Abstract
Acinetobacter PobR and PcaU are transcriptional activators that closely resemble each other in primary structure, DNA-binding sites, metabolic modulators, and physiological function. PobR responds to the inducer-metabolite p-hydroxybenzoate and activates transcription of pobA, the structural gene for the enzyme that converts p-hydroxybenzoate to protocatechuate. This compound, differing from p-hydroxybenzoate only in that it contains an additional oxygen atom, binds to PcaU and thereby specifically activates transcription of the full set of genes for protocatechuate catabolism. Particular experimental attention has been paid to PobR and PcaU from Acinetobacter strain ADP1, which exhibits exceptional competence for natural transformation. This trait allowed selection of mutant strains in which pobR function had been impaired by nucleotide substitutions introduced by PCR replication errors. Contrary to expectation, the spectrum of amino acids whose substitution led to loss of function in PobR shows no marked similarity to the spectrum of amino acids conserved by the demand for continued function during evolutionary divergence of PobR, PcaU, and related proteins. Surface plasmon resonance was used to determine the ability of mutant PobR proteins to bind to DNA in the pobA-pobR intergenic region. Deleterious mutations that strongly affect DNA binding all cluster in and around the PobR region that contains a helix-turn-helix motif, whereas mutations causing defects in the central portion of the PobR primary sequence do not seem to have a significant effect on operator binding. PCR-generated mutations allowing PobR to mimic PcaU function invariably caused a T57A amino acid substitution, making the helix-turn-helix sequence of PobR more like that of PcaU. The mutant PobR depended on p-hydroxybenzoate for its activity, but this dependence could be relieved by any of six amino acid substitutions in the center of the PobR primary sequence. Independent mutations allowing PcaU to mimic PobR activity were shown to be G222V amino acid substitutions in the C terminus of the 274-residue protein. Together, the analyses suggest that PobR and PcaU possess a linear domain structure similar to that of LysR transcriptional activators which largely differ in primary structure.
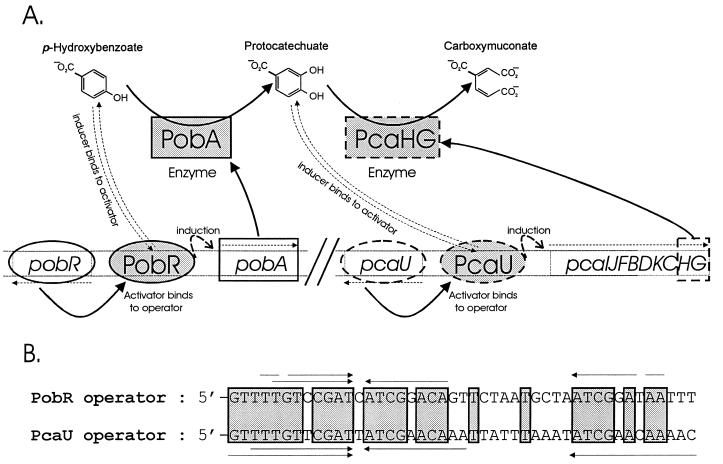
An early documentation of the physiological plasticity of bacteria was demonstration of specific patterns of induction in response to the chemically similar growth substrates p-hydroxybenzoate and protocatechuate (44). More recent investigations have elucidated molecular mechanisms underlying the specific inducible response of Acinetobacter to these compounds. As summarized in Fig. 1A, the transcriptional activator PobR elicits transcription of pobA in response to p-hydroxybenzoate (8). Action of PobA upon this compound produces protocatechuate, which triggers the action of PcaU, the transcriptional activator of the complete set of genes required for catabolism of protocatechuate (17). In light of the tight specificity of their controls, it is remarkable that PobR and PcaU are similar in many respects. The amino acid sequences of the proteins are 54% identical (17), and as shown in Fig. 1B, the operators to which the proteins bind are markedly similar (17).
FIG. 1.
Similar mechanisms govern transcriptional activation of p-hydroxybenzoate and protocatechuate catabolism in Acinetobacter. (A) Rectangles represent enzymes, and ovals represent transcriptional activators. Proteins are shaded, and genes are not. Genes and proteins associated with PobR are circumscribed by solid lines, and those associated with PcaU are surrounded by dashed lines. The curved arrows at the top represent the enzymatic conversions of p-hydroxybenzoate to protocatechuate and of protocatechuate to carboxymuconate. Carboxymuconate is a toxic metabolite, and strains blocked in its metabolism can be used to select secondary mutations blocking catabolism of either p-hydroxybenzoate (8, 15, 23) or protocatechuate (16). As indicated by the curved dashed arrows, p-hydroxybenzoate and protocatechuate act upon the respective activators PobR and PcaU to exert specific control over gene transcription (9, 17). PobA, the enzyme that acts upon p-hydroxybenzoate, is induced in response to interaction of the compound to PobR bound to an operator upstream from pobA (9). By an analogous mechanism, protocatechuate triggers expression of genes encoding its catabolism by interaction with PcaU bound to an operator upstream from the pcaIJFBDKCHG operon, which encodes genes for protocatechuate catabolism (17). Dashed horizontal arrows indicate directions of transcription. Curved solid arrows pointing from gene to protein indicate formation of the translated gene product. (B) Similar nucleotide sequences in the PobR and PcaU operators (17). Horizontal arrows indicate inverted repetitions in nucleotide sequences, and shaded boxes mark nucleotide residues that are identical in the aligned operator sequences.
Investigation of pobR and pcaU has been facilitated by the remarkable competence of Acinetobacter strain ADP1 for natural transformation (22), and this trait facilitated isolation of 89 mutants in which pobR function had been impaired by nucleotide substitutions caused by errors during PCR amplification of the gene (23). The mutations were distributed widely throughout pobR, and the mutant strains exhibited a range of phenotypes: some were null, some were leaky, some were heat sensitive, and others were cold sensitive. In this report, we describe how some of these mutations influence the ability of PobR to bind to its DNA target, the PobR operator. As might be expected, these mutations are clustered in and around nucleotide sequences encoding an apparent helix-turn-helix region, a DNA-binding motif found in many regulatory proteins.
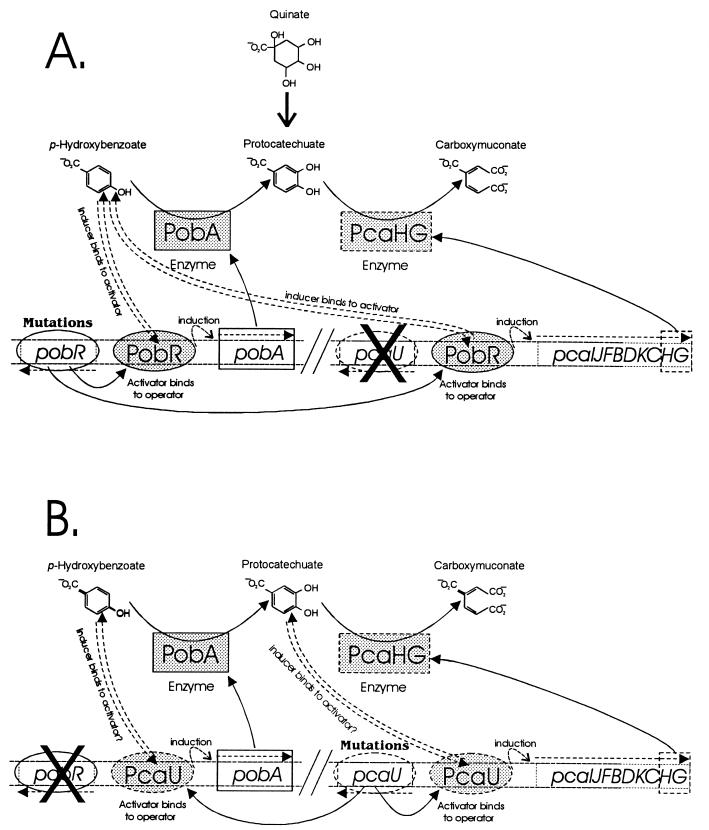
An alternative approach to understanding regulatory proteins is investigation of amino acid substitutions that allow gain rather than loss of function. This was made possible by selection of mutant strains in which PCR mutagenesis of pobR produced a protein altered so that it could activate pca gene expression. As indicated in Fig. 2A, proteins emerging from such a selection required p-hydroxybenzoate for their activity. This requirement was relieved by a limited subset of PCR-generated mutations within the central region of pobR. It also proved possible to select strains in which PCR-generated nucleotide substitutions had modified pcaU so that it formed a protein that mimicked PobR function (Fig. 2B). As described here, the selection invariably produced strains containing a single amino acid substitution in the C-terminal region of PcaU, remote from the helix-turn-helix motif segment that would be predicted to be most directly associated with DNA binding.
FIG. 2.
