Abstract
Sulfotransferases are ubiquitous enzymes that transfer a sulfo group from the universal cofactor donor 3′-phosphoadenosine 5′-phosphosulfate to a broad range of acceptor substrates. In humans, the cytosolic sulfotransferases are involved in the sulfation of endogenous compounds such as steroids, neurotransmitters, hormones, and bile acids as well as xenobiotics including drugs, toxins, and environmental chemicals. The Golgi associated membrane-bound sulfotransferases are involved in post-translational modification of macromolecules from glycosaminoglycans to proteins. The sulfation of small molecules can have profound biologic effects on the functionality of the acceptor, including activation, deactivation, or enhanced metabolism and elimination. Sulfation of macromolecules has been shown to regulate a number of physiologic and pathophysiological pathways by enhancing binding affinity to regulatory proteins or binding partners. Over the last 25 years, crystal structures of these enzymes have provided a wealth of information on the mechanisms of this process and the specificity of these enzymes. This review will focus on the general commonalities of the sulfotransferases, from enzyme structure to catalytic mechanism as well as providing examples into how structural information is being used to either design drugs that inhibit sulfotransferases or to modify the enzymes to improve drug synthesis.
SIGNIFICANCE STATEMENT
This manuscript honors Dr. Masahiko Negishi’s contribution to the understanding of sulfotransferase mechanism, specificity, and roles in biology by analyzing the crystal structures that have been solved over the last 25 years.
Introduction
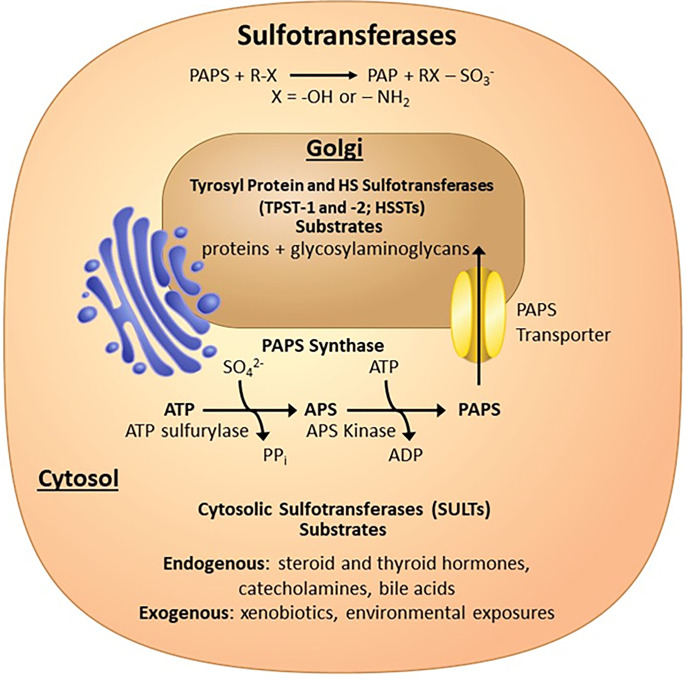
The sulfated products of reactions catalyzed by the sulfotransferase family of enzymes are involved in many physiologic and pathophysiological processes (Gamage et al., 2006; Bishop et al., 2007; Duffel, 2010 updated 2016; Lindahl et al., 2015; Coughtrie, 2016; Langford et al., 2017). Vertebrate sulfotransferases use 3′-phosphoadenosine 5′-phosphosulfate (PAPS) as the universal sulfo donor. PAPS is synthesized by one of two PAPS synthases, which are bifunctional enzymes that use two ATP molecules to produce one molecule of PAPS (Fig. 1) (Lyle et al., 1994; Xu et al., 2000). The ATP sulfurylase domain displaces pyrophosphate from ATP with inorganic sulfate to produce adenosine 5′-phosphosulfate (APS). APS is subsequently phosphorylated at the 3′ hydroxyl by the APS kinase domain to produce PAPS.
Fig. 1.
Sulfotransferases covered in this review. Sulfotransferases transfer a sulfo group (SO3) from PAPS, which is generated in the cytosol by the bifunctional PAPS synthases to many different types of acceptor substrates. The SULT enzymes in the cytosol sulfate endogenous and exogenous small molecules, whereas the Golgi-associated sulfotransferase sulfate macromolecules such as proteins and glycosaminoglycans.
Sulfation by cytosolic sulfotransferases (SULTs) of small molecules on hydroxyl or amine moieties is one of the major pathways for the detoxification of xenobiotics and elimination of endogenous small molecules such as hormones, neurotransmitters, and bile acids from the body (Gamage et al., 2006; Duffel, 2010 updated 2016; Coughtrie, 2016). Sulfation of these compounds typically improves water solubility, enhancing elimination. However, sulfation can also result in biologically active compounds. Dehydroepiandrosterone and estrone sulfate can act as circulating intermediates for the biosynthesis of hormones, whereas pregnenolone sulfate can regulate neurotransmitter receptors (Mueller et al., 2015). As such, sulfation/desulfation is a critical process that modulates steroidogenesis and hormone action in various tissues (Mueller et al., 2015). Inhibition of sulfotransferases due to environmental exposures to chemicals such as polychlorinated biphenols and flame retardants may result in disruption of proper endocrine homeostasis (Kester et al., 2000, 2002; Hamers et al., 2008).
In addition to aiding in the elimination of dietary compounds and environmental toxins, SULTs are major Phase II drug metabolizing enzymes, not only involved in their elimination, but also in the activation of prodrugs. Sulfation of minoxidil, for example, is critical for its hair growth effects, whereas sulfation of oxamniquine activates the drug to treat schistosomiasis (Buhl et al., 1990; Pica-Mattoccia et al., 2006; Valentim et al., 2013).
In humans, there are 13 SULT genes encoding 14 proteins, with two of the proteins resulting from alternative splicing (Coughtrie, 2016). These proteins are between 284 to 365 amino acids in length. Historical nomenclature for the SULTs was based upon the original substrate with which they were identified (e.g., EST estrogen sulfotransferase, PST phenol sulfotransferase, and AST aryl sulfotransferase), but as it became clear that many of these enzymes catalyzed substrates with different functional groups and exhibited overlapping specificity, a more systematic nomenclature was needed (Blanchard et al., 2004; Duffel, 2010 updated 2016). In brief, in the current nomenclature, the first numeral after the SULT defines the family (45% sequence identity) and the following capital Arabic letter defines the subfamily (60%), with the final numeral defining the isoform. Splice variants are then assigned a lowercase letter (ex. SULT2B1b).
Tyrosyl protein sulfotransferases (TPSTs) are Type-II integral membrane proteins localized in the trans-Golgi network that are responsible for sulfation of the tyrosine side chain of acceptor proteins, representing one of the major post-translational modifications (Fig. 1) (Moore, 2003). Tyrosine sulfation can have a number of biologic consequences including effecting circulation half-life, proteolytic processing of bioactive peptides, and protein-protein interactions impacting many processes including blood coagulation, inflammation, and viral infection (Leyte et al., 1991; Pouyani and Seed, 1995; Farzan et al., 1999; Moore, 2003). Humans contain two TPSTs (TPST-1 and -2) that share 64% sequence identity and are 370 and 377 amino acids in length, respectively (Niehrs and Huttner, 1990; Beisswanger et al., 1998; Ouyang et al., 1998). These enzymes have broad and slightly different substrate specificities (Mishiro et al., 2006).
There are at least 37 Golgi-associated sulfotransferases in humans, a large number of which are involved in the sulfation of the glycosaminoglycans (GAGs)—heparan sulfate, chondroitin sulfate, and dermatan sulfate (Langford et al., 2017; Zerbino et al., 2018). These GAGs are found attached to a select group of specific proteins or lipids and are found ubiquitously on the cell surface and in the extracellular matrix (Bishop et al., 2007; Lindahl and Li, 2009; Iozzo and Schaefer, 2015; Lindahl et al., 2015). As such, they play important roles in how cells communicate with their surrounding environment through interactions with protein binding partners. These GAGs are involved in a long list of physiologic and pathophysiological processes including embryonic development, blood coagulation, inflammation, bacterial and viral infection, neurodegenerative diseases, and cancer (Bishop et al., 2007; Lindahl and Li, 2009; Shi et al., 2021). GAGs are made up of linear repeating disaccharide units that can be modified by deacetylation, epimerization, and sulfation. This review will focus on heparan sulfate sulfotransferases due to the availability of structural information. The sulfation of specific hydroxyls and amines on the GAGs is carried out by unique sulfotransferases and appears to be a somewhat ordered process (Multhaupt and Couchman, 2012). In general, heparan sulfates (HS) are synthesized through chain elongation by a set of glycosyltransferases (exotosin 1 and 2) onto a common linker region to produce repeating disaccharides units of N-acetylglucosamine (GlcNAc) and glucuronic acid (GlcA). The bifunctional N-deacetylase/N-sulfotransferase (4 isoforms in humans) can deacetylate and subsequently sulfate the glucosamine at the 2-amine position. This becomes a good substrate for the C5-epimerase, which can convert adjacent GlcAs into iduronic acids (IdoA). Although the 2-O-sulfotranerase (1 isoform) can sulfate the 2-hydroxyl on GlcA, sulfation on IdoA appears to be the preferred substrate and is much more prevalent in biology (Rong et al., 2001; Bethea et al., 2008; Liu et al., 2014). Sulfation by both the 6-O-sulfotransferases (3 isoforms) and the 3-O-sulfotransfereases (7 isoforms) occurs at the 6- and 3-hydroxyl groups of glucosamines, respectively. Whereas some sulfotransferase isoforms appear to have similar substrate specificities, others are very distinct. The extent and types of modification in a given tissue or cell type are spatially and temporally regulated and are determined by which isoforms are present and expressed.
Historical Prospective
In 1876, the relevance of sulfotransferases in medicine was first reported when E. Baumann determined that the conjugate of carbolic acid, a common antiseptic used in surgery at the time, could be absorbed by the skin and secreted in the urine as phenol sulphuric ester (Baumann, 1876; Folin and Denis, 1915). Recognizing the importance and relevance of sulfation in chondroitin sulfate, heparin, steroids, and phenol detoxification, Robbins and Lipmann reported in 1956 that the biologic sulfo donor was indeed PAPS, which could be produced by a liver enzyme with the addition of ATP (Robbins and Lipmann, 1956; Lipmann, 1958). They referred to the enzymes that must use PAPS for the sulfo donor as sulfokinases. It was around this same time period that the first tyrosine-O-sulfate was found in a peptide from fibrinogen (Bettelheim, 1954). By this time, HS had already been identified as an effective anticoagulant and had been introduced as a widely available therapeutic (Howell and Holt, 1918; Lim, 2017). It wasn’t until 1987 that the first cytosolic sulfotransferase was unknowingly cloned as an androgen-repressible rat liver protein (Chatterjee et al., 1987). The cloning of the first Golgi sulfotransferases involved in HS biosynthesis occurred in 1992, and the TPSTs followed in 1998 (Hashimoto et al., 1992; Ouyang et al., 1998).
Prior to the first crystal structures, sequence alignments of the SULTs were used to identify critical residues in the functionality of these enzymes (Roche et al., 1991; Khan et al., 1993; Weinshilboum and Otterness, 1994). These alignments revealed two highly conserved regions including an N-terminal region YPKSGTXW and a carboxy terminus GXXGXXK reminiscent of the GXXXXGK[TS] P-loop structures found to bind ATP in kinases (Saraste et al., 1990). In 1997, the Negishi laboratory at the National Institute of Environmental Health Sciences at the National Institutes of Health published the first crystal structure of a sulfotransferase (mouse estrogen sulfotransferase or mSULT1E1), ushering in a new era of understanding of the molecular function of these enzymes (Kakuta et al., 1997). This structure revealed that both regions indeed bound to the PAPS; however, it was the amino terminal region that was structurally related to the P-loop and cradled the 5′-phosphate, whereas the C-terminal region bound to the 3′-phosphate unique to PAPS (Fig. 2). The structure also revealed a conservation of a central scaffold as well as substrate positioning similar to that of the adenylate kinase family of enzymes, suggesting a common mechanism between sulfation and phosphorylation. This analogy can be extended to desulfation, since sulfatases display conservation in structure and activity to that of alkaline phosphatase (Bond et al., 1997; O'Brien and Herschlag, 1998). Crystal structures currently exist for 12 of the 14 human SULTs, representatives from each of the vertebrate HS sulfotransferases as well as both TPSTs (Table 1).
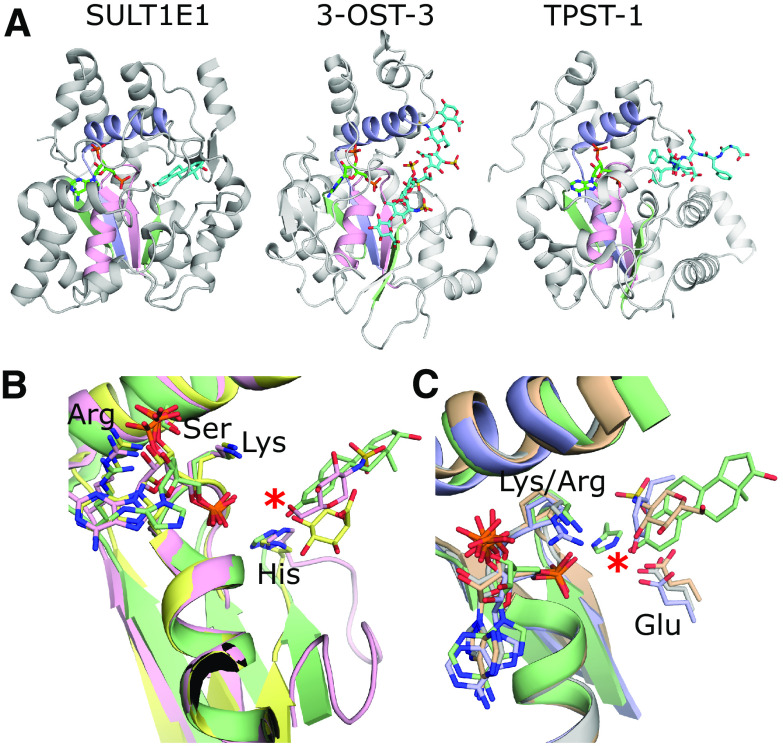
Fig. 2.
Representative structures of the different types of sulfotransferases and their PAPS binding sites. (A) From left to right: crystal structure of hSULT1E1 in complex with PAP and estradiol [protein data bank (PDB) code 4JVL) (Gosavi et al., 2013); crystal structure of 3-OST-3 with PAP and an 8mer NS2S heparan sulfate bound (PDB code 6XL8) (Wander et al., 2021); crystal structure of TPST-1 with PAP and a polypeptide substrate bound (PDB code 5WRI) (Tanaka et al., 2017). The PAP is colored green and acceptor substrates are cyan. The strand-loop-helix containing the PSB loop is colored pink, whereas the strand-loop-helix containing the 3′-phosphate binding motif (3′SB) is colored light purple. The remaining two strands making up the central β-sheet are colored light green. (B) Comparisons of the PAP binding motifs and catalytic residues. The SULTs including SULT1E1 (green) and heparan sulfotransferases 2-OST (yellow) and 6-OST (pink) all use a lysine on the PSB loop and histidines for the proposed catalytic base (PDB codes 4JVL, 4NDZ, and 5T0A, respectively) (Gosavi et al., 2013; Liu et al., 2014; Xu et al., 2017b). The acceptor hydroxyl on the substrates all superimpose well, supporting a conserved mechanism. (C) Comparisons of the PAP binding motifs and catalytic residues of the sulfotransferase domain of SULT1E1 (green), NDST-1 (gray), 3-OST-3 (wheat), and TPST-1 (light purple) (PDB codes: 4JVL, NST1, 6XL8, and 5WRI, respectively) (Kakuta et al., 1999; Gosavi et al., 2013; Tanaka et al., 2017; Wander et al., 2021). NDST-1, 3-OST-3, and TPST-1 appear to use a conserved glutamate for their catalytic base, as opposed to the SULTs, 2-OST, and 6-OST. However, the PAPs and acceptor hydroxyls on the substrates all superimpose perfectly, supporting a conserved inline reaction mechanism for all the vertebrate sulfotransferases. For panels B and C, only the acceptor saccharide of the substrate is shown for 3-OST-3 and 2-OST and acceptor tyrosine of the peptide for TPST-1. No substrate is present in the crystal structure of NST-1.
Table 1.
