Abstract
Viral infection can be used to uncover host regulatory mechanisms controlling both cell response and homeostasis. Using cell biological, biochemical and genetic tools, we reveal that influenza A virus (IAV) infection induces global transcriptional defects at the 3’-end of active host genes and RNA polymerase II (RNAPII) run-through into extragenic regions. This effect induces the biogenesis of aberrant RNAs (3’-extensions and host gene fusions) which ultimately causes global transcriptional downregulation of physiological transcripts, an effect that impacts antiviral response and virulence. This phenomenon occurs with multiple strains of IAV and it is dependent on influenza NS1 protein. Mechanistically, pervasive RNAPII run-through can be modulated by SUMOylation of an intrinsically disordered region (IDR) of NS1 expressed by the 1918 pandemic IAV. Our data identify a general strategy used by IAV to suppress host gene expression and indicate that polymorphisms in IDRs of viral proteins can determine the outcome of an infection.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Introduction
Due to their fast sampling of the evolutionary space, comparative analysis of virus-host interaction networks can provide us with valuable information about evolutionary constraints governing protein-protein interaction principles. The analysis of proteins from different viruses (or of the same protein from different strains) can guide the study of human proteins and ultimately reveal universal principles and regulatory mechanism controlling cellular functions.
Influenza A virus (IAV) is a negative-sense single-stranded RNA virus. The IAV genome is organized in 8 segments, short ‘mini RNA chromosomes” of negative-sense (non-coding) polarity that are packaged inside virions after the virus completes a life cycle in infected cells. IAV, like any other virus, is an obligate parasite as it requires cellular functions to multiply and disseminate. However, unlike most other RNA viruses, it replicates in the nucleus, a feature that evolutionarily ‘pushed’ a number of viral proteins to adapt and coopt (or interfere) with host chromatin-based regulatory processes in the infected cell1,2.
All IAV encode for a non-structural protein called NS1, whose function is to antagonize host antiviral response3. NS1-mediated host-antagonism occurs through multiple mechanisms, including inhibition of host-sensing of the virus4 and suppression of host functions that are detrimental to the virus such as host translation5 and inflammatory gene expression1,6. NS1 protein sequences differ in different strains. Only a few domains of NS1 have been associated to being pathogenic determinants in multiple strains, such as the RNA binding domain7 and the C-terminal domain8. Interestingly, the C-terminus is among the most divergent sequences within the NS1 protein, and it is unstructured1,9,10. These features are reminiscent of IDR, or unstructured domains often referred to as SLIMs (short linear modules), peptide motifs, or linear domains11. Pathogens often use IDRs to promote novel and evolvable interactions to adapt to new hosts and to enhance transmission and virulence12.
In this manuscript, we used Influenza 1918 virus13,14, a pandemic IAV that caused the worst pandemic to date15, as a case study to identify novel feature of IAV-host interaction and adaptation mediated by IDRs. We show that 1918 IAV encodes a NS1 that contains a unique C-terminal domain composed of a small ubiquitin-like modifier (SUMO) site embedded in a PDZ-Binding Domain (PDZBD) consensus sequence. We show that this extended SUMO consensus or SUP (SUMO inside PDZBD) domain predicts site-specific SUMOylation of 1918 NS1 during infection. Using viral engineering, we developed a virus expressing unmodified 1918 NS1 and a mutant expressing a constitutively SUMOylated NS1. These viruses promoted the discovery that, IAV elicits global deregulation of RNAPII transcription termination by impairing 3’-end cleavage and termination. This effect is augmented by NS1 SUMOylation, which increases partitioning of NS1 in nuclear granules containing 3’-end cleavage factors. Termination defects lead to RNAPII travelling through intergenic regions, causing formation of aberrant mRNAs, which ultimately results in global gene downregulation. To determine whether this event is dependent on NS1 and specific to 1918 IAV, we analyzed host transcriptional responses to non-pandemic IAV. Overall, our results indicate that IAV-induced 3’-end termination defects is a general feature of IAV infection that is dependent on NS1 expression modulated by post-translational modification of the unstructured region of NS1. Our data support the idea that study of viral polymorphic proteins can reveal important molecular events at play during infection and provide novel insights about how regulatory event occurring at steady state can be influenced by host and viral genetics.
Results
A unique domain present in the NS1 of 1918 influenza virus
Influenza A virus (IAV) is a major threat to human health. Along with its structural proteins, IAV encodes the non-structural protein NS1, which is an antagonist of cellular anti-viral responses16,17. We surveyed highly pathogenic IAV strains for the presence of unique protein domains and found a unique sequence present in the C-terminal domain of NS1 from the Brevig/mission strain (Fig. 1A and Supplementary Fig. 1A). This strain is responsible for the pandemic influenza that occurred in 191814. Interestingly, different strains of IAV bear divergent NS1 C-termini (NS1 tail) (Fig. 1B, Supplementary Fig. 1A and 1B), and NS1 tails have been linked to host tropism and virulence8,18. The 1918 NS1 (hereafter referred to as NS1) C-terminus ends with the sequence: 226IKSEV230 (Fig. 1A). We realize that this sequence is a SUMOylation consensus site (ψ K x E) embedded in a PDZBD (Fig. 1A). We examined whether this domain predicts a functional acceptor site for SUMO conjugation. As shown in Fig. 1C, the K227 site is the only one modified by SUMO, while the only other canonical SUMO consensus in position 68–71 is not. To check whether the 1918-like domain can confer SUMOylation to a non-cognate NS1, we generated chimeric NS1s bearing the residues 1–225 of H3N2 NS1 (A/New York/739/1994) and residues 226–230 of 1918 or 1918-like sequences from avian isolates. As shown in Fig. 1D, the tail-swapping confers SUMOylation in trans. We then assessed whether NS1 is modified during infection. To do so, we generated, using reverse genetics19, a chimeric virus with the first 7 segments of A/Puerto Rico/8/1934 H1N1 strain (PR8) and the 8th segment of the 1918 strain (reassortant 7+1, hereafter referred to as NS1 virus). We used this virus to infect A549 human epithelial cells. Immunoprecipitation (IP) of NS1 followed by Western blotting (WB) using anti-NS1 and anti-SUMO2–3 antibodies revealed that NS1 is SUMOylated during infection (Fig. 1E). This experiment also suggests that NS1 SUMOylation is controlled by SUMO de-conjugating enzymes, as shown by the loss of NS1 modification upon omission of the SUMO-protease inhibitor N-ethylmaleimide (NEM) from the cell extracts. Overall, our results indicate that the C-terminus of NS1 from the 1918 strain is modified by SUMO within the consensus sequence embedded in a PDZBD. The modified NS1 domain is short, present in an unstructured region of the protein1,9, and can be transferred to other substrates to confer SUMO conjugation. All these features are characteristics of SLIMs and peptide domains20. We therefore refer to this as SUP SLIM or SUP peptide domain.
Fig. 1. NS1 from 1918 pandemic influenza virus is SUMOylated in its unique C-terminal domain.
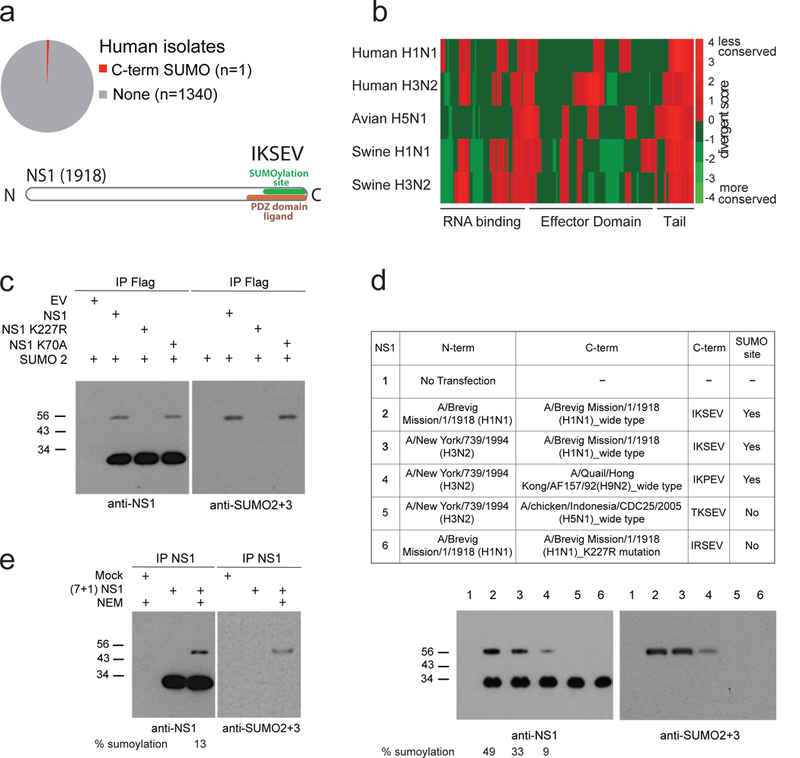
a, Venn diagram of influenza virus NS1s bearing a SUMO site in the C-terminal domain (tail). Bottom: amino acidic sequence of the NS1 tail from A/Brevig Mission/1/1918 (H1N1) bearing SUMOylation site and PDZ domain ligand site. 1918 NS1 C-termini is unique (1 out 1341 H1N1 from human isolates). b, Conservation plot of amino acidic sequences of NS1 among different viral isolates and hosts. The color-coding is the difference between the conservation score and average value of the score across the protein. Protein domains of NS1 are shown at the bottom. c, Ectopic expression of SUMO2 and the indicated NS1 (WT NS1 and the SUMO-consensus mutants at positions K70 and K227) in A549 cells. Immunoprecipitation (IP) with anti-Flag antibody, and Western Blotting (WB) analysis with anti-NS1 and anti-SUMO2/3 antibodies are shown. d, IP with anti-NS1 antibody and WB with anti-NS1 and anti-SUMO2/3 antibodies in A549 cells transfected with the indicated NS1s. The tail swapping of H3N2 NS1 was performed using the sequences from the indicated viruses, all of human origins but Hong Kong 1992 and Indonesia 2005 which are avian viruses. e, IP and WB with anti NS1- antibody of whole cellular extract from A549 cells infected with the reassortant NS1 virus bearing the segment 8 encoding for 1918 NS1 (see Figure 2B). NEM: N-Ethylmaleimide, a SUMO peptidase inhibitor
Rescue and analysis of 1918 NS1-SUMO fusion virus mutant
The observation that the PDZBD is modified by a bulky modification (molecular weight of SUMO is > 10 KDa) invites two possible scenarios: either SUMO conjugation alters PDZBD-PDZ protein interaction or the dual domain could simply predict functional SUMO sites that confer additional properties to substrates. To discriminate between these two possibilities, we undertook biochemical analyses as well as structural modeling. First, leveraging the fact that the PDZBD is at the C-terminus of NS1, we engineered a NS1 covalently fused with SUMO. This strategy has been used to identify ubiquitin-dependent mechanisms in histone biology21 as it avoids the confounding effect of unmodified substrates since post-translational modifications often occur only in a fraction of total proteins. We named this configuration NS1-SUMO (Fig. 2A). We also made a mutant of the lysine 227 that cannot undergo SUMO-conjugation (NS1-KR) (Fig. 2A). These mutations did not change NS1 nuclear localization (Supplementary Fig. 2A). Using Bio-Layer Interferometry (BLI), we then determined the dissociation constant (Kd) of the interaction between a prototypical PDZ domain-containing protein, PSD95, and the three NS1 variants. Our results show that the Kd values of the three NS1 with PSD95 are in the same order of magnitude (1.60, 2.30 and 0.63 μM, respectively) (Fig. 2A). These results were corroborated by pull-down assays (Supplementary Fig. 2B) and by modeling of NS1 binding with three PDZ proteins (PSD95, GIPC2, and DVL1) (Supplementary Fig. 2C-E and Supplementary Table 1). Overall, our results indicate that NS1 retains PDZ interactions independently of its conjugation by SUMO and suggest that SUMOylation of NS1 may have functions other than altering PDZ-PDZ interactions.
Fig. 2. 1918 NS1 wt and mutant viruses.
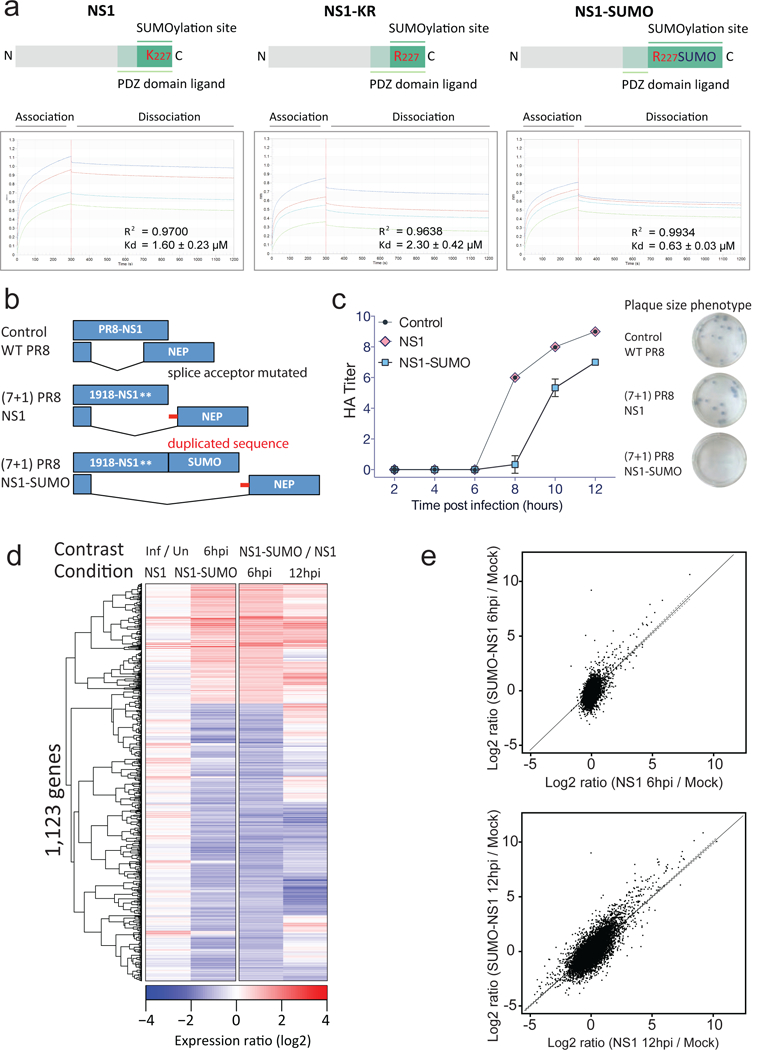
a, Top: schematic representations of the recombinant NS1, NS1-KR and NS1-SUMO proteins used for Bio-layer interferometry (BLI). Bottom: spectra showing BLI measurements of the interactions between recombinant PSD95 and NS1, NS1-KR and NS1-SUMO. Kd and R2 are shown. b, Schematic representation of WT PR8, the reassortant virus (7+1) PR8 NS1 bearing 7 segments of WT PR8 and the Segment 8 of the 1918 Influenza virus, and the reassortant virus (7+1) PR8 NS1-SUMO bearing the Segment 8 of the 1918 Influenza virus encoding a fusion protein between NS1 and SUMO2. c, Growth curves of the indicated viruses after infection of MDCK cells at MOI=1 (n=2). Plotted values represent one of two independent experiments. Plaque size from a representative experiment is shown (n=2). Error bars correspond to mean ± s.e.m. d, Hierarchical clustering of genes with significant changes in expression (FDR q<0.001) between A549 cells infected with NS1 or NS1-SUMO virus at indicated time (hpi). Rows show the log-fold change in expression (IDs not shown and color key at bottom), and columns represent different experimental conditions (labeled at the top and bottom of the panel). e, Scatterplot of cellular gene expression changes in A549 cells at 6 hours (upper panel) or 12 hours (lower panel) post-infection with PR8 NS1 or PR8 NS1-SUMO virus, relative to mock- infected (Mock) cells (log2 ratio). Solid and dotted lines correspond to the regression line and 95% confidence interval, respectively.
This prompted us to study the effect of SUMOylation of the 1918 SUP domain, and in an effort to probe the intrinsic role of SUMOylation during infection we then engineered two viruses respectively expressing NS1 and NS1-SUMO (hereafter referred to as NS1 virus and NS1-SUMO virus) (Fig. 2B). We predicted that the rescue of this virus would be safe as fusion of SUMO to NS1 would result in a loss-of-function with respect to virulence, similarly to other NS1 fusion viruses22. This prediction turned out to be correct, as demonstrated by hemagglutination (HA) assays (Fig. 2C) and severely impaired plaque formation (Fig. 2C, right panel). Plaque-sequencing indicated that the delayed growth of the NS1-SUMO virus at low multiplicity of infection (MOI) was associated with a reversion to its wild-type virotype (Supplementary Fig. 2F). The reduced virulence of NS1-SUMO virus may be driven by formation of many defective particles, impaired assembly, inefficient cell-to-cell spreading or by indirect effect caused by the fact that SUMOylation controls NS1 levels (Supplementary Fig. 3A), which impeded the rescue of the NS1-KR mutant23.
Despite having reduced pathogenicity, the NS1-SUMO virus still retained the capability to transiently infect at high MOIs, allowing us to monitor early events that are independent of virus budding, egress and re-infection of nearby cells. We thus compared NS1 and NS1-SUMO viruses in experiments conducted at high MOI. A549 cells were infected with NS1 or NS1-SUMO viruses at a MOI of 3, and total RNA was extracted at 6 and 12 hours post-infection (hpi). RNA-sequencing (RNA-Seq) analysis showed that NS1 and NS1-SUMO RNA level were comparable (Supplementary Fig. 3B). Both viruses caused altered host gene expression of roughly 1000 shared genes (Fig. 2D and 2E), including upregulation of genes that are part of the host defense to infection, consistent with the high induction of cellular response genes upon 1918 infection24. Notably, infection caused the downregulation of most transcriptionally active genes, which accounted for approximately two thirds of genes whose expression was altered during infection (Fig. 2D). Compared to the NS1 virus, the NS1-SUMO virus displayed a higher magnitude of gene expression changes in both induced and suppressed genes at both 6 and 12 hpi (Fig. 2D). Importantly, both viruses upregulate and down regulated the same set of genes (Fig. 2E). Overall, these results suggest that NS1 controls the magnitude of induction and suppression at host genes. This prompted the investigation of how NS1 can achieve this effect during infection.
SUMOylation controls NS1 oligomeric assembly and partitioning in RNA granules
Since NS1 binds RNA25 and forms polymers in vitro26,27, we investigated the putative role of NS1 in the interaction with RNA granules. These compartments are nucleated by aggregation-prone proteins28 interacting with RNA-binding proteins29, and are regulated by post-translational modifications such as phosphorylation30, SUMOylation31 and possibly many more. SUMO enhances the valency and strength of interaction between scaffold and client proteins of RNA granules31. We thus performed Semi-Denaturating Detergent Agarose Gel Electrophoresis (SDD-AGE), which allows the resolution of high molecular weight (HMW) complexes formed as a result of protein multimerization, a feature that can influence partitioning into RNA granules. Results from this assay indicate that NS1 forms HMW complexes during infection, and that this feature is enhanced by SUMO-fusion (Fig. 3A). We then took advantage of the recent discovery that the synthetic compound biotinylated isoxazole (B-isox) can selectively precipitate RNA granule components32. We synthesized B-isox (Supplementary Fig. 4A and 4B) and performed selective precipitation from total cell extracts derived from control and infected cells. Three conclusions could be drawn from the results of this analysis. First, we confirmed previous findings indicating that B-isox treatment induces the selective precipitation of RNA granule proteins in a dose-dependent manner (Supplementary Fig. 4C, 4D and Supplementary Table 2)32. Second, quantitative mass-spectrometry analysis indicated that infection affects RNA granule composition (Supplementary Table 3). Third, SUMOylation of NS1 increases NS1’s partitioning in RNA granules (Fig. 3B) and interaction with proteins involved in mRNA processing (Fig. 3C, left panel). This was further supported by the validation experiments, which showed NS1-SUMO-dependent enrichment with the cleavage and polyadenylation specificity factor (CPSF) and absence of enrichment of other proteins known to be present in RNA granules (FUS and EWS) (Fig. 3C, right panel, and Supplementary Table 3). Since NS1 from other strains are known to antagonize antiviral host gene expression1,3,16,17 and recent findings indicate that phase-separated RNA granules play a key role in controlling RNA polymerase II (RNAPII)30,33–36, we therefore investigated the relationship between NS1 expression and gene suppression by looking at RNAPII activity and mRNA levels upon infection with 1918 viruses and non-pandemic strains proficient and deficient for NS1.
