Abstract
Most cancers exhibit aneuploidy, but its functional significance in tumor development is controversial. Here, we describe ReDACT (Restoring Disomy in Aneuploid cells using CRISPR Targeting), a set of chromosome engineering tools that allow us to eliminate specific aneuploidies from cancer genomes. Using ReDACT, we created a panel of isogenic cells that have or lack common aneuploidies, and we demonstrate that trisomy of chromosome 1q is required for malignant growth in cancers harboring this alteration. Mechanistically, gaining chromosome 1q increases the expression of MDM4 and suppresses p53 signaling, and we show that TP53 mutations are mutually-exclusive with 1q aneuploidy in human cancers. Thus, tumor cells can be dependent on specific aneuploidies, raising the possibility that these “aneuploidy addictions” could be targeted as a therapeutic strategy.
INTRODUCTION
Chromosome copy number changes, otherwise known as aneuploidy, are a ubiquitous feature of tumor genomes (1, 2). While the pervasiveness of aneuploidy in cancer has been known for over a century (3, 4), the role of aneuploidy in tumor development has remained controversial (5–8). Chromosome gains have been proposed to serve as a mechanism for increasing the dosage of tumor-promoting genes that are found within altered regions (9, 10). However, proof of this hypothesis is lacking, and it has alternately been suggested that aneuploidy could arise as a result of the loss of checkpoint control that frequently occurs in advanced malignances (11). Indeed, individuals with Down syndrome, which is caused by the triplication of chromosome 21, have a markedly decreased risk of developing most solid cancers, suggesting that in certain cases aneuploidy may actually have tumor-suppressive properties (12).
Our ability to directly interrogate the role of aneuploidy in cancer has historically been limited by the experimental difficulties involved in manipulating entire chromosome arms. Over the past 40 years, cancer researchers have used the standard tools of molecular genetics, including gene overexpression, knockdown, and mutagenesis, to develop a deep understanding of many individual oncogenes and tumor suppressors (13, 14). For instance, the biological functions of genes like KRAS and TP53 were elucidated in part by creating and analyzing isogenic cell lines that express or lack these genes (15, 16). However, existing approaches for single-gene manipulations are insufficient to interrogate the chromosome-scale changes that affect hundreds of genes simultaneously. The consequences of eliminating specific aneuploid chromosomes from human cancer cells have not previously been established.
Studies of individual cancer driver genes led to the discovery of a phenomenon called “oncogene addiction”, in which loss or inhibition of a single oncogene is sufficient to induce cancer regression (17). For example, mutations in KRAS cause the development of pancreas cancer, and genetically ablating KRAS in a “KRAS-addicted” pancreas tumor blocks growth and triggers apoptosis (18). Previous cancer genome sequencing projects have revealed that the aneuploidy patterns observed in human tumors are non-random, and specific events like the gain of chromosome 1q and 8q occur more often than expected by chance (1, 19). We speculated that these recurrent aneuploidies could themselves represent a type of cancer “addiction”, analogous to the concept of oncogene addictions. To investigate this hypothesis, we developed a set of computational and functional techniques to facilitate the analysis of cancer aneuploidy.
RESULTS
Specific chromosome gains recurrently occur early in cancer development
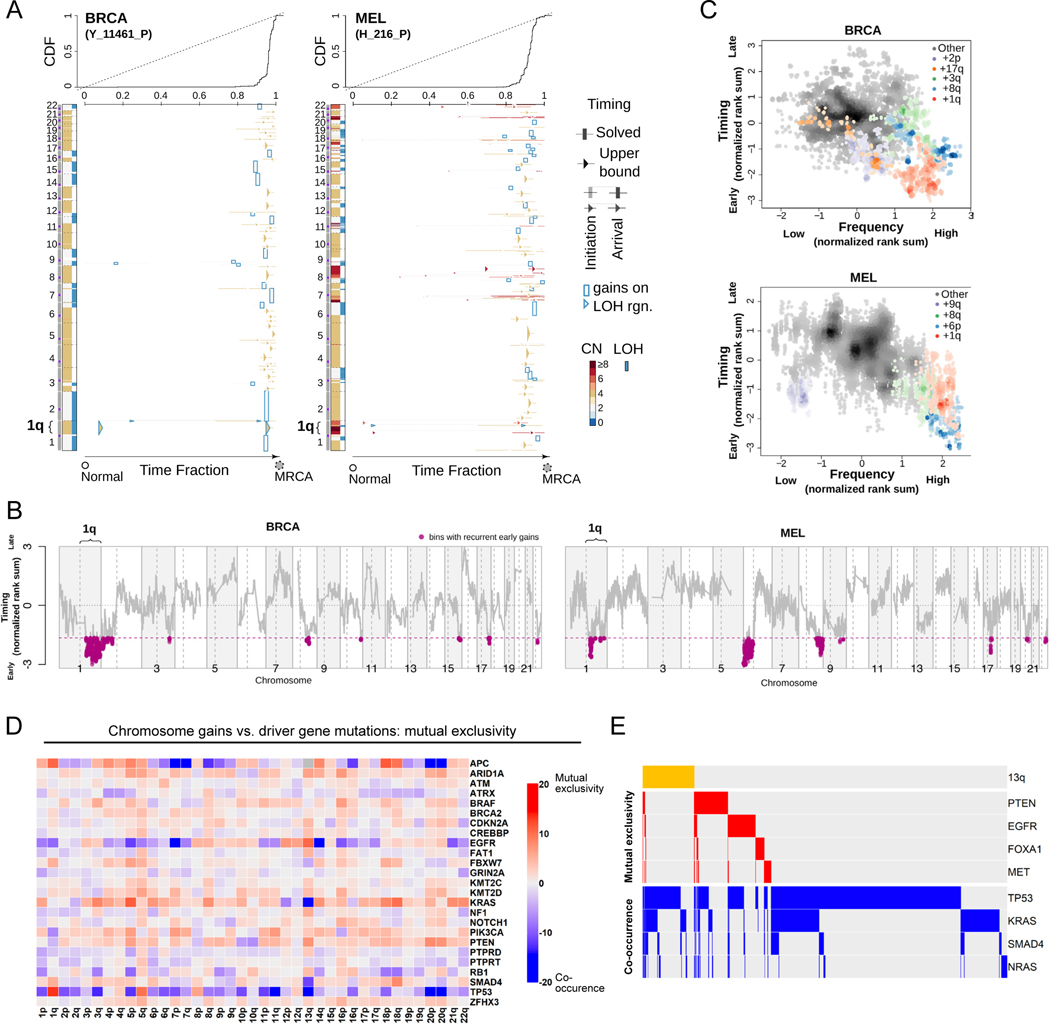
We recently established a computational approach to leverage multi-sample tumor sequencing data to determine the relative timing of somatic copy number alterations (SCNAs) in cancer evolution (20). We applied this tool to investigate the timing of aneuploidy events in a cohort of patients with breast cancer (BRCA) or melanoma (MEL)(21, 22). We modeled the relative timing of SCNAs in whole genome sequences from these tumor samples by assuming that (1) somatic point mutations accumulate over time at a rate that is proportional to DNA copy number, and (2) the multiplicity of early point mutations increases with copy number gains. We found that specific chromosome copy number changes are consistently observed early in tumor development (Fig. 1A-B). Notably, we observed that chromosome 1q gains are recurrently the first copy number alterations that occurs in breast cancer evolution, and these gains are also among the first alterations in melanoma evolution. In general, we found that common aneuploidies arose earlier in tumor development than less-common aneuploidies, in agreement with the assumption that early somatic alterations are likely to be fitness-driving events (Fig. 1C)(23). However, the correlation between frequency and timing was not maintained across all chromosomes. For instance, in breast cancer, chromosome 8q gains and chromosome 1q gains occurred with similar frequencies, but we found that 1q gains consistently arose earlier during tumor development than 8q gains. We conclude that, as previously observed with oncogenic point mutations (24), specific chromosome gains occur in a defined temporal order, and we speculate that aneuploidies that are consistently gained early during tumorigenesis may enhance cancer fitness.
Figure 1. Specific chromosome gains arise early in tumor development and are mutually exclusive with driver gene mutations.
(A) The inferred timing of somatic copy number gains in the evolution of two tumors. A breast tumor is shown on the left and a melanoma on the right. Copy number (CN) states along the genome are shown on the left in each panel and color coded. The plot visualizes the time fraction of somatic evolution from germline to the most recent common ancestor (MRCA) of the patient tumor sample. For each copy number segment, the inferred timing is shown as a rectangle (exactly solved timing) or an arrow (upper bounds of timing when the timing solutions are not unique) with the same color-coding as its CN. The top panel shows the cumulative distribution (CDF) of the timing. Genome doubling (GD) can be observed as the punctuated gains occurring in a narrow time window, and chromosome 1q gains appear to be extremely early and preceding GD in these two tumors.
(B) Recurrent early gains of chromosome 1q in BRCA (n = 38 tumor samples) and MEL (n = 37 tumor samples). For each tumor type, we converted the timing of gains into ranks for genomic bins within a patient and computed the rank sums across patients for each bin. The normalized rank sums for each genomic bin are shown for BRCA and MEL. The large negative values indicate recurrent early initiating gains. We used the normalized rank sums to test against the null hypothesis (no regions show recurrent early gain across patients). Bins from chromosome 1q reject this null for both tumor types (with 90% confidence level).
(C) The timing of a gain compared to the frequency of its occurrence in BRCA (n = 38 tumor samples) and MEL (n = 37 tumor samples). The points on the plots show the timing of gain of a genomic bin versus its frequency of copy number gain. Colors represent chromosomal arms, and color darkness indicates the density of points. Both the timings and frequencies were transformed into normalized rank sums (see Methods). In total, 15 out of 21 BRCA patients and 24 out of 37 MEL patients exhibited arm-scale gains of chromosome 1q.
(D) A pan-cancer analysis of mutual exclusivity between mutations in 25 commonly-mutated cancer genes and chromosome arm gain events. The complete results of this analysis are included in Table S1.
(E) Mutual exclusivity and co-occurrence patterns between one representative chromosome gain (+13q, orange bars at the top), and point mutations in several different cancer driver genes.
Specific chromosome gains are associated with altered mutational patterns and cancer progression
In instances where two oncogenes converge to activate the same pathway, cancers frequently acquire mutations in either gene but not both (25). If chromosome gains play an oncogene-like role supporting cancer growth, then specific aneuploidies may also be expected to exhibit mutual exclusivity with individual oncogenic mutations. To investigate this possibility, we calculated patterns of mutual exclusivity between chromosome arm gains and mutations across 23,544 cancer patients (26, 27). We detected several hundred instances in which aneuploidies and mutations co-occur less often than expected by chance both within individual cancer types and in a pan-cancer analysis (Fig. 1D-E, S1, and Table S1). For instance, KRAS mutations are mutually exclusive with chromosome 18q gains in pancreatic cancer, while BRAF mutations are mutually exclusive with chromosome 20q gains in colorectal cancer (Fig. S1 and Table S1). These results are consistent with our hypothesis that specific chromosome gains can play an oncogene-like role in cancer, thereby making the acquisition of certain oncogenic mutations redundant in the presence of that aneuploidy.
High levels of aneuploidy are generally associated with poor cancer patient outcomes (28–30). However, it is less clear whether specific copy number changes drive tumor progression, or whether the aneuploid state itself represents a universal risk factor. We calculated the association between patient outcome and copy number gains affecting every chromosome band across 10,884 patients and 33 cancer types from The Cancer Genome Atlas (TCGA)(31). We discovered that certain copy number alterations were commonly prognostic across multiple cancer types, particularly gains affecting chromosome 1q (Fig. S2A-C and Table S2A). The strong association between 1q gains and disease progression was robust to the inclusion of multiple clinical variables, including patient age, sex, tumor stage, and tumor grade (Fig. S2D and Table S2B). 1q copy number correlated with hallmarks of aggressive disease in genetically-diverse cancer types, including with Gleason score in prostate adenocarcinoma and with thrombocytopenia in acute myeloid leukemia (Fig. S2E). We performed a similar analysis for cancer-associated mutations, and we found that the only gene for which mutations were prognostic in more than four cancer types was TP53 (Fig. S2A and S2D). These results illustrate that specific chromosome gain events, particularly involving regions of chromosome 1q, are robust pan-cancer markers for the risk of disease progression.