Mutations altering the specificity of transcriptional activation by PobR and PcaU. (A) A knockout mutation in pcaU impairs expression of the pca operon including PcaHG. PCR mutagenesis can alter PobR so that it assumes the function of PcaU. The altered PobR still requires p-hydroxybenzoate for activity because this compound is required for growth of the cells with either quinate or protocatechuate. Secondary mutations relieve PobR of the demand for p-hydroxybenzoate and allow the activator to elicit pca gene expression. (B) PCR mutagenesis of pcaU creates mutations allowing PcaU to replace the function of an inactivated PobR.
MATERIALS AND METHODS
Strain and culture conditions.
Mineral medium (23) supplemented with 10 mM succinate was routinely used for growth of Acinetobacter strains in tubes on a gyratory shaker, or on plates (solidified with 1.8% [wt/vol] agar), at 37 or 22°C. Mutations causing defects in the naturally transformable Acinetobacter strain ADP1 were prepared and sequenced in an earlier investigation. Where indicated, p-hydroxybenzoate (5 mM), quinate (5 mM), or protocatechuate (3 mM) was used as the carbon and energy source. Luria-Bertani (LB) broth was used for growth of Escherichia coli strains. Ampicillin was added to a concentration of 80 μg/ml for selection of resistance in E. coli; kanamycin was used at a concentration of 50 μg/ml for both E. coli and Acinetobacter.
Recombinant DNA techniques.
All recombinant DNA techniques were performed as described previously (24, 25) and according to Sambrook et al. (40). Isolation of bacterial chromosomal DNA, to be used as template DNA in PCRs with Taq polymerase, was done with Instagene DNA purification matrix (Bio-Rad) as recommended by the supplier. A scaled-down version of the protocol described before (16) was used for isolation of chromosomal template DNA to be used with Pfu polymerase. Restriction enzymes were obtained from New England Biolabs, Inc. PCR primers were custom synthesized (Keck Biotechnology Resource Laboratory, Yale University).
Taq polymerase (Boehringer Mannheim) and Pfu polymerase (Stratagene) were used as indicated by the suppliers for amplification of DNA fragments. Standard PCRs were carried out with 200 nM each primer, 200 μM each deoxynucleoside triphosphate (dNTP), 50 to 100 ng of chromosomal template DNA, and 0.5 U of polymerase in a final volume of 50 μl. The standard thermocycle protocol consisted of a total of 30 cycles, with a denaturation step at 94°C, primer annealing at 58°C (all primers), and elongation at 72°C. For generation of template DNA for sequencing, unincorporated primers and dNTPs were removed from PCR mixtures by using GeneClean Glassmilk as described by the supplier (Bio 101, Inc.).
Transformation-facilitated mutagenesis.
PCR for transformation-facilitated mutagenesis was performed as described above except that the number of cycles was increased to 35. Mutagenesis of pobR was performed as in a previous study (23) with primers pob1 (5′-GCAGTTGACCGAGTAGTAATCCCG-3′) and pob2 (5′-GAAAACTGTCCACTCCGATTCC-3′), which generated a 1,434-bp product. For mutagenesis of pcaU, primers pcaU1 (5′GATAACTCCAATGTGCATCTAGC-3′) and pcaU4 (5′-GATGAATCAGATCGATATGGCAA-3′) were used to generate a 1,460-bp product. Strains of Acinetobacter were transformed with PCR DNA as described before (23) or with slight modification. Routinely, 10 μl of an unpurified PCR product was added to 500 μl of an early-exponential-phase culture. After growth for an additional 3 to 16 h, the transformation mixture was plated directly onto selective medium.
Sequence analysis of mutations.
Sequence analysis was performed as described before (23), with Taq-amplified chromosomal DNA as the template in cycle sequence reactions, using an ABI PRISM dye terminator cycle sequencing kit with Amplitaq DNA polymerase (-FS) as recommended by the supplier (Perkin-Elmer). DNA fragments were denatured at 95°C for 2 min prior to electrophoresis on a denaturing 6% polyacrylamide gel in an ABI 373 automated sequencer (Perkin-Elmer ABI), linked to an Apple PowerMac, equipped with appropriate sequencing software (Perkin-Elmer ABI). Sequences were analyzed using the program DNASTAR (Lasergene).
Overproduction of wild-type and mutant PobR in E. coli.
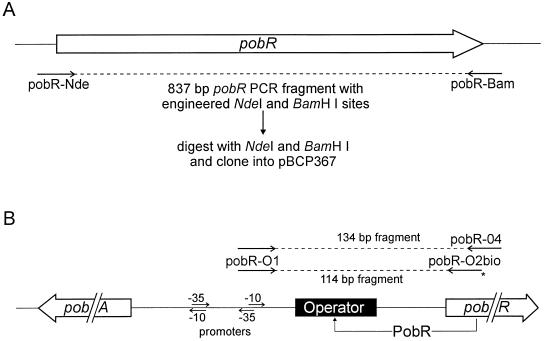
Expression vector pBCP367 (47) was used for generation of PobR overexpression plasmids. This vector contains an NdeI site (CATATG) that allows cloning of a gene with its own ATG translation initiation codon inserted directly downstream of both the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible trc promoter and a plasmid-derived ribosome-binding site. The availability of a unique BamHI site downstream of NdeI permitted forced cloning of PCR-amplified pobR alleles for overproduction of PobR in E. coli (Fig. 3A). Wild-type and mutant alleles of Acinetobacter pobR were amplified by PCR with the high-fidelity polymerase Pfu (Stratagene), using chromosomal DNA as the template and with primers pobR-Nde and pobR-Bam (Fig. 3A). Primer pobR-Nde (5′-GGATTTGAATCATATGGAACAACATCACCAATAC-3′) overlaps with the first seven codons of the 816-bp pobR open reading frame (in italics) and carries an engineered (two nucleotide substitutions; doubly underlined) NdeI restriction site (underlined). The second primer, pobR-Bam (5′-TCTAGGATCCAAATTATACCAAATTACGCAG-3′), carries an engineered (two nucleotide substitutions) BamHI recognition site and anneals at the end of pobR (in italics). The 837-bp fragment formed after PCR amplification was separated from Pfu polymerase, unincorporated primers, and dNTPs and digested with NdeI and BamHI; the resulting fragments were gel purified and ligated into NdeI/BamHI-digested pBCP367. Transformants of E. coli DH5α (19) were selected for vector-encoded ampicillin resistance, and the nucleotide sequence of each cloned pobR gene was verified. Plasmid pZR85 contains the cloned wild-type pobR, and plasmids pZR110 through pZR130 carry mutant pobR alleles.
FIG. 3.
(A) Construction of PobR overproduction plasmids. (B) Generation of PCR fragments, including the PobR operator in the pobA-pobR intergenic region, for use as DNA-binding probes in BIAcore experiments. The biotin label coupled at the 5′ end to primer pobR-O2bio is depicted as an asterisk.
For overproduction of wild-type or mutant PobR in E. coli, fresh overnight cultures of E. coli DH5α with appropriate pobR expression plasmids were diluted 25-fold into fresh LB medium (25 ml in a 200-ml Erlenmeyer flask), and the cultures were grown for about 2.5 h at 37°C on a gyratory shaker until they had reached an optical density at 540 nm of 1.5 (±0.1). IPTG was added to a final concentration of 200 μg/ml and the cultures were further incubated for 1 h. After cooling on ice, the cells were pelleted by centrifugation; after being washed once in ice-cold HEPES-buffered saline (HBS) (10 mM HEPES [pH 7.5], 3.4 mM EDTA, 150 mM NaCl, 0.05% [vol/vol] Tween 20), the cells were resuspended in 2 ml of ice-cold HBS and frozen at −70°C overnight. Thawed cells were sonicated on ice with a Braun Sonifier with a microtip at maximum output (four bursts of 30 s), and the sonicated cell suspensions were centrifuged at 15,000 × g for 20 min. Cleared supernatant was collected as cell extract and stored at −70°C until further use. Protein concentration in cell extracts was determined according to Bradford (3). For analysis of PobR overexpression, cell extracts (normalized with respect to total protein) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% acrylamide gel, and proteins were visualized by staining with Coomassie brilliant blue as described by Sambrook et al. (40). Based on relative intensity of staining, the concentration of PobR in each sample was estimated to be 20 to 30% of the total protein content of the cell extract.
Surface plasmon resonance detection of PobR operator binding.