Fig. 3. SUMOylation of NS1 induces oligomeric assembly and increases pervasive RNAPII termination defects at host transcripts.
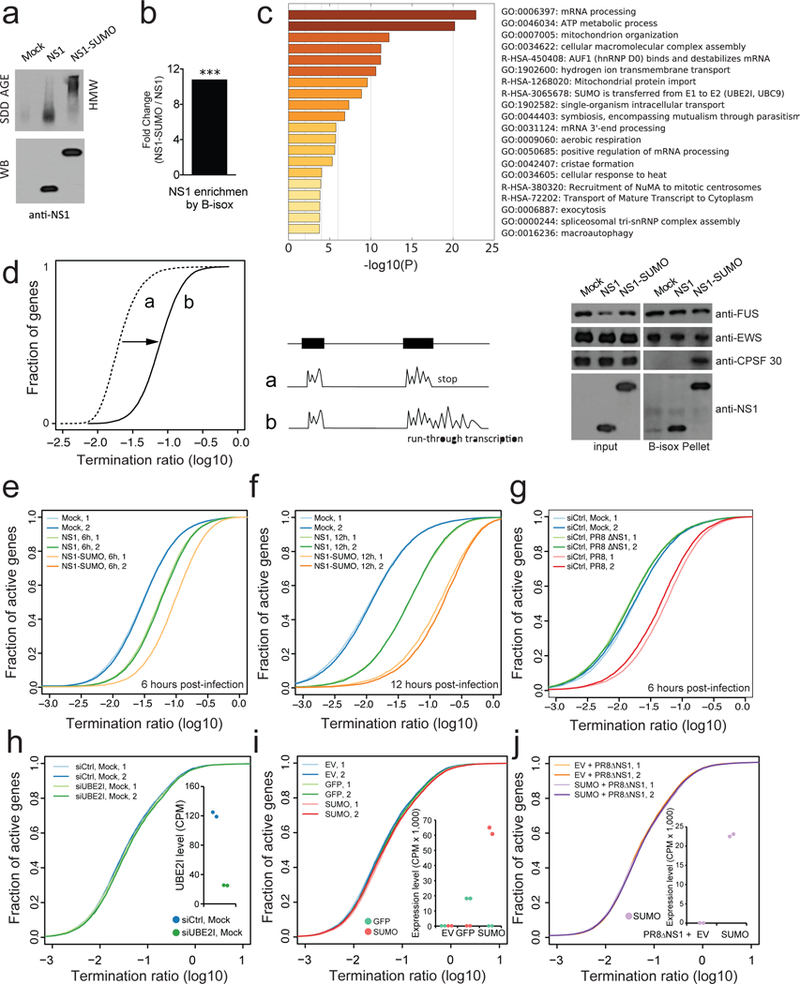
a, SDD-AGE (top) and WB (bottom) analysis on NS1 and NS1-SUMO virus infected lysates of A549. Mock: uninfected control. b, B-isox enrichment of NS1 from A549 lysate infected by NS1 and NS1-SUMO virus as analyzed by mass-spectrometry. ***, p value: 1.98*10−3. c, GO enrichment analysis of biological processes of B-isox enriched proteins during NS1 and NS1-SUMO infection (left panel), WB of the input and B-isox precipitates from NS1 and NS1-SUMO virus infected A549 lysates by the indicated antibodies (right panel). FUS and EWS are controls for resident RNA granule proteins. d, Schematic of the approach used to detect a relative increase in transcript levels in 3’ end gene-flanking regions. For each gene, a 3-prime transcript ratio (Termination ratio, TR) was calculated as the average number of total RNA-Seq reads per bp in 5-kilobase 3’ gene-flanking regions, divided by that in exonic regions. A global relative increase in 3’ transcript levels results in a horizontal transformation of the cumulative TR plot, as indicated. e-f, TR plots at termination region of active genes (RPKM > 1 in 50% of samples) in uninfected A549 cells and 6 hpi (e) or 12 hpi (f) with NS1 or NS1-SUMO virus. Data is shown for two replicates in each condition (n=2). g, TR plots at termination region of active genes (RPKM > 1 in 50% of samples) in uninfected A549 cells and 6 hpi with PR8 or ∆NS1 virus. Data is shown for two replicates in each condition. h-j, TR plots at termination region of uninfected A549 cells depleted for Ubiquitin Conjugating Enzyme E2 (siUBE2I) and control (siCtrl) (h), transfected with either empty vector (EV), or a vector expressing GFP or SUMO (i), and infected with PR8ΔNS1 A459 cells transfected with an empty vector (EV) or a vector expressing SUMO (j). The insets show the RNA expression levels of UBE2I in siCtrl or siUBE2I treated cells (h) and of GFP and/or SUMO in each condition (i-j) as counts per million sequenced reads (CPM). Results are shown for two biological replicates.
SUMOylation of NS1 enhances suppression of the host transcriptional response via interference with RNAPII termination
Due to the known role of CPSF complex in 3’-end cleavage and termination of RNAPII37, we first analyzed genome-wide RNAPII travelling ratios (TRs) at transcriptional termination sites, namely termination ratio, at both 6 and 12 hpi (Fig. 3D). This analysis indicated that downregulation of gene expression upon infection is associated with a genome-wide deregulation of RNAPII 3’-end cleavage and termination. This was evidenced by an increase in run-through transcription, as judged by high levels of RNA after the 3’-end at infection-regulated host genes at both 6 (Fig. 3E) and 12 hpi (Fig. 3F). Notably, this effect was evident during NS1 virus infection and was magnified upon NS1-SUMO virus infection (Fig. 3E and 3F). To check whether run-through transcription is also a feature of non-1918 viruses and dependent on NS1 expression, we used WT and NS1-deficient PR8 virus (hereafter referred to as dNS1 virus)6. Comparison between termination ratios induced by these two viruses indicated that while WT PR8 virus infection causes RNAPII run-through transcription, this effect was abrogated in dNS1 virus infected cells (Fig. 3G). Importantly, downregulation of Ube2L, an enzyme required for SUMOylation of substrate proteins, did not induce RNAPII termination defects (Fig. 3H). Additionally, overexpression of GFP or SUMO in uninfected cells did not induce RNAPII run-through (Fig. 3I), and similar run-through was seen in infection with WT H5N1 IAV and a modified virus encoding a GFP-fused NS1 (Supplementary Fig. 4E). Lastly, overexpression of SUMO in dNS1 virus-infected cells did not affect RNAPII at termination region (Fig. 3J). Overall, these results indicate that genome-wide deregulation of RNAPII termination is 1) a feature of IAV infection, 2) dependent on NS1 expression and 3) controlled by SUMOylation of the 1918 NS1 SUP.
Transcriptional shut-off during infection
RNAPII run-through occurring during infection can be functionally relevant if it is linked to gene repression of factors involved in the orchestration of the antiviral response. To explore this, we analyzed the relationship between the change in run-through transcription and expression changes in NS1 vs NS1-SUMO virus-infected cells by cross-comparing transcript density at 3’-end of genes and percentage of gene repression. While virtually all genes experienced increased run-through (positive signal > 0 of all gene curves in Supplementary Fig. 5A in NS1-SUMO vs. NS1 conditions), the effect was exacerbated at genes with decreased expression in NS1-SUMO. This implicates a key role for run-through transcription in down-regulating constitutively expressed genes important for homeostasis of cell division (e.g., CDC25A and MYC) and metabolism (e. g., PDK4 and RANBP6), apoptosis (e. g., MCL1 and SOX4) and cell defense (e.g., NFKBIA and CXCL1) (Supplementary Table 4, and representative gene-browser tracks, Supplementary Fig. 5B). Although some upregulated genes enriched for host antiviral functions (e.g. IFIT1 and IFIT2) experienced run-through, their induction exceeded any negative impact of the increased run-through transcription seen in NS1-SUMO virus-infected cells. Notably, RNAPII run-through upon infection was universally seen at all transcriptionally active genes but four (LY6E, APOL1, DEFB1, and IFI6, Supplementary Table 4), possibly suggesting unique features of their 3’-end. Overall, our data demonstrate that IAV infection can cause a global effect on gene expression by de-regulating RNAPII termination. This effect undermines the activity of many cell defense genes and constitutively expressed genes that play a key role in cell defense. It is modulated by a post-translational modification of a viral antagonist, and may represent a general way by which viruses antagonize hosts via inducing a state of transcriptional shut-off in the infected cells.
Mechanism of run-through transcription
The defect in RNAPII cleavage and increased run into distal extragenic regions after the 3’-end of genes observed during viral infection could be the result of deregulated termination and/or other regulatory events that indirectly affect termination. To test whether this effect could be driven by inhibition of CPSF, we first reanalyzed published RNA-Seq datasets in controls and cells depleted for CPSF73, an essential subunit of the CPSF complex38. Our analysis indicates that loss of function of CPSF activity induces RNAPII termination defects at all active genes (Fig. 4A), recapitulating the effect caused by NS1 during infection. Since CPSF also plays a key role in alternative splicing39–41, we analyzed datasets taken from cells treated with the splicing inhibitors spliceostatin and isoginkgetin42. Splicing inhibition induced pervasive RNAPII run-through, as evidenced by the increase of run-through ratio in treated cells (Fig. 4B). These data suggest that RNAPII run-through upon infection could be caused by pervasive defects in splicing rather than impaired 3’-end cleavage and termination. We thus analyzed genome-wide splicing efficiencies upon infection with NS1 and NS1-SUMO viruses. As shown in Fig. 4C-E, global splicing defects were detected only at 12 hpi, while deregulation of termination was already evident at 6 hpi (Fig. 3E). No increase of splicing defects was evident in genes with multiple exons compared with mono-exonic genes (Supplementary Fig. 6A). To gain further support to the idea that transcription termination defects precede splicing defects, we performed Iso-sequencing (Iso-seq), a method that allows to detect and define all splicing isoforms generated by a given gene on a genome-wide scale through long read sequencing (see Methods). Iso-seq of mRNA from A549 cells that were mock-infected or infected with WT or NS1-SUMO virus indicated that infection does not cause a pervasive impact on isoforms expression at 6 hpi. Despite this, infection induces some ‘non-physiological’ transcripts poorly detected in mock infected cells. These RNA biotypes, potentially generated from interference with mRNA maturation, are increased in NS1-SUMO infection (Fig. 4F, left panel). Among the affected genes, NS1-SUMO infection has a stronger effect than NS1 infection in generating intron retention and RNAPII run-through even of poly A transcripts. Furthermore, analysis of long reads mapping to the viral segment 8 demonstrated that, during infection with NS1-SUMO, all the mRNAs were full-length (including the SUMO-coding region) rather than truncated (Fig. 4F, right panel).
