Abstract
New work reveals differences in oogenic gene expression between parthenogenetic and sexually reproducing Drosophila mercatorum strains. Recapitulating those changes in D. melanogaster oocytes induced parthenogenesis in this normally sexually reproducing species, providing molecular insight into how these reproductive modes arise.
Most animals derive from the union of an egg and a sperm. Combining genetic material from two parents into a single offspring contributes to genetic diversity and allows for generation of potentially advantageous allelic combinations and removal of disadvantageous alleles/combinations by recombination. Given the advantages of biparental reproduction, it is not surprising to find cellular mechanisms that render it obligatory. Such mechanisms include genomic imprinting in mammals1 and the need for sperm to provide important molecules that ‘activate’ eggs to start embryogenesis. For example, sperm-derived phospholipase C raises calcium levels in mammalian eggs, thereby activating them2, and sperm organelles such as centrioles are needed for embryonic divisions in many taxa (e.g.3).
However, as with many biological phenomena, there is immense variation in reproductive strategies. In bees and their relatives, females develop from the standard egg-meets-sperm situation, but males develop from unfertilized eggs that initiate development and make their own centrioles de novo4. In ‘gynogenetic’ species, such as the crucian carp5, female progeny develop from eggs fertilized by sperm from another species. The eggs are activated, but the heterospecific sperm’s genome is not incorporated into that of the offspring. Conversely, progeny in ‘androgenetic’ species have paternally derived genomes, again by exploiting gametes from the opposite sex6.
A particularly intriguing method of reproduction is parthenogenesis. Here, progeny develop from a female’s oocytes with no involvement of a male. Parthenogenesis occurs in numerous insect species (e.g.7,8) as well as in reptiles, birds, and fishes (e.g.9,10). Although parthenogenesis lacks some of the genome-diversifying advantages of biparental inheritance, it has its own advantages: it allows progeny production without finding and mating with a male — an expedient strategy if animals are very dispersed. Some species are obligatorily parthenogenetic, others obligatorily sexually reproducing. So-called ‘facultatively parthenogenetic’ species avail themselves of the benefits of each reproductive mode, generating biparental offspring when males are available and conditions warrant, and undertaking parthenogenesis when they are not.
Despite its existence in many taxa, mechanisms of parthenogenesis and its regulation have remained mysterious. We know that parthenogenesis often produces diploid progeny from diploid mothers by mechanisms ranging from suppression of meiosis to fusion of meiotic products; the latter can also give rise to higher-ploidy offspring. But, while previous studies have associated candidate loci or chromosomal regions with parthenogenesis, the molecular mechanisms that control the switch between the production of biparental vs. parthenogenetic progeny remain unknown. A new paper by Sperling et al.11 in this issue of Current Biology takes the first molecular steps towards solving this mystery, by taking advantage of the existence of parthenogenetic, sexually reproducing, and facultatively parthenogenetic strains within Drosophila mercatorum8.
To search for a parthenogenesis-promoting gene(s) or genomic region(s), Sperling et al. sequenced and compared genomes of the different D. mercatorum strains. The genomes’ contents and karyotypes were remarkably similar across strains, apart from some inversions in one chromosome arm; no obvious ‘parthenogenesis gene(s)’ jumped out. The authors then wondered whether a parthenogenesis-regulating gene might differ in expression in the germline of parthenogenetic vs. sexually reproducing strains. Accordingly, they determined and compared the transcriptomes of mature oocytes between strains. They observed expression differences, often in conserved genes with known functions. Though many of those genes were not obviously connected to parthenogenesis, some — namely cell-cycle regulators and centriole and spindle factors — hinted at a role in early development.
The authors then took a brave leap. They hypothesized that the differential expression of these genes might underlie parthenogenetic ability and decided to test this with Drosophlia melanogaster, a species that is simple to genetically manipulate in the lab. D. melanogaster is normally considered obligately sexually reproducing, although an early report documented that some wild-caught strains show a low level of parthenogenesis12. Sperling et al. tested whether altering the expression of genes discovered as differentially expressed in D. mercatorum oocytes could induce parthenogenesis in D. melanogaster. They used publicly available as well as ‘homemade’ strains to increase or decrease expression of the candidate genes in the D. melanogaster germline, looking for cases where unmated females laid unfertilized eggs that developed to adulthood.