Wild-type Acinetobacter PobR binds to the intergenic region between pobR and the divergently transcribed pobA in a 35-bp region containing inverted sequence repetitions (9). This region was independently amplified by PCR, using Taq polymerase and chromosomal DNA of Acinetobacter as the template, with two sets of primers. (i) A 114-bp DNA fragment was generated with a standard primer annealing in the pobA-pobR intergenic region (pobR-O1 [5′-AAAGACATCTATAATCAAGGCTC-3′]) and a 5′-biotinylated primer that anneals near the start of pobR (pobR-O2bio [5′-biotin-GCAAGGTATTGGTGATGTTGTTC-3′]) (Fig. 3B). The resulting biotinylated PCR fragment was used for immobilization of the operator onto a sensor chip (see below), to be used as the probe in determining DNA-binding characteristics of PobR proteins. (ii) A second operator fragment was generated via PCR amplification with primers pobR-O1 and pobR-04 (5′-CTTCACTTGAATGGGGATGTGC-3′). The resulting 134-bp fragment (Fig. 3B) is not labeled with biotin and was used as free operator DNA in competition experiments to determine the specificity of the association signal. The PCR fragments were purified on a 12% polyacrylamide gel, eluted with a buffer containing 10 mM Tris HCl (pH 7.5), 0.5 mM EDTA, and 200 mM NaCl, after precipitated, and then taken up in 10 mM Tris HCl (pH 8.0)–1 mM EDTA–300 mM NaCl. The concentration of the PCR-amplified operator DNA fragments in these solutions was estimated by relative UV-induced ethidium bromide fluorescence of samples run on a 12% acrylamide gel. A BIAcore (Pharmacia Biosensor) apparatus was used as recommended by the supplier for semiquantitative surface plasmon resonance detection of binding of wild-type PobR and mutant PobR proteins in cell extracts of E. coli (see above for overproduction of PobR in E. coli) to the 114-bp operator fragment immobilized on a dextran-streptavidin-coated sensor chip (sensor chip SA; Pharmacia Biosensor). Routinely, between 40 and 80 ng of biotinylated operator DNA (1 to 2 pmol) was injected (in HBS with 0.5 M NaCl) at a flow rate of 5 μl/min (for 9 min) into the flow cell of a sensor chip for binding of the operator DNA to the chip. This resulted in a net increase in relative response units of between 1,100 and 1,700.
After immobilization of DNA on the chip, cell extracts of E. coli strains overexpressing wild-type or mutant PobR were injected to determine binding of the PobR proteins. Routinely, the E. coli cell extract that was injected contained 1.0 μg of total protein (corresponding to an estimated 2 pmol of PobR protein) in HBS. To reduce nonspecific effects of protein binding, excess (4 μg) poly(dI-dC) · poly(dI-dC) (Sigma) was added as a nonspecific competitor DNA. All cell extract samples were passed over the chip with immobilized operator DNA at a flow rate of 5 μl/min for a period of 9 min (allowing PobR-operator association), followed by a flow of HBS for 10 to 15 min to monitor dissociation of PobR-operator complexes. Prior to injection of a new sample, undissociated protein-operator complexes were dissolved by repeated injections of 0.05% SDS for 2 min. All BIAcore analyses were carried out at 22°C. BIAcore-generated data were collected at 0.5-s intervals and a response-versus-time curve (sensorgram) was generated with the BIAlogue software packet (Pharmacia Biosensor) installed on a personal computer linked to the BIAcore.
RESULTS
Distribution of PCR-generated mutations in Acinetobacter pobR.
In a previous study (23), the transcriptional regulator gene pobR was used to illustrate the advantage of combining PCR mutagenesis with natural transformation to obtain Acinetobacter mutants impaired in growth with p-hydroxybenzoate as the sole carbon and energy source. In this investigation, 57 such regulatory mutations were analyzed in detail by phenotypic characterization and by sequencing the corresponding mutant pobR allele. Most of the mutations caused single amino acid substitutions in PobR; three mutations altered the length of the PobR C terminus (Table 1). The mutations impeded growth with p-hydroxybenzoate in one of four ways: 23 of the mutants exhibited a null phenotype, 9 were heat sensitive, 16 were cold sensitive, and 9 were leaky (Table 1).
TABLE 1.
Mutations causing phenotypically detectable pobR defectsa
| Mutant strain | Genotype | Substitution
|
Phenotype (growth with p-hydroxybenzoate) | |
|---|---|---|---|---|
| Nucleotide | Amino Acid | |||
| ADP1410 | pobR1410 | T770G | J257Stop | Null |
| ADP1411 | pobR1411 | A65C | Y22S | Null |
| ADP1412 | pobR1412b | A168T | R56S | Leaky |
| ADP1413 | pobR1413 | T634C | S212P | Null |
| ADP1114 | pobR1414 | T242A | L81H | Null |
| ADP1415 | pobR1415b | A294T | L98F | Cold sensitive |
| ADP1416 | pobR1416b | C476G | T159S | Cold sensitive |
| ADP1417 | pobR1417b | T359A | V120E | Null |
| ADP1418 | pobR1418 | T499C | S167P | Heat sensitive |
| ADP1419 | pobR1419b | A200T | K67I | Cold sensitive |
| ADP1420 | pobR1420b | T456A | N152K | Null |
| ADP1421 | pobR1421 | C305A | A102E | Null |
| ADP1422 | pobR1422b | C457T | R153C | Cold sensitive |
| ADP1423 | pobR1423 | T383A | V128E | Null |
| ADP1424 | pobR1424b | T722A | M241K | Null |
| ADP1425 | pobR1425b | T238C | W80R | Cold sensitive |
| ADP1426 | pobR1426 | T800A | L267Q | Heat sensitive |
| ADP1427 | pobR1427 | T204A | F68L | Cold sensitive |
| ADP1428 | pobR1428 | C677A | P226H | Cold sensitive |
| ADP1429 | pobR1429b | A191T | K64M | Cold sensitive |
| ADP1430 | pobR1430 | A163C | S55R | Heat sensitive |
| ADP1431 | pobR1431 | T274C | Y92H | Null |
| ADP1432 | pobR1432 | T590C | F197S | Heat sensitive |
| ADP1433c | pobR1433 | A815C | Stop272S | Cold sensitive |
| ADP1434 | pobR1434 | T236C | F79S | Null |
| ADP1435 | pobR1435 | T64G | Y22D | Null |
| ADP1436 | pobR1436 | A298G | K100E | Null |
| ADP1437 | pobR1437 | A230T | H77L | Leaky |
| ADP1438 | pobR1438 | T680G | V227G | Null |
| ADP1440d | pobR1440 | ΔT699 | T234Shift | Null |
| ADP1441 | pobR1441 | T69G | I23M | Cold sensitive |
| ADP1442 | pobR1442 | G179A | R60Q | Null |
| ADP1448 | pobR1448 | T77C | L26S | Null |
| ADP1449 | pobR1449 | T108A | F36L | Null |
| ADP1450 | pobR1450 | C482T | T161I | Leaky |
| ADP1451 | pobR1451 | T659C | L220P | Null |
| ADP1452 | pobR1452e | T795A | N265K | Cold sensitive |
| ADP1453 | pobR1453 | T64A | Y22N | Null |
| ADP1454 | pobR1454 | G450T | L150F | Null |
| ADP1456 | pobR1456 | C245T | T82I | Null |
| ADP1458 | pobR1458 | A67C | I23L | Cold sensitive |
| ADP1459 | pobR1459 | A84T | K28N | Cold sensitive |
| ADP1460 | pobR1460 | A290C | H98P | Leaky |
| ADP1461 | pobR1461 | A224G | D75G | Leaky |
| ADP1462 | pobR1462 | G110T | G37V | Null |
| ADP1463 | pobR1463 | T478C | S160P | Null |
| ADP1464 | pobR1464 | G472T | A158S | Cold sensitive |
| ADP1467 | pobR1467 | T497A | L166H | Heat sensitive |
| ADP1468 | pobR1468 | T203C | F68S | Leaky |
| ADP1470 | pobR1470 | A318T | L106F | Leaky |
| ADP1471 | pobR1471 | T443G | M148R | Cold sensitive |
| ADP1472 | pobR1472 | T653A | L218Q | Cold sensitive |
| ADP1473 | pobR1473 | C250A | R84S | Heat sensitive |
| ADP1477 | pobR1477 | A470T | H157L | Heat sensitive |
| ADP1478 | pobR1478 | T209C | L69S | Heat sensitive |
| ADP1479 | pobR1479e | A793G | N265D | Leaky |
| ADP1480 | pobR1480 | C569A | T190N | Leaky |
The mutant strains were isolated as part of an earlier investigation.
The PobR protein encoded by the gene was overproduced by E. coli (Table 2; Fig. 5) and studied with respect to operator-binding characteristics (Fig. 6).
Mutation of the pobR translation termination codon in ADP1433 results in gain of two amino acids (S272 and F273).
Due to a frameshift mutation in pobR codon 234, PobR1440 lacks 38 carboxy-terminal wild-type amino acid residues but contains 48 additional carboxy-terminal amino acid residues (T234LQHLIVCHKPIGCSHSILSIKFYLYYAILRMSCVIWYNFNHRYLLSY-carboxy terminus).
The mutant alleles pobR1452 and pobR1479 carry different mutations that alter the asparagine at position 265, only 6 amino acids from the carboxy terminus of PobR.
