Summary
Arabidopsis RESISTANCE TO POWDERY MILDEW 8.1 (RPW8.1) is an important tool for engineering broad‐spectrum disease resistance against multiple pathogens. Ectopic expression of RPW8.1 leads to enhanced disease resistance with cell death at leaves and compromised plant growth, implying a regulatory mechanism balancing RPW8.1‐mediated resistance and growth. Here, we show that RPW8.1 constitutively enhances the expression of transcription factor WRKY51 and activates salicylic acid and ethylene signalling pathways; WRKY51 in turn suppresses RPW8.1 expression, forming a feedback regulation loop. RPW8.1 and WRKY51 are both induced by pathogen infection and pathogen−/microbe‐associated molecular patterns. In ectopic expression of RPW8.1 background (R1Y4), overexpression of WRKY51 not only rescues the growth suppression and cell death caused by RPW8.1, but also suppresses RPW8.1‐mediated broad‐spectrum disease resistance and pattern‐triggered immunity. Mechanistically, WRKY51 directly binds to and represses RPW8.1 promoter, thus limiting the expression amplitude of RPW8.1. Moreover, WRKY6, WRKY28 and WRKY41 play a role redundant to WRKY51 in the suppression of RPW8.1 expression and are constitutively upregulated in R1Y4 plants with WRKY51 being knocked out (wrky51 R1Y4) plants. Notably, WRKY51 has no significant effects on disease resistance or plant growth in wild type without RPW8.1, indicating a specific role in RPW8.1‐mediated disease resistance. Altogether, our results reveal a regulatory circuit controlling the accumulation of RPW8.1 to an appropriate level to precisely balance growth and disease resistance during pathogen invasion.
Keywords: RPW8.1, WRKY51, immunity, growth, balance
Introduction
Plant broad‐spectrum disease resistance confers resistance against multiple species of pathogens or many races/isolates of the same pathogen. Identification and utilization of genes that confer broad‐spectrum resistance is the most effective and environmentally friendly method to reduce the loss of crops, vegetables and fruits caused by diverse pathogens. In recent years, many genes conferring broad‐spectrum immunity have been identified and used in breeding programmes. In crops, certain resistance (R) genes have been identified to confer broad‐spectrum immunity. Most R genes encode proteins belonging to the family of nucleotide‐binding site‐leucine rich repeat (NBS‐LRR) immune receptors (NLRs) that recognize pathogen‐secreted effectors leading to activation of effector‐triggered immunity (ETI) (Cui et al., 2015). For example, Pi50, Pigm, Pizh, Pi2 and Pi9 are well‐known NBS‐LRR‐encoding genes in rice conferring broad‐spectrum resistance against Magnaporthe oryzae, the causative agent of rice blast disease (Deng et al., 2017; Xie et al., 2019). Similarly, Xa21, Xa7, Xa23, Xa41(t) and Xa47(t) are R genes conferring broad‐spectrum resistance against Xanthomonas oryzae pv. oryzae (Xoo), the causative agent of rice bacterial blight (Chen et al., 2021; Chen and Ronald, 2011).
Besides R genes, some defence regulators are also involved in the regulation of broad‐spectrum immunity. In rice, more than 50 broad‐spectrum defence regulators have been characterized. For example, OsMYB30 improves bsr‐d1‐mediated broad‐spectrum blast resistance by activating lignin biosynthesis genes to strengthen cell walls (Li et al., 2017). OsWRKY45 mediates blast resistance conferred by coiled‐coil (CC)‐NB‐LRR protein Pb1 (Inoue et al., 2013). Rice cysteine‐rich‐receptor‐like kinases OsCRK6 and OsCRK10 contribute to OsNPR1 (non‐expressor of pathogenesis‐related genes 1)‐mediated resistance to Xoo (Chern et al., 2016). In barley, mlo is a nonspecific recessive gene conferring broad‐spectrum disease resistance to powdery mildew. Because of its excellence in conferring broad‐spectrum immunity, the resistant mlo allele has been widely used in most high‐yield European spring barley for more than four decades (Dreiseitl, 2020).
A series of plant receptor‐like kinases (RLKs) and receptor‐like proteins (RLPs) act as pattern recognition receptors (PRRs), which recognize pathogen−/microbe‐associated molecular patterns (PAMPs/MAMPs) or damage‐associated molecular patterns (DAMPs) to induce basal, non‐host resistance against multiple pathogens, contributing to broad‐spectrum disease resistance. For example, Pi‐d2, encoding a receptor‐like kinase protein, confers gene‐for‐gene resistance to multiple races of M. oryzae (Chen et al., 2006). Bph3, a locus encoding plasma membrane‐localized lectin receptor kinases (OsLecRK1‐OsLecRK3), enhances resistance to brown planthopper (BPH) and white‐backed planthopper in susceptible rice varieties (Liu et al., 2015). Xa21, encoding a leucine‐rich‐repeats serine/threonine kinase, confers high levels of resistance to most races of Xoo (Ercoli et al., 2022). In tomato, the I‐3 gene, encoding an S‐receptor‐like kinase (SRLK), confers resistance against multiple races of Fusarium oxysporum f. sp. lycopersici (Fol) when incorporated into cultivated tomato from a wild tomato species (Catanzariti et al., 2015). In apple, Vf genes, encoding receptor‐like proteins, represent the best‐studied apple scab R genes (Malnoy et al., 2008); HcrVf1 and HcrVf2, derived from wild species Malus floribunda 821, confer scab resistance when expressed in a susceptible cultivar (Belfanti et al., 2004; Malnoy et al., 2008).
RESISTANCE TO POWDERY MILDEW 8 (RPW8) is an Arabidopsis thaliana locus identified from wild‐type accession Ms‐0 containing two naturally polymorphic, dominant R genes, RPW8.1 and RPW8.2, which confer resistance to powdery mildew (Orgil et al., 2007; Xiao et al., 2001). Both RPW8.1 and RPW8.2 are atypical R genes encoding proteins containing a CC domain homologous to the CC domain of CC‐NBS‐LRR type R proteins (Xiao et al., 2001). A typical RPW8 domain is present in the N‐termini of a subclass of NLRs termed helper NLRs (hNLRs), which are genetically required for the immune signalling of diverse sensor NLRs that directly or indirectly recognize their cognate effectors (Jubic et al., 2019). For example, NRG1, a hNLR containing a RPW8 domain, is required for the hypersensitive response (HR) cell death and full oomycete resistance (Castel et al., 2019), suggesting an important role for RPW8 genes and RPW8 domains in NLR‐mediated immunity.
Transgenic lines ectopically expressing RPW8.1 from its native promoter in Col‐gl background (R1Ys) showed greatly enhanced broad‐spectrum resistance to powdery mildew fungus, downy mildew oomycete (Ma et al., 2014) and Pseudomonas syringae bacterium (Li et al., 2018) in Arabidopsis. Studies also reveal that RPW8.1 boosts basal defence responses against multiple pathogens in Arabidopsis, including callose deposition, production of reactive oxygen species (ROS), expression of defence‐related genes and hypersensitive response‐like cell death with elevated H2O2 accumulation (Li et al., 2018). Moreover, transgenic rice lines expressing RPW8.1 display significantly enhanced broad‐spectrum resistance against fungal pathogen M. oryzae and bacterial pathogen Xoo (Li et al., 2018). These results demonstrate that RPW8.1 could be used as an important tool for engineering broad‐spectrum disease resistance in plants.
Plant innate immune system contains multiple layers of defence responses to fight against the invasion of diverse pathogens. However, excessive, or prolonged defence responses usually impede growth resulting in yield penalties (Bergelson and Purrington, 1996; Nelson et al., 2018). Conversely, inactivation of immune responses easily leads to great yield loss upon pathogen invasion (Shi et al., 2013). Therefore, controlling defence responses at a precise level and time is important for plants to maintain normal growth in complicated environments. Plants have developed various regulatory mechanisms to precisely control the growth‐immunity trade‐off. In recent years, several proteins have been identified as key regulators of growth‐immunity trade‐offs in plants. In Arabidopsis, BRASSINAZOLE‐RESISTANT1 (BZR1) mediates suppression of immune responses when fast growth is required (Lozano‐Duran et al., 2013). HOMOLOGUE OF BRASSINOSTEROID ENHANCED EXPRESSION 2 (BEE2) INTERACTING WITH IBH1 (HBI1), a bHLH transcription factor, negatively regulates a subset of genes involved in immunity and in turn, PTI signals repress HBI1 transcription to mediate a trade‐off between growth and PTI (Fan et al., 2014). Conversely, transcription factor WRKY45 functions to enhance immunity but suppresses growth in rice (Goto et al., 2015; Goto et al., 2016). Therefore, limiting the expression levels of defence genes to an appropriate level is critical for plants to maintain normal growth. Breeders have selected genes that balance growth and immunity to improve disease resistance without causing yield penalty. In rice, the Pigm (Pyricularia‐Gumei) locus precisely controls a yield‐immunity trade‐off. PigmS (Pigm Susceptible) interacts with PigmR (Pigm Resistant) to attenuate the immune response and yield penalty conferred by PigmR, thus maintaining yield under blast disease conditions (Deng et al., 2017). The Ideal Plant Architecture 1 (IPA1) protein improves both yield and blast disease resistance by maintaining an optimal balance between growth and immunity via switching DNA binding specificity upon phosphorylation or dephosphorylation at a critical residue in the DNA binding domain (Wang et al., 2018).
The transgenic R1Y lines ectopically expressing RPW8.1 with its native promoter exhibited a stunted morphology and spontaneous lesions in leaves but enhanced disease resistance (Li et al., 2018; Ma et al., 2014). In contrast, the wild‐type accessions expressing RPW8.1, such as Wa‐1 and Ms‐0, show no cell death (Orgil et al., 2007), indicating the existence of a regulatory mechanism to control RPW8.1 to an appropriate level avoiding excessive cell death and growth inhibition in wild‐type accessions. In this study, we found that WRKY51 (At5g64810) was constitutively up‐regulated in RPW8.1‐expressing R1Y lines but down‐regulated in RPW8.1 mutant lines. We generated transgenic lines to overexpress or mutate WRKY51 in an RPW8.1‐expressing line (R1Y4) or wild‐type accession and examined their disease resistance and defence responses. Our results reveal a regulatory circuit mediated by WRKY51, WRKY6, WRKY28 and WRKY41 that limits RPW8.1 expression to avoid excessive defence responses and fitness costs.
