Abstract
Dyslipidemia has been associated with depression, but individual lipid species associated with depression remain largely unknown. The temporal relationship between lipid metabolism and the development of depression also remains to be determined. We studied 3721 fasting plasma samples from 1978 American Indians attending two exams (2001–2003, 2006–2009, mean ~5.5 years apart) in the Strong Heart Family Study. Plasma lipids were repeatedly measured by untargeted liquid chromatography-mass spectrometry (LC-MS). Depressive symptoms were assessed using the 20-item Center for Epidemiologic Studies for Depression (CES-D). Participants at risk for depression were defined as total CES-D score ≥16. Generalized estimating equation (GEE) was used to examine the associations of lipid species with incident or prevalent depression, adjusting for covariates. The associations between changes in lipids and changes in depressive symptoms were additionally adjusted for baseline lipids. We found that lower levels of sphingomyelins and glycerophospholipids and higher level of lysophospholipids were significantly associated with incident and/or prevalent depression. Changes in sphingomyelins, glycerophospholipids, acylcarnitines, fatty acids and triacylglycerols were associated with changes in depressive symptoms and other psychosomatic traits. We also identified differential lipid networks associated with risk of depression. The observed alterations in lipid metabolism may affect depression through increasing the activities of acid sphingomyelinase and phospholipase A2, disturbing neurotransmitters and membrane signaling, enhancing inflammation, oxidative stress, and lipid peroxidation, and/or affecting energy storage in lipid droplets or membrane formation. These findings illuminate the mechanisms through which dyslipidemia may contribute to depression and provide initial evidence for targeting lipid metabolism in developing preventive and therapeutic interventions for depression.
INTRODUCTION
Major depressive disorder (MDD) is a debilitating mental disorder that affects over 300 million people worldwide and a major contributor to the overall global burden of disease [1]. American Indians suffer a higher rate of depression compared to Caucasians and African-Americans [2, 3]. Currently, the diagnosis of MDD relies solely on clinical symptoms, which results in a high rate of misdiagnosis [4]. In addition, about one-third of patients failed to respond to antidepressants [5], highlighting the pressing need of novel biomarkers for accurate diagnosis, prognosis and drug response monitoring. Clinically, depressive symptoms are often accompanied by and associated with increased risk of metabolic disorders, such as dyslipidemia, impaired glucose metabolism, obesity, diabetes, and heart diseases [6–8]. Epidemiological studies have reported that depressive patients had lower levels of total cholesterol [9, 10] and low-density lipoprotein (LDL) cholesterol [11] compared to healthy controls, although results are mixed [12–18]. Together, these findings strongly suggest that metabolic dysregulation may underlie depression and its related phenotypes [19, 20]. However, as routine blood lipids provide only a narrow snapshot of lipid metabolism, it fails to capture the complexity of the blood lipidome that includes hundreds to thousands of individual lipid species (both endogenous and exogenous) in a blood sample.
Lipidomics can identify and quantify up to thousands of individual lipid species in a biological sample, and is well suited for characterizing perturbed lipid metabolism that precedes the onset of depressive symptoms. In support of this, several studies have reported associations of altered blood lipid species, such as fatty acids, phosphatidylcholines (PCs), phosphatidylethanolamines (PEs), phosphatidylinositols (PIs), sphingolipids, cholesterol esters (CEs), diacylglycerols (DAGs), and triacylglycerols (TAGs), with depression in various populations [21–31]. However, previous studies were mostly limited by small sample size and a low coverage of the blood lipidome. Importantly, prior studies measured blood lipids only once (baseline), which did not reflect the temporal relationship between change in the blood lipidome and development of depression. Potential confounding and comorbid conditions were not adequately controlled either in previous research.
Here we report findings from the first large-scale longitudinal lipidomic profiling of risk for depressive symptoms in 3721 fasting plasma samples from 1978 American Indians attending two clinical exams (~5.5 year apart) in the Strong Heart Family Study. Our primary goals are to (1) identify individual lipid species associated with risk of depression beyond known clinical factors, and (2) examine whether longitudinal changes in plasma lipids are associated with changes in depressive symptoms and other psychosomatic traits.
MATERIALS AND METHODS
Study participants
All participants are American Indians in the Strong Heart Family Study (SHFS, 2001-ongoing), a family-based prospective study designed to identify genetic, metabolic and behavioral factors for cardiovascular disease (CVD) and its risk factors in American Indians [32–34]. Briefly, 2786 tribal members (aged 14 and older) in three geographic regions (Arizona, North/South Dakota, Oklahoma) were initially examined in 2001–2003 and re-examined in 2006–2009 (mean 5.5 years apart) using the same protocols. Information for demography, lifestyle, family history and medical records was collected at each visit.
Assessment of clinical factors
Fasting plasma glucose, insulin, and lipoproteins were measured by standard laboratory methods as previously described [35, 36]. Body mass index (BMI) was calculated as body weight in kilograms divided by the square of height in meters. Information regarding previous/current medical conditions, education, smoking, alcohol consumption, and family history of diseases was collected during the personal interview. For cigarette smoking, participants were classified as current smokers and non-current smokers (former and never smokers combined). Alcohol consumption was determined by self-reported history of alcohol intake, the type of alcoholic beverages consumed, frequency of alcohol consumption, and average quantity consumed per day and per week. Participants were classified as current drinkers and non-current drinkers (former and never drinkers). Information on use of lipid-lowering drugs and anti-depressants was collected at each visit. Hypertension was defined as blood pressure ≥140/90 mmHg or use of antihypertensive medications. Type 2 diabetes was defined as fasting plasma glucose ≥126 mg/dL or use of hypoglycemic drugs. Cardiovascular disease (CVD) was defined as any definite or possible fatal or non-fatal myocardial infarction, coronary heart disease, stroke, or heart failure, as previously described [32, 37, 38]. Chronic kidney disease (CKD) was defined as estimated glomerular filtration rate (eGFR) < 60 mL / min /1.73m2 or a history of dialysis or kidney transplant [39, 40]. In this study, we defined presence of chronic diseases if participants had any of the four chronic conditions, including type 2 diabetes, hypertension, CVD, or CKD. Those without any of these conditions were defined as absence of chronic diseases. A binary variable (yes/no) was included as a covariate in the statistical models. High sensitivity C-reactive protein (CRP) was measured by an ELISA method using purified CRP and anti-CRP antibodies from Calbiochem [41–43]. Plasma fibrinogen was measured using the Clauss method as previously described [44]. The coefficient of variations (CVs) for the CRP and fibrinogen assays were less than 4% and 3%, respectively [43]. All participants provided informed consents. The SHFS protocols were approved by the Institutional Review Boards of each participating institution and tribes.
Assessment of depressive symptoms and other psychosomatic measures
Current depressive symptoms were assessed using the 20-item Center for Epidemiologic Studies for Depressive symptoms Scale (CES-D) [45]. The questionnaire asks participants to rate how often over the past week they experienced symptoms associated with depression such as depressed mood, feeling of guilt or worthlessness, feeling of helplessness or hopelessness, poor appetite, and sleep disturbance. Each of the 20 items rates severity of depressive symptoms on a 4-point scale, with 0 indicating rarely or none, and 3 indicating most or all of the time. The total score for all 20 items ranges from 0 to 60, with a higher score indicating a greater frequency of depressive experiences [45]. Individuals with an increased risk for depression were defined as those with total CES-D score ≥16 [45, 46]. Incident depression was identified if participants had CES-D score <16 at baseline but developed new depression by end of study follow-up. We also collected information for quality of life, perceived stress and social support for all participants (Supplementary Methods). Missing values were imputed by multiple imputation [47, 48] via the R package mice [49].
Lipidomic data acquisition, processing and normalization
Methods for blood sample collection, data acquisition, processing and normalization of the lipidomic data in the SHFS have been described previously [50]. Briefly, fasting plasma samples were first extracted using a modified liquid-liquid extraction method (cold methanol/ MTBE/water) [51], and then subjected to liquid chromatography–mass spectrometry (LC-MS) on both positive and negative ion modes. An in-house cloud-based software (LC-BinBase) was used to process raw data with peak detection and deconvolution algorithms adapted from the MS-DIAL software [52]. Results of lipid peak intensity were manually checked against raw data files. Peaks and features with 50% missing values across all samples were excluded. Batch effect was corrected by the SERRF software [53]. After stringent quality control, we obtained 1809 lipids (579 known, 1230 unknown) in 3977 samples (1983 participants at baseline, 1994 participants at follow-up). The coefficient of variation of the BioIVT and NIST plasma quality control samples was 9% and 16%, respectively. Relative abundances of duplicated samples were highly correlated (r > 0.95). After further excluding lipids with multimodal distribution (n = 267) [54], and participants with outliers or missing covariates, a total of 1542 lipids (518 known, 1024 unknown, Table S1) in 1978 participants (1922 at baseline, 1799 at follow-up) were included in the final data analysis.
Statistical analysis
Figure S1 illustrates the procedures for participants’ selection and statistical analyses. Prior to data analysis, lipidomic data were further standardized to zero mean and unit variance. Other continuous variables (e.g., age, BMI) were similarly scaled.