Representative structures of the human SULT, TPST, and HSSTs
| Sulfotransferase | Acceptor | Nucleotide | PDB Code | Res Å | Reference |
|---|---|---|---|---|---|
| Human Cytosolic Sulfotransferases (SULTs) | |||||
| SULT1A1 | p-nitrophenol | PAP | 1LS6 | 1.9 | (Gamage et al., 2003) |
| estradiol | PAP | 2D06 | 2.3 | (Gamage et al., 2005) | |
| p-nitrophenol | PAP | 3QVU | 2.5 | (Alcolombri et al., 2011) | |
| 3-cyanoumbelliferone | PAP | 3QVV | 2.35 | (Alcolombri et al., 2011) | |
| 2-napthol | PAP | 3U3K | 2.36 | (Berger et al., 2011) | |
| 3-cyano-7-hydroxycoumarin | PAP | 3U3M, 3U3O | 2.3, 2.0 | (Berger et al., 2011) | |
| p-nitrophenol | PAP | 3U3R | 2.36 | (Berger et al., 2011) | |
| — | PAP | 3U3J | 2.70 | (Berger et al., 2011) | |
| — | PAP | 4GRA | 2.56 | (Cook et al., 2013a) | |
| SULT1A1*3 | — | PAP | 1Z28 | 2.3 | (Lu et al., 2010) |
| SULT1A2 | PAP | 1Z29 | 2.4 | (Lu et al., 2010) | |
| SULT1A3 | Dopamine | PAP | 2A3R | 2.6 | (Lu et al., 2005) |
| — | — | 1CJM | 2.4 | (Bidwell et al., 1999) | |
| SULT1B1 | Resveratrol | PAP | 3CKL | 2.0 | TBPf |
| — | PAP | 2Z5F | 2.1 | (Dombrovski et al., 2006) | |
| SULT1C2a | — | PAP | 3BFX | 1.8 | (Dombrovski et al., 2006) |
| SULT1C3d | — | PAP | 2H8K | 3.2 | (Allali-Hassani et al., 2007) |
| — | PAP | 2REO | 2.65 | TBP | |
| SULT1C4b | Pentrachlorophenol | PAP | 2GWH | 1.80 | (Allali-Hassani et al., 2007) |
| — | — | 2AD1 | 2.00 | (Allali-Hassani et al., 2007) | |
| SULT1E1 | Estradiol | PAP | 4JVL | 1.94 | (Gosavi et al., 2013) |
| Tetrabromobisphenol A | PAP | 4JVM | 1.99 | (Gosavi et al., 2013) | |
| 3,5,3′,5′-tetrachloro-biphenyl-4-4’-diol | PAP | 1G3M | 1.7 | (Shevtsov et al., 2003) | |
| 3-hydroxylbromodiphenyl ether | PAP | 4JVN | 2.05 | (Gosavi et al., 2013) | |
| — | PAPS | 1HY3 | 1.80 | (Pedersen et al., 2002) | |
| SULT2A1 | Androsterone | — | 1OV4 | 2.70 | (Chang et al., 2004) |
| DHEAg | — | 1J99 | 1.99 | (Rehse et al., 2002) | |
| (3Beta,5alpha)-3-Hydroxyandrostan-17-one | — | 2QP32QP4 | 2.63.0 | (Lu et al., 2008) | |
| Lithocholic acid | PAP | 3F3Y | 2.2 | TBP | |
| PAPS | 4IFB | 2.3 | TBP | ||
| — | PAP | 1EFH | 2.4 | (Pedersen et al., 2000) | |
| SULT2B1a | — | PAP | 1Q1Q | 2.91 | (Lee et al., 2003) |
| SULT2B1b | DHEA | PAP | 1Q22 | 2.5 | (Lee et al., 2003) |
| Pregnenolone | PAP | 1Q20 | 2.3 | (Lee et al., 2003) | |
| — | PAP | 1Q1Z | 2.4 | (Lee et al., 2003) | |
| SULT4A1 | — | — | 1ZD1 | 2.24 | (Allali-Hassani et al., 2007) |
| Tyrosyl Protein Sulfotransferase (TPSTs) | |||||
| TPST-1 | DFEDYEFD | PAP | 5WRI | 1.60 | (Tanaka et al., 2017) |
| EEEEEAYGWMDF | PAP | 5WRJ | 2.33 | (Tanaka et al., 2017) | |
| TPST-2 | EDFEDYEFD | 3AP1 | 1.9 | (Teramoto et al., 2013) | |
| EDFEDYEFD | PAP | 3AP2 | 2.4 | TBP | |
| — | PAP | 3AP3 | 3.5 | (Teramoto et al., 2013) | |
| Heparan Sulfate Sulfotransferases (HSSTs) | |||||
| NDST-1 (sulfotransferase catalytic domain) | — | PAP | 1NST | (Kakuta et al., 1999) | |
| 2-OSTc | GlcA-GlcNAc-GlcA-GlcNS-IdoAh-GlcNS-GlcA-pNP | PAP | 4NDZ | 3.45 | (Liu et al., 2014) |
| — | PAP | 3F5F | 2.65 | (Bethea et al., 2008) | |
| 6-OST-3d | GlcNS-GlcA-GlcNS-IdoA2S-GlcNS-GlcA-pNP | PAP | 5T05 | 1.95 | (Xu et al., 2017b) |
| GlcNS-GlcA-GlcNS-GlcA-GlcNS-GlcA-pNP | PAP | 5T03 | 2.1 | (Xu et al., 2017b) | |
| GlcA-GlcNS-GlcA-GlcNS-GlcA-GlcNS-GlcA-pNP | PAP | 5T0A | 1.95 | (Xu et al., 2017b) | |
| 3-OST-1e | GlcNAc6S-GlcA-GlcNS6S-IdoA2S-GlcNS6S-GlcA-UA | PAP | 3UAN | 1.84 | (Moon et al., 2012) |
| — | PAP | 1VKJ | 2.5 | (Edavettal et al., 2004) | |
| 3-OST-1 | — | PAP | 1ZRH | 2.1 | TBP |
| 3-OST-3 | GlcNAc-GlcA-GlcNS-Ido2S-GlcNS-Ido2S-GlcNS-GlcA-pNP | PAP | 6XL8 | 2.34 | (Wander et al., 2021) |
| GlcNAc6S-GlcA-GlcNS6S-Ido2S-GlcNS6S-Ido2S-GlcNS6S-GlcA-pNP | PAP | 6XKG | 1.55 | (Wander et al., 2021) | |
| δUA2S-IdoA2S-GlcNS6S-IdoA2S | PAP | 1T8U | 1.95 | (Moon et al., 2004) | |
| — | PAP | 1T8T | 1.85 | (Moon et al., 2004) | |
| 3-OST-5 | — | PAP | 3BD9 | 2.3 | (Xu et al., 2008) |
aIn PDB under SULT1C1.
bIn PDB under SULT1C2.
c Gallus gallus.
d Danio rerio.
e Mus musculus.
fTBP (To be published).
gdehydroepiandrosterone.
hacceptor saccharide is in bold.
In this review, we will delve into what has been learned from crystal structures of the various sulfotransferase families (SULTs, HSSTs, and TPSTs) regarding conservation of structure and mechanism, determinants of specificity, and how this information is being used to design and engineer new pharmacological tools.
Key Advances in Understanding
Conservation of Structure and Mechanism among Sulfotransferases
The crystal structures of SULTs, heparan sulfate sulfotransferases (HSSTs), and TPSTs all share many central commonalities with respect to structure and function. All three families of sulfotransferases contain a conserved core structure consisting of 4–5 β-strands flanked on both faces with α-helixes (Fig. 2A) (Negishi et al., 2001; Teramoto et al., 2013; Gunal et al., 2019). The need for binding specificity to PAPS dictates structural conservation throughout the sulfotransferases. From the structural analysis of mouse estrogen sulfotransferase (mSULT1E1), two conserved motifs were identified (Fig. 2) (Kakuta et al., 1997). The first strand-loop-helix motif is analogous to the P-loop structures found in the uridylate kinase family that cradles the 5′-phosphate of the PAPS, forming an extensive hydrogen bonding network via backbone amide interactions with the loop [referred to as the termed phosphosulfate binding (PSB)-loop] (Kakuta et al., 1998a). Central to the loop is a conserved basic residue, usually a lysine but sometimes an arginine (such as in the TPSTs) that hydrogen bonds to the 5′-phosphate (Fig. 2, B–C) (Teramoto et al., 2013; Valentim et al., 2013). The second region, termed 3′-sulfate binding (SB), consists of a strand from the central β-sheet and an α-helix that runs across the top of the PSB loop that can form interactions with both the donor and acceptor substrates (Fig. 2) (Kakuta et al., 1998a). This region is involved in binding of the 3′-phosphate of the 3′-phosphoadenosine 5′-phosphate (PAP) via sidechain interactions with a conserved arginine from the β-strand and a serine residue from the 3′SB α-helix (Fig. 2, B–C). The SULTs contain an additional conserved region of sequence GXXGXXK that, as mentioned, was proposed to interact with the PAPS based on its similarity in sequence to P-loops (Marsolais and Varin, 1995). The structure of mSULT1E1 revealed that indeed this region interacted with the PAPS, but via the 3′-phosphate, and perhaps plays other critical roles in function, as discussed later.
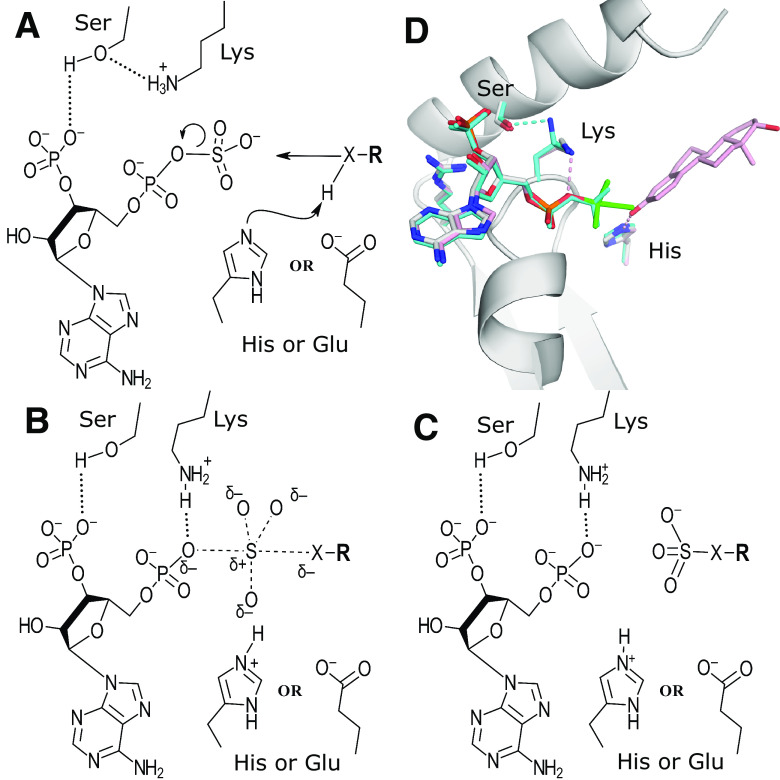
The relative position of the substrate acceptor with respect to the position of the donor suggested an inline displacement mechanism for the sulfo group transfer (Fig. 3, A–C) (Kakuta et al., 1997). This was further supported by a cocrystal structure of mSULT1E1 with vanadate and estradiol, as well as mSULT1E1 with PAPS alone (Fig. 3D) (Kakuta et al., 1998b; Pedersen et al., 2002). Based on the structural comparisons to uridylate kinase, it was hypothesized that the reaction would proceed via an SN2-like associative reaction mechanism whereby the acceptor nucleophile could be primed by a catalytic base, allowing for nucleophilic attack on the sulfur of PAPS (Müller-Dieckmann and Schulz, 1994; Kakuta et al., 1997). The reaction would proceed through a trigonal bipyramidal transition state, with the leaving oxygen on the PAP and the incoming nucleophile in the axial positions (Fig. 3B). This transition state was proposed to be mimicked by the crystal structure of mSULT1E1 in the presence of PAP and vanadate (Fig. 3D) (Kakuta et al., 1998b). In the mSULT1E1/PAPS structure, the position of the conserved lysine on the PSB loop is found in a different orientation than when PAP is bound (Fig. 3D) (Pedersen et al., 2002). Here, the lysine is found hydrogen bonding with the conserved serine from the 3′SB loop rather than with the 5′-phosphate of the PAP. It was proposed that this interaction discourages PAPS hydrolysis in the absence of acceptor substrate (Pedersen et al., 2002). Once the acceptor substrate is bound, the lysine was proposed to undergo a conformational change to hydrogen bond with the phospho-sulfo bridging oxygen, functioning as a catalytic acid to encourage dissociation of the PAP leaving group. For the SULTs, a conserved histidine was identified within hydrogen bonding distance of the acceptor hydroxyl position that could function as a catalytic base to deprotonate the acceptor group (Fig. 2B, Fig. 3, A–C) (Kakuta et al., 1997). Mutations of the conserved His108, Lys48, and Ser138 in mSULT1E1 all greatly reduced or eliminated detectable activity, supporting the proposed mechanism (Kakuta et al., 1998b; Pedersen et al., 2002).
Fig. 3.
Catalytic mechanism of the sulfotransferases. (A–C) Proposed catalytic mechanism of the PAPS-dependent sulfotransferases. (D) Common architecture of the sulfotransferase PAPS binding site including the PSB-loop and 3′SB strand-loop-helix from the crystal structure of mSULT1E1 (PDB code 1BO6) with PAP (white) and a vanadate molecule (green) in a trigonal bipyramidal arrangement thought to mimic the transition state. Also shown are the proposed catalytic base (His) and acid (Lys) (Kakuta et al., 1998b). Superimposed onto this structure are PAPS (cyan) and estradiol (pink) from two different structures of human SULT1E1 (PDB codes 1HY3 and 4JVL, respectively) with their equivalent His and Lys residues (Pedersen et al., 2002; Gosavi et al., 2013). When PAPS is present, the lysine forms a hydrogen bond with a conserved serine from the 3′SB binding region but is bound to the 5′-phosphate when PAP is present. These structures, combined with comparisons to uridylate kinase, shaped the hypothesis for the reaction mechanism shown in panels (A–C).
One year after solving the structure of mSULT1E1, the Negishi laboratory solved the structure of the sulfotransferase domain (NST-1) of the bifunctional enzyme N-deacetylase/N-sulfotransferase isoform 1 (NDST-1) (Kakuta et al., 1999). This structure confirmed that conservation of the PAPS binding core for the sulfotransferases exists outside of the SULTs, including the lysine from the PSB-loop and the arginine and serine from the 3′PB motif, but lacking the conserved proposed histidine base (Fig. 2C) (Kakuta et al., 1999). The proposed catalytic histidine was later found to be conserved in Golgi HS 2-O-sulfotransferase (2-OST) and the 6-OST-1, but not in NDST-1, the 3-OSTs, or the TPSTs (Fig. 2, B–C) (Kakuta et al., 1999; Edavettal et al., 2004; Moon et al., 2004, 2012; Bethea et al., 2008; Xu et al., 2008, 2017b; Teramoto et al., 2013; Liu et al., 2014; Tanaka et al., 2017). In place of the histidine, these enzymes have a glutamate emanating from nonconserved structural elements within hydrogen bonding distance of the acceptor amine and hydroxyl atoms, respectively (Fig. 2C) (Kakuta et al., 1999; Edavettal et al., 2004; Moon et al., 2004, 2012; Xu et al., 2008; Teramoto et al., 2013; Tanaka et al., 2017). Mutations of these proposed alternative catalytic base residues also greatly reduced activity (Kakuta et al., 2003; Edavettal et al., 2004; Teramoto et al., 2013; Xu et al., 2017b). Noteworthy is the fact that, although the base may differ between different members of the phylogenetic family tree, the relative orientations of the acceptor to the leaving group PAP are highly conserved, supporting an inline transfer mechanism.
Interestingly, analysis using kinetic isotope effects and linear free-energy relationships on the SULTs suggests a mostly dissociative SN1-like reaction mechanism (Chapman et al., 2003; Hoff et al., 2006). This conclusion is supported by recent crystallographic analysis of the mouse SULT2A8 (Teramoto et al., 2021). This enzyme sulfates the 7α position of bile acids. In this structure, the catalytic histidine is replaced with a leucine. Mutations to nonconserved His48 and Glu237, within proximity to the acceptor 7α-OH, show a greater impact on KM rather than Vmax, indicating greater importance in binding than catalysis. These results suggest in sulfotransferases that the proposed catalytic base is required less for catalysis and more for binding and likely proper positioning. It is plausible that the ratio of dissociative to associative behavior of the sulfotransferases may vary within the family.