Results
RPW8.1 boosts WRKY51 expression and the activation of salicylic acid and ethylene signalling pathways
In this study, we try to explore how plants control the expression of RPW8.1 to balance the growth and disease resistance. As RPW8 locus contains RPW8.1 and RPW8.2, we first explored whether RPW8.2 affected the expression of RPW8.1. We mutated RPW8.2 (rpw8.2) in a RPW8‐contained accession Wa‐1 (Orgil et al., 2007) using the CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 technology (Figure S1a–c). Intriguingly, RPW8.1 mRNA level was decreased in rpw8.2 (Figure S1d), indicating that RPW8.2, at least, did not suppress RPW8.1 expression and there existed other mechanisms controlling RPW8.1 expression.
To explore how plants control RPW8.1‐mediated cell death and growth penalty, we conducted RNA‐seq experiments to compare the transcriptomic profiles of a transgenic line expressing RPW8.1‐YFP from the native RPW8.1 promoter in Col‐gl background (R1Y4) and the Col‐gl control that does not contain the RPW8 locus (Table S1). We found that 1197 genes were constitutively up‐regulated in R1Y4 (Table S2), including 16 WRKYs (Table S3). WRKY51 displayed the highest constitutive elevation among the 16 WRKYs 1 week post‐appearance of the spontaneous cell death (Figure S2a) and constitutively greatly up‐regulated (by 10 to 30‐fold) in R1Y4 and R1Y5 when the spontaneous cell death appeared (Figure 1a), indicating that WRKY51 was constitutively elevated by RPW8.1.
Figure 1.
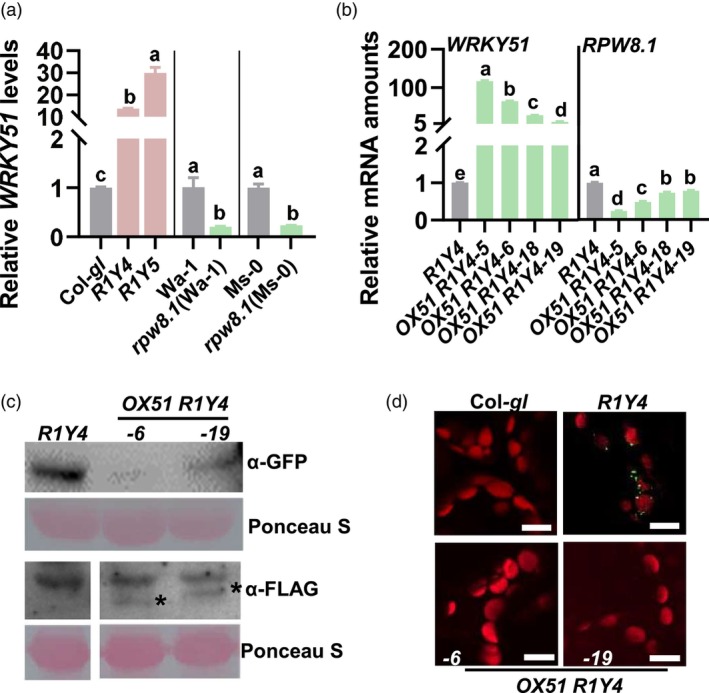
WRKY51 is up‐regulated by RPW8.1 and feedback‐suppresses RPW8.1 expression. (a) Reverse‐transcription quantitative polymerase chain reaction (RT‐qPCR) show the relative mRNA levels of WRKY51 in native promoter‐expressed RPW8.1 transgenic lines (R1Y4 & R1Y5) in Col‐gl background and rpw8.1 mutants in Ms‐0 and Wa‐1 background. (b) mRNA levels of WRKY51 and RPW8.1 in the transgenic lines OX51 R1Y4 (35S‐expressed WRKY51 and native promoter‐expressed RPW8.1) and the R1Y4 control. (c) Western blots for RPW8.1 protein amount in the presence or absence of WRKY51 overexpression. Protein was extracted from five‐week‐old plants for immunoblot analysis using antibodies against green fluorescent protein (GFP) (for RPW8.1) and Flag (for WRKY51) individually. ‘*’ indicates WRKY51‐Flag bands. (d) Subcellular localization of RPW8.1‐YFP (yellow fluorescent protein) in OX51 R1Y4 and R1Y4. Representative confocal images are acquired from leaves of five‐week‐old plants. YFP‐tagged RPW8.1 protein was pseudo‐colored green, and auto‐fluorescent chloroplasts were pseudo‐colored red. Scale bar = 10 μm. For (a) and (b), data are shown as mean ± SD (n = 3 independent samples). The letters above bars indicate significant differences at P < 0.01 determined by one‐way ANOVA followed by post hoc Tukey HSD analysis.
We generated RPW8.1 mutants (rpw8.1) using the CRISPR/Cas9 technology in RPW8‐contained accessions Ms‐0 and Wa‐1(Figures S3 and S4). Compared to wild‐type plants, rpw8.1 plants expressed significantly decreased mRNA levels (by over threefold) of WRKY51 (Figure 1a). Moreover, the other 15 WRKYs were also constitutively suppressed in the rpw8.1 mutant compared to the Ms‐0 control except WRKY58 (Figure S2b). These results indicate that the up‐regulation of WRKY51 requires RPW8.1.
We also examined the expression of these WRKYs in RPW8.2‐expressing line R2Y3 to detect the effect of RPW8.2 on WRKYs. Different from that in R1Y4, seven of the 16 tested WRKYs were slightly and constitutively elevated in R2Y3, but their relative mRNA levels were less than 2‐fold of those in the Col‐gl control (Figure S2c). Moreover, WRKY51 in R2Y3 was significantly suppressed (Figure S2c). These results suggest that the up‐regulation of WRKY51 is independent of RPW8.2.
It was reported that RPW8.1 enhances SA signalling and ethylene signalling (Xiao et al., 2003; Xiao et al., 2005; Zhao et al., 2021). Moreover, Gene Ontology (GO) pathway analysis showed that the up‐regulated genes in R1Y4 are most enriched in SA‐related pathways: ‘salicylic acid biosynthetic process’, ‘salicylic acid metabolic process’ and ‘response to salicylic acid’ (Table S4; Figure S5a), confirming the prominent role of SA in RPW8.1‐mediated immunity. We then tested whether the hormone signals were involved in RPW8.1‐enhanced WRKY51 expression by examining the marker genes of the two signalling pathways (PR1 and PR2 for SA, ERF1 and ERF2 for ethylene) in plants with or without RPW8.1. Consistently, the mRNA levels of these marker genes were greatly elevated in R1Y4 compared to the Col‐gl control without hormone treatment, whereas suppressed in rpw8.1 compared to the Ms‐0 control (Figure S5b,c). However, these marker genes and WRKY51 were not largely induced by benzothiadiazole (BTH) and ethrel (ETH) treatment in R1Y4 (Figure S5d) but greatly induced to similar levels comparable to Ms‐0 control in rpw8.1 12 or 24 h post‐treatment (Figure S5d,e). Based on these data, we deduced that RPW8.1 functioned upstream of SA and ETH to enhance WRKY51 expression.
WRKY51 suppresses RPW8.1 expression
To explore whether WRKY51 was involved in RPW8.1‐mediated growth penalty and resistance, we generated transgenic lines overexpressing FLAG‐tagged WRKY51 using the 35S promoter in the R1Y4 background (OX51 R1Y4). We obtained more than 20 independent lines and examined WRKY51 mRNA and protein levels in four independent lines. All tested OX51 R1Y4 lines displayed markedly elevated WRKY51 mRNA and protein (probed with FLAG antibody) levels in comparison with R1Y4 control (Figure 1b,c). Surprisingly, OX51 R1Y4 showed significantly decreased (down to 20%) mRNA and protein (probed with GFP antibody) levels of RPW8.1 compared with R1Y4 control (Figure 1b,c). Consistent with this result, the YFP signals from RPW8.1‐YFP in OX51 R1Y4 were greatly reduced compared to that in R1Y4 (Figure 1d). These results indicate that WRKY51 feedback suppresses the expression of RPW8.1.
WRKY51 rescues RPW8.1‐suppressed plant growth and cell death
We then looked at the effects of WRKY51 on RPW8.1‐suppressed plant growth and biomass. R1Y4 showed obviously stunted plant stature and cell death‐caused necrotic lesions in leaves compared to Col‐gl control (Figure 2a). Intriguingly, the stunted morphology, necrotic lesions in leaves and decreased biomass caused by RPW8.1 in R1Y4 were completely rescued by WRKY51 overexpression in OX51 R1Y4, which showed normal stature and biomass, as well as leaves without cell death like Col‐gl control (Figure 2a,b). Consistently, although RPW8.1 RNA levels were gradually increased in both OX51 R1Y4 and R1Y4 control during development, they remained relatively suppressed due to WRKY51 overexpression, showing lower levels in OX51 R1Y4 than in R1Y4 control from day 30 when the cell death‐necrotic lesions appeared in R1Y4 (Figure 2c). In contrast, WRKY51 RNA levels were remarkably elevated in OX51 R1Y4 lines than in Col‐gl and R1Y4 control from day 30 (Figure 2d), but were suppressed in rpw8.1 from day 20 compared to the Ms‐0 control (Figure S6). These results indicate that, during development, RPW8.1 induces the expression of WRKY51, which in turn suppresses RPW8.1 expression to alleviate RPW8.1‐mediated cell death and growth penalty.
Figure 2.
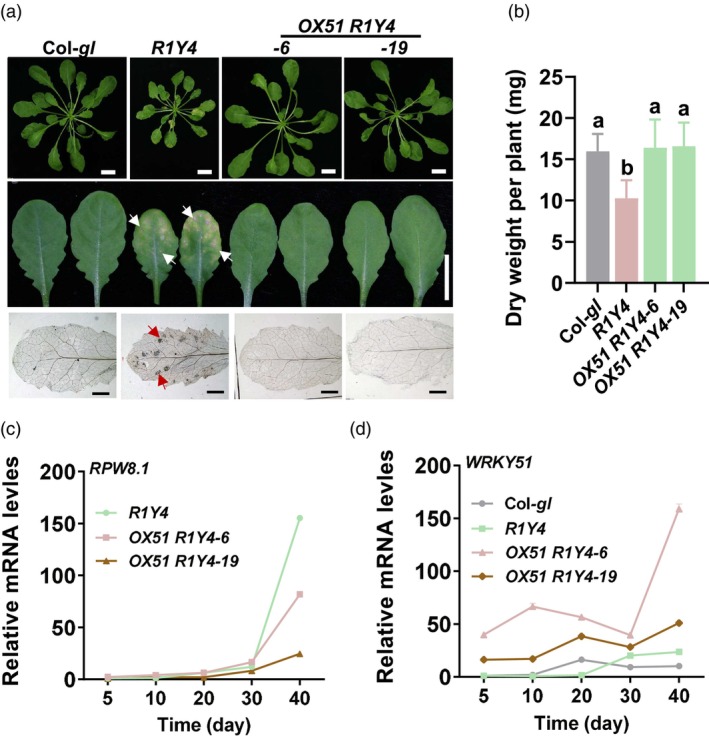
WRKY51 rescues RPW8.1‐suppressed plant growth. (a) Representative five‐week‐old plants and leaves of R1Y4, OX51 R1Y4 and Col‐gl control. The white arrows indicate necrotic lesions and the red arrows indicate cell death nearby the necrotic lesions. Cell death was stained with trypan blue in the lower panel. Scale bar: 1 cm for upper and middle panels and 1 mm for the lower panel. (b) Dry weights of R1Y4, OX51 R1Y4 and Col‐gl control. Data are shown as mean ± SD (n = 6 independent samples). The letters above bars indicate significant differences at P < 0.01 determined by one‐way ANOVA followed by post hoc Tukey HSD analysis. (c‐d) Reverse‐transcription quantitative polymerase chain reaction (RT‐qPCR) show the relative mRNA levels of RPW8.1 and WRKY51 in the indicated lines during the growth period. Data are shown as mean ± SD, n = 3 independent samples.
WRKY51 and RPW8.1 were simultaneously induced by pathogens and PAMPs
WRKY51 was constitutively up‐regulated in RPW8.1 lines (Figure 1a), in which PTI responses and disease resistance were boosted compared with Col‐gl control (Li et al., 2018). Intriguingly, RNA‐seq data showed that WRKY51 is responsive to PAMPs chitin and flg22 (Table S1). Based on these results, we predicted that pathogens could induce the expression of WRKY51 and RPW8.1 simultaneously to limit the expression amplitude of RPW8.1. We first examined the expression of WRKY51 and RPW8.1 in wild‐type Ms‐0 following pathogen inoculation or PAMPs treatment. The expression of both genes was induced by pathogens (powdery mildew or P. syringae) and PAMPs (flg22 and chitin) (Figure 3a), indicating that pathogen infection could induce WRKY51 and RPW8.1 expression simultaneously.
Figure 3.
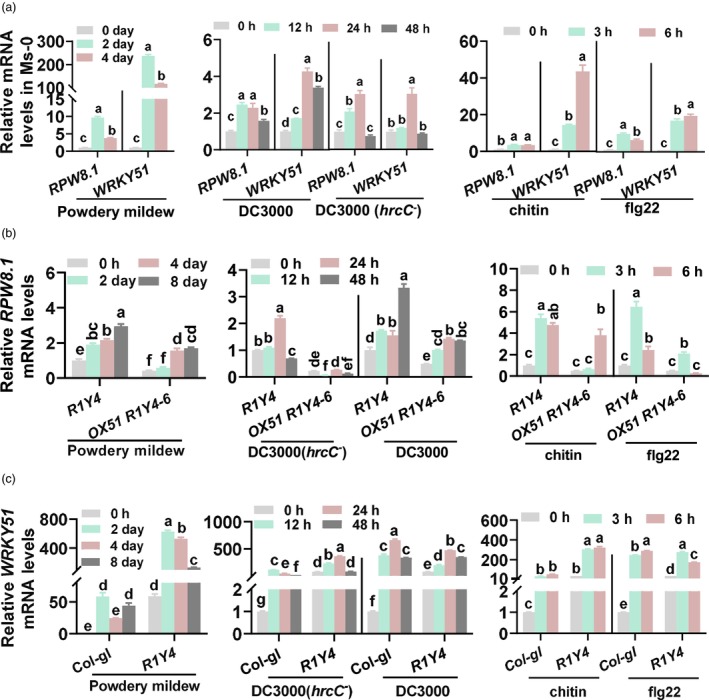
WRKY51 and RPW8.1 were simultaneously induced by pathogens and PAMPs. (a) Reverse‐transcription quantitative polymerase chain reaction (RT‐qPCR) show the relative mRNA levels of RPW8.1 and WRKY51 induced by pathogens (powdery mildew and Pseudomonas syringae) or PMAPs (flg22 and chitin) in Ms‐0 background expressing RPW8.1. (b) RT‐qPCR show the relative mRNA levels of RPW8.1 induced by pathogens or PMAPs in OX51 R1Y4 (containing 35S‐WRKY51 and native promoter‐RPW8.1) and R1Y4 (containing native promoter‐expressed RPW8.1) control. (c) RT‐qPCR show the relative mRNA levels of WRKY51 induced by pathogens or PMAPs in R1Y4 and Col‐gl control. In (a), (b) and (c), data are shown as mean ± SD (n = 3). The letters above bars indicate significant differences at P < 0.01 determined by one‐way ANOVA followed by post hoc Tukey HSD analysis.
We then explored WRKY51 expression in R1Y4 and Col‐gl control, and RPW8.1 expression in OX51 R1Y4 and R1Y4 control. Again, WRKY51 and RPW8.1 RNA levels were induced in all the examined lines (Figure 3b,c). Two points are worth of noting. First, RPW8.1 was less induced in OX51 R1Y4 than in R1Y4 by pathogens and PAMPs (Figure 3b), implying that WRKY51 may decrease RPW8.1 expression during pathogen inoculation. Second, WRKY51 RNA levels were significantly induced to higher levels in R1Y4 than in Col‐gl control by chitin and powdery mildew, but not by flg22 and P. syringae, at both time points (Figure 3c), implying that RPW8.1 has an enhancement effect on WRKY51 RNA expression and that this effect has a probable preference toward fungi (chitin and powdery mildew) than bacteria (flg22 and P. syringae).
WRKY51 suppresses RPW8.1‐boosted disease resistance and PTI‐responses
RPW8.1 expression enhances resistance to fungal pathogen powdery mildew and bacterial pathogen P. syringae (Li et al., 2018; Ma et al., 2014). We then assessed whether WRKY51 could affect RPW8.1‐mediated resistance. OX51 R1Y4 supported significantly more bacterial and fungal growth than R1Y4 control with completely or partially compromised RPW8.1‐mediated immunity, restoring susceptibility to pathogens to disease levels comparable to wild‐type Col‐gl (Figure 4a‐c). These results were consistent with the suppressed RPW8.1 expression in OX51 R1Y4 lines, and indicate that pathogen‐induced WRKY51 suppresses RPW8.1‐mediated disease resistance.
Figure 4.
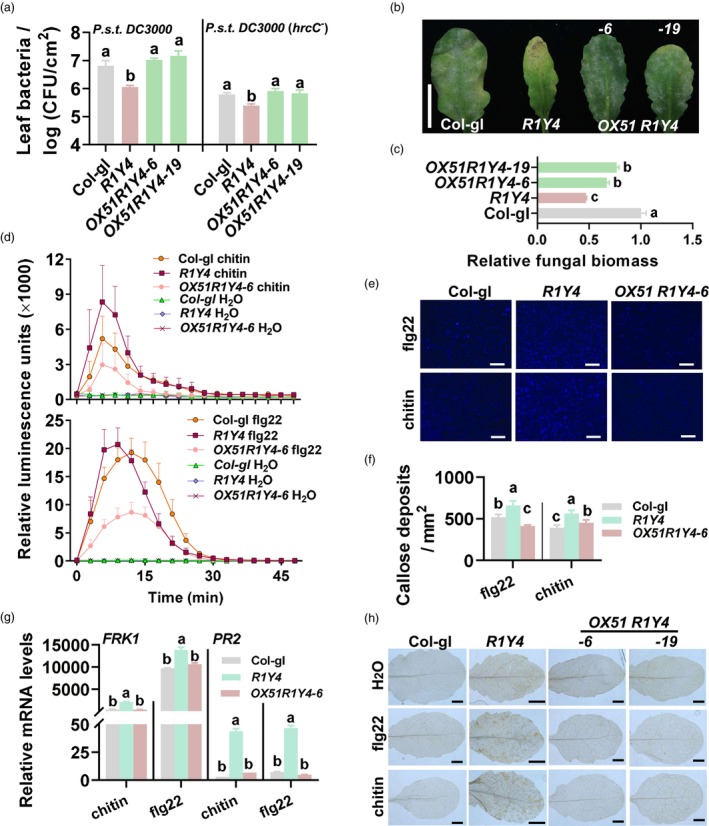
WRKY51 suppresses RPW8.1‐mediated resistance and PAMPs‐triggered immune responses. (a) Bacterial growth in OX51 R1Y4 (containing 35S:WRKY51 and native promoter‐expressed RPW8.1), R1Y4 (containing native promoter‐expressed RPW8.1) and Col‐gl control 3 days post‐inoculation (dpi) with Pseudomonas syringae DC3000 or DC3000(hrcC − ). (b) Disease symptoms of indicated lines 10 dpi with powdery mildew. Scale bar = 1 cm. (c) Fungal biomass of powdery mildew from (b). Fungal biomass was examined by quantitative polymerase chain reaction (qPCR) and shown as the ratio of powdery mildew GDSL‐like lipase DNA against Arabidopsis actin2 DNA. (d) Reactive oxygen species (ROS) bursts triggered by chitin and flg22 in indicated lines. (e) Callose depositions triggered by chitin and flg22 in OX51 R1Y4, R1Y4 and Col‐gl. Callose depositions stained by aniline blue appear as bright spots. Scale bars = 0.1 mm. (f) Quantitation of callose depositions in (f). (g) Reverse‐transcription qPCR (RT‐qPCR) show the relative mRNA levels of FRK1 and PR2 in indicated lines (3 h post‐treatment (hpt) for FRK1 and 12 hpt for PR2) treated with chitin or flg22. Data were normalized to that of Col‐gl at 0 hpt. (h) H2O2 accumulation in indicated lines 48 hpt with or without chitin or flg22 treatment. 3, 3‐diaminobezidin (DAB) was used to stain H2O2 (reddish‐brown). Scale bar = 1 mm. For a–h, data are shown as mean ± SD (n = 3 independent samples for c and h; n = 4 for a, d and e; n = 6 for g). The letters above bars indicate significant differences at P < 0.01 determined by one‐way ANOVA followed by post hoc Tukey HSD analysis.
