Abstract
An epoxide hydrolase from Rhodococcus erythropolis DCL14 catalyzes the hydrolysis of limonene-1,2-epoxide to limonene-1,2-diol. The enzyme is induced when R. erythropolis is grown on monoterpenes, reflecting its role in the limonene degradation pathway of this microorganism. Limonene-1,2-epoxide hydrolase was purified to homogeneity. It is a monomeric cytoplasmic enzyme of 17 kDa, and its N-terminal amino acid sequence was determined. No cofactor was required for activity of this colorless enzyme. Maximal enzyme activity was measured at pH 7 and 50°C. None of the tested inhibitors or metal ions inhibited limonene-1,2-epoxide hydrolase activity. Limonene-1,2-epoxide hydrolase has a narrow substrate range. Of the compounds tested, only limonene-1,2-epoxide, 1-methylcyclohexene oxide, cyclohexene oxide, and indene oxide were substrates. This report shows that limonene-1,2-epoxide hydrolase belongs to a new class of epoxide hydrolases based on (i) its low molecular mass, (ii) the absence of any significant homology between the partial amino acid sequence of limonene-1,2-epoxide hydrolase and amino acid sequences of known epoxide hydrolases, (iii) its pH profile, and (iv) the inability of 2-bromo-4′-nitroacetophenone, diethylpyrocarbonate, 4-fluorochalcone oxide, and 1,10-phenanthroline to inhibit limonene-1,2-epoxide hydrolase activity.
Epoxides are highly reactive compounds which readily react with numerous biological compounds, including proteins and nucleic acids. Consequently, epoxides are cytotoxic, mutagenic, and potentially carcinogenic, and there is considerable interest in biological degradation mechanisms for these compounds.
In bacteria, epoxides are formed during the metabolism of alkenes (23) and halohydrins (15, 26, 34, 49). Enzymes belonging to a large number of enzyme classes, including dehydrogenases (17), lyases (21), carboxylases (1, 43), glutathione S-transferases (6, 8), isomerases (24), and hydrolases (7, 19, 44), are involved in the microbial degradation of epoxides.
Epoxide hydrolases are enzymes catalyzing the addition of water to epoxides forming the corresponding diol. This group of enzymes has been extensively studied in mammals, while only limited information is available on bacterial epoxide hydrolases. Three functions for epoxide hydrolases are recognized (42). In bacteria, epoxide hydrolases are involved in the degradation of several hydrocarbons, including 1,3-dihalo-2-propanol (34), 2,3-dihalo-1-propanol (15, 26), epichlorohydrin (46), propylene oxide (16), 9,10-epoxy fatty acids (30, 36), trans-2,3-epoxysuccinate (2), and cyclohexene oxide (14). Other epoxide hydrolases, such as microsomal and cytosolic epoxide hydrolase from mammals (for reviews, see references 4, 8, and 44), are involved in the detoxification of epoxides formed due to the action of P-450-dependent monooxygenases (8). Epoxide hydrolases are also involved in biosynthesis of hormones, such as leukotrienes and juvenile hormone (40, 45), and plant cuticular elements (11). Remarkably, the bacterial and eukaryotic epoxide hydrolases described so far form a homogeneous group of enzymes belonging to the α/β-hydrolase fold superfamily (10, 38).
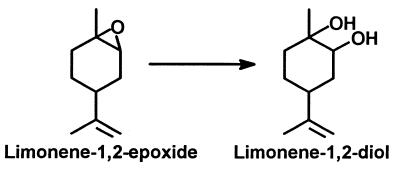
Rhodococcus erythropolis DCL14, a gram-positive bacterium, is able to grow on both (+)- and (−)-limonene as the sole source of carbon and energy (47). Cells grown on limonene contained a novel epoxide hydrolase that does not belong to the α/β-hydrolase fold superfamily. This limonene-1,2-epoxide hydrolase converts limonene-1,2-epoxide to limonene-1,2-diol (p-menth-8-ene-1,2-diol [Fig. 1]). In this report, we describe the purification and characterization of this enzyme and show that limonene-1,2-epoxide hydrolase belongs to a novel class of epoxide hydrolases.
FIG. 1.
Reaction catalyzed by limonene-1,2-epoxide hydrolase.
MATERIALS AND METHODS
Isolation of strain DCL14.
R. erythropolis DCL14 was isolated from an enrichment culture containing a sediment sample (10 g) from a ditch in Reeuwijk, The Netherlands, diluted in 30 ml of mineral salts medium (pH 7.0) (24) in the presence of 1 mM (−)-dihydrocarveol as the carbon and energy source. After incubation of this culture for 2 weeks on a shaker at 30°C and two successive transfers into fresh medium, samples of the enrichments were plated onto agar plates with mineral salts medium. These plates were incubated in a desiccator in which (+)-limonene was supplied via the gas phase. Colonies that developed were isolated and checked for purity by plating on yeast extract-glucose plates. R. erythropolis DCL14 (CIMW 0387B) is maintained at the Division of Industrial Microbiology, Wageningen, The Netherlands.
Growth conditions.
R. erythropolis DCL14 was subcultured once a month and grown at 30°C on a yeast extract-glucose agar plate for 2 days, after which the plates were stored at room temperature. Cultures were grown in 5-liter Erlenmeyer flasks containing 1 liter of mineral salts medium with 0.01% (vol/vol) carbon source and fitted with rubber stoppers. The flasks were incubated at 30°C on a horizontal shaker oscillating at 1 Hz with an amplitude of 10 cm. After growth was observed, the concentration of the toxic substrates was increased with steps of 0.01% (vol/vol) until a total of 0.1% (vol/vol) carbon source had been added.
