Abstract
Deubiquitylating enzymes (DUBs) maintain relative homeostasis of the cellular ubiquitome by removing the post‐translational modification ubiquitin moiety from substrates. Numerous DUBs have been demonstrated specificity for cleaving a certain type of ubiquitin linkage or positions within ubiquitin chains. Moreover, several DUBs perform functions through specific protein–protein interactions in a catalytically independent manner, which further expands the versatility and complexity of DUBs’ functions. Dysregulation of DUBs disrupts the dynamic equilibrium of ubiquitome and causes various diseases, especially cancer and immune disorders. This review summarizes the Janus‐faced roles of DUBs in cancer including proteasomal degradation, DNA repair, apoptosis, and tumor metastasis, as well as in immunity involving innate immune receptor signaling and inflammatory and autoimmune disorders. The prospects and challenges for the clinical development of DUB inhibitors are further discussed. The review provides a comprehensive understanding of the multi‐faced roles of DUBs in cancer and immunity.
Keywords: cancer, deubiquitylating enzymes (DUBs), immunity, therapeutic approach
This review presents the fundamental understanding of deubiquitinating enzymes (DUBs) and then outlines the mechanisms and multi‐role of DUBs in determining the occurrence and development of tumor and immune disorders. DUBs in the crosstalk between cancer and immune response is then presented. The therapeutic potential and clinical translation of DUB inhibitors are also summarized.
1. Introduction
Protein ubiquitination is a dynamic and reversible posttranslational modification (PTM), wherein one or more ubiquitin protein (Ub) with 76 amino acids is covalently attached to a substrate protein. This modification occurs on a diverse range of cellular proteins in numerous cellular processes.[ 1 ] Ubiquitin is coded by four different genes in Homo sapiens: ubiquitin A‐52 residue ribosomal protein fusion product 1 (UBA52) and ribosomal protein S27a (UBA80) code for a single ubiquitin fused at the C‐terminus to the ribosomal proteins L40 and S27a, respectively. Ubiquitin B (UBB), and ubiquitin C (UBC) genes are polyubiquitin precursor presented with tandem repeats.[ 2 ] The ubiquitination process is a cascade reaction involving three types of enzymes: Ub‐activating enzymes (E1s), Ub‐conjugating enzymes (E2s), and Ub ligase enzymes (E3s).[ 3 ] Ub is activated by E1 in an ATP‐dependent manner by forming a thioester bond between the Cys of its active site and glycine C‐terminal carboxyl group of Ub. Subsequently, Ub is bound to E2 through transthiolation and then covalