Abstract
Molecular phylogenetic surveys have recently revealed an ecologically widespread crenarchaeal group that inhabits cold and temperate terrestrial and marine environments. To date these organisms have resisted isolation in pure culture, and so their phenotypic and genotypic characteristics remain largely unknown. To characterize these archaea, and to extend methodological approaches for characterizing uncultivated microorganisms, we initiated genomic analyses of the nonthermophilic crenarchaeote Cenarchaeum symbiosum found living in association with a marine sponge, Axinella mexicana. Complex DNA libraries derived from the host-symbiont population yielded several large clones containing the ribosomal operon from C. symbiosum. Unexpectedly, cloning and sequence analysis revealed the presence of two closely related variants that were consistently found in the majority of host individuals analyzed. Homologous regions from the two variants were sequenced and compared in detail. The variants exhibit >99.2% sequence identity in both small- and large-subunit rRNA genes and they contain homologous protein-encoding genes in identical order and orientation over a 28-kbp overlapping region. Our study not only indicates the potential for characterizing uncultivated prokaryotes by genome sequencing but also identifies the primary complication inherent in the approach: the widespread genomic microheterogeneity in naturally occurring prokaryotic populations.
Molecular phylogenetic surveys of mixed microbial populations have revealed the existence of many new lineages undetected by classical microbiological approaches (7, 25). Furthermore, quantitative rRNA hybridization experiments demonstrate that some of these novel prokaryotic groups represent major components of natural microbial communities. These molecular phylogenetic approaches have altered current views of microbial diversity and ecology and have demonstrated that traditional cultivation techniques may recover only a small, skewed fraction of naturally occurring microbes. However, phylogenetic identification using single gene sequences provides a limited perspective on other biological properties, particularly for novel lineages only distantly related to cultivated and characterized organisms. Consequently, additional approaches are necessary to better characterize ecologically abundant and potentially biotechnologically useful microorganisms, many of which resist cultivation attempts.
Nonthermophilic members of the kingdom Crenarchaeota are one of the more abundant, widespread, and frequently recovered prokaryotic groups revealed by molecular phylogenetic approaches. These microorganisms were originally detected in high abundance in temperate ocean waters and polar seas (6, 8, 10, 22, 23, 28). Representatives have now been reported to exist in terrestrial environments (2, 15, 19, 37) and freshwater lake sediments (14, 20, 31), indicating a widespread distribution. The ecological distribution of these organisms was initially surprising, since their closest cultivated relatives are all thermophilic or hyperthermophilic. No representative of this new archaeal group has yet been obtained in pure culture, and so the phenotypic and metabolic properties of these organisms as well as their impact on the environment and global nutrient cycling remain unknown. Since growth temperature and habitat characteristics vary so widely between nonthermophilic and the hyperthermophilic Crenarchaeota, these groups are likely to differ greatly with respect to specific physiology and metabolism.
To gain a better perspective on the genetic and physiological characteristics of nonthermophilic crenarchaeotes, we began a genomic study of Cenarchaeum symbiosum. This archaeon lives in specific association with the marine sponge Axinella mexicana off the coast of California (28), allowing access to relatively large amounts of biomass from this species. Our approach differs in several respects from now standard genomic characterization of cultivated organisms, and also from comparable studies of uncultivated obligate parasites or symbionts. C. symbiosum has not been completely physically separated from the tissues of its metazoan host. Therefore, its genetic material needs to be identified within the context of complex genomic libraries that contain significant amounts of eucaryotic DNA, as well as DNA derived from members of the domain Bacteria.
In the course of our study, we identified the presence of at least two major variants or strains of C. symbiosum that coexist inside the sponge tissues. This complexity of the C. symbiosum population was not detected in initial studies based solely on direct sequencing of PCR-amplified small-subunit (SSU) rRNA genes (28). This natural variation would also have been lost upon isolation of a pure culture. One component of our genomic analysis involved the sequencing and comparison of large, overlapping chromosomal regions from the two dominant naturally co-occurring C. symbiosum variants. The variability and distribution of the variants within different sponge individuals were also investigated.
MATERIALS AND METHODS
Enrichment for C. symbiosum cells from sponge tissue, DNA extraction, and preparation of fosmid libraries.
Preparation of archaeal cells for the first fosmid (16) library has been previously described (28). A small individual of A. mexicana was incubated in calcium- and magnesium-free artificial seawater (ASW) containing pronase (0.25 mg/ml); the tissue was then homogenized and enriched for archaeal cells by differential centrifugation. For the second library, prepared from a different sponge individual, this cell fraction was further incubated for 1 h at 4°C in 10 mM Tris-HCl (pH 8)–200 mM EDTA. This additional incubation step was found to increase the lysis of sponge cells, which resulted in an enhanced separation of archaeal and eucaryotic cells in the Percoll gradient. The cells were then pelleted and subsequently purified on a 15% Percoll (Sigma) cushion in ASW. Archaeal cells banded in the light, upper fraction after centrifugation at 2,500 rpm in a Beckman SS34 rotor. This cell fraction was washed in ASW and resuspended in TE buffer (10 mM Tris-HCl [pH 8], 0.1 mM EDTA). Quantitative hybridization experiments using a domain-specific oligonucleotide (6) indicated that 25 to 30% of the total rRNA from this fraction was derived from archaea. DNA extraction, preparation of the fosmid libraries, and PCR-based screening were performed as previously described (28, 36). The first fosmid library yielded 7 unique C. symbiosum rRNA operon-containing clones out of a total 10,236 recombinant fosmids (0.07%). The second fosmid library yielded eight unique C. symbiosum rRNA operon-containing clones out of 2,100 recombinants (0.38%).
Fosmid sequencing.
Small (1- to 2-kbp)-insert plasmid libraries were prepared by cloning partial restriction enzyme digests of purified fosmids. Plasmids were sequenced by using Applied Biosystems Inc. (ABI; Foster City, Calif.) Prism dye terminator FS reaction mix. Direct sequencing from fosmids was used for gap filling and resequencing to ensure accuracy. Fosmid sequencing was performed by using DNA from a single 3-ml overnight culture purified on an Autogen 740 automated plasmid isolation system. Each reaction consisted of one preparation of DNA directly resuspended by the addition of 16 μl of H2O, 8 μl of oligonucleotide primer (1.4 pmol/μl), and 16 μl of ABI Prism dye terminator FS reaction mix. Cycle sequencing was performed with a 3-min preincubation at 96°C followed by 25 cycles of the sequence 96°C for 20 s-50°C for 20 s-60°C for 4 min and a 5-min postcycling incubation at 60°C. Sequencing reaction products were analyzed on ABI 377 sequencers.
Direct sequencing of PCR fragments.
PCRs with two archaeon-specific 16S rDNA primers (21F and 958R [6], one biotinylated) were used to amplify a 950-bp fragment from total nucleic acids of 16 different sponge individuals. Primers 21F and 459R-LSU (CTTTCCCTCACGGTA) were used to amplify the 16S-23S spacer region from fosmids. The PCR products were purified and sequenced as described previously (28), with primer 519R for 16S rDNA and primer SP23rev (CTA TTG CCG TCT TTA CACC) for the spacer region.
rRNA hybridization.
Two oligonucleotides specific for each variant type were designed from the 23S rDNA gene sequences (positions 283 to 303, E. coli numbering) of fosmids 101G10 and 60A5. The probes differ by three point mutations: L-St-C.symA-283-a-A-19 (variant A), ACACTTCAACTATTTCCTG; and L-St-C.symB-283-a-A-19 (variant B), ACACTTTGACTATTTCGTG. Nucleic acids from sponges (300 ng) and controls (fosmids 101G10 and 60A5, 50 ng of each) were denatured, bound to nylon membranes (Hybond-N; Amersham), hybridized with the labeled probes (22), and washed at 41.5°C. Hybridization was analyzed by autoradiography.
RFLP analysis of PCR fragments.
Primers 21F (6) and 459R-LSU for amplification of 2.2 kbp of the ribosomal operon, primers GSAT810F (GAATCCGCCCCCGACTATCTT) and 16S37REV (CATGGCTTAGTATCAATC) for amplification of the 16S RNA-glutamate semialdehyde aminotransferase (GSAT) region (2.2 kbp), and primers Cenpol357F (ACITACAACGGIGACGAYTTTGA) and Cenpol735R (CACCCCGAARTAGTTYTTYTT) for an internal DNA polymerase fragment (of 1,134 bp) were used in PCRs with 5 ng of purified fosmids. The PCR products were cut with TaqI and HpaII (16S-23S RNA), HaeIII and RsaI (GSAT-16S RNA), or HaeIII and AvaII (polymerase) and analyzed on 2% agarose gels. If the pattern did not exactly match but closely resembled the restriction fragment length polymorphism (RFLP) of either type A or B, it was denoted by a lowercase letter (a or b [Table 1]), meaning that at least three of four or three of five bands created by restriction digest appear identical in size to the ones from either type A or B.
TABLE 1.
Analysis of polymorphism at four distinct loci in different fosmids
| Fosmida | Patternb
|
|||||
|---|---|---|---|---|---|---|
| 16S RNAc | 16S-23S spacerd | 16S-GSATe
|
DNA Pole
|
|||
| HaeIII | RsaI | HaeIII | AvaII | |||
| 101G10 | A | A | A | A | A | A |
| 60A5 | B | B | B | B | B | B |
| 15A5 | B | B | — | — | b | b |
| 43H4 | A | — | — | — | A | A |
| 60H6 | A | A | — | — | a/b | B |
| 69H2 | A | — | — | — | A | A |
| 87F4 | B | — | — | — | b | a/b |
| C1H5 | A | A | A | A | ||
| C4H1 | A | A | A | A | ||
| C4H9 | A | A | A | A | A | B |
| C7D4 | A | A | A | A | A | A |
| C8B8 | B | B | B | B | B | b |
| C15A3 | A | A | A | A | ||
| C17D2 | B | — | b | B | B | b |
| C20B5 | A | A | a | a/b | ||
The first seven fosmids were isolated from a first library; the last eight fosmids (prefix C) are from a second library.
A or B, pattern identical to that of either 101G10 (=A) or 60A5 (=B); a or b, pattern similar to that of either A or B (see Materials and Methods); —, not determined. Fosmids C1H5, C4H1, C15A3, and C20B5 did not yield PCR products with polymerase (Pol)-specific primers.
Partial sequence (101G10 through 87F4) or RFLP analysis (C1H5 through C20B5).
Partial sequence.
RFLP analysis of PCR products.
Nucleotide sequence accession numbers.
The sequences described in this report have been deposited in GenBank under accession no. AF083072 (fosmid 101G10) and AF083071 (fosmid 60A5).
RESULTS
Isolation and comparison of fosmid clones from two environmental libraries.
We constructed two environmental fosmid libraries from tissue preparations of the A. mexicana-C. symbiosum association that were enriched for archaeal cells. These libraries yielded 15 unique C. symbiosum rRNA operon-containing fosmids. Partial sequence and RFLP analysis of the SSU RNA genes of all 15 fosmids revealed the presence of two variants, termed A and B, that differed by only two point mutations over a 590-bp region. Southern blotting of restriction digests of entire fosmids also confirmed the presence of two classes of rRNA operon-containing clones (data not shown). A total of 10 clones were identified as variant A, and 5 clones represented variant B (Table 1). The A/B variant recovery ratios were 4:3 in the first library and 6:2 in the second library.
We determined the complete sequences of two fosmids, both containing an rRNA operon, which corresponded to the two variant types. The insert of fosmid 101G10 (designated variant A) was 32,998 bp and is syntenic over ca. 28 kbp with the 42,432-bp insert of fosmid 60A5 (variant B). Analysis of the common 28-kbp region is shown in Fig. 1. The large-subunit (LSU) and SSU rRNA genes of the two variants were 99.2 and 99.3% identical, respectively. Protein coding regions were highly similar in both nucleic acid and deduced amino acid sequences (Fig. 1; Table 2). The data provide strong evidence that these genomic clones are derived from two very closely related but distinct strains, as opposed to representing two rRNA operon regions originating from the same organism. This conclusion is consistent with the observation that all crenarchaeotes characterized to date contain only one rRNA operon (11).
FIG. 1.
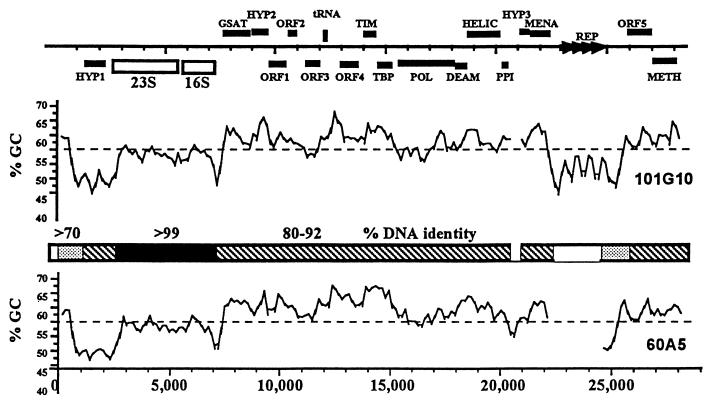
Gene map of related 28-kbp regions from two fosmids of C. symbiosum. Sequences were aligned over their entire length. The same genes and gene order were found on both fosmid 101G10 and fosmid 60A5 (for abbreviations of depicted ORFs, see the footnote to Table 2). G+C plots for both fosmids are shown underneath (calculated within a window size of 400 bp and an increment of 150 bp). A bar depicts different levels of DNA identity between the two aligned sequences (see text for more details). Insertions/deletions were found predominantly in intergenic regions. The two largest insertions, one found between PPI and HYP3 (hypothetical 03) in fosmid 60A5 and the REP (repeat) element between MenA and ORF 5 in fosmid 101G10, are shown as gaps in the G+C plots. POL, DNA polymerase; DEAM, deaminase; METH, methylase.
TABLE 2.
Comparison of overlapping coding sequences from fosmid 101G10 and fosmid 60A5
| Gene namea | Functional category | % Identity
|
|
|---|---|---|---|
| Nucleotide | Amino acid | ||
| Hypothetical 01 | Unknown | 81.4 | 76.6 |
| 23S | Translation | 99.16 | |
| 16S | Translation | 99.3 | |
| GSAT | Heme biosynthesis | 83.2 | 83.8 |
| Hypothetical 02 | Unknown | 83.4 | 81.4 |
| ORF 01 | Unknown | 83.3 | 85.7 |
| ORF 02 | Unknown | 89.9 | 95.2 |
| ORF 03 | Unknown | 87.9 | 86.7 |
| tRNATyr | Translation | 99.2 | |
| ORF 04 | Unknown | 87.8 | 88.1 |
| TIM | Glycolysis | 80.9 | 83.3 |
| TBP | Transcription | 83.4 | 86.3 |
| DNA polymerase | Replication/repair | 89.0 | 93.9 |
| dCMP deaminase | Pyrimidine synthesis | 85.7 | 89.8 |
| RNA helicase (ATP dependent) | Translation | 86.1 | 92.2 |
| PPI | Chaperone | 88.4 | 92.5 |
| Hypothetical 03 | Unknown | 91.5 | 92.4 |
| MenA | Menaquinone biosynthesis | 86 | 89.4 |
| ORF 05 | Unknown | 87.5 | 90.6 |
| Methylase | Restriction/modification | 86.4 | 87.5 |
Hypothetical, ORF with similarity to proteins of unknown function from the databases; ORF, open reading frame identified by similarity between both fosmids, including upstream promoter sequence; TIM, triosephosphate isomerase; PPI, peptidylprolyl cis,trans-isomerase.