Cells for enzyme purification were grown fed-batch in a fermentor with a working volume of 2.0 liters at 28°C. (+)-Limonene was supplied via the gas phase by passing the airflow (300 ml/min) into the fermentor through a bubble column containing (+)-limonene. Every day, 1.5 liters of the culture was harvested, after which the working volume was immediately increased to 2.0 liters. Cells were collected by centrifugation (4°C, 10 min at 16,000 × g) and washed with 50 mM potassium phosphate buffer (pH 7.0). The pellet was resuspended in 7 ml of this buffer and stored at −20°C until used.
Preparation of cell extract.
Two aliquots of frozen cell suspension (7 ml) were thawed and disrupted by sonication (20 min, 30% duty cycle, output control 2.3) with a Branson Sonifier 250. Cell debris was removed by centrifugation at 20,000 × g for 20 min. The supernatant was used as the cell extract. Protein was determined by the method of Bradford (12), with bovine serum albumin as the standard.
Purification of limonene-1,2-epoxide hydrolase.
All purification steps were performed at 4°C and pH 7.0. If necessary, the pooled fractions were concentrated by ultrafiltration with an Amicon ultrafiltration unit using a membrane with a molecular weight cutoff of 10,000 under nitrogen at a pressure of 4 bar.
Step 1: gel filtration.
The cell extract was applied onto a Sephacryl S300 (Pharmacia) column (2.5 by 98 cm) equilibrated with 10 mM potassium phosphate buffer (flow rate, 0.75 ml/min; collected fraction volume, 7.5 ml). Fractions containing limonene-1,2-epoxide hydrolase were pooled.
Step 2: hydroxyapatite.
The pooled fractions from the gel filtration step were applied to a hydroxyapatite (Bio-Rad) column (5 by 6 cm) equilibrated with 10 mM potassium phosphate buffer (flow rate, 0.3 ml/min; collected fraction volume, 3 ml). The column was washed with 50 ml of the same buffer, and subsequently the enzyme was eluted with a 10 to 500 mM linear gradient of potassium phosphate (total volume, 400 ml). Limonene-1,2-epoxide hydrolase eluted at a potassium phosphate concentration of 100 mM. Active fractions were pooled.
Step 3: anion-exchange chromatography.
The pooled fractions from the hydroxyapatite step were applied onto a DEAE-Sepharose CL-6B (Pharmacia) column (2.5 by 31 cm) equilibrated with 25 mM potassium phosphate buffer. The column was washed with 100 ml of the same buffer (flow rate, 0.75 ml/min; collected fraction volume, 7.5 ml), and the enzyme was eluted with a 0 to 1 M linear gradient of NaCl in the same buffer (total volume, 1 liter). Limonene-1,2-epoxide hydrolase eluted at an NaCl concentration of 260 mM. Fractions exhibiting limonene-1,2-epoxide hydrolase activity were pooled and concentrated.
Determination of molecular weight.
The molecular weight of the denatured protein was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). An SDS–20% polyacrylamide gel was prepared by the method of Laemmli (28). Proteins were stained with Coomassie brilliant blue G. A Pharmacia low-molecular-weight calibration kit containing phosphorylase b (94,000), bovine serum albumin (67,000), ovalbumin (43,000), carbonic anhydrase (30,000), soybean trypsin inhibitor (20,100), and α-lactalbumin (14,400) was used for the estimation of the molecular weight.
The molecular weight of the native protein was determined by gel filtration on a Sephacryl S300 column as described under step 1 of the purification procedure. Aldolase (158,000), bovine serum albumin (67,000), ovalbumin (43,000), and chymotrypsinogen A (25,000) were used as the reference proteins.
Assay of limonene-1,2-epoxide hydrolase activity.
Limonene-1,2-epoxide hydrolase activity was determined by monitoring (+)-limonene-1,2-epoxide degradation by gas chromatography (GC). The reaction mixtures consisted of cell extract (10 to 100 μl) and 2 ml of a freshly prepared 2.5 mM (+)-limonene-1,2-epoxide solution in 50 mM potassium phosphate (pH 7.0) in 15-ml vials fitted with Teflon Mininert valves (Supelco Inc.) preventing evaporation of limonene-1,2-epoxide. The vials were placed in a shaking water bath (30°C); after 5, 10, and 15 min, a vial was removed from the water bath and the reaction was terminated by the addition of 1 ml of ethylacetate. The vials were vigorously shaken to enable quantitative extraction of the terpenes. The ethylacetate layer was pipetted in a microcentrifuge tube and centrifuged (3 min, 15,000 × g) to achieve separation of the two layers, and then 1 μl of the ethylacetate layer was analyzed by GC. The effects of inhibitors and ions were studied by adding 1 mM effector to limonene-1,2-epoxide hydrolase. This mixture was preincubated at 30°C for 15 min, after which the limonene-1,2-epoxide hydrolase activity was determined as described above.
The substrate specificity of limonene-1,2-epoxide hydrolase was tested by incubating different amounts of limonene-1,2-epoxide hydrolase with 5 mM epoxide in potassium phosphate buffer (pH 7.0) for 1 h at 30°C. The samples were extracted with ethylacetate and analyzed by GC. Epoxide degradation was corrected for chemical hydrolysis. With styrene oxide, cis-2,3-epoxybutane, epichlorohydrin, and 1,2-epoxyhexane, substrate degradation was followed by analyzing the headspace of these incubations by GC. The incubations with indene oxide and cis-stilbene oxide (tested at 1 mM because of the low solubility of these compounds) were extracted with hexane, and the hexane phase was analyzed by high-pressure liquid chromatography (HPLC).
Analytical methods.
