activity facilitates liver regeneration upon acute CCl4 intoxication in mice. Arch. Toxicol. 2016;90:347–357. doi: 10.1007/s00204-014-1436-y. [DOI] [PubMed] [Google Scholar]
- 189.Masuda A., Nakamura T., Abe M., Iwamoto H., Sakaue T., Tanaka T., Suzuki H., Koga H., Torimura T. Promotion of liver regeneration and anti-fibrotic effects of the TGF-β receptor kinase inhibitor galunisertib in CCl4-treated mice. Int. J. Mol. Med. 2020;46:427–438. doi: 10.3892/ijmm.2020.4594. [DOI] [PubMed] [Google Scholar]
- 190.Chen L., Tao F., Zhang Y., Shu C., Xiang W., Yang L., Chen X., Hong Y., Chen B., Li K., et al. Islet-cell autoantigen 69 accelerates liver regeneration by downregulating Tgfbr1 and attenuating Tgfβ signaling in mice. FEBS Lett. 2020;594:2881–2893. doi: 10.1002/1873-3468.13859. [DOI] [PubMed] [Google Scholar]
- 191.Bird T.G., Müller M., Boulter L., Vincent D.F., Ridgway R.A., Lopez-Guadamillas E., Lu W.Y., Jamieson T., Govaere O., Campbell A.D., et al. TGFβ inhibition restores a regenerative response in acute liver injury by suppressing paracrine senescence. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aan1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Heo S.H., Jeong E.S., Lee K.S., Seo J.H., Lee W.K., Choi Y.K. Knockout of krüppel-like factor 10 suppresses hepatic cell proliferation in a partially hepatectomized mouse model. Oncol. Lett. 2017;13:4843–4848. doi: 10.3892/ol.2017.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ling N., Ying S.Y., Ueno N., Shimasaki S., Esch F., Hotta M., Guillemin R. Pituitary FSH is released by a heterodimer of the beta-subunits from the two forms of inhibin. Nature. 1986;321:779–782. doi: 10.1038/321779a0. [DOI] [PubMed] [Google Scholar]
- 194.Bloise E., Ciarmela P., Dela Cruz C., Luisi S., Petraglia F., Reis F.M. Activin A in Mammalian Physiology. Physiol. Rev. 2019;99:739–780. doi: 10.1152/physrev.00002.2018. [DOI] [PubMed] [Google Scholar]
- 195.Li F., Long Y., Yu X., Tong Y., Gong L. Different Immunoregulation Roles of Activin A Compared With TGF-β. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.921366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Date M., Matsuzaki K., Matsushita M., Tahashi Y., Sakitani K., Inoue K. Differential regulation of activin A for hepatocyte growth and fibronectin synthesis in rat liver injury. J. Hepatol. 2000;32:251–260. doi: 10.1016/s0168-8278(00)80070-7. [DOI] [PubMed] [Google Scholar]
- 197.Manavski Y., Abel T., Hu J., Kleinlützum D., Buchholz C.J., Belz C., Augustin H.G., Boon R.A., Dimmeler S. Endothelial transcription factor KLF2 negatively regulates liver regeneration via induction of activin A. Proc. Natl. Acad. Sci. USA. 2017;114:3993–3998. doi: 10.1073/pnas.1613392114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chen L., Zhang W., Liang H.F., Zhou Q.F., Ding Z.Y., Yang H.Q., Liu W.B., Wu Y.H., Man Q., Zhang B.X., Chen X.P. Activin A induces growth arrest through a SMAD- dependent pathway in hepatic progenitor cells. Cell Commun. Signal. 2014;12:18. doi: 10.1186/1478-811X-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lv X., Chen J., He J., Hou L., Ren Y., Shen X., Wang Y., Ji T., Cai X. Gasdermin D-mediated pyroptosis suppresses liver regeneration after 70% partial hepatectomy. Hepatol. Commun. 2022;6:2340–2353. doi: 10.1002/hep4.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gao X., Zhao P., Hu J., Zhu H., Zhang J., Zhou Z., Zhao J., Tang F. MicroRNA-194 protects against chronic hepatitis B-related liver damage by promoting hepatocyte growth via ACVR2B. J. Cell Mol. Med. 2018;22:4534–4544. doi: 10.1111/jcmm.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Herrera B., Dooley S., Breitkopf-Heinlein K. Potential roles of bone morphogenetic protein (BMP)-9 in human liver diseases. Int. J. Mol. Sci. 2014;15:5199–5220. doi: 10.3390/ijms15045199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Urist M.R. Bone: formation by autoinduction. Science. 