Prospective association analysis.
Among 1248 participants free of depression at baseline, we constructed multivariate generalized estimating equation (GEE) models to identify baseline lipids predictive of risk for depression. The model accounted for family relatedness and adjusted for age, sex, study center, BMI, a composite variable indicating the presence or absence of chronic diseases (i.e., type 2 diabetes, hypertension, CVD or chronic kidney disease), and use of alcohol, anti-depressants or lipid-lowering drugs at baseline. For the purpose of replication, we randomly split the total sample into discovery (n = 875, 141 cases) and replication (n = 37,359 cases). The analysis was first conducted in the discovery sample, followed by replication of top hits (P < 0.05) in the remaining sample. Results from both stages were then combined by fixed-effects meta-analysis. Lipids with raw P < 0.05 in the discovery stage, and those with false discovery rate (FDR)-adjusted P (i.e., q-value [55, 56]) < 0.05 in the replication and meta-analysis were considered statistically significant. The incremental predictive value of lipids over clinical factors was assessed by area under the receiver operating characteristic curve (AUROC), net reclassification improvement (NRI) and integrated discrimination improvement (IDI) [57].
Cross-sectional association analysis.
The association between lipid species and prevalent depression at baseline (n = 1922) or follow-up (n = 1799) was separately examined by GEE, adjusting for same covariates. Results were then combined by fixed-effects meta-analysis.
Repeated measurement analysis.
To examine the temporal relationship between change in plasma lipidome and the development of depressive symptoms, we regressed the change in total CES-D score (dependent variable) on the change in a specific lipid (independent variable), adjusting for same covariates plus baseline lipid and CES-D score. The longitudinal associations of lipids with other psychosomatic measures, including physical composite scale (PCS), mental health composite scale (MCS), perceived stress scale (PSS), and social support scale (SSS), were similarly examined. This analysis included 1542 lipids (518 known) from 1248 participants who were free of depression and attended clinical exams at both time points.
Differential lipid network analysis.
To identify lipid networks (i.e., sets of lipids that are highly correlated) associated with risk of depression, we constructed lipid modules (subnetworks) using the Weighted Correlation Network Analysis (WGCNA) [58]. This was achieved by building signed weighted lipid co-regulation networks using all 1542 baseline lipids among incident cases and non-cases, separately. Lipid features were hierarchically clustered, and those with a high topological overlap similarity were grouped into a same module. Differential modular analysis was performed to dissect intra-module difference (i.e., difference of connectivity among lipids within a module) between cases and non-cases. To further quantify the intra-module difference between cases and non-cases, we calculated modular differential connectivity (MDC) [59, 60], i.e., total connectivity for any pair of lipids within a module from cases minus that of the same lipid pairs from non-cases. Gain of connectivity (GOC) was defined if MDC > 0 and loss of connectivity (LOC) if MDC < 0. Statistical significance of MDC was evaluated by 1000 permutations [61, 62].
Lipid enrichment and pathway analysis.
Lipid enrichment analysis was performed using the lipid ontology (LION) program [63]. Metabolic pathways involved in the differential networks were annotated using the MetaboAnalyst 5.0 [64]. A pathway impact score, defined as the sum of importance measures of identified lipids divided by the total sum of importance measures of all lipids in a specific pathway, was assigned to each metabolic pathway. Multiple testing for these analyses was controlled at q < 0.05.
Sensitivity analysis.
To examine the potential impact of bulk lipids (e.g., low-density lipoprotein (LDL), high-density lipoprotein (HDL), total cholesterol, triglycerides) on our results, we additionally adjusted for baseline bulk lipids in the statistical models. To examine whether and how missing/imputed data in CES-D affected our results, we performed additional analyses by excluding participants with missing information for one or more items in the 20-item CES-D questionnaire. We also conducted ordinal GEE regression to examine whether lipids significantly associated with status of depression (total CES-D score ≥16) were also associated with severity of depressive symptoms, defined as total CES-D score <10 (none), 10–15 (mild), 16–24 (moderate), and >24 (severe). Information for severity of depressive symptoms in the SHFS is shown in Figure S2.
RESULTS
Characteristics of participants
Prevalence of depression at baseline and follow-up was 27.9% and 26.1%, respectively. As expected, depression was more prevalent in women than in men (baseline: 31.7% vs. 21.7%, P = 2.50 × 10−6; follow-up: 30.0% vs. 19.3%, P = 7.36 × 10−7. Individuals who developed incident depression (cases) were significantly younger, more likely to be females and current smokers, were less educated, and had lower levels of LDL, total cholesterol and systolic blood pressure and a higher level of eGFR at baseline (Table 1). More details for the discovery and replication samples used in the prospective association analysis are described in the Table S2.
Table 1.
Characteristics of American Indian participants according to their status of incident depression by the end of 5-year follow-up in the Strong Heart Family Study (n = 1248).
| Characteristics | All (n = 1248) | Cases (n = 200) | Non-cases (n = 1048) | P-value |
|---|---|---|---|---|
| Age (years) | 41.4 ± 14.0 | 38.1 ± 13.5 | 42.0 ± 14.0 | <0.001 |
| Female, n (%) | 751 (60.2%) | 133 (66.5%) | 618 (59.0%) | 0.042 |
| BMI (kg/m2) | 31.5 ±7.0 | 31.8 ±7.5 | 31.4 ±6.9 | 0.503 |
| Current smoking, n (%) | 461 (36.9%) | 90 (45.0%) | 371 (35.4%) | 0.012 |
| Current drinking, n (%) | 748 (59.9%) | 127 (63.5%) | 621 (59.3%) | 0.308 |
| Education higher than high school, n (%) | 551 (44.2%) | 65 (32.5%) | 486 (46.4%) | <0.001 |
| Hypertension, n (%) | 375 (30.0%) | 57 (28.5%) | 318 (30.3%) | 0.605 |
| Type 2 diabetes, n (%) | 212 (17.0%) | 39 (19.5%) | 173 (16.5%) | 0.580 |
| Chronic kidney disease, n (%) | 11 (0.9%) | 3 (1.5%) | 8 (0.8%) | 0.318 |
| Cardiovascular disease, n (%) | 13 (1.0%) | 2 (1.0%) | 11 (1.0%) | 0.945 |
| Presence of chronic diseases, n (%) | 469 (37.6%) | 76 (38.0%) | 393 (37.5%) | 0.894 |
| LDL cholesterol (mg/dL) | 103.4 ±30.7 | 96.7 ±31.0 | 104.7 ±30.5 | 0.002 |
| HDL cholesterol (mg/dL) | 52.0 ± 14.6 | 52.3 ± 15.3 | 52.0 ± 14.4 | 0.798 |
| Total cholesterol (mg/dL) | 187.2 ±35.2 | 180.3 ±35.4 | 188.5 ±35.0 | 0.008 |
| Triglycerides (mg/dL) | 164.3 ± 123.2 | 162.4 ±113.1 | 164.6 ± 125.1 | 0.814 |
| Fasting plasma glucose (mg/dL) | 106.8 ±41.1 | 107.9 ±51.6 | 106.6 ±38.8 | 0.709 |
| Systolic blood pressure (mmHg) | 123.3 ± 15.7 | 121.3 ± 15.4 | 123.7 ± 15.7 | 0.048 |
| Diastolic blood pressure (mmHg) | 77.7 ± 10.7 | 76.4 ± 10.8 | 78.0 ± 10.6 | 0.070 |
| eGFR (mL/min/1.73m2) | 112.9 ± 17.7 | 116.0 ±20.0 | 112.3 ± 17.2 | 0.028 |
| C-reactive protein (mg/L) | 6.7 ± 10.4 | 7.9 ± 13.5 | 6.5 ± 9.7 | 0.119 |
| Fibrinogen (mg/dL) | 377.8 ±87.1 | 377.2 ±91.1 | 377.9 ± 86.4 | 0.914 |
| Use of lipid-lowering drugs, n (%) | 43 (3.4%) | 6 (3.0%) | 37 (3.5%) | 0.698 |
| Use of antidepressants, n (%) | 36 (2.9%) | 7 (3.5%) | 29 (2.8%) | 0.571 |
Continuous variables were expressed as mean ± standard deviation and qualitative variables were expressed as n (%). P-values were obtained by generalized estimating equation (GEE) model accounting for family relatedness. Cases: participants who developed incident depression over 5-year follow-up; Non-cases: participants who remained to be free of depression over 5-year follow-up. Presence of chronic diseases was defined as participants who had one or more of these conditions (hypertension, type 2 diabetes, chronic kidney disease, cardiovascular disease), while absence was defined as those without any of these conditions.
BMI body mass index, LDL low-density lipoprotein, HDL high-density lipoprotein, eGFR estimated glomerular filtration rate.