All epoxides except indene oxide and cis-stilbene oxide were analyzed by chiral GC on fused silica cyclodextrin capillary columns (30-m length, 0.25-mm internal diameter, 0.25-μm film coating; Supelco, Zwijndrecht, The Netherlands). GC was performed on a Chrompack CP9000 gas chromatograph equipped with a flame ionization detector, using N2 as the carrier gas. The detector and injector temperatures were 250 and 200°C, respectively, and the split ratio was 1:50. For limonene-1,2-epoxide, 1-methylcyclohexene oxide, and cyclohexene oxide, an α-DEX 120 column was used at oven temperatures of 100, 70, and 70°C, respectively. Indene oxide and cis-stilbene oxide were analyzed by HPLC using a Chiralcel OB column as described by Zhang et al. (50).
Electroelution of limonene-1,2-epoxide hydrolase was performed with gel slices from an unstained nondenaturing 20% polyacrylamide gel, using a Hoefer GE 200 Gel Eluter (180 min at 100 V; Hoefer Pharmacia Biotech Inc.). The N terminus of limonene-1,2-epoxide hydrolase was determined by the Eiwitsequencerfaciliteit Leiden, Vakgroep Medische Biochemie, Sylvius Laboratoria, Leiden, The Netherlands. The metal composition of limonene-1,2-epoxide hydrolase (95 μM) was determined by inductively coupled plasma mass spectrometry (ICP-MS) using a Perkin-Elmer Elan 6000. The IPC-MS detection limit was 1 μg of metal/liter.
Sources of chemicals.
(+)- and (−)-limonene-1,2-epoxide and (+)-limonene were purchased from Acros; cyclohexene oxide was purchased from Aldrich. Limonene-1,2-diol was prepared by acid (H2SO4) hydrolysis of limonene-1,2-epoxide (39). 1-Methylcyclohexene oxide was prepared as described before (14). Indene oxide was prepared as described by Gagis et al. (20). All other chemicals were of the highest purity commercially available.
RESULTS
Induction of limonene-1,2-epoxide hydrolase activity in R. erythropolis.
R. erythropolis DCL14 was grown on different carbon sources, and the limonene-1,2-epoxide hydrolase activity was determined (Table 1). Growth on monoterpenes resulted in a 10- to 100-fold increase in limonene-1,2-epoxide hydrolase activity. Notably, growth on the (+)-isomer(s) of the terpenes resulted in a limonene-1,2-epoxide hydrolase activity higher than that found after growth of the cells on the (−)-isomer(s) of the same compound (Table 1). Cells for further experiments were grown on (+)-limonene.
TABLE 1.
Limonene-1,2-epoxide hydrolase activity in cell extracts of R. erythropolis DCL14 grown in batch cultures on various carbon sources
| Growth substrate | Sp acta (nmol · min−1 · [mg of protein]−1) |
|---|---|
| (+)-Limonene | 795 |
| (−)-Limonene | 185 |
| (+)-Limonene-1,2-epoxide | 705 |
| (−)-Limonene-1,2-epoxide | 315 |
| (+)-Limonene-1,2-diol | 1,450 |
| (−)-Limonene-1,2-diol | 300 |
| (+)-Carvone | 185 |
| (−)-Carvone | 150 |
| Succinate | 15 |
| Ethanol | 15 |
Activity with (+)-limonene-1,2-epoxide.
Purification of limonene-1,2-epoxide hydrolase.
The limonene-1,2-epoxide hydrolase activity was present in the 100,000 × g supernatant of cell extract. Storage of cell extract at room temperature for 1 month did not result in any apparent loss of limonene-1,2-epoxide hydrolase activity.
The purification scheme for limonene-1,2-epoxide hydrolase is presented in Table 2. Limonene-1,2-epoxide hydrolase was purified 53-fold, with an overall yield of 60%. From Table 2, it can be calculated that limonene-1,2-epoxide hydrolase represents 2% of the total soluble cellular protein of (+)-limonene-grown cells. The absorption spectrum of this colorless protein gave no indication of the presence of a prosthetic group. The results of the ICP-MS analysis indicate that limonene-1,2-epoxide hydrolase does not contain a metal ion as a cofactor. Limonene-1,2-epoxide hydrolase could be stored at −20°C for 6 months without loss of activity.
TABLE 2.
Purification of limonene-1,2-epoxide hydrolase from (+)-limonene-grown cells of R. erythropolis DCL14
| Purification step | Total protein (mg) | Sp acta (nmol · min−1 · [mg of protein]−1) | Purification (fold) | Yield (%) |
|---|---|---|---|---|
| Cell extract | 248 | 1,590 | 1 | 100 |
| Gel filtration | 17.8 | 21,800 | 13.6 | 98 |
| Hydroxyapatite | 3.94 | 72,900 | 45.5 | 72 |
| Anion exchange | 2.75 | 85,100 | 53.2 | 59 |
Activity with (+)-limonene-1,2-epoxide.
SDS-PAGE revealed one distinct band, corresponding to a protein with a molecular mass of 17 kDa (Fig. 2). Activity determinations with this protein electroeluted from a nondenaturating polyacrylamide gel revealed that the stained protein band indeed represented limonene-1,2-epoxide hydrolase (not shown). Gel filtration revealed a molecular mass of about 15 kDa, indicating that the native enzyme is a monomer.
FIG. 2.