We next tested whether WRKY51 suppressed RPW8.1‐boosted PTI hallmarks, including reactive oxygen species (ROS) bursts, callose deposition, defence‐related gene expression and H2O2 accumulation. We found that these PTI hallmarks were affected by WRKY51 overexpression. First, chitin and flg22 treatments induced higher ROS bursts and more callose depositions in R1Y4 than in Col‐gl control. In contrast, chitin and flg22 induced ROS bursts in OX51 R1Y4 to levels even lower than those in Col‐gl control (Figure 4d). Second, callose depositions after induction were lower in OX51 R1Y4 than in R1Y4 at levels like that in Col‐gl control (Figure 4e,f). Third, R1Y4 showed higher RNA levels of two PTI‐related marker genes, FRK1 and PR2, than Col‐gl control upon chitin and flg22 treatments, whereas OX51 R1Y4 showed RNA levels comparable to those in Col‐gl control (Figure 4g). Fourth, flg22 and chitin induced less hydrogen peroxide (H2O2) accumulation in OX51 R1Y4 than in R1Y4 as revealed by 3,3′‐diaminobenzidine (DAB) staining (Figure 4h). These results indicate that PAMPs‐induced WRKY51 compromises RPW8.1‐boosted PTI responses by suppressing PAMPs‐induced expression of RPW8.1.
WRKY51 directly binds to the RPW8.1 promoter and represses its activity
As WRKY51 encodes a transcription factor, we then tested whether WRKY51 protein suppresses RPW8.1 expression via repressing the activity of the RPW8.1 promoter by co‐expressing RFP‐HA‐tagged WRKY51 (35S:WRKY51‐HA‐RFP) with a reporter construct containing YFP‐tagged RPW8.1 driven by the RPW8.1 promoter (R8.1 P :R8.1‐YFP) containing 1137 bp sequence before the translational start site (ATG) in Nicotiana benthamiana. As shown in Figures 5a,b, the RPW8.1‐YFP fluorescence intensity and protein level driven by the RPW8.1 promoter were greatly reduced in the presence of WRKY51‐HA‐RFP compared with HA‐YFP, indicating that WRKY51 suppressed RPW8.1 promoter activity.
Figure 5.
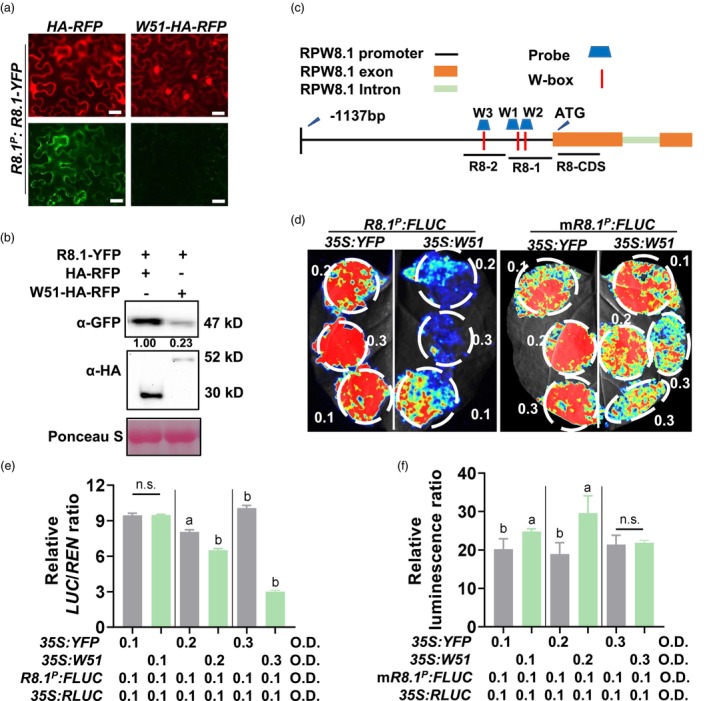
WRKY51 represses the activity of RPW8.1 promoter. (a‐b) Fluorescence intensity (a) and protein amounts (b) of YFP‐fused RPW8.1 (R8.1‐YFP), RFP‐fused WRKY51‐HA (W51‐HA‐RFP) and HA‐RFP. The RPW8.1 promoter‐expressed R8.1‐YFP construct was transiently co‐expressed with construct 35S:HA‐RFP or 35S:WRKY51‐HA‐RFP in Nicotiana benthamiana leaves via Agrobacterium‐mediated infiltration. Scale bar = 20 μm. (c) Distribution of fragments and WRKY protein‐binding W‐boxes in RPW8.1 promoter. Orange rectangles indicate exons; green rectangle indicates intron; black line indicates promoter region. Short red lines indicate W‐boxes. R8‐1, R8‐2 and R8‐CDS indicate the fragments used for DAP‐qPCR experiments. Blue trapezoids indicate the probes (W1, W2 and W3) containing W‐boxes used in EMSA. ATG: translational start site. (d) Images of firefly luciferase (FLUC) signals expressed from wild‐type RPW8.1 promoter (R8.1 P ) or RPW8.1 promoter carrying mutated W‐boxes (mR8.1 P ). The R8.1 P :FLUC and mR8.1 P :FLUC reporter construct were individually co‐expressed with construct 35S:YFP or 35S:WRKY51 (35S:W51) in N. benthamiana leaves via Agrobacterium‐mediated infiltration. LUC light signals were imaged 40 h post‐infiltration with Agrobacterium at indicated concentrations. Renilla luciferase (RLUC) was co‐expressed as an internal reference to normalize the transfection efficiency. (e‐f) Quantitation of firefly luminescence from (d) with different combinations of Agrobacterium concentrations. Luminescence signals were detected using a Promega GloMax 96 Luminometer. Data are shown as mean ± SD (n = 3 independent samples). The letters above bars indicate significant differences at P < 0.01 determined by one‐way ANOVA followed by post hoc Tukey HSD analysis. ‘n.s.’ indicate no significant differences.
Because WRKY proteins are known to bind to the W‐box (TTGACC/T) (Rushton et al., 2010), we analysed the RPW8.1 promoter sequence and found three putative W boxes (TTGACC/T) at −137 to −132 bp, −163 to −158 bp and −274 to −269 bp (Figure 4c). We then tested the cis‐element on the RPW8.1 promoter for WRKY51‐mediated suppression employing a luciferase reporter assay system in N. benthamiana. To this end, we co‐expressed WRKY51 (35S:WRKY51) with a luciferase reporter driven by wild‐type RPW8.1 promoter (R8.1 P :FLUC) or by a mutated RPW8.1 promoter (mR8.1 P :FLUC) in which the three W‐boxes were mutated. A YFP construct (35S:YFP) was used as a negative control. Renilla luciferase (35S:RLUC) was co‐expressed as an internal reference to normalize transfection efficiency. Compared to the YFP control, WRKY51 greatly reduced luciferase expression from the RPW8.1 promoter at two higher concentrations of the WRKY51 construct, but did not affect, or even enhance luciferase expression from the mutated promoter (Figure. 5d–f), suggesting that WRKY51 might bind to the W box and repressed RPW8.1 promoter activity.
We next conducted DNA affinity purification (DAP)‐qPCR and electrophoretic mobility shift assay (EMSA) to validate the binding of WRKY51 to the RPW8.1 promoter. We selected three fragments with or without putative W‐boxes: R8‐1 (−206 ~ −49 bp with W1 and W2, two W‐boxes), R8‐2 (−350 ~ −187 bp with W3, one W‐box) and R8‐CDS (22 ~ 169 bp without a W‐box) (Figure 5c). R8‐CDS is localized in exon one and used as a negative control. We conducted DAP‐qPCR using a GST‐tagged WRKY51 protein expressed in E. coli. The DAP‐qPCR results showed that R8‐1 was significantly highly enriched in the DNA pulled down by GST‐WRKY51 compared to the GST control, whereas R8‐2 and R8‐CDS were not enriched (Figure 6a), indicating that WRKY51 preferentially binds to the W‐box in fragment R8‐1. To confirm the binding of WRKY51 to the W boxes, we amplified a 58‐bp fragment containing W1, a 36‐bp fragment containing W2 located near ATG site and a 30‐bp fragment containing W3 located far away from ATG site. We also amplified mutated W1 and W2, in which the TTGAC core sequences were mutated to TAAAA. These wild‐type probes were tagged with biotin at their 5′‐termini (W‐biotin). Untagged wild‐type and mutant W1 and W2 fragments were used as wild‐type (W) and mutant (Wm) competitors, respectively, to compete for binding to WRKY51. Consistent with the DAP‐qPCR results, EMSA results showed that GST‐WRKY51 bound to W1‐biotin moderately and W2‐biotin strongly causing a shift‐up, respectively. Moreover, the shift‐up band was greatly reduced by the addition of unlabeled wild‐type competitors, but not by mutated competitors (Figure 6b,c). In contrast, GST‐WRKY51 did not cause a shift‐up when incubated with W3‐biotin (Figure 6d). These results indicate that WRKY51 directly binds to W1 and W2 boxes, but not to W3, in the RPW8.1 promoter.
Figure 6.