The SULTs are believed to employ a sequential mechanism, whereby binding of both substrates occurs prior to product release (Leyh, 1993). Although the binding was originally thought to be through an ordered mechanism, recent more thorough analysis of the reaction, taking into account the formation of a dead-end product complexes, suggests the reaction may proceed via a random sequential mechanism (Zhang et al., 1998; Wang et al., 2014a, 2014b). These inhibitory dead-end complexes are present in many crystal structures and contain both the PAP product and the acceptor substrate bound (Table 1). The catalytic mechanism for SULT2A1 has been eloquently described to contain eight enzyme forms and 22 rate constants (Wang et al., 2014a). Their highly conserved sequence and structural features suggest this mechanism is maintained for many SULTs. Although the reaction may proceed regardless of which substrate binds first, positive or negative cooperativity may regulate substrate binding, suggesting a quasi-ordered mechanism may be a better description for some SULTs, depending on the enzyme and substrates examined (Tibbs et al., 2015). One example of this has been reported for SULT2A1 (Cook et al., 2012). Although SULT2A1 in the presence or absence of PAP does not demonstrate cooperative binding with dehyroepiandrosterone, it does display negative cooperativity for Raloxifene in the presence of PAP (Cook et al., 2012). This has been suggested to be due to conformational changes associated with nucleotide binding to the SULT that restricts access of larger substrates such as Raloxifene to the acceptor binding site after PAPS binds. Thus, cellular levels of PAPS may contribute to a dynamic specificity of the enzyme (Tibbs et al., 2015).
Although sharing the inline transfer geometry with the SULTs, an ordered mechanism as opposed to a bi-bi reaction mechanism has also been suggested for 6-OST-3, based on lack of binding to a substrate N-sulfoheparosan in the absence of PAPS (Sterner et al., 2014). Interestingly, based on liquid chromatography mass spectrometry analysis, TPSTs were originally suggested to proceed through a two-site ping-pong reaction mechanism, whereby the PAPS and substrate bind independently, and the reaction involves a covalent sulfo-histidine intermediate of the sulfotransferase prior to transfer of the sulfo group to the acceptor tyrosine substrate residue (Danan et al., 2010). However, structural comparisons of the TPST-2 ternary complex structure with bound PAP and substrate peptide to those of the SULTs and HSSTs, along with the lack of a histidine proximal to the active site, support an inline transfer. Additionally, like the 6-OST-3, PAPS/PAP was required for binding of protein to peptide column, suggesting an ordered sequential mechanism of substrate binding (Niehrs and Huttner, 1990).
Substrate Specificity of SULTs
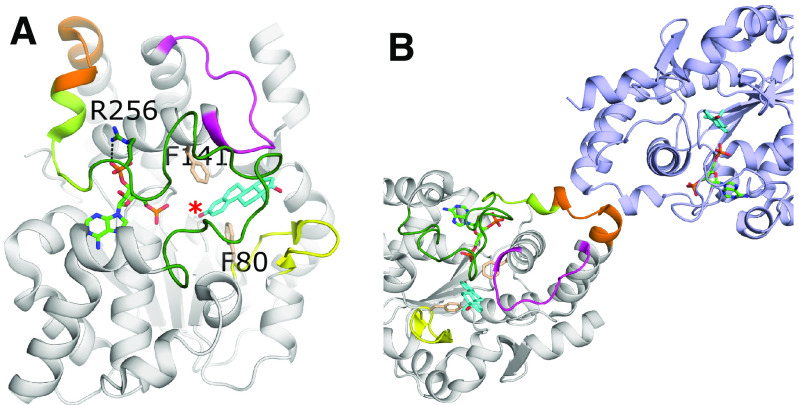
Although the SULTs, TPSTs and HSSTs all show conservation in nucleotide binding and catalytic mechanism, they can vary greatly in acceptor substrate recognition, due to the wide ranges in shape, size, and charge of substrates. The acceptor substrates of sulfotransferases can range in size from small phenolic compounds to very large protein or proteoglycan substrates. Modes of recognition and substrate orientation inside the binding pocket can also vary within the families themselves. Due to the smaller, often hydrophobic nature of the substrates, the SULTs’ acceptor substrate binding pockets are typically buried within the core of the protein by three loops (Fig. 4A) (Tibbs et al., 2015). Originally termed phenol sulfotransferases, the human SULT1 family possess a substrate gate consisting of aromatic residues (Phe80 and Phe141 in hSULT1E1) with the one exception being SULT1B1, which contains a methionine at the first position (figure 7 in Tibbs et al., 2015). These residues lie above and below the aromatic plane of the substrate and select for a phenolic acceptor, yet the rest of the pocket can accommodate diverse structural features (Fig. 4A) (Petrotchenko et al., 1999; Duffel, 2010 updated 2016). The SULT2 family were originally referred to as hydroxysteroid or bile acid sulfotransferases, but were later reclassified with the more general term alcohol sulfotransferases due to the developing knowledge of their broad specificity (Lyon and Jakoby, 1980; Duffel, 2010 updated 2016). The residues at the substrate gate differ in the SULT2 family, allowing for nonaromatic acceptors such as dehydroepiandrosterone. A thorough analysis on specificity overlap among and within the SULTs has been previously reviewed in fine detail (Duffel, 2010 updated 2016; Dong et al., 2012; Coughtrie, 2016). In general, the acceptor specificity is largely determined by three loops with variable sequences that define and restrict the entrance to the substrate binding pocket. These loops are flexible and often disordered in SULT structures, particularly in the absence of substrate (Tibbs et al., 2015). In human SULTs, loop 1 is ∼9–10 residues and highly variable in the SULT1 family and is 5–6 residues shorter in the SULT2 and SULT4 families (Dong et al., 2012; Tibbs et al., 2015). It has been postulated that a smaller loop 1 for the SULT2s allows the binding of larger substrates compared with SULT1 family members (Dong et al., 2012). Loop 2 is located after the SB-helix and can contribute to substrate interactions (Fig. 4A). Loop 3, also referred to as “lid” or “cap”, is the largest of the loops and covers both the PAPS and acceptor substrate binding sites when substrates are bound and may regulate binding cooperativity for certain substrates such as Raloxifene to SULT2A1, as mentioned previously (Cook et al., 2012; Leyh et al., 2013). Ordering of loop 3 is due in part to PAPS binding, as the conserved GXXGXXK is located at the C-terminus of this loop and, along with the preceding conserved arginine, forms interactions with the 3′-phosphate of the PAPS (Fig. 4A). In fact, there are relatively few crystal structures of sulfotransferases without nucleotides present. Binding of the nucleotide generally improves the thermostability and increases the likelihood of crystallization by decreasing surface heterogeneity via limiting loop 3 flexibility (Tibbs et al., 2015).
Fig. 4.
Substrate binding and dimerization of SULTS. (A) A representative SULT crystal structure, SULT1E1 with PAP (green) and estradiol (cyan) bound (PDB code 4JVL) (Gosavi et al., 2013). Shown are Loops 1 (yellow), 2 (magenta), and 3 (green) that contribute to substrate specificity and binding. The SULT-specific GXXGXXK region that connects loop 3 to the dimerization domain (orange) is colored lemon. The acceptor hydroxyl of the estradiol is designated with a red asterisk. Also shown are highly conserved aromatic residues (wheat) found in the SULT1 enzymes that contribute to selectivity for phenolic compounds. (B) Dimer of SULT1E1. The dimerization domain consists of a small seven residue motif (264-270 in SULT1E1, orange) that is conserved in human SULTs. The other protomer in the dimer is shown in light purple.
Single amino acid differences between isoforms SULT1A1 and SULT1A2 as well as SULT1A1 and SULT1A3 have been suggested to dictate specificity among these isoforms (Dajani et al., 1998; Lu et al., 2010; Dong et al., 2012). As well, loops 1–3 display structural plasticity within single isoforms to accommodate different acceptor substrates, accentuating the need for structures with multiple acceptors bound to better interrogate these enzymes for design of specific inhibitors (Dong et al., 2012).
Crystal structures of human SULTs revealed a conserved KXXXTVXXXE dimerization motif that presents an unusually small protomer interface (Fig. 4B) (Petrotchenko et al., 2001; Weitzner et al., 2009). The N-terminal lysine of this motif overlaps with the C-terminal lysine of the GXXGXXK sequence involved in 3′-phosphate binding at the end of loop 3 (Fig. 4). The dimerization motif is located distal from the acceptor binding site and appears important for enzyme stability, based on studies with SULT1A1 and 1B1, but may also regulate function (Lu et al., 2009; Tibbs and Falany, 2016). Half-site reactivity has been demonstrated for SULT1E1, SULT1A1, and SULT2A1, suggesting that substrate binding at one active site may influence binding to the other protomer (Sun and Leyh, 2010; Wang et al., 2014a, 2014b). The proximity of the dimerization domain to residues involved in PAP binding suggests a potential mode of regulation described in detail by Tibbs et al. (2015). However, this theme may not be universal, as SULT1B1 forms homodimers but does not display half-site reactivity (Tibbs and Falany, 2016). Homo- and heterodimers of SULTs have been described in vivo (Heroux and Roth, 1988; Kiehlbauch et al., 1995). Heterodimers could provide a mechanism by which substrates of one SULT could affect sulfation of another. Also, SULT4A1 alone has been hypothesized to play important roles in neuronal development by heterodimerizing with and regulating the activity of other SULTs (Idris et al., 2020).
Substrate Specificity of TPSTs
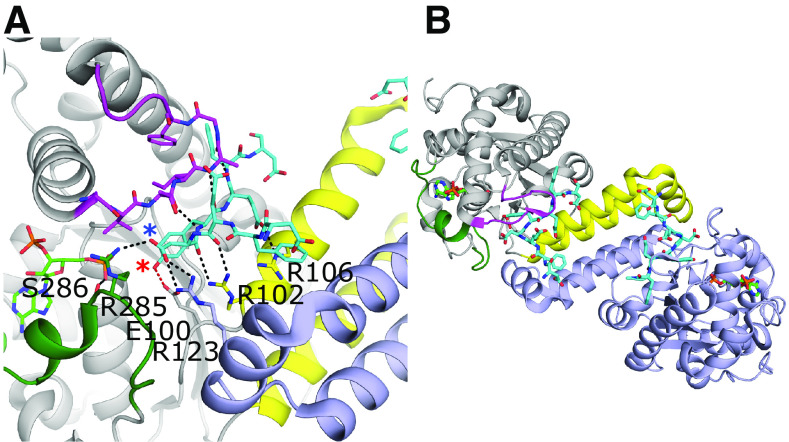
The TPSTs have broad, overlapping substrate specificities and are believed to sulfate up to 1% of all tyrosine residues in the eukaryotic proteome (Baeuerle and Huttner, 1985; Hille et al., 1990). The human TPSTs -1 and -2 bind to intrinsically disordered sequences containing a tyrosine flanked by acidic residues within five amino acids on both the N- and C- terminal sides (Hortin et al., 1986; Rosenquist and Nicholas, 1993; Teramoto et al., 2013). The crystal structures of the catalytic domains of TPST-1 and -2 reveal that the tyrosine acceptor is buried deep within a positively charged binding pocket, necessitating the peptide to take on an intrinsically unfolded conformation (Fig. 5A) (Teramoto et al., 2013; Tanaka et al., 2017). The peptide adopts a sharply bent configuration with the bend’s apex immediately N-terminal (-1 position) to the tyrosine acceptor (0 position). For TPST-1 binding to a substrate peptide, the tyrosine and aspartate at the -1 position appear to be the most critical residues for binding (Fig. 5A) (Tanaka et al., 2017). Like the human SULTs, the TPSTs form functional dimers; however, the dimerization interface is very different. For TPSTs, the dimerization interface buries ∼25% of the surface area and is located at the acceptor substrate binding pocket (Fig. 5) (Teramoto et al., 2013; Tanaka et al., 2017). Although not critical for catalysis, dimerization appears to have a role in substrate binding (Teramoto et al., 2013). Structurally located at the position of loop 1 in the SULTs are three helices that form the majority of the dimer interface (Fig. 5B). Heterodimers between TPST-1 and -2 have been reported in vivo, likely due to the high sequence conservation of this region (Hartmann-Fatu et al., 2015). This region also contributes to acceptor substrate binding interactions at each active site. Two conserved arginines (Arg102 and Arg106) from the first α-helix, shown to be important for binding in TPST2, interact with the substrate in the same protomer binding site, although Arg123 (not critical for catalysis from the other protomer) also simultaneously interacts with the substrate (Fig. 5A) (Teramoto et al., 2013). SULT-equivalent loop 2 from TPST forms β-sheet like interactions with the substrate peptide when present and is disordered in the absence of acceptor substrate (Teramoto et al., 2013). Although the extent of the conformational flexibility of “loop 3” in TPST remains unknown, it appears to be involved in dimer formation, exclusion of water from the active site, and PAPS binding via a serine residue that interacts with the 5′- phosphate (Fig. 5A) (Teramoto et al., 2013). It has been suggested that the presence of PAPS enhances peptide binding by organizing the substrate binding site (Niehrs and Huttner, 1990). The ability to bind and sulfonate a variety of peptide sequences may also make the enzyme susceptible to nonproductive binding, as demonstrated in the crystal structure of TPST-1, where the Gastrin peptide is interpreted to be present in both a productive and a nonproductive binding orientation (Tanaka et al., 2017).
Fig. 5.
Substrate binding and dimerization of TPST-1. (A) Substrate binding site of TPST-1 (PDB code 5WRI) (Tanaka et al., 2017). Residues Arg102 and Arg106 from the α-helix bundle and loop 2 (magenta) contribute significantly to substrate binding. Residues from loop 3 (green) form interactions with both the acceptor (Arg285) and donor substrates (Ser286). Arg123 from the other protomer forms a nonessential interaction with the substrate. The acceptor tyrosine is positioned for catalysis and forms a hydrogen bond with the proposed catalytic base Glu100 (red dashed line). The acceptor hydroxyl on the tyrosine and the Asp at the -1 position of the substrate are designated with red and blue asterisks, respectively. (B) Dimer interface of TPST-1. One protomer is colored light purple, whereas the other is colored gray with the equivalent to loops 1, 2, and 3 of the SULTs colored in yellow, magenta, and dark green, respectively.
Substrate Specificity of HSSTs
HSSTs are highly specific for which functional group becomes sulfated and on the types of modifications (sulfation or epimerization) on the HS that can be accommodated within the active site. Unlike the SULTs and TPSTs, these enzymes use an open binding cleft to bind the large, often anionic polysaccharide acceptor substrates. Historically, the study of these enzymes was hindered by the inability to obtain homogeneous substrates. However, with the advent of chemoenzymatic synthesis, which utilizes glycosyltransferase and sulfotransferase enzymes to produce homogenous HS with specific lengths, sequences, sulfation, and epimerization modifications, cocrystal structures with acceptor substrates bound with chicken 2-OST, zebrafish 6-OST-3, and 3-OST-1 and -3 (mouse and human, respectively) have been obtained (Moon et al., 2004, 2012; Liu et al., 2014; Xu et al., 2017b; Wander et al., 2021). This structural information has provided a wealth of information into features dictating the specificity between families and isoforms within families.
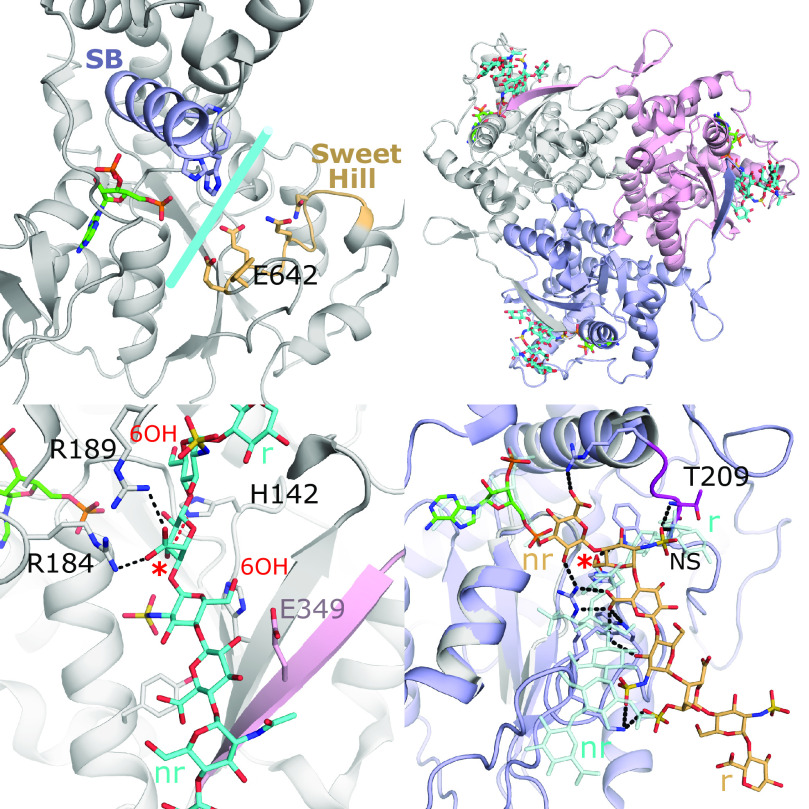
Despite lacking an acceptor substrate, the crystal structure of the sulfotransferase domain of NDST-1 (NST-1) revealed a large open cleft that extends across the PAPS binding site to accommodate the acceptor substrate binding (Fig. 6A) (Kakuta et al., 1999). The NDSTs’ sulfotransferase domains are most similar to that of the 3-OSTs (discussed later), with NST-1 sharing 28% sequence identity in the catalytic domain (Edavettal et al., 2004; Moon et al., 2004; Xu et al., 2008). Comparing NST-1 to the crystal structure of 3-OST-1 revealed the acceptor binding cleft to be less positively charged than that of the 3-OST-1, consistent with its substrate containing a reduced anionic charge due to its role earlier in the HS maturation process (Edavettal et al., 2004). To determine residues involved in substrate binding, an acceptor substrate was modeled into the active site (Kakuta et al., 2003). Mutations were then made to residues lining the cleft on the 3′SB helix as well as a loop coined “sweet hill”, with a unique insert of residues (640–649) on the opposite side of the cleft. Residues from this motif, including the proposed catalytic base Glu642, were shown to be important for activity (Kakuta et al., 2003).