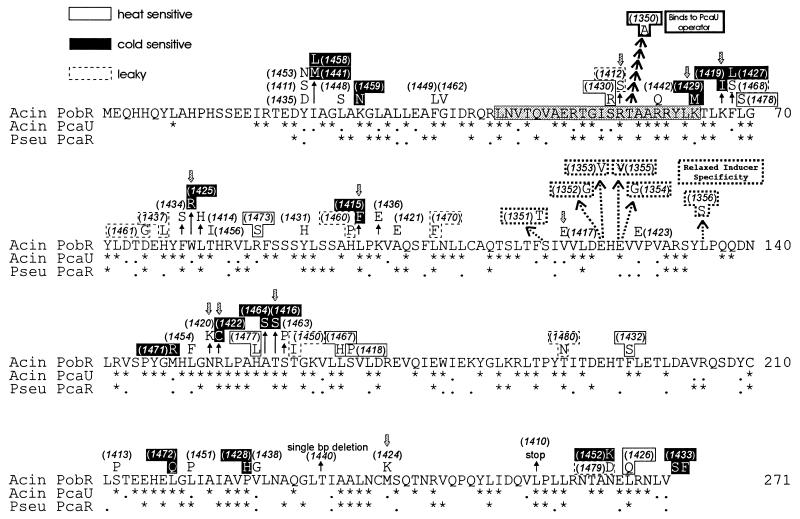
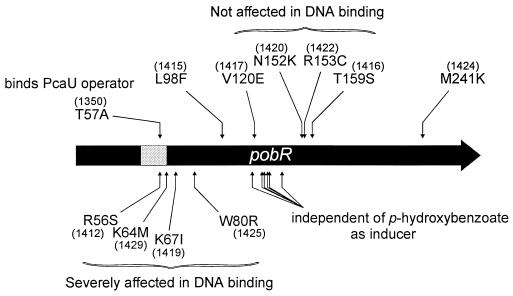
Distribution of the mutations in the PobR primary structure is shown in Fig. 4. Multiple substitutions occurred at four positions in the amino acid sequence. Different amino acid substitutions of Y22 and I23 resulted in indistinguishable phenotypes, whereas varied substitutions of F68 and N265 led to different phenotypes. Otherwise, the mutations causing varied phenotypes are widely distributed throughout the PobR primary sequence. Also shown in Fig. 4 is an amino acid alignment of PobR with two closely related homologs that regulate different steps in the β-ketoadipate pathway: Acinetobacter PcaU (17) and Pseudomonas putida PcaR (39). Amino acid residues identified genetically in this study as being important for PobR activity are not predominantly those that one would predict based on conservation within this family of regulators.
FIG. 4.
Alignment of the amino acid sequence of Acinetobacter PobR (Acin PobR) with those of Acinetobacter PcaU (Acin PcaU) and P. putida PcaR (Pseu PcaR). Only identical residues (as asterisks) and similar residues (dots) are represented. A shaded rectangle encompasses the potential DNA-binding helix-turn-helix motif (8). The effects in PobR of the nucleotide substitutions in the pobR mutants listed in Tables 1 and Table 3 are shown; the numbers in parentheses correspond to the pobR alleles (Tables 1 and 3). The phenotypes of the strains containing these mutations, with respect to growth on 4-hydroxybenzoate as the sole carbon source, are indicated as follows: null (no growth at all; not boxed), heat sensitive (little or no growth at 37°C; boxed), cold sensitive (little or no growth at 22°C; black-shaded box), and leaky (slow growth at both 22 and 37°C; boxed with a dashed line). Downward-pointing grey arrows mark the mutant PobR proteins that have been overproduced in E. coli (Table 2; Fig. 5) and tested with respect to operator binding. Six consecutive arrows represent the six independent mutations giving rise to pobR1350 (boxed with dark outline). The six different mutations leading to relaxation of inducer specificity are marked by dashed arrows, and the boxes surrounding the mutations are dashed.
Identification of PobR residues involved in DNA binding.
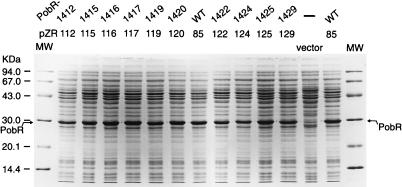
Acinetobacter PobR binds in the 134-bp pobA-pobR intergenic region to a 35-bp interval containing inverted DNA repeats (9) (Fig. 1). PobR negatively regulates its own expression and activates transcription of pobA, the structural gene for p-hydroxybenzoate hydroxylase (8, 9). In this study, PobR amino acid residues involved in DNA binding were identified by qualitative surface plasmon resonance detection in a BIAcore apparatus. Ten of the mutant pobR alleles generated by PCR mutagenesis (23) were cloned into expression vector pBCP367 (47) (Fig. 3), and the encoded PobR mutant proteins were overproduced in E. coli (Table 2). In cell extracts of these E. coli hosts, PobR appeared as a major protein band of the expected size (29 kDa) on Coomassie blue-stained SDS-polyacrylamide gels (Fig. 5), indicating its presence as soluble protein. The mutant alleles were chosen to include a range of phenotypes. The 10 mutants were also chosen so as to sample a range of locations in the pobR gene; included were two strains with mutations in the N-terminal helix-turn-helix motif and three strains with mutations in the conserved region centered on A158 (Fig. 4). Originally, an additional eight pobR alleles (pobR1410, pobR1411, pobR1413, pobR1414, pobR1421, pobR1423, pobR1427, and pobR1428 [Table 1; Fig. 4]) were cloned in pBCP367. Unexpectedly, however, cell extracts of the eight respective E. coli strains lacked detectable soluble PobR protein. Since these mutant PobR proteins were detectable in whole-cell extracts (on SDS-PAGE) and were produced in amounts comparable to the first 10 mutant proteins, their absence in cell extracts suggests that the encoded proteins form inclusion bodies in E. coli. This is striking since they all differ from the wild-type protein by just a single amino acid. Possibly, the latter eight amino acid substitutions individually lead to a destabilization of the PobR secondary structure, resulting in aggregation of the mutant proteins in the E. coli cytosol.
TABLE 2.
Binding of wild-type and mutant PobR proteins to the PobR operator as determined by surface resonance detection
| PobR overproduction plasmid | pobR allele cloned in pBCP367 | PobR mutation | Operator binding | Dissociation from operator |
|---|---|---|---|---|
| pZR85 | pobR (wild type) | None | Normal | Normal |
| pZR112 | pobR1412 | R56S | None observed | None observed |
| pZR115 | pobR1415 | L98F | Normal | Increased |
| pZR116 | pobR1416 | T159S | Normal | Normal |
| pZR117 | pobR1417 | V120E | Normal | Normal |
| pZR119 | pobR1419 | K67I | Reduced | Increased |
| pZR120 | pobR1420 | N152K | Normal | Normal |
| pZR122 | pobR1422 | R153C | Normal | Normal |
| pZR124 | pobR1424 | M241K | Normal | Decreased |
| pZR125 | pobR1425 | W80R | Reduced | None observed |
| pZR129 | pobR1429 | K64M | None observed | None observed |
FIG. 5.
Coomassie blue-stained SDS-polyacrylamide gel of cell extract of E. coli DH5α strains carrying plasmid pBCP367 (vector; control without pobR), pZR85 (overexpressing wild-type pobR), or plasmids pzR112 through -129 (overexpressing mutant pobR genes). Overproduction of PobR was induced for 1 h with IPTG (see Materials and Methods), and cell extract corresponding to 15 μg of total protein was loaded for each sample. The band corresponding to PobR is indicated. MW, molecular weight markers (Pharmacia).
The PobR operator was amplified by PCR, biotin labeled, and immobilized on the BIAcore sensor chip (Fig. 3). Cell extract containing soluble PobR protein (Fig. 5) was passed over the sensor chip, and binding to the operator was monitored directly by surface plasmon resonance detection. Cell extract derived from an E. coli strain carrying the empty vector served as the reference in these experiments. Wild-type PobR supplied in cell extract of E. coli DH5α(pZR85) bound specifically to the operator DNA immobilized on the sensor chip surface: the binding signal was reduced dramatically by addition of increasing amounts of competing nonbiotinylated operator to the cell extract (Fig. 6A). Addition of p-hydroxybenzoate, the coinducer of pobA expression, did not affect the association or dissociation characteristics of wild-type PobR and therefore was not added in the BIAcore experiments shown in Fig. 6 (up to 1 mM p-hydroxybenzoate was tested). This finding confirms the previous observation with DNA gel mobility shift and DNase I footprinting experiments (9) that p-hydroxybenzoate is not required for binding of PobR to its operator.
FIG. 6.