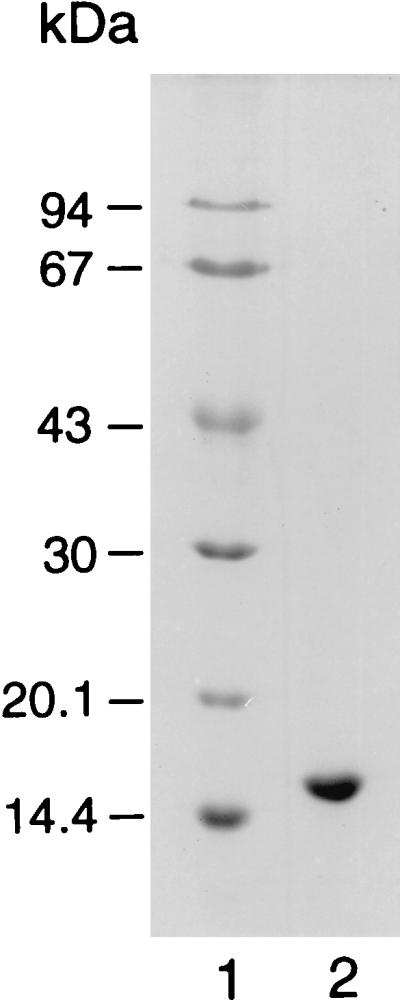
SDS-PAGE of limonene-1,2-epoxide hydrolase from R. erythropolis DCL14. Lane 1; molecular weight markers, lane 2; 20 μg of limonene-1,2-epoxide hydrolase.
N-terminal amino acid sequence.
The N-terminal amino acid sequence of limonene-1,2-epoxide hydrolase was determined to be Thr-Ser-Lys-Ile-Glu-Gln-Pro-Arg-Trp-Ala-Ser-Lys-Asp-Ser-Ala-Ala-Gly-Ala-Ala-Ser-Thr-Pro-Asp-Glu-Lys-Ile-Val-Leu-Glu-Phe-Met-Asp-Ala-Leu-Thr-Ser-Asn-Asp-Ala-Ala-Lys-Leu-Ile-Glu-Tyr-Phe-Ala-Glu-Asp-Thr. Comparison of this amino acid sequence with entries in the databases by using the BLAST search program revealed no substantial homology with any other protein. The highest similarity (38%) was with checkpoint protein RAD24 from Saccharomyces cerevisiae (SWISS-PROT accession no. P32641).
Temperature and pH optimum and temperature stability of the purified enzyme.
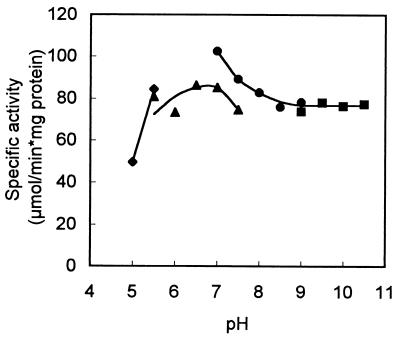
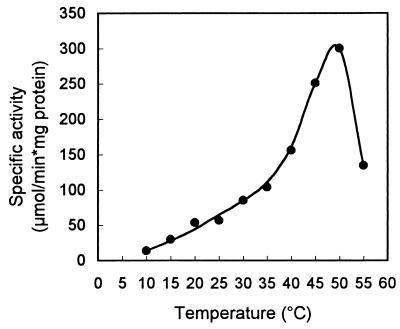
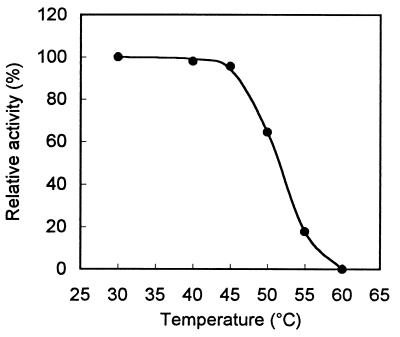
The enzyme has a very broad pH optimum, peaking at around pH 7 (Fig. 3). Phosphate buffer seemed to slightly inhibit limonene-1,2-epoxide hydrolase activity. Below pH 5 the activity could no longer be measured accurately, as chemical hydrolysis of the substrate overtook the biological hydrolysis rate. The temperature optimum of the enzyme was 50°C (Fig. 4). From the data presented in Fig. 4, we calculated an activation energy of 51.8 kJ/mol for limonene-1,2-epoxide hydrolase. The temperature stability of limonene-1,2-epoxide hydrolase is shown in Fig. 5. At temperatures over 45°C, rapid inactivation of the enzyme was observed.
FIG. 3.
Effect of pH on limonene-1,2-epoxide hydrolase activity. Specific activity was determined with (+)-limonene-1,2-epoxide at 30°C. ⧫, 50 mM citrate buffer; ▴, 50 mM potassium phosphate buffer; •, 50 mM Tris-HCl buffer; ■, 50 mM glycine-NaOH buffer.
FIG. 4.
Effect of temperature on limonene-1,2-epoxide hydrolase activity. Specific activity was determined with (+)-limonene-1,2-epoxide in 50 mM potassium phosphate buffer, pH 7.0.
FIG. 5.
Temperature stability of limonene-1,2-epoxide hydrolase (50 mM potassium phosphate buffer, pH 7.0). The enzyme was incubated for 15 min at the indicated temperature, after which the specific activity was determined at 30°C.
Inhibitors and metal ions.
We tested a variety of enzyme inhibitors for the ability to inhibit limonene-1,2-epoxide hydrolase activity: EDTA, 1,10-phenanthroline, α,α′-dipyridyl, nitrilotriacetate, SDS, iodoacetamide, iodoacetate, p-chloromercuribenzoate, dithiothreitol, cysteine, 2-mercaptoethanol, glutathione and phenylhydrazine, hydroxylamine, potassium cyanate, N-ethylmaleimide, semicarbazide, diethylpyrocarbonate, 2-bromo-4′-nitroacetophenone, and 2-bromo-4′-methyl-acetophenone (all at 1 mM). None of these compounds inhibited limonene-1,2-epoxide hydrolase activity, nor did 4-fluorochalcone oxide and 4-phenylchalcone oxide, which are competitive inhibitors of mammalian soluble epoxide hydrolase (33).