In protein coding regions, the DNA identity of the two contigs ranged from 80.9% (triosephosphate isomerase) to 91.5% (hypothetical 03) (Table 2). Within intergenic regions, the identity dropped to 70 to 86%, and small insertions or deletions were found frequently. The high similarity in coding regions and upstream sequences aided in the identification of genes, start codons, and putative transcriptional promoter motifs (see below).
Although both sequences could be aligned unambiguously over most of the overlapping region, four large insertions/deletions ranging in size from 142 to 1,994 bp were identified between positions 20500 and 25800. The longest insertion contained a repetitive element of 1,784 bp, which was found in variant A between menA and open read frame (ORF) 05. It was composed of a threefold direct repeat of 575 bp (REP in Fig. 1), with repeats exhibiting only minor sequence variation (95.8 to 98.7% identity). A segment of 56 bp at the start of this repeat is also found adjacent to the 3′ terminus of the third direct repeat. No obvious structural or sequence similarities to known repeats or mobile genetic elements from other organisms were identified within the repeat sequence. Its occurrence in only one variant and its relatively low G+C content relative to the rest of the fragment suggest that it may have been acquired by horizontal transfer from a different genetic context.
Analysis of the 28-kbp genomic fragment from C. symbiosum.
The average G+C contents in the fosmid inserts were 55.6% for variant A and 57.1% for variant B in protein coding regions but were significantly lower in the gene for the hypothetical protein 01 and in the repetitive element of variant A (Fig. 1). Genes appear as densely packed in C. symbiosum as they are in other sequenced archaeal genomes (4, 17, 34). The rRNA operon is composed of the genes for 16S and 23S RNAs separated by spacer of 131 bp. This organization is typical of crenarchaeotes and differs from that of rRNA operons of euryarchaeotes, which usually contain 5S RNA and tRNA genes (11). Another stable RNA gene, coding for tRNATyr, is found separate from the rRNA operon. This tRNA contains a 45 bp intron in the vicinity of the anticodon loop. Of the 17 predicted protein-encoding genes, 9 show significant matches to genes of assigned function from the public databases. Three are homologous to hypothetical proteins from other organisms (Table 1). In a number of cases, the highest similarity of derived amino acid sequences is with known archaeal proteins. In particular, DNA polymerase, TATA box-binding protein (TBP), and triosephosphate isomerase gene sequences could be analyzed in greater detail because several archaeal homologs are known. The DNA polymerase shares highest overall similarity with the crenarchaeal homologs from the extreme thermophiles Sulfolobus acidocaldarius and Pyrodictium occultum (54 and 53%, respectively) and exhibits all conserved motifs of B-(α-)type DNA polymerases and 3′-5′-exonuclease motifs, both indicative of archaeal polymerases. A more detailed phylogenetic analysis and biochemical characterization of the C. symbiosum polymerase has been published elsewhere (32). The TBP is similar to other known archaeal TBPs and is N-terminally truncated with respect to the eucaryal homologs. It shows 49% amino acid similarity with TBP from Pyrococcus woesii. The triosephosphate isomerase represents the first such protein sequence reported for a crenarchaeote and has known archaeal signature sequences and deletions which distinguish archaeal triosephosphate isomerase genes from their eucaryal and eubacterial homologues (data not shown). We identified an ATP-dependent RNA helicase that is highly similar in sequence to homologues found in the complete genome sequences of three euryarchaeotes (4, 17, 34). GSAT, also detected in an rRNA operon containing genomic fragment of a planktonic marine crenarchaeote (36), is also present.