This investigation would have been impossible in any other insect, but even in D. melanogaster it was not easy. Expecting induced parthenogenesis to be rare, the authors screened tens of thousands of female flies to find conditions that switched on parthenogenesis. Excitingly, 16 genes, when manipulated to echo the expression in parthenogenetic D. mercatorum, converted D. melanogaster eggs to parthenotes at low rates: these included cell-cycle or centriole regulatory genes. As a proxy for manipulating many of the latter, the authors focused on Polo kinase, a known centriole regulator13. Increasing polo expression, and thus modulation of its targets, in D. melanogaster eggs resulted in parthenogenesis, but at a low (0.1%) rate. The authors then tested whether manipulating other candidates in combination with increased Polo could increase the rate of D. melanogaster parthenogenesis.
Combining increased polo dosage with knockouts of fatty acid desaturases (desat1/2) increased parthenogenesis; 0.6% of unfertilized eggs developed to adulthood. Parthenogenesis increased further (to 1.4% surviving adults) upon the addition of an extra copy of Myc, a transcriptional regulator of cell cycle/proliferative genes14, and a homolog of one of the original Yamanaka pluripotency factors15. This is, to our knowledge, the first identification of genetic changes that can trigger parthenogenesis and modulate its rate.
How could these genes trigger parthenogenetic development? The transition from differentiated oocyte to early embryo normally involves at least two distinct processes. First, eggs must be activated by a calcium-triggered event that relieves meiotic arrest and alters the egg’s transcriptome, proteome, and membranes/envelopes to support development (e.g.16). Second, the activated eggs must begin embryogenesis by generating a zygotic nucleus from egg- and sperm-derived pronuclei, making centrioles if necessary, and undertaking mitotic divisions. While egg activation and the initiation of embryo development are often tightly coupled, they are separate in some species. In particular, egg activation in numerous insects including D. melanogaster is independent of fertilization; it is induced instead by physical forces that eggs experience as they move through the reproductive tract17,18. Thus, to trigger parthenogenetic development, a D. melanogaster egg only needs what is necessary to start embryo development.