Compared to individuals without depression, those with prevalent depression at either baseline or follow-up were significantly younger, more likely to be females, had higher BMI, more likely to be current smokers, were less educated, and had lower levels of LDL, total cholesterol and systolic blood pressure and a higher level of eGFR (Table S3). The distributions of CES-D scales between imputed and original non-missing data did not differ significantly (Fig. S3). Fig. S4 A–H shows the distributions of physical composite scale (PCS), mental health composite scale (MCS), perceived stress scale (PSS), and social support scale (SSS) at baseline and follow-up. Total CES-D score and other psychosomatic measures were highly correlated (Fig. S4 I–G). Missing rates for CES-D and other psychosomatic measures are shown in Table S4.
Baseline plasma lipids predicted incident depression beyond clinical factors
Prospective association analysis identified 96 lipids (24 known) significantly associated with incident depression in the discovery stage (Fig. 1, Table S5). Of the 24 known lipids, 21 lipids including 7 sphingomyelins (SMs), 5 phosphatidylcholines (PCs), 3 phosphatidylethanolamines (PEs), 2 phosphatidylinositols (PIs), and 4 ether-phospholipids were inversely, while three lipids including FA(15:1), LPC(20:1), and LPC(p-18:0)/LPC(o-18:1) were positively associated with risk of depression. These 24 lipids were also significantly associated with bulk lipids (e.g., HDL, LDL, total cholesterol, and triglycerides) and other clinical measures, such as BMI, fasting plasma glucose, blood pressure, inflammatory markers (i.e., C-reactive protein, fibrinogens) and kidney function (e.g., eGFR) (Fig. S5). Five (out of 24) lipids, including PC(40:6) A, SM(d36:3) A, SM(d36:3) B, SM(d40:2) B, and SM(d41:2) B, were confirmed in the replication sample (Fig. 1, Table S6, Fig. S6). Nine lipids, including PC(40:6) A, PE(16:0/22:5), and 7 sphingomyelins (SMs), were inversely associated with incident depression in the meta-analysis (Table S7). As shown in Fig. 2, additional inclusion of he top six lipids (i.e., SM(d36:3) A, SM(d32:2) B, SM(d40:2) B, SM(d41:2) B, SM(d39:1) B, and PC(40:6) A) significantly improved risk prediction over clinical factors (AUROC increased from 0.575 to 0.627, P = 0.035; continuous NRI = 0.417, PNRI = 0.001; IDI = 0.016, PIDI = 0.001). These results suggest that the newly identified lipid species may have potential clinical utility in risk prediction for depression.
Fig. 1. Baseline plasma lipids associated with incident depression (mean follow-up: 5.5 years).
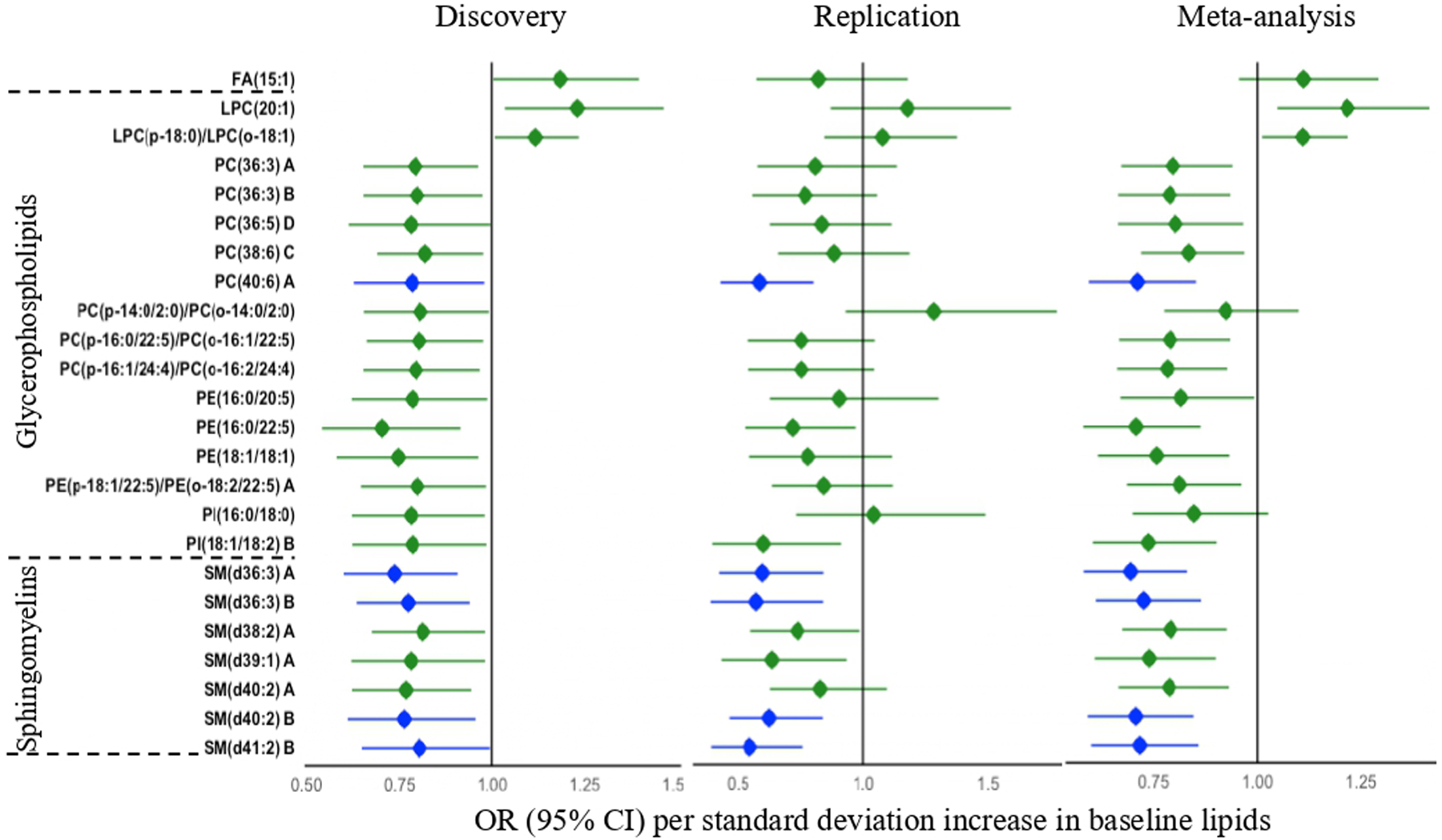
Odds ratios (ORs) were obtained by GEE, adjusting for age, sex, study center, BMI, a composite variable indicating the presence or absence of chronic diseases (i.e., type 2 diabetes, hypertension, cardiovascular disease or chronic kidney disease), and use of alcohol, anti-depressants or lipid-lowering drugs at baseline. Only known lipids significantly associated with incident depression in the discovery stage are shown. Lipids confirmed in the replication stage are highlighted in blue. The letter A, B, C, or D in the name of lipids indicates isomers.
Fig. 2. Incremental value of the identified plasma lipids in risk prediction for depression.