Loss of trisomy 1q blocks malignant growth in human cancers
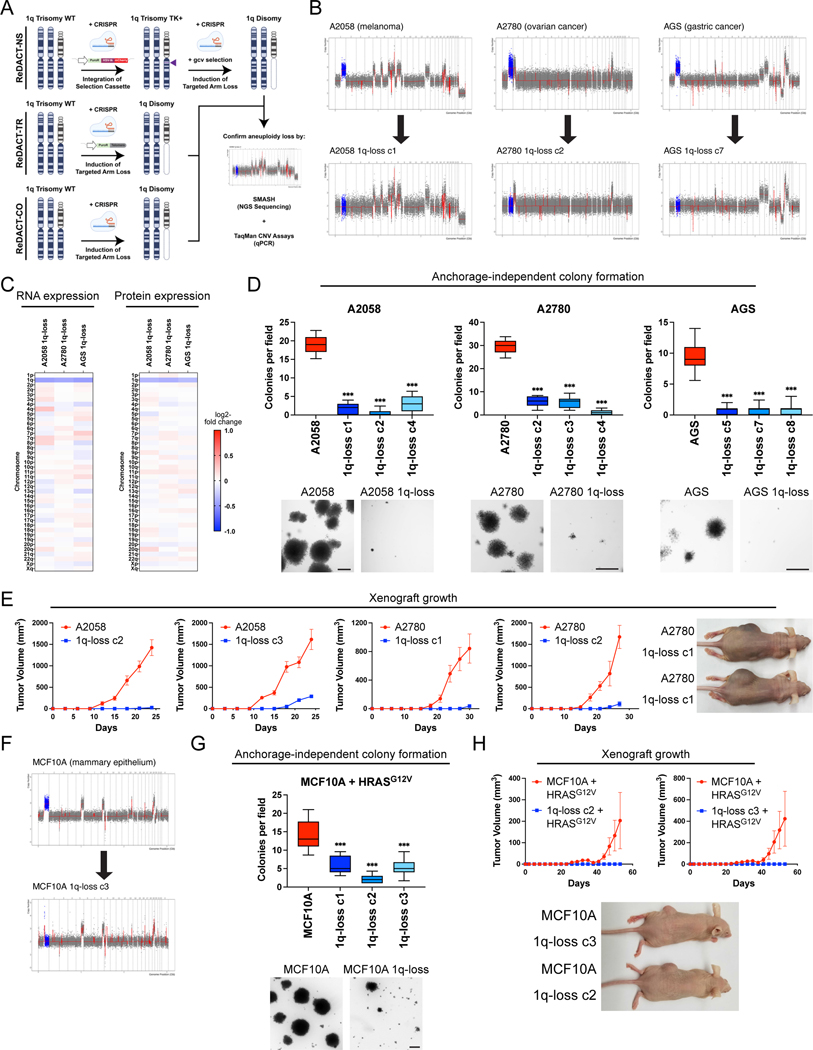
The computational analyses described above highlighted several similarities between chromosome copy number gains and driver mutations, raising the possibility that these aneuploidies could represent oncogene-like cancer addictions. The oncogene addiction paradigm was first established by developing genetic techniques to eliminate individual genes from established cancer cell lines (16, 17). In order to conduct comparable assays with aneuploidy, we created a set of approaches collectively called ReDACT (Restoring Disomy in Aneuploid cells using CRISPR Targeting) (Fig. 2A). In the first approach, called ReDACT-NS (Negative Selection), we integrate a copy of herpesvirus thymidine kinase (HSV-TK) onto an aneuploid chromosome of interest. Then, the cells are transfected with a gRNA that cuts between the integrant and the centromere and treated with ganciclovir, which is toxic to cells that express HSV-TK (32). Loss of the aneuploid chromosome harboring HSV-TK allows cells to survive ganciclovir selection. In the second approach, called ReDACT-TR (Telomere Replacement), cells are co-transfected with a gRNA that cuts near the centromere of an aneuploid chromosome along with a cassette encoding ~100 repeats of the human telomere seed sequence (33). CRISPR cleavage coupled with integration of the telomeric seed sequence leads to loss of an aneuploid chromosome arm and formation of a de novo telomere. In the third approach, called ReDACT-CO (CRISPR Only), we took advantage of prior reports demonstrating that in rare circumstances CRISPR cleavage by itself is sufficient to trigger chromosome loss (34, 35), and we transfected cells with a gRNA targeting an aneuploid chromosome arm without any other selection markers. We successfully applied all three approaches to create clones derived from human cell lines that had lost specific aneuploid chromosomes.
Figure 2. Phenotypic effects of losing chromosome 1q-aneuploidy.
(A) Chromosomal engineering strategies for the targeted deletion of chromosome arms: (1) ReDACT-NS: using CRISPR-Cas9 homology-directed repair, we integrated a positive-negative selection cassette encoding a fluorescent reporter, a positive selection marker, and a negative selection marker (HSV thymidine kinase) at a centromere-proximal region on chromosome 1q. We induced arm loss by generating a dsDNA break centromere-proximal to the cassette with Cas9, and isolated clonal populations of cells that were ganciclovir-resistant. (2) ReDACT-TR: We induced arm loss by generating a dsDNA break at a centromere-proximal location with Cas9 while providing cells with an ectopic telomere seed sequence for repair. (3) ReDACT-CO: We induced arm loss by generating a dsDNA break at a centromere-proximal location with Cas9, and isolated clonal populations of cells. For all three approaches, we screened clonal populations of cells for targeted chromosome loss through TaqMan CNV assays and validated their karyotypes through SMASH sequencing.
(B) Representative SMASH karyotypes of the 1q-disomic clones generated from the 1q-trisomic cancer cell lines A2780, AGS, and A2058. Chromosome 1q is highlighted in blue. A complete list of aneuploidy-loss clones and how they were generated is included in Table S3.
(C) 1q-disomic clones display decreased RNA expression and protein expression of genes encoded on chromosome 1q. RNA expression data were obtained through bulk RNA-seq and represent the average expression of genes by chromosome arm across multiple 1q-disomic clones for each cell line. Protein expression data were obtained through mass spectrometry, and representative data from one 1q-disomic clone are shown for each cell line. Data are log2 transformed, normalized to the parental cell line, and adjusted so that the mean expression across all chromosomes is 0.
(D) 1q-disomic clones exhibit decreased anchorage-independent growth. The micrographs display representative images of colony formation for 1q-trisomic and 1q-disomic clones.
(E) 1q-disomic clones exhibit impaired xenograft growth in vivo. 1q-trisomic and 1q-disomic cells were injected contralaterally and subcutaneously into immunocompromised mice. The graphs display the mean ± SEM for each trial. Representative mice are shown on the right.
(F) SMASH karyotype of a 1q-disomic clone generated from the mammary epithelial cell line MCF10A. Chromosome 1q is highlighted in blue.
(G) 1q-disomic MCF10A clones transduced with HRASG12V exhibit decreased anchorage-independent growth relative to 1q-trisomic MCF10A cells.
(H) 1q disomic MCF10A clones transduced with HRASG12V clones exhibit impaired xenograft growth in vivo. 1q-trisomic and 1q-disomic cells were injected contralaterally and subcutaneously into immunocompromised mice. The graphs display the mean ± SEM for each trial. Representative mice are shown below.
For anchorage-independent growth assays in D and G, the boxplots represent the 25th, 50th, and 75th percentiles of colonies per field, while the whiskers represent the 10th and 90th percentiles. Unpaired t-test, n = 15 fields of view, data from representative trial (n ≥ 2 total trials). Representative images are shown below. Scale bars = 250 μm.
**p < 0.005, ***p < 0.0005
We first focused on aneuploidies of chromosome 1q, as we found that 1q gains were an early event in multiple cancer types and were strongly associated with disease progression (Fig. 1 and S2). We targeted the 1q trisomy in the A2058 melanoma cell line, AGS gastric cancer cell line, and A2780 ovarian cancer cell line. We generated multiple independent derivatives of each line in which a single copy of chromosome 1q had been eliminated, thereby producing cell lines that were disomic rather than trisomic for chromosome 1q. We verified loss of the 1q trisomy and the absence of any other chromosome copy number changes using SMASH-Seq, a sequencing-based approach to determine DNA copy number (36), and by G-banding analysis of metaphase spreads (Fig. 2B, S3-S4, and Table S3). Loss of the 1q trisomy decreased the expression of genes encoded on chromosome 1q by an average of 26% at the RNA level and 21% at the protein level (Fig. 2C). These results suggest that chromosome loss causes a substantial downregulation of genes encoded on an aneuploid chromosome, though these effects can be buffered to some extent by cellular dosage compensation (37).
Next, we tested whether losing the 1q trisomy affects malignant growth in cancer cells. Toward that end, we quantified anchorage-independent colony formation, an in vitro proxy for malignant potential (38), in the 1q-trisomic and 1q-disomic cells. While 1q-trisomic A2058, A2780, and AGS cells displayed robust colony formation, multiple independent 1q-disomic clones derived from each cell line exhibited minimal anchorage-independent growth (Fig. 2D). We then performed contralateral subcutaneous injections with each cell line to test whether aneuploidy-loss affected tumor growth in vivo. Consistent with our colony formation assays, we observed that 1q-trisomic A2058 and A2780 cells rapidly formed large tumors, while 1q-disomic cells displayed minimal tumor growth (Fig. 2E). At the end of these assays, the trisomic cells had formed tumors that were on average 25-fold larger than the tumors formed by the 1q-disomic cells. For the AGS cancer cell line, neither the trisomic nor the disomic cells formed tumors following subcutaneous injection (Fig. S5). Finally, we performed proliferation assays to measure the doubling time of the 1q-trisomic and the 1q-disomic cells in culture (Fig. S6A-C). The aneuploidy-loss clones divided more slowly in vitro compared to the 1q-trisomic cells, though the difference in doubling time (~35%) was substantially less than the differences observed in the soft agar and xenograft assays. In total, these results suggest that multiple human cancer cell lines are dependent on the presence of a third copy of chromosome 1q to support malignant growth, and elimination of this aneuploid chromosome compromises their tumorigenic potential. Furthermore, we note that this phenotypic pattern, in which aneuploidy loss causes a moderate effect on in vitro doubling but a severe effect on anchorage-independent growth and xenograft formation, resembles the previously-reported consequences of eliminating bona fide oncogene addictions (16, 39).
Loss of trisomy 1q prevents malignant transformation
We discovered that 1q gains were commonly the first copy number alteration to occur during breast tumor development (Fig. 1A-C). We therefore hypothesized that, in addition to being required for cancer growth, aneuploidy of chromosome 1q may directly promote cellular transformation. To test this, we performed chromosome engineering in MCF10A, an immortal but non-tumorigenic mammary epithelial cell line. SMASH-Seq revealed that this cell line harbors a trisomy of chromosome 1q, and we successfully applied ReDACT-CO and ReDACT-TR to generate derivatives of MCF10A with two copies rather than three copies of 1q (Fig. 2F, S3D, and Table S3). We then attempted to transform the 1q-trisomic and 1q-disomic cells by transducing them with a retrovirus encoding the HRASG12V oncogene. HRASG12V expression was sufficient to transform trisomic MCF10A, as these cells were able to form colonies in soft agar and grow as xenografts in nude mice (Fig. 2G-H). In contrast, 1q-disomic MCF10A clones expressing HRASG12V exhibited impaired colony formation and were unable to produce tumors in vivo, demonstrating that loss of the trisomic chromosome prevented cellular transformation. These results are consistent with our finding that 1q gains are an early event during breast cancer development and demonstrate that specific aneuploidies can cooperate with oncogenes to transform non-malignant cells.
Robust anchorage-independent growth in human cancer cell lines subjected to CRISPR cutting and ganciclovir selection
In order to confirm that our findings were a specific consequence of aneuploidy loss, we generated and tested a series of control clones subjected to various CRISPR manipulations that did not induce loss of the 1q trisomy (Fig. S7 and S8A). These control clones included:
Cell lines harboring a CRISPR-mediated integration of the HSV-TK cassette that were not subjected to selection for 1q-loss
Cell lines in which the HSV-TK cassette was deleted using two gRNAs followed by ganciclovir selection
Cell lines transfected with a 1q-targeting gRNA in which the lesion was repaired without inducing chromosome loss
Cell lines transfected with a gRNA targeting the non-coding Rosa26 locus
Cell lines in which dual CRISPR guides were used to generate segmental deletions of genes encoding olfactory receptors on chromosome 1q
Cell lines in which CRISPR was used to delete a terminal segment on chromosome 1q, eliminating the telomere and decreasing the copy number of 26 out of 968 protein-coding genes on the chromosome
We performed SMASH-Seq on each control clone that we generated and we confirmed that each clone retained an extra copy of chromosome 1q (Fig. S7). We then measured the effects of these manipulations on anchorage-independent growth. We found that every control clone maintained the ability to form colonies in soft agar, with some variability between independent clones. Across the three cancer cell lines and 37 different control clones, we observed that the 1q-disomic clones exhibited worse anchorage-independent growth than every control clone that we generated (Fig. S8B-E). These results indicate that the deficiencies in malignant growth exhibited by the 1q-disomic clones are not a result of our use of CRISPR or ganciclovir selection.
Eliminating different cancer aneuploidies produces distinct phenotypic consequences
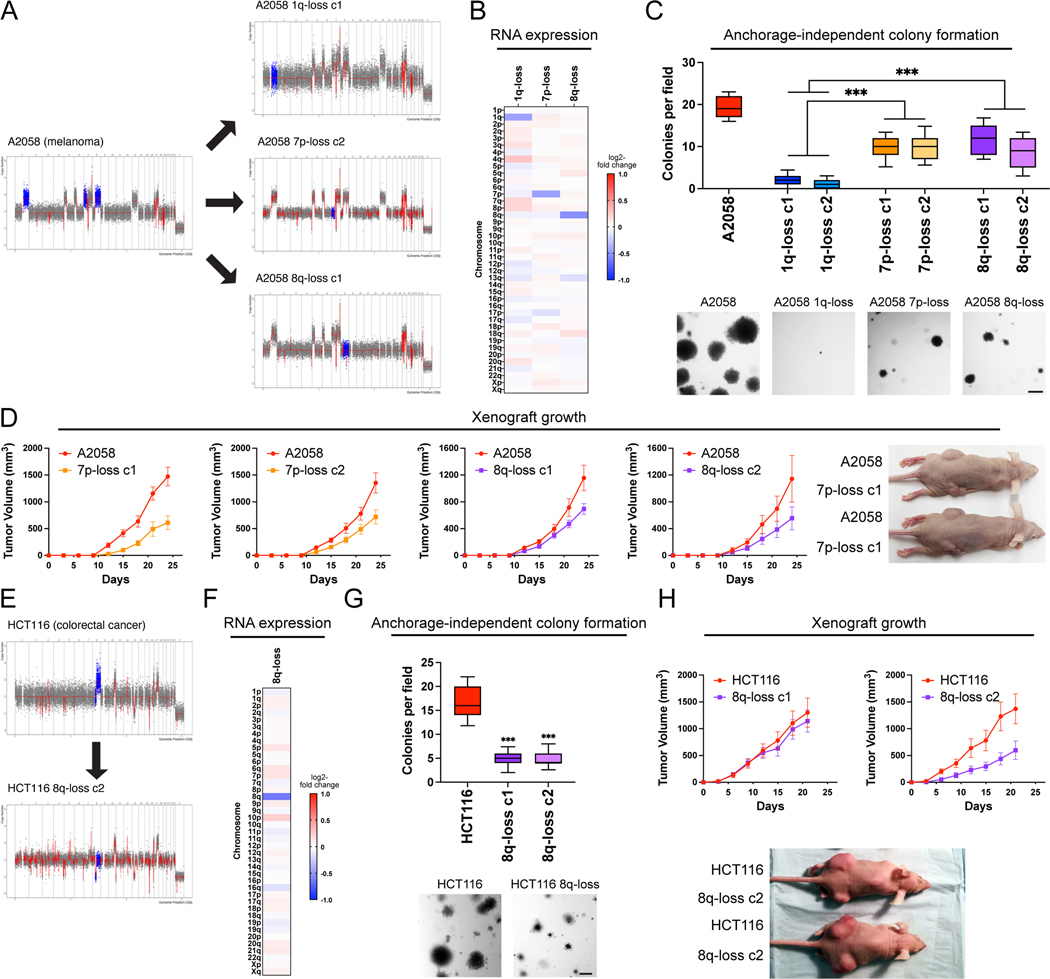
To further investigate the consequences of inducing aneuploidy loss, we used ReDACT to eliminate the trisomy of either chromosome 7p or 8q from A2058 melanoma cells. SMASH-Seq confirmed the desired aneuploidy-loss events without other karyotypic changes (Fig. 3A, S9, and Table S3). As expected, loss of either trisomy 7p or trisomy 8q resulted in a decrease in the expression of genes encoded on the affected chromosomes (Fig. 3B). 7p-disomic and 8q-disomic clones exhibited impaired anchorage-independent growth compared to a panel of control clones, though this defect was not as severe as the defect observed among A2058 1q-disomic clones (Fig. 3C and S10). In vitro doubling times were also closer to wild-type levels for 7p-disomic and 8q-disomic cells compared to 1q-disomic cells (Fig. S6D). Finally, we performed subcutaneous injections of the 7p-disomic and the 8q-disomic cells in nude mice, and we found that loss of either the 7p or the 8q trisomy resulted in a moderate decrease in tumor growth (Fig. 3D). At the end of the assay, the wild-type tumors were on average two-fold larger than the tumors formed by either 7p-disomic or 8q-disomic cells, compared to a 30-fold difference between A2058 wild-type and 1q-disomic tumors (Fig. 2E). In total, these results indicate that A2058 melanoma cells exhibit a greater degree of “addiction” to the 1q trisomy compared to the trisomies of chromosome 7p or 8q.