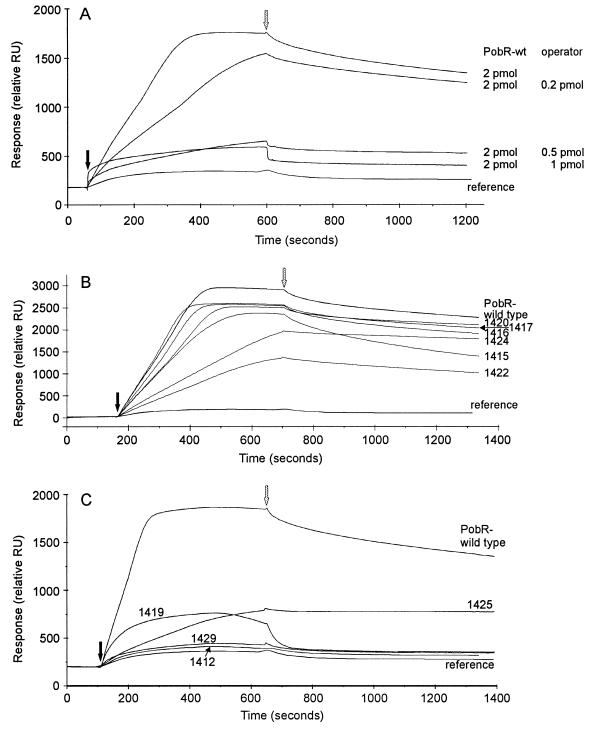
BIAcore sensorgram (relative response versus time) traces derived from the PobR-operator binding experiments. The response is given in relative response units (RU). Black arrows mark the time of injection of the cell extract into the flowcell (onset of the association phase), grey arrows indicate the time of replacement of the cell extract by HBS (onset of the dissociation phase). The reference trace shows the response observed after injection of cell extract of the E. coli strain with the vector (pBCP367) alone (no PobR protein [Fig. 5]). PobR numerals next to the traces correspond to the mutant allele designations given in Table 2. (A) Competition experiment showing how the addition of free specific competitor DNA (operator) together with wild-type PobR protein (PobR-wt) affects binding of PobR to the immobilized operator on the chip surface. The amount of injected PobR protein and free operator DNA is next to each trace. (B) Response observed after injection of cell extract of E. coli strains with overproduced PobR proteins that appear to exhibit wild-type binding properties. (C) Responses observed after injection of cell extract of E. coli strains with overproduced PobR proteins that exhibit abnormal binding properties.
As judged by the shape of the association curves shown in Fig. 6B, the operator-binding ability of some PobR mutants is indistinguishable from that of the wild type. The apparent reduction in DNA binding of PobR1422(R153C) and PobR1424(M241K) that might be inferred by their association curve slopes may be caused partly by the relatively low concentration of these mutant PobR proteins in their respective E. coli cell extracts (Fig. 5). The dissociation curves shown in Fig. 6B reveal that two mutant proteins differ from the wild type: PobR1424(M241K) dissociates less readily and PobR1415(L98F) dissociates more readily than wild-type PobR. Figure 6C depicts properties of four PobR mutant proteins that are severely affected in DNA binding. Mutants PobR1412(R56S) and PobR1429(K64M) completely fail to bind the operator. PobR1419(K67I) is also severely impaired, apparently mainly due to a strongly enhanced dissociation of the regulator-operator complex. PobR1425(W80R) seems to show strongly reduced operator association, but the protein remains bound and its dissociation from the complex is barely detectable.
Importance of the PobR C-terminal region.
Amino acid conservation between PobR and its homologs in the β-ketoadipate pathway extends almost to the last residue (Fig. 4). Two mutations that result in 15 amino acids lost (PobR1410) or 2 amino acids gained (PobR1433) at the C terminus confer, respectively, null and cold-sensitive phenotypes (Fig. 4). The frameshift mutation altering the C-terminal 38 residues (PobR1440) abolishes growth with p-hydroxybenzoate (Fig. 4; Table 1). Implying that the conserved PobR C-terminal region may (directly or indirectly) play a role in DNA binding, the null mutation in PobR1424 (M241K [Fig. 4]) produces a protein that appears to bind relatively tightly to its operator in vitro (Fig. 6B).
Importance of the PobR N-terminal helix-turn-helix region.
A potential helix-turn-helix motif consisting of amino acid residues 43 to 64 was identified near the N terminus of PobR (8) (Fig. 4). In the second helix of the PobR helix-turn-helix, there are three arginyl residues conserved with its two closest homologs, PcaU and PcaR, that appear to be essential for PobR function (Fig. 4). The PCR-generated PobR1442 (R60Q [Fig. 4]) and the previously identified (8) spontaneous mutation R61H both completely prevent growth with p-hydroxybenzoate. The PCR-generated mutation R56S, although it produces a leaky phenotype (PobR1412 [Fig. 4]), abolishes DNA binding in vitro (Fig. 6C). The helix-turn-helix motif also includes the other PobR mutation assayed in this study that eliminates DNA binding, K64M (PobR1429), and is flanked by the three remaining mutations that severely affect binding, most significantly W80R in the tightly binding PobR1425 (Fig. 4; Table 2). Immediately adjacent to the helix-turn-helix region, PobR R40 is intolerant of amino acid substitutions (23), consistent with its conservation in the PobR family (Fig. 4).
The amino acid substitution T57A in the PobR helix-turn-helix appears to broaden operator-binding specificity.
Acinetobacter PobR and PcaU are homologous transcriptional regulators that govern sequential steps in the β-ketoadipate pathway (8, 17) (Fig. 1). The two proteins are 54% identical at the amino acid level (Fig. 4), recognize similar effector molecules (Fig. 1), and bind to operator sites containing nearly identical inverted DNA repeats (9, 17) (Fig. 1). A potential helix-turn-helix motif can be identified near the N terminus in both proteins (9, 17). Despite these similarities, the functions of pobR and pcaU do not appear to overlap. Knockout mutations in one gene are not complemented by the wild-type copy of the other gene. The similarity of PobR and PcaU tempted us to use PCR mutagenesis to isolate gain-of-function mutations that would confer on one regulatory protein some of the activity of its homolog (Fig. 2). We anticipated that such mutations could highlight the specificity determinants in each protein.
Selection of gain-of-function pobR mutations required a starting strain in which inactivation of pcaU prevented growth with protocatechuate. Since pcaU is not absolutely required for growth with protocatechuate (17), strain ADP1349 was constructed by transforming the pcaU1::ΩSmrSpcr insertion from ADP92 (17) into strain ADP6338 (16) carrying the leaky ΔpcaH7 mutation which produces a four-amino-acid deletion in the β-subunit of protocatechuate 3,4-dioxygenase. The combination in ADP1349 of mutations in both the regulatory gene and in one of the structural genes in the pathway for protocatechuate degradation completely blocks growth with protocatechuate either when provided directly or when produced intracellularly from either p-hydroxybenzoate or quinate (Fig. 2).
To select mutants in which PobR had gained PcaU activity, the pobR gene was amplified by PCR with Taq polymerase, and the PCR DNA was directly used to transform ADP1349. Transformation reactions were plated onto mineral agar medium containing p-hydroxybenzoate as the sole carbon and energy source. Selection for growth with p-hydroxybenzoate was consistently successful, and the pobR gene was sequenced from six transformants, each independently derived from different PCRs. All six strains contained the same mutation (pobR1350 [Table 3; Fig. 4]) causing a T57A amino acid substitution in the PobR helix-turn-helix motif likely to be involved in DNA binding. Because the selection demanded growth with p-hydroxybenzoate, the mutant PobR was constrained to maintain its wild-type function (Fig. 2), and this may in part explain why only one specific amino acid substitution was recovered. The T57A substitution produces a stretch of six contiguous amino acids identical between the PobR and PcaU proteins (Fig. 4), including helix-turn-helix positions known to be directly involved in DNA sequence recognition (30). Therefore, T57 in PobR is likely to be a key contributor to DNA sequence-specific binding, possibly by a direct interaction with the one base pair that differs between the half-sites of the inverted DNA repeats common to the PobR and PcaU operators (17) (Fig. 1). Consistent with the PobR1350 mutant being specifically a gain of function in DNA binding, strain ADP1350(pobR1350) containing this mutation cannot grow with either quinate or protocatechuate unless p-hydroxybenzoate is added at effector concentrations (100 μM).
TABLE 3.
Mutations in pobR that suppress inactivation of PcaU
| Strain | Primary mutation | Suppressor mutation(s) | pobR mutation (no. of independent isolates) | Mutation(s) in PobR | Growth on protocatechuate | p-hydroxybenzoate needed as effector |
|---|---|---|---|---|---|---|
| ADP1349 | pcaU11::Ω/ΔpcaH7 | None | None | No | No | |
| ADP1350 | pcaU1::Ω/ΔpcaH7 | pobR1350 | A169G (6) | T57A | Yes | Yes |
| ADP1351 | pcaU1::Ω/ΔpcaH7 | pobR1350, pobR1351 | A169G, T352A (1) | T57A, S118T | Yes | No |
| ADP1352 | pcaU1::Ω/ΔpcaH7 | pobR1350, pobR1352 | A169G, A371G (3) | T57A, E124G | Yes | No |
| ADP1353 | pcaU1::Ω/ΔpcaH7 | pobR1350, pobR1353 | A169G, A371T (1) | T57A, E124V | Yes | No |
| ADP1354 | pcaU1::Ω/ΔpcaH7 | pobR1350, pobR1354 | A169G, A377G (2) | T57A, E126G | Yes | No |
| ADP1355 | pcaU1::Ω/ΔpcaH7 | pobR1350, pobR1355 | A169G, A377T (1) | T57A, E126V | Yes | No |
| ADP1356 | pcaU1::Ω/ΔpcaH7 | pobR1350, pobR1356 | A169G, T404C (2) | T57A, L135S | Yes | No |
Mutations in PobR amino acid residues 118 to 135 define a domain where single substitutions relax dependence of PobR activity on p-hydroxybenzoate.