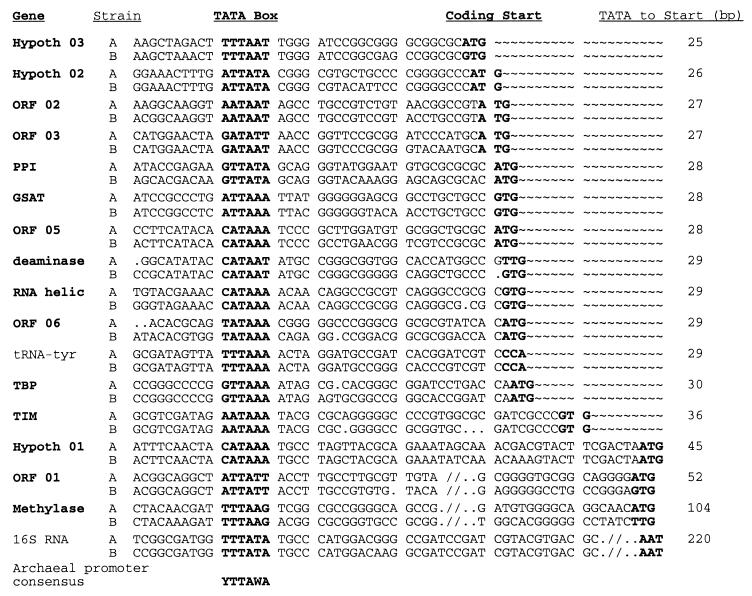
The high conservation between the two chromosomal segments is not entirely confined to coding regions but also extends into adjacent upstream sequences. Due to this upstream similarity, and also because the average G+C content of the sequences is relatively high, it was possible to readily identify putative transcriptional (A+T-rich) promoter elements. A signature corresponding to the consensus of the archaeal TATA box-like element ([C/T]TTA[T/A]A) (13) was identified upstream of nearly all genes (Fig. 2). The exceptions were the genes encoding MenA and DNA polymerase, which are located immediately downstream of other ORFs and may therefore be transcribed as polycistronic mRNAs. In vivo and in vitro studies of other archaea have shown that initiation of transcription occurs consistently 24 to 28 bp downstream from the central T of this motif (13, 26). For 12 of the protein-encoding genes, the promoter element was found 25 to 30 bp upstream of the ORF (Fig. 2), suggesting that transcriptional initiation occurs near or at the translational start codon.
FIG. 2.
Alignment of the promoter regions of 17 genes identified in both the A and B variants of C. symbiosum. The distance from the TATA box to the start codon is indicated at the right. The TATA box consensus sequence is shown below the alignment.
Distribution of C. symbiosum variants in host-associated natural populations.
The unexpected finding of two distinct but highly related genomic variants of C. symbiosum in libraries derived from two different sponge isolates led us to investigate whether this variation occurred consistently in other samples. Sequence analysis of 590 bp of the 16S rRNA gene revealed two variant positions (175 and 183.7, E. coli numbering [Table 3]). These signature nucleotides were used to determine the presence of the variants in natural populations of A. mexicana by direct sequencing of 16S rDNA PCR products from 16 different sponge individuals collected from different locations and at different times. In 15 cases, U/C ambiguities were found at the signature positions, indicating the presence of both variants (Table 3). Only one sponge (s4) yielded an unambiguous sequence identical to that of variant A, but variant B was detected in this individual by another criterion. The second approach detected variation in LSU rRNA, using oligonucleotides uniquely specific for each variant type. In the majority of host individuals examined, the presence of both LSU rRNA variants was observed (Table 3), again suggesting that the specific association of C. symbiosum with its host typically involves the presence of both variants.
TABLE 3.
Detection of C. symbiosum variants in natural populations of A. mexicana
| A. mexicana individual or isolated DNA sourcea | Variation in 16S rDNA positionsb
|
Variation in 23S rRNA hybridizationc
|
||
|---|---|---|---|---|
| 175 | 183.7 | Variant type A | Variant type B | |
| Fosmid 101G10 from s12 | U | U | + | − |
| Fosmid 60A5 from s12 | C | C | − | + |
| s12 | Y | Y | + | + |
| s1 | —d | — | + | + |
| s2 | — | — | + | + |
| s3 | Y | Y | + | + |
| s4 | U | U | + | w |
| s5 | Y | Y | — | — |
| s6 | Y | Y | + | + |
| s7 | — | — | + | w |
| s8 | Y | Y | + | + |
| s9 | Y | Y | + | w |
| s10 | — | — | + | + |
| s11 | Y | Y | + | + |
| s13 | — | — | + | + |
| s14 | — | — | − | w |
| s16 | — | — | + | + |
| s17 | — | — | − | w |
| s18 | Y | Y | − | w |
| s19 | — | — | + | + |
| s20 | — | — | + | + |
| s21 | — | — | + | + |
| s22 | — | — | + | + |
| s23 | — | — | + | + |
| s24 | — | — | + | + |
| s25 | — | — | + | + |
| s26 | — | — | + | + |
| s27 | — | — | + | + |
| s28 | — | — | + | + |
| s29 | — | — | + | − |
| s30 | — | — | + | + |
| hs1 | — | — | + | + |
| hs2 | — | — | + | + |
| hs3 | Y | Y | + | w |
| hs4 | Y | Y | + | w |
| hs5 | Y | Y | + | + |
| hh1 | — | — | w | w |
| hh2 | Y | Y | + | + |
| hh3 | Y | Y | + | + |
| Aq1 | Y | Y | — | — |
| Aq2 | Y | Y | — | — |
| Aq3 | — | — | + | + |
Prefixes: s, Naples Reef; hs, Haskle Reef; hh, Hermit Hole; Aq, captive sponge.
Y, direct sequence of PCR product yields C and U at the same position.
+, positive; −, negative; w, weakly positive.
—, not determined.