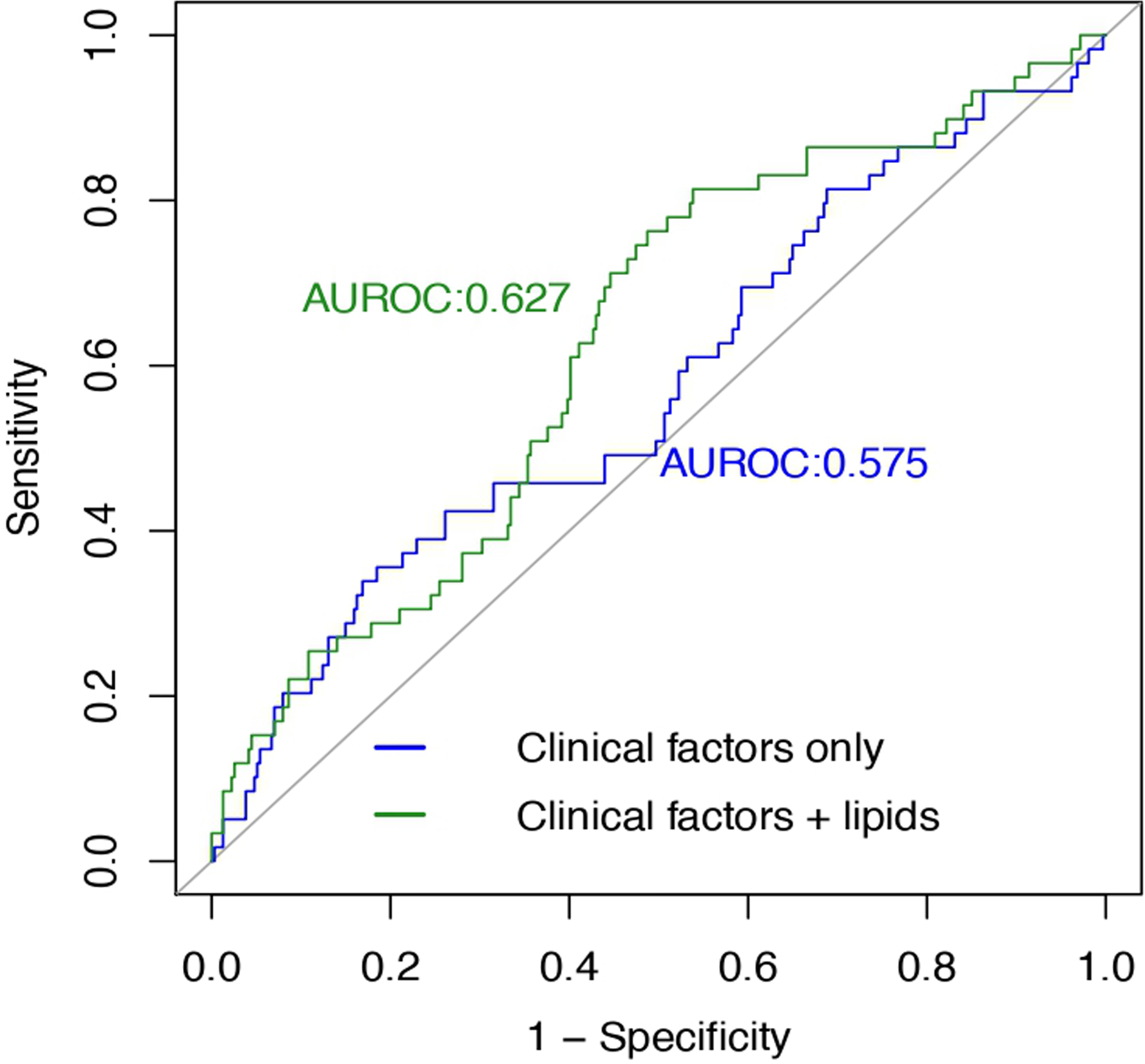
Model 1: clinical factors only, including age, sex, study center, BMI, a composite variable indicating the presence or absence of chronic diseases (i.e., type 2 diabetes, hypertension, cardiovascular disease or chronic kidney disease), and use of alcohol, anti-depressants or lipid-lowering drugs at baseline. Model 2: clinical factors + 6 significant lipids, including SM(d36:3) A, SM(d32:2) B, SM(d40:2) B, SM(d41:2) B, SM(d39:1) B, and PC(40:6) A. Compared to the model that included clinical factors only (model 1), additional inclusion of plasma lipids (model 2) significantly increases risk prediction for depression. AUROC increases from 0.575 to 0.627 (P = 0.035); continuous NRI = 0.417, PNRI = 0.001; IDI = 0.016, PIDI = 0.001.
Altered plasma lipids associated with prevalent depression
After correction for multiple testing (q < 0.05), 66 lipids (23 known) were significantly associated with prevalent depression at baseline (Fig. 3, Table S8). The 23 known lipids largely belong to sphingomyelins (SMs). Of these, 19 lipids were inversely (ORs: 0.72–0.84) whereas 4 lipids were positively (ORs: 1.10–1.19) associated with prevalent depression. In contrast, 175 lipids (62 known) were significantly associated with prevalent depression at follow-up (Fig. 3, Table S9). The 62 known lipids largely belong to phosphatidylcholines (PCs), ether-phosphatidylcholines (PC-P/PC-O), and sphingomyelins (SMs). Of these, 59 lipids were inversely (ORs: 0.71–0.92) associated, whereas 3 lipids were positively associated with prevalent depression (ORs: 1.15–1.18). Decreased levels of 14 lipids were significantly associated with prevalent depression at both time points (Fig. 3). Multiple lipids, such as SM(d39:1) A, SM(d40:2) B, and SM(d41:2) B, were also significantly associated with incident depression. Meta-analysis combining results at both time points revealed significant associations of 333 lipids (114 known) with prevalent depression at q < 0.05 (Table S10).
Fig. 3. Plasma lipids associated with prevalent depression at baseline or follow-up.
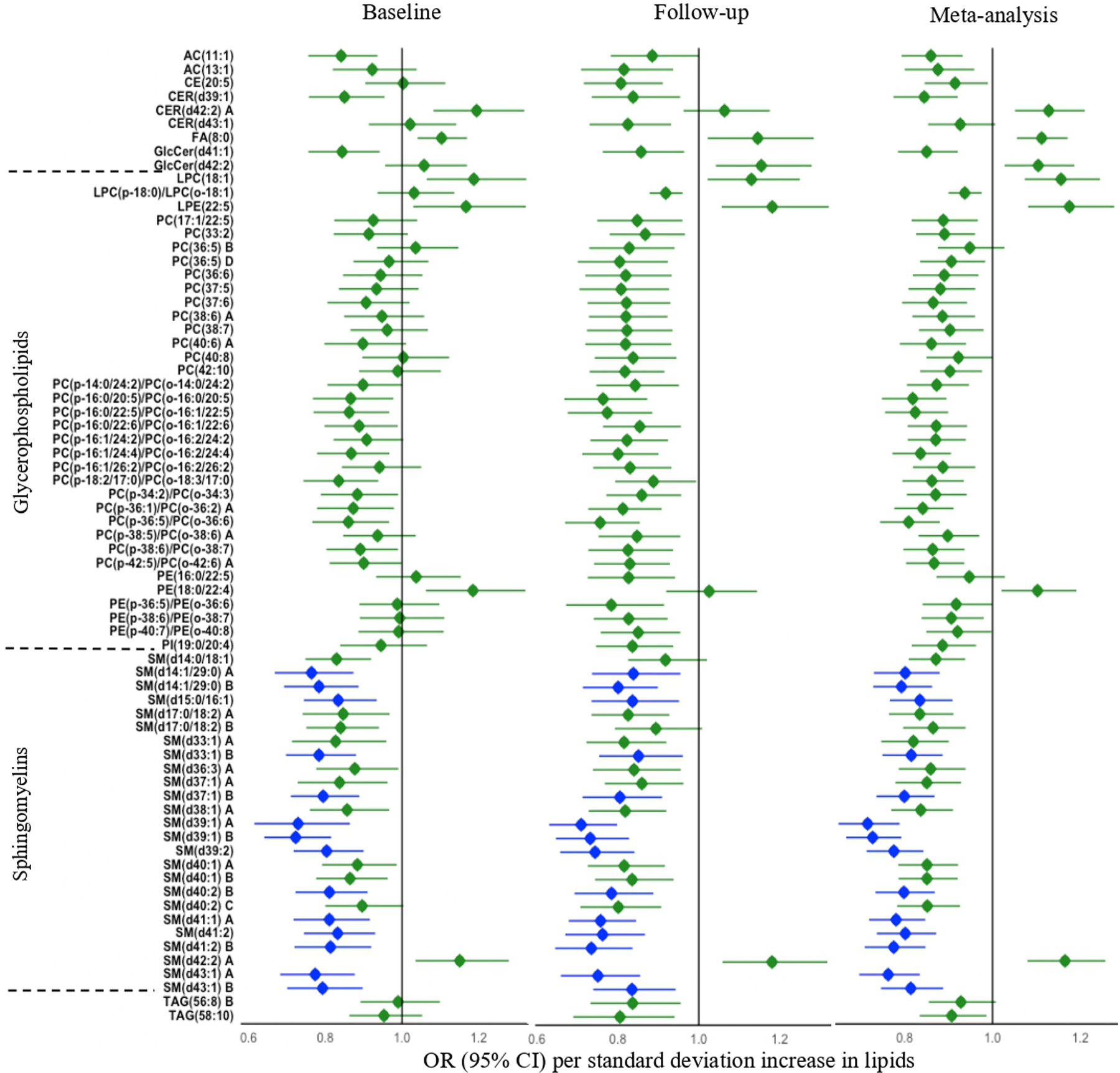
Odds ratios (ORs) were obtained by GEE, adjusting for age, sex, study center, BMI, a composite variable indicating the presence or absence of chronic diseases (i.e., diabetes, hypertension, cardiovascular disease or chronic kidney disease), and use of alcohol, anti-depressants or lipid-lowering drugs. Only known lipids with q < 0.05 are shown. The blue highlighted lipids indicate statistically significant at both baseline and follow-up. The letter A, B, C, or D in the name of lipids indicates isomers.
Longitudinal changes in fasting plasma lipid species associated with change in depressive symptoms
After correction for covariates and multiple testing (q < 0.05), longitudinal changes in 223 lipids (51 known) were significantly associated with changes in CES-D score or other psychosomatic measures (e.g., physical composite scale (PCS), mental health composite scale (MCS), perceived stress scale (PSS), and social support scale (SSS)) (Figs. 4, S7–8 and Table S11). The 51 known lipids largely belong to acylcarnitines (ACs), fatty acids (FAs), ether-phosphatidylcholines (PC-P/PC-O), phosphatidylinositols (PIs), sphingomyelins (SMs), and triacylglycerols (TAGs). To ease of data interpretation, we plotted Fig. 5 to summarize the directions of associations and possible biological pathways for the identified lipids in this study.
Fig. 4. Manhattan plots displaying the longitudinal associations of change in plasma lipids with change in psychosomatic measures between baseline and follow-up.