Figure 3. Variable degrees of addiction to aneuploidy of chromosome 1q, 7p, and 8q.
(A) Representative SMASH karyotypes of the 1q-disomic, 7p-disomic, and 8q-disomic clones generated from the melanoma cell line A2058. Trisomy of chromosomes 1q, 7p, and 8q are highlighted in blue in the parental cell line on the left, and the respective targeted chromosome loss is highlighted in blue in the derived clones on the right. A complete list of aneuploidy-loss clones and how they were generated is included in Table S3.
(B) 1q-disomic, 7p-disomic, and 8q-disomic clones in A2058 exhibit decreased RNA expression of genes encoded on the targeted chromosome. RNA expression data were obtained through bulk RNA-seq and represent the average expression of genes by chromosome arm across multiple aneuploidy-loss clones for each targeted chromosome. Data are log2 transformed, normalized to the parental cell line, and adjusted so that the mean expression across all chromosomes is 0.
(C) 7p-disomic and 8q-disomic clones in A2058 exhibit a milder deficit in anchorage-independent growth as compared to 1q-disomic clones. The micrographs display representative images of colony formation for the indicated cell lines.
(D) 7p-disomic and 8q-disomic clones in A2058 exhibit a moderate defect in xenograft growth. Wild-type (7p-trisomic and 8q-trisomic) cells and either 7p-disomic or 8q-disomic cells were injected contralaterally and subcutaneously into immunocompromised mice. The graphs display the mean ± SEM for each trial. Representative mice are shown on the right.
(E) SMASH karyotype of an 8q-disomic clone generated from the colorectal cancer cell line HCT116. Chromosome 8q is highlighted in blue.
(F) 8q-disomic clones in HCT116 exhibit decreased RNA expression of genes encoded on chromosome 8q. RNA expression data were obtained through bulk RNA-seq and represent the average expression of genes by chromosome arm across multiple aneuploidy-loss clones for each cell line. Data are log2 transformed, normalized to the parental cell line, and adjusted so that the mean expression across all chromosomes is 0.
(G) 8q-disomic clones in HCT116 exhibit decreased anchorage-independent growth. The micrographs display representative images of colony formation for the indicated cell lines.
(H) 8q-disomic clones in HCT116 exhibit variable xenograft growth. 8q-trisomic and 8q-disomic cells were injected contralaterally and subcutaneously into immunocompromised mice. The graphs display the mean ± SEM for each trial. Representative mice are shown below the graphs.
For anchorage-independent growth assays in C and G, boxes represent the 25th, 50th, and 75th percentiles of colonies per field, while the whiskers represent the 10th and 90th percentiles. Unpaired t-test, n = 15 fields of view, data from representative trial (n ≥ 2 total trials). Representative images are shown below. Scale bars = 250 μm.
***p < 0.0005
To explore the consequences of losing chromosome 8q aneuploidy in a distinct cancer type, we eliminated the 8q trisomy from the colorectal cancer cell line HCT116 (Fig. 3E-F, S9C and Table S3). Consistent with our observations in A2058, loss of the 8q trisomy decreased but did not fully prevent anchorage-independent growth in HCT116 (Fig. 3G). We then tested xenograft formation in the HCT116 8q-disomic cells, and we observed that one 8q-disomic clone exhibited a moderate defect in tumor growth while a second clone was able to form tumors at levels comparable to the trisomic parental line (Fig. 3H). These results demonstrate that eliminating aneuploid chromosomes has variable effects, depending on the identity of the chromosome and the genetic background of the cancer.
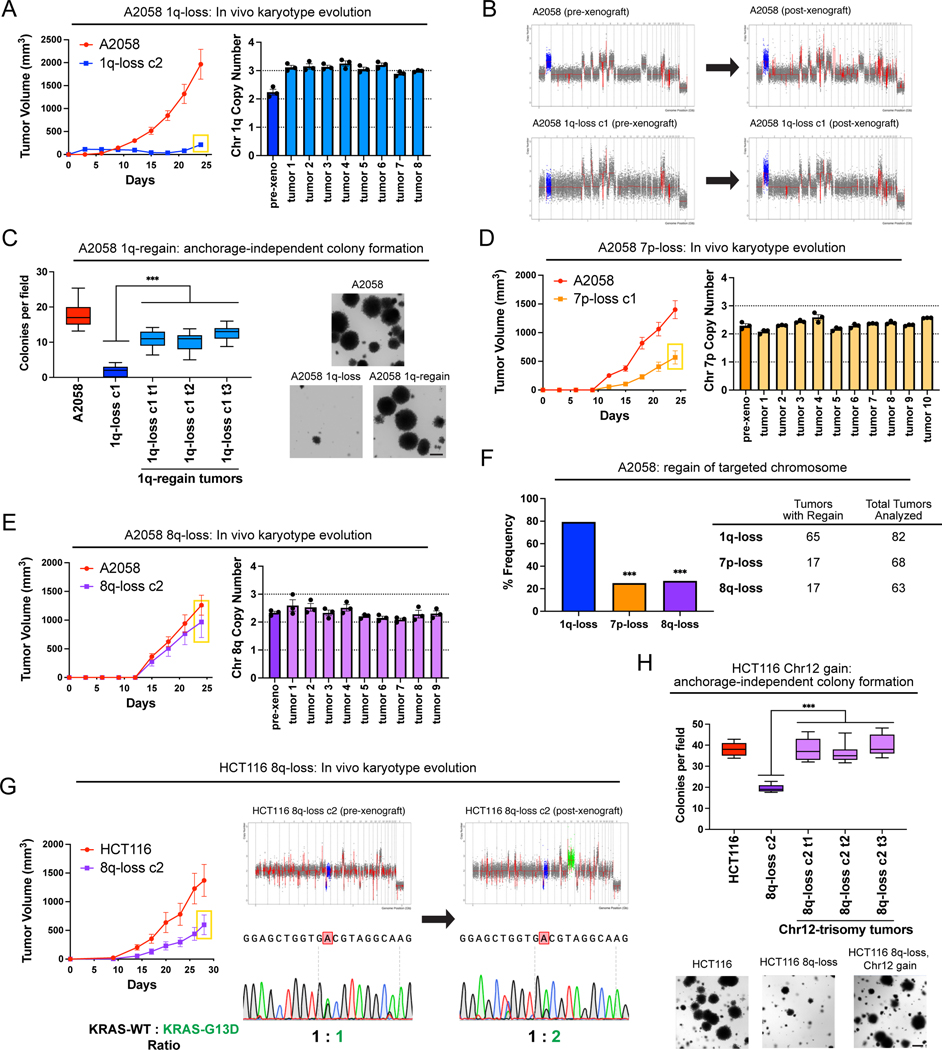
Karyotype evolution and 1q trisomy restoration after aneuploidy loss
A hallmark of oncogene addictions is that loss or inhibition of a driver oncogene results in strong and rapid selection to re-establish oncogenic signaling (40). For instance, when EGFR-driven lung cancers are treated with an EGFR inhibitor, these cancers evolve to acquire specific mutations, including EGFRT790M (which restores EGFR activity) and KRASG13D (which activates a parallel oncogenic pathway)(41). We sought to investigate whether elimination of an “aneuploidy addiction” would also result in evolutionary pressure to restore the lost aneuploidy. We injected 1q-disomic A2058 cells into nude mice and then determined the copy number of chromosome 1q in the resulting xenografts using qPCR (Fig. 4A). We discovered that 65 out of 82 1q-disomic xenografts re-acquired an extra copy of chromosome 1q, demonstrating strong selective pressure to regain the initial 1q aneuploidy. We subjected 20 of these post-xenograft clones to SMASH-Seq, and we found that chromosome 1q-regain was the only detectable chromosome-scale copy number change (Fig. 4B and S11A). No gross karyotypic changes were observed when the parental 1q-trisomic cells were grown as xenografts (Fig. 4B and S11B). If loss of the chromosome 1q trisomy compromises malignant potential, then we would expect that regaining 1q aneuploidy would restore cell fitness. Consistent with this, we found that cells that had re-acquired the 1q trisomy exhibited increased clonogenicity compared to 1q-disomic cells when grown under anchorage-independent conditions (Fig. 4C).
Figure 4. Cancers rapidly recover chromosome 1q aneuploidy.
(A) A2058 1q-disomic cells frequently evolve to recover a third copy of chromosome 1q during xenograft growth.
(B) Representative SMASH karyotypes of A2058 wildtype and 1q-disomic tumors. The initial karyotypes for these lines prior to the xenograft assay are shown on the left, and karyotypes of tumors following the xenograft assay are shown on the right. Chromosome 1q is highlighted in blue.
(C) 1q-disomic clones that have evolved to regain 1q trisomy following xenograft growth exhibit increased anchorage-independent growth relative to the pre-xenograft 1q-disomic parental cells.
(D) Variable evolution of 7p-disomic cells to recover a third copy of chromosome 7p during xenograft growth.
(E) Variable evolution of 8q-disomic cells to recover a third copy of chromosome 8q during xenograft growth.
(F) Regain of trisomy 1q occurs more frequently than regain of trisomy 7p or trisomy 8q. Tumors were classified as exhibiting regain if the mean copy number of the targeted chromosome was ≥ 2.5, as determined through TaqMan copy number assays. n = 213 tumors, chi-squared test.
(G) HCT116 8q-disomic clones evolve to gain a copy of chromosome 12 during xenograft assays, resulting in the acquisition of an extra copy of the KRASG13D allele. Cell lines were rederived from tumors harvested at the endpoint of xenograft assays, and subjected to SMASH karyotyping and Sanger sequencing of KRAS. The xenograft growth curve is shown on the left, and representative SMASH karyotype profiles and Sanger sequencing chromatograms pre- and post-xenograft are shown on the right. Chromosome 8q is highlighted in blue and chromosome 12 is highlighted in green.
(H) 8q-disomic clones that have evolved to acquire trisomy of chromosome 12 following xenograft growth exhibit increased anchorage-independent growth relative to the pre-xenograft 8q-disomic parental cells.
For copy number profiling in A, D, and E, cell lines were rederived from tumors at the endpoint of the xenograft assays, and chromosome copy number was determined through TaqMan copy number assays. Mean ± SEM, n = 3 probes on targeted chromosome, data from representative trials are shown (n = 2 total trials). The corresponding xenograft assays are shown on the left.
For the anchorage-independent growth assays in C and H, the boxes represent the 25th, 50th, and 75th percentiles of colonies per field, while the whiskers represent the 10th and 90th percentiles. Unpaired t-test, n = 15 fields of view, data from representative trial (n ≥ 2 total trials). Representative images are shown on the right. Scale bars = 250 μm.
***p < 0.0005
Next, we assessed karyotype evolution following in vivo growth of A2058 7p-disomic and 8q-disomic clones. Interestingly, 17 out of 68 7p-disomic xenografts and 17 out of 63 8q-disomic xenografts were found to exhibit 7p and 8q trisomy regain, respectively (Fig. 4D-E). These rates of chromosome re-gain were significantly lower than the rates that we observed for chromosome 1q (P < 0.0001, chi-square test; Fig. 4F). These results suggest that there is moderate selective pressure to restore 7p and 8q trisomies and stronger selective pressure to restore 1q trisomy in A2058.