1965;150:893–899. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- 203.García de Vinuesa A., Abdelilah-Seyfried S., Knaus P., Zwijsen A., Bailly S. BMP signaling in vascular biology and dysfunction. Cytokine Growth Factor Rev. 2016;27:65–79. doi: 10.1016/j.cytogfr.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 204.Sánchez-Duffhues G., Hiepen C., Knaus P., Ten Dijke P. Bone morphogenetic protein signaling in bone homeostasis. Bone. 2015;80:43–59. doi: 10.1016/j.bone.2015.05.025. [DOI] [PubMed] [Google Scholar]
- 205.Choi T.Y., Khaliq M., Tsurusaki S., Ninov N., Stainier D.Y.R., Tanaka M., Shin D. Bone morphogenetic protein signaling governs biliary-driven liver regeneration in zebrafish through tbx2b and id2a. Hepatology. 2017;66:1616–1630. doi: 10.1002/hep.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Katagiri T., Watabe T. Bone Morphogenetic Proteins. Cold Spring Harb. Perspect. Biol. 2016;8 doi: 10.1101/cshperspect.a021899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tsugawa D., Oya Y., Masuzaki R., Ray K., Engers D.W., Dib M., Do N., Kuramitsu K., Ho K., Frist A., et al. Specific activin receptor-like kinase 3 inhibitors enhance liver regeneration. J. Pharmacol. Exp. Ther. 2014;351:549–558. doi: 10.1124/jpet.114.216903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Cui B., Yang L., Zhao Y., Lu X., Song M., Liu C., Yang C. HOXA13 promotes liver regeneration through regulation of BMP-7. Biochem. Biophys. Res. Commun. 2022;623:23–31. doi: 10.1016/j.bbrc.2022.07.018. [DOI] [PubMed] [Google Scholar]
- 209.Breitkopf-Heinlein K., Meyer C., König C., Gaitantzi H., Addante A., Thomas M., Wiercinska E., Cai C., Li Q., Wan F., et al. BMP-9 interferes with liver regeneration and promotes liver fibrosis. Gut. 2017;66:939–954. doi: 10.1136/gutjnl-2016-313314. [DOI] [PubMed] [Google Scholar]
- 210.Do N., Zhao R., Ray K., Ho K., Dib M., Ren X., Kuzontkoski P., Terwilliger E., Karp S.J. BMP4 is a novel paracrine inhibitor of liver regeneration. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;303:G1220–G1227. doi: 10.1152/ajpgi.00105.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Xu C.P., Ji W.M., van den Brink G.R., Peppelenbosch M.P. Bone morphogenetic protein-2 is a negative regulator of hepatocyte proliferation downregulated in the regenerating liver. World J. Gastroenterol. 2006;12:7621–7625. doi: 10.3748/wjg.v12.i47.7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Liu Z., Kuna V.K., Xu B., Sumitran-Holgersson S. Wnt ligands 3a and 5a regulate proliferation and migration in human fetal liver progenitor cells. Transl. Gastroenterol. Hepatol. 2021;6:56. doi: 10.21037/tgh.2020.01.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jeng L.B., Lee S.G., Soin A.S., Lee W.C., Suh K.S., Joo D.J., Uemoto S., Joh J., Yoshizumi T., Yang H.R., et al. Efficacy and safety of everolimus with reduced tacrolimus in living-donor liver transplant recipients: 12-month results of a randomized multicenter study. Am. J. Transplant. 2018;18:1435–1446. doi: 10.1111/ajt.14623. [DOI] [PubMed] [Google Scholar]
- 214.Saliba F., Duvoux C., Dharancy S., Dumortier J., Calmus Y., Gugenheim J., Kamar N., Salamé E., Neau-Cransac M., Vanlemmens C., et al. Early Switch From Tacrolimus to Everolimus After Liver Transplantation: Outcomes at 2 Years. Liver Transpl. 2019;25:1822–1832. doi: 10.1002/lt.25664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Karin M., Clevers H. Reparative inflammation takes charge of tissue regeneration. Nature. 2016;529:307–315. doi: 10.1038/nature17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Solhi R., Lotfinia M., Gramignoli R., Najimi M., Vosough M. Metabolic hallmarks of liver regeneration. Trends Endocrinol. Metab. 2021;32:731–745. doi: 10.1016/j.tem.2021.06.002. [DOI] [PubMed] [Google Scholar]