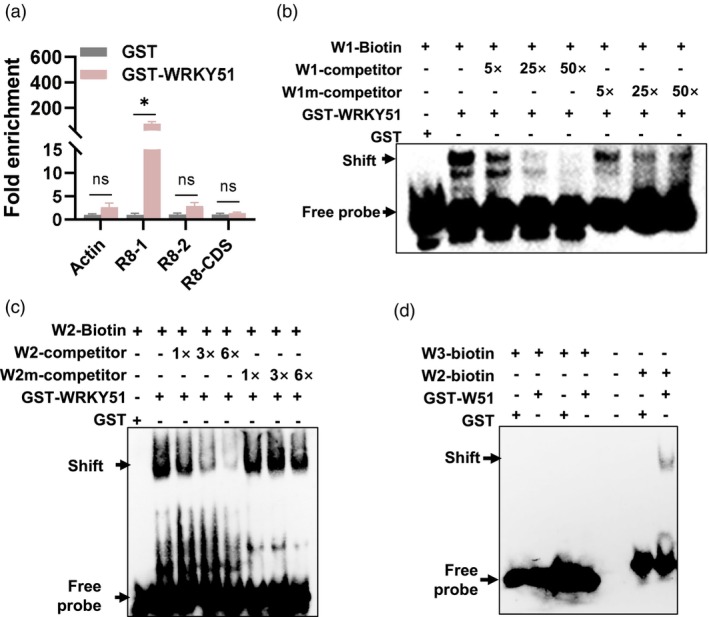
WRKY51 directly binds to the RPW8.1 promoter. (a) Amounts of R8‐1, R8‐2 and R8‐CDS DNA pulled down by GST‐WRKY51 protein or GST control in DNA affinity purification‐quantitative polymerase chain reaction (DAP‐qPCR). PCR product amounts were normalized to actin2, R8‐1, R8‐2, R8‐CDS DNA pulled down by GST, respectively. Actin2 and R8‐CDS serve as negative controls. Data are shown as mean ± SD (n = 3 independent samples). The star above a bar indicates significant differences at P < 0.05 determined by a one‐way ANOVA followed by post hoc Tukey HSD analysis. (b–d) Binding of WRKY51 protein to probes W1, W2 and W3 in EMSA. W‐biotin: W probes labelled with biotin. Unlabeled W1 and W2 were used as competitors in fold excess to probes W1‐biotin or W2‐biotin as indicated. Wm‐competitors: competitors with mutated W‐boxes. (−) indicates absence and (+) indicates presence of the reagent.
WRKY6, WRKY28 and WRKY41 play a role redundant to WRKY51 in the suppression of RPW8.1
To further study the role of WRKY51 on RPW8.1‐controlled disease resistance and growth, we created homozygous w51 R1Y4 lines by crossing a T‐DNA insertional mutant of WRKY51 (w51, SALK_022198C) with R1Y4 (Figure S7a,b). Compared to R1Y4, w51 R1Y4 expressed significantly decreased WRKY51 levels and elevated RPW8.1 mRNA levels (Figure S7c). However, the protein levels and YFP intensity of RPW8.1 were almost unchanged in w51 R1Y4 plants (Figure S7d). Moreover, w51 R1Y4 displayed a similar phenotype and biomass as R1Y4 with necrotic lesions and cell death on leaves (Figure 7e,f). Subsequently, w51 R1Y4 plants displayed the same resistance levels against P. syringae and powdery mildew and supported the same pathogen growth levels as R1Y4 (Figure S8a–c). Moreover, chitin‐ and flg22‐induced callose deposition and H2O2 accumulation in w51 R1Y4 were indistinguishable from those in R1Y4 control (Figure S8d–f). These results imply the existence of other genes that are redundant to WRKY51 in regulating RPW8.1 expression.
Figure 7.
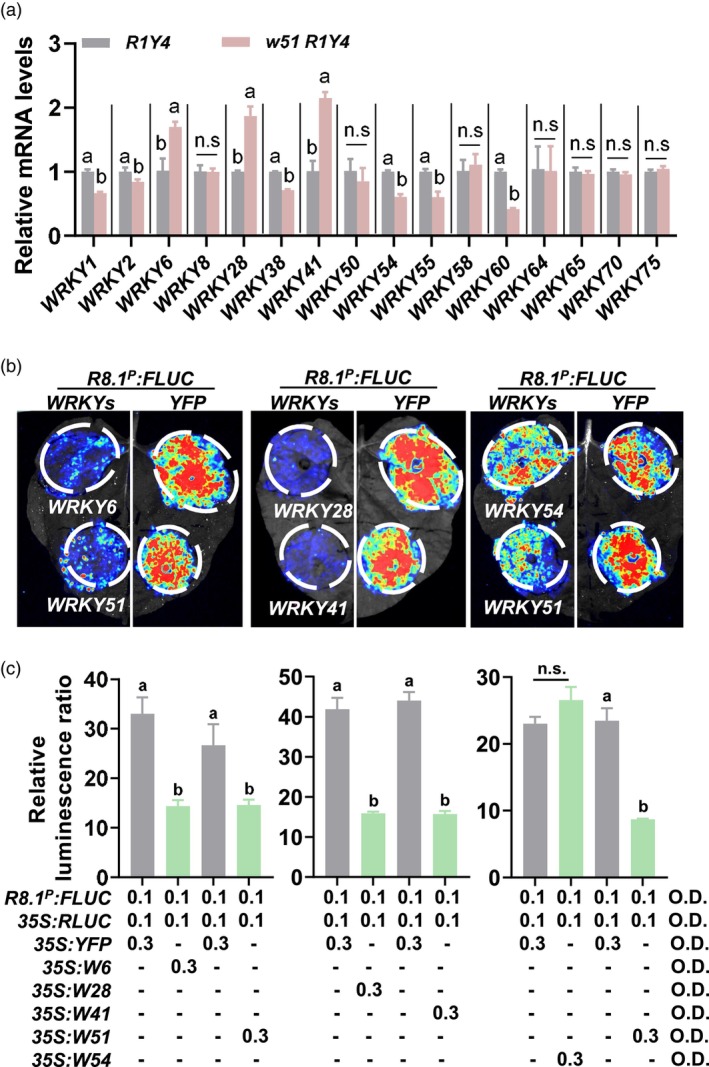
WRKY6, WRKY28 and WRKY41 repress the activity of RPW8.1 promoter. (a) Reverse‐transcription quantitative polymerase chain reaction (RT‐qPCR) show the relative mRNA levels of 16 WRKYs in w51 R1Y4 (containing wrky51 and native promoter‐expressed RPW8.1) line and R1Y4 (containing native promoter‐expressed RPW8.1) control. The letters above bars indicate significant differences at P < 0.01 determined by one‐way ANOVA followed by post hoc Tukey HSD analysis. (b) Images of firefly luciferase (FLUC) signals expressed from the RPW8.1 promoter (R8.1 P ). The R8.1 P :FLUC reporter construct was transiently co‐expressed with construct 35S:YFP or 35S:WRKYs in Nicotiana benthamiana leaves via Agrobacterium‐mediated infiltration. LUC light signals were imaged 40 h post‐infiltration with different combinations of Agrobacterium. Renilla luciferase (RLUC) was co‐expressed as an internal reference to normalize the transfection efficiency. (c) Quantitation of firefly luminescence from (b) with different combinations of Agrobacterium concentrations. Luminescence signals were detected using a Promega GloMax 96 Luminometer. For a and c, data are shown as mean ± SD (n = 3 independent samples). The letters above bars indicate significant differences at P < 0.01 determined by one‐way ANOVA followed by post hoc Tukey HSD analysis. ‘n.s.’ indicate no significant differences.
We then tested the expression of RPW8.1‐boosted WRKYs in w51 R1Y4. Intriguingly, WRKY6, WRKY28 and WRKY41 mRNA levels were constitutively elevated in w51 R1Y4 compared to those in R1Y4 control (Figure 7a), implying a compensatory role of these WRKYs for loss of WRKY51. To test the role of these three WRKYs in the regulation of RPW8.1, we co‐expressed these WRKYs (35S:WRKYs) individually with R8.1 P :FLUC. Compared to the YFP control, WRKY6, WRKY28 and WRKY41 each greatly reduced luciferase expression from the RPW8.1 promoter, whereas WRKY54, a control WRKY gene not up‐regulated in w51 R1Y4, failed to significantly suppress luciferase expression from the RPW8.1 promoter (Figure. 7b,c). These results suggest that WRKY6, WRKY28 and WRKY41 play a role redundant to WRKY51 in repressing the activity of RPW8.1 promoter.
Regulation of growth and disease resistance by WRKY51 depends on RPW8.1
To test whether WRKY51 affects non‐RPW8.1‐regulated growth and disease resistance, we overexpressed WRKY51 in wild‐type Col‐gl accession (OX51) and generated WRKY51 knockout mutant (w51) in Col‐gl background by crossing a wrky51 mutant (w51, SALK_022198C) with Col‐gl (Figure 8a). Both OX51 and w51 lines displayed a normal morphology and dry weight like wild‐type Col‐gl (Figure 8b,c). Upon inoculation with virulent and non‐virulent P. syringae and powdery mildew individually, both OX51 and w51 displayed a similar disease symptom and supported a similar pathogen biomass as Col‐gl control (Figure 8d–f). Consistently, PAMPs chitin and flg22 induced similar levels of callose deposition and H2O2 accumulation in OX51 and w51 as in Col‐gl control (Figure 8g–i). These results indicate that WRKY51‐mediated regulation of growth and disease resistance depends on RPW8.1.
Figure 8.