Fig. 4. Relationship between RNAPII run-through and splicing defect.
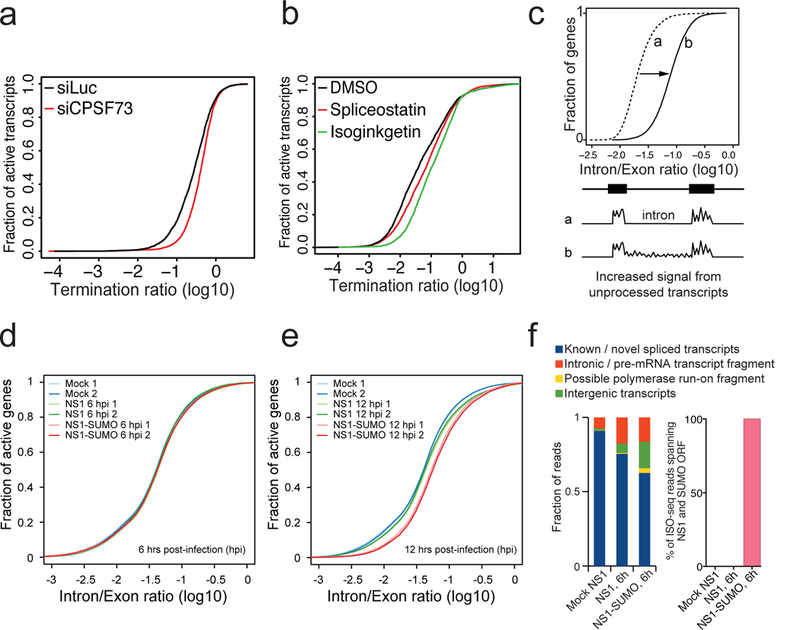
a, TR plots at termination region of uninfected cells depleted for CPSF73, one subunit of the CPSF complex, (siCPSF73) and control (siLuc). b, TR plots at termination region of uninfected cells treated with the indicated inhibitors and control (DMSO). c, Schematic of the approach used to detect a relative accumulation of unprocessed intronic transcripts. Analogous to the TR plots, an intron/exon transcript ratio was calculated by dividing the average read coverage per basepair in 5,000 bp intronic regions directly flanking an upstream exon, by the average read coverage of the upstream exon. An increase in unprocessed intronic transcripts results in horizontal transformation of the cumulative intron/exon ratio plots, as indicated. d-e, Cumulative Intron/Exon ratio distributions of active genes (RPKM > 1 in 50% of samples) in uninfected A549 cells and 6 hpi with NS1 or NS1-SUMO virus (d), and at 12 hpi with NS1 or NS1-SUMO virus (e). Data is shown for two replicates in each condition. (f) Classification of all IsoSeq circular consensus (CCS) reads based on comparisons with GENCODE v38 transcript annotations using cuffcompare. (g) Percentage of reads containing full length RNA from the segment 8 in cells infected with NS1-SUMO virus.
Overall, our data on total RNA seq and Iso-Seq indicate that RNAPII run-through during IAV infection (and the downregulation of most active genes) is due to interference with 3’-end cleavage and termination, which in turn causes a defect in splicing at a later time as infection progresses, and support the notion that defective RNAPII termination can cause delay in RNAPII initiation and splicing fidelity43–46.
Finally, downregulation of genes that experience RNAPII run-through in NS1-SUMO virus vs NS1 virus infected cells is likely to be caused by both delayed/reduced initiation and the direct effect of run-through RNAPII into neighboring genes, as suggested by the linear relationship between each gene increased run-through in cells infected with NS1-SUMO virus vs NS1 virus and changes in gene expression (Supplementary Fig. 6B).
Lastly, the fact that chemical inhibition of splicing causes RNAPII run-through, albeit to a less dramatic level than during infection, indicates interconnection between these two biological processes. This is further supported by evidences showing that last-exon splicing and RNAPII termination are mechanistically and evolutionarily linked events39,47,48.
Overall, our data demonstrate that chemical or virus-induced interference with co-transcriptional events can generate pervasive RNAPII run-through.
Discussion
By sampling sequences from highly pathogenic viruses, we identified the presence in the C-terminal region of NS1 from 1918 pandemic IAV of a novel peptide domain constituted of a SUMO site embedded in a PDZ binding domain. The SUP domain confers SUMOylation of NS1 during infection. The rescue of a constitutively SUMOylated NS1 virus indicated that, during infection, SUMO enhances NS1 oligomeric assembly and association with nuclear ribonucleoprotein complexes controlling RNAPII activity. NS1-SUMO infection causes pervasive RNAPII run-through into downstream extragenic DNA and nearby genes.
This result prompted us to verify whether infection with other IAV strains displayed any evidence of transcription termination defects. Indeed, infection with WT PR8 virus induced RNAPII run-through, an effect also recently recapitulated by two independent studies that mapped nascent RNA –seq49 and chromatin profiling of RNAPII50 (Heinz et al, Cell, in press) during IAV infection with multiple strains. Unlike WT PR8, NS1-deficient virus did not induce RNAPII run through. This provides genetic evidence for the role of NS1 in inducing aberrant RNA biogenesis and suggests that different influenza strains have evolved to target factors controlling termination. On this note, it is important to emphasize that, despite different NS1s bind to transcription regulators (like CPSF30) in vitro with different strength, recent evidences suggest that cognate interaction (from viral protein of the same viral strain) between viral polymerase and NS1 can increase interaction strengths and inhibitory activity during infection51.
Based on this and our results, it is tempting to speculate that all IAVs have evolved to inhibit 3’-end formation through different strategies – like by using direct binding via viral protein or viral complex to host factors or by affecting the microenvironment in which termination occurs. This rationale is reinforced by the knowledge that pathogen-derived effector proteins tend to target similar key event controlling cell functions52–56.
Effect of RNAPII run-through in gene induction and gene repression
While most genes affected by RNAPII run-through were downregulated, some host antiviral genes were upregulated despite being affected by run-through. This is possibly due to unique regulatory sequences and events that control their 3’-ends or by the fact that their extremely high induction level and burst rate allows to partially bypass/escape termination inhibition. Overall, our results implicate deregulated RNAPII termination in suppression of active genes and the establishment of a transcriptional shut off. Many viruses cause such effect by means of different strategies of host interference57–61. We suggest that different mechanisms of host transcriptional shut off are linked to virus-specific requirements for their life cycle. For example, IAV requires high levels of host gene transcription (or initiation events) in the first hours of infection to mediate cap-snatching and to coordinate viral mRNA production and viral protein expression2. As such, gene suppression needs to be in sync with viral replication. Overall, our result suggests that virus-induced RNAPII run-through serves to kinetically control host gene expression to maximize fitness. We show that this effect is achieved by NS1 and modulated by SUMOylation in the case of 1918 IAV. Constitutive NS1 SUMOylation is associated with an increased degree of RNAPII run-through at most active loci and concomitantly with gene suppression. Interestingly, NS1-SUMO induces also higher levels of upregulation of some host response genes (which partially escape downregulation). These results suggest that SUMOylation skews NS1 activity toward the control of RNAPII termination and, as seen in the NS1-SUMO virus, constitutive SUMOylation might limit other NS1 host antagonist function. This is supported by the limited infectivity in multicycle growth of the NS1-SUMO virus, which cannot undergo dynamic control of this post-translational modification. In this respect, it is important to consider that SUMOylation can be a signal for NS1 degradation. Several transcription factors are known to function transiently on chromatin and undergo rapid turnover62. NS1 may have adopted such feature to provide a titratable strategy to interfere with host transcription. Overall, our data suggest that 1918 IAV uses dynamic regulation of post-translational modifications to switch between various host-antagonizing strategies. On this note, we show that polymorphism and damaging-mutations in SUMO-modifying enzymes exist in humans (Supplementary Fig. 6C and Supplementary Table 5) and they could affect host response to 1918-like IAVs.
Evolutionary considerations
Sequence analysis of pathogen-derived polypeptides can provide us with valuable information about sequences governing protein-protein interaction principles12,63. Proteins encoded by viruses, especially the ones that have evolved to suppress the host response or co-opt cellular signaling to increase pathogen fitness (viral antagonists) display multivalent interaction surfaces that allow simultaneous binding with many host proteins or/and nucleic acids8,64,65.
This functional pleiotropy can be achieved by evolving peptide domains and IDRs66–69. There are key defining features of pathogen-derived linear domains and IDRs: 1) they are located at the C- or N-terminus of a protein70; 2) they are enriched in viral proteins that reside in the nucleus of infected cells63, and 3) they contain post-translational modification sites71. The SUP domain in 1918 NS1 fits all these parameters.
Most IDRs arise de novo via convergent evolution11,72,73, and the 1918 SUP is not conserved in other influenza viruses. This suggests that it either evolved independently or was co-opted via viral mimicry. Previous work has identified how a circulating strain of IAV (H3N2) employed molecular mimicry to evolve histone-like sequences1. Despite the fact that the 1918 IAV circulated for a short period of time, the NS1 SUP domain may have undergone high levels of mutagenesis and fixation in different strains. Intriguingly, the NS1 tail sequence of different strains can predict the host species from which the virus has been isolated18. Understanding the molecular mechanisms underlying this correlation can be informative in the context of surveillance and zoonosis74. Notably, the only three sequences that contain the NS1 SUP domain have been isolated from avian species, which are key host reservoirs linked to acquisition of increased pathogenicity and pandemic potentials75,76. Lastly, our work shows that despite being close to the centennial of its insurgence, 1918 influenza research can still instruct us about biological principles controlling pathogens and humans.