To take advantage of the selective growth conditions still available with the PobR1350 mutant, a further round of PCR mutagenesis was performed. Strain ADP1349 was again transformed, this time with PCR-amplified DNA containing the pobR1350 mutation, and recombinants were selected on plates containing quinate as the sole carbon source. Quinate, a metabolite upstream of protocatechuate in the β-ketoadipate pathway (10, 11) (Fig. 2), was used rather than protocatechuate because of the latter’s relative toxicity and instability. Recombinants which could grow with quinate or protocatechuate were readily obtained, and pobR from 10 independently generated mutant strains was sequenced.
All 10 strains maintained the original pobR1350 mutation retained the ability to grow with p-hydroxybenzoate and, in addition, had acquired a mutation causing an amino acid substitution in one of four residues clustered near the middle of the PobR primary sequence (Table 3; Fig. 4). The tight clustering and the fact that three of the substitutions were recovered more than once (Table 3) suggest that they define a specific functional domain. Unlike the previous round of mutagenesis, only in one case, E124G(PobR1352), did the mutation substitute the PobR residue with the equivalent PcaU residue. Instead, in four of the six different mutations, an acidic residue, either E124 or E126, was replaced by an uncharged residue, either G or V (Fig. 4). The opposite amino acid substitution was found in the only PCR-generated loss-of-function mutations in this region of PobR, the two null mutations V120E (PobR1417) and V128E (PobR1423 [Fig. 4]). Unlike PobR1423 which appeared to form a protein aggregate in E. coli, PobR1417 was successfully overproduced and showed wild-type binding of the PobR operator in vitro (Fig. 6B). Given this phenotype, the V120E mutation may prevent PobR1417 from responding to p-hydroxybenzoate, thereby locking the protein in an inactive conformation, the opposite effect of the gain-of-function mutations adjacent in the PobR primary sequence (Fig. 4) which confer activity that is independent of p-hydroxybenzoate. As expected with a two-step evolution of PobR during the two rounds of PCR mutagenesis, first to a broadened operator-binding specificity and second to a relaxed coinducer dependence, no recombinants were obtained by attempts to endow PobR with PcaU activity in a single step by demanding growth with quinate after a single round of mutagenesis.
A single amino acid substitution in PcaU (G222V) confers PobR activity.
To complete the symmetric strategy depicted in Fig. 2, we used transformation-facilitated PCR mutagenesis to obtain gain-of-function mutations allowing PcaU to replace PobR. The choice of starting strain ADP607 (10) for this selection was straightforward: the deletion removing the 59 C-terminal amino acids of PobR in this strain completely abolishes growth with p-hydroxybenzoate (9). Transformation of ADP607 with PCR-amplified pcaU DNA readily produced recombinants that grew with p-hydroxybenzoate. Sequencing of pcaU from six independently generated mutant strains revealed the same mutation in each case (pcaU1300 [Table 4]). This mutation caused an amino acid substitution, G222V, located not in the helix-turn-helix domain of PcaU but near the C terminus in a residue equivalent in position to PobR G219 (Fig. 4).
TABLE 4.
pcaU mutation that suppresses inactivation of PobR
| Strain | Primary mutation | Suppressor mutation | pcaU mutation (no. of independent isolates) | Mutation in PcaU | Growth on p-hydroxybenzoate |
|---|---|---|---|---|---|
| ADP607 | ΔpobR-quiA | None | None | None | No |
| ADP1300 | ΔpobR-quiA | pcaU1300 | G665T (six isolates) | G222V | Yes |
DISCUSSION
Selection in the laboratory and selection during evolutionary history.
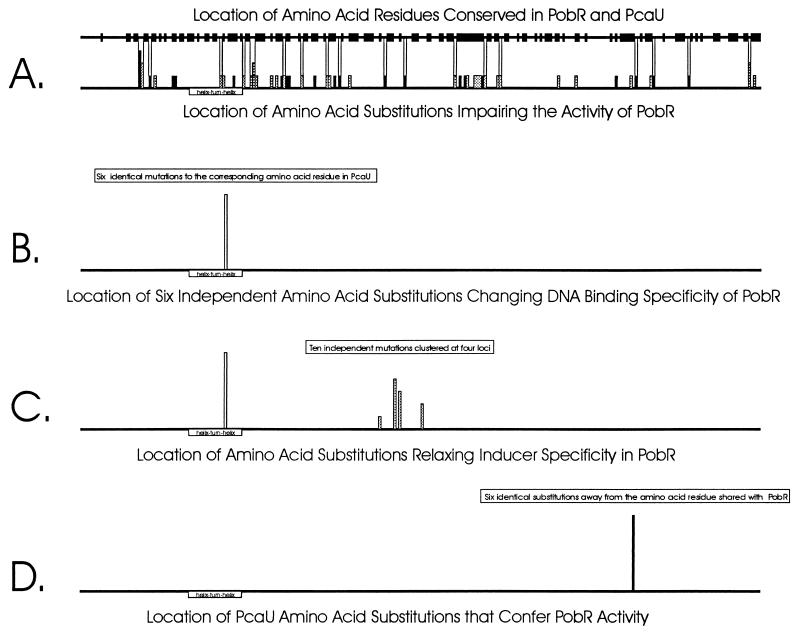
Nucleotide substitutions introduce variations in protein structure that are played upon by selection. Throughout evolutionary history, those variations that impair protein function are likely to have been lost. Therefore amino acid residues conserved within the aligned primary sequences of divergent proteins may be regarded as essential or at least valuable contributors to function. In this investigation, direct selection for loss of PobR function was imposed, and it might have been anticipated that most PCR-generated mutations would alter amino acids that had been conserved during the evolutionary divergence of PobR and PcaU. As shown in Fig. 7A, this prediction is not particularly accurate. About half (29 of 54) of the amino acid substitutions selected on the basis of loss of PobR function occurred at positions where the PobR and PcaU sequences resisted change during their evolutionary divergence. The other mutations leading to loss of function are located at positions where change, presumably associated with either maintenance or acquisition of function, was accepted during evolution of the proteins.
FIG. 7.
Locations of amino acid substitutions within the primary sequences of PobR and PcaU. The height of each rectangle indicates the frequency with which a selected mutation was observed at that position. (A) Selection for loss of function after PCR mutagenesis compared with selection for retention of function during evolution. Null mutations are indicated by dark rectangles, and mutations allowing some residual activity are indicated by shaded rectangles. Vertical lines connect the sites of PCR-generated mutations with loci that accepted change during evolutionary divergence of PobR and PcaU. (B) Selection for PcaU transcriptional activator function yielded the same mutation six times after PCR mutagenesis of pobR. (C) Demand for PcaU function in the absence of p-hydroxybenzoate produced PCR-generated mutants carrying amino acid substitutions at four different loci in the middle of the PobR primary sequence. (D) PCR mutagenesis of pcaU followed by selection for PobR function invariably yielded strains carrying the same amino acid substitution in the carboxy-terminal region of PcaU.
In contrast to the general demand for loss of function in PobR (Fig. 7A), more rigorous selection was imposed by the requirement that PobR activate pca gene transcription, and the greater rigor of the selection is reflected in the specificity of the response. As summarized in Fig. 7B, the full range of PCR-generated mutations yielded only one amino acid substitution that met the demand, T57A in the helix-turn-helix motif of PobR. Somewhat greater genetic flexibility was demonstrated in response to the demand for relaxed inducer specificity so that PobR could effectively mimic PcaU function (Fig. 7C), but severe selective constraints are reflected in the failure to recover any PobR variants that had achieved this function after a single round of PCR mutagenesis. Such success was achieved by the demand that PcaU mimic PobR function (Fig. 7D). Intriguingly, repeated selections produced a protein with the same amino acid substitution not in the helix-turn-helix portion but in the C-terminal portion of PcaU (Fig. 7D).
Linear organization of PobR functional domains.
Based on amino acid sequence alignment, PobR is a member of a small family of bacterial transcriptional regulators, all of which contain an N-terminal helix-turn-helix motif (8, 17, 21, 39, 43, 45). The distribution of mutations affecting DNA binding detected in this study emphasizes the functional importance of the helix-turn-helix region and suggests that the first 100 of the 271 amino acid residues of PobR constitute the DNA-binding domain (Fig. 4 and 8). The PobR C-terminal region, particularly in light of comparisons to members of the LysR family discussed below, may function indirectly in DNA binding by mediating PobR multerimization. Mutations that do not significantly affect DNA binding are found toward the middle of the PobR primary sequence, both in the conserved region centered on A158 (Fig. 4 and 8) and in the region where mutation can alleviate dependence of PobR activity on the coinducer p-hydroxybenzoate (Fig. 4 and 8). Further work is needed to more precisely define the coinducer response domain and to characterize the mutations that alter coinducer response, either directly, by allowing a new effector (most likely protocatechuate) to bind, or indirectly, by producing a constitutively active protein.
FIG. 8.