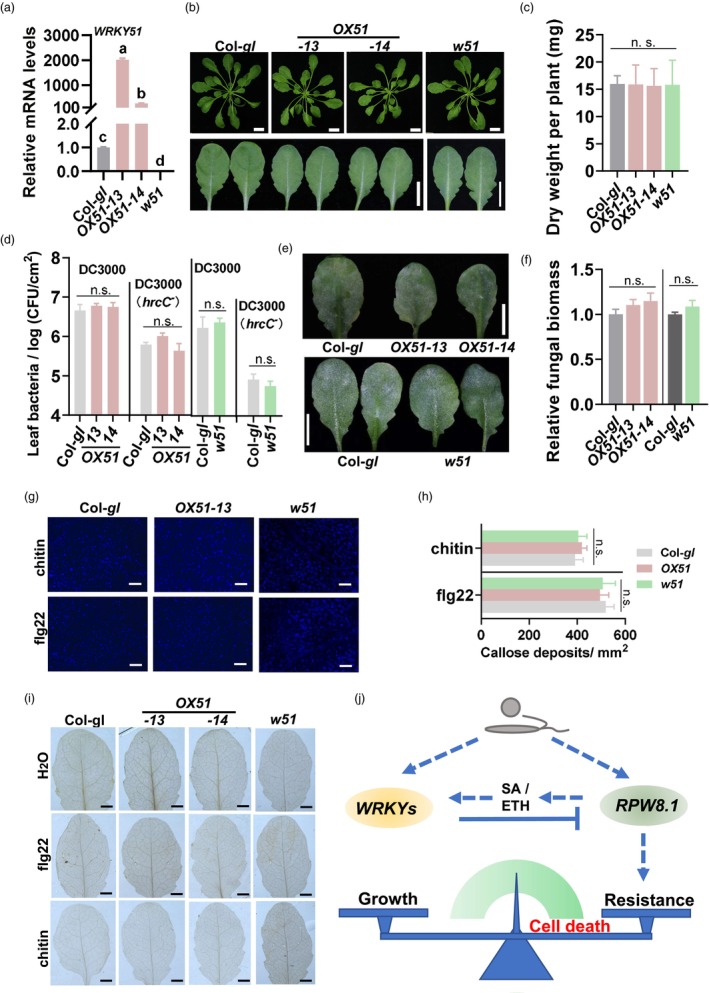
Regulation of growth and disease resistance by WRKY51 depends on RPW8.1. (a) Reverse‐transcription quantitative polymerase chain reaction (RT‐qPCR) show the relative mRNA levels of WRKY51 in OX51, w51 and Col‐gl control. (b) Morphology of five‐week‐old plants and leaves of indicated lines. Scale bar = 1 cm. (c) Dry weights of five‐week‐old seedlings in (b). (d) Bacterial growth in indicated lines 3 days post‐inoculation (dpi) with Pseudomonas syringae tomato DC3000 and DC3000(hrcC −). (e) Disease symptoms of indicated lines seven dpi with powdery mildew. Scale bars = 1 cm. (f) Fungal biomass of powdery mildew in (e). Fungal biomass was examined by quantitative PCR and shown as the ratio of powdery mildew GDSL‐like lipase DNA against Arabidopsis actin2 DNA. (g) PAMPs‐triggered callose depositions in leaves of indicated lines 18 h post‐inoculation (hpi) as stained by aniline blue. Scale bars = 0.1 mm. (h) Quantitation of callose depositions induced by chitin or flg22 in (g). (i) DAB‐stained H2O2 in leaves of indicated lines treated with or without chitin or flg22 for 48 h. Scale bars = 1 mm. For a, c, d, f and h, data are shown as mean ± SD (n = 3 independent samples for a, cand f; n = 4 for d; n = 6 for h). Letters above bars indicate significant differences at P < 0.01 determined by one‐way ANOVA followed by post hoc Tukey HSD analysis. ‘n.s.’ indicates no significant difference. (j) A model for the regulatory circuit of WRKYs and RPW8.1 that controls immune response amplitude. Pathogens trigger expression of WRKYs and RPW8.1 simultaneously. On the one hand, elevated levels of WRKYs suppress pathogen‐induced expression of RPW8.1 leading to reduced cell death and decreased immune responses. On the other hand, RPW8.1 also boosts expression of WRKYs via SA and ethylene signalling pathway to further limit cell death and immune responses. Thus, plants have evolved a feedback‐regulatory circuit where RPW8.1 enhances expression of WRKYs, which further limits RPW8.1 expression to ensure attenuation of cell death and the balance of growth and immunity.
Discussion
Here, we report a regulatory circuit controlling the expression of RPW8.1 with or without pathogen invasion. RPW8.1 constitutively enhances WRKY51 expression, which feedback‐suppresses RPW8.1 expression to an appropriate level avoiding excessive defence responses and cell death. During pathogen invasion, pathogens simultaneously trigger the expression of RPW8.1 and WRKY51. On the one hand, the increased RPW8.1 level boosts defence responses leading to broad‐spectrum disease resistance; on the other hand, the accumulated WRKY51 directly binds to the RPW8.1 promoter and suppresses RPW8.1 expression to avoid excessive cell death and growth inhibition. Moreover, WRKY6, WRKY28 and WRKY41, which are also constitutively up‐regulated by RPW8.1, play a redundant role to WRKY51 to suppress RPW8.1 expression. Therefore, plants have evolved a feedback‐regulatory circuit to avoid excessive RPW8.1‐mediated defence responses via enhancing the expression of WRKYs (Figure 8j). As a result, plants balance the trade‐off between growth and immunity to avoid growth penalties caused by excessive defence responses. These findings provide new insight into the tradeoffs of RPW8.1‐mediated immunity with growth and resources for the application of RPW8.1 to engineer broad disease resistance without growth penalty.
RPW8.1 enhances expression of WRKYs associated with the activation of SA and ethylene signalling
We demonstrate that WRKY51 is induced by PAMPs and pathogens, as well as by RPW8.1 (Figures 1 and 3). It is intriguing that RPW8.1 up‐regulates a gene that directly represses its transcription in turn. WRKY51 was boosted in RPW8.1‐expressing lines with or without pathogen invasion. However, how RPW8.1 up‐regulates WRKY51 expression remains elusive. RPW8.1 is localized near chloroplasts, whereas WRKY51 is localized in the nucleus. RPW8.1 encodes an atypical R protein, whose biochemical function remains unknown. RPW8.1 binds to and stabilizes 1‐aminocyclopropane‐1‐carboxylate oxidase 4 (ACO4), an ACC oxidase converting ACC into ethylene; ectopic expression of RPW8.1 leads to higher ethylene production and activation of ethylene signalling (Zhao et al., 2021). Moreover, RPW8.1 recruits salicylic acid to activate powdery mildew resistance and HR (Xiao et al., 2005). In this study, we found that WRKY51 is induced by BTH and ETH in Col‐gl, R1Y4 and rpw8.1 plants, implying that RPW8.1 enhances the expression of WRKY51 via downstream SA and ethylene‐mediated pathways.
WRKY51 specifically regulates RPW8.1‐mediated disease resistance
In this study, we identified WRKY51 as a key regulator of RPW8.1 that controls the amplification of defence responses to an appropriate level to avoid excessive cell death. Intriguingly, WRKY51 specifically participates in RPW8.1‐mediated disease resistance to balance the trade‐off between resistance and growth. Overexpression and knockout of WRKY51 in non‐RPW8.1 accessions neither affect disease resistance nor penalize plant growth (Figure 8). It is possible that the effects of WRKY51 are limited to RPW8.1‐mediated immunity and growth inhibition because WRKY51 directly binds to the RPW8.1 promoter and controls RPW8.1 expression.
It was reported that WRKY50 and WRKY51 proteins repress JA signalling, and WRKY50 and WRKY51 knockout mutations restore both JA‐inducible PDF1.2 expression and basal resistance to Botrytis cinerea (Gao et al., 2011). WRKY51 is a component of a novel JAV1‐JAZ8‐WRKY51 (JJW) complex that suppresses JA biosynthesis in the absence of an insect attack; once plants are attacked by insects, the JJW complex is disintegrated, resulting in the activation of JA biosynthesis to enhance plant defence responses against insects (Yan et al., 2018). As WRKY51 is up‐regulated in R1Y4 plants, it may be worthwhile to test whether the expression of JA biosynthesis and signalling‐related genes were suppressed in R1Y4. Nevertheless, Gene Ontology results show that the RPW8.1‐upregulated genes are enriched in JA biosynthetic process and JA‐mediated signalling pathway (Table S4), implying that the regulation of RPW8.1‐mediated disease resistance and plant growth by WRKY51 is probably independent of JJW complex‐mediated signalling.
WRKY51 collaborates with other factors to control activity of the RPW8.1 promoter
Knockout of WRKY51 alone has no obvious effects on RPW8.1, as w51 R1Y4 and R1Y4 plants display similar phenotypes, RPW8.1 levels, disease resistance levels and defence responses (Figures S7 and S8). Thus, WRKY51 knockout has no clear effects on RPW8.1‐mediated disease resistance, suggesting the existence of other redundant genes playing a similar role such as WRKY51 in modulating RPW8.1. Consistently, the expression levels of WRKY6, WRKY28 and WRKY41 were constitutively elevated in the R1Y4 line (compared to Col‐gl), and especially in the w51 R1Y4 line (Figure 7a). Promoter activity analysis reveals that these WRKYs, like WRKY51, can repress RPW8.1 expression. Moreover, other TFs also have been identified as trans‐suppressors of RPW8.1. Transient expression of ethylene response factors ORA59, ERF6 and ERF016 significantly suppress the promoter activity of RPW8.1; ORA59 directly binds to the T/A‐rich motif in the RPW8.1 promoter (Zhao et al., 2021). Mutations of these TF genes in R1Y4 result in more severe cell death and ion leakage, as well as more H2O2 accumulation upon pathogen infection, supporting their suppressor roles in RPW8.1‐mediated immune responses (Zhao et al., 2021). In summary, WRKY51 collaborates with other factors to control activity of the RPW8.1 promoter.
Materials and methods
Plant materials and growth conditions
Arabidopsis accessions Col‐gl (containing a glabrous mutation in Col‐0 background), Ms‐0, Wa‐1 and a transgenic line R1Y4 expressing RPW8.1‐YFP in Col‐gl background (Ma et al., 2014) were used. Col‐gl and R1Y4 were used to generate WRKY51 overexpressing lines (abbreviated as OX51 R1Y4 and OX51) and WRKY51 mutant lines (abbreviated as w51 R1Y4 and w51). Arabidopsis plants were planted in a growth room with 23 °C temperature, 70% relative humidity and a 10−/14‐h day/night light setup.
Plant biomass assay
Five‐week‐old plants were collected and dried in a 42 °C oven overnight. The weight of plants was analysed with three replications (three plants in each replication). Data were analysed by the one‐way ANOVA followed by post hoc Tukey HSD analysis with significant differences at P < 0.01.
Generation of transgenic plants
To generate transgenic lines overexpressing WRKY51, the sequence of WRKY51 was amplified from Col‐gl cDNA and cloned into binary vector 35S‐pCAMBIA1300. The constructs were introduced into Col‐gl or R1Y4 via Agrobacterium‐mediated floral dipping (Clough and Bent, 1998) to generate transgenic plants OX51 and OX51 R1Y4, respectively. To generate mutants wrky51, a wrky51 mutant (SALK_022198C) was crossed with R1Y4 and Col‐gl, respectively. Homozygous w51 in Col‐gl background and R1Y4 background (w51R1Y4) were obtained by screening F2 individuals with specific primers w51‐F and T‐DNA primer LB (Table S5). We construct rpw8.1 mutants using the CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 technology in Ms‐0. The two RPW8.1 guide RNA (gRNA) sequences listed in Table S2 were screened by the Cas‐OFFinder system (http://www.rgenome.net/cas‐offinder/) to avoid potential off‐target sites with the screen parameters to allow less than three base‐pair mismatches and one DNA/RNA bulge. For the assembly of two gRNAs, the two target sites were incorporated into PCR primers, respectively. The PCR fragment was amplified from pCBC‐DT1T2 for dicot targets with four shorter primers, among which two forward or two reverse primers were partially overlapping. The purified PCR fragment, together with the pHEE401E vector, was used to set up restriction‐ligation reactions using BsaI and T4 Ligase (New England Biolabs). The reaction was incubated in a thermocycler for 5 h at 37 °C, 5 min at 50 °C and 10 min at 80 °C. The vector was transformed into Agrobacterium strain GV3101 for Arabidopsis transformation.