Materials and Methods
Reagents
Chemical synthesis of B-isox: 5-(2-thienyl)-3-isoxazolecarboxylic acid (763109–71-3), N-hydroxysuccinimide (HOSu, 6066–82-6), N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDC·HCl, 25952–53-8), 6-amino-1-hexanol (4048–33-3), biotin (B4639), and 4-(dimethylamino)pyridine (DMAP, 1122–58-3) from Sigma-Aldrich. B-isox mediated precipitation of proteins: Halt™ Protease Inhibitor Cocktail (78438), Dynabeads™ MyOne™ Streptavidin C1 (65001), SilverQuest™ Silver Staining Kit (LC6070), and GelCode™ Blue Stain Reagent (24590) from Thermo Fisher Scientific. Expression and purification of recombinant proteins: isopropyl β-D-1-thiogalactopyranoside (IPTG, I6758), cOmpleteTM, EDTA-free Protease Inhibitor Cocktail Tablets (11873580001) from Sigma-Aldrich, Ni-NTA Agarose (30210, QIAGEN), and Nanosep® Centrifugal Devices with Omega™ Membrane, 3K (OD003C33, Pall Corporation). Bio-layer interferometry (BLI): EZ-LinkTM Sulfo-NHS-LC-Biotin (21327, Thermo Fisher Scientific) and Dip and ReadTM Streptavidin (SA) Biosensors (18–5021, ForteBio). Immunofluorescence (IF): Lipofectamine 2000 (11668027) and DAPI (D1306) from Thermo Fisher Scientific. Pull down assay: anti-FLAG® M2 Affinity Gel (A2220, Thermo Fisher Scientific). Proteasome inhibition: MG132 (M7449, Sigma-Aldrich). Directional RNA-Sequencing (RNA-Seq): Ribo-Zero Gold rRNA Removal Kit (Human/Mouse/Rat) (MRZG12324, illumina), Agencourt AMPure XP (A63882, Beckman Coulter Life Sciences), TruSeq Directional Library Prep Kit (RS-122–2203, illumina), and BluePippin 2% M1 gels (BEF2010, Sage Scientific). Antibodies: anti-NS1 from Dr. Adolfo Garcia-Sastre’s lab. Anti-FLAG (F7425, Sigma-Aldrich), anti-SUMO 2/3 (07–2167, Millipore), anti-FUS (A300–302A), anti-EWS (A300–418A) and anti-CPSF30 (A301–585A-M) from Bethyl Laboratories. Plasmids: pcDNA5 vectors encoding indicated genes were used to express corresponding recombinant proteins in mammalian system. pET28c vectors encoding indicated genes were used to express recombinant proteins in bacterial system. A549 cells (adenocarcinomic human alveolar basal epithelial cells), HEK293T cells (human embryonic kidney cells) and MDCK (Madin-Darby canine kidney cells) were originally obtained from the American Type Culture Collection (ATCC). The cell cultures were tested and guaranteed to be mycoplasma-free.
Conservation analysis
All complete NS1 amino acid sequences were downloaded from the NCBI Influenza Virus Database for Human and Swine H1N1 and H3N2 and Avian H5N1. A multiple alignment was made of all sequences in the 5 strains and for all sequences together as well. The multiple alignments were performed using multialign in MATLAB. For each group consensus sequence and conservation score was defined using seqconsensus in MATLAB. The average score was plotted across a 10 residue sliding window for each group of proteins and then normalized so the maximum and minimum score was consistent across groups. These values were plotted in the accompanying heatmap.
Modeling of PDZ/peptide/SUMO complexes
Initial models of the PDZ domains with NS1 peptides were generated using MODELLER 9.12 using the crystal structures of the PDZ domains or homology models when needed (supplementary Table 1) and the sequences of the peptides77. Seven residues from NS1 peptide were modeled (sequence: RTIKSEV). The crystal structure of the Par-6 PDZ domain in complex with the Pals1 peptide was used as a template peptide conformation in all NS1 complexes78. The initial PDZ/peptide complexes were then refined using the FlexPepDock server with default parameters79. The top-scoring model from FlexPepDock for each complex was selected and the SUMO conjugated version was modeled by bonding the C-terminal glycine residue of the SUMO structure to the lysine residue in the peptide80. The possible spatial orientations of the conjugated SUMO domain were explored by varying the conformation of the lysine side chain in the peptides.
Transfection, Infection, and Western Blot
For transfected samples, 70% confluent A549 cells were transfected with the vectors encoding the indicated genes by using Lipofectamine for 2 days. The cells were washed by cold PBS twice, scraped and suspended in the lysis buffer containing 20 mM Tris·HCl (pH=7.5), 150 mM NaCl, 5 mM MgCl2, 20mM β-mercaptoethanol, 0.5% NP-40, 10% glycerol, 20 mM NEM and 2X Halt protease inhibitor. The cells were then lysed at 4 °C by using BIORUPTOR® 300 for 10 cycles of 30 seconds Time ON and 30 seconds Time OFF at high level of sonication. For infected samples, 100% confluent A549 cells were infected by indicated viruses at indicated MOI and collected at different time points. The cells were lysed as described above. Crude lysates were analyzed by Western Blot.
Semi-denaturating detergent agarose gel electrophoresis (SDD-AGE)
The infected A549 lysates were prepared as described above and centrifuged at 2, 000 rcf at 4 °C for 2 minutes. The supernatants were taken out carefully and incubated with 4X SDD-AGE loading buffer containing 2X TAE buffer, 20% glycerol, 8% SDS and 1% bromophenol blue at room temperature for 5 minutes. The samples were loaded on 1.5% agarose gel containing 0.1% SDS. The gel was running in 1X TAE buffer with 0.1% SDS at a constant voltage of 40 V at 4 °C for 3 hours. Capillary transfer and Western Blot were processed81.
Purification of recombinant proteins
Recombinant proteins His tagged NS1, NS1-KR, NS1-SUMO and PSD95 were overexpressed in E. Coli BL21(DE3), OD600 nm=0.6, with the induction of IPTG at 1 mM and the incubation at 30 °C overnight. Bacteria were harvested, washed by PBS twice and suspended in lysis buffer containing 50 mM NaH2PO4, 300 mM NaCl and 10 mM imidazole at pH 8.0 complemented with EDTA-free protease inhibitor. Bacteria mixture were placed on ice and sonicated by Misonix 3000 for 6 cycles of 10 seconds Pulse “ON” and 10 seconds Pulse “OFF” at Initial Output Level 10. The lysate was centrifuged at 15, 000 rpm at 4 °C for 15 minutes. The soluble fraction of the lysate was mixed with Ni-NTA resin and gently rotated at 4 °C overnight. The Ni-NTA agarose was fully washed with buffer containing 50 mM NaH2PO4, 300 mM NaCl and 20 mM imidazole at pH 8.0. The proteins were finally eluted by elution buffer containing 50 mM NaH2PO4, 300 mM NaCl and 250 mM imidazole at pH 8.0. The purified proteins were concentrated by Nanosep (3K) and the concentrations were determined by NanoDrop 1000 Spectrophotometer. The purity of recombinant proteins was confirmed by SDS-PAGE and staining by Coomassie Blue.
BLI
Recombinant PSD95 was biotinylated by reacting with Sulfo-NHS-LC-Biotin with molar coupling ratio of 1:1. Extra biotin reagent was fully removed through buffer exchange for 4 times after biotinylation reaction was completed. Stock of biotinylated PSD95 was diluted to 20 ug/mL in PBS and stocks of NS1, NS1-KR and NS1-SUMO were serially diluted to 10, 5, 2.5 and 1.25 uM in PBS, respectively. BLI assay was performed on a ForteBio OctetRED 96. SA biosensors were washed first by PBS for 60 seconds, immobilized subsequently with biotinylated PSD95 (20 ug/mL) for 300 seconds, washed for the second time by PBS for 60 seconds, associated respectively with blank (PBS), NS1, NS1-KR and NS1-SUMO at serial diluted concentrations (10, 5, 2.5 and 1.25 μM) for 300 seconds and dissociated finally by washing with PBS for 900 seconds. All the steps were performed with agitation at 1, 000 rpm at 30 °C. BLI data was processed and analyzed by using software Data Analysis 8.2. Steady state analyses eventually generated Kd and R2 value82.
Pull-down assay
Purified FLAG tagged NS1, NS1-KR and NS1-SUMO proteins were incubated with anti-FLAG agarose in PBS by rotation at 4 °C for 1 hour. The beads were fully washed with washing buffer containing 50 mM Tris–HCl (pH 7.5), 200 mM NaCl, 1 mM EDTA and 0.2% NP40 followed by being washed with the same buffer without NP40. NS1, NS1-KR and NS1-SUMO bound agarose were respectively incubated with purified His tagged PSD95 in PBS by rotation at 4 °C for 1 hour (agarose washed by PBS was included as a control). The beads were fully washed with washing buffer. Bound proteins were then eluted by 3x FLAG peptide (0.25 mg/mL) by shaking agarose at 1, 000 rpm at 4 °C for 1 hour. The input and eluted proteins were run on SDS-PAGE and stained by Coomassie Blue.
Analysis of proteins stability
A549 cells were infected with NS1 and NS1-SUMO virus for 12 hours and sequentially treated with mg132 (50 nM) or cycloheximide (CHX, 50 ug/mL) for 0/4/8/24 hours. Cells were lysed and Western Blot was processed.
IF
The FLAG tagged GFP, NS1, NS1-KR and NS1-SUMO transfected A549 lysates were washed, fixed, permeabilized and stained with anti-GFP, anti-FLAG and DAPI. Images were observed under microscope, Zeiss Axio Imager Z2M and processed by Software, Zen.
Rescue of recombinant influenza viruses
Recombinant NS1 and NS1-SUMO viruses were prepared by plasmid transfection as previously described83. Two viruses rescued contained 7 segments from strain A/Puerto Rico/8/34 (PR8). For the NS1-SUMO version, a modified plasmid encoding the NS segment from strain A/Brevig Mission/1/1918 in which the two viral proteins (NS1 and NEP) are encoded in separated open reading frames was used. Plasmid pDZ_NS1 encoded the original amino acid sequence, with 3 silent changes to allow differentiation. The plasmid pDZ_NS1-SUMO contained the SUMO2 coding sequence as a fusion at the end of the NS1 open reading frame. Rescued viruses were amplified in embryonated chicken eggs and the NS segment was confirmed by sequencing. All work using infectious viruses containing sequences from A/Brevig Mission/1/1918 was performed in strict accordance with CDC guidelines for biosafety level 3 (BSL3) agents at the BSL3 laboratory of the Icahn School of Medicine at Mount Sinai, NY.
Single cycle growth curve
Confluent monolayers of MDCK cells growing in 6 well plates were infected in triplicate with the indicated viruses at a multiplicity of infection of 1 PFU per cell. After 1 h adsorption the inoculum was aspirated and replaced by 2 mL of infection medium (MEM supplemented with 1% antibiotics, 0.3% BSA and 1 ug/mL of TPCK treated trypsin). At the indicated times 200 uL of supernatant were collected for titration and replaced by 200 uL of fresh infection medium. Supernatants were titrated by hemaglutination assay using turkey red blood cells83.