We also examined the possibility of an even greater diversity of variants, as opposed to a symbiont population composed strictly of two variant types. Since the rRNA spacer region displays greater heterogeneity between the two variant types than the SSU rRNA sequence, we PCR amplified and sequenced the variable spacer region (containing 10 distinguishing signature nucleotides) from 11 unique rRNA operon-containing fosmids. In all cases, we found a sequence identical to one or the other variant type (i.e., type A [101G10] or B [60A5] [Table 1]). We then amplified fragments from less highly conserved regions, i.e., an 1,150-bp fragment covering the 5′ end of the GSAT gene and 16S gene and an internal fragment of 1,134 bp from the DNA polymerase gene. RFLP patterns of these fragments revealed that all fosmids analyzed could again be assigned to either the A or B type, but slight variations were also detected (lowercase letters in Table 1), suggesting that both variants exhibit further microheterogeneity which is detectable only in protein coding and intergenic regions.
DISCUSSION
We chose C. symbiosum as a representative of the nonthermophilic crenarchaeotes to begin characterization of this newly detected, ecologically widespread but phenotypically uncharacterized lineage. The specific association of C. symbiosum with the marine sponge A. mexicana provides a tractable experimental system, allowing access to these novel uncultivated microorganisms in an enriched form.
Environmental genomic analysis reveals a heterogenous population of C. symbiosum.
Initial studies suggested that only one specific archaeal phylotype was associated with the sponge (28). The analysis presented here reveals heterogeneity at the subspecies level in C. symbiosum (see below), as a result of the higher resolution and more comprehensive nature of genome characterization compared to phylogenetic surveys based on a single genetic locus (e.g., SSU rRNA). We found two highly similar phylotypes (A and B) in two independently created libraries from the symbiotic association (Table 1) and have detected these consistently in nearly all sponge individuals analyzed (Table 3). By extending analyses outside the rRNA operon, we detected further divergence in less conserved protein coding and intergenic regions (Table 1; Fig. 1). Over the 28-kbp region analyzed, the variants showed >99.2% identity in their rRNA genes, approximately 87.8% overall DNA identity, an average of 91.6% similarity in ORF amino acid sequence, and complete colinearity of protein-encoding regions. Our data therefore suggest that the sponge-associated archaeal population consists of two major types, represented by fosmids 101G10 and 60A5, whose similarity is so great that by standard criteria (e.g., rRNA and genomic DNA similarity [38]) they could be considered different strains of a single species, C. symbiosum. Both variants also display further microheterogeneity, which is not evident on the rRNA level (Table 1).
Generally, the concept of symbiosis implies a specific association between a particular symbiont and a particular host. Very few studies, however, have analyzed the degree of diversity within single symbiotic populations (21, 29, 30, 35) in single host individuals. Our finding of two distinguishable variants in the archaeal-metazoan symbiosis indicates that both variants are concurrently stably maintained in the sponge host. Their close phylogenetic relationship and the extremely high similarity of protein-encoding regions, combined with evidence for further microheterogeneity of the two rRNA variants, strongly suggests that a population of strains or variants of C. symbiosum is coevolving in the context of the sponge-archaeon association. If this is the case, then transmission of the symbionts to the next host generation probably involves a population of archaeal cells that is large enough to maintain the diversity that we observed within each particular host individual. The mode of transmission of C. symbiosum, as well as its physical distribution within the sponge tissues, is under investigation in our laboratory. It is also possible that there is another environmental reservoir of C. symbiosum and that A. mexicana is selectively infected with closely related strains.
Contemporary surveys of natural microbial assemblages now routinely use cloning and analysis of phylogenetically informative gene sequences from mixed populations. Many novel and previously undetected microbial groups, including C. symbiosum and other previously uncultivated archaea, have been discovered by using this approach. A consistent pattern that has emerged in virtually all such studies among widely disparate taxa is the recovery of highly related but distinct rRNA gene clades or clusters (1, 6, 9, 12). Several explanations have been suggested for the high genetic diversity found in rRNA gene surveys, including PCR or sequencing artifacts, variation within rRNA operons in a single chromosome, or variation in rRNA genes among highly related, co-occurring strains (1, 9). The environmental distributions of such highly related rRNA genes (9), as well as the expression of rRNA variation within individual cells (1), indicates that much of the naturally occurring rRNA gene microheterogeneity may be due to authentic organismal genetic diversity. Our results tend to corroborate these findings, since the highly similar, syntenic chromosomal DNA fragments that we analyzed appear to be derived from naturally co-occurring but closely related strains.
How are such highly related sympatric strains, nearly indistinguishable by SSU rRNA sequence, stably maintained in their natural habitat? Previous studies of diversity at the level of protein-encoding genes indicate that ecologically distinct bacterial populations, through the action of purging selection, form distinctive clusters at all genetic loci examined (5). Furthermore, ecologically distinct species of highly related strains, indistinguishable by SSU rRNA sequences, do fall into separate sequence similarity clusters when their protein-encoding genes are compared. Palys et al., on the basis of theoretical considerations and empirical data, conclude that in cases where sympatric species form distinct similarity clusters, they must certainly show ecological differences (27). A good example of this is a recent report of naturally co-occurring closely related Proclorococcus 16S rRNA variants, each adapted for optimal growth at different light intensities (24). One explanation of our data that is consistent with this hypothesis is that the symbiotic variants occupy different compartments within their sponge host, occupying different niches that allow their stable transmission. Another distinct possibility is the evolution of a metabolic interdependence between the two strains, requiring the cotransmission of both variants to maintain the symbiosis. A third possibility is that the differences between the strains are currently neutral, and although one is expected to dominate eventually, there is a period of time when both coexist.