Properties of mutations within the linear amino acid sequence of PobR. Mutations that have been tested with respect to their effect on DNA binding are indicated along with those that convert PobR into the regulator of both pobA and the pca operon. The N-terminal helix-turn-helix motif is shaded grey. Numerals in parentheses correspond to the PobR alleles (Fig. 4). The amino acid substitutions L98F and M241K have relatively subtle effects on DNA binding in vitro (Fig. 6; Table 2).
Comparison of the organization of functional domains in PobR with that in LysR family regulators.
It has been noted (9) that the proposed model for regulation by PobR has characteristics typical of the large LysR family of bacterial transcriptional regulators (20, 41). Not unexpectedly then, the linear domain organization of PobR presented here is also similar to that found in the LysR family (41), particularly the well-characterized member NahR (5, 42), regulating two operons for the catabolism of the aromatic compound naphthalene and its metabolic intermediate salicylate. NahR mutations eliminating DNA binding cluster in and around the N-terminal helix-turn-helix motif, but as in PobR, the C terminus of the protein is also essential: a nonsense mutation resulting in loss of nine amino acid residues from the NahR C terminus abolishes DNA binding (42). Most nearby amino acid substitutions that eliminate DNA binding generate NahR proteins that are not trans dominant, suggesting that the C terminus may be indirectly involved in DNA binding by mediating multerimization (41, 42).
PobR amino acids 118 to 135 define a domain where mutation relaxes dependence of PobR activity on its effector p-hydroxybenzoate. This domain in the 271-amino-acid PobR is remarkably similar in position to the region in the 300-amino-acid residue NahR where mutation allows the binding of new effectors, i.e., residues 116 to 169, although several mutations in this region also increase the basal level of NahR activity (5). The one mutation in NahR outside the above-mentioned domain that alters coinducer response does so by increasing the general responsiveness of the NahR protein to a range of effectors (5). This mutation at NahR amino acid residue 248 in the putative multerimization domain may be analogous to the single mutation at PcaU amino acid residue 222 that confers PobR activity.
Distribution in the β-ketoadipate pathway of regulators from the PobR and LysR families.
In Acinetobacter strain ADP1, the two branches of the β-ketoadipate pathway are encoded by genes in two supraoperonic clusters, separated on the chromosome by approximately 280 kb (18) and each governed by regulatory proteins from distinct families: conversion of p-hydroxybenzoate to citric acid cycle intermediates is governed by the PobR family regulators PobR (8) and PcaU (17), whereas the equivalent enzymatic steps in the benzoate branch of the pathway are regulated by LysR family members BenM (6) and CatM (38). This same branch specificity has been noted (39) for the P. putida regulators PcaR and CatR. Given the similarity of PobR to LysR family members discussed above, this strict branch specificity in Acinetobacter may reflect more than historical accident.
Recent work has revealed new layers of regulation within the β-ketoadipate pathway of P. putida (28) and Acinetobacter strain ADP1 (14) that result in sequential use of available carbon and energy sources. Branch specificity of regulatory proteins may facilitate negative interactions between branches, such as that allowing the preferential growth with benzoate over p-hydroxybenzoate, seen in both organisms (14, 28). At the same time, having multiple members from one family of regulators confined to one branch of the pathway may facilitate coordinate control of each supraoperonic cluster, either directly in response to environmental signals or indirectly, mediated by more globally active regulatory proteins. The regulatory organization of the p-hydroxybenzoate branch of the β-ketoadipate pathway in Agrobacterium tumefaciens, with members of three regulatory families clustered in 4.2 kb, genes for two of which overlap at their 3′ ends, indicates that other regulatory solutions are possible (31). Comparison of these different solutions should give insight into the evolutionary relationship between the organisms (4, 29, 31) and the strategies they have employed to adapt to individual ecological niches (29, 31).
Implications of PcaU and PobR gain-of-function mutations.
A single amino acid substitution in PcaU allows it to functionally replace PobR and activate PobA expression. This mutation near the PcaU C terminus is far from the domain containing the helix-turn-helix that is expected to mediate DNA sequence-specific binding, implying that wild-type PcaU can already bind the PobR operator, albeit not productively. Supporting this possibility is the fact that substitution at one position of the PobR helix-turn-helix with the equivalent PcaU residue appears to allow the PobR protein to bind both the PobR and PcaU operators.
Overlapping activation by related regulatory proteins at their DNA targets (cross talk) has been detected in many systems (2, 12, 13, 26, 27, 32, 48), including regulation of Acinetobacter catA (6, 38) by BenM and CatM. It is unclear if the potential interaction of PcaU and PobR at the PobR operator is an important component of pobA regulation or is only an evolutionary vestige reflecting the homology between the two regulatory proteins. Nevertheless, this “regulatory noise” (7) probably underlies the ease of isolation of a PcaU mutant that could functionally replace PobR and underscores another potential adaptive significance of branch-specific regulators from one protein family: the ready availability of a backup (7) if one protein is lost by mutation. This may be especially relevant to pobR, a preferred target in Acinetobacter for IS1236 insertion (15), and exactly such a situation has already been documented with two regulators from the LysR family (27).
In contrast to the evolutionary stability provided by regulator gene families, individual members exhibit remarkable plasticity. The ease of isolation of PobR mutants whose activity was independent of p-hydroxybenzoate suggests that acquiring a new effector profile would not be difficult, as was seen with mutagenesis of NahR (5). This regulatory plasticity which has been exploited over evolutionary time can also be exploited to overcome regulatory bottlenecks during the engineering of new biochemical pathways (33, 35–37, 46). In Acinetobacter, the combination of a positive selection for mutants blocked in the β-ketoadipate pathway with PCR mutagenesis and natural transformation is a distinct advantage in attempts both to understand what has happened over evolutionary time and to explore what might have happened by directed evolution in the laboratory.
ACKNOWLEDGMENTS
This research was supported by grants from the Army Research Office, the National Science Foundation, and the General Reinsurance Corporation.
R. Kok was supported by research funds from K. Hellingwerf during the conclusion of this project.
Footnotes
Publication 18 from the Biological Transformation Center in the Yale Biospherics Institute.
REFERENCES
- 1.Averhoff B A, Gregg-Jolly L A, Elsemore D A, Ornston L N. Genetic analysis of supraoperonic clustering by use of natural transformation in Acinetobacter calcoaceticus. J Bacteriol. 1992;174:200–204. doi: 10.1128/jb.174.1.200-204.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belitsky B R, Sonenshein A L. Altered transcription activation specificity of a mutant form of Bacillus subtilis GltR, a LysR family member. J Bacteriol. 1997;179:1035–1043. doi: 10.1128/jb.179.4.1035-1043.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Cánovas J L, Ornston L N, Stanier R Y. Evolutionary significance of metabolic control systems. Science. 1967;156:1695–1699. doi: 10.1126/science.156.3783.1695. [DOI] [PubMed] [Google Scholar]
- 5.Cebolla A, Sousa C, de Lorenzo V. Effector specificity mutants of the transcriptional activator NahR of naphthalene degrading Pseudomonas define protein sites involved in binding of aromatic inducers. J Biol Chem. 1997;272:3986–3992. doi: 10.1074/jbc.272.7.3986. [DOI] [PubMed] [Google Scholar]
- 6.Collier L S, Neidle E L. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Characterization of BenM, a LysR-type transcriptional activator regulating benzoate degradation in Acinetobacter calcoaceticus, abstr. K-127; p. 557. [Google Scholar]
- 7.de Lorenzo V, Pérez-Martin J. Regulatory noise in prokaryotic promoters: how bacteria learn to respond to novel environmental signals. Mol Microbiol. 1996;19:1177–1184. doi: 10.1111/j.1365-2958.1996.tb02463.x. [DOI] [PubMed] [Google Scholar]