It is not simple to come up with a model for how Polo, Myc, and Desat2 levels could promote parthenogenesis. Sperling et al. propose that each of these genes contributes in a different way (Figure 1). Higher Myc may ‘prime’ the parthenogenetic egg for mitotic divisions by ensuring abundant cell-cycle gene products in the egg, thereby supporting later proliferation. Increased amounts of Polo kinase, which has roles in mitotic entry and whose activity may be regulated during egg activation19, are suggested to drive the centriole biogenesis needed for parthenogenetic embryos to undertake mitosis. The authors’ images of Polo puncta at sites of centriole formation support this model. How decreasing Desat1/2 levels promotes parthenogenesis seems more mysterious. The authors suggest it might alter membrane fluidity, allowing polar body nuclei to fuse or engage in mitosis.
Figure 1. Inducing parthenogenesis in D. melanogaster.
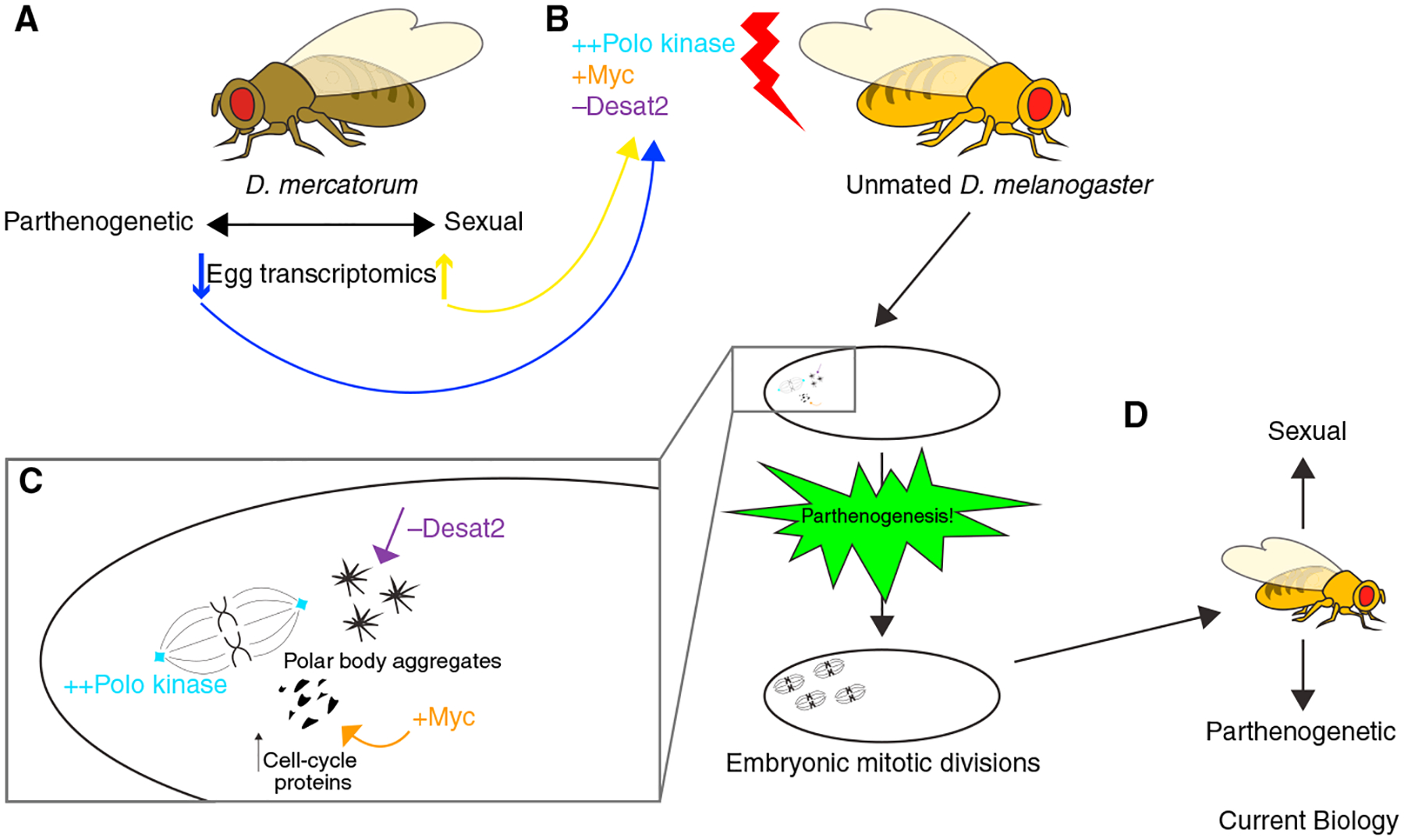
(A) Sperling et al.11 performed RNA sequencing analysis to compare transcriptomes of sexually reproducing and parthenogenetic (obligatory and facultative) D. mercatorum eggs. (B) They mimicked the differential expression of some of these genes in the germline of female D. melanogaster, a normally sexually reproducing species. (C) Unfertilized eggs from D. melanogaster females overexpressing Polo kinase and Myc, and deficient for desaturase 2 (Desat2), became parthenogenetic. The authors suggest that these genetic manipulations promote centriole biogenesis (Polo), ‘prime’ the embryo with abundant cell-cycle proteins to increase proliferative capacity (Myc), and allow polar bodies to participate in mitosis by altering membrane fluidity (Desat2). (D) Adult flies that develop from these parthenogenetic embryos can reproduce both sexually and by parthenogenesis.
Sperling et al.’s exciting results will motivate many fascinating future studies of cell, developmental, reproductive, and evolutionary questions. For example, although a few parthenogenetic D. melanogaster embryos can develop to adulthood (and are fertile, either by sexual or parthenogenetic reproduction), many arrest development very early, around the time of the initiation of mitotic divisions. Among embryos that proceed beyond that point, the authors saw intriguing deviations from stereotypical spindle distribution and organization. D. melanogaster sperm normally provide centrioles for the embryos3. Do the spindle abnormalities in the later-arresting parthenogenetic embryos reflect inefficiencies in de novo centriole generation or function?
It will also be interesting to know whether additional ‘ingredients’ improve the ‘recipe’ for parthenogenesis (a pinch of extra Polo and Myc, a little less Desat2). Does parthenogenesis efficiency in D. melanogaster increase if the expression of additional candidates is manipulated along with these three genes? The full molecular instructions will be important to know for their significance to fundamental and applied reproductive/developmental biology and to compare to gene expression changes in other situations in which non-dividing cells are induced to proliferate, such as cancer initiation or tissue regeneration.