Fig. 6.
Substrate binding of the heparan sulfate sulfotransferases. (A) Crystal structure of the sulfotransferase domain of NDST-1 (PDB code 1NST) with PAP shown (Kakuta et al., 1999). The regions colored in light purple (α-helix from 3′SB motif) and wheat (“Sweet Hill” region, including Glu642, the catalytic base) have been shown to be important for substrate binding and lie along an open cleft across the active site, which is similar to that seen in 3-OST-3 (Fig. 2A). (B) Crystal structure of the 2-OST trimer with protomers shown in white, light purple, and pink (PDB code 4NZD). PAP and the octasaccharide substrate bound to each active site are shown in green and cyan, respectively. Of note, the C-terminal residues of one protomer extend into the active site of the other (Liu et al., 2014). (C) Active site of 2-OST suggests how the enzyme accommodates both IdoA and GlcA in the active site by supporting binding of a 4C1 acceptor sugar conformation that relies on Arg189. The positions of the 6-OH as shown would likely exclude 6S moieties, due to steric conflict on the reducing end and possible electrostatic repulsion with Glu349 from another protomer on the GlcNS on the nonreducing side. The acceptor hydroxyl of the octasaccharide is designated with a red asterisk, whereas the reducing and nonreducing ends are labeled r and nr, respectively. (D) Crystal structure of zf6-OST-3 (PDB code 5T0A) active site (light purple) with PAP (green) and bound heptamer (wheat) substrate (Xu et al., 2017b). Hydrogen bonds with substrate are shown in black dashed lines. The acceptor hydroxyl is designated with a red asterisk. 2-OST (gray) is superimposed with its substrate (transparent cyan). The superposition reveals 6-OST binds its substrate with opposite polarity, relative to the active site, as compared to 2-OST and 3-OSTs. In 6-OST, a loop including Thr209 (magenta) blocks the cleft found in the other heparan sulfotransferases and forms interactions with the N-sulfo moiety on the acceptor glucosamine.
The product of the N-deacetylase/N-sulfotransferase and C5-epimerization becomes the substrate for 2-OST. To obtain the crystal structure of chicken 2-OST (92% sequence identity to human), 2-OST was crystallized as an maltose binding protein fusion to enhance solubility and likelihood of crystallization (Bethea et al., 2008). Unlike the other HSSTs that appear to function as monomers, the structure revealed that 2-OST exists as a homotrimer with ∼24% of each protomer’s surface area buried at the interface with the C-terminus of one molecule contributing to the substrate binding pocket of the other (Fig. 6, B–C). Removal of this tail disrupts trimer formation and greatly diminishes activity, supporting an important role for trimerization (Bethea et al., 2008). The active sites of each protomer are located ∼50 Å apart and are believed to function independently of one another (Liu et al., 2014). The structure revealed extensive interactions with five saccharides in the bound heptasaccharide substrate. The N-sulfo groups on the GlcNSs flanking the acceptor uronic acid form multiple interactions, suggesting a GlcNS-containing pentasaccharide represents the minimum binding motif required for activity (Liu et al., 2014). The structure also revealed that sulfation on the 6-OH of the adjacent GlcNS on the reducing end would result in significant steric clashes, whereas 6S on the nonreducing side could create electrostatic repulsion with the Glu349 on the neighboring protomer (Fig. 6C). Taken together, the structures support the central dogma that 2-O-sulfation occurs after N-sulfation and before 6-O-sulfation. Although 2-OST can transfer the sulfo group to both GlcA and IdoA saccharides, in a heptascaccharide with a mixture of GlcA or IdoA flanked by GlcNS, IdoA2S was produced 10:1 over GlcA2S (Liu et al., 2014). IdoA is typically found in 1C4 or 2S0, whereas GlcA adopts the 4C1 conformation, suggesting that each would need to bind differently to the active site. However, the crystal structure, combined with mutagenesis, revealed that Arg189 may act to stabilize IdoA in the unexpected 4C1 conformation, allowing both IdoA and GlcA to use the same active site in similar conformations (Fig. 6C) (Liu et al., 2014). In contrast, a recent molecular modeling study suggests that for longer endogenous substrates, the IdoA would be in the preferred 1C4 conformation and that Arg189 sterically excludes GlcA, generating the preference for IdoA substrate (Gesteira et al., 2021).
Like 2-OST, the zebrafish 6-OST isoform 3 (zf6-OST-3; > 70% sequence identity to human isoforms) was crystallized as a maltose binding protein fusion (Xu et al., 2017b). Three crystal structures were obtained with three oligosaccharides (a hepta- and two hexasaccarides) comprised of different sequences that bound in similar conformations. Although the 6-OSTs share many features with the other HSSTs, the orientation of substrate binding is completely different, with the substrate oriented almost perpendicular to that observed in the 2-OST and 3-OST crystal structures (Fig. 6D). In all three 6-OST structures, the nonreducing end GlcNS is positioned with the 6-hydroxyl in the catalytic position. The structures revealed that the reducing end of the 2-OST and 3-OST cleft is occluded by the loop immediately after the 3′SB helix, structurally equivalent to that of loop 2 in the SULTs (Fig. 6D). The 6-OSTs are the most promiscuous of the HSSTs with 6-OST-1 requiring only a trisaccharide composed of two glucosamines (GlcNS or GlcNAc) and a central uronic acid (GlcA, IdoA, or IdoA2S) (Liu and Liu, 2011). The lack of specificity is explained by the crystal structure, which shows that the majority of the interactions are with the acceptor GlcNS and the -GlcA-GlcNS- on its reducing side (Xu et al., 2017b). The NS on the acceptor GlcNS forms multiple interactions with the occluding “loop 2,” consistent with the preference of GlcNS substrate over GlcNAc. The lack of strict specificity by these enzymes may be important to generate a larger pool of diverse HS. 6-OSTs are capable of sulfating on adjacent GlcNS-GlcA/IdoAS repeat sequences. Although all the substrates in the crystal structures bind across the reducing end of the binding interface, one of the substrates has an additional GlcA on the nonreducing side of the acceptor GlcNS, suggesting the path of the nonreducing end, if it were present, would extend across the PAPS binding site (Fig. 6D).
Humans have three isoforms of 6-OST, and the extent of specificity differences between them has been up for debate (Habuchi et al., 2000). Mapping the conserved residues of the three human isoforms onto the zf6-OST-3 revealed that all residues interacting with the substrate were conserved. This, combined with recent results of activity between the isoforms on different homogeneous substrates, supports similar specificities between the isoforms (Xu et al., 2017b).
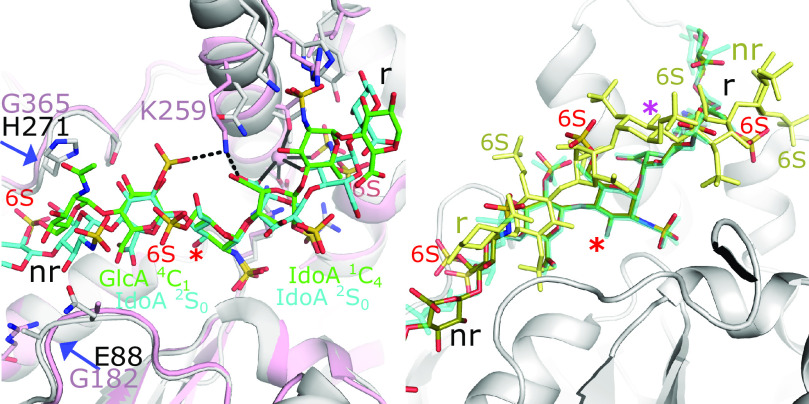
The 3-OSTs appear to have the strictest specificity requirements of all the HSSTs (Shworak et al., 1999; Liu and Pedersen, 2007). Of the seven human isoforms, the specificity of three (3-OST-1, 3-OST-3, and 3-OST-5) have been well characterized and examined structurally (Xu et al., 2008; Moon et al., 2004, 2012; Wander et al., 2021). 3-OST-1 is responsible for producing the pentasaccharide sequence GlcNS6S-GlcA-GlcNS6S3S-IdoA2S-GlcNS6S with strong anticoagulant activity (Liu et al., 1996; Liu and Pedersen, 2007). Alternatively, 3-OST-3 is involved in the production of the HS co-entry receptor for HSV-1, with the petasaccharide sequence GlcNS6S-IdoA2S-GlcNS6S3S-IdoA2S-GlcNS6S (Shukla et al., 1999). 3-OST-5 is more promiscuous and can generate both substrates (Xia et al., 2002). Crystal structures exist for all three of these enzymes in binary complexes with PAP, and acceptor substrate-bound structures exist for 3-OST-1 and 3-OST-3 (Edavettal et al., 2004; Moon et al., 2004, 2012; Xu et al., 2008; Wander et al., 2021). Major differences in substrate specificities exist between the isoforms. 3-OST-1 prefers substrates that are 6-O-sulfated and contain a GlcA on the nonreducing side of the acceptor GlcNS, whereas 3-OST-3 prefers substrates lacking 6S that contain Ido2S saccharides flanking the acceptor GlcNS on both sides (Wang et al., 2017). Although substrates for both enzymes occupy the similar substrate cleft as found in NST and 2-OST, the conformations of the uronic acids differ in 3-OST-1 and -3, resulting in different interactions with the enzyme (Fig. 7A). For 3-OST-1 binding, the GlcA and IdoA2S are in the 4C1 and 1C4 conformation, respectively (Moon et al., 2012). For 3-OST-3, both IdoA2Ss are found in the 2S0 conformation (Fig. 7A) (Wander et al., 2021). Lys259 in 3-OST-3, not conserved in 3-OST-1, along with a Na+ ion binding site on the reducing side have been suggested to contribute to 3-OST-3 specificity (Fig. 7A) (Moon et al., 2004; Wander et al., 2021). In addition, Lys259 may help to alleviate charge repulsions between the sulfo group of the Ido2S and the carboxylate group of the Ido2S on the nonreducing and reducing sides of the acceptor GlcNS, respectively. 3-OST-1 lacks these features, contributing to its inability to accommodate a IdoA2S on the nonreducing side of the acceptor.
Fig. 7.
Substrate binding to 3-OST-1 and 3-OST-3 active sites. (A) Crystal structure of 3-OST-3 (pink) with bound NS2S 8mer oligosaccharide (cyan) (PDB code 6XL8) superimposed with 3-OST-1 (gray) with bound heptasaccharide (green) (PDB code 3UAN) (Moon et al., 2012; Wander et al., 2021). Residue side chains that differ between the two isoforms lining the substrate binding pocket are drawn in stick. Hydrogen bonds are depicted with dashed black lines and interactions with the Na+ ion (pink) involved in substrate binding to 3-OST-3 are shown in solid black lines. The positions of the two uronic acids flanking the acceptor glucosamine, the acceptor 3-OH on the glucosamine, and the reducing (r) and nonreducing (nr) ends of the oligosaccharide are labeled. Residues associated with the nonreducing end substrate "gate” and labeled and denoted with blue arrows. (B) Crystal structure of 3-OST-3 with the productive binding mode of NS2S6S containing 8mer bound to one protomer (olive) with the position of the NS2S6S containing 8mer in the nonproductive mode superimposed (all yellow) (PDB code 6XK6) (Wander et al., 2021). The nonproductive binding mode has reversed polarity with respect to the active site and the acceptor substrate is not in position for catalysis (magenta asterisk). The correct position for the acceptor 3OH hydroxyl is designated with a red asterisk. The NS2S substrate from the 3-OST-3 structure (PDB code 6XL8) is also superimposed (cyan) and displays very similar binding to productive positioning of the NS2S6S.
Based on the structures, a substrate “gate” on the nonreducing side of the substrate binding cleft has been identified (Fig. 7A) (Xu et al., 2008). Having a 4C1 GlcA (3-OST-1) versus a 2S0 IdoA2S (3-OST-3) on the nonreducing side of the acceptor GlcNS results in a different trajectory of the oligosaccharide through the gate for each substrate, suggesting that the different sequences for the isoforms at the gate also contribute to specificity, which has been supported with mutagenesis studies (Xu et al., 2008; Wander et al., 2021).
Whereas the 6S on two of the three ordered GlcNS6S saccharides in the 3-OST-1 heptaasaccharide substrate forms interactions with the enzyme, the 6S on the acceptor glucosamine is solvent-exposed (Moon et al., 2012). Due to the decrease in activity for 3-OST-3 on 6-O-sulfated substrates, it was originally hypothesized that 6S would generate steric and/or electrostatic clashes while binding to 3-OST-3. However, a crystal structure at 1.55 Å with two 3-OST-3 molecules in the asymmetric unit demonstrated that a 6S-containing octasaccharide was easily accommodated and, surprisingly, could bind in the same conformation as the octasaccharide lacking 6S (Fig. 7B) (Wander et al., 2021). In contrast, the other 3-OST-3 molecule in the asymmetric unit bound the 6S-containing octasaccharide in a nonproductive manner, binding with the opposite polarity across the binding cleft (Fig. 7B, yellow molecule). The structural information led to further analysis revealing that the 6S-containing substrate bound with 10-fold higher affinity and displayed a mix of product and substrate inhibition contributing to the perceived specificity difference (Wander et al., 2021).
Understanding Disease through Sulfotransferase Structures
Missense genetic polymorphisms in sulfotransferases have been linked to differential metabolism of hormones and drugs by the SULTs as well as effecting activity of the HSSTs (Tornberg et al., 2011; Reuter et al., 2014; Schneeberger et al., 2020; Kurogi et al., 2021). These polymorphisms have been linked to diseases and conditions including malaria, cancer, congenital ichthyosis, idiopathic hypogonadotropic hypogonadism, and other developmental disorders including neurologic, skeletal, and renal abnormalities (Table 2). Crystal structure analysis allows for hypothesizing of how these changes may affect function. Although some of the polymorphisms are located distally from the active site and may reflect changes in the protein stability or interactions with binding partners, many of these mutations have been linked to the substrate binding loops 2 and 3 for SULTs 1A1, 1B1, 1E1, and 2B1 and could impact substrate binding and/or catalysis (Chung et al., 2009; Cohen et al., 2009; Kinnersley et al., 2016; Heinz et al., 2017). Variants associated with disease also have been found lining the substrate binding clefts of NDST-1, 2-OST, and 6-OST-1 (Najmabadi et al., 2011; Tornberg et al., 2011; Reuter et al., 2014; Schneeberger et al., 2020). Disease-associated variants are also found clustered within the highly conserved 3′SB and 5′PSB strand-loop-helix motifs associated with PAPS and acceptor substrate binding (Reuter et al., 2014; Tibbs et al., 2018; Schneeberger et al., 2020). One of the variants for NDST-1 involves the proposed catalytic base, whereas the R189S of 2-OST involves the residue proposed to dictate specificity for IdoA over GluA (Reuter et al., 2014; Schneeberger et al., 2020). The R189S mutation would likely not inhibit GlcA binding, but would lose reactivity to IdoA, as suggested by the R189A in vitro studies (Liu et al., 2014).
Table 2.