- 8.DiMarco A A, Averhoff B, Ornston L N. Identification of the transcriptional activator pobR and characterization of its role in the expression of pobA, the structural gene for p-hydroxybenzoate hydroxylase in Acinetobacter calcoaceticus. J Bacteriol. 1993;175:4499–4506. doi: 10.1128/jb.175.14.4499-4506.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiMarco A A, Ornston L N. Regulation of p-hydroxybenzoate hydroxylase synthesis by PobR bound to an operator in Acinetobacter calcoaceticus. J Bacteriol. 1994;176:4277–4284. doi: 10.1128/jb.176.14.4277-4284.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsemore D A, Ornston L N. The pca-pob supraoperonic cluster of Acinetobacter calcoaceticus contains quiA, the structural gene for quinate-shikimate dehydrongenase. J Bacteriol. 1994;176:7659–7666. doi: 10.1128/jb.176.24.7659-7666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elsemore D A, Ornston L N. Unusual ancestry of dehydratases associated with quinate catabolism in Acinetobacter calcoaceticus. J Bacteriol. 1995;177:5971–5978. doi: 10.1128/jb.177.20.5971-5978.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett M, Walsh T, Guay G, Bennett P. GcvA, a LysR-type transcriptional regulator protein, activates expression of the cloned Citrobacter freundii ampC β-lactamase gene in Escherichia coli: cross-talk between DNA-binding proteins. Microbiology. 1995;141:419–430. doi: 10.1099/13500872-141-2-419. [DOI] [PubMed] [Google Scholar]
- 13.Fernández S, Shingler V, de Lorenzo V. Cross-regulation by XylR and DmpR activators of Pseudomonas putida suggests that transcriptional control of biodegradative operons evolves independently of catabolic genes. J Bacteriol. 1994;176:5052–5058. doi: 10.1128/jb.176.16.5052-5058.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaines G L, III, Smith L, Neidle E L. Novel nuclear magnetic resonance spectroscopy methods demonstrate preferential carbon source utilization by Acinetobacter calcoaceticus. J Bacteriol. 1996;178:6833–6841. doi: 10.1128/jb.178.23.6833-6841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerischer U, D’Argenio D A, Ornston L N. IS1236, a newly discovered member of the IS3 family, exhibits varied patterns of insertion into the Acinetobacter calcoaceticus chromosome. Microbiology. 1996;142:1825–1831. doi: 10.1099/13500872-142-7-1825. [DOI] [PubMed] [Google Scholar]
- 16.Gerischer U, Ornston L N. Spontaneous mutations in pcaH and -G, structural genes for protocatechuate 3,4-dioxygenase in Acinetobacter calcoaceticus. J Bacteriol. 1995;177:1336–1347. doi: 10.1128/jb.177.5.1336-1347.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerischer U, Segura A, Ornston L N. PcaU, transcriptional activator of genes for protocatechuate utilization in Acinetobacter. J Bacteriol. 1998;180:1512–1524. doi: 10.1128/jb.180.6.1512-1524.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gralton E M, Campbell A L, Neidle E L. Directed introduction of DNA cleavage sites to produce a high-resolution genetic and physical map of the Acinetobacter sp. strain ADP1 (BD413) chromosome. Microbiology. 1997;143:1345–1357. doi: 10.1099/00221287-143-4-1345. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 20.Henikoff S, Haughn G W, Calvo J M, Wallace J C. A large family of bacterial activator proteins. Proc Natl Acad Sci USA. 1988;85:6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. Regulation of pectinolysis in Erwinia chrysanthemi. Annu Rev Microbiol. 1996;50:213–257. doi: 10.1146/annurev.micro.50.1.213. [DOI] [PubMed] [Google Scholar]
- 22.Juni E, Janick A. Transformation of Acinetobacter calcoaceticus (Bacterium anitratum) J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kok R G, D’Argenio D A, Ornston L N. Combining localized PCR-mutagenesis and natural transformation in direct genetic analysis of a transcriptional regulator gene, pobR. J Bacteriol. 1997;179:4270–4276. doi: 10.1128/jb.179.13.4270-4276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kok R G, van Thor J J, Nugteren-Roodzant I M, Brouwer M B W, Egmond M R, Nudel C B, Vosman B, Hellingwerf K J. Characterization of the extracellular lipase, LipA, of Acinetobacter calcoaceticus BD413 and sequence analysis of the cloned structural gene. Mol Microbiol. 1995;15:803–818. doi: 10.1111/j.1365-2958.1995.tb02351.x. [DOI] [PubMed] [Google Scholar]
- 25.Kok R G, van Thor J J, Nugteren-Roodzant I M, Vosman B, Hellingwerf K J. Characterization of lipase-deficient mutants of Acinetobacter calcoaceticus BD413: identification of a periplasmic lipase chaperone essential for the production of extracellular lipase. J Bacteriol. 1995;177:3295–3307. doi: 10.1128/jb.177.11.3295-3307.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leahy J G, Johnson G R, Olsen R H. Cross-regulation of toluene monooxygenases by the transcriptional activators TbmR and TbuT. Appl Environ Microbiol. 1997;63:3736–3739. doi: 10.1128/aem.63.9.3736-3739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leveau J H, van der Meer J R. The tfdR gene product can successfully take over the role of the insertion element-inactivated TfdT protein as a transcriptional activator of the tfdCDEF gene cluster, which encodes chlorocatechol degradation in Ralstonia eutropha JMP134(pJP4) J Bacteriol. 1996;178:6824–6832. doi: 10.1128/jb.178.23.6824-6832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nichols N N, Harwood C S. Repression of 4-hydroxybenzoate transport and degradation by benzoate: a new layer of regulatory control in the Pseudomonas putida β-ketoadipate pathway. J Bacteriol. 1995;177:7033–7040. doi: 10.1128/jb.177.24.7033-7040.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ornston L N, Parke D. The evolution of induction mechanisms in bacteria: insights derived from study of the β-ketoadipate pathway. Curr Top Regul. 1997;12:209–262. doi: 10.1016/b978-0-12-152812-6.50011-1. [DOI] [PubMed] [Google Scholar]
- 30.Pabo C O, Sauer R T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 31.Parke D. Acquisition, reorganization, and merger of genes: novel management of the β-ketoadipate pathway in Agrobacterium tumefaciens. FEMS Microbiol Lett. 1997;146:3–12. [Google Scholar]
- 32.Parsek M R, McFall S M, Shinabarger D L, Chakrabarty A M. Interaction of two LysR-type regulatory proteins CatR and ClcR with heterologous promoters: functional and evolutionary implications. Proc Natl Acad Sci USA. 1994;91:12393–12397. doi: 10.1073/pnas.91.26.12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavel H, Forsman M, Shingler V. An aromatic effector specificity mutant of the transcriptional regulator DmpR overcomes the growth constraints of Pseudomonas sp. strain CF600 on para-substituted methylphenols. J Bacteriol. 1994;176:7550–7557. doi: 10.1128/jb.176.24.7550-7557.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peeters B P H, de Boer J H, Bron S, Venema G. Structural plasmid instability in Bacillus subtilis: effect of direct and inverted repeats. Mol Gen Genet. 1988;212:450–458. doi: 10.1007/BF00330849. [DOI] [PubMed] [Google Scholar]
- 35.Ramos J L, Stolz A, Reineke W, Timmis K N. Altered effector specificities in regulators of gene expression: TOL plasmid xylS mutants and their use to engineer expansion of the range of aromatics degraded by bacteria. Proc Natl Acad Sci USA. 1986;83:8467–8471. doi: 10.1073/pnas.83.22.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramos, J. L., A. Wasserfallen, K. Rose, and K. N. Timmis. Redesigning metabolic routes: manipulation of TOL plasmid pathway for catabolism of alkylbenzoates. Science 235:593–596. [DOI] [PubMed]
- 37.Rojo F, Pieper D H, Engesser K-H, Knackmuss H-J, Timmis K N. Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science. 1987;238:1395–1398. doi: 10.1126/science.3479842. [DOI] [PubMed] [Google Scholar]
- 38.Romero-Arroyo C E, Schell M A, Gaines III G L, Neidle E L. catM encodes a LysR-type transcriptional activator regulating catechol degradation in Acinetobacter calcoaceticus. J Bacteriol. 1995;177:5891–5898. doi: 10.1128/jb.177.20.5891-5898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romero-Steiner S, Parales R E, Harwood C S, Houghton J E. Characterization of the pcaR regulatory gene from Pseudomonas putida, which is required for the complete degradation of p-hydroxybenzoate. J Bacteriol. 1994;176:5771–5779. doi: 10.1128/jb.176.18.5771-5779.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 42.Schell M A, Brown P H, Raju S. Use of saturation mutagenesis to localize probable functional domains in the NahR protein, a LysR-type transcription activator. J Biol Chem. 1990;265:3844–3850. [PubMed] [Google Scholar]
- 43.Smith C P, Chater K F. Structure and regulation of controlling sequences for the Streptomyces coelicolor glycerol operon. J Mol Biol. 1988;204:569–580. doi: 10.1016/0022-2836(88)90356-7. [DOI] [PubMed] [Google Scholar]
- 44.Stanier R Y. Simultaneous adaptation: a new technique for the study of metabolic pathways. J Bacteriol. 1951;177:339–348. doi: 10.1128/jb.54.3.339-348.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sunnarborg A, Klumpp D, Chung T, LaPorte D C. Regulation of the glyoxylate bypass operon: cloning and characterization of iclR. J Bacteriol. 1990;172:2642–2649. doi: 10.1128/jb.172.5.2642-2649.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Timmis K N, Steffan R J, Unterman R. Designing microorganisms for the treatment of toxic wastes. Annu Rev Microbiol. 1994;48:525–557. doi: 10.1146/annurev.mi.48.100194.002521. [DOI] [PubMed] [Google Scholar]
- 47.Velterop J S, Dijkhuizen M A, van’t Hof R, Postma P W. A versatile vector for controlled expression of genes in Escherichia coli and Salmonella typhimurium. Gene. 1995;153:63–65. doi: 10.1016/0378-1119(94)00790-y. [DOI] [PubMed] [Google Scholar]
- 48.von Lintig J, Kreusch D, Schröder J. Opine-regulated promoters and LysR-type regulators in the nopaline (noc) and octopine (occ) catabolic regions of Ti plasmids of Agrobacterium tumefaciens. J Bacteriol. 1994;176:495–503. doi: 10.1128/jb.176.2.495-503.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]