Plaque phenotype
MDCK monolayers growing in 6 well plates were infected with serial dilutions of the indicated virus. After 1 hour adsorption the inoculum was aspirated and cells were overlaid with medium containing 0.6% agar and trypsin (1 ug/mL). After 2 days cells were fixed with 4% formaldehyde and plaques visualized with a monoclonal antibody against the viral protein NP.
Analysis of infectious viral progeny
Supernatants from a 24 hours infection were analyzed by plaque assay as described above. Isolated visible plaques were suspended in PBS and inoculated onto fresh MDCK cells. When cytopathic effect was observed, viral RNA was purified from the supernatant and amplified by RT-PCR using primers specific for the NS segment. The amplified product was purified from an agarose gel and sequenced using the same primers.
RT-PCR
A549 cells were lysed with Trizol reagent (Ambion, 15596026) followed by RNA extraction, and DNase digestion. cDNA was synthesized using high capacity cDNA Reverse transcription kit (Applied Biosystems, 4368813) according to manufacture protocol. Primers: b-actin forward, 5′-ACCTTCTACAATGAGCTGCG-3′; b-actin reverse, 5′-CCTGGATAGCAACGTACATGG-3′; GAPDH forward, 5′-TCTGACGCTGACTGGTTAGT; GAPDH reverse, 5’-GAGGGCACAGAAAGCAATAGAG; CDC25A forward, 5’-GTGGGATGGCCTTCAGATT; CDC25A reverse, 5’-CCATCAAGAACTAGGCAGAGAG.
Chemical synthesis of B-isox
B-isox was chemically synthesized according to the protocol as previously reported with minor modification84. A solution of 5-(2-thienyl)-3-isoxazolecarboxylic acid (a, 200 mg, 1 eq.), HOSu (239 mg, 2 eq.) and EDC·HCl (397 mg, 2 eq.) in 5 mL aqueous THF was stirring at room temperature overnight to produce chemical intermediate b. 6-amino-1-hexanol (243 mg, 2 eq.) was added into the mixture and reaction was processing by stirring at room temperature for additional 5 hours. The product c was then purified by applying CombiFlash® Purification Systems with gradient elution of MeOH and DCM. Chemical intermediate c (100 mg, 1 eq.) were next mixed with biotin (85 mg, 1 eq.), DMAP (6.7 mg, 0.2 eq.) and EDC·HCl (128 mg, 2 eq.) in 5 mL DCM. The reaction was stirring at room temperature for 3 days. The final aim product B-isox (d) was obtained with purity ≥ 98% through employing CombiFlash® Purification Systems with gradient elution of MeOH and DCM. Total yield was 11%. B-isox was characterized by HRMS. Calculation for C24H33N4O5S2, [M+H]+ and C48H65N8O10S4, [2M+H]+, were 521.1892 and 1041.3706, respectively, 521.2162 and 1041.3704 were found.
B-isox mediated precipitation
Six 15 cm dishes of 100% confluent HEK293T cells were lysed as described above. The crude lysates were centrifuged at 15, 000 rpm at 4 °C for 10 minutes to produce 6 supernatants (1 mL). Four supernatants (800 uL) were treated with B-isox (0, 10, 30, 100 uM) and two supernatants were treated with B-isox (0 and 100 uM) and streptavidin agarose as controls. The mixtures were rotated gently at 4 °C for 1 hour. The mixtures were spun at 15, 000 rpm for 10 minutes and the supernatants were removed. The pellets were washed by cold lysis buffer (20 mM Tris·HCl (pH=7.5), 150 mM NaCl, 5 mM MgCl2, 20mM β-mercaptoethanol, 0.5% NP-40, 10% glycerol, 20 mM NEM and 2X Halt protease inhibitor) twice and dissolved in 100 uL 2% SDS. The samples were boiled in 4X SDS loading buffer and run on SDS-PAGE. The gel was stained by silver staining. A549 cells were infected by Mock, NS1 and NS1-SUMO viruses at MOI of 3 for 12 hours and the infected lysates were precipitated by 100 uM B-isox as described above. The precipitated samples were analyzed by Western Blot.
Preparation of peptides for mass spectrometry (MS)
B-isox precipitated samples of A549 lysates infected by Mock, NS1 and NS1-SUMO viruses were separated by SDS-PAGE. Gel bands for each sample were excised and then diced into small sections (1 mm2). Gel pieces were dehydrated in 25 mM NH4HCO3 / 50% acetonitrile (ACN) for 10 minutes with vortexing in two consecutive rounds and then dried under vacuum centrifugation. Slices were rehydrated in 15 mM tris(2-carboxyethyl)phosphine (TCEP) / 25 mM NH4HCO3 and the samples were incubated for 20 minutes. Freshly prepared 1 M iodoacetamide / 25 mM NH4HCO3 was then added to attain a final concentration of 50 mM iodoacetamide and the samples were incubated in the dark for 20 minutes. The supernatants were removed and 25 mM NH4HCO3 was added to cover gel pieces. The samples were vortexed for 10 minutes, the supernatants were removed and 25 mM NH4HCO3 / 50% ACN was added. Following incubation with vortexing for 5 minutes, supernatants were removed and the gel slices were dried under vacuum centrifugation. A total of 0.5 ug trypsin in NH4HCO3 was added with enough volume to cover the gel slices and samples were incubated at 37 °C for digestion overnight. The supernatants containing extracted peptides were collected in separate lo-bind tubes and enough 50% ACN / 5% formic acid (FA) were added to the remaing gel slices. Samples were vortexed for 10 minutes and the supernatants were combined with the previous extractions. Repeat this step by using 100% CAN and the supernatants were combined with the previous combinations. Peptides were dried under vacuum centrifugation and suspended in 10 L of 3.0% ACN, 0.1% FA prior to MS analysis.
Protein identification by liquid chromatography tandem mass spectrometry (LC-MS/MS)
Digested peptides were subjected to LC-MS/MS analysis using an Easy-nLC 1000 coupled to a dual-pressure linear ion trap (Velos Pro) Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Online LC separation was performed using a 75 mm x 25 cm fused silica IntegraFrit capillary packed with 1.9 mm Reprosil-Pur C18 AQ reversed-phase resin. Peptides were eluted by a gradient of 5% to 30% ACN in 0.1% FA in 160 minutes delivered at a flow rate of 300 nL/minute. For each cycle, one full MS scan (150–1500 m/z, resolution of 120, 000) in the Orbitrap was followed by 20 data-dependent MS/MS scans fragmented by normalized collision energy (setting of 35%) and acquired in the linear ion trap. Target ions already acquired in MS/MS scans were dynamically excluded for 20 seconds. Raw MS files were analyzed by MaxQuant version 1.3.0.385 and MS/MS spectra searched by the Andromeda search engine86 against a database containing reviewed SwissProt human and influenza protein sequences (20, 194 in total)87. All runs were analyzed simultaneously to maximize the “match between runs” algorithm available on Maxquant. Multiplicity was set to 1 (as recommended for label free experiments) and a false discovery rate imposed to 0.01 for peptide and protein identification.
Statistical relative quantification of proteins and enrichment analysis
Normalization of raw peptide intensities, protein level abundance inference, and differential expression analysis (log2 fold change and p-values) were calculated using the open source R package MSstats version 3.3.1088. To ensure reproducibility, data was filtered to select for proteins identified in both biological replicas in at least one of the conditions compared. Differential expression values for proteins available in only one condition were estimated by imputation of the intensity values by random sampling from the lowest 20 normalized intensity values from the conditions where proteins were not detected.
Gene Ontology (GO) Enrichment Analysis of biological processes was performed using Panther89. Data analysis and integration was carried out using the R language for statistical computing and graphics.
Directional RNA-Seq
One ug of DNase-treated RNA was depleted of rRNAs using the Ribo-Zero Gold rRNA Removal Kit (Human/Mouse/Rat) according to the manufacturer’s instructions and purified post-depletion with 1.6X volume AMPure XP beads. Barcoded directional RNA-Seq libraries were then prepared using the TruSeq Directional Library Prep kit (Illumina), per kit instructions. PCR products were purified with 1.8X volume AMPure XP beads and fragments of 300–500 bp were size-selected using BluePippin 2% M1 gels (Sage Scientific). Afterwards, libraries were sequenced on the Illumina HiSeq 2500 platform in a 100 bp single-end read run format.
RNA-seq
After adapter removal with cutadapt90 and base quality trimming to remove 3’ read sequences if more than 20 bases with Q < 20 were present, paired-end reads were mapped to the human (hg38) reference genome using STAR91 and gene count summaries were generated using featureCounts92. Raw fragment (i.e. paired-end read) counts were then combined into a numeric matrix, with genes in rows and experiments in columns and used as input for differential gene expression analysis with the Bioconductor Limma package93 after multiple filtering steps to remove low-expressed genes. First, gene counts were converted to FPKM (fragments per kb per million reads) using the RSEM package94 with default settings in strand specific mode and only genes with expression levels above 1 FPKM in at least 50% of samples were retained for further analysis. Additional filtering removed genes with less than 50 total reads across all samples or less than 200 nucleotides in length. Finally, normalization factors were computed on the filtered data matrix using the weighted trimmed mean of M-values (TMM) method, followed by voom95 mean-variance transformation in preparation for Limma linear modeling. Data was fitted to a design matrix containing all sample groups and pairwise comparisons were performed between sample groups.
Termination ratio and intron/exon ratio calculation
To quantify an increase in transcription beyond transcription termination sites at the 3’ end of genes, we calculated a measure called the termination ratio (TR) that compares the ratio between the average read coverage in 3’ gene-flanking regions and the average read coverage per basepair (bp) of exonic sequence. We defined the 3’ flanking region between +1 to +5, 000 bp relative to most distal 3’ end of annotated transcripts for known genes. Analogously, to assess the accumulation of unprocessed transcripts that could be the result of a global defect in splicing we also calculated an intron/exon transcript density ratio for all introns. This measure was defined as the average read coverage per bp in 5, 000 bp intronic regions directly flanking an upstream exon, divided by the average read coverage of the upstream exon. Transcript densities in 3’ gene and exon-flanking were calculated after excluding any regions overlapping annotated genes. The termination and intron/exon transcript density ratios are analogous to the traveling ratio as described previously96.