Polymorphisms of human sulfotransferases with potential clinical significance
| Gene | Reference SNP Cluster ID | Impact | Amino Acid Change | Location in Structure |
|---|---|---|---|---|
| SULT1A1 | rs1042008 | Increased risk of oral squamous cell carcinoma (Chung et al., 2009) | His149Tyr | Loop 2 |
| rs1801030 | Endometrial cancer (Rebbeck et al., 2006) | Met223Vala | Pre-loop 3 | |
| SULT1A2 | rs4987024 | Inversely associated with bladder cancer risk (Figueroa et al., 2008) | Tyr62PheTyr62Cys | Away from active site |
| SULT1B1 | rs11569729 | Higher odds ratio in glioblastoma gliomas (Kinnersley et al., 2016) | Thr261Met | Loop 3 |
| SULT1C4 | Rs1402467 | Increased relapse rate in acute myeloblastic leukemia (Monzo et al., 2006) | Asp5Glua | N-terminus not in structure |
| SULT1E1 | Increased risk of breast cancer (Cohen et al., 2009) | His224Gln | Pre-loop 3 | |
| SULT2B1 | rs140526640 | May relate to autosomal-recessive congenital ichthyosis (Youssefian et al., 2019) | Glu78Lysa | 5′PSB-loop helix Buried H-bond to Arg100 |
| rs1303127476 | May relate to autosomal-recessive congenital ichthyosis (Youssefian et al., 2019) | Arg100Trpa | Buried H-bond to Glu78 | |
| rs1114167424 | May cause autosomal-recessive congenital ichthyosis (Heinz et al., 2017) | Pro149Arg | 3′SB loop | |
| rs1052131 | Associated with esophageal squamous cell carcinoma risk (Hong et al., 2019) | Asp316Glu | C-terminus not ordered | |
| rs762765702 | May cause autosomal-recessive ichthyosis (Heinz et al., 2017) | Arg274Gln | Loop 3, binds 3′- phosphate of PAPS | |
| NDST1 | rs606231456 | Significant overlap in both demonstrated and apparent intellectual disability, muscular hypotonia, epilepsy, and postnatal growth deficiency (Najmabadi et al., 2011; Reuter et al., 2014) | Arg709Gln | On 3′SB helix |
| rs606231457 | Significant overlap in both demonstrated and apparent intellectual disability, muscular hypotonia, epilepsy, and postnatal growth deficiency (Reuter et al., 2014). | Glu642Asp | Catalytic base on Sweet Hill | |
| rs606231458 | Associated with intellectual disability, muscular hypotonia, epilepsy, and postnatal growth deficiency (Reuter et al., 2014) | Phe640Leu | On Sweet Hill | |
| rs606231459 | Associated with intellectual disability, muscular hypotonia, epilepsy, and postnatal growth deficiency (Reuter et al., 2014) | Gly611Ser | 5′PSB-loop | |
| HS2ST1b | rs758990524 | Associated with a syndromic phenotype comprising neurologic, skeletal, and renal abnormalities (Schneeberger et al., 2020) | Asp165Tyr | 3′SB loop |
| rs1651972168 | Associated with a syndromic phenotype comprising neurologic, skeletal, and renal abnormalities (Schneeberger et al., 2020) | Phe176Ser | 3′SB helix. Lines substrate binding pocket | |
| rs1651973144 | Associated with a syndromic phenotype comprising neurologic, skeletal, and renal abnormalities (Schneeberger et al., 2020). | Arg189Ser | Binds acceptor saccharide. Determinant residue for IdoA sulfation | |
| HS3ST3A1 | rs60532842 | Associated with P. falciparum parasitaemia (Atkinson et al., 2012) | Ala85Ser | N-terminal of sulfotransferase domain |
| HS3ST3B1c | rs9906590 | Associated with P. falciparum parasitaemia (Atkinson et al., 2012; Nguyen et al., 2018) | Glu363LysGlu363Gln | Away from active site |
| rs62056073 | Associated with P. falciparum parasitaemia (Atkinson et al., 2012; Nguyen et al., 2018) | Ile196ValIle196Phe | Away from active site | |
| rs62636623 | Associated with P. falciparum parasitaemia (Atkinson et al., 2012; Nguyen et al., 2018) | Gly83ArgGly83Trp | N-terminal of sulfotransferase domain | |
| HS6ST1d | rs780352591 | Found in hypogonadotropic hypogonadism patients (Tornberg et al., 2011) | Arg306Trp1 | Away from active site. |
| rs201307896 | Found in hypogonadotropic hypogonadism patients (Tornberg et al., 2011) | Arg306Glne | Away from active site | |
| rs761325768 | Found in hypogonadotropic hypogonadism patients (Tornberg et al., 2011) | Arg323Glne | Lines non-reducing end of substrate binding pocket | |
| rs199538589 | Found in hypogonadotropic hypogonadism patients (Tornberg et al., 2011; Cangiano et al., 2019; Choi et al., 2015) | Arg382Trpe | Extended C-terminal tail away from active site | |
| Found in hypogonadotropic hypogonadism patients (Tornberg et al., 2011) | Met404Vale | Extended C-terminal tail away from active site | ||
| HS6ST2d | rs866919041 | Associated with X-linked intellectual disability and severe myopia in two male twins (Paganini et al., 2019). | Gly306CysGly306Arg | Disordered loop away from active site |
List was obtained by utilizing the NSBI dbSNP search with clinical significance (likely pathogenic, pathogenic, and pathogenic likely pathogenic) and function class (missense).
anot found in NSBI dbSNP using listed criteria.
bHS2ST analysis based on crystal structure of chicken 2-OST PDB code 4NDZ.
cH3ST3B1 structural analysis based on structure of equivalent residue in H3ST3A1 PDB code 6XL8.
dHS6ST1 and HS6ST2 structural analysis based on crystal structure of zebrafish 6-OST-3 PDB id code 5T0A.
eNumbering of Arg306, Arg323, Arg382, and Met404 are listed as Arg296, Arg313, Arg372, and Met294 in Tornberg et al. (2011).
ST Structures in Molecular Modeling
The value of sulfotransferase crystal structures has extended beyond basic understanding of the enzymatic functions, serving as a starting point for in silico calculations to predict substrates and inhibitors and better understand how conformational dynamics affect substrate binding. Early work applied crystal structures and homology models to an adaptation of the three-dimensional quantitative structure-activity relationship, referred to as comparative molecular field analysis, to better understand enzyme kinetics and help predict specificity of rat ASTIV and SULT1A3 (Sharma and Duffel, 2002, 2005; Sipilä et al., 2003). In silico docking based on crystal structures was used to determine that PAPS binding can regulate binding of larger substrates such as Raloxifene via conformational dynamics of loop 3 (Cook et al., 2012). Quantitive structure-activity relationship was later combined with molecular dynamics (MD) to consider both ligand and protein flexibility to successfully identify ligands of isoforms SULT1A1, SULT1A3, and SULT1E1 (Martiny et al., 2013). Others have used MD to study thermal stability and conformational flexibility associated with ligand binding in SULT2A1 (Zhao et al., 2015, 2016; Zhou et al., 2019; Zhu et al., 2019). Utilizing molecular dynamics and an FDA-approved small molecule drug database, Cook et al. (2013b) were able to accurately predict known substrates and identify novel substrates for SULT1A1 and SULT2A1. Further MD analysis of SULT1A1 revealed a molecular clamp by two aromatic residues that reposition to “sandwich”, in a catalytically relevant position, the phenolic substrates that display enhanced affinities for binding in the presence of saturating nucleotide (Cook et al., 2015b). In contrast, substrates unaffected by nucleotide binding tend to “wander” in the binding pocket, rarely sampling a catalytically relevant position (Cook et al., 2015b). Analysis using MD simulations was also used to understand substrate inhibition differences between mouse and human SULT1E1, suggesting that a subpocket within the binding pocket of hSULT1E1 could accommodate the substrate in a noncatalytic binding mode (Stjernschantz et al., 2010). A recent approach in MD applied to SULT1A1 that includes the use of excited normal modes has been used to sample more conformational space than classic MD and may provide more insight into substrate and/or inhibitor binding (Dudas et al., 2021).
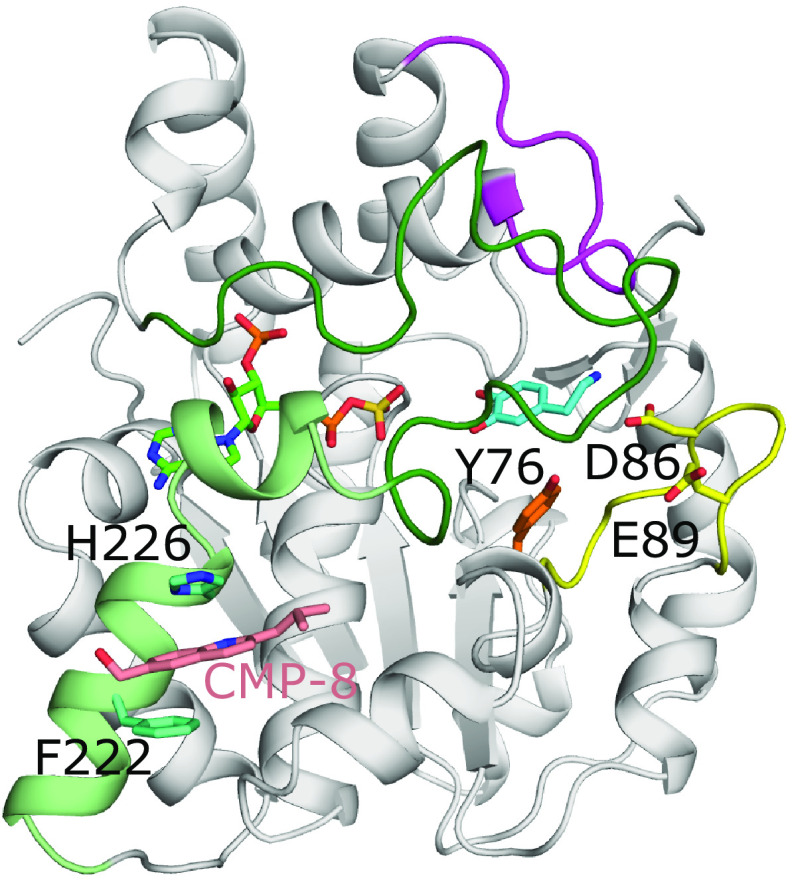
Molecular docking experiments have also been used to better understand how allosteric interactions can regulate sulfotransferase function. SULT1A1 and 1A3 have been shown to contain two allosteric binding sites (Fig. 8) (Cook et al., 2015a, 2017). Utilizing the crystal structure of SULT1A1 as a starting model of SULT1A3, monoamine neurotransmitter metabolites were screened against the known catechin-binding site of SULT1A3 predicting tetrahydrobiopterin, an essential cofactor in monoamine neurotransmitter biosynthesis, as a potential allosteric regulator of SULT1A3 activity. When tested, tetrahydrobiopterin had a Ki = 23nM for SULT1A3, but with no detectable affinity for SULT1A1, SULT1E1, SULT2A1, or SULT2B1 (Cook et al., 2017). Binding to the catechin-binding site, located at the entrance to the substrate binding pocket between loops 1 and 3, was confirmed through mutagenesis and spin-labeled triangulation NMR experiments (Cook et al., 2017). Darrah et al. (2019) identified a SULT1A3 specific inhibitor which also inhibited the sulfotransferase activity at nanomolar concentrations (Ki = ∼34nM). Using spin-labeled triangulation NMR, combined with distance constraint molecular dynamics of models derived from crystal structures, a novel allosteric site specific to SULT1A3 was identified and confirmed by mutagenesis of surrounding residues (Darrah et al., 2019). This site is located outside the active site prior to loop 3, in a region disordered in the crystal structure of SULT1A3 when no substrates are present, that must open and close during catalysis to bind and release substrate (Fig. 8) (Bidwell et al., 1999; Wang et al., 2014a). A subsequent study identified two other structurally similar compounds that also inhibited with nanomolar Kis and were specific to SULT1A3 when compared with SULT1A1, SULT1E1, and SULT2A1. These compounds also inhibited SULT1A3 in cultured cells (Cook et al., 2021). Further molecular dynamics analysis supported the assessment that these compounds inhibit via loop 3 stabilization (Cook et al., 2021).
Fig. 8.
The allosteric binding sites of SULT1A3. Model of SULT1A3 with bound PAPS (green) and inhibitor CMP-8 (coral) bound to an allosteric site via aromatic stacking with His226 and Phe222 (model found at: https://www.modelarchive.org/doi/10.5452/ma-qtj80) (Darrah et al., 2019). The position of the acceptor substrate dopamine (cyan) was superimposed from PDB coordinates 2A3R (Lu et al., 2005). Also shown are residues Tyr76 (orange), Asp86, and Glu89 (both yellow) that compose the catechin-binding allosteric site that binds the monoamine neurotransmitter cofactor tetrahydrobiopterin (Cook et al., 2017). Loops 1, 2, and 3 are colored yellow, magenta, and dark green, respectively, as seen in SULT1E1 (Fig. 4). In the crystal structure of SULT1A3 without substrate present, residues 216-261 are disordered. This comprises loop 3 and the two helices (light green) N-terminal to loop 3 that make up one of the allosteric binding sites (PDB code 1CJM) (Bidwell et al., 1999).
Modeling and molecular dynamics studies of HS substrates using the crystal structure of NST-1 with bound PAP has provided insight into residues that are involved in substrate recognition and influence catalysis (Gorokhov et al., 2000; Kakuta et al., 2003; Gesteira et al., 2013). Molecular dynamics was also used to provide information on global dynamics, residues involved in binding, and a possible route for HS during modification by NDST-1 and 2-OST (Gesteira and Coulson-Thomas, 2018). Conformational dynamics has also been investigated for TPST-2 (Singh et al., 2016a, 2016b).
In addition to these dynamic studies, quantum mechanics calculations as well as Quantum mechanics/molecular mechanics studies have been performed on SULT1E1, 3-OST-3, 2-OST, and 6-OST to better understand the underlying catalytic mechanisms (Bartolotti et al., 1999; Lin et al., 2006; Sousa et al., 2016; Gesteira et al., 2021). Quantum mechanics/molecular mechanics has also been applied to investigate the potential endocrine disrupting effects of hydroxylated bromodiphenyl ethers from environmental exposures via inhibition of SULT1A1 (Ma et al., 2021).
Future Directions: Utilizing Sulfotransferase Structures for the Development of New Pharmacological Tools
Crystal structures can be powerful aids in the development of novel therapeutics. The Leyh group has used structural information on the closing of loop 3 in the presence of PAPS to design compounds that can interact with receptors but cannot be effectively sulfated by SULTs, which should increase the half-life of compounds in vivo (Cook et al., 2016). Such work could be useful for improved birth control bioavailability or treatment of Parkinson’s disease (Cook et al., 2016). Another example is the prodrug Oxamniquine used to treat schistosomiasis, which becomes activated through sulfation by cytosolic sulfotransferases. Resistance has emerged due to sulfotransferase variants. Crystal structures of these helminth sulfotransferases are being evaluated to help design new broad-spectrum oxamniquine derivatives that kill both S. mansoni and S hematobium species (Valentim et al., 2013; Guzman et al., 2020).
Historically, the design of effective therapeutic inhibitors has been hampered by inability to obtain a high degree of specificity for individual SULTs. The recent discovery of a selective allosteric site in SULT1A3 has changed that (Cook et al., 2021). SULT1A3 is the isoform responsible for sulfating 80% of serotonin levels in the brain, resulting in decreased levels of the active neurotransmitter. Regulating unmodified serotonin levels is important for the treatment of depressive disorders. Molecular dynamics of SULT1A3 structural models, in combination with spin-labeled triangulation NMR, appears to have been critical for determining the allosteric binding pocket specific for SULT1A3. Taking advantage of ways to specifically inhibit SULT1A3 should greatly aid current and future therapeutics studies (Cook et al., 2021).
High throughput screening has recently been used to search for inhibitors of the Golgi-resident TPST and GAG sulfotransferases (Cheung et al., 2017; Byrne et al., 2018a, 2018b). Positive hits were evaluated by docking to crystal structures to validate their ability to bind to the active site (Byrne et al., 2018a, 2018b). Crystal structures of the TPSTs and HS 2-OST are also being used for structure-based design of nucleotide analog inhibitors (Kershaw et al., 2019). Such initial inhibitors could be used to improve specificity to TPSTs or 2-OST to probe function or potentially modulate disease states such as viral infection and inflammation (Byrne et al., 2018a, 2018b; Kershaw et al., 2019).
A trending method for generation of pharmacological tools to probe physiologic and pathophysiological pathways and potential therapeutics is utilization of a chemoenzymatic approach to synthesize homogeneous GAG oligosaccharides. This method makes use of recombinant glycosyltransferases and sulfotransferases to efficiently produce the desired product, in contrast to tediously purifying heterogeneous compounds from source (Xu et al., 2011, 2017a; Zhang et al., 2020). Currently this process is being used for the development of improved anticoagulant and autoinflammatory therapeutics to treat acute liver failure due to acetaminophen overdose (Xu et al., 2014, 2017a; Arnold et al., 2020; Wang et al., 2021).