A CES-D score; B physical composite scale (PCS); C mental health composite scale (MCS). X-axis: lipid classes; Y-axis: −log10 P. Different colors represent different lipid categories. The dashed line represents significance level at q = 0.05. The letter A or B in the name of lipids represents isomers.
Fig. 5. Directions of associations between plasma lipid species and depression in the SHFS.
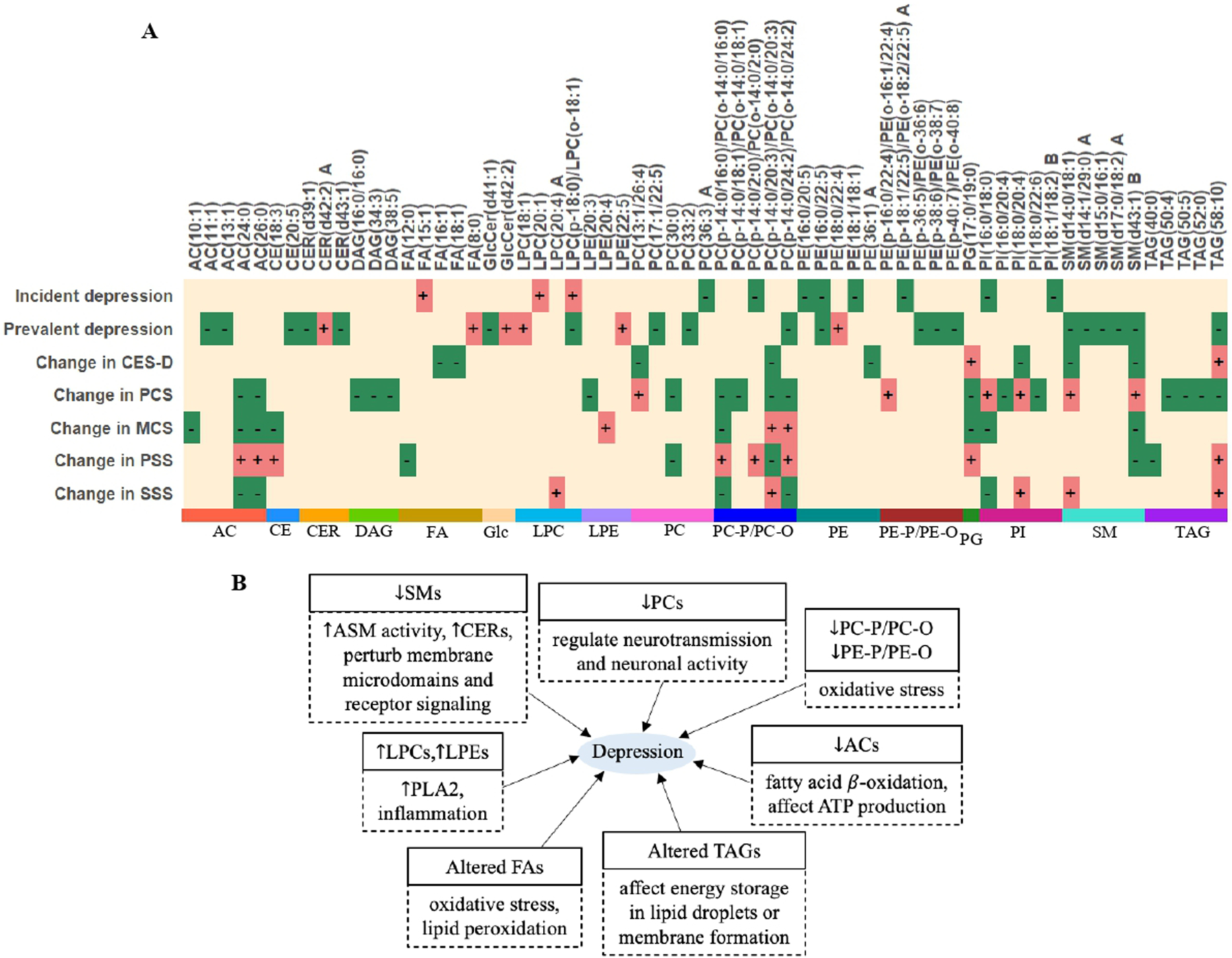
A Summary of the directions of the associations between plasma lipid species and depression-related phenotypes in each analysis. “+” denotes positive association (regression coefficient) between lipid and each trait; “−” denotes inverse association (regression coefficient) between lipid and each trait. Only top 5 known lipids in each class are shown. The letter A or B in the name of lipids represents isomers. B Diagram illustrating the relationship between altered lipid metabolism and depression. Each box contains two panels. The upper panel indicates the direction of association (regression coefficient) between lipid species and depression in the current study. The lower panel indicates the possible metabolic pathways through which altered lipid metabolism may affect depression. “↓” denotes lower plasma lipids in individuals with depression compared to non-depressed individuals. “↑” denotes higher plasma lipids (or enhanced activities) in individuals with depression compared to non-depressed individuals. Abbreviations: PCS physical composite scale, MCS mental health composite scale, PSS perceived stress scale, SSS social support scale, ASM activity of acid sphingomyelinase, PLA2 phospholipase A2, ATP adenosine triphosphate, AC acylcarnitine, CE cholesterol ester, CER ceramide, DAG diacylglycerol, FA fatty acid, Glc glycosylceramide, LPC lysophosphatidylcholine, LPE lysophosphatidylethanolamine, PC phosphatidylcholine, PC-P/PC-O ether-phosphatidylcholine, PE phosphatidylethanolamine, PE-P/PE-O ether-phosphatidylethanolamine, PG phosphatidylglycerol, PI phosphatidylinositol, SM sphingomyelin, TAG triacylglycerol.
Differential lipid networks for risk of depression
Three lipid networks exhibited significant difference in lipid connectivity between individuals who developed incident depression (cases) and those who did not (Figs. S9–13). Specifically, compared to non-cases, incident cases exhibited gain of connectivity (GOC) for lipids in the magenta module (modular differential connectivity (MDC) = 36.6, P = 0.015, q = 0.07) and the green module (MDC = 118.2, P = 0.018, q = 0.07) (Figs. S11–13). Hub lipids in these two GOC modules included CER(d38:1), CER(d39:1), CER(d40:1), PC(36:1), PC(38:2), PC(38:3), and PC(38:4) B. In contrast, incident cases exhibited loss of connectivity (LOC) for lipids the red module (MDC = −91.5, P = 0.021, q = 0.070) compared to non-cases (Figs. S11–13). Hub lipids in this LOC module included AC(12:1), AC(14:1), AC(14:2), and AC(16:1). These differentially expressed lipids are enriched in pathways related to endoplasmic reticulum, plasma membrane, and lipid-mediated signaling, glycerophospholipid metabolism, and biosynthesis of unsaturated fatty acids (Fig. S14).
Results from sensitivity analyses
The observed associations of lipid species with incident or prevalent depression were largely unchanged after further adjustments for bulk lipids (Tables S12–14). After excluding individuals with missing data on the CES-D questionnaire, 8 individual lipid species (out of 24 detected in the data including both non-missing and imputed data) in the prospective analysis and majority of the individual lipid species (63 out of 99 lipids at baseline, 109 out of 132 lipids at follow-up) in the cross-sectional analysis remained significant (Tables S12–14). There was no change in the identified lipid classes before and after excluding participants with missing data on the CES-D survey. Finally, most lipids associated with prevalent depression (total CES-D ≥ 16) were similarly associated with severity of depressive symptoms (Table S15).
DISCUSSION
In this first large-scale longitudinal profiling of plasma lipidome for depression, we have several important findings. First, several lipid species were significantly associated with depression. Specifically, we found that lower baseline levels of unsaturated sphingomyelins (e.g., SM(d36:3) A, SM(d40:2) B, SM(d41:2) B), phosphatidylcholines (e.g., PC(36:3) A, PC(38:6) C, PC(40:6) A), phosphatidylethanolamines (e.g., PE(16:0/20:5), PE(16:0/22:5), PE(18:1/18:1)), phosphatidylinositols (e.g., PI(16:0/18:0), PI(18:1/18:2) B), and ether-phospholipids (e.g., PC(p-14:0/2:0)/PC(o-14:0/2:0), PC(p-16:1/24:4)/PC(o-16:2/24:4)), and higher baseline levels of lysophospholipids (e.g., LPC(20:1), LPC(p-18:0)/LPC(o-18:1)) were significantly associated with an increased risk of depression. Six individual lipids, including SM(d36:3) A, SM(d32:2) B, SM(d40:2) B, SM(d41:2) B, SM(d39:1) B, and PC(40:6) A, significantly improved risk prediction for depression above and over clinical factors. Cross-sectional analysis revealed that, compared to nondepressed individuals, depressed individuals had lower levels of unsaturated sphingomyelins, phosphatidylcholines, ether-phospholipids (e.g., PCP/PC-O), acylcarnitines, and cholesterol esters, and higher levels of unsaturated phosphatidylethanolamines, lysophosphatidylcholines, lysophosphatidylethanolamines, and saturated fatty acid (FA(8:0)). Second, repeated measurement analysis revealed that changes in multiple lipid species were significantly associated with changes in depressive symptoms and other psychosomatic traits, after adjusting for clinical factors and baseline lipids. Specifically, changes in unsaturated sphingomyelins (e.g., SM(d30:1) B, SM(d39:1) B, SM(d41:2)) and fatty acids (e.g., FA(16:1), FA(18:1), FA(18:2)) were inversely, whereas changes in saturated acylcarnitines (e.g., AC(24:0), AC(26:0)) were positively associated with depressive symptoms. These findings suggest that disturbed lipid metabolism may accompany the development of depression. Third, we identified differential lipid networks and hub lipids associated with risk of depression. These hub lipids may represent important therapeutic targets in future investigations. Together, our results unraveled substantial perturbations in lipid metabolism and distinct lipidomic signatures associated with depressive symptoms among American Indians.