The metal salts CuSO4, CoSO4, BaCl2, NiCl2, CaCl2, CdCl2, HgCl2, AgNO3, ZnCl2, MgSO, MnSO4 Fe(II)SO4, and Fe(III)3(SO4)2 (all at 1 mM) and NaCl, KCl, NH4Cl, CsCl, and LiCl (all at 10 mM) did not affect limonene-1,2-epoxide hydrolase activity.
Substrate specificity.
Limonene-1,2-epoxide hydrolase has a rather narrow substrate specificity. Of the compounds tested, only limonene-1,2-epoxide, 1-methylcyclohexene oxide, cyclohexene oxide, and indene oxide were substrates for limonene-1,2-epoxide hydrolase (Table 3). The enzyme showed the highest activity with (+)-limonene-1,2-epoxide.
TABLE 3.
Substrate specificity of limonene-1,2-epoxide hydrolasea
| Substrate | Relative activityb |
|---|---|
| (+)-Limonene-1,2-epoxide | 100 |
| (−)-Limonene-1,2-epoxide | 27 |
| 1-Methylcyclohexene oxide | 47 |
| Cyclohexene oxide | 4.0 |
| Indene oxide | 57 |
Limonene-1,2-epoxide hydrolase showed less than 0.25% of the (+)-limonene-1,2-epoxide hydrolysis rate with (−)-α-pinene oxide, cyclopentene oxide, cyclooctene oxide, cyclododecene oxide, cyclohexene sulfide, 3-oxo-cyclohexene oxide, 1-oxaspiro[2.5]octane, styrene oxide, trans-β-methylstyrene oxide, cis-stilbene oxide, epichlorohydrin, cis-2,3-epoxybutane, and 1,2-epoxyhexane.
Relative to 100% activity with (+)-limonene-1,2-epoxide.
DISCUSSION
This report describes the purification and characterization of a novel epoxide hydrolase from R. erythropolis DCL14. Limonene-1,2-epoxide hydrolase is induced when R. erythropolis DCL14 is grown on monoterpenes (Table 1), reflecting its function in the limonene degradation pathway of the bacterium (47). So far, very few enzymes involved in microbial degradation of monoterpenes have been characterized (48).
Epoxide hydrolases form a remarkably homogeneous group of enzymes. They belong to the α,β-hydrolase fold superfamily (10, 38), based on the observation that they show low but significant sequence similarity with haloalkane dehalogenase from Xanthobacter autotrophicus GJ10, of which the three-dimensional structure has been solved (49). Epoxide hydrolases do not contain a prosthetic group. Their catalytic activity depends on a catalytic triad consisting of Asp, His, and Asp(Glu) residues (5, 38). On nucleophilic attack by the catalytic Asp the epoxide ring opens, resulting in the formation of a covalent hydroxy ester intermediate (3). This covalent intermediate was recently visualized in elegant studies by Müller et al. (32). Subsequently, a proton is abstracted from a water molecule by the His-Asp(Glu) pair, resulting in hydrolysis of the ester bond and in release of a corresponding diol.
In bacteria, only soluble epoxide hydrolases have been found (25, 35), while in eukaryotes, α/β-hydrolase folded epoxide hydrolases are either soluble or membrane bound (9, 11, 44). Bacterial epoxide hydrolases have a subunit mass of 35 to 40 kDa, and both homodimeric and monomeric enzymes have been described (25, 31, 35). Plant epoxide hydrolases have similar subunit masses (32 to 40 kDa [11, 27, 41]) and the only plant epoxide purified to homogeneity was a homodimer (11). Soluble epoxide hydrolases from mammals are homodimers and are much larger than bacterial epoxide hydrolases (subunit mass of 55 to 62 kDa [44]). Mammalian membrane-bound epoxide hydrolases are thought to be dodecamers with a subunit mass of 46 to 53 kDa (8).
Two epoxide hydrolases which do not belong to the α/β-hydrolase fold superfamily have been described (10, 44): leukotriene A4 hydrolase and cholesterol-epoxide hydrolase.
Leukotriene A4 hydrolase is an enzyme involved in the production of the hormone derivatives of archidonic acid. This cytosolic enzyme has been purified from several mammalian sources and is a monomer of 68 to 70 kDa (44). The amino acid sequence of leukotriene A4 hydrolase revealed that this enzyme belongs to the metallohydrolase superfamily (29). It contains 1 mol of catalytic Zn2+ per mol of enzyme (22). Remarkably, leukotriene A4 hydrolase is also a highly efficient aminopeptidase (37).
Cholesterol-epoxide hydrolase is an enzyme involved in the conversion of epoxides arising from cholesterol during lipid peroxidation in mammals. It is a membrane-bound protein which has not yet been purified to homogeneity (32). The molecular mass of this enzyme is unknown. In contrast to the α/β-epoxide hydrolases, there is no evidence that a covalent intermediate is formed in the course of epoxide hydrolysis, suggesting a completely different reaction mechanism (32).
Limonene-1,2-epoxide hydrolase from R. erythropolis DCL14 clearly belongs to a separate class of epoxide hydrolases. It is a cytoplasmic enzyme with an unprecedented low molecular mass of 17 kDa (Fig. 2). This low mass distinctly sets this enzyme apart from the α/β-hydrolase fold class of epoxide hydrolases. It is estimated that a minimal molecular mass of 25 kDa is necessary to accommodate the reaction mechanism as used by the α/β-hydrolase folded epoxide hydrolases (2a).