We then sought to determine whether we could observe evolutionary pressure to restore chromosome 1q aneuploidy when 1q-disomic cells were grown in vitro. Toward that end, we passaged A2058, A2780, and AGS 1q-trisomic and 1q-disomic cancer cell lines for thirty days in culture and then we assessed their karyotypes. Similar to our in vivo results, we uncovered multiple instances in which 1q-disomic cells independently regained an extra copy of chromosome 1q over the course of the assay (Fig. S12). We reasoned that there were two possible sources for the regained chromosome: the third copy of 1q could result from the missegregation of an endogenous copy of chromosome 1q, or the disomic cell line could have been out-competed by an exogenous population of trisomic cells (for instance, from metastatic colonization when 1q-disomic and 1q-trisomic xenografts were grown in the same mouse). To differentiate between these possibilities, we identified SNPs on 1q that were heterozygous in the parental cell line and that exhibited loss-of-heterozygosity (LOH) following 1q-elimination. We reasoned that if LOH was maintained following 1q-regain, then that would be evidence for an endogenous missegregation event, while the reappearance of heterozygosity would be evidence of an exogenous cell population (Fig. S13A). Sanger sequencing analysis revealed that both potential causes of 1q-regain were observed during these evolution experiments (Fig. S13B-C). In both cases, 1q-regain correlated with restored anchorage-independent growth relative to the 1q-disomic clones, further verifying the link between 1q copy number and cell fitness (Fig. S13D-E).
Finally, we assessed karyotype evolution in xenografts produced by 8q-disomic HCT116 cells (Fig. 4G). We found that 0 out of 13 tumors regained the trisomy of chromosome 8q, but 7 out of 13 tumors gained a de novo trisomy of chromosome 12. HCT116 cells are driven by a heterozygous KRASG13D mutation (16), and KRAS is encoded on chromosome 12. Sanger sequencing analysis revealed that every chromosome 12-trisomic tumor had amplified the copy of chromosome 12 harboring the mutant KRASG13D allele (Fig. 4G). Increasing dosage of mutant KRAS has previously been associated with enhanced tumor fitness (42), and we observed that these chromosome 8q-disomic/chromosome 12-trisomic cells exhibited superior anchorage-independent growth relative to the chromosome 8q-disomic/chromosome 12-disomic pre-xenograft population (Fig. 4H). In total, these results suggest that aneuploidy loss creates strong selective pressure for karyotype evolution, and the effects of aneuploidy loss can be suppressed in cis (by regaining the lost chromosome) or in trans (by acquiring a beneficial secondary alteration).
Chromosome 1q aneuploidy suppresses p53 signaling by increasing MDM4 expression
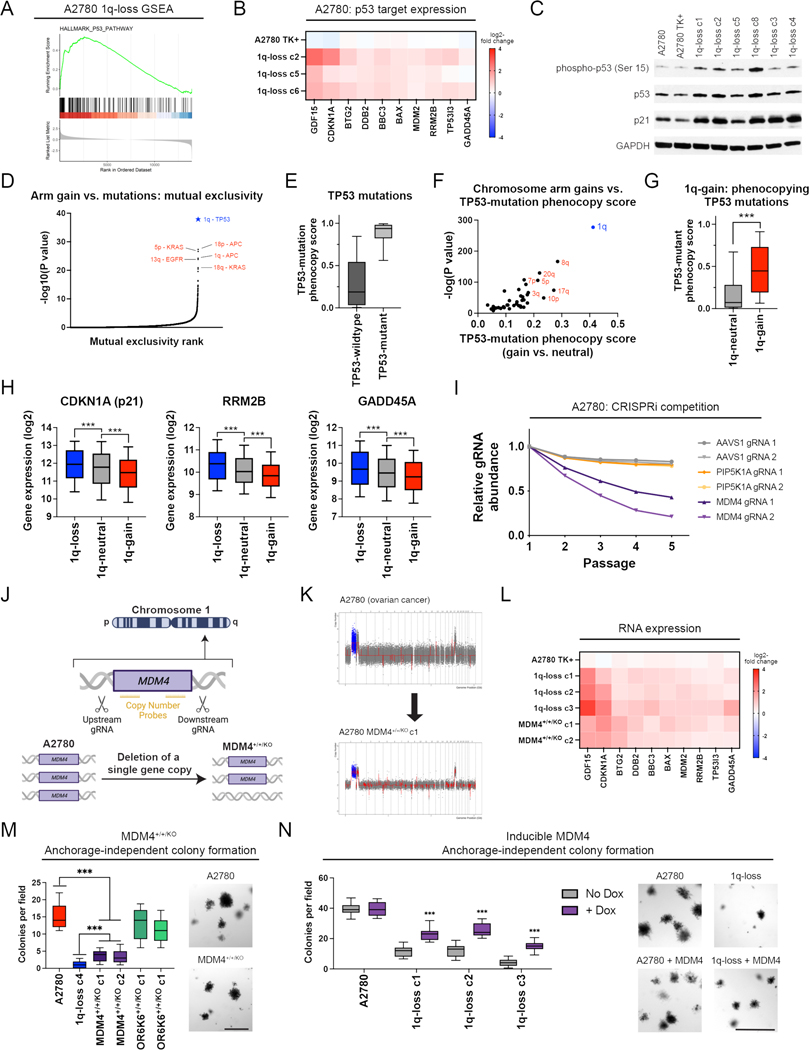
We sought to uncover the biological mechanism underlying the addiction to chromosome 1q aneuploidy. RNA-seq analysis revealed that elimination of the 1q trisomy caused upregulation of tumor suppressor p53 target genes in A2780 and MCF10A, which are both wild-type for TP53 (Fig. 5A-B and Fig. S14A-B). Western blotting confirmed that 1q-disomic clones exhibited increased phosphorylation of p53 at serine-15 and increased expression of the canonical p53 target p21 (Fig. 5C)(43). These results were not a by-product of CRISPR mutagenesis, as A2780 cells harboring a CRISPR-mediated integration of HSV-TK did not display evidence of p53 activation (Fig. 5B-C). Additionally, 1q-disomic A2780 and MCF10A cells exhibited a delay in the G1 phase of the cell cycle and increased senescence-associated beta-galactosidase staining, both of which are associated with p53-mediated tumor suppression (Fig. S14C-F)(44). These results suggest that the chromosome 1q trisomy inhibits p53 signaling, and elimination of this trisomy antagonizes malignant growth at least in part by triggering p53 activation.
Figure 5. A single extra copy of MDM4 suppresses p53 signaling and contributes to the 1q trisomy addiction.
(A) GSEA analysis of A2780 RNA-seq data reveals upregulation of the p53 pathway in the 1q-disomic clones, relative to the parental trisomy.
(B) A heatmap displaying the upregulation of 10 p53 target genes in A2780 1q-disomic clones. The TK+ clone indicates a clone that harbors the CRISPR-mediated integration of the HSV-TK transgene but that was not treated to induce chromosome 1q-loss.
(C) Western blot analysis demonstrating activation of p53 signaling in 1q-disomic clones. GAPDH was analyzed as a loading control. The TK+ clone indicates a clone that harbors the CRISPR-mediated integration of the HSV-TK transgene but that was not treated to induce chromosome 1q-loss.
(D) A waterfall plot highlighting the most-significant instances of mutual exclusivity between chromosome arm gains and mutations in cancer-associated genes. The complete dataset for mutual exclusivity and co-occurrence is included in Table S1.
(E) Boxplots displaying the TP53-mutation phenocopy signature (45) in cancers from TCGA, split based on whether the cancers harbor a non-synonymous mutation in TP53.
(F) A scatterplot comparing the association between chromosome arm gains and the TP53-mutation phenocopy signature (45) in TP53-wildtype cancers from TCGA. Cancers with chromosome 1q gains are highlighted in blue.
(G) Boxplots displaying the TP53-mutation phenocopy signature (45) in cancers from TCGA, split based on whether tumors harbor a gain of chromosome 1q. Only TP53-wildtype cancers are included in this analysis.
(H) Boxplots displaying the expression of three p53 target genes – CDKN1A (p21), RRM2B, and GADD45A – in cancers from TCGA split based on the copy number of chromosome 1q. Only TP53-wildtype cancers are included in this analysis.
(I) A CRISPRi competition assay demonstrates that gRNAs targeting MDM4 drop out over time in A2780 cells. In contrast, gRNAs targeting AAVS1 and PIP5K1A, another gene encoded on chromosome 1q, exhibit minimal depletion.
(J) A schematic displaying the strategy for using paired CRISPR gRNAs to delete a single copy of MDM4 in a cell line with a trisomy of chromosome 1q.
(K) SMASH karyotype demonstrating maintenance of the chromosome 1q trisomy in an MDM4+/+/KO clone. Chromosome 1q is highlighted in blue.
(L) 1q-disomic clones and MDM4+/+/KO clones in A2780 exhibit comparable upregulation of p53 transcriptional targets, as determined through TaqMan gene expression assays.
(M) MDM4+/+/KO clones exhibit decreased anchorage-independent growth relative to the MDM4+/+/+ parental cell line.
(N) Induction of MDM4 cDNA in 1q-disomic clones in A2780 increases anchorage-independent growth.
For the graphs in E, G, H, M, and N, the boxplots represent the 25th, 50th, and 75th percentiles of the indicated data, while the whiskers represent the 10th and 90th percentiles of the indicated data. For the soft agar experiments in M and N, the data are from n = 15 fields of view, and a representative trial is shown (n ≥ 2 total trials). Scale bars = 250 μm.
***p < 0.0005
To explore whether p53 inhibition is a common consequence of chromosome 1q gains, we examined our prior analysis of aneuploidy-mutation mutual exclusivity in cancer genomes (Table S1). Out of 14,383 aneuploidy-gene mutation pairs, the single strongest instance of mutual exclusivity was between 1q gains and TP53 mutations (Fig. 5D). Next, we applied a classification algorithm capable of predicting cancers that lack p53 function based on their transcriptional profiles (45). As expected, cancers from TCGA with non-synonymous TP53 mutations scored higher with this classifier than cancers with wild-type TP53 (Fig. 5E). Considering only tumors with wild-type TP53, we calculated the association between the p53 status classifier and every possible chromosome arm gain in TCGA. Across all chromosomes, 1q gains exhibited the strongest correlation with the p53-loss signature (Fig. 5F-G). Among tumors with wild-type TP53, gains of chromosome 1q were associated with lower expression of the p53 target genes CDKN1A (p21), GADD45A, and RRM2B (Fig. 5H)(43). In total, these results indicate that gaining chromosome 1q phenocopies the effects of p53 mutations and suppresses p53 activity in human tumors.
We sought to discover the gene(s) on chromosome 1q responsible for inhibiting p53 signaling. We noted that MDM4, a negative regulator of p53 activity, is located on 1q32 (46). MDM4 expression increased with chromosome 1q copy number and higher MDM4 expression correlated with the p53-loss transcriptional signature (Fig. S15A-B). To uncover whether MDM4 is directly responsible for the 1q-aneuploidy addiction observed in A2780, we first used CRISPR-interference (CRISPRi) to downregulate MDM4 expression without fully ablating it (47). In A2780 competition assays, we observed that downregulating MDM4 impaired cell fitness relative to A2780 cells in which AAVS1 or PIP5K1A, an unrelated gene on chromosome 1q, were downregulated (Fig. 5I)(48). Next, we used a two-guide strategy to delete a single copy of MDM4 in an otherwise trisomic background (Fig. 5J-K and S15C-D). We found that the subsequent A2780 MDM4+/+/KO clones downregulated MDM4 and upregulated p53 target genes, comparable to the effects observed in cells lacking the entire 1q trisomy (Fig. 5L and S15E). We then tested the colony formation ability of MDM4+/+/KO clones, and we discovered that losing a single copy of MDM4 decreased anchorage-independent growth (Fig. 5M). Subsequently, we performed the converse experiment: we cloned MDM4 cDNA under the control of a doxycycline-inducible promoter and transduced the construct into both 1q-trisomic and 1q-disomic cells. We found that moderate (1.7-fold) overexpression of MDM4 was sufficient to cause an increase in anchorage-independent growth in the 1q-disomic cells, while this same treatment did not affect the 1q-trisomic cells (Fig. 5N and S15F).
Finally, to investigate the role of p53 as a mediator of 1q-aneuploidy addiction from an orthogonal approach, we used CRISPR to delete the TP53 gene in A2780 1q-disomic and 1q-trisomic cells (Fig. S16A-B). We discovered that loss of TP53 rescued the G1 delay and enhanced anchorage-independent growth in 1q-disomic cells (Fig. S16C-D). The magnitude of the increase in colony formation was significantly greater in the 1q-disomic cells compared to the 1q-trisomic cells (4-fold vs. 1.5-fold; P < 0.0001, t-test)(Fig. S16D). In total, these results indicate that chromosome 1q gains are a mechanism by which TP53-wildtype cancers can suppress p53 activity, and this suppression occurs due to the increased expression of MDM4.
BCL9 promotes the growth of 1q-aneuploid cancers via a p53-independent mechanism
We noted that the deletion of TP53 and the overexpression of MDM4 in 1q-disomic clones did not fully restore anchorage-independent growth to 1q-trisomic levels (Fig. 5N and S16D). We therefore hypothesized that there were additional dosage-sensitive genes encoded on chromosome 1q that promote the fitness of 1q-aneuploid cancers. To uncover these genes, we assembled a panel of 16 1q genes that have previously been associated with tumorigenesis, including MDM4 as a positive control, and we conducted CRISPRi competition assays to assess the effects of downregulating each gene in 1q-trisomic A2780 cells (Fig. S17A). gRNAs targeting six genes, including MDM4, exhibited a mean depletion >1.75-fold in cellular competitions (Fig. S17B). We cloned cDNAs for these six genes into doxycycline-inducible vectors and transduced them into A2780 1q-disomic cells. We found that overexpression of three of these genes increased anchorage-independent growth: MDM4, the anti-apoptotic gene MCL1, and the Wnt/β-catenin signaling gene BCL9 (Fig. S17C). We then conducted two further analyses to test whether the effects of the latter two genes were independent of the MDM4/p53 pathway. First, we co-transduced 1q-disomic cells with vectors to overexpress both MDM4 and BCL9 or MDM4 and MCL1, and second, we assessed the effects of overexpressing BCL9 or MCL1 in TP53-KO 1q-disomic cells. We found that MCL1 had no effect on anchorage-independent growth in 1q-disomic cells that lacked TP53, and co-expressing MCL1 and MDM4 in p53-wildtype 1q-disomic cells did not increase clonogenicity beyond the effects of expressing MCL1 alone (Fig. S17D-E). As both MCL1 and TP53 control apoptosis, we speculate that the overexpression of MCL1 and MDM4 are to some extent epistatic with one another (49).