Transcript isoform sequencing (Iso-seq)
RNA QC was performed according to Qubit (Qubit RNA HS Assay Kit, Thermo Fisher) and Agilent RNA Pico Kit protocols to assess quantity and quality (RIN integrity) of the samples, respectively. Ribosomal RNA depletion was performed according to the Thermo Fisher Scientific RiboMinus Transcriptome Isolation Kit, human/mouse. Magnetic beads were prepared by re-suspending the RiboMinus Magnetic Beads by thoroughly vortexing. For each sample, 250μl of the bead suspension was pipetted into a sterile tube. The tube were placed on a magnetic rack for 1 minute, the supernatant was removed, discarded and replaced with 250μl DEPC-treated water. The tube were placed on a magnetic rack for 1 minute, the supernatant was removed, discarded and replaced with, 250 μL Hybridization Buffer. The tube were placed on a magnetic rack for 1 minute, the supernatant was removed, discarded and replaced and resuspended with 100 μL Hybridization Buffer. The tubes were incubated at 37°C until use. Hybridization was performed by combining 20 μL of Poly (A)-tailed total RNA, 4 μL RIboMinus probe (100 pmol/μL) and 100 μL hybridization buffer. The tubes were incubated at 37°C for 5 minutes. The tubes were placed on ice for 30 seconds and centrifuged. 124 μL of the samples were transferred to the prepared RiboMinus Magnetic beads and mixed. The tubes were incubated at 37°C for 15 minutes. The tubes were placed on a magnetic rack for 1 minute. The supernatant was removed and transferred to a sterile tube. RNA QC was then repeated on each sample.
Isoform generation was performed according to the Isoform Sequencing (Iso-Seq ™) Template Preparation for Sequel Systems. The first strand synthesis was performed by addition of 1–3.5 μL 3’ SMART CDS Primer IIA, 1–3.5 μL RNA sample, and 0–2.5 μL Nuclease free water; for a total of 4.5 μL volume. The tubes were mixed by pipetting, centrifuged briefly, and incubated on a hot lid thermal cycler at 72°C for 3 minutes; slow ramp to 42°C at 0.1°C/sec for 2 minutes. During the incubation step the Master mix was prepared by addition of 2 μL 5X Frist strand buffer, 0.25 μL 100mM DTT, 1 μL 10mM dNTP, 1 μL 12 μM SMARTer II A Oligonucleotide, 0.25 μL RNase Inhibitor, 1 μL SMARTScribe Reverse Transcriptase, for a total volume of 5.5 μL per reaction. The master mix was heated at 42°C for 1 minute. Aliquots of 5.5 μL of master mix were added each reaction tube, and mixed gently by pipetting, and spun briefly. The tubes were incubated at 42°C for 90 minutes. The reaction was terminated by heating the tubes at 70°C for 10 minutes. The first strand reaction products were diluted by adding 90 μL of PacBio Elution buffer (EB).
cDNA amplification was performed as follows 24 × 50 μL PCR reactions were set up. Per sample, PCR master mix was prepared by addition of 240 μL 5X PrimeSTAR GXL Buffer, 240 μL filuted first-strand cDNA, 96 μL 10mM each dNTP Mix, 16 μL 5’ PCR primer II A, 576 μL nuclease free water and 24 μL 1.25U/μl PrimeSTAR GXL DNA polymerase. The samples were incubated at 98°C for 30 seconds; followed by 15 cycles: 98°C for 10 seconds, 65°C for 15 seconds, 68°C for 10 minutes; with a final extension time of 68°C for 5 minutes.
PCR products purification:
The 12 × 50 μL PCR reactions were pooled and the sample purified using 1X AMPure PacBio (PB), fraction 1. The remaining 12 × 50 μL PCR reactions were pooled and the sample purified using 0.4X AMPure PacBio, fraction 2, following the manufacture’s protocol. The Beads purifications were repeated on fractions 1 and fraction 2, 1X and 0.4X volume, respectively. cDNA QC was performed using Qubit (Qubit dsDNA HS Assay Kit, Thermo Fisher), and the quality accessed using Agilent cDNA 12K Kit, following the manufacturers’ instructions. Equal molar quantities of the two fractions per sample were pooled and preceded to SMRTbell Template preparation.
SMRTbell libraries were prepared following manufacturer’s protocol. First, DNA damage repair was performed, followed by end repair, blunt ligation of SMRTbell adaptors and Exonucleases III and IV digestion of any unligated material. SMRTbells were purified using 1X AMPure PacBio (PB) beads and eluted in 30 μL of PacBio Elution Buffer. The sample was transferred to a fresh tube and the concentration was determined using Qubit (Qubit dsDNA HS Assay Kit, Thermo Fisher), and the quality accessed using Agilent cDNA 12K Kit or HS Kit, following the manufacturers’ instructions.
For sequencing, SMRTbell libraries were annealed to sequencing primer v3 sequenced using 2.1 chemistry and 10 hour movies on the Sequel system according to manufacturer’s instructions. Further information on the usage of long read-sequencing can be found in 97 and at https://www.pacb.com/smrt-science/smrt-sequencing/.
Statistics and Reproducibility
EBayes adjusted P-values of RNA-seq data were corrected for multiple testing using the Benjamin-Hochberg (BH) method and used to select genes with significant expression differences. One representative western blot data of two independent experiments is shown in the main text. T-test with MSstats was applied to calculate P value in the proteomic analysis88. Growth curve experiments were performed two times and results of one representative experiment are shown in the main text.
Supplementary Material
Acknowledgments
We thank all members of the Marazzi and García-Sastre Lab, Dr. Jesse Bloom and Dr. Alberto Kornblihtt for the valuable discussion and suggestions of the manuscript. We thank Medicinal Chemistry Core, Integrated Screening Core, Microscopy CoRE, and the Global Health and Emerging Pathogens Institute (GHEPI) at Icahn School of Medicine at Mount Sinai. H.v.B., I.M. and A.G.-S., are partially supported by HHSN272201400008C - Center for Research on Influenza Pathogenesis (CRIP), a NIAID-funded Center of Excellence for Influenza Research and Surveillance (CEIRS). I.M. is supported in part by The Department of Defense W911NF-14–1-0353 I.M. and H.v.B. are supported by NIH grant 1R01AN3663134. I.M., R.A.A., S.C., N.K., and A.G.-S. are all supported by the NIH grant U19AI106754 FLUOMICS. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention/the Agency for Toxic Substances and Disease Registry. This work was supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Footnotes
Competing Financial Interests Statement
The authors declare no competing financial interests.
Data availability
RNA-Seq data associated with this study is available through the Gene Expression Omnibus (GEO) data repository, accession number: GSE103604.
References
- 1.Marazzi I, Ho JS, Kim J, Manicassamy B, Dewell S, Albrecht RA, Seibert CW, Schaefer U, Jeffrey KL, Prinjha RK, Lee K, García-Sastre A, Roeder RG & Tarakhovsky A Suppression of the antiviral response by an influenza histone mimic. Nature 483, 428–433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rialdi A, Hultquist J, Jimenez-Morales D, Peralta Z, Campisi L, Fenouil R, Moshkina N, Wang ZZ, Laffleur B, Kaake RM, McGregor MJ, Haas K, Pefanis E, Albrecht RA, Pache L, Chanda S, Jen J, Ochando J, Byun M, Basu U, García-Sastre A, Krogan N, van Bakel H & Marazzi I The RNA exosome syncs IAV-RNAPII transcription to promote viral ribogenesis and infectivity. Cell 169, 679–692 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayllon J & García-Sastre A The NS1 protein: a multitasking virulence factor. Curr. Top Microbiol. Immunol. 386, 73–107 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, Farzan M, Inoue S, Jung JU & García-Sastre A Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5, 439–449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S, Min JY, Krug RM & Sen GC Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349, 13–21 (2006). [DOI] [PubMed] [Google Scholar]
- 6.García-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P & Muster T Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252, 324–320 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Donelan NR, Basler CF & García-Sastre A A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol. 77, 13257–13266 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson D, Hossain MJ, Hickman D, Perez DR & Lamb RA A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc. Natl. Acad. Sci. USA 105, 4381–4386 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrillo B, Choi JM, Bornholdt ZA, Sankaran B, Rice AP & Prasad BV The influenza A virus protein NS1 displays structural polymorphism. J. Virol. 88, 4113–4122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hale BG Conformational plasticity of the influenza A virus NS1 protein. J. Gen. Virol. 95, 2099–2105 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Davey NE, Van Roey, K., Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H & Gibson TJ Attributes of short linear motifs. Mol. Biosyst. 8, 268–281 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Gitlin L, Hagai T, LaBarbera A, Solovey M & Andino R Rapid evolution of virus sequences in intrinsically disordered protein regions. PLoS Pathog. 10, e1004529 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taubenberger JK & Morens DM The pathology of influenza virus infections. Annu. Rev. Pathol. 3, 499–522 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taubenberger JK & Kash JC Insights on influenza pathogenesis from the grave. Virus Res. 162, 2–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taubenberger JK, Baltimore D, Doherty PC, Markel H, Morens DM, Webster RG & Wilson IA Reconstruction of the 1918 influenza virus: unexpected rewards from the past. MBio. 3, e00201–e00212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hale BG, Randall RE, Ortín J & Jackson D The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89, 2359–2376 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Krug RM Functions of the influenza A virus NS1 protein in antiviral defense. Curr. Opin. Virol. 12, 1–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, Rakestraw KM, Webster RG, Hoffmann E, Krauss S, Zheng J, Zhang Z & Naeve CW Large-scale sequence analysis of avian influenza isolates. Science 311, 1576–1580 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Neumann G, Whitt MA & Kawaoka Y A decade after the generation of a negative-sense RNA virus from cloned cDNA - what have we learned? J. Gen. Virol. 83, 2635–2662 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Van Roey K, Uyar B, Weatheritt RJ, Dinkel H, Seiler M, Budd A, Gibson TJ & Davey NE Short linear motifs: ubiquitous and functionally diverse protein interaction modules directing cell regulation. Chem. Rev. 114, 6733–6778 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH & Verma IM BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477, 179–184 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B & García-Sastre A Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. USA 107, 11531–11536 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reardon S US suspends risky disease research. Nature 514, 411–412 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Geiss GK, Salvatore M, Tumpey TM, Carter VS, Wang X, Basler CF, Taubenberger JK, Bumgarner RE, Palese P, Katze MG & García-Sastre A Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99, 10736–10741 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatada E & Fukuda R Binding of influenza A virus NS1 protein to dsRNA in vitro. J. Gen. Virol. 73, 3325–3329 (1992). [DOI] [PubMed] [Google Scholar]