Genomic analysis of C. symbiosum reveals typical features of Archaea.
The sequence analysis of 28 kbp of contiguous DNA from two C. symbiosum variants reveals many features typical for Archaea, and particularly for Crenarchaeota, confirming the phylogenetic affiliation inferred from analysis of the SSU rRNA sequence (28): the rRNA gene order, spacer region, and structure are most similar to those found in the hyperthermophilic Crenarchaeota. The GSAT gene, which is located directly upstream of the ribosomal operon, was found in the same relative location on a fosmid derived from a planktonic marine crenarchaeote (36). Deduced amino acid sequences of proteins all share highest overall similarity with archaeal proteins, whenever known homologues are available.
The findings of a TBP gene and of promoter elements that follow the archaeal TATA box consensus suggest a typical archaeal transcription mechanism. Interestingly, most of the C. symbiosum promoters that we identified were located such that transcription initiation must occur close to the translational start codon, allowing no space for a ribosomal binding site in an untranslated mRNA leader. A similar observation has been made for 30 of the predicted 100 strong and medium promoters from 156-kbp sequence of Sulfolobus solfataricus (33). Transcription initiation at or near the translational start codons has been mapped for some genes in Halobacterium salinarium (3) and S. solfataricus (18), and alternative mechanisms for initial mRNA-ribosome contact in Archaea have been hypothesized (3).
The analysis of 28 kbp from C. symbiosum has identified genes indicative of several metabolic pathways (Table 2). As our study progresses, we expect to gain more critical information on the physiological potential of the organism. We have already identified overlapping fosmids that represent ca. 90 kbp of the C. symbiosum genome (unpublished data). While serving as a model for the development of environmental genomic approaches to characterize uncultivated organisms, the C. symbiosum genome analysis also highlights the complications inherent in such studies that arise from the widespread genomic heterogeneity in natural populations, even those occupying a well-defined symbiotic niche. Our study represents a departure from pure-culture genomics. This approach provides a clearer view of the characteristics of naturally occurring genomic variability.
Environmental genomics has the potential to elucidate the physiologies of organisms that have resisted cultivation in the laboratory. The true test of our understanding of the field of genomics will be our ability to infer an organism’s physiology solely from its genome sequence.
ACKNOWLEDGMENTS
We thank Shane Andersen and Chris Gottschalk for their expert diving and collecting assistance; Kathy Foltz for use of aquarium facilities; Anna Lenox, Bill Young, and Monnette Aujay for DNA sequencing; and Jeff Stein for comments on the manuscript.
This work was supported by Diversa Corporation and by NSF grants OCE95-29804 and OPP94-18442 to E.E.D. C.S. was supported by a fellowship from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Amann R, Snaidr J, Wagner M, Ludwig W, Schleifer K H. In situ visualization of high genetic diversity in a natural microbial community. J Bacteriol. 1996;178:3496–3500. doi: 10.1128/jb.178.12.3496-3500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bintrim S B, Donohue T J, Handelsman J, Roberts G P, Goodman R M. Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown J W, Daniels C J, Reeve J N. Gene structure, organization, and expression in archaebacteria. Crit Rev Microbiol. 1989;16:287–337. doi: 10.3109/10408418909105479. [DOI] [PubMed] [Google Scholar]
- 4.Bult C, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrman J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 5.Cohan F M. The effects of rare but promiscuous genetic exchange on evolutionary divergence in prokaryotes. Am Nat. 1994;143:956–986. doi: 10.1016/0169-5347(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 6.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong E F. Marine microbial diversity: the tip of the iceberg. Trends Biotechnol. 1997;15:2–9. doi: 10.1016/S0167-7799(97)01044-5. [DOI] [PubMed] [Google Scholar]
- 8.DeLong E F, Wu K Y, Prezelin B B, Jovine R V M. High abundance of Archaea in Antarctic marine picoplankton. Nature. 1994;371:695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- 9.Field K G, Gordon D, Wright T, Rappe M, Urbach E, Vergin K, Giovannoni S J. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuhrmann J A, McCallum K, Davis A A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 11.Garrett R A, Dalgaard J, Larsen N, Kjems J, Mankin A S. Archaeal rRNA operons. Trends Biochem Sci. 1991;16:22–26. doi: 10.1016/0968-0004(91)90011-j. [DOI] [PubMed] [Google Scholar]