In contrast, BCL9 expression enhanced anchorage-independent growth in TP53-KO cells and BCL9-MDM4 co-expression resulted in more colony formation compared to BCL9 alone (Fig. S17D-E). BCL9 encodes an adaptor protein that binds to nuclear β-catenin and enhances β-catenin–mediated transcriptional activity (50). We found that eliminating the trisomy of chromosome 1q reduced the expression of BCL9 as well as AXIN2 and LGR5, which are canonical targets of the Wnt/β-catenin pathway (51)(Fig. S18A). GSEA revealed a general decrease in the expression of transcripts associated with the Wnt/β-catenin pathway in 1q-disomic vs. 1q-trisomic cells (Fig. S18B). Ectopic overexpression of BCL9 increased the expression of AXIN2 and LGR5 (Fig. S18C). In human cancers, chromosome 1q gains were associated with higher levels of BCL9 expression, and high BCL9 was associated with the upregulation of AXIN2 and LGR5 (Fig. S18D-E). Mutations in CTNNB1, which encodes β-catenin, were over-represented in cancers with 1q gains (Fig. S18F). Finally, 1q aneuploidy was associated with higher β-catenin activity in several cancer types, including ovarian cancer, hepatocellular carcinoma, and rectal adenocarcinoma (Fig. S18G). In total, these results indicate that chromosome 1q aneuploidy can enhance oncogenic Wnt/β-catenin signaling via the upregulation of BCL9, and increased BCL9 expression promotes cancer cell fitness in a p53-independent manner.
Aneuploidy addictions create collateral therapeutic vulnerabilities
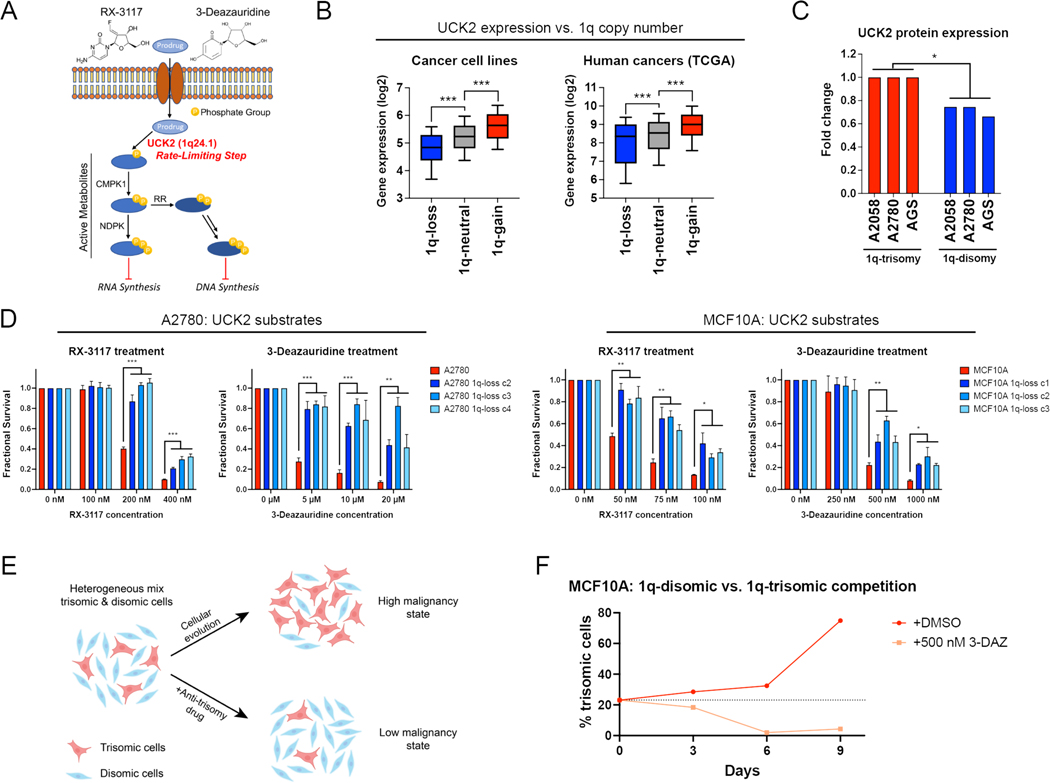
The oncogene addiction hypothesis is the conceptual foundation for the use of targeted therapies in cancer (40). Correspondingly, we sought to uncover whether aneuploidy addictions could also represent a therapeutic vulnerability for certain cancers. We noted that chromosome 1q harbors the UCK2 gene, which encodes a pyrimidine salvage kinase that controls a rate-limiting step in the activation of certain toxic nucleotide analogs, including RX-3117 and 3-deazauridine (Fig. 6A)(52, 53). We found that UCK2 is overexpressed in human cancers that contain extra copies of chromosome 1q, and elimination of the chromosome 1q trisomy consistently decreased UCK2 protein expression in our engineered cell lines (Fig. 6B-C). We therefore investigated whether gaining chromosome 1q could create a collateral sensitivity to UCK2-dependent nucleotide analogs.
Figure 6. Gaining chromosome 1q increases sensitivity to UCK2 substrates.
(A) A schematic of the metabolism of two pyrimidine analogs, RX-3117 and 3-deazauridine. UCK2, a kinase encoded on chromosome 1q, phosphorylates these compounds to produce cytotoxic derivatives that can poison DNA and RNA synthesis.
(B) Boxplots displaying the expression of UCK2 from the cancer cell line encyclopedia (left) and TCGA (right), divided based on the copy number of chromosome 1q. The boxplots represent the 25th, 50th, and 75th percentiles of the indicated data, while the whiskers represent the 10th and 90th percentiles of the indicated data. Data were analyzed using unpaired t-tests; n = 10,331 samples from TCGA and 942 samples from CCLE.
(C) Expression of UCK2 protein in cancer cell lines with 1q trisomies or following aneuploidy-elimination.
(D) Cellular sensitivity of A2780 and MCF10A treated with different concentrations of RX-3117 or 3-deazauridine. Mean ± SEM, data from representative trials are shown (n ≥ 3 total trials).
(E) A schematic displaying cellular competition between trisomic and disomic cells. Under normal conditions, certain trisomies enhance cellular fitness, allowing these cells to overtake the population and enhance malignant growth (top). However, treatment with an “anti-trisomy” compound could selectively impair the growth of the aneuploid cells, keeping the population in a low-malignant state (bottom).
(F) A cellular competition between fluorescently-labeled MCF10A 1q-trisomic and unlabeled 1q-disomic cells. These cells were mixed at a ratio of 20% to 80% and then cultured in either DMSO or 3-deazauridine. While the trisomic cells quickly dominate the population in drug-free medium, treatment with 3-deazauridine prevents the outgrowth of the 1q-trisomic subpopulation. Data from representative trial are shown (n = 2 total trials).
*p < 0.05, ** p < 0.005, *** p < 0.0005
First, as the mechanism of many cancer drugs is poorly understood (54, 55), we sought to verify that the cytotoxicity of RX-3117 and 3-deazauridine requires UCK2 expression. We used CRISPR to delete UCK2 in the haploid HAP1 cell line, and we confirmed that UCK2-knockout cells were highly resistant to both compounds (Fig. S19A-C). Next, we tested the effects of RX-3117 and 3-deazauridine in our engineered 1q-trisomic and 1q-disomic cell lines. We found that A2780 and MCF10A cells harboring a trisomy of chromosome 1q were more sensitive to both compounds compared to isogenic cells containing two copies of chromosome 1q (Fig. 6D). This effect was specific for UCK2 substrates, as the 1q-trisomic cells did not exhibit greater sensitivity to UCK2-independent nucleotide poisons and other cancer drugs (Fig. S19D). Furthermore, deletion of a single copy of UCK2 in 1q-trisomic A2780 cells was sufficient to decrease sensitivity to RX-3117, while ectopic overexpression of UCK2 cDNA in 1q-disomic A2780 cells was sufficient to increase sensitivity to RX-3117 (Fig. S19E-H). However, we did not detect any difference in RX-3117 sensitivity between 1q-trisomic and 1q-disomic A2058 and AGS cells (Fig. S20A-B). As A2780 and MCF10A harbor wild-type TP53 while AGS and A2058 exhibit compromised p53 signaling, we speculate that TP53 status or the expression of other related proteins may also influence the response to UCK2 substrates.
To determine whether 1q copy number changes generated without using ReDACT could also increase sensitivity to UCK2 substrates, we transiently treated near-diploid DLD1 colon cancer cells with an inhibitor of the spindle checkpoint kinase Mps1, and we isolated a clone that harbored a trisomy of chromosome 1 (Fig. S20C-D). DLD1 trisomy-1 cells were significantly more sensitive to RX-3117 compared to the parental DLD1 cells (P < 0.005; t-test; Fig. S20E). Finally, RX-3117 and 3-deazauridine have been screened across the NCI-60 cell line panel, and we found that higher UCK2 expression correlates with greater sensitivity to both compounds (Fig. S20F)(56). In total, these results indicate that 1q gains induce a collateral sensitivity to certain nucleotide analogs by increasing the expression of UCK2.
We hypothesized that we could use the greater sensitivity of 1q-trisomic cells to UCK2 substrates to re-direct cellular evolution away from aneuploidy and towards a disomic state with lower malignant potential (Fig. 6E). We mixed fluorescently-labeled 1q-trisomic and 1q-disomic MCF10A cells at a ratio of 20:80 and then co-cultured the two cell populations. After nine days of growth in drug-free medium, the trisomic cells had expanded to comprise 75% of the culture, but when the same cell populations were grown in the presence of 500 nM 3-deazauridine, the trisomic population decreased to comprise only 4% of the final culture (Fig. 6F). We conclude that trisomy-selective compounds can be used to manipulate cellular evolution to prevent the outgrowth of malignant aneuploid cells in a pre-malignant setting.
Finally, we investigated whether we could generalize this approach to identify compounds that exhibit selective toxicity against other aneuploidies. To nominate drugs that could be used to target chromosome 7p, we assessed the PRISM dataset of 4,518 compounds tested against 578 cancer cell lines (57). We calculated the correlation between the expression of every gene encoded on chromosome 7p and the sensitivity to each drug (Fig. S21A). One of the strongest relationships that we found was between expression of the gene AHR, which encodes a ligand-activated transcription factor, and sensitivity to the drug CGS-15943 (Fig. S21B). It has previously been reported that CGS-15943 binds to AHR and causes it to upregulate the expression of pro-apoptotic genes, providing a potential mechanistic explanation for this result (Fig. S21C)(58). AHR expression was upregulated in human cancer cell lines and tumors that are trisomic for chromosome 7p, while eliminating the 7p trisomy in our A2058 cell line model decreased the expression of AHR (Fig. S21D). Consistent with our hypothesis, we found that A2058 cells with a 7p trisomy were moderately but significantly more sensitive to CGS-15943 compared to 7p-disomic A2058 cells (P < 0.0005; t-test; Fig. S21E). We conclude that by using a combination of computational and experimental approaches, we can uncover compounds that exhibit greater activity toward cancer cells with specific aneuploidies.
DISCUSSION
In this work, we eliminated endogenous aneuploidies from established cancer cell lines, and we revealed that the removal of trisomic chromosomes compromises cancer-like growth. We posit that these phenotypes are due specifically to the loss of the aneuploid chromosome and are not a by-product of CRISPR selection or the elimination of point mutations encoded on the targeted chromosome (discussed in more detail in the Supplementary Text)(59). Due to the similarity between our observations and the previously-described oncogene addiction phenomenon (17), we suggest that in certain circumstances cancers may also be “addicted” to the aneuploidy found in their genomes. We speculate that during tumor evolution, certain aneuploidies can provide context-specific benefits that enhance tumorigenesis. For instance, we showed that chromosome 1q gains are an early event during cancer development, and we demonstrated that MDM4 and BCL9 are dosage-sensitive genes on 1q that enhance malignant growth. In cells that already harbor TP53 mutations or in cancer types that are not driven by WNT signaling, the beneficial effects of gaining chromosome 1q may be outweighed by the detrimental effects of overexpressing hundreds of other 1q genes.
MDM4 and many other genes have tumor-promoting properties when highly overexpressed (60, 61). For instance, MDM4 is focally amplified in ~1% of cancers in TCGA, and strong overexpression of MDM4 via retrovirus immortalizes primary cells and renders them sensitive to Ras-mediated transformation (62). Here, we demonstrated that a single extra copy of MDM4 is sufficient to suppress the expression of p53 target genes and promote oncogenic growth. Our results are consistent with a recent report showing that low-level overexpression of MDM4 can enhance fitness in hematopoietic cell competitions (63). The overlap between single-copy dosage-sensitive genes like MDM4 and genes found to have tumor-promoting properties when highly overexpressed is at present unknown. Furthermore, our work demonstrates that MDM4 overexpression alone is insufficient to fully recapitulate the oncogenic effects of 1q aneuploidy, and we established that BCL9 is a second dosage-sensitive 1q gene that enhances fitness in a TP53-independent manner. As arm-length aneuploidy is more common in tumor genomes than focal gene amplifications (19), we expect that recurrently-gained chromosomes harbor multiple independent loci like MDM4 and BCL9 that cooperate to drive tumor development.
Finally, our results raise the exciting possibility that “aneuploidy addictions” may represent a therapeutic vulnerability in cancer. Previous attempts to target aneuploidy have focused on phenotypes that are shared across highly-aneuploid cells, such as alterations in spindle geometry (64, 65). Here, we sought to develop an approach to take advantage of the genes that are encoded on an aneuploid chromosome, thereby allowing chromosome-specific targeting. In particular, we hypothesized that the overexpression of specific genes – for instance, drug-importer pumps or enzymes required for a prodrug’s activation – could sensitize cancers to compounds that are otherwise better-tolerated in euploid tissue. We demonstrated that gaining chromosome 1q creates a collateral vulnerability to the nucleotide analogs RX-3117 and 3-deazauridine due to the overexpression of the kinase UCK2. Notably, RX-3117 has been tested in phase 2A clinical trials, but without the use of any genomic biomarkers for patient selection (66). High UCK2 expression has been proposed as a potential sensitivity biomarker for RX-3117, and we speculate that patients whose tumors harbor gains of chromosome 1q may exhibit strong responses due to the constitutive overexpression of UCK2 (67). More broadly, compounds whose anti-cancer function is enhanced by genes encoded on aneuploid chromosomes could be used to direct cellular evolution away from certain aneuploidies and toward the lower-malignancy diploid state.