The N-terminal amino acid sequence of limonene-1,2-epoxide hydrolase does not show homology with known amino acid sequences of α/β-hydrolase folded epoxide hydrolases (10, 38). At first sight, α/β-hydrolase folded epoxide hydrolases do not show any obvious sequence similarity (10). However, several regions which are highly conserved have been identified in the amino acid sequences of α/β-hydrolase folded epoxide hydrolases (10). There was also no homology between the partial amino acid sequence of limonene-1,2-epoxide hydrolase and the amino acid sequence of leukotriene A4 hydrolase (29). Remarkably, no substantial homology of limonene-1,2-epoxide hydrolase with any other protein present in the databases was detected.
The imidazole-modifying compounds 2-bromo-4′-nitroacetophenone and diethylpyrocarbonate did not affect limonene-1,2-epoxide hydrolase activity. This suggests that unlike in the α/β-hydrolase folded epoxide hydrolases, no catalytic histidine is involved in limonene-1,2-epoxide hydrolase (5, 18). The involvement of histidine in the mechanism of α/β-folded epoxide hydrolases has also been deduced from the slope of the pH optimum of this enzyme (9). Limonene-1,2-epoxide hydrolase behaved differently in this respect, and the activity remained constant at pH values of >8 (Fig. 3). Also 4-fluoro- and 4-phenylchalcone oxide, compounds which selectively inhibit soluble α/β-hydrolase folded epoxide hydrolases (33), and 1,10-phenanthroline, an inhibitor of leukotriene A4 hydrolase activity (22), did not inhibit limonene-1,2-epoxide hydrolase activity. These results with inhibitors indicate that limonene-1,2-epoxide hydrolase uses a reaction mechanism that is completely different from those used by known epoxide hydrolases.
In general, epoxides are hydrolyzed chemically under both acidic and basic conditions (13). However, limonene-1,2-epoxide is much more stable under basic than under neutral conditions (unpublished results). An overall base-catalyzed reaction mechanism, as used by the α/β-folded epoxide hydrolases, would therefore be unfavorable for the hydrolysis of limonene-1,2-epoxide. In this respect, it seems understandable that R. erythropolis DCL14 has evolved another type of epoxide hydrolase, in view of its specialized function in the limonene degradation pathway (reference 46 and unpublished results). The specialized function of limonene-1,2-epoxide hydrolase in limonene degradation is also reflected by the narrow substrate range of this enzyme (Table 3), in contrast to the broad substrate specificity of other epoxide hydrolases (35, 38, 44).
In conclusion, based on (i) the molecular mass, (ii) the partial amino acid sequence, (iii) the pH profile, and (iv) the effect of different inhibitors on the activity of limonene-1,2-epoxide hydrolase, limonene-1,2-epoxide hydrolase from R. erythropolis DCL14 clearly belongs to a separate class of epoxide hydrolases.
ACKNOWLEDGMENTS
This work was supported by grant BIO4-CT95-0049 from the European Community.
We thank Martin de Wit for technical assistance. David Leak (Department of Biochemistry, Imperial College, London, England) is acknowledged for sending us a sample of 1-methylcyclohexene oxide. We thank Michael Arand (Institute of Toxicology, University of Mainz, Mainz, Germany) for carefully reading the manuscript and sending us a sample of 4-fluorochalcone oxide.
REFERENCES
- 1.Allen J R, Ensign S A. Purification to homogeneity and reconstitution of the individual components of the epoxide carboxylase multiprotein enzyme complex from Xanthobacter strain Py2. J Biol Chem. 1997;272:32121–32128. doi: 10.1074/jbc.272.51.32121. [DOI] [PubMed] [Google Scholar]
- 2.Allen R H, Jakoby W B. Tartaric acid metabolism. IX. Synthesis with tartrate epoxidase. J Biol Chem. 1969;244:2078–2084. [PubMed] [Google Scholar]
- 2a.Arand, M. Personal communication.