- 217.Zhang L., Li Y., Wang Y., Qiu Y., Mou H., Deng Y., Yao J., Xia Z., Zhang W., Zhu D., et al. mTORC2 Facilitates Liver Regeneration Through Sphingolipid-Induced PPAR-α-Fatty Acid Oxidation. Cell. Mol. Gastroenterol. Hepatol. 2022;14:1311–1331. doi: 10.1016/j.jcmgh.2022.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Huang W., Ma K., Zhang J., Qatanani M., Cuvillier J., Liu J., Dong B., Huang X., Moore D.D. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312:233–236. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 219.Belenguer G., Mastrogiovanni G., Pacini C., Hall Z., Dowbaj A.M., Arnes-Benito R., Sljukic A., Prior N., Kakava S., Bradshaw C.R., et al. RNF43/ZNRF3 loss predisposes to hepatocellular-carcinoma by impairing liver regeneration and altering the liver lipid metabolic ground-state. Nat. Commun. 2022;13:334. doi: 10.1038/s41467-021-27923-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Li X.L., Man K., Ng K.T., Lee T.K., Lo C.M., Fan S.T. Insulin in UW solution exacerbates hepatic ischemia/reperfusion injury by energy depletion through the IRS-2/SREBP-1c pathway. Liver Transpl. 2004;10:1173–1182. doi: 10.1002/lt.20240. [DOI] [PubMed] [Google Scholar]
- 221.Ding H.R., Wang J.L., Tang Z.T., Wang Y., Zhou G., Liu Y., Ren H.Z., Shi X.L. Mesenchymal Stem Cells Improve Glycometabolism and Liver Regeneration in the Treatment of Post-hepatectomy Liver Failure. Front. Physiol. 2019;10:412. doi: 10.3389/fphys.2019.00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Jung Y.S. Metabolism of Sulfur-Containing Amino Acids in the Liver: A Link between Hepatic Injury and Recovery. Biol. Pharm. Bull. 2015;38:971–974. doi: 10.1248/bpb.b15-00244. [DOI] [PubMed] [Google Scholar]
- 223.Nelsen C.J., Rickheim D.G., Timchenko N.A., Stanley M.W., Albrecht J.H. Transient expression of cyclin D1 is sufficient to promote hepatocyte replication and liver growth in vivo. Cancer Res. 2001;61:8564–8568. [PubMed] [Google Scholar]
- 224.Mead J.E., Braun L., Martin D.A., Fausto N. Induction of replicative competence ("priming") in normal liver. Cancer Res. 1990;50:7023–7030. [PubMed] [Google Scholar]
- 225.He Z.Y., Lou K.H., Zhao J.H., Zhang M., Zhang L.C., Li J., Yu H.F., Zhang R.P., Wei-Yan H. Resina Draconis Reduces Acute Liver Injury and Promotes Liver Regeneration after 2/3 Partial Hepatectomy in Mice. Evid. Based. Complement. Alternat. Med. 2020;2020 doi: 10.1155/2020/2305784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tiberio G.A.M., Tiberio L., Benetti A., Cervi E., Montani N., Dreano M., Garotta G., Cerea K., Steimberg N., Pandolfo G., et al. IL-6 Promotes compensatory liver regeneration in cirrhotic rat after partial hepatectomy. Cytokine. 2008;42:372–378. doi: 10.1016/j.cyto.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 227.Bou Saleh M., Louvet A., Ntandja-Wandji L.C., Boleslawski E., Gnemmi V., Lassailly G., Truant S., Maggiotto F., Ningarhari M., Artru F., et al. Loss of hepatocyte identity following aberrant YAP activation: A key mechanism in alcoholic hepatitis. J. Hepatol. 2021;75:912–923. doi: 10.1016/j.jhep.2021.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Konishi T., Schuster R.M., Lentsch A.B. Proliferation of hepatic stellate cells, mediated by YAP and TAZ, contributes to liver repair and regeneration after liver ischemia-reperfusion injury. Am. J. Physiol. Gastrointest. Liver Physiol. 2018;314:G471–G482. doi: 10.1152/ajpgi.00153.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Verboven E., Moya I.M., Sansores-Garcia L., Xie J., Hillen H., Kowalczyk W., Vella G., Verhulst S., Castaldo S.A., Algueró-Nadal A., et al. Regeneration Defects in Yap and Taz Mutant Mouse Livers Are Caused by Bile Duct Disruption and Cholestasis. Gastroenterology. 2021;160:847–862. doi: 10.1053/j.gastro.2020.10.035. [DOI] [PubMed] [Google Scholar]