In line with previous studies reporting negative associations of specific sphingomyelins with depression [22, 24, 29], we also found that lower baseline levels of several long-chain unsaturated sphingomyelins (e.g., SM(d36:3) A, SM(d40:2) B, SM(d41:2) B) were associated with an increased risk of depression in American Indians. In addition, lower levels of multiple sphingomyelins (e.g., SM(d15:0/16:1), SM(d33:1) B, SM(d39:1) B, SM(d39:2), SM(d40:2) B, SM(d41:2), SM(d43:1) A) were significantly associated with prevalent depression at both baseline and follow-up. Moreover, longitudinal changes in sphingomyelins (e.g., SM(d39:1) B, SM(d41:2), SM(d43:1) B) were inversely associated with change in CES-D score and other psychosomatic phenotypes. Decreased sphingomyelins may affect depressive symptoms through increasing the activity of acid sphingomyelinase (ASM), an enzyme that catalyzes the breakdown of sphingomyelin into ceramide and phosphorylcholine [65]. Indeed, patients with depression exhibited higher ASM activity than healthy controls [66], and overexpression of ASM in mouse brain resulted in depression-like phenotypes [67]. Moreover, antidepressants inhibited the activity of ASM both in vitro and in vivo [68]. Inhibition of ASM has been considered a promising therapeutic approach for depression [69].
In addition to sphingomyelins, decreased glycerophospholipids, such as phosphatidylcholines (PCs) and ether-phosphatidylcholines (PC-P/PC-O), were associated with both incident and prevalent depression in American Indians. Moreover, changes in phosphatidylcholines, phosphatidylethanolamines, phosphatidylinositols, and phosphatidylglycerol were associated with changes in CES-D score and other psychometric traits, including quality of life, perceived stress and social support. Further, many glycerophospholipids identified in our study were previously associated with depression in human and/or animal studies. For example, epidemiological studies [21–23, 31] have shown that depression was associated with decreased phosphatidylcholines (e.g., PC(32:0), PC(36:1)), phosphatidylinositols (e.g., PI (36:3), PI(38:5)), ether-phosphatidylcholines (e.g, PC-O 36:2, PC-O 36:4), and ether-phosphatidylethanolamines (e.g., PE-O 36:5, PE-O 36:6) in plasma or serum samples. Animal models with depressive symptoms exhibited altered glycerophospholipids, such as phosphatidylcholines, phosphatidylethanolamines and phosphatidylinositols as well as ether-phospholipids (e.g., PE-O/PE-P, PC-O/PC-P) [29, 30, 70]. Moreover, use of antidepressants can change these lipid species in brain or blood samples of animal models [71–74]. Because glycerophospholipids are the important structural and functional components of plasma membranes, they play crucial roles in regulating neurotransmission and neuronal activity. Disturbed glycerophospholipids may affect neurotransmitters and signaling pathways, thereby contributing to depression [75].
Besides phosphatidylcholines, phosphatidylethanolamines, phosphatidylinositols, and ether-phospholipids, elevated plasma lysophosphatidylcholines (e.g., LPC(16:1), LPC(18:1)) and lysophosphatidylethanolamines (e.g., LPE(18:0), LPE(22:5)) were also significantly associated with prevalent depression. These observations are consistent with previous findings in human and animal studies [21, 24, 30]. The elevated lysophospholipids may result from the increased activity of phospholipase A2 (PLA2), which promotes inflammation, thereby contributing to depression [21, 22, 75]. Other changes included decreased ether-phospholipids, such as ether-phosphatidylcholines (PC-P/PC-O) or ether-phosphatidylethanolamines (PE-P/PE-O), which are well-known endogenous antioxidants [76]. This could be related to increased oxidative stress observed in individuals with depression [77, 78]. Together, our results corroborate and strengthen the notion that altered sphingolipids (e.g., sphingomyelins, ceramides) and glycerophospholipids (e.g., PCs, PEs, PIs, LPCs, LPEs, and plasmalogens) are implicated in the pathogenesis of depressive symptoms [21–24, 29].
Our longitudinal lipidomic analysis for the first time revealed the temporal relationship between changes in 51 individual lipid species, largely phosphatidylcholines, phosphatidylethanolamines, phosphatidylinositols, sphingomyelins, unsaturated fatty acids, acylcarnitines, and triacylglycerols, and changes in depressive symptoms as well as other psychosomatic traits. Altered sphingomyelins may promote the generation of ceramides via stimulating acid sphingomyelinase (ASM) activity, which can perturb membrane microdomains and receptor signaling with implications for brain function [29, 75]. Altered metabolism in fatty acids may contribute to depression through enhancing oxidative stress and lipid peroxidation [27, 79]. Acylcarnitines are involved in fatty acid β-oxidation [30]. It has been shown that elevated medium- and long-chain acylcarnitines resulted in incomplete β-oxidation and reduced adenosine triphosphate (ATP) production, thereby contributing to depression [30, 31, 80]. The observed associations between changes in phosphatidylcholines, phosphatidylethanolamines, and lysophosphatidylethanolamines and changes in psychosomatic traits could be attributed to their effects on inflammation and oxidized stress [21, 22, 30]. Altered triacylglycerols may contribute to depression through affecting energy storage in lipid droplets or membrane formation [19, 23].
Several limitations of our study should be noted. First, although we included a large number of American Indians and have internally validated our results, we did not have an external validation due to lack of comparable cohorts with available lipidomic data. Second, we used the CES-D scale but not diagnostic interview for depression diagnosis, albeit CES-D has been widely used in large-scale epidemiological studies [81]. Also, as most epidemiological studies that examine participants during a specific time period (in general about every 4–5 years), data and sample collection in the Strong Heart Study were pre-selected. The optimal time interval between measurements is uncertain. Third, despite the large number of lipids detected, many lipids are unknown or isomers. Additional experiments are needed to characterize these unknowns if considered of interest. Fourth, although our statistical models controlled many known clinical factors, residual confounding cannot be entirely ruled out. Finally, the observational nature of our study precludes any causal inference. In addition, the potential clinical utilities of the identified lipids in disease diagnosis, prognosis and drug response monitoring awaits further investigation. However, our study has several strengths. First, to our knowledge, the current study represents the first and by far the largest longitudinal lipidomic analysis of depressive symptoms in any racial/ethnic groups. Second, we conducted both prospective and cross-sectional association analysis, repeated measurement analysis, and network analysis. Many lipids were consistently detected in different statistical models, signifying the robustness of our findings. Moreover, our analyses adjusted for a comprehensive list of covariates including comorbid conditions, bulk lipids and use of lipid-lowering medications and antidepressants.
In summary, in a large-scale longitudinal lipidomic profiling of depressive symptoms, we demonstrated that disturbed lipid metabolism precedes the onset and accompanies the development of depression. Our findings provide novel insights into the role of dyslipidemia in depression and offer potential for targeting lipid metabolism in preventive and therapeutic interventions for depression.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Strong Heart Study (SHS) participants, the Indian Health Service (IHS) facilities, and the participating tribes for their extraordinary cooperation and involvement, which has contributed to the success of the SHS. The content expressed in this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the IHS.
FUNDING
This study was supported by the National Institute of Health (NIH) grant R01DK107532. The Strong Heart Study (SHS) has been funded in whole or in part with federal funds from the National Heart, Lung, and Blood Institute, National Institute of Health, Department of Health and Human Services, under contract numbers 75N92019D00027, 75N92019D00028, 75N92019D00029, and 75N92019D00030. The study was previously supported by research grants: R01HL109315, R01HL109301, R01HL109284, R01HL109282, and R01HL109319 and by cooperative agreements: U01HL41642, U01HL41652, U01HL41654, U01HL65520, and U01HL65521.
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41380-023-01948-w.
DATA AVAILABILITY
The phenotype data used in this study can be requested through the Strong Heart Study (https://strongheartstudy.org/). The lipidomic data can be obtained from the corresponding author upon a reasonable request.