Based on crystal structures of sulfotransferases engaged with oligosaccharide substrates, some of these enzymes have been engineered via site-directed mutagenesis to alter their specificities to improve control of the chemoenzymatic pathway (Xu et al., 2008, 2017b; Liu et al., 2012, 2014; Yi et al., 2020). Particularly, a triple mutation of zf6-OST-3 designed based on the crystal structures allows for sulfation only on the nonreducing terminal glucosamine, providing fine-tuned control of 6-O-sulfation within the chemoenzymatic synthesis pathway (Yi et al., 2020). Cytosolic sulfotransferases are also being investigated for their ability to generate sulfated products for biologic reagents and drug discovery (Lamb et al., 2006; Shi et al., 2012; Shimohira et al., 2017; Matsumura et al., 2018; Xie et al., 2020).
Historically, two obstacles to using sulfotransferases for product development were product inhibition by PAP and the cost of PAPS. Recycling PAP efficiently circumvents both obstacles by utilizing the cytosolic sulfotransferase rat ASTIV to regenerate PAPS from PAP and p-nitrophenyl sulfate (Burkart et al., 1999, 2000). In concert with production of the desired sulfated product, recycling of PAP reduces both the need for additional PAPS and reduces PAP inhibition by keeping the PAP concentration low (Burkart et al., 1999, 2000; Chen et al., 2005; Datta et al., 2020). Recent work has sought to improve upon the efficiency of this approach by re-engineering ASTIV (Zhou et al., 2019). Information obtained from the crystal structure of SULT1A1 was used to choose the loop sequence between Lys81 and Thr95 for molecular evolution that identified the double mutant L89S/E90L to have improved efficiency. These residues were then further subjected to site-saturation mutagenesis to systematically test for improved efficiency. Zhou et al. (2019) identified the variant L89M/E90Q, which has a 2-fold lower KM and a 10-fold higher catalytic efficacy than the wild-type enzymes. This mutant could prove useful to improve chemoenzymatic synthesis by sulfotransferases. Thus, combining engineered SULTs with engineered GAG sulfotransferases could be useful in the discovery and production of pharmacological tools for probing pathways and designing therapeutics.
Conclusion
Crystal structures of the SULTs, HSSTs, and TPSTs have provided a crucial foundation of structural information to better understand the mechanisms and specificities of these enzymes. These structures are proving useful for in silico substrate and inhibitor screening from chemical databases as well as for inhibitor design. Mutational engineering based on the sulfotransferase structures has proven successful in altering specificity and kinetic properties. Understanding how the sulfated products of these enzymes interact with their downstream receptors and targets will likely stimulate an interactive, iterative process of redesigning the specificity of the sulfotransferases to enhance specificity of the products with their targets. The design of inhibitors along with the use of sulfotransferases in synthesizing compounds should enhance the production of pharmacological tools for probing pathways and the design of novel therapeutics.
Acknowledgments
The authors thank J. Krahn and L. Perera for critical reading of the manuscript and Dr. Masahiko Negishi for his mentoring and friendship as well as unwavering support of structural biology at the National Institutes of Health, National Institute of Environmental Health Sciences.
Abbreviations
- APS
adenosine 5′-phosphosulfate
- GAG
glycosaminoglycan
- GlcA
glucuronic acid
- GlcNAc
N-acetylglucosamine
- HSST
heparan sulfate sulfotransferase
- IdoA
Iduronic acid
- MD
molecular dynamics
- NDST-1
N-deacetylase/N-sulfotransferase isoform 1
- NST-1
sulfotransferase domain of NDST-1
- OST
O-sulfotransferase
- PAP
3′-phosphoadenosine 5′-phosphate
- PAPS
3′-phosphoadenosine 5′-phosphosulfate
- PDB
protein data bank
- PSB
phosphosulfate binding
- SB
sulfate binding
- SULT
cytosolic sulfotransferase
- TPST
Tyrosyl protein sulfotransferase
Authorship Contributions
Performed data analysis: L.C. Pedersen, Yi.
Wrote or contributed to the writing of the manuscript: L.C. Pedersen, Yi, L.G. Pedersen, Kaminski.
Footnotes
This work was supported by the Division of Intramural Research of the National Institutes of Health, National Institutes of Environmental Health Sciences [Grant 1ZIA-ES102645] (to L.C.P).
References
- Alcolombri U, Elias M, Tawfik DS (2011) Directed evolution of sulfotransferases and paraoxonases by ancestral libraries. J Mol Biol 411:837–853. [DOI] [PubMed] [Google Scholar]
- Allali-Hassani APan PWDombrovski LNajmanovich RTempel WDong ALoppnau PMartin FThornton JEdwards AM, et al. (2007) Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol 5:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold KXu YSparkenbaugh EMLi MHan XZhang XXia KPiegore MZhang FZhang X, et al. (2020) Design of anti-inflammatory heparan sulfate to protect against acetaminophen-induced acute liver failure. Sci Transl Med 12:eaav8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson A, Garnier S, Afridi S, Fumoux F, Rihet P (2012) Genetic variations in genes involved in heparan sulphate biosynthesis are associated with Plasmodium falciparum parasitaemia: a familial study in Burkina Faso. Malar J 11:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle PA, Huttner WB (1985) Tyrosine sulfation of yolk proteins 1, 2, and 3 in Drosophila melanogaster. J Biol Chem 260:6434–6439. [PubMed] [Google Scholar]
- Bartolotti L, Kakuta Y, Pedersen L, Negishi M, Pedersen L (1999) A quantum mechanical study of the transfer of biological sulfate. J Mol Struc-Theochem 461:105–111. [Google Scholar]
- Baumann E (1876) Ueber sulfosäuren im harn. Dtsch Chem Ges:54–58. [Google Scholar]
- Beisswanger R, Corbeil D, Vannier C, Thiele C, Dohrmann U, Kellner R, Ashman K, Niehrs C, Huttner WB (1998) Existence of distinct tyrosylprotein sulfotransferase genes: molecular characterization of tyrosylprotein sulfotransferase-2. Proc Natl Acad Sci USA 95:11134–11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger I, Guttman C, Amar D, Zarivach R, Aharoni A (2011) The molecular basis for the broad substrate specificity of human sulfotransferase 1A1. PLoS One 6:e26794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethea HN, Xu D, Liu J, Pedersen LC (2008) Redirecting the substrate specificity of heparan sulfate 2-O-sulfotransferase by structurally guided mutagenesis. Proc Natl Acad Sci USA 105:18724–18729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelheim FR (1954) Tyrosine-o-sulfate in a peptide from fibrinogen. J Am Chem Soc 76:2838–2839. [Google Scholar]
- Bidwell LM, McManus ME, Gaedigk A, Kakuta Y, Negishi M, Pedersen L, Martin JL (1999) Crystal structure of human catecholamine sulfotransferase. J Mol Biol 293:521–530. [DOI] [PubMed] [Google Scholar]
- Bishop JR, Schuksz M, Esko JD (2007) Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446:1030–1037. [DOI] [PubMed] [Google Scholar]
- Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW (2004) A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics 14:199–211. [DOI] [PubMed] [Google Scholar]
- Bond CS, Clements PR, Ashby SJ, Collyer CA, Harrop SJ, Hopwood JJ, Guss JM (1997) Structure of a human lysosomal sulfatase. Structure 5:277–289. [DOI] [PubMed] [Google Scholar]
- Buhl AE, Waldon DJ, Baker CA, Johnson GA (1990) Minoxidil sulfate is the active metabolite that stimulates hair follicles. J Invest Dermatol 95:553–557. [DOI] [PubMed] [Google Scholar]
- Burkart MD, Izumi M, Chapman E, Lin C-H, Wong C-H (2000) Regeneration of PAPS for the enzymatic synthesis of sulfated oligosaccharides. J Org Chem 65:5565–5574. [DOI] [PubMed] [Google Scholar]
- Burkart MD, Izumi M, Wong CH (1999) Enzymatic regeneration of 3′-phosphoadenosine-5′-phosphosulfate using aryl sulfotransferase for the preparative enzymatic synthesis of sulfated carbohydrates. Angew Chem Int Ed Engl 38:2747–2750. [DOI] [PubMed] [Google Scholar]
- Byrne DPLi YNgamlert PRamakrishnan KEyers CEWells CDrewry DHZuercher WJBerry NGFernig DG, et al. (2018a) New tools for evaluating protein tyrosine sulfation: tyrosylprotein sulfotransferases (TPSTs) are novel targets for RAF protein kinase inhibitors. Biochem J 475:2435–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne DPLi YRamakrishnan KBarsukov ILYates EAEyers CEPapy-Garcia DChantepie SPagadala VLiu J, et al. (2018b) New tools for carbohydrate sulfation analysis: heparan sulfate 2-O-sulfotransferase (HS2ST) is a target for small-molecule protein kinase inhibitors. Biochem J 475:2417–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cangiano B, Duminuco P, Vezzoli V, Guizzardi F, Chiodini I, Corona G, Maggi M, Persani L, Bonomi M (2019) Evidence for a common genetic origin of classic and milder adult-onset forms of isolated hypogonadotropic hypogonadism. J Clin Med 8:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HJ, Shi R, Rehse P, Lin SX (2004) Identifying androsterone (ADT) as a cognate substrate for human dehydroepiandrosterone sulfotransferase (DHEA-ST) important for steroid homeostasis: structure of the enzyme-ADT complex. J Biol Chem 279:2689–2696. [DOI] [PubMed] [Google Scholar]
- Chapman E, Bryan MC, Wong CH (2003) Mechanistic studies of beta-arylsulfotransferase IV. Proc Natl Acad Sci USA 100:910–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee B, Majumdar D, Ozbilen O, Murty CVR, Roy AK (1987) Molecular cloning and characterization of cDNA for androgen-repressible rat liver protein, SMP-2. J Biol Chem 262:822–825. [PubMed] [Google Scholar]
- Chen J, Avci FY, Muñoz EM, McDowell LM, Chen M, Pedersen LC, Zhang L, Linhardt RJ, Liu J (2005) Enzymatic redesigning of biologically active heparan sulfate. J Biol Chem 280:42817–42825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung ST, Miller MS, Pacoma R, Roland J, Liu J, Schumacher AM, Hsieh-Wilson LC (2017) Discovery of a small-molecule modulator of glycosaminoglycan sulfation. ACS Chem Biol 12:3126–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JHBalasubramanian RLee PHShaw NDHall JEPlummer LBuck CLKottler MLJarzabek KWołczynski S, et al. (2015) Expanding the spectrum of founder mutations causing isolated gonadotropin-releasing hormone deficiency. J Clin Endocrinol Metab 100:E1378–E1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YT, Hsieh LL, Chen IH, Liao CT, Liou SH, Chi CW, Ueng YF, Liu TY (2009) Sulfotransferase 1A1 haplotypes associated with oral squamous cell carcinoma susceptibility in male Taiwanese. Carcinogenesis 30:286–294. [DOI] [PubMed] [Google Scholar]
- Cohen S, Laitman Y, Kaufman B, Milgrom R, Nir U, Friedman E (2009) SULT1E1 and ID2 genes as candidates for inherited predisposition to breast and ovarian cancer in Jewish women. Fam Cancer 8:135–144. [DOI] [PubMed] [Google Scholar]
- Cook I, Cacace M, Wang T, Darrah K, Deiters A, Leyh TS (2021) Small-molecule control of neurotransmitter sulfonation. J Biol Chem 296:100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook I, Wang T, Almo SC, Kim J, Falany CN, Leyh TS (2013a) The gate that governs sulfotransferase selectivity. Biochemistry 52:415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook I, Wang T, Falany CN, Leyh TS (2012) A nucleotide-gated molecular pore selects sulfotransferase substrates. Biochemistry 51:5674–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook I, Wang T, Falany CN, Leyh TS (2013b) High accuracy in silico sulfotransferase models. J Biol Chem 288:34494–34501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook I, Wang T, Falany CN, Leyh TS (2015a) The allosteric binding sites of sulfotransferase 1A1. Drug Metab Dispos 43:418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook I, Wang T, Leyh TS (2015b) Sulfotransferase 1A1 substrate selectivity: a molecular clamp mechanism. Biochemistry 54:6114–6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook I, Wang T, Leyh TS (2017) Tetrahydrobiopterin regulates monoamine neurotransmitter sulfonation. Proc Natl Acad Sci USA 114:E5317–E5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook I, Wang T, Wang W, Kopp F, Wu P, Leyh TS (2016) Controlling sulfuryl-transfer biology. Cell Chem Biol 23:579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughtrie MWH (2016) Function and organization of the human cytosolic sulfotransferase (SULT) family. Chem Biol Interact 259 (Pt A):2–7. [DOI] [PubMed] [Google Scholar]
- Dajani R, Hood AM, Coughtrie MWH (1998) A single amino acid, glu146, governs the substrate specificity of a human dopamine sulfotransferase, SULT1A3. Mol Pharmacol 54:942–948. [DOI] [PubMed] [Google Scholar]
- Danan LM, Yu Z, Ludden PJ, Jia W, Moore KL, Leary JA (2010) Catalytic mechanism of Golgi-resident human tyrosylprotein sulfotransferase-2: a mass spectrometry approach. J Am Soc Mass Spectrom 21:1633–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah K, Wang T, Cook I, Cacace M, Deiters A, Leyh TS (2019) Allosteres to regulate neurotransmitter sulfonation. J Biol Chem 294:2293–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta P, Fu L, He W, Koffas M, Dordick J, Linhardt R (2020) Expression of enzymes for 3′-phosphoadenosine-5′-phosphosulfate (PAPS) biosynthesis and their preparation for PAPS synthesis and regeneration. Appl Microbiol Biot 104:7067–7078. [DOI] [PubMed] [Google Scholar]
- Dombrovski L, Dong A, Bochkarev A, Plotnikov AN (2006) Crystal structures of human sulfotransferases SULT1B1 and SULT1C1 complexed with the cofactor product adenosine-3′- 5′-diphosphate (PAP). Proteins 64:1091–1094. [DOI] [PubMed] [Google Scholar]
- Dong D, Ako R, Wu B (2012) Crystal structures of human sulfotransferases: insights into the mechanisms of action and substrate selectivity. Expert Opin Drug Metab Toxicol 8:635–646. [DOI] [PubMed] [Google Scholar]
- Dudas B, Toth D, Perahia D, Nicot AB, Balog E, Miteva MA (2021) Insights into the substrate binding mechanism of SULT1A1 through molecular dynamics with excited normal modes simulations. Sci Rep 11:13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffel MW(2010. updated 2016) Sulfotransfearses, in Comprehensive Toxicology (McQueen CA ed) pp 367–384, Elsevier, Oxford. [Google Scholar]
- Edavettal SC, Lee KA, Negishi M, Linhardt RJ, Liu J, Pedersen LC (2004) Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1. J Biol Chem 279:25789–25797. [DOI] [PubMed] [Google Scholar]
- Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard NP, Gerard C, Sodroski J, Choe H (1999) Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96:667–676. [DOI] [PubMed] [Google Scholar]
- Figueroa JDMalats NGarcía-Closas MReal FXSilverman DKogevinas MChanock SWelch RDosemeci MLan Q, et al. (2008) Bladder cancer risk and genetic variation in AKR1C3 and other metabolizing genes. Carcinogenesis 29:1955–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folin O, Denis W (1915) The excretion of free and conjugated phenols and phenol derivatives. J Biol Chem 22:309–320. [Google Scholar]
- Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME (2006) Human sulfotransferases and their role in chemical metabolism. Toxicol Sci 90:5–22. [DOI] [PubMed] [Google Scholar]
- Gamage NU, Duggleby RG, Barnett AC, Tresillian M, Latham CF, Liyou NE, McManus ME, Martin JL (2003) Structure of a human carcinogen-converting enzyme, SULT1A1. Structural and kinetic implications of substrate inhibition. J Biol Chem 278:7655–7662. [DOI] [PubMed] [Google Scholar]