Pathogen infection
Five‐week‐old Arabidopsis plants were syringe‐infiltrated with the virulent bacterial strain P. syringae DC3000 at the concentration of OD600 = 0.0005, or the mutant strain DC3000(hrcC − ) at OD600 = 0.002. Bacterial biomass was determined as previously described 3 days post‐inoculation (dpi) (Li et al., 2010).
The Golovinomyces cichoracearum UCSC1 powdery mildew strain was maintained on pad4 sid2 plants for the generation of fresh inocula. Photos of mildew disease phenotypes were captured at 10 dpi. Relative fungal biomass of powdery mildew was determined by the relative ratio of DNA levels of the powdery mildew GDSL‐like lipase gene against the Arabidopsis actin2 gene via quantitative polymerase chain reaction (qPCR) (Wessling and Panstruga, 2012).
Gene expression assay
Five‐week‐old Arabidopsis plants were pretreated with PAMPs chitin and flg22 or inoculated with pathogens. PAMPs‐treated leaves were collected at 0, 3 and 6 hpi. P. syringae‐inoculated leaves were collected at 0, 12, 24, 48 hpi. Powdery mildew‐inoculated leaves were collected at 0, 2, 4, 8 dpi. Total RNAs were extracted using TRIzol reagent (Invitrogen, Shanghai, China) for gene expression assay by reverse transcription‐quantitative polymerase chain reaction (RT‐qPCR). Total RNAs were used as templates for reverse transcription using the NovoScript® Plus All‐in‐one 1st Strand cDNA Synthesis SuperMix (gDNA Purge) (Novoprotein, Suzhou, China). RT‐qPCRs were conducted using SYBR mix (Bioground Biotech, Chongqing, China) and gene‐specific primers (Table S2). Arabidopsis actin2 (ACT2) gene is used as the internal reference to normalize expression levels.
Reactive oxygen species (ROS) burst
Leaves of four‐week‐old plants were sliced into strips and about 10 mm2 strips were incubated in 200 μL water in a 96‐well plate for 12 h. Then ROS bursts were triggered by adding 1 μM flg22 or 20 μg/ mL chitin in 200 μL buffer containing 20 mM luminol, 10 μg/ mL horseradish peroxidase (Sigma‐Aldrich Shanghai Trading Co Ltd, Shanghai, China) and determined as relative luminescence units using a GloMax96 Microplate Luminometer (Promega Biotech Co., Ltd, Beijing, China) for about 45 min.
Western blot assays
Total protein samples were extracted with protein extraction buffer (0.25 M Tris–HCL (pH 6.8), 4% sodium dodecyl sulfate, 0.1% bromophenol blue, 40% glycerol); polyclonal antibodies against GFP (BBI Life Science, Shanghai, China), HA (Roche, Shanghai, China), Flag (Sigma‐Aldrich Shanghai Trading Co Ltd, Shanghai, China) were individually used with the Clarity Western ECL Substrate System (Bio‐Rad, Chengdu, China) to detect protein levels.
Callose Staining
Leaves of four‐week‐old plants were infiltrated with 1 μM flg22 or 20 μg/ mL chitin and incubated for 12 h. Then the leaves were discolored and stained with 0.01% aniline blue for 30 min (Hauck et al., 2003). Callose depositions were captured with a fluorescence microscope under a UV channel (340 to 380 nm) (Zeiss imager A2.0, Germany) and calculated by using the Image J software (Zhang et al., 2007).
H2O2 accumulation
Leaves of four‐week‐old plants were collected and submerged in 0.5 mg/mL 3,3′‐diaminobenzidine (DAB) (Sigma‐Aldrich Shanghai Trading Co Ltd, Shanghai, China) solution for staining overnight. Then the leaves were decolored with 95% alcohol at 65 °C and pictured under a stereomicroscope (OLYMPUS SZX16, Japan).
Dual‐luciferase reporter system
The 1137‐bp promoter sequence before the transcription start site of RPW8.1 was amplified with specific primers (Table S2) and cloned into binary vector pCAMBIA1300 to express firefly luciferase (FLUC), creating construct R8.1 P :LUC, via homologous recombination (Zhao et al., 2021). The plasmid was introduced into Agrobacterium strain GV3101. An Aagrobacterium strain expressing Renilla luciferase (RLUC) driven by the 35S promoter was used as an internal reference to normalize the transfection efficiency. The Agrobacterium strains were syringe‐infiltrated into N. benthamiana leaves with an Agrobacterium strain containing 35S:WRKY51 for transient expression assay. We observed the results at 40 hpi. Leaves were sprayed with 0.15 mg/ ml D‐luciferin potassium salt (BioVision, USA) containing 0.01% silwet L‐77 for imaging. Intensity of firefly luminescence was detected by a low‐light cooled charge‐coupled device imaging apparatus (Bio‐rad, Chengdu, China). The relative intensity of firefly luminescence was normalized to the Renilla luminescence intensity using a dual‐luciferase reporter Kit (Beyotime, China) and measured in a GloMax96 Microplate Luminometer following the manufacturer's instruction.
Electrophoretic mobility shift assay (EMSA)
Biotin‐labelled probes were prepared by a company (Sangon Biotech, Shanghai, China). For the binding assay, purified GST‐WRKY51 protein was incubated with 1.5 nM specific probes in binding buffer (10 mM Tris, 0.2 mM EDTA, 20 mM KCl, 1% glycerol, 0.02% TritonX‐100). For the competition assay, cold competitors (unlabelled probes) were added to the reaction at a concentration of 5×, 25×, or 50× of the labelled probe. The samples were incubated at 25 °C for 20 min. Then the reaction mixtures were separated on a 6% native polyacrylamide gel and transferred to a nylon membrane (Amersham Hybond™‐N+, GE, America). Later steps followed the instructions of the Chemiluminescent EMSA Kit (Beyotime, China). Luminescence intensity was detected using a low‐light cooled charge‐coupled device imaging apparatus. The probe sequences are listed in Table S2.
DNA affinity purification (DAP)‐qPCR
The experiment was conducted as described before (Li et al., 2017) with some modifications. Generally, we sonicated the R1Y4 genomic DNA (5 μg) into 200 to 500‐bp fragments and added 10 μL sonicated DNA sample to 500 μL F‐buffer (50 mM Tris–HCl pH = 8.0, 1 mM EDTA, 100 mM KCl, 5% glycerol, 0.1% Triton, 100 mM DTT) containing purified GST‐WRKY51 or GST protein with 50 μL pre‐washed Pierce™ Glutathione Agarose resins (ThermoFisher, Shanghai, China). The DNA‐protein‐agarose mixture was incubated at 4 °C overnight. We collected the agarose resins by spinning at 700 g for 5 min at 4 °C and washed five times with 1× PBS + Triton X‐100 (0.005%) and two times with 1× PBS. Beads were then resuspended in 25 μL EB (10 mM Tris–HCl, pH = 8.5) and the samples were heated for 10 min at 98 °C. Samples were spun and the supernatants were kept for further qPCR analysis. The equivalent of 1 μL of immunoprecipitated DNA was then amplified by real‐time PCR reactions using gene‐specific primers. PCR products were normalized to actin2.
Author contributions
Y. L. and W‐M. Wang conceived the experimental design, and together with X‐M. Y., J‐H. Z., X‐Y. X., J‐F. S., H. S., Z‐W. H., L. X., Y‐J. L. and Y. Zhu carried out the experiments; G‐B. L., S‐X. Z., J‐L. L., S‐H. B., H. W., Z‐X. Z., J‐W. Z., J. F. and Y‐Y. Huang analysed the data; C. L. and M. Pu carried out the planting; Y. L. and W‐M. Wang wrote the paper.
Conflict of interest
No conflict of interest in this study.
Supporting information
Figure S1 Mutation of RPW8.2 leads to decreased expression of RPW8.1.
Figure S2. RPW8.1 constitutively boosts the expression of WRKYs.
Figure S3. rpw8.1 mutant in Ms‐0 background.
Figure S4. rpw8.1 mutant in Wa‐1 background.
Figure S5. RPW8.1 activates SA and ethylene signalling pathways, which enhances the expression of WRKY51.
Figure S6. WRKY51 is suppressed in rpw8.1 mutant.
Figure S7. WRKY51 knockout has no major effects on RPW8.1‐mediated growth inhibition.
Figure S8. WRKY51 knockout has no major effects on RPW8.1‐mediated disease resistance and PTI responses.
Table S1 mRNA reads in R1Y4 and the Col‐gl with or without PAMPs treatment.
Table S2. Genes up‐regulated in R1Y4 compared to Col‐gl control.
Table S3. WRKYs up‐regulated in R1Y4 compared to Col‐gl control.
Table S4. GO analysis of the up‐regulated genes in R1Y4.
Table S5. Primers used in this study.
Appendix S1 Supporting Information.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 32121003, 32172417), the Department of Science and Technology of Sichuan Province (2022JDTD0023), the Open Research Fund of State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China (SKL‐ZY202209, SKL‐ZD202202, SKL‐ZY202205, SKL‐KF202225), the Open Research Fund of State Key Laboratory of Hybrid Rice (Hunan Hybrid Rice Research Center, 2021KF07) and the Yunnan Basic Research Special Youth Project (202201 AU070010).
Contributor Information
Wen‐Ming Wang, Email: j316wenmingwang@sicau.edu.cn.
Yan Li, Email: liyan_rice@sicau.edu.cn.