- 3.Arand M, Grant D F, Beetham J K, Friedberg T, Oesch F, Hammock B D. Sequence similarity of mammalian epoxide hydrolases to the bacterial haloalkane dehalogenase and other related proteins. Implication for the potential catalytic mechanism of enzymatic epoxide hydrolysis. FEBS Lett. 1994;338:251–256. doi: 10.1016/0014-5793(94)80278-5. [DOI] [PubMed] [Google Scholar]
- 4.Arand M, Hinz W, Müller F, Hänel K, Winkler L, Mecky A, Knehr M, Dürk H, Wagner H, Ringhoffer M, Oesch F. Structure and mechanism of soluble epoxide hydrolase and its relation to microsomal epoxide hydrolase. In: Oesch F, Hengstler J, editors. Control mechanisms of carcinogenesis. Berlin, Germany: Springer Verlag; 1996. pp. 116–134. [Google Scholar]
- 5.Arand M, Wagner H, Oesch F. Asp333, Asp495, and His523 form the catalytic triad of rat soluble epoxide hydrolase. J Biol Chem. 1996;271:4223–4229. doi: 10.1074/jbc.271.8.4223. [DOI] [PubMed] [Google Scholar]
- 6.Arca P, Hardisson C, Suárez J E. Purification of a glutathione S-transferase that mediates fosfomycin resistance in bacteria. Antimicrob Agents Chemother. 1990;34:844–848. doi: 10.1128/aac.34.5.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Archer I V J. Epoxide hydrolases as asymmetric catalysts. Tetrahedron. 1997;53:15617–15662. [Google Scholar]
- 8.Armstrong R N. Enzyme-catalyzed detoxication reactions: mechanisms and stereochemistry. Crit Rev Biochem. 1987;22:39–88. doi: 10.3109/10409238709082547. [DOI] [PubMed] [Google Scholar]
- 9.Armstrong R N, Levin W, Jerina D M. Hepatic microsomal epoxide hydrolase. Mechanistic studies of the hydration of K-region arene oxides. J Biol Chem. 1980;255:4698–4705. [PubMed] [Google Scholar]
- 10.Beetham J K, Grant D, Arand M, Garbarino J, Kiyosue T, Pinot F, Oesch F, Belknap W R, Shinozaki K, Hammock B D. Gene evolution of epoxide hydrolases and recommended nomenclature. DNA Cell Biol. 1995;14:61–71. doi: 10.1089/dna.1995.14.61. [DOI] [PubMed] [Google Scholar]
- 11.Blée E, Schuber F. Occurrence of fatty acid epoxide hydrolases in soybean (Glycine max). Purification and characterization of the soluble form. Biochem J. 1992;282:711–714. doi: 10.1042/bj2820711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 13.Buchanan J G, Sable H Z. Stereoselective epoxide cleavages. In: Thyagarajan B S, editor. Organic transformations. New York, N.Y: John Wiley & Sons; 1972. pp. 1–95. [Google Scholar]
- 14.Carter S F, Leak D J. The isolation and characterisation of a carbocyclic epoxide-degrading Corynebacterium sp. Biocatal Biotransform. 1995;13:111–129. [Google Scholar]
- 15.Castro C E, Bartnicki E W. Biodehalogenation. Epoxidation of halohydrins, epoxide opening, and transhalogenation by a Flavobacterium sp. Biochemistry. 1968;7:3213–3218. doi: 10.1021/bi00849a025. [DOI] [PubMed] [Google Scholar]
- 16.de Bont J A M, van Dijken J P, van Ginkel K G. The metabolism of 1,2-propanediol by the propylene oxide utilizing bacterium Nocardia A60. Biochim Biophys Acta. 1982;714:465–470. [Google Scholar]
- 17.de Bont J A M, Harder W. Metabolism of ethylene by Mycobacterium E20. FEMS Microbiol Lett. 1978;3:89–93. [Google Scholar]
- 18.DuBois G C, Appella E, Levin W, Lu A Y H, Jerina D M. Hepatic microsomal epoxide hydrase. Involvement of a histidine at the active site suggests a nucleophilic mechanism. J Biol Chem. 1978;253:2932–2939. [PubMed] [Google Scholar]
- 19.Faber K, Mischitz M, Kroutil W. Microbial epoxide hydrolases. Acta Chem Scand. 1996;50:249–258. [Google Scholar]
- 20.Gagis A, Fusco A, Benedict J T. A stereochemical study of the ring opening of indene oxide by benzoic acid. J Org Chem. 1972;37:3181–3182. [Google Scholar]
- 21.Griffiths E T, Harries P C, Jeffcoat R, Trudgill P W. Purification and properties of α-pinene oxide lyase from Nocardia sp. strain P18.3. J Bacteriol. 1987;169:4980–4983. doi: 10.1128/jb.169.11.4980-4983.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haeggström J Z, Wetterholm A, Shapiro R, Vallee B L, Samuelsson B. Leukotriene A4 hydrolase: a zinc metalloenzyme. Biochem Biophys Res Commun. 1990;172:965–970. doi: 10.1016/0006-291x(90)91540-9. [DOI] [PubMed] [Google Scholar]
- 23.Hartmans S, de Bont J A M, Harder W. Microbial metabolism of short-chain unsaturated hydrocarbons. FEMS Microbiol Rev. 1989;63:235–264. doi: 10.1016/0168-6445(89)90034-x. [DOI] [PubMed] [Google Scholar]
- 24.Hartmans S, Smits J P, van der Werf M J, Volkering F, de Bont J A M. Metabolism of styrene oxide and 2-phenylethanol in the styrene-degrading Xanthobacter strain 124X. Appl Environ Microbiol. 1989;55:2850–2855. doi: 10.1128/aem.55.11.2850-2855.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs M H J, van den Wijngaard A J, Pentenga M, Janssen D B. Characterization of the epoxide hydrolase from an epichlorohydrin-degrading Pseudomonas sp. Eur J Biochem. 1991;202:1217–1222. doi: 10.1111/j.1432-1033.1991.tb16493.x. [DOI] [PubMed] [Google Scholar]
- 26.Kasai N, Tsujimura K, Unoura K, Suzuki T. Degradation of 2,3-dichloro-1-propanol by a Pseudomonas sp. Agric Biol Chem. 1990;54:3185–3190. [Google Scholar]