MATERIALS & METHODS
The identities of all cell lines used in this study were confirmed using STR (short tandem repeat) profiling. CRISPR plasmids were cloned as described in (68). Chromosome copy number analysis was performed as described in (36). Complete methods are provided in the supplementary materials (59).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dr. Tobias Cantz (Hannover Medical School) for providing the TK plasmids used in this work. We thank Peter Andrews (CSHL) for assistance with SMASH-Seq. We thank Yale Flow Cytometry, especially Chao Wang and Lesley Devine, for their assistance with single-cell sorting. We thank Al Mennone and the Yale Center for Advanced Light Microscopy Facility for their assistance with soft agar imaging. We thank the Yale Center for Genome Analysis for performing SMASH-Seq. We thank the Yale Center for Research Computing, specifically Robert Bjornson, for guidance and assistance in computations run on the Farnam and Ruddle clusters. We thank the Yale Animal Resources Center Staff for assistance with mouse experiments. Copy number timing analysis conducted in the Sun Lab uses the computing resources of the Minnesota Supercomputing Institute. The timing analysis was prepared using limited access datasets obtained from the Cancer Genome Project from the Wellcome Sanger Institute and does not necessarily reflect the opinions of the provider institution. Part of the BRCA sequencing data was originally generated by research led by Dr. Masahito Kawazu. We also thank the International Cancer Genome Consortium (ICGC) for providing access to the MEL dataset.
Funding.
Research in the Sheltzer Lab is supported by NIH grants R01CA237652 and R01CA276666, Department of Defense grant W81XWH-20-1-068, an American Cancer Society Research Scholar Grant, a Breast Cancer Alliance Young Investigator Award, a Damon Runyon-Rachleff Innovation Award, a sponsored research agreement from Ono Pharmaceuticals, and a sponsored research agreement from Meliora Therapeutics. Research in the Liu Lab is supported by NIH grant R01GM137031. Yale Flow Cytometry is supported in part by an NCI Cancer Center Support Grant # NIH P30 CA016359. This work was performed with assistance from the CSHL Flow Cytometry, Microscopy, Animal, and Sequencing Technologies & Analysis Shared Resources, which are supported in part by the Cancer Center Support Grant 5P30CA045508.
J.C.S. is a co-founder of Meliora Therapeutics, a member of the advisory board of Surface Ventures, and an employee of Google, Inc. This work was performed outside of her affiliation with Google and used no proprietary knowledge or materials from Google. J.M.S. has received consulting fees from Merck, Pfizer, Ono Pharmaceuticals, and Highside Capital Management, is a member of the advisory boards of Tyra Biosciences, BioIO, and the Chemical Probes Portal, and is a co-founder of Meliora Therapeutics.
Footnotes
Competing Interests. All other authors declare that they have no competing interests.
Data and materials availability.
The parental cell lines used in this work are available from various commercial sources (Table S4). The plasmids used to induce chromosome loss have been deposited at Addgene (Table S5). The code used to perform the TCGA survival analysis is available at https://github.com/joan-smith/comprehensive-tcga-survival (31). The code used to perform the mutual exclusivity analysis is available at github.com/sheltzer-lab/aneuploidy-addictions (27). The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE PXD037956127. RNA-seq data have been deposited at GSE222379.
References and notes
- 1.Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, Schumacher SE, Wang C, Hu H, Liu J, Lazar AJ, Cherniack AD, Beroukhim R, Meyerson M, Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell. 33, 676–689.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon DJ, Resio B, Pellman D, Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet 13, 189–203 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Boveri T, Concerning the Origin of Malignant Tumours by Theodor Boveri. Translated and annotated by Henry Harris. J. Cell Sci 121, 1–84 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Hardy PA, Zacharias H, Reappraisal of the Hansemann-Boveri hypothesis on the origin of tumors. Cell Biol. Int 29, 983–992 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Vasudevan A, Schukken KM, Sausville EL, Girish V, Adebambo OA, Sheltzer JM, Aneuploidy as a promoter and suppressor of malignant growth. Nat. Rev. Cancer 21, 89–103 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Sheltzer JM, Amon A, The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends Genet. TIG 27, 446–453 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheltzer JM, Ko JH, Replogle JM, Habibe Burgos NC, Chung ES, Meehl CM, Sayles NM, Passerini V, Storchova Z, Amon A, Single-chromosome Gains Commonly Function as Tumor Suppressors. Cancer Cell. 31, 240–255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weaver BAA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW, Aneuploidy Acts Both Oncogenically and as a Tumor Suppressor. Cancer Cell. 11, 25–36 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ, Cumulative Haploinsufficiency and Triplosensitivity Drive Aneuploidy Patterns to Shape the Cancer Genome. Cell. 155, 948–962 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sack LM, Davoli T, Li MZ, Li Y, Xu Q, Naxerova K, Wooten EC, Bernardi RJ, Martin TD, Chen T, Leng Y, Liang AC, Scorsone KA, Westbrook TF, Wong K-K, Elledge SJ, Profound Tissue Specificity in Proliferation Control Underlies Cancer Drivers and Aneuploidy Patterns. Cell. 173, 499–514.e23 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zimonjic D, Brooks MW, Popescu N, Weinberg RA, Hahn WC, Derivation of Human Tumor Cells in Vitro without Widespread Genomic Instability. Cancer Res. 61, 8838–8844 (2001). [PubMed] [Google Scholar]
- 12.Hasle H, Friedman JM, Olsen JH, Rasmussen SA, Low risk of solid tumors in persons with Down syndrome. Genet. Med 18, 1151–1157 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Bister K, Discovery of oncogenes: The advent of molecular cancer research. Proc. Natl. Acad. Sci. U. S. A 112, 15259–15260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Loughlin TA, Gilbert LA, Functional Genomics for Cancer Research: Applications In Vivo and In Vitro. Annu. Rev. Cancer Biol 3, 345–363 (2019). [Google Scholar]
- 15.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B, Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 282, 1497–1501 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T, Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 260, 85–88 (1993). [DOI] [PubMed] [Google Scholar]
- 17.Weinstein IB, Addiction to Oncogenes--the Achilles Heal of Cancer. Science. 297, 63–64 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Waters AM, Der CJ, KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med 8 (2018), doi: 10.1101/cshperspect.a031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang C-Z, Wala J, Mermel CH, Sougnez C, Gabriel SB, Hernandez B, Shen H, Laird PW, Getz G, Meyerson M, Beroukhim R, Pan-cancer patterns of somatic copy number alteration. Nat. Genet 45, 1134–1140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Xia Y, Mills L, Nikolakopoulos AN, Maeser N, Sheltzer JM, Sun R, Evolving copy number gains promote tumor expansion and bolster mutational diversification (2022), p. 2022.06.14.495959, , doi: 10.1101/2022.06.14.495959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yates LR, Knappskog S, Wedge D, Farmery JHR, Gonzalez S, Martincorena I, Alexandrov LB, Van Loo P, Haugland HK, Lilleng PK, Gundem G, Gerstung M, Pappaemmanuil E, Gazinska P, Bhosle SG, Jones D, Raine K, Mudie L, Latimer C, Sawyer E, Desmedt C, Sotiriou C, Stratton MR, Sieuwerts AM, Lynch AG, Martens JW, Richardson AL, Tutt A, Lønning PE, Campbell PJ, Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell. 32, 169–184.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, Patch A-M, Kakavand H, Alexandrov LB, Burke H, Jakrot V, Kazakoff S, Holmes O, Leonard C, Sabarinathan R, Mularoni L, Wood S, Xu Q, Waddell N, Tembe V, Pupo GM, De Paoli-Iseppi R, Vilain RE, Shang P, Lau LMS, Dagg RA, Schramm S-J, Pritchard A, Dutton-Regester K, Newell F, Fitzgerald A, Shang CA, Grimmond SM, Pickett HA, Yang JY, Stretch JR, Behren A, Kefford RF, Hersey P, Long GV, Cebon J, Shackleton M, Spillane AJ, Saw RPM, López-Bigas N, Pearson JV, Thompson JF, Scolyer RA, Mann GJ, Whole-genome landscapes of major melanoma subtypes. Nature. 545, 175–180 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Paterson C, Clevers H, Bozic I, Mathematical model of colorectal cancer initiation. Proc. Natl. Acad. Sci 117, 20681–20688 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C, Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med 7, 283ra54–283ra54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cisowski J, Bergo MO, What makes oncogenes mutually exclusive? Small GTPases. 8, 187–192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen B, Fong C, Luthra A, Smith SA, DiNatale RG, Nandakumar S, Walch H, Chatila WK, Madupuri R, Kundra R, Bielski CM, Mastrogiacomo B, Donoghue MTA, Boire A, Chandarlapaty S, Ganesh K, Harding JJ, Iacobuzio-Donahue CA, Razavi P, Reznik E, Rudin CM, Zamarin D, Abida W, Abou-Alfa GK, Aghajanian C, Cercek A, Chi P, Feldman D, Ho AL, Iyer G, Janjigian YY, Morris M, Motzer RJ, O’Reilly EM, Postow MA, Raj NP, Riely GJ, Robson ME, Rosenberg JE, Safonov A, Shoushtari AN, Tap W, Teo MY, Varghese AM, Voss M, Yaeger R, Zauderer MG, Abu-Rustum N, Garcia-Aguilar J, Bochner B, Hakimi A, Jarnagin WR, Jones DR, Molena D, Morris L, Rios-Doria E, Russo P, Singer S, Strong VE, Chakravarty D, Ellenson LH, Gopalan A, Reis-Filho JS, Weigelt B, Ladanyi M, Gonen M, Shah SP, Massague J, Gao J, Zehir A, Berger MF, Solit DB, Bakhoum SF, Sanchez-Vega F, Schultz N, Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell. 185, 563–575.e11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagenson RA, sheltzer-lab/aneuploidy-addictions: v1.0.0 (2023), doi: 10.5281/zenodo.7859759. [DOI] [Google Scholar]
- 28.Stopsack KH, Whittaker CA, Gerke TA, Loda M, Kantoff PW, Mucci LA, Amon A, Aneuploidy drives lethal progression in prostate cancer. Proc. Natl. Acad. Sci 116, 11390–11395 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hieronymus H, Murali R, Tin A, Yadav K, Abida W, Moller H, Berney D, Scher H, Carver B, Scardino P, Schultz N, Taylor B, Vickers A, Cuzick J, Sawyers CL, Tumor copy number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. eLife. 7, e37294 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukow DA, Sausville EL, Suri P, Chunduri NK, Wieland A, Leu J, Smith JC, Girish V, Kumar AA, Kendall J, Wang Z, Storchova Z, Sheltzer JM, Chromosomal instability accelerates the evolution of resistance to anti-cancer therapies. Dev. Cell 56, 2427–2439.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith J, joan-smith/comprehensive-tcga-survival: Genome-wide identification and analysis of prognostic features in human cancers (2022), doi: 10.5281/zenodo.6632067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fillat C, Carrio M, Cascante A, Sangro B, Suicide Gene Therapy Mediated by the Herpes Simplex Virus Thymidine Kinase Gene / Ganciclovir System: Fifteen Years of Application. Curr. Gene Ther. 3, 13–26 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Uno N, Hiramatsu K, Uno K, Komoto S, Kazuki Y, Oshimura M, CRISPR/Cas9-induced transgene insertion and telomere-associated truncation of a single human chromosome for chromosome engineering in CHO and A9 cells. Sci. Rep 7, 12739 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuccaro MV, Xu J, Mitchell C, Marin D, Zimmerman R, Rana B, Weinstein E, King RT, Palmerola KL, Smith ME, Tsang SH, Goland R, Jasin M, Lobo R, Treff N, Egli D, Allele-Specific Chromosome Removal after Cas9 Cleavage in Human Embryos. Cell. 183, 1650–1664.e15 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Papathanasiou S, Markoulaki S, Blaine LJ, Leibowitz ML, Zhang C-Z, Jaenisch R, Pellman D, Whole chromosome loss and genomic instability in mouse embryos after CRISPR-Cas9 genome editing. Nat. Commun 12, 5855 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Andrews P, Kendall J, Ma B, Hakker I, Rodgers L, Ronemus M, Wigler M, Levy D, SMASH, a fragmentation and sequencing method for genomic copy number analysis. Genome Res. 26, 844–851 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schukken KM, Sheltzer JM, Extensive protein dosage compensation in aneuploid human cancers. Genome Res. 32, 1254–1270 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin SI, Freedman VH, Risser R, Pollack R, Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc. Natl. Acad. Sci 72, 4435–4439 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O’Hagan R, Pantginis J, Zhou H, Horner JW, Cordon-Cardo C, Yancopoulos GD, DePinho RA #, Essential role for oncogenic Ras in tumour maintenance. Nature. 400, 468–472 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Torti D, Trusolino L, Oncogene addiction as a foundational rationale for targeted anti-cancer therapy: promises and perils. EMBO Mol. Med 3, 623–636 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westover D, Zugazagoitia J, Cho BC, Lovly CM, Paz-Ares L, Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann. Oncol 29, i10–i19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgess MR, Hwang E, Mroue R, Bielski CM, Wandler AM, Huang BJ, Firestone AJ, Young A, Lacap JA, Crocker L, Asthana S, Davis EM, Xu J, Akagi K, Beau MML, Li Q, Haley B, Stokoe D, Sampath D, Taylor BS, Evangelista M, Shannon K, KRAS Allelic Imbalance Enhances Fitness and Modulates MAP Kinase Dependence in Cancer. Cell. 168, 817–829.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fischer M, Census and evaluation of p53 target genes. Oncogene. 36, 3943–3956 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugrue MM, Shin DY, Lee SW, Aaronson SA, Wild-type p53 triggers a rapid senescence program in human tumor cells lacking functional p53. Proc. Natl. Acad. Sci 94, 9648–9653 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fito-Lopez B, Salvadores M, Alvarez M-M, Supek F, Prevalence, causes and impact of TP53-loss phenocopying events in human tumors (2022), p. 2022.11.01.514743, , doi: 10.1101/2022.11.01.514743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karni-Schmidt O, Lokshin M, Prives C, The Roles of MDM2 and MDMX in Cancer. Annu. Rev. Pathol. Mech. Dis 11, 617–644 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, Weissman JS, Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife. 5, e19760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Girish V, Sheltzer JM, A CRISPR Competition Assay to Identify Cancer Genetic Dependencies. Bio-Protoc. 10, e3682–e3682 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leu JI-J, Dumont P, Hafey M, Murphy ME, George DL, Mitochondrial p53 activates Bak and causes disruption of a Bak–Mcl1 complex. Nat. Cell Biol 6, 443–450 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Mani M, Carrasco DE, Zhang Y, Takada K, Gatt ME, Dutta-Simmons J, Ikeda H, Diaz-Griffero F, Pena-Cruz V, Bertagnolli M, Myeroff LL, Markowitz SD, Anderson KC, Carrasco DR, BCL9 Promotes Tumor Progression by Conferring Enhanced Proliferative, Metastatic, and Angiogenic Properties to Cancer Cells. Cancer Res. 69, 7577–7586 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nusse R, Clevers H, Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 169, 985–999 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Van Rompay AR, Norda A, Lindén K, Johansson M, Karlsson A, Phosphorylation of uridine and cytidine nucleoside analogs by two human uridine-cytidine kinases. Mol. Pharmacol 59, 1181–1186 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Sarkisjan D, Julsing JR, Smid K, de Klerk D, van Kuilenburg ABP, Meinsma R, Lee YB, Kim DJ, Peters GJ, The Cytidine Analog Fluorocyclopentenylcytosine (RX-3117) Is Activated by Uridine-Cytidine Kinase 2. PLOS ONE. 11, e0162901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin A, Giuliano CJ, Palladino A, John KM, Abramowicz C, Yuan ML, Sausville EL, Lukow DA, Liu L, Chait AR, Galluzzo ZC, Tucker C, Sheltzer JM, Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med 11 (2019), doi: 10.1126/scitranslmed.aaw8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin A, Sheltzer JM, Discovering and validating cancer genetic dependencies: approaches and pitfalls. Nat. Rev. Genet 21, 671–682 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Luna A, Elloumi F, Varma S, Wang Y, Rajapakse VN, Aladjem MI, Robert J, Sander C, Pommier Y, Reinhold WC, CellMiner Cross-Database (CellMinerCDB) version 1.2: Exploration of patient-derived cancer cell line pharmacogenomics. Nucleic Acids Res. 49, D1083–D1093 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corsello SM, Nagari RT, Spangler RD, Rossen J, Kocak M, Bryan JG, Humeidi R, Peck D, Wu X, Tang AA, Wang VM, Bender SA, Lemire E, Narayan R, Montgomery P, Ben-David U, Garvie CW, Chen Y, Rees MG, Lyons NJ, McFarland JM, Wong BT, Wang L, Dumont N, O’Hearn PJ, Stefan E, Doench JG, Harrington CN, Greulich H, Meyerson M, Vazquez F, Subramanian A, Roth JA, Bittker JA, Boehm JS, Mader CC, Tsherniak A, Golub TR, Discovering the anticancer potential of non-oncology drugs by systematic viability profiling. Nat. Cancer 1, 235–248 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Donnell EF, Jang HS, Liefwalker DF, Kerkvliet NI, Kolluri SK, Discovery and Mechanistic Characterization of a Select Modulator of AhR-regulated Transcription (SMAhRT) with Anti-cancer Effects. Apoptosis Int. J. Program. Cell Death. 26, 307–322 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Girish et al. Supplemental Material.