It is also tempting to wonder whether manipulating analogous genes in eggs of other organisms could induce parthenogenesis, although success is uncertain given potential technical barriers and biological differences (e.g. imprinting in mammals, but see20). The results of such studies would interest not only developmental, cell, and evolutionary biologists but also people looking to reproduce without the need for sperm-meets-egg: same-sex couples who desire biologically related offspring, or people who can produce eggs and would like a biological child without a partner or with a partner who is infertile due to sperm defects.
Sperling et al.’s important findings bring a new understanding of how parthenogenesis can be induced. Now that we know some of the genes involved, it will be intriguing to determine what causes their germline gene expression differences between sexually reproducing and parthenogenetic species or strains. Did the changes arise individually and accumulate with selection? Or is there an upstream regulator — intrinsic or extrinsic — that controls them all? The answers will illuminate the genesis of this aspect of the amazing diversity in reproductive strategies across the animal kingdom.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Surani MA, Barton SC, and Norris ML (1984). Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308, 548–550. [DOI] [PubMed] [Google Scholar]
- 2.Swann K, and Lai FA (2016). Egg activation at fertilization by a soluble sperm protein. Physiol. Rev 96, 127–149. [DOI] [PubMed] [Google Scholar]
- 3.Blachon S, Khire A, and Avidor-Reiss T (2014). The origin of the second centriole in the zygote of Drosophila melanogaster. Genetics 197, 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tram U, and Sullivan W (2000). Reciprocal inheritance of centrosomes in the parthenogenetic hymenopteran Nasonia vitripennis. Curr. Biol 10, 1413–1419. [DOI] [PubMed] [Google Scholar]
- 5.Yamashita M, Onozato H, Nakanishi T, and Nagahama Y (1990). Breakdown of the sperm nuclear envelope is a prerequisite for male pronucleus formation: direct evidence from the gynogenetic crucian carp Carassius auratus langsdorfii. Dev. Biol 137, 155–160. [DOI] [PubMed] [Google Scholar]
- 6.Schwander T, and Oldroyd BP (2016). Androgenesis: where males hijack eggs to clone themselves. Philos. Trans. R. Soc. Lond. B Biol. Sci 371, 20150534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sperling AL, and Glover DM (2023). Parthenogenesis in dipterans: a genetic perspective. Proc. Biol. Sci 290, 20230261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Markow TA (2013). Parents without partners: Drosophila as a model for understanding the mechanisms and evolution of parthenogenesis. G3 3, 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryder OA, Thomas S, Judson JM, Romanov MN, Dandekar S, Papp JC, Sidak-Loftis LC, Walker K, Stalis IH, Mace M, et al. (2021). Facultative parthenogenesis in California condors. J. Hered 112, 569–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Booth W, Levine BA, Corush JB, Davis MA, Dwyer Q, De Plecker R, and Schuett GW (2023). Discovery of facultative parthenogenesis in a new world crocodile. Biol. Lett 19, 20230129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sperling AL, Fabian DK, Garrison E, and Glover DM (2023). A genetic basis for facultative parthenogenesis in Drosophila. Curr. Biol 33, 3545–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stalker HD (1954). Parthenogenesis in Drosophila. Genetics 39, 4–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, and Jallepalli PV (2009). Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell 17, 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Gutierrez L, Delgado MD, and Leon J (2019). MYC oncogene contributions to release of cell cycle brakes. Genes 10, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi K, and Yamanaka S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- 16.Aviles-Pagan EE, and Orr-Weaver TL (2018). Activating embryonic development in Drosophila. Semin. Cell Dev. Biol 84, 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heifetz Y, Yu J, and Wolfner MF (2001). Ovulation triggers activation of Drosophila oocytes. Dev. Biol 234, 416–424. [DOI] [PubMed] [Google Scholar]
- 18.Went DF, and Krause G (1974). Egg activation in Pimpla turionellae (Hym.). Naturwissenschaften 61, 407–408. [DOI] [PubMed] [Google Scholar]
- 19.Whitfield ZJ, Chisholm J, Hawley RS, and Orr-Weaver TL (2013). A meiosis-specific form of the APC/C promotes the oocyte-to-embryo transition by decreasing levels of the Polo kinase inhibitor matrimony. PLoS Biol. 11, e1001648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei Y, Yang CR, and Zhao ZA (2022). Viable offspring derived from single unfertilized mammalian oocytes. Proc. Natl. Acad. Sci. USA 119, e2115248119. [DOI] [PMC free article] [PubMed] [Google Scholar]