- 26.Nemeroff ME, Qian XY & Krug RM The influenza virus NS1 protein forms multimers in vitro and in vivo. Virology 212, 422–428 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Bornholdt ZA & Prasad BV X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus. Nature 456, 985–988 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T & Taylor JP Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y, Protter DS, Rosen MK & Parker R Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL & McKnight SL Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R & Rosen MK Compositional control of phase-separated cellular bodies. Cell 166, 651–663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R & McKnight SL Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harlen KM & Churchman LS The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 18, 263–273 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Heyn P, Salmonowicz H, Rodenfels J & Neugebauer KM Activation of transcription enforces the formation of distinct nuclear bodies in zebrafish embryos. RNA Biol. 14, 752–760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hnisz D, Shrinivas K, Young RA, Chakraborty AK & Sharp PA A phase separation model for transcriptional control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu H, Yu D, Hansen AS, Ganguly S, Liu R, Heckert A, Darzacq X & Zhou Q Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richard P & Manley JL Transcription termination by nuclear RNA polymerases. Genes Dev. 23, 1247–1269 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlackow M, Nojima T, Gomes T, Dhir A, Carmo-Fonseca M & Proudfoot NJ Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol. Cell 65, 25–38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinson HG An active role for splicing in 3’-end formation. Wiley Interdiscip. Rev. RNA 2, 459–470 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Misra A, Ou J, Zhu LJ & Green MR Global promotion of alternative internal exon usage by mRNA 3’ end formation factors. Mol. Cell 58, 819–831 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Misra A & Green MR From polyadenylation to splicing: dual role for mRNA 3’ end formation factors. RNA Biol. 13, 259–264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tseng CK, Wang HF, Burns AM, Schroeder MR, Gaspari M & Baumann P Human telomerase RNA processing and quality control. Cell Rep. 13, 2232–2243 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Orphanides G & Reinberg D A unified theory of gene expression. Cell 108, 439–451 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Perales R & Bentley D “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell 36, 178–191 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E & Jensen TH Crosstalk between mRNA 3’ end processing and transcription initiation. Mol. Cell 40, 410–422 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Gu B, Eick D & Bensaude O CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res. 41, 1591–1603 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niwa M, Rose SD & Berget SM In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 4, 1552–1559 (1990). [DOI] [PubMed] [Google Scholar]
- 48.Dye MJ & Proudfoot NJ Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell 3, 371–378 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Bauer DLV, Tellier M, Martinez-Alonso M, Nojima T, Proudfoot NJ, Murphy S & Fodor E Influenza Virus Mounts a Two-Pronged Attack on Host RNA Polymerase II Transcription. Cell Rep. 23, 2119–2129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heinz, in press. [Google Scholar]
- 51.Kuo RL & Krug RM Influenza a virus polymerase is an integral component of the CPSF30-NS1A protein complex in infected cells. J. Virol. 83, 1611–1616 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pichlmair A, Kandasamy K, Alvisi G, Mulhern O, Sacco R, Habjan M, Binder M, Stefanovic A, Eberle CA, Goncalves A, Bürckstümmer T, Müller AC, Fauster A, Holze C, Lindsten K, Goodbourn S, Kochs G, Weber F, Bartenschlager R, Bowie AG, Bennett KL, Colinge J & Superti-Furga G Viral immune modulators perturb the human molecular network by common and unique strategies. Nature 487, 486–490 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, Pevzner SJ, Abderazzaq F, Byrdsong D, Carvunis AR, Chen AA, Cheng J, Correll M, Duarte M, Fan C, Feltkamp MC, Ficarro SB, Franchi R, Garg BK, Gulbahce N, Hao T, Holthaus AM, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Roecklein-Canfield J, Johannsen E, Barabási AL, Beroukhim R, Kieff E, Cusick ME, Hill DE, Münger K, Marto JA, Quackenbush J, Roth FP, DeCaprio JA & Vidal M Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487, 491–495 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marazzi I & Garcia-Sastre A Interference of viral effector proteins with chromatin, transcription, and the epigenome. Curr. Opin. Microbiol. 26, 123–129 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Heaton NS, Moshkina N, Fenouil R, Gardner TJ, Aguirre S, Shah PS, Zhao N, Manganaro L, Hultquist JF, Noel J, Sachs D, Hamilton J, Leon PE, Chawdury A, Tripathi S, Melegari C, Campisi L, Hai R, Metreveli G, Gamarnik AV, García-Sastre A, Greenbaum B, Simon V, Fernandez-Sesma A, Krogan NJ, Mulder LCF, van Bakel H, Tortorella D, Taunton J, Palese P & Marazzi I Targeting Viral Proteostasis Limits Influenza Virus, HIV, and Dengue Virus Infection. Immunity 44, 46–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marazzi I, Greenbaum BD, Low DHP & Guccione E Chromatin dependencies in cancer and inflammation. Nat. Rev. Mol. Cell Biol. 19, 245–261 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Maldonado E, Cabrejos ME, Banks L & Allende JE Human papillomavirus-16 E7 protein inhibits the DNA interaction of the TATA binding transcription factor. J. Cell Biochem. 85, 663–669 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Dasgupta A Targeting TFIIH to inhibit host cell transcription by Rift Valley Fever Virus. Mol. Cell 13, 456–458 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Di Valentin E, Bontems S, Habran L, Jolois O, Markine-Goriaynoff N, Vanderplasschen A, Sadzot-Delvaux C & Piette J Varicella-zoster virus IE63 protein represses the basal transcription machinery by disorganizing the pre-initiation complex. Biol. Chem. 386, 255–267 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Kundu P, Raychaudhuri S, Tsai W & Dasgupta A Shutoff of RNA polymerase II transcription by poliovirus involves 3C protease-mediated cleavage of the TATA-binding protein at an alternative site: incomplete shutoff of transcription interferes with efficient viral replication. J. Virol. 79, 9702–9713 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fraser KA & Rice SA Herpes simplex virus immediate-early protein ICP22 triggers loss of serine 2-phosphorylated RNA polymerase II. J. Virol. 81, 5091–5101 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geng F, Wenzel S & Tansey WP Ubiquitin and proteasomes in transcription. Annu. Rev. Biochem. 81, 177–201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hagai T, Azia A, Babu MM & Andino R Use of host-like peptide motifs in viral proteins is a prevalent strategy in host-virus interactions. Cell Rep. 7, 1729–1739 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daugherty MD & Malik HS Rules of engagement: molecular insights from host-virus arms races. Annu. Rev. Genet. 46, 677–700 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Garamszegi S, Franzosa EA & Xia Y Signatures of pleiotropy, economy and convergent evolution in a domain-resolved map of human-virus protein-protein interaction networks. PLoS Pathog. 9, e1003778 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dyson HJ, and Wright PE Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Uversky VN & Dunker AK Understanding protein non-folding. Biochim. Biophys. Acta 1804, 1231–1264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Babu MM, Kriwacki RW & Pappu RV Structural biology. Versatility from protein disorder. Science 337, 1460–1461 (2012). [DOI] [PubMed] [Google Scholar]
- 69.Tompa P, Davey NE, Gibson TJ & Babu MM A million peptide motifs for the molecular biologist. Mol. Cell 55, 161–169 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Coletta A, Pinney JW, Solís DY, Marsh J, Pettifer SR & Attwood TK Low-complexity regions within protein sequences have position-dependent roles. BMC Syst. Biol. 4, 43 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beltrao P, Albanese V, Kenner LR, Swaney DL, Burlingame A, Villen J, Lim WA, Fraser JS, Frydman J & Krogan NJ Systematic functional prioritization of protein posttranslational modifications. Cell 150, 413–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP & Morgan DO Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mosca R, Pache RA & Aloy P The role of structural disorder in the rewiring of protein interactions through evolution. Mol. Cell. Proteomics 11, 014969 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schrauwen EJ & Fouchier RA Host adaptation and transmission of influenza A viruses in mammals. Emerg. Microbes Infect. 3, e9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller MS & Palese P Peering into the crystal ball: influenza pandemics and vaccine efficacy. Cell 157, 294–299 (2014). [DOI] [PubMed] [Google Scholar]
- 76.Taft AS, Ozawa M, Fitch A, Depasse JV, Halfmann PJ, Hill-Batorski L, Hatta M, Friedrich TC, Lopes TJ, Maher EA, Ghedin E, Macken CA, Neumann G & Kawaoka Y Identification of mammalian-adapting mutations in the polymerase complex of an avian H5N1 influenza virus. Nat Commun. 6, 7491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 77.Eswar N, Eramian D, Webb B, Shen MY & Sali A Protein structure modeling with MODELLER. Methods Mol. Biol. 426, 145–159 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Penkert RR, DiVittorio HM & Prehoda KE Internal recognition through PDZ domain plasticity in the Par-6-Pals1 complex. Nat. Struct. Mol. Biol. 11, 1122–1127 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.London N, Raveh B, Cohen E, Fathi G & Schueler-Furman O Rosetta FlexPepDock web server-high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 39, W249–253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reverter D & Lima CD A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure 12, 1519–1531 (2004). [DOI] [PubMed] [Google Scholar]
- 81.Halfmann R & Lindquist S Screening for Amyloid Aggregation by Semi-Denaturing Detergent-Agarose Gel Electrophoresis. J. Vis. Exp. 17, e838 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shah NB and Duncan TM Bio-layer interferometry for measuring kinetics of protein-protein interactions and allosteric ligand effects. J. Vis. Exp. 84, e51383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martínez-Sobrido L & García-Sastre A Generation of recombinant influenza virus from plasmid DNA. J. Vis. Exp. 42, 2057 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R & McKnight SL Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 86.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV & Mann M Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011). [DOI] [PubMed] [Google Scholar]
- 87.The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Choi M, Chang CY, Clough T, Broudy D, Killeen T, MacLean B & Vitek O MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526 (2014). [DOI] [PubMed] [Google Scholar]
- 89.Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D & Thomas PD PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways and data analysis tool enhancements. Nucleic Acids Res. 45, D183–189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martin M Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011). [Google Scholar]
- 91.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M & Gingeras TR STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liao Y, Smyth GK & Shi W Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 93.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W & Smyth GK Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li B & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Law CW, Chen Y, Shi W & Smyth GK Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 25, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA & Young RA C-myc regulates transcriptional pause release. Cell 141, 432–445 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rhoads A & Au K. F. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13, 278–89 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