REFERENCES
- 1.Friedrich MJ. Depression is the leading cause of disability around the world. JAMA. 2017;317:1517. [DOI] [PubMed] [Google Scholar]
- 2.Wulsin LR, Evans JC, Vasan RS, Murabito JM, Kelly-Hayes M, Benjamin EJ. Depressive symptoms, coronary heart disease, and overall mortality in the framingham heart study. Psychosom Med. 2005;67:697–702. [DOI] [PubMed] [Google Scholar]
- 3.Gaffey AE, Cavanagh CE, Rosman L, Wang K, Deng Y, Sims M, et al. Depressive symptoms and incident heart failure in the jackson heart study: differential risk among black men and women. J Am Heart Assoc. 2022;11:e022514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell AJ, Vaze A, Rao S. Clinical diagnosis of depression in primary care: a meta-analysis. Lancet. 2009;374:609–19. [DOI] [PubMed] [Google Scholar]
- 5.Fawcett J, Barkin RL. Efficacy issues with antidepressants. J Clin Psychiatry. 1997;58:32–39. [PubMed] [Google Scholar]
- 6.Vancampfort D, Correll CU, Wampers M, Sienaert P, Mitchell AJ, De Herdt A, et al. Metabolic syndrome and metabolic abnormalities in patients with major depressive disorder: a meta-analysis of prevalences and moderating variables. Psychol Med. 2014;44:2017–28. [DOI] [PubMed] [Google Scholar]
- 7.Pan A, Hu FB. Response to comment on: Pan et al. Bidirectional association between depression and metabolic syndrome: a systematic review and meta-analysis of epidemiological studies. Diabetes Care. 2013;36:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan A, Sun Q, Okereke OI, Rexrode KM, Hu FB. Depression and risk of stroke morbidity and mortality: a meta-analysis and systematic review. JAMA. 2011;306:1241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin JY, Suls J, Martin R. Are cholesterol and depression inversely related? A meta-analysis of the association between two cardiac risk factors. Ann Behav Med. 2008;36:33–43. [DOI] [PubMed] [Google Scholar]
- 10.Olusi SO, Fido AA. Serum lipid concentrations in patients with major depressive disorder. Biol Psychiatry. 1996;40:1128–31. [DOI] [PubMed] [Google Scholar]
- 11.Persons JE, Fiedorowicz JG. Depression and serum low-density lipoprotein: a systematic review and meta-analysis. J Affect Disord. 2016;206:55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreira FP, Jansen K, Cardoso TA, Mondin TC, Magalhães PVDS, Kapczinski F, et al. Metabolic syndrome in subjects with bipolar disorder and major depressive disorder in a current depressive episode: population-based study: metabolic syndrome in current depressive episode. J Psychiatr Res. 2017;92:119–23. [DOI] [PubMed] [Google Scholar]
- 13.van Reedt Dortland AK, Giltay EJ, van Veen T, van Pelt J, Zitman FG, Penninx BW. Associations between serum lipids and major depressive disorder: results from the Netherlands Study of Depression and Anxiety (NESDA). J Clin Psychiatry. 2010;71:729–36. [DOI] [PubMed] [Google Scholar]
- 14.Nakao M, Yano E. Relationship between major depression and high serum cholesterol in Japanese men. Tohoku J Exp Med. 2004;204:273–87. [DOI] [PubMed] [Google Scholar]
- 15.Ledochowski M, Murr C, Sperner-Unterweger B, Neurauter G, Fuchs D. Association between increased serum cholesterol and signs of depressive mood. Clin Chem Lab Med. 2003;41:821–4. [DOI] [PubMed] [Google Scholar]
- 16.Brown SL, Salive ME, Harris TB, Simonsick EM, Guralnik JM, Kohout FJ. Low cholesterol concentrations and severe depressive symptoms in elderly people. BMJ. 1994;308:1328–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacka FN, Pasco JA, McConnell S, Williams LJ, Kotowicz MA, Nicholson GC, et al. Self-reported depression and cardiovascular risk factors in a community sample of women. Psychosomatics. 2007;48:54–59. [DOI] [PubMed] [Google Scholar]
- 18.Ergün UG, Uguz S, Bozdemir N, Güzel R, Burgut R, Saatçi E, et al. The relationship between cholesterol levels and depression in the elderly. Int J Geriatr Psychiatry. 2004;19:291–6. [DOI] [PubMed] [Google Scholar]
- 19.Walther A, Cannistraci CV, Simons K, Durán C, Gerl MJ, Wehrli S, et al. Lipidomics in major depressive disorder. Front Psychiatry. 2018;9:459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ong KL, Morris MJ, McClelland RL, Maniam J, Allison MA, Rye KALipids. lipoprotein distribution and depressive symptoms: the multi-ethnic study of atherosclerosis. Transl Psychiatry. 2016;6:e962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Zheng P, Zhao X, Zhang Y, Hu C, Li J, et al. Discovery and validation of plasma biomarkers for major depressive disorder classification based on liquid chromatography-mass spectrometry. J Proteome Res. 2015;14:2322–30. [DOI] [PubMed] [Google Scholar]
- 22.Demirkan A, Isaacs A, Ugocsai P, Liebisch G, Struchalin M, Rudan I, et al. Plasma phosphatidylcholine and sphingomyelin concentrations are associated with depression and anxiety symptoms in a Dutch family-based lipidomics study. J Psychiatr Res. 2013;47:357–62. [DOI] [PubMed] [Google Scholar]
- 23.Kim EY, Lee JW, Lee MY, Kim SH, Mok HJ, Ha K, et al. Serum lipidomic analysis for the discovery of biomarkers for major depressive disorder in drug-free patients. Psychiatry Res. 2018;265:174–82. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Li J, Zheng P, Zhao X, Zhou C, Hu C, et al. Plasma lipidomics reveals potential lipid markers of major depressive disorder. Anal Bioanal Chem. 2016;408:6497–507. [DOI] [PubMed] [Google Scholar]
- 25.Knowles EEM, Huynh K, Meikle PJ, Göring HHH, Olvera RL, Mathias SR, et al. The lipidome in major depressive disorder: shared genetic influence for ether-phosphatidylcholines, a plasma-based phenotype related to inflammation, and disease risk. Eur Psychiatry. 2017;43:44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang T, Balasubramanian R, Yao Y, Clish CB, Shadyab AH, Liu B, et al. Associations of depression status with plasma levels of candidate lipid and amino acid metabolites: a meta-analysis of individual data from three independent samples of US postmenopausal women. Mol Psychiatry. 2021;26:3315–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X, Liu L, Lan X, Cohen D, Zhang Y, Ravindran AV, et al. Polyunsaturated fatty acids metabolism, purine metabolism and inosine as potential independent diagnostic biomarkers for major depressive disorder in children and adolescents. Mol Psychiatry. 2019;24:1478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Z, Zhao P, Long Z, Li J, Yang G, Zhang Q, et al. Biomarker screening for antenatal depression in women who underwent caesarean section: a matched observational study with plasma lipidomics. BMC Psychiatry. 2019;19:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira TG, Chan RB, Bravo FV, Miranda A, Silva RR, Zhou B, et al. The impact of chronic stress on the rat brain lipidome. Mol Psychiatry. 2016;21:80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen S, Wei C, Gao P, Kong H, Jia Z, Hu C, et al. Effect of Allium macrostemon on a rat model of depression studied by using plasma lipid and acylcarnitine profiles from liquid chromatography/mass spectrometry. J Pharm Biomed Anal. 2014;89:122–9. [DOI] [PubMed] [Google Scholar]
- 31.Mocking RJT, Naviaux JC, Li K, Wang L, Monk JM, Bright AT, et al. Metabolic features of recurrent major depressive disorder in remission, and the risk of future recurrence. Transl Psychiatry. 2021;11:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee ET, Welty TK, Fabsitz R, Cowan LD, Le NA, Oopik AJ, et al. The Strong Heart Study. A study of cardiovascular disease in American Indians: design and methods. Am J Epidemiol. 1990;132:1141–55. [DOI] [PubMed] [Google Scholar]
- 33.Howard BV, Welty TK, Fabsitz RR, Cowan LD, Oopik AJ, Le NA, et al. Risk factors for coronary heart disease in diabetic and nondiabetic Native Americans. The Strong Heart Study. Diabetes. 1992;41:4–11. [DOI] [PubMed] [Google Scholar]
- 34.North KE, Howard BV, Welty TK, Best LG, Lee ET, Yeh JL, et al. Genetic and environmental contributions to cardiovascular disease risk in American Indians: the strong heart family study. Am J Epidemiol. 2003;157:303–14. [DOI] [PubMed] [Google Scholar]
- 35.Welty TK, Lee ET, Yeh J, Cowan LD, Go O, Fabsitz RR, et al. Cardiovascular disease risk factors among American Indians. The strong heart study. Am J Epidemiol. 1995;142:269–87. [DOI] [PubMed] [Google Scholar]
- 36.Howard BV, Lee ET, Yeh JL, Go O, Fabsitz RR, Devereux RB, et al. Hypertension in adult American Indians. The strong heart study. Hypertension. 1996;28:256–64. [DOI] [PubMed] [Google Scholar]
- 37.Lee ET, Cowan LD, Welty TK, Sievers M, Howard WJ, Oopik A, et al. All-cause mortality and cardiovascular disease mortality in three American Indian populations, aged 45–74 years, 1984–8. The strong heart study. Am J Epidemiol. 1998;147:995–1008. [DOI] [PubMed] [Google Scholar]