- Gamage NU, Tsvetanov S, Duggleby RG, McManus ME, Martin JL (2005) The structure of human SULT1A1 crystallized with estradiol. An insight into active site plasticity and substrate inhibition with multi-ring substrates. J Biol Chem 280:41482–41486. [DOI] [PubMed] [Google Scholar]
- Gesteira TF, Coulson-Thomas VJ (2018) Structural basis of oligosaccharide processing by glycosaminoglycan sulfotransferases. Glycobiology 28:885–897. [DOI] [PubMed] [Google Scholar]
- Gesteira TF, Marforio TD, Mueller JW, Calvaresi M, Coulson-Thomas VJ (2021) Structural determinants of substrate recognition and catalysis by heparan sulfate sulfotransferases. ACS Catal 11:10974–10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteira TF, Pol-Fachin L, Coulson-Thomas VJ, Lima MA, Verli H, Nader HB (2013) Insights into the N-sulfation mechanism: molecular dynamics simulations of the N-sulfotransferase domain of NDST1 and mutants. PLoS One 8:e70880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorokhov A, Perera L, Darden TA, Negishi M, Pedersen LC, Pedersen LG (2000) Heparan sulfate biosynthesis: a theoretical study of the initial sulfation step by N-deacetylase/N-sulfotransferase. Biophys J 79:2909–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosavi RA, Knudsen GA, Birnbaum LS, Pedersen LC (2013) Mimicking of estradiol binding by flame retardants and their metabolites: a crystallographic analysis. Environ Health Perspect 121:1194–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günal S, Hardman R, Kopriva S, Mueller JW (2019) Sulfation pathways from red to green. J Biol Chem 294:12293–12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman M, Rugel A, Tarpley RS, Cao X, McHardy SF, LoVerde PT, Taylor AB (2020) Molecular basis for hycanthone drug action in schistosome parasites. Mol Biochem Parasitol 236:111257. [DOI] [PubMed] [Google Scholar]
- Habuchi H, Tanaka M, Habuchi O, Yoshida K, Suzuki H, Ban K, Kimata K (2000) The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for hexuronic acid adjacent to the targeted N-sulfoglucosamine. J Biol Chem 275:2859–2868. [DOI] [PubMed] [Google Scholar]
- Hamers T, Kamstra JH, Sonneveld E, Murk AJ, Visser TJ, Van Velzen MJ, Brouwer A, Bergman A (2008) Biotransformation of brominated flame retardants into potentially endocrine-disrupting metabolites, with special attention to 2,2′,4,4′-tetrabromodiphenyl ether (BDE-47). Mol Nutr Food Res 52:284–298. [DOI] [PubMed] [Google Scholar]
- Hartmann-Fatu C, Trusch F, Moll CN, Michin I, Hassinen A, Kellokumpu S, Bayer P (2015) Heterodimers of tyrosylprotein sulfotransferases suggest existence of a higher organization level of transferases in the membrane of the trans-Golgi apparatus. J Mol Biol 427 (6 Pt B):1404–1412. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Orellana A, Gil G, Hirschberg CB (1992) Molecular cloning and expression of rat liver N-heparan sulfate sulfotransferase. J Biol Chem 267:15744–15750. [PubMed] [Google Scholar]
- Heinz L, Kim GJ, Marrakchi S, Christiansen J, Turki H, Rauschendorf MA, Lathrop M, Hausser I, Zimmer AD, Fischer J (2017) Mutations in SULT2B1 cause autosomal-recessive congenital ichthyosis in humans. Am J Hum Genet 100:926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heroux JA, Roth JA (1988) Physical characterization of a monoamine-sulfating form of phenol sulfotransferase from human platelets. Mol Pharmacol 34:194–199. [PubMed] [Google Scholar]
- Hille A, Braulke T, von Figura K, Huttner WB (1990) Occurrence of tyrosine sulfate in proteins–a balance sheet. 1. Secretory and lysosomal proteins. Eur J Biochem 188:577–586. [DOI] [PubMed] [Google Scholar]
- Hoff RH, Czyryca PG, Sun M, Leyh TS, Hengge AC (2006) Transition state of the sulfuryl transfer reaction of estrogen sulfotransferase. J Biol Chem 281:30645–30649. [DOI] [PubMed] [Google Scholar]
- Hong W, Guo F, Yang M, Xu D, Zhuang Z, Niu B, Bai Q, Li X (2019) Hydroxysteroid sulfotransferase 2B1 affects gastric epithelial function and carcinogenesis induced by a carcinogenic agent. Lipids Health Dis 18:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortin G, Folz R, Gordon JI, Strauss AW (1986) Characterization of sites of tyrosine sulfation in proteins and criteria for predicting their occurrence. Biochem Biophys Res Commun 141:326–333. [DOI] [PubMed] [Google Scholar]
- Howell WH, Holt E (1918) Two new factors in blood coagulation - heparin and pro- antithrombin. Am J Physiol 47:328–341. [Google Scholar]
- Idris M, Mitchell DJ, Gordon R, Sidharthan NP, Butcher NJ, Minchin RF (2020) Interaction of the brain-selective sulfotransferase SULT4A1 with other cytosolic sulfotransferases: effects on protein expression and function. Drug Metab Dispos 48:337–344. [DOI] [PubMed] [Google Scholar]
- Iozzo RV, Schaefer L (2015) Proteoglycan form and function: a comprehensive nomenclature of proteoglycans. Matrix Biol 42:11–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta Y, Li L, Pedersen LC, Pedersen LG, Negishi M (2003) Heparan sulphate N-sulphotransferase activity: reaction mechanism and substrate recognition. Biochem Soc Trans 31:331–334. [DOI] [PubMed] [Google Scholar]
- Kakuta Y, Pedersen LG, Carter CW, Negishi M, Pedersen LC (1997) Crystal structure of estrogen sulphotransferase. Nat Struct Biol 4:904–908. [DOI] [PubMed] [Google Scholar]
- Kakuta Y, Pedersen LG, Pedersen LC, Negishi M (1998a) Conserved structural motifs in the sulfotransferase family. Trends Biochem Sci 23:129–130. [DOI] [PubMed] [Google Scholar]
- Kakuta Y, Petrotchenko EV, Pedersen LC, Negishi M (1998b) The sulfuryl transfer mechanism. Crystal structure of a vanadate complex of estrogen sulfotransferase and mutational analysis. J Biol Chem 273:27325–27330. [DOI] [PubMed] [Google Scholar]
- Kakuta Y, Sueyoshi T, Negishi M, Pedersen LC (1999) Crystal structure of the sulfotransferase domain of human heparan sulfate N-deacetylase/ N-sulfotransferase 1. J Biol Chem 274:10673–10676. [DOI] [PubMed] [Google Scholar]
- Kershaw NM, Byrne DP, Parsons H, Berry NG, Fernig DG, Eyers PA, Cosstick R (2019) Structure-based design of nucleoside-derived analogues as sulfotransferase inhibitors. RSC Advances 9:32165–32173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kester MHABulduk STibboel DMeinl WGlatt HFalany CNCoughtrie MWHBergman ASafe SHKuiper GGJM, et al. (2000) Potent inhibition of estrogen sulfotransferase by hydroxylated PCB metabolites: a novel pathway explaining the estrogenic activity of PCBs. Endocrinology 141:1897–1900. [DOI] [PubMed] [Google Scholar]
- Kester MHABulduk Svan Toor HTibboel DMeinl WGlatt HFalany CNCoughtrie MWHSchuur AGBrouwer A, et al. (2002) Potent inhibition of estrogen sulfotransferase by hydroxylated metabolites of polyhalogenated aromatic hydrocarbons reveals alternative mechanism for estrogenic activity of endocrine disrupters. J Clin Endocrinol Metab 87:1142–1150. [DOI] [PubMed] [Google Scholar]
- Khan AS, Taylor BR, Chung K, Etheredge J, Gonzales R, Ringer DP (1993) Genomic structure of rat liver aryl sulfotransferase IV-encoding gene. Gene 137:321–326. [DOI] [PubMed] [Google Scholar]
- Kiehlbauch CC, Lam YF, Ringer DP (1995) Homodimeric and heterodimeric aryl sulfotransferases catalyze the sulfuric acid esterification of N-hydroxy-2-acetylaminofluorene. J Biol Chem 270:18941–18947. [DOI] [PubMed] [Google Scholar]
- Kinnersley BKamatani YLabussière MWang YGalan PMokhtari KDelattre JYGousias KSchramm JSchoemaker MJ, et al. (2016) Search for new loci and low-frequency variants influencing glioma risk by exome-array analysis. Eur J Hum Genet 24:717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurogi K, Rasool MI, Alherz FA, El Daibani AA, Bairam AF, Abunnaja MS, Yasuda S, Wilson LJ, Hui Y, Liu MC (2021) SULT genetic polymorphisms: physiological, pharmacological and clinical implications. Expert Opin Drug Metab Toxicol 17:767–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb SS, Patel T, Koteva KP, Wright GD (2006) Biosynthesis of sulfated glycopeptide antibiotics by using the sulfotransferase StaL. Chem Biol 13:171–181. [DOI] [PubMed] [Google Scholar]
- Langford R, Hurrion E, Dawson PA (2017) Genetics and pathophysiology of mammalian sulfate biology. J Genet Genomics 44:7–20. [DOI] [PubMed] [Google Scholar]
- Lee KA, Fuda H, Lee YC, Negishi M, Strott CA, Pedersen LC (2003) Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of pregnenolone and 3′-phosphoadenosine 5′-phosphate. Rationale for specificity differences between prototypical SULT2A1 and the SULT2BG1 isoforms. J Biol Chem 278:44593– 44599. [DOI] [PubMed] [Google Scholar]
- Leyh TS (1993) The physical biochemistry and molecular genetics of sulfate activation. Crit Rev Biochem Mol Biol 28:515–542. [DOI] [PubMed] [Google Scholar]
- Leyh TS, Cook I, Wang T (2013) Structure, dynamics and selectivity in the sulfotransferase family. Drug Metab Rev 45:423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyte A, van Schijndel HB, Niehrs C, Huttner WB, Verbeet MP, Mertens K, van Mourik JA (1991) Sulfation of Tyr1680 of human blood coagulation factor VIII is essential for the interaction of factor VIII with von Willebrand factor. J Biol Chem 266:740–746. [PubMed] [Google Scholar]
- Lim GB (2017) Milestone 1: discovery and purification of heparin. Nat Rev Cardiol. [DOI] [PubMed] [Google Scholar]
- Lin P, Yang WT, Pedersen LC, Negishi M, Pedersen LG (2006) Searching for the minimum energy path in the sulfuryl transfer reaction catalyzed by human estrogen sulfotransferase: role of enzyme dynamics. Int J Quantum Chem 106:2981–2998. [Google Scholar]
- Lindahl U, Couchman J, Kimata K, Esko JD (2015) Proteoglycans and Sulfated Glycosaminoglycans, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Lindahl U, Li JP (2009) Interactions between heparan sulfate and proteins-design and functional implications. Int Rev Cell Mol Biol 276:105–159. [DOI] [PubMed] [Google Scholar]
- Lipmann F (1958) Biological sulfate activation and transfer. Science 128:575–580. [DOI] [PubMed] [Google Scholar]
- Liu C, Sheng J, Krahn JM, Perera L, Xu Y, Hsieh PH, Dou W, Liu J, Pedersen LC (2014) Molecular mechanism of substrate specificity for heparan sulfate 2-O-sulfotransferase. J Biol Chem 289:13407–13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Moon AF, Sheng J, Pedersen LC (2012) Understanding the substrate specificity of the heparan sulfate sulfotransferases by an integrated biosynthetic and crystallographic approach. Curr Opin Struct Biol 22:550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Pedersen LC (2007) Anticoagulant heparan sulfate: structural specificity and biosynthesis. Appl Microbiol Biotechnol 74:263–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Shworak NW, Fritze LMS, Edelberg JM, Rosenberg RD (1996) Purification of heparan sulfate D-glucosaminyl 3-O-sulfotransferase. J Biol Chem 271:27072–27082. [DOI] [PubMed] [Google Scholar]
- Liu R, Liu J (2011) Enzymatic placement of 6-O-sulfo groups in heparan sulfate. Biochemistry 50:4382–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Li H, Zhang J, Li M, Liu MY, An X, Liu MC, Chang W (2010) Crystal structures of SULT1A2 and SULT1A1 *3: insights into the substrate inhibition and the role of Tyr149 in SULT1A2. Biochem Biophys Res Commun 396:429–434. [DOI] [PubMed] [Google Scholar]
- Lu JH, Li HT, Liu MC, Zhang JP, Li M, An XM, Chang WR (2005) Crystal structure of human sulfotransferase SULT1A3 in complex with dopamine and 3′-phosphoadenosine 5′-phosphate. Biochem Biophys Res Commun 335:417–423. [DOI] [PubMed] [Google Scholar]
- Lu LY, Chiang HP, Chen WT, Yang YS (2009) Dimerization is responsible for the structural stability of human sulfotransferase 1A1. Drug Metab Dispos 37:1083–1088. [DOI] [PubMed] [Google Scholar]
- Lu LY, Hsieh YC, Liu MY, Lin YH, Chen CJ, Yang YS (2008) Identification and characterization of two amino acids critical for the substrate inhibition of human dehydroepiandrosterone sulfotransferase (SULT2A1). Mol Pharmacol 73:660–668. [DOI] [PubMed] [Google Scholar]
- Lyle S, Stanczak J, Ng K, Schwartz NB (1994) Rat chondrosarcoma ATP sulfurylase and adenosine 5′-phosphosulfate kinase reside on a single bifunctional protein. Biochemistry 33:5920–5925. [DOI] [PubMed] [Google Scholar]
- Lyon ES, Jakoby WB (1980) The identity of alcohol sulfotransferases with hydroxysteroid sulfotransferases. Arch Biochem Biophys 202:474–481. [DOI] [PubMed] [Google Scholar]
- Ma G, Geng L, Lu Y, Wei X, Yu H (2021) Investigating the molecular mechanism of hydroxylated bromdiphenyl ethers to inhibit the thyroid hormone sulfotransferase SULT1A1. Chemosphere 263:128353. [DOI] [PubMed] [Google Scholar]
- Marsolais F, Varin L (1995) Identification of amino acid residues critical for catalysis and cosubstrate binding in the flavonol 3-sulfotransferase. J Biol Chem 270:30458–30463. [DOI] [PubMed] [Google Scholar]
- Martiny VY, Carbonell P, Lagorce D, Villoutreix BO, Moroy G, Miteva MA (2013) In silico mechanistic profiling to probe small molecule binding to sulfotransferases. PLoS One 8:e73587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura E, Nakagawa A, Tomabechi Y, Ikushiro S, Sakaki T, Katayama T, Yamamoto K, Kumagai H, Sato F, Minami H (2018) Microbial production of novel sulphated alkaloids for drug discovery. Sci Rep 8:7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiro E, Sakakibara Y, Liu MC, Suiko M (2006) Differential enzymatic characteristics and tissue-specific expression of human TPST-1 and TPST-2. J Biochem 140:731–737. [DOI] [PubMed] [Google Scholar]
- Monzo MBrunet SUrbano-Ispizua ANavarro APerea GEsteve JArtells RGranell MBerlanga JRibera JM, et al. ; CETLAM (2006) Genomic polymorphisms provide prognostic information in intermediate-risk acute myeloblastic leukemia. Blood 107:4871–4879. [DOI] [PubMed] [Google Scholar]
- Moon A, Edavettal SC, Krahn JX, Munoz EM, Negishi M, Linhardt RJ, Liu J, Pedersen LC (2004) Structural analysis of the sulfotransferase (3-o-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor of herpes simplex virus 1. J Biol Chem 279:45185–45193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AF, Xu Y, Woody SM, Krahn JM, Linhardt RJ, Liu J, Pedersen LC (2012) Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin. Proc Natl Acad Sci USA 109:5265–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KL (2003) The biology and enzymology of protein tyrosine O-sulfation. J Biol Chem 278:24243–24246. [DOI] [PubMed] [Google Scholar]
- Mueller JW, Gilligan LC, Idkowiak J, Arlt W, Foster PA (2015) The regulation of steroid action by sulfation and desulfation. Endocr Rev 36:526–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Dieckmann H-J, Schulz GE (1994) The structure of uridylate kinase with its substrates, showing the transition state geometry. J Mol Biol 236:361–367. [DOI] [PubMed] [Google Scholar]
- Multhaupt HA, Couchman JR (2012) Heparan sulfate biosynthesis: methods for investigation of the heparanosome. J Histochem Cytochem 60:908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmabadi HHu HGarshasbi MZemojtel TAbedini SSChen WHosseini MBehjati FHaas SJamali P, et al. (2011) Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 478:57–63. [DOI] [PubMed] [Google Scholar]
- Negishi M, Pedersen LG, Petrotchenko E, Shevtsov S, Gorokhov A, Kakuta Y, Pedersen LC (2001) Structure and function of sulfotransferases. Arch Biochem Biophys 390:149–157. [DOI] [PubMed] [Google Scholar]
- Nguyen NT, Vivès RR, Torres M, Delauzun V, Saesen E, Roig-Zamboni V, Lortat-Jacob H, Rihet P, Bourne Y (2018) Genetic and enzymatic characterization of 3-O-sulfotransferase SNPs associated with Plasmodium falciparum parasitaemia. Glycobiology 28:534–541. [DOI] [PubMed] [Google Scholar]