References
- Belfanti, E. , Silfverberg‐Dilworth, E. , Tartarini, S. , Patocchi, A. , Barbieri, M. , Zhu, J. , Vinatzer, B.A. et al. (2004) The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivated variety. Proc. Natl. Acad. Sci. U. S. A. 101, 886–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson, J. and Purrington, C.B. (1996) Surveying patterns in the cost of resistance in plants. Am. Nat. 148, 536–558. [Google Scholar]
- Castel, B. , Ngou, P.M. , Cevik, V. , Redkar, A. , Kim, D.S. , Yang, Y. , Ding, P. et al. (2019) Diverse NLR immune receptors activate defence via the RPW8‐NLR NRG1. New Phytol. 222, 966–980. [DOI] [PubMed] [Google Scholar]
- Catanzariti, A.M. , Lim, G.T.T. and Jones, D.A. (2015) The tomato I‐3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytol. 207, 106–118. [DOI] [PubMed] [Google Scholar]
- Chen, X. and Ronald, P.C. (2011) Innate immunity in rice. Trends Plant Sci. 16, 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Shang, J. , Chen, D. , Lei, C. , Zou, Y. , Zhai, W. , Liu, G. et al. (2006) A B‐lectin receptor kinase gene conferring rice blast resistance. Plant J. 46, 794–804. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Liu, P. , Mei, L. , He, X. , Chen, L. , Liu, H. , Shen, S. et al. (2021) Xa7, a new executor R gene that confers durable and broad‐spectrum resistance to bacterial blight disease in rice. Plant Commun 2, 100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern, M. , Xu, Q. , Bart, R.S. , Bai, W. , Ruan, D. , Sze‐To, W.H , Canlas, P.E. et al. (2016) A Genetic Screen Identifies a Requirement for Cysteine‐Rich‐Receptor‐Like Kinases in Rice NH1 (OsNPR1)‐Mediated Immunity. PLoS Genet. 12, e1006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cui, H. , Tsuda, K. and Parker, J.E. (2015) Effector‐triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. [DOI] [PubMed] [Google Scholar]
- Deng, Y. , Zhai, K. , Xie, Z. , Yang, D. , Zhu, X. , Liu, J. , Wang, X. et al. (2017) Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965. [DOI] [PubMed] [Google Scholar]
- Dreiseitl, A. (2020) Specific Resistance of Barley to Powdery Mildew, p. 11. Genes (Basel): Its Use and Beyond. A Concise Critical Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercoli, M.F. , Luu, D.D. , Rim, E.Y. , Shigenaga, A. , Teixeira De Araujo, A., Jr. , Chern, M. , Jain, R. et al. (2022) Plant immunity: Rice XA21‐mediated resistance to bacterial infection. Proc. Natl. Acad. Sci. U. S. A. 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, M. , Bai, M.Y. , Kim, J.G. , Wang, T. , Oh, E. , Chen, L. , Park, C.H. et al. (2014) The bHLH transcription factor HBI1 mediates the trade‐off between growth and pathogen‐associated molecular pattern‐triggered immunity in Arabidopsis. Plant Cell, 26, 828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Q.M. , Venugopal, S. , Navarre, D. and Kachroo, A. (2011) Low oleic acid‐derived repression of jasmonic acid‐inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 155, 464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, S. , Sasakura‐Shimoda, F. , Suetsugu, M. , Selvaraj, M.G. , Hayashi, N. , Yamazaki, M. , Ishitani, M. et al. (2015) Development of disease‐resistant rice by optimized expression of WRKY45. Plant Biotechnol. J. 13, 753–765. [DOI] [PubMed] [Google Scholar]
- Goto, S. , Sasakura‐Shimoda, F. , Yamazaki, M. , Hayashi, N. , Suetsugu, M. , Ochiai, H. and Takatsuji, H. (2016) Development of disease‐resistant rice by pathogen‐responsive expression of WRKY45. Plant Biotechnol. J. 14, 1127–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck, P. , Thilmony, R. and He, S.Y. (2003) A Pseudomonas syringae type III effector suppresses cell wall‐based extracellular defense in susceptible Arabidopsis plants. Proc. Natl. Acad. Sci. U. S. A. 100, 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, H. , Hayashi, N. , Matsushita, A. , Xinqiong, L. , Nakayama, A. , Sugano, S. , Jiang, C.J. et al. (2013) Blast resistance of CC‐NB‐LRR protein Pb1 is mediated by WRKY45 through protein‐protein interaction. Proc. Natl. Acad. Sci. U. S. A. 110, 9577–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubic, L.M. , Saile, S. , Furzer, O.J. , El Kasmi, F. and Dangl, J.L. (2019) Help wanted: helper NLRs and plant immune responses. Curr. Opin. Plant Biol. 50, 82–94. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Zhang, Q. , Zhang, J. , Wu, L. , Qi, Y. and Zhou, J.M. (2010) Identification of microRNAs involved in pathogen‐associated molecular pattern‐triggered plant innate immunity. Plant Physiol. 152, 2222–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Zhu, Z. , Chern, M. , Yin, J. , Yang, C. , Ran, L. , Cheng, M. et al. (2017) A natural allele of a transcription factor in rice confers broad‐spectrum blast resistance. Cell, 170, e115. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Zhang, Y. , Wang, Q.X. , Wang, T.T. , Cao, X.L. , Zhao, Z.X. , Zhao, S.L. et al. (2018) RESISTANCE TO POWDERY MILDEW8.1 boosts pattern‐triggered immunity against multiple pathogens in Arabidopsis and rice. Plant Biotechnol. J. 16, 428–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Wu, H. , Chen, H. , Liu, Y. , He, J. , Kang, H. , Sun, Z. et al. (2015) A gene cluster encoding lectin receptor kinases confers broad‐spectrum and durable insect resistance in rice. Nat. Biotechnol. 33, 301–305. [DOI] [PubMed] [Google Scholar]
- Lozano‐Duran, R. , Macho, A.P. , Boutrot, F. , Segonzac, C. , Somssich, I.E. and Zipfel, C. (2013) The transcriptional regulator BZR1 mediates trade‐off between plant innate immunity and growth. Elife 2, e00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X.F. , Li, Y. , Sun, J.L. , Wang, T.T. , Fan, J. , Lei, Y. , Huang, Y.Y. et al. (2014) Ectopic expression of RESISTANCE TO POWDERY MILDEW8.1 confers resistance to fungal and oomycete pathogens in Arabidopsis. Plant Cell Physiol. 55, 1484–1496. [DOI] [PubMed] [Google Scholar]
- Malnoy, M. , Xu, M. , Borejsza‐Wysocka, E. , Korban, S.S. and Aldwinckle, H.S. (2008) Two receptor‐like genes, Vfa1 and Vfa2, confer resistance to the fungal pathogen Venturia inaequalis inciting apple scab disease. Mol. Plant Microbe Interact. 21, 448–458. [DOI] [PubMed] [Google Scholar]
- Nelson, R. , Wiesner‐Hanks, T. , Wisser, R. and Balint‐Kurti, P. (2018) Navigating complexity to breed disease‐resistant crops. Nat. Rev. Genet. 19, 21–33. [DOI] [PubMed] [Google Scholar]
- Orgil, U. , Araki, H. , Tangchaiburana, S. , Berkey, R. and Xiao, S. (2007) Intraspecific genetic variations, fitness cost and benefit of RPW8, a disease resistance locus in Arabidopsis thaliana. Genetics, 176, 2317–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J. , Somssich, I.E. , Ringler, P. and Shen, Q.J. (2010) WRKY transcription factors. Trends Plant Sci. 15, 247–258. [DOI] [PubMed] [Google Scholar]
- Shi, H. , Shen, Q. , Qi, Y. , Yan, H. , Nie, H. , Chen, Y. , Zhao, T. et al. (2013) BR‐SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell, 25, 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Zhou, L. , Shi, H. , Chern, M. , Yu, H. , Yi, H. , He, M. et al. (2018) A single transcription factor promotes both yield and immunity in rice. Science, 361, 1026–1028. [DOI] [PubMed] [Google Scholar]
- Wessling, R. and Panstruga, R. (2012) Rapid quantification of plant‐powdery mildew interactions by qPCR and conidiospore counts. Plant Methods, 8, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S. , Ellwood, S. , Calis, O. , Patrick, E. , Li, T. , Coleman, M. and Turner, J.G. (2001) Broad‐spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science, 291, 118–120. [DOI] [PubMed] [Google Scholar]
- Xiao, S. , Brown, S. , Patrick, E. , Brearley, C. and Turner, J.G. (2003) Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid‐dependent amplification circuit is required for hypersensitive cell death. Plant Cell, 15, 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, S. , Calis, O. , Patrick, E. , Zhang, G. , Charoenwattana, P. , Muskett, P. , Parker, J.E. et al. (2005) The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. Plant J. 42, 95–110. [DOI] [PubMed] [Google Scholar]
- Xie, Z. , Yan, B.X. , Shou, J.Y. , Tang, J. , Wang, X. , Zhai, K.R. , Liu, J.Y. et al. (2019) A nucleotide‐binding site‐leucine‐rich repeat receptor pair confers broad‐spectrum disease resistance through physical association in rice. Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20180308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, C. , Fan, M. , Yang, M. , Zhao, J. , Zhang, W. , Su, Y. , Xiao, L. et al. (2018) Injury Activates Ca(2+)/Calmodulin‐Dependent Phosphorylation of JAV1‐JAZ8‐WRKY51 Complex for Jasmonate Biosynthesis. Mol. Cell 70, e137. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Shao, F. , Li, Y. , Cui, H. , Chen, L. , Li, H. , Zou, Y. et al. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP‐induced immunity in plants. Cell Host Microbe 1, 175–185. [DOI] [PubMed] [Google Scholar]
- Zhao, Z.X. , Feng, Q. , Liu, P.Q. , He, X.R. , Zhao, J.H. , Xu, Y.J. , Zhang, L.L. et al. (2021) RPW8.1 enhances the ethylene‐signaling pathway to feedback‐attenuate its mediated cell death and disease resistance in Arabidopsis. New Phytol. 229, 516–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Mutation of RPW8.2 leads to decreased expression of RPW8.1.
Figure S2. RPW8.1 constitutively boosts the expression of WRKYs.
Figure S3. rpw8.1 mutant in Ms‐0 background.
Figure S4. rpw8.1 mutant in Wa‐1 background.
Figure S5. RPW8.1 activates SA and ethylene signalling pathways, which enhances the expression of WRKY51.
Figure S6. WRKY51 is suppressed in rpw8.1 mutant.
Figure S7. WRKY51 knockout has no major effects on RPW8.1‐mediated growth inhibition.
Figure S8. WRKY51 knockout has no major effects on RPW8.1‐mediated disease resistance and PTI responses.
Table S1 mRNA reads in R1Y4 and the Col‐gl with or without PAMPs treatment.
Table S2. Genes up‐regulated in R1Y4 compared to Col‐gl control.
Table S3. WRKYs up‐regulated in R1Y4 compared to Col‐gl control.
Table S4. GO analysis of the up‐regulated genes in R1Y4.
Table S5. Primers used in this study.
Appendix S1 Supporting Information.