- 12.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 13.Hain J, Reiter W D, Huedepohl U, Zillig W. Elements of an archaeal promoter defined by mutational analysis. Nucleic Acids Res. 1992;20:5423–5428. doi: 10.1093/nar/20.20.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershberger K L, Barns S M, Reysenbach A L, Dawson S C, Pace N R. Wide diversity of Crenarchaeota. Nature. 1996;384:420. doi: 10.1038/384420a0. [DOI] [PubMed] [Google Scholar]
- 15.Jurgens G, Lindstroem K, Saano A. Novel group within the kingdom Crenarchaeota from boreal forest soil. Appl Environ Microbiol. 1997;63:803–805. doi: 10.1128/aem.63.2.803-805.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim U-J, Shizuya H, de Jong P J, Birren B, Simon M I. Stable propagation of cosmid sized human DNA inserts in an F factor based vector. Nucleic Acids Res. 1992;20:1083–1085. doi: 10.1093/nar/20.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 18.Klenk H P, Schleper C, Schwass V, Brudler R. Nucleotide sequence, transcription and phylogeny of the gene encoding the superoxide dismutase of Sulfolobus acidocaldarius. Biochim Biophys Acta. 1993;1174:95–98. doi: 10.1016/0167-4781(93)90099-y. [DOI] [PubMed] [Google Scholar]
- 19.Kudo Y, Shibata S, Miyaki T, Tono T, Oyaizu H. Peculiar archaea found in Japanese paddy soils. Biosci Biotechnol Biochem. 1997;61:917–920. [PubMed] [Google Scholar]
- 20.MacGregor B J, Moser D P, Alm E W, Nealson K H, Stahl D. Crenarchaeota in Lake Michigan sediment. Appl Environ Microbiol. 1997;63:1178–1181. doi: 10.1128/aem.63.3.1178-1181.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Romero E, Caballero-Mellado J. Rhizobium phylogenies and bacterial genetic diversity. Crit Rev Plant Sci. 1996;15:113–140. [Google Scholar]
- 22.Massana R, Murray A E, Preston C M, DeLong E F. Vertical distribution and phylogenetic characterization of marine planktonic archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McInerney J O, Wilkinson M, Patching J W, Embley T M, Powell R. Recovery and phylogenetic analysis of novel archaeal rRNA sequences from a deep-sea deposit feeder. Appl Environ Microbiol. 1995;61:1646–1648. doi: 10.1128/aem.61.4.1646-1648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore L R, Rocap G, Chisholm S W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 25.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 26.Palmer J R, Daniels C J. In vivo definition of an archaeal promoter. J Bacteriol. 1995;177:1844–1849. doi: 10.1128/jb.177.7.1844-1849.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palys T, Nakamura L K, Cohan F M. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int J Syst Bacteriol. 1997;47:1145–1156. doi: 10.1099/00207713-47-4-1145. [DOI] [PubMed] [Google Scholar]
- 28.Preston C M, Wu K Y, Molinski T F, DeLong E F. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc Natl Acad Sci USA. 1996;93:6241–6246. doi: 10.1073/pnas.93.13.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rowan R, Knowlton N. Intraspecific diversity and ecological zonation in coral-algal symbiosis. Proc Natl Acad Sci USA. 1995;92:2850–2853. doi: 10.1073/pnas.92.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruby E G. Lessons from a cooperative bacterial-animal association: the Vibrio fischeri-Eupreymna scolopes light organ symbioses. Annu Rev Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 31.Schleper C, Holben W, Klenk H P. Recovery of crenarchaeotal ribosomal DNA sequences from freshwater-lake sediments. Appl Environ Microbiol. 1997;63:321–323. doi: 10.1128/aem.63.1.321-323.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schleper C, Swanson R V, Mathur E J, DeLong E F. Characterization of a DNA polymerase from the uncultivated psychrophilic archaeon Cenarchaeum symbiosum. J Bacteriol. 1997;179:7803–7811. doi: 10.1128/jb.179.24.7803-7811.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sensen C W, Klenk H P, Singh R K, Allard G, Chan C C, Liu Q Y, Penny S L, Young F, Schenk M E, Gaasterland T, Doolittle W F, Ragan M A, Charlebois R L. Organizational characteristics and information content of an archaeal genome: 156 kb of sequence from Sulfolobus solfataricus P2. Mol Microbiol. 1996;22:175–191. doi: 10.1111/j.1365-2958.1996.tb02666.x. [DOI] [PubMed] [Google Scholar]
- 34.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Souza V, Eguiarte L, Avila G, Cappello R, Gallardo C, Montoya J, Pinero D. Genetic structure of Rhizobium etli biovar phaseoli associated with wild and cultivated bean plants (Phaseolus vulgaris and Phaseolus coccineus) in Morelos, Mexico. Appl Environ Microbiol. 1994;60:1260. doi: 10.1128/aem.60.4.1260-1268.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stein J L, Marsh T L, Wu K Y, Shizuya H, DeLong E F. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol. 1996;178:591–599. doi: 10.1128/jb.178.3.591-599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueda T, Suga Y, Matsuguchi T. Molecular phylogenetic analysis of a soil microbial community. Eur J Soil Sci. 1995;46:415–421. [Google Scholar]
- 38.Wayne L, Brenner D J, Colwell R R, Grimont P A D, Kandler O, Krichevsky M I, Moore L H, Moore W E C, Murray R G E, Stackebrandt E, Starr M P, Truper H G. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]