- Niehrs C, Huttner WB (1990) Purification and characterization of tyrosylprotein sulfotransferase. EMBO J 9:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien PJ, Herschlag D (1998) Sulfatase activity of E-coli alkaline phosphatase demonstrates a functional link to arylsulfatases, an evolutionarily related enzyme family. J Am Chem Soc 120:12369–12370. [Google Scholar]
- Ouyang Yb, Lane WS, Moore KL (1998) Tyrosylprotein sulfotransferase: purification and molecular cloning of an enzyme that catalyzes tyrosine O-sulfation, a common posttranslational modification of eukaryotic proteins. Proc Natl Acad Sci USA 95:2896–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganini LHadi LAChetta MRovina DFontana LColapietro PBonaparte EPezzani LMarchisio PTabano SM, et al. (2019) A HS6ST2 gene variant associated with X-linked intellectual disability and severe myopia in two male twins. Clin Genet 95:368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen LC, Petrotchenko E, Shevtsov S, Negishi M (2002) Crystal structure of the human estrogen sulfotransferase-PAPS complex: evidence for catalytic role of Ser137 in the sulfuryl transfer reaction. J Biol Chem 277:17928–17932. [DOI] [PubMed] [Google Scholar]
- Pedersen LC, Petrotchenko EV, Negishi M (2000) Crystal structure of SULT2A3, human hydroxysteroid sulfotransferase. FEBS Lett 475:61–64. [DOI] [PubMed] [Google Scholar]
- Petrotchenko EV, Doerflein ME, Kakuta Y, Pedersen LC, Negishi M (1999) Substrate gating confers steroid specificity to estrogen sulfotransferase. J Biol Chem 274:30019–30022. [DOI] [PubMed] [Google Scholar]
- Petrotchenko EV, Pedersen LC, Borchers CH, Tomer KB, Negishi M (2001) The dimerization motif of cytosolic sulfotransferases. FEBS Lett 490:39–43. [DOI] [PubMed] [Google Scholar]
- Pica-Mattoccia L, Carlini D, Guidi A, Cimica V, Vigorosi F, Cioli D (2006) The schistosome enzyme that activates oxamniquine has the characteristics of a sulfotransferase. Mem Inst Oswaldo Cruz 101 (Suppl 1):307–312. [DOI] [PubMed] [Google Scholar]
- Pouyani T, Seed B (1995) PSGL-1 recognition of P-selectin is controlled by a tyrosine sulfation consensus at the PSGL-1 amino terminus. Cell 83:333–343. [DOI] [PubMed] [Google Scholar]
- Rebbeck TRTroxel ABWang YWalker AHPanossian SGallagher SShatalova EGBlanchard RBunin GDeMichele A, et al. (2006) Estrogen sulfation genes, hormone replacement therapy, and endometrial cancer risk. J Natl Cancer Inst 98:1311–1320. [DOI] [PubMed] [Google Scholar]
- Rehse PH, Zhou M, Lin SX (2002) Crystal structure of human dehydroepiandrosterone sulphotransferase in complex with substrate. Biochem J 364:165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter MSMusante LHu HDiederich SSticht HEkici ABUebe SWienker TFBartsch OZechner U, et al. (2014) NDST1 missense mutations in autosomal recessive intellectual disability. Am J Med Genet A 164A:2753–2763. [DOI] [PubMed] [Google Scholar]
- Robbins PW, Lipmann F (1956) Identification of enzymatically active sulfate as adenosine-3′-phosphate-5′-phosphosulfate. J Am Chem Soc 78:2652–2653. [Google Scholar]
- Roche P, Debellé F, Maillet F, Lerouge P, Faucher C, Truchet G, Dénarié J, Promé J-C (1991) Molecular basis of symbiotic host specificity in Rhizobium meliloti: nodH and nodPQ genes encode the sulfation of lipo-oligosaccharide signals. Cell 67:1131–1143. [DOI] [PubMed] [Google Scholar]
- Rong J, Habuchi H, Kimata K, Lindahl U, Kusche-Gullberg M (2001) Substrate specificity of the heparan sulfate hexuronic acid 2-O-sulfotransferase. Biochemistry 40:5548–5555. [DOI] [PubMed] [Google Scholar]
- Rosenquist GL, Nicholas HB Jr (1993) Analysis of sequence requirements for protein tyrosine sulfation. Protein Sci 2:215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste M, Sibbald PR, Wittinghofer A (1990) The P-loop–a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci 15:430–434. [DOI] [PubMed] [Google Scholar]
- Schneeberger PEvon Elsner LBarker ELMeinecke PMarquardt IAlawi MSteindl KJoset PRauch AZwijnenburg PJG, et al. (2020) Bi-allelic pathogenic variants in HS2ST1 cause a syndrome characterized by developmental delay and corpus callosum, skeletal, and renal abnormalities. Am J Hum Genet 107:1044–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Duffel MW (2002) Comparative molecular field analysis of substrates for an aryl sulfotransferase based on catalytic mechanism and protein homology modeling. J Med Chem 45:5514–5522. [DOI] [PubMed] [Google Scholar]
- Sharma V, Duffel MW (2005) A comparative molecular field analysis-based approach to prediction of sulfotransferase catalytic specificity. Methods Enzymol 400:249–263. [DOI] [PubMed] [Google Scholar]
- Shevtsov S, Petrotchenko EV, Pedersen LC, Negishi M (2003) Crystallographic analysis of a hydroxylated polychlorinated biphenyl (OH-PCB) bound to the catalytic estrogen binding site of human estrogen sulfotransferase. Environ Health Perspect 111:884–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi D, Sheng A, Chi L (2021) Glycosaminoglycan-protein interactions and their roles in human disease. Front Mol Biosci 8:639666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Munger C, Kalan L, Sulea T, Wright GD, Cygler M (2012) Sulfonation of glycopeptide antibiotics by sulfotransferase StaL depends on conformational flexibility of aglycone scaffold. Proc Natl Acad Sci USA 109:11824–11829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimohira T, Kurogi K, Hashiguchi T, Liu MC, Suiko M, Sakakibara Y (2017) Regioselective production of sulfated polyphenols using human cytosolic sulfotransferase-expressing Escherichia coli cells. J Biosci Bioeng 124:84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG (1999) A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13–22. [DOI] [PubMed] [Google Scholar]
- Shworak NW, Liu J, Petros LM, Zhang L, Kobayashi M, Copeland NG, Jenkins NA, Rosenberg RD (1999) Multiple isoforms of heparan sulfate D-glucosaminyl 3-O-sulfotransferase. Isolation, characterization, and expression of human cdnas and identification of distinct genomic loci. J Biol Chem 274:5170–5184. [DOI] [PubMed] [Google Scholar]
- Singh W, Karabencheva-Christova TG, Black GW, Sparagano O, Christov CZ (2016a) Conformational flexibility influences structure-function relationships in tyrosyl protein sulfotransferase-2. RSC Advances 6:11344–11352. [Google Scholar]
- Singh W, Karabencheva-Christova TG, Sparagano O, Black GW, Petrov PY, Christov CZ (2016b) Dimerization and ligand binding in tyrosylprotein sulfotransferase-2 are influenced by molecular motions. RSC Advances 6:18542–18548. [Google Scholar]
- Sipilä J, Hood AM, Coughtrie MW, Taskinen J (2003) CoMFA modeling of enzyme kinetics: K(m) values for sulfation of diverse phenolic substrates by human catecholamine sulfotransferase SULT1A3. J Chem Inf Comput Sci 43:1563–1569. [DOI] [PubMed] [Google Scholar]
- Sousa RP, Fernandes PA, Ramos MJ, Brás NF (2016) Insights into the reaction mechanism of 3-O-sulfotransferase through QM/MM calculations. Phys Chem Chem Phys 18:11488–11496. [DOI] [PubMed] [Google Scholar]
- Sterner E, Li L, Paul P, Beaudet JM, Liu J, Linhardt RJ, Dordick JS (2014) Assays for determining heparan sulfate and heparin O-sulfotransferase activity and specificity. Anal Bioanal Chem 406:525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stjernschantz E, Reinen J, Meinl W, George BJ, Glatt H, Vermeulen NP, Oostenbrink C (2010) Comparison of murine and human estrogen sulfotransferase inhibition in vitro and in silico–implications for differences in activity, subunit dimerization and substrate inhibition. Mol Cell Endocrinol 317:127–140. [DOI] [PubMed] [Google Scholar]
- Sun M, Leyh TS (2010) The human estrogen sulfotransferase: a half-site reactive enzyme. Biochemistry 49:4779–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Nishiyori T, Kojo H, Otsubo R, Tsuruta M, Kurogi K, Liu MC, Suiko M, Sakakibara Y, Kakuta Y (2017) Structural basis for the broad substrate specificity of the human tyrosylprotein sulfotransferase-1. Sci Rep-Uk 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramoto TFujikawa YKawaguchi YKurogi KSoejima MAdachi RNakanishi YMishiro-Sato ELiu MCSakakibara Y, et al. (2013) Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction. Nat Commun 4:1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramoto T, Nishio T, Kurogi K, Sakakibara Y, Kakuta Y (2021) The crystal structure of mouse SULT2A8 reveals the mechanism of 7α-hydroxyl, bile acid sulfation. Biochem Biophys Res Commun 562:15–20. [DOI] [PubMed] [Google Scholar]
- Tibbs ZE, Falany CN (2016) An engineered heterodimeric model to investigate SULT1B1 dependence on intersubunit communication. Biochem Pharmacol 115:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbs ZE, Guidry AL, Falany JL, Kadlubar SA, Falany CN (2018) A high frequency missense SULT1B1 allelic variant (L145V) selectively expressed in African descendants exhibits altered kinetic properties. Xenobiotica 48:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbs ZE, Rohn-Glowacki KJ, Crittenden F, Guidry AL, Falany CN (2015) Structural plasticity in the human cytosolic sulfotransferase dimer and its role in substrate selectivity and catalysis. Drug Metab Pharmacokinet 30:3–20. [DOI] [PubMed] [Google Scholar]
- Tornberg JSykiotis GPKeefe KPlummer LHoang XHall JEQuinton RSeminara SBHughes VVan Vliet G, et al. (2011) Heparan sulfate 6-O-sulfotransferase 1, a gene involved in extracellular sugar modifications, is mutated in patients with idiopathic hypogonadotrophic hypogonadism. Proc Natl Acad Sci USA 108:11524–11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentim CLCioli DChevalier FDCao XTaylor ABHolloway SPPica-Mattoccia LGuidi ABasso ATsai IJ, et al. (2013) Genetic and molecular basis of drug resistance and species-specific drug action in schistosome parasites. Science 342:1385–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wander R, Kaminski AM, Xu Y, Pagadala V, Krahn JM, Pham TQ, Liu J, Pedersen LC (2021) Deciphering the substrate recognition mechanisms of the heparan sulfate 3-O-sulfotransferase-3. RSC Chemical Biology 2:1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Cook I, Falany CN, Leyh TS (2014a) Paradigms of sulfotransferase catalysis: the mechanism of SULT2A1. J Biol Chem 289:26474–26480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Cook I, Leyh TS (2014b) 3′-Phosphoadenosine 5′-phosphosulfate allosterically regulates sulfotransferase turnover. Biochemistry 53:6893–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Arnold K, Dhurandhare VM, Xu Y, Liu J (2021) Investigation of the biological functions of heparan sulfate using a chemoenzymatic synthetic approach. RSC Chem Biol 2:702–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hsieh P-H, Xu Y, Thieker D, Chai EJE, Xie S, Cooley B, Woods RJ, Chi L, Liu J (2017) Synthesis of 3-O-sulfated oligosaccharides to understand the relationship between structures and functions of heparan sulfate. J Am Chem Soc 139:5249–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinshilboum R, Otterness D (1994) Sulfotransferase Enzymes, in Conjugation—Deconjugation Reactions in Drug Metabolism and Toxicity (Kauffman FC ed) pp 45–78, Springer Berlin Heidelberg, Berlin, Heidelberg. [Google Scholar]
- Weitzner B, Meehan T, Xu Q, Dunbrack RL Jr (2009) An unusually small dimer interface is observed in all available crystal structures of cytosolic sulfotransferases. Proteins 75:289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia G, Chen J, Tiwari V, Ju W, Li JP, Malmstrom A, Shukla D, Liu J (2002) Heparan sulfate 3-O-sulfotransferase isoform 5 generates both an antithrombin-binding site and an entry receptor for herpes simplex virus, type 1. J Biol Chem 277:37912–37919. [DOI] [PubMed] [Google Scholar]
- Xie LXiao DWang XWang CBai JYue QYue HLi YMolnár IXu Y, et al. (2020) Combinatorial biosynthesis of sulfated benzenediol lactones with a phenolic sulfotransferase from fusarium graminearum PH-1. MSphere 5:e00949-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Moon AF, Song D, Pedersen LC, Liu J (2008) Engineering sulfotransferases to modify heparan sulfate. Nat Chem Biol 4:200–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YCai CChandarajoti KHsieh PHLi LPham TQSparkenbaugh EMSheng JKey NSPawlinski R, et al. (2014) Homogeneous low-molecular-weight heparins with reversible anticoagulant activity. Nat Chem Biol 10:248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YChandarajoti KZhang XPagadala VDou WHoppensteadt DMSparkenbaugh EMCooley BDaily SKey NS, et al. (2017a) Synthetic oligosaccharides can replace animal-sourced low-molecular weight heparins. Sci Transl Med 9:eaan5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Masuko S, Takieddin M, Xu H, Liu R, Jing J, Mousa SA, Linhardt RJ, Liu J (2011) Chemoenzymatic synthesis of homogeneous ultralow molecular weight heparins. Science 334:498–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Moon AF, Xu S, Krahn JM, Liu J, Pedersen LC (2017b) Structure based substrate specificity analysis of heparan sulfate 6-O-sulfotransferases. ACS Chem Biol 12:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZHOtterness DMFreimuth RRCarlini EJWood TCMitchell SMoon EKim UJXu JPSiciliano MJ, et al. (2000) Human 3′-phosphoadenosine 5′-phosphosulfate synthetase 1 (PAPSS1) and PAPSS2: gene cloning, characterization and chromosomal localization. Biochem Biophys Res Commun 268:437–444. [DOI] [PubMed] [Google Scholar]
- Yi LXu YKaminski AMChang XPagadala VHorton MSu GWang ZLu GConley P, et al. (2020) Using engineered 6-O-sulfotransferase to improve the synthesis of anticoagulant heparin. Org Biomol Chem 18:8094–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssefian LVahidnezhad HSaeidian AHTouati ASotoudeh SMahmoudi HMansouri PDaneshpazhooh MAghazadeh NHesari KK, et al. (2019) Autosomal recessive congenital ichthyosis: genomic landscape and phenotypic spectrum in a cohort of 125 consanguineous families. Hum Mutat 40:288–298. [DOI] [PubMed] [Google Scholar]
- Zerbino DRAchuthan PAkanni WAmode MRBarrell DBhai JBillis KCummins CGall AGirón CG, et al. (2018) Ensembl 2018. Nucleic Acids Res 46 (D1):D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Varlamova O, Vargas FM, Falany CN, Leyh TS (1998) Sulfuryl transfer: the catalytic mechanism of human estrogen sulfotransferase. J Biol Chem 273:10888–10892. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lin L, Huang H, Linhardt RJ (2020) Chemoenzymatic synthesis of glycosaminoglycans. Acc Chem Res 53:335–346. [DOI] [PubMed] [Google Scholar]
- Zhao L, Zhang P, Long S, Wang L, Jin H, Han W, Tian P (2016) Investigating the substrate binding mechanism of sulfotransferase 2A1 based on substrate tunnel analysis: a molecular dynamics simulation study. J Mol Model 22:176. [DOI] [PubMed] [Google Scholar]
- Zhao L, Zhang P, Long S, Wang L, Tian P (2015) The impact of ligands on the structure and flexibility of sulfotransferases: a molecular dynamics simulation study. J Mol Model 21:190. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Li Q, Xu R, Wang B, Du G, Kang Z (2019) Secretory expression of the rat aryl sulfotransferases IV with improved catalytic efficiency by molecular engineering. 3 Biotech 9:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Qi R, Liu Y, Zhao L, Han W (2019) Mechanistic insights into the effect of ligands on structural stability and selectivity of sulfotransferase 2A1 (SULT2A1). ACS Omega 4:22021–22034. [DOI] [PMC free article] [PubMed] [Google Scholar]