- 60.Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS, A census of amplified and overexpressed human cancer genes. Nat. Rev. Cancer 10, 59–64 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Hüllein J, Słabicki M, Rosolowski M, Jethwa A, Habringer S, Tomska K, Kurilov R, Lu J, Scheinost S, Wagener R, Huang Z, Lukas M, Yavorska O, Helfrich H, Scholtysik R, Bonneau K, Tedesco D, Küppers R, Klapper W, Pott C, Stilgenbauer S, Burkhardt B, Löffler M, Trümper LH, Hummel M, Brors B, Zapatka M, Siebert R, Kreuz M, Keller U, Huber W, Zenz T, MDM4 Is Targeted by 1q Gain and Drives Disease in Burkitt Lymphoma. Cancer Res. 79, 3125–3138 (2019). [DOI] [PubMed] [Google Scholar]
- 62.Danovi D, Meulmeester E, Pasini D, Migliorini D, Capra M, Frenk R, de Graaf P, Francoz S, Gasparini P, Gobbi A, Helin K, Pelicci PG, Jochemsen AG, Marine J-C, Amplification of Mdmx (or Mdm4) Directly Contributes to Tumor Formation by Inhibiting p53 Tumor Suppressor Activity. Mol. Cell. Biol 24, 5835–5843 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sebert M, Gachet S, Leblanc T, Rousseau A, Bluteau O, Kim R, Ben Abdelali R, Sicre de Fontbrune F, Maillard L, Fedronie C, Murigneux V, Bellenger L, Naouar N, Quentin S, Hernandez L, Vasquez N, Da Costa M, Prata PH, Larcher L, de Tersant M, Duchmann M, Raimbault A, Trimoreau F, Fenneteau O, Cuccuini W, Gachard N, Auger N, Tueur G, Blanluet M, Gazin C, Souyri M, Langa Vives F, Mendez-Bermudez A, Lapillonne H, Lengline E, Raffoux E, Fenaux P, Adès L, Forcade E, Jubert C, Domenech C, Strullu M, Bruno B, Buchbinder N, Thomas C, Petit A, Leverger G, Michel G, Cavazzana M, Gluckman E, Bertrand Y, Boissel N, Baruchel A, Dalle J-H, Clappier E, Gilson E, Deriano L, Chevret S, Sigaux F, Socié G, Stoppa-Lyonnet D, de Thé H, Antoniewski C, Bluteau D, Peffault de Latour R, Soulier J, Clonal hematopoiesis driven by chromosome 1q/MDM4 trisomy defines a canonical route toward leukemia in Fanconi anemia. Cell Stem Cell. 30, 153–170.e9 (2023). [DOI] [PubMed] [Google Scholar]
- 64.Quinton RJ, DiDomizio A, Vittoria MA, Kotýnková K, Ticas CJ, Patel S, Koga Y, Vakhshoorzadeh J, Hermance N, Kuroda TS, Parulekar N, Taylor AM, Manning AL, Campbell JD, Ganem NJ, Whole-genome doubling confers unique genetic vulnerabilities on tumour cells. Nature. 590, 492–497 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cohen-Sharir Y, McFarland JM, Abdusamad M, Marquis C, Bernhard SV, Kazachkova M, Tang H, Ippolito MR, Laue K, Zerbib J, Malaby HLH, Jones A, Stautmeister L-M, Bockaj I, Wardenaar R, Lyons N, Nagaraja A, Bass AJ, Spierings DCJ, Foijer F, Beroukhim R, Santaguida S, Golub TR, Stumpff J, Storchová Z, Ben-David U, Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition. Nature. 590, 486–491 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salgia N, Pal SK, Chung V, Tagawa ST, Picus J, Babiker HM, Gupta S, Wadlow RC, Poore J, Peterson C, Benaim E, Activity of RX-3117, an oral antimetabolite nucleoside, in subjects with advanced urothelial cancer: Preliminary results of a phase IIa study. J. Clin. Oncol 37, 455–455 (2019). [Google Scholar]
- 67.El Hassouni B, Infante J, Mantini G, Ricci C, Funel N, Giovannetti E, Peters GJ, Uridine Cytidine Kinase 2 as a Potential Biomarker for Treatment with RX-3117 in Pancreatic Cancer. Anticancer Res. 39, 3609–3614 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Giuliano CJ, Lin A, Girish V, Sheltzer JM, Generating Single Cell–Derived Knockout Clones in Mammalian Cells with CRISPR/Cas9. Curr. Protoc. Mol. Biol 128, e100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, Vaimberg EW, Goodale A, Root DE, Piccioni F, Doench JG, Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun 9 (2018), doi: 10.1038/s41467-018-07901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Di Tommaso P, Chatzou M, Floden EW, Barja PP, Palumbo E, Notredame C, Nextflow enables reproducible computational workflows. Nat. Biotechnol 35, 316–319 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Andrews PA, Iossifov I, Kendall J, Marks S, Muthuswamy L, Wang Z, Levy D, Wigler M, MUMdex: MUM-based structural variation detection. bioRxiv, 078261 (2016). [Google Scholar]
- 72.Wickham H, Averick M, Bryan J, Chang W, McGowan LD, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen TL, Miller E, Bache SM, Müller K, Ooms J, Robinson D, Seidel DP, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H, Welcome to the Tidyverse. J. Open Source Softw 4, 1686 (2019). [Google Scholar]
- 73.Mayo P, Hartshorne T, Li K, McMunn-Gibson C, Spencer K, Schnetz-Boutaud N, Curr. Protoc. Hum. Genet, in press, doi: 10.1002/0471142905.hg0213s67. [DOI] [PubMed] [Google Scholar]
- 74.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinforma. Oxf. Engl 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu G, Hu E, Gao C-H, enrichplot: Visualization of Functional Enrichment Result (2023), , doi: 10.18129/B9.bioc.enrichplot. [DOI] [Google Scholar]
- 76.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G, clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. Camb. Mass 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y, Mi Y, Mueller T, Kreibich S, Williams EG, Van Drogen A, Borel C, Frank M, Germain P-L, Bludau I, Mehnert M, Seifert M, Emmenlauer M, Sorg I, Bezrukov F, Bena FS, Zhou H, Dehio C, Testa G, Saez-Rodriguez J, Antonarakis SE, Hardt W-D, Aebersold R, Multi-omic measurements of heterogeneity in HeLa cells across laboratories. Nat. Biotechnol 37, 314–322 (2019). [DOI] [PubMed] [Google Scholar]
- 78.Collins BC, Hunter CL, Liu Y, Schilling B, Rosenberger G, Bader SL, Chan DW, Gibson BW, Gingras A-C, Held JM, Hirayama-Kurogi M, Hou G, Krisp C, Larsen B, Lin L, Liu S, Molloy MP, Moritz RL, Ohtsuki S, Schlapbach R, Selevsek N, Thomas SN, Tzeng S-C, Zhang H, Aebersold R, Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat. Commun 8, 291 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mehnert M, Li W, Wu C, Salovska B, Liu Y, Combining Rapid Data Independent Acquisition and CRISPR Gene Deletion for Studying Potential Protein Functions: A Case of HMGN1. Proteomics. 19, e1800438 (2019). [DOI] [PubMed] [Google Scholar]
- 80.Bruderer R, Bernhardt OM, Gandhi T, Xuan Y, Sondermann J, Schmidt M, Gomez-Varela D, Reiter L, Optimization of Experimental Parameters in Data-Independent Mass Spectrometry Significantly Increases Depth and Reproducibility of Results. Mol. Cell. Proteomics MCP 16, 2296–2309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bruderer R, Muntel J, Müller S, Bernhardt OM, Gandhi T, Cominetti O, Macron C, Carayol J, Rinner O, Astrup A, Saris WHM, Hager J, Valsesia A, Dayon L, Reiter L, Analysis of 1508 Plasma Samples by Capillary-Flow Data-Independent Acquisition Profiles Proteomics of Weight Loss and Maintenance. Mol. Cell. Proteomics MCP 18, 1242–1254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bruderer R, Bernhardt OM, Gandhi T, Miladinović SM, Cheng L-Y, Messner S, Ehrenberger T, Zanotelli V, Butscheid Y, Escher C, Vitek O, Rinner O, Reiter L, Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteomics MCP 14, 1400–1410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tsou C-C, Avtonomov D, Larsen B, Tucholska M, Choi H, Gingras A-C, Nesvizhskii AI, DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods 12, 258–264, 7 p following 264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eggenschwiler R, Moslem M, Fráguas MS, Galla M, Papp O, Naujock M, Fonfara I, Gensch I, Wähner A, Beh-Pajooh A, Mussolino C, Tauscher M, Steinemann D, Wegner F, Petri S, Schambach A, Charpentier E, Cathomen T, Cantz T, Improved bi-allelic modification of a transcriptionally silent locus in patient-derived iPSC by Cas9 nickase. Sci. Rep 6, 38198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mansour SL, Thomas KR, Capecchi MR, Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 336, 348–352 (1988). [DOI] [PubMed] [Google Scholar]
- 86.Li LB, Chang K-H, Wang P-R, Hirata RK, Papayannopoulou T, Russell DW, Trisomy Correction in Down Syndrome Induced Pluripotent Stem Cells. Cell Stem Cell. 11, 615–619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vasudevan A, Baruah PS, Smith JC, Wang Z, Sayles NM, Andrews P, Kendall J, Leu J, Chunduri NK, Levy D, Wigler M, Storchová Z, Sheltzer JM, Single-Chromosomal Gains Can Function as Metastasis Suppressors and Promoters in Colon Cancer. Dev. Cell 52, 413–428.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kluesner MG, Nedveck DA, Lahr WS, Garbe JR, Abrahante JE, Webber BR, Moriarity BS, EditR: A Method to Quantify Base Editing from Sanger Sequencing. CRISPR J. 1, 239–250 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho Y-J, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao M-S, DeMichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M, The landscape of somatic copy-number alteration across human cancers. Nature. 463, 899–905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, Goodale A, Lee Y, Ali LD, Jiang G, Lubonja R, Harrington WF, Strickland M, Wu T, Hawes DC, Zhivich VA, Wyatt MR, Kalani Z, Chang JJ, Okamoto M, Stegmaier K, Golub TR, Boehm JS, Vazquez F, Root DE, Hahn WC, Tsherniak A, Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet 49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kawazu M, Kojima S, Ueno T, Totoki Y, Nakamura H, Kunita A, Qu W, Yoshimura J, Soda M, Yasuda T, Hama N, Saito-Adachi M, Sato K, Kohsaka S, Sai E, Ikemura M, Yamamoto S, Ogawa T, Fukayama M, Tada K, Seto Y, Morishita S, Hazama S, Shibata T, Yamashita Y, Mano H, Integrative analysis of genomic alterations in triple-negative breast cancer in association with homologous recombination deficiency. PLoS Genet. 13, e1006853 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maeser N, Khan A, Sun R, Somatic variant detection from multi-sampled genomic sequencing data of tumor specimens using the ith.Variant pipeline. STAR Protoc. 4, 101927 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Purdom E, Ho C, Grasso CS, Quist MJ, Cho RJ, Spellman P, Methods and challenges in timing chromosomal abnormalities within cancer samples. Bioinforma. Oxf. Engl 29, 3113–3120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Azzalini A, Menardi G, Clustering via Nonparametric Density Estimation: The R Package pdfCluster. J. Stat. Softw 57, 1–26 (2014).25400517 [Google Scholar]