- 27.Kiyosue T, Beetham J K, Pinot F, Hammock B D, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of an Arabidopsis cDNA for a soluble epoxide hydrolase gene that is inducible by auxin and water stress. Plant J. 1994;6:259–269. doi: 10.1046/j.1365-313x.1994.6020259.x. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Malfroy B, Kado-Fong H, Gros C, Giros B, Schwartz J-C, Hellmiss R. Molecular cloning and amino acid sequence of rat kidney aminopeptidase M: a member of a super family of zinc-metallohydrolases. Biochem Biophys Res Commun. 1989;161:236–241. doi: 10.1016/0006-291x(89)91586-6. [DOI] [PubMed] [Google Scholar]
- 30.Michaels B C, Ruettinger R T, Fulco A J. Hydration of 9,10-epoxypalmitic acid by a soluble enzyme from Bacillus megaterium. Biochem Biophys Res Commun. 1980;92:1189–1195. doi: 10.1016/0006-291x(80)90412-x. [DOI] [PubMed] [Google Scholar]
- 31.Mischitz M, Faber K, Willetts A. Isolation of a highly enantioselective epoxide hydrolase from Rhodococcus sp. NCIMB 11216. Biotechnol Lett. 1995;17:893–898. [Google Scholar]
- 32.Müller F, Arand M, Frank H, Seidel A, Hinz W, Winkler L, Hänel K, Blée E, Beetham J K, Hammock B D, Oesch F. Visualization of a covalent intermediate between microsomal epoxide hydrolase, but not cholesterol epoxide hydrolase, and their substrates. Eur J Biochem. 1997;245:490–496. doi: 10.1111/j.1432-1033.1997.00490.x. [DOI] [PubMed] [Google Scholar]
- 33.Mullin C A, Hammock B D. Chalcone oxides—potent selective inhibitors of cytosolic epoxide hydrolase. Arch Biochem Biophys. 1982;216:423–439. doi: 10.1016/0003-9861(82)90231-4. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura T, Nagasawa T, Yu F, Watanabe I, Yamada H. Resolution and some properties of enzymes involved in enantioselective transformation of 1,3-dichloro-2-propanol to (R)-3-chloro-1,2-propanediol by Corynebacterium sp. strain N-1074. J Bacteriol. 1992;174:7613–7619. doi: 10.1128/jb.174.23.7613-7619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakamura T, Nagasawa T, Yu F, Watanabe I, Yamada H. Purification and characterization of two epoxide hydrolases from Corynebacterium sp. strain N-1074. Appl Environ Microbiol. 1994;60:4630–4633. doi: 10.1128/aem.60.12.4630-4633.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niehaus W G, Kisic A, Torkelson A, Bednarczyk D J, Schroepfer G J. Stereospecific hydration of cis- and trans-9,10-epoxyoctadecanoic acids. J Biol Chem. 1970;245:3802–3809. [PubMed] [Google Scholar]
- 37.Orning L, Gierse J K, Fitzpatrick F A. The bifunctional enzyme leukotriene-A4 hydrolase is an arginine aminopeptidase of high efficiency and specificity. J Biol Chem. 1994;269:11269–11273. [PubMed] [Google Scholar]
- 38.Rink R, Fennema M, Smids M, Dehmel U, Janssen D B. Primary structure and catalytic mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J Biol Chem. 1997;272:14650–14657. doi: 10.1074/jbc.272.23.14650. [DOI] [PubMed] [Google Scholar]
- 39.Royals E E, Leffingwell J C. Reactions of the limonene 1,2-oxides. I. The stereospecific reactions of the (+)-cis- and (+)-trans-limonene 1,2-oxides. J Org Chem. 1966;31:1937–1944. [Google Scholar]
- 40.Samuelsson B, Funk C D. Enzymes involved in the biosynthesis of leukotriene B4. J Biol Chem. 1989;264:19469–19472. [PubMed] [Google Scholar]
- 41.Stapleton A, Beetham J K, Pinot F, Garbarino J E, Rockhold D R, Friedman M, Hammock B D, Belknap W R. Cloning and expression of soluble epoxide hydrolase from potato. Plant J. 1994;6:251–258. doi: 10.1046/j.1365-313x.1994.6020251.x. [DOI] [PubMed] [Google Scholar]
- 42.Swaving J, de Bont J A M. Microbial transformation of epoxides. Enzyme Microb Technol. 1998;22:19–26. [Google Scholar]
- 43.Swaving J, de Bont J A M, Westphal A, de Kok A. A novel type of pyridine nucleotide-disulfide oxidoreductase is essential for NAD+- and NADPH-dependent degradation of epoxyalkanes by Xanthobacter strain Py2. J Bacteriol. 1996;178:6644–6646. doi: 10.1128/jb.178.22.6644-6646.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas H, Timms C W, Oesch F. Epoxide hydrolases: molecular properties, induction, polymorphisms and function. In: Ruckpaul K, Rein H, editors. Frontiers in biotransformation. Vol. 2. Berlin, Germany: Akademie-Verlag; 1990. pp. 278–337. [Google Scholar]
- 45.Touhara K, Prestwich G D. Juvenile hormone epoxide hydrolase. Photoaffinity labelling, purification, and characterization from tobacco hornworm eggs. J Biol Chem. 1993;268:19604–19609. [PubMed] [Google Scholar]
- 46.van den Wijngaard A J, Janssen D B, Witholt B. Degradation of epichlorohydrin and halohydrins by bacterial cultures isolated from freshwater sediment. J Gen Microbiol. 1989;135:2199–2208. [Google Scholar]
- 47.van der Werf M J, de Bont J A M. Screening for microorganisms converting limonene into carvone. Stud Org Chem. 1998;53:231–234. [Google Scholar]
- 48.van der Werf M J, de Bont J A M, Leak D J. Opportunities in microbial biotransformation of monoterpenes. Adv Biochem Eng Biotechnol. 1997;55:147–177. [Google Scholar]
- 49.Verschueren K H G, Franken S M, Rozeboom H J, Kalk K H, Dijkstra B W. Refined X-ray structure of haloalkane dehalogenase at pH 6.2 and pH 8.2 and implications for the reaction mechanism. J Mol Biol. 1993;232:856–872. doi: 10.1006/jmbi.1993.1436. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J, Reddy J, Roberge C, Senanayake C, Greasham, R. R, Chartrain M. Chiral bio-resolution of racemic indene oxide by fungal epoxide hydrolases. J Ferment Bioeng. 1995;80:244–246. [Google Scholar]