- 38.Howard BV, Lee ET, Cowan LD, Devereux RB, Galloway JM, Go OT, et al. Rising tide of cardiovascular disease in American Indians. The strong heart study. Circulation. 1999;99:2389–95. [DOI] [PubMed] [Google Scholar]
- 39.Levey AS, Coresh J, Balk E, Kausz AT, Levin A, Steffes MW, et al. National Kidney Foundation practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Ann Intern Med. 2003;139:137–47. [DOI] [PubMed] [Google Scholar]
- 40.Lucove J, Vupputuri S, Heiss G, North K, Russell M. Metabolic syndrome and the development of CKD in American Indians: the strong heart study. Am J Kidney Dis. 2008;51:21–28. [DOI] [PubMed] [Google Scholar]
- 41.Macy EM, Hayes TE, Tracy RP. Variability in the measurement of C-reactive protein in healthy subjects: implications for reference intervals and epidemiological applications. Clin Chem. 1997;43:52–58. [PubMed] [Google Scholar]
- 42.Best LG, Zhang Y, Lee ET, Yeh JL, Cowan L, Palmieri V, et al. C-reactive protein as a predictor of cardiovascular risk in a population with a high prevalence of diabetes: the strong heart study. Circulation. 2005;112:1289–95. [DOI] [PubMed] [Google Scholar]
- 43.National Heart, Lung and Blood Institute. The strong heart study: cardiovascular disease in American Indians. Phase V operations manual. Strong Heart Study Coordinating Center; 2006. https://strongheartstudy.org/Research/Research-Overview/Phase-V. [Google Scholar]
- 44.Clauss A Rapid physiological coagulation method in determination of fibrinogen. Acta Haematol. 1957;17:237–46. [DOI] [PubMed] [Google Scholar]
- 45.Radloff LS. The CES-D Scale: a self-report depression scale for research in the general population. Appl Psychological Meas. 1977;1:385–401. [Google Scholar]
- 46.Schroevers MJ, Sanderman R, van Sonderen E, Ranchor AV. The evaluation of the Center for Epidemiologic Studies Depression (CES-D) scale: depressed and positive affect in cancer patients and healthy reference subjects. Qual Life Res. 2000;9:1015–29. [DOI] [PubMed] [Google Scholar]
- 47.Schafer JL, Olsen MK. Multiple imputation for multivariate missing-data problems: a data analyst’s perspective. Multivar Behav Res. 1998;33:545–71. [DOI] [PubMed] [Google Scholar]
- 48.Pedersen AB, Mikkelsen EM, Cronin-Fenton D, Kristensen NR, Pham TM, Pedersen L, et al. Missing data and multiple imputation in clinical epidemiological research. Clin Epidemiol. 2017;9:157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Buuren S, Groothuis-Oudshoorn K. MICE: Multivariate Imputation by Chained Equations in R. J Stat Softw. 2011;45:1–67. [Google Scholar]
- 50.Miao G, Zhang Y, Huo Z, Zeng W, Zhu J, Umans JG, et al. Longitudinal plasma lipidome and risk of type 2 diabetes in a large sample of american indians with normal fasting glucose: the strong heart family study. Diabetes Care. 2021;44:2664–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cajka T, Smilowitz JT, Fiehn O. Validating quantitative untargeted lipidomics across nine liquid chromatography-high-resolution mass spectrometry platforms. Anal Chem. 2017;89:12360–8. [DOI] [PubMed] [Google Scholar]
- 52.Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods. 2015;12:523–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan S, Kind T, Cajka T, Hazen SL, Tang WHW, Kaddurah-Daouk R, et al. Systematic error removal using random forest for normalizing large-scale untargeted lipidomics data. Anal Chem. 2019;91:3590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hartigan JA, Hartigan PM.The dip test of unimodality. Ann Statist.1985;13:70–84. [Google Scholar]
- 55.Storey JD. A direct approach to false discovery rates. J R Stat Soc Ser B (Stat Methodol). 2002;64:479–98. [Google Scholar]
- 56.Storey John D, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci. 2003;100:9440–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pencina MJ, D’Agostino RB, Vasan RS. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med. 2008;27:157–72. [DOI] [PubMed] [Google Scholar]
- 58.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinforma. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput Biol. 2008;4:e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput Biol. 2011;7:e1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peschel S, Müller CL, von Mutius E, Boulesteix AL, Depner M. NetCoMi: network construction and comparison for microbiome data in R. Brief Bioinform. 2021;22:bbaa290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Molenaar MR, Jeucken A, Wassenaar TA, van de Lest CHA, Brouwers JF, Helms JB. LION/web: a web-based ontology enrichment tool for lipidomic data analysis. Gigascience. 2019;8:giz061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49:W388–W396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneider PB, Kennedy EP. Sphingomyelinase in normal human spleens and in spleens from subjects with Niemann-Pick disease. J Lipid Res. 1967;8:202–9. [PubMed] [Google Scholar]
- 66.Kornhuber J, Medlin A, Bleich S, Jendrossek V, Henkel AW, Wiltfang J, et al. High activity of acid sphingomyelinase in major depression. J Neural Transm (Vienna). 2005;112:1583–90. [DOI] [PubMed] [Google Scholar]
- 67.Zoicas I, Schumacher F, Kleuser B, Reichel M, Gulbins E, Fejtova A, et al. The forebrain-specific overexpression of acid sphingomyelinase induces depressive-like symptoms in mice. Cells. 2020;9:1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beckmann N, Sharma D, Gulbins E, Becker KA, Edelmann B. Inhibition of acid sphingomyelinase by tricyclic antidepressants and analogons. Front Physiol. 2014;5:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kornhuber J, Gulbins E. New molecular targets for antidepressant drugs. Pharm (Basel). 2021;14:894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Faria R, Santana MM, Aveleira CA, Simões C, Maciel E, Melo T, et al. Alterations in phospholipidomic profile in the brain of mouse model of depression induced by chronic unpredictable stress. Neuroscience. 2014;273:1–11. [DOI] [PubMed] [Google Scholar]
- 71.Lee LH, Tan CH, Shui G, Wenk MR, Ong WY. Role of prefrontal cortical calcium independent phospholipase A2 in antidepressant-like effect of maprotiline. Int J Neuropsychopharmacol. 2012;15:1087–98. [DOI] [PubMed] [Google Scholar]
- 72.Lee LH, Shui G, Farooqui AA, Wenk MR, Tan CH, Ong WY. Lipidomic analyses of the mouse brain after antidepressant treatment: evidence for endogenous release of long-chain fatty acids? Int J Neuropsychopharmacol. 2009;12:953–64. [DOI] [PubMed] [Google Scholar]
- 73.Xue SS, Zhou CH, Xue F, Liu L, Cai YH, Luo JF, et al. The impact of repetitive transcranial magnetic stimulation and fluoxetine on the brain lipidome in a rat model of chronic unpredictable stress. Prog Neuropsychopharmacol Biol Psychiatry. 2020;102:109946. [DOI] [PubMed] [Google Scholar]
- 74.Hamilton PJ, Chen EY, Tolstikov V, Peña CJ, Picone JA, Shah P, et al. Chronic stress and antidepressant treatment alter purine metabolism and beta oxidation within mouse brain and serum. Sci Rep. 2020;10:18134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Müller CP, Reichel M, Mühle C, Rhein C, Gulbins E, Kornhuber J. Brain membrane lipids in major depression and anxiety disorders. Biochim Biophys Acta. 2015;1851:1052–65. [DOI] [PubMed] [Google Scholar]
- 76.Dean JM, Lodhi IJ. Structural and functional roles of ether lipids. Protein Cell. 2018;9:196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu Y, Wang C, Klabnik JJ, O’Donnell JM. Novel therapeutic targets in depression and anxiety: antioxidants as a candidate treatment. Curr Neuropharmacol. 2014;12:108–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu T, Zhong S, Liao X, Chen J, He T, Lai S, et al. A meta-analysis of oxidative stress markers in depression. PLoS One. 2015;10:e0138904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Assies J, Mocking RJ, Lok A, Ruhé HG, Pouwer F, Schene AH. Effects of oxidative stress on fatty acid- and one-carbon-metabolism in psychiatric and cardiovascular disease comorbidity. Acta Psychiatr Scand. 2014;130:163–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.MahmoudianDehkordi S, Ahmed AT, Bhattacharyya S, Han X, Baillie RA, Arnold M, et al. Alterations in acylcarnitines, amines, and lipids inform about the mechanism of action of citalopram/escitalopram in major depression. Transl Psychiatry. 2021;11:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lewinsohn PM, Seeley JR, Roberts RE, Allen NB. Center for Epidemiologic Studies Depression Scale (CES-D) as a screening instrument for depression among community-residing older adults. Psychol Aging. 1997;12:277–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The phenotype data used in this study can be requested through the Strong Heart Study (https://strongheartstudy.org/). The lipidomic data can be obtained from the corresponding author upon a reasonable request.