- 95.Anaconda | The World’s Most Popular Data Science Platform. Anaconda, (available at https://www.anaconda.com/). [Google Scholar]
- 96.Smith JC, Sheltzer JM, Systematic identification of mutations and copy number alterations associated with cancer patient prognosis. eLife. 7, e39217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith JC, Sheltzer JM, Genome-wide identification and analysis of prognostic features in human cancers. Cell Rep. 38, 110569 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Caesar-Johnson SJ, Demchok JA, Felau I, Kasapi M, Ferguson ML, Hutter CM, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, (Julia) Zhang J, Chudamani S, Liu J, Lolla L, Naresh R, Pihl T, Sun Q, Wan Y, Wu Y, Cho J, DeFreitas T, Frazer S, Gehlenborg N, Getz G, Heiman DI, Kim J, Lawrence MS, Lin P, Meier S, Noble MS, Saksena G, Voet D, Zhang H, Bernard B, Chambwe N, Dhankani V, Knijnenburg T, Kramer R, Leinonen K, Liu Y, Miller M, Reynolds S, Shmulevich I, Thorsson V, Zhang W, Akbani R, Broom BM, Hegde AM, Ju Z, Kanchi RS, Korkut A, Li J, Liang H, Ling S, Liu W, Lu Y, Mills GB, Ng K-S, Rao A, Ryan M, Wang J, Weinstein JN, Zhang J, Abeshouse A, Armenia J, Chakravarty D, Chatila WK, de Bruijn I, Gao J, Gross BE, Heins ZJ, Kundra R, La K, Ladanyi M, Luna A, Nissan MG, Ochoa A, Phillips SM, Reznik E, Sanchez-Vega F, Sander C, Schultz N, Sheridan R, Sumer SO, Sun Y, Taylor BS, Wang J, Zhang H, Anur P, Peto M, Spellman P, Benz C, Stuart JM, Wong CK, Yau C, Hayes DN, Parker JS, Wilkerson MD, Ally A, Balasundaram M, Bowlby R, Brooks D, Carlsen R, Chuah E, Dhalla N, Holt R, Jones SJM, Kasaian K, Lee D, Ma Y, Marra MA, Mayo M, Moore RA, Mungall AJ, Mungall K, Robertson AG, Sadeghi S, Schein JE, Sipahimalani P, Tam A, Thiessen N, Tse K, Wong T, Berger AC, Beroukhim R, Cherniack AD, Cibulskis C, Gabriel SB, Gao GF, Ha G, Meyerson M, Schumacher SE, Shih J, Kucherlapati MH, Kucherlapati RS, Baylin S, Cope L, Danilova L, Bootwalla MS, Lai PH, Maglinte DT, Berg DJVD, Weisenberger DJ, Auman JT, Balu S, Bodenheimer T, Fan C, Hoadley KA, Hoyle AP, Jefferys SR, Jones CD, Meng S, Mieczkowski PA, Mose LE, Perou AH, Perou CM, Roach J, Shi Y, Simons JV, Skelly T, Soloway MG, Tan D, Veluvolu U, Fan H, Hinoue T, Laird PW, Shen H, Zhou W, Bellair M, Chang K, Covington K, Creighton CJ, Dinh H, Doddapaneni H, Donehower LA, Drummond J, Gibbs RA, Glenn R, Hale W, Han Y, Hu J, Korchina V, Lee S, Lewis L, Li W, Liu X, Morgan M, Morton D, Muzny D, Santibanez J, Sheth M, Shinbro E, Wang L, Wang M, Wheeler DA, Xi L, Zhao F, Hess J, Appelbaum EL, Bailey M, Cordes MG, Ding L, Fronick CC, Fulton LA, Fulton RS, Kandoth C, Mardis ER, McLellan MD, Miller CA, Schmidt HK, Wilson RK, Crain D, Curley E, Gardner J, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton C, Shelton T, Sherman M, Thompson E, Yena P, Bowen J, Gastier-Foster JM, Gerken M, Leraas KM, Lichtenberg TM, Ramirez NC, Wise L, Zmuda E, Corcoran N, Costello T, Hovens C, Carvalho AL, de Carvalho AC, Fregnani JH, Longatto-Filho A, Reis RM, Scapulatempo-Neto C, Silveira HCS, Vidal DO, Burnette A, Eschbacher J, Hermes B, Noss A, Singh R, Anderson ML, Castro PD, Ittmann M, Huntsman D, Kohl B, Le X, Thorp R, Andry C, Duffy ER, Lyadov V, Paklina O, Setdikova G, Shabunin A, Tavobilov M, McPherson C, Warnick R, Berkowitz R, Cramer D, Feltmate C, Horowitz N, Kibel A, Muto M, Raut CP, Malykh A, Barnholtz-Sloan JS, Barrett W, Devine K, Fulop J, Ostrom QT, Shimmel K, Wolinsky Y, Sloan AE, Rose AD, Giuliante F, Goodman M, Karlan BY, Hagedorn CH, Eckman J, Harr J, Myers J, Tucker K, Zach LA, Deyarmin B, Hu H, Kvecher L, Larson C, Mural RJ, Somiari S, Vicha A, Zelinka T, Bennett J, Iacocca M, Rabeno B, Swanson P, Latour M, Lacombe L, Têtu B, Bergeron A, McGraw M, Staugaitis SM, Chabot J, Hibshoosh H, Sepulveda A, Su T, Wang T, Potapova O, Voronina O, Desjardins L, Mariani O, Roman-Roman S, Sastre X, Stern M-H, Cheng F, Signoretti S, Berchuck A, Bigner D, Lipp E, Marks J, McCall S, McLendon R, Secord A, Sharp A, Behera M, Brat DJ, Chen A, Delman K, Force S, Khuri F, Magliocca K, Maithel S, Olson JJ, Owonikoko T, Pickens A, Ramalingam S, Shin DM, Sica G, Meir EGV, Zhang H, Eijckenboom W, Gillis A, Korpershoek E, Looijenga L, Oosterhuis W, Stoop H, van Kessel KE, Zwarthoff EC, Calatozzolo C, Cuppini L, Cuzzubbo S, DiMeco F, Finocchiaro G, Mattei L, Perin A, Pollo B, Chen C, Houck J, Lohavanichbutr P, Hartmann A, Stoehr C, Stoehr R, Taubert H, Wach S, Wullich B, Kycler W, Murawa D, Wiznerowicz M, Chung K, Edenfield WJ, Martin J, Baudin E, Bubley G, Bueno R, Rienzo AD, Richards WG, Kalkanis S, Mikkelsen T, Noushmehr H, Scarpace L, Girard N, Aymerich M, Campo E, Giné E, Guillermo AL, Bang NV, Hanh PT, Phu BD, Tang Y, Colman H, Evason K, Dottino PR, Martignetti JA, Gabra H, Juhl H, Akeredolu T, Stepa S, Hoon D, Ahn K, Kang KJ, Beuschlein F, Breggia A, Birrer M, Bell D, Borad M, Bryce AH, Castle E, Chandan V, Cheville J, Copland JA, Farnell M, Flotte T, Giama N, Ho T, Kendrick M, Kocher J-P, Kopp K, Moser C, Nagorney D, O’Brien D, O’Neill BP, Patel T, Petersen G, Que F, Rivera M, Roberts L, Smallridge R, Smyrk T, Stanton M, Thompson RH, Torbenson M, Yang JD, Zhang L, Brimo F, Ajani JA, Gonzalez AMA, Behrens C, Bondaruk J, Broaddus R, Czerniak B, Esmaeli B, Fujimoto J, Gershenwald J, Guo C, Lazar AJ, Logothetis C, Meric-Bernstam F, Moran C, Ramondetta L, Rice D, Sood A, Tamboli P, Thompson T, Troncoso P, Tsao A, Wistuba I, Carter C, Haydu L, Hersey P, Jakrot V, Kakavand H, Kefford R, Lee K, Long G, Mann G, Quinn M, Saw R, Scolyer R, Shannon K, Spillane A, Stretch J, Synott M, Thompson J, Wilmott J, Al-Ahmadie H, Chan TA, Ghossein R, Gopalan A, Levine DA, Reuter V, Singer S, Singh B, Tien NV, Broudy T, Mirsaidi C, Nair P, Drwiega P, Miller J, Smith J, Zaren H, Park J-W, Hung NP, Kebebew E, Linehan WM, Metwalli AR, Pacak K, Pinto PA, Schiffman M, Schmidt LS, Vocke CD, Wentzensen N, Worrell R, Yang H, Moncrieff M, Goparaju C, Melamed J, Pass H, Botnariuc N, Caraman I, Cernat M, Chemencedji I, Clipca A, Doruc S, Gorincioi G, Mura S, Pirtac M, Stancul I, Tcaciuc D, Albert M, Alexopoulou I, Arnaout A, Bartlett J, Engel J, Gilbert S, Parfitt J, Sekhon H, Thomas G, Rassl DM, Rintoul RC, Bifulco C, Tamakawa R, Urba W, Hayward N, Timmers H, Antenucci A, Facciolo F, Grazi G, Marino M, Merola R, de Krijger R, Gimenez-Roqueplo A-P, Piché A, Chevalier S, McKercher G, Birsoy K, Barnett G, Brewer C, Farver C, Naska T, Pennell NA, Raymond D, Schilero C, Smolenski K, Williams F, Morrison C, Borgia JA, Liptay MJ, Pool M, Seder CW, Junker K, Omberg L, Dinkin M, Manikhas G, Alvaro D, Bragazzi MC, Cardinale V, Carpino G, Gaudio E, Chesla D, Cottingham S, Dubina M, Moiseenko F, Dhanasekaran R, Becker K-F, Janssen K-P, Slotta-Huspenina J, Abdel-Rahman MH, Aziz D, Bell S, Cebulla CM, Davis A, Duell R, Elder JB, Hilty J, Kumar B, Lang J, Lehman NL, Mandt R, Nguyen P, Pilarski R, Rai K, Schoenfield L, Senecal K, Wakely P, Hansen P, Lechan R, Powers J, Tischler A, Grizzle WE, Sexton KC, Kastl A, Henderson J, Porten S, Waldmann J, Fassnacht M, Asa SL, Schadendorf D, Couce M, Graefen M, Huland H, Sauter G, Schlomm T, Simon R, Tennstedt P, Olabode O, Nelson M, Bathe O, Carroll PR, Chan JM, Disaia P, Glenn P, Kelley RK, Landen CN, Phillips J, Prados M, Simko J, Smith-McCune K, VandenBerg S, Roggin K, Fehrenbach A, Kendler A, Sifri S, Steele R, Jimeno A, Carey F, Forgie I, Mannelli M, Carney M, Hernandez B, Campos B, Herold-Mende C, Jungk C, Unterberg A, von Deimling A, Bossler A, Galbraith J, Jacobus L, Knudson M, Knutson T, Ma D, Milhem M, Sigmund R, Godwin AK, Madan R, Rosenthal HG, Adebamowo C, Adebamowo SN, Boussioutas A, Beer D, Giordano T, Mes-Masson A-M, Saad F, Bocklage T, Landrum L, Mannel R, Moore K, Moxley K, Postier R, Walker J, Zuna R, Feldman M, Valdivieso F, Dhir R, Luketich J, Pinero EMM, Quintero-Aguilo M, Carlos Gilberto Carlotti J, Santos JSD, Kemp R, Sankarankuty A, Tirapelli D, Catto J, Agnew K, Swisher E, Creaney J, Robinson B, Shelley CS, Godwin EM, Kendall S, Shipman C, Bradford C, Carey T, Haddad A, Moyer J, Peterson L, Prince M, Rozek L, Wolf G, Bowman R, Fong KM, Yang I, Korst R, Rathmell WK, Fantacone-Campbell JL, Hooke JA, Kovatich AJ, Shriver CD, DiPersio J, Drake B, Govindan R, Heath S, Ley T, Tine BV, Westervelt P, Rubin MA, Lee JI, Aredes ND, Mariamidze A, Hu H, An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 173, 400–416.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW, Cancer Genome Landscapes. Science. 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.MSK-IMPACT: A Targeted Test for Mutations in Both Rare and Common Cancers | Memorial Sloan Kettering Cancer Center, (available at https://www.mskcc.org/msk-impact). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The parental cell lines used in this work are available from various commercial sources (Table S4). The plasmids used to induce chromosome loss have been deposited at Addgene (Table S5). The code used to perform the TCGA survival analysis is available at https://github.com/joan-smith/comprehensive-tcga-survival (31). The code used to perform the mutual exclusivity analysis is available at github.com/sheltzer-lab/aneuploidy-addictions (27). The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE PXD037956127. RNA-seq data have been deposited at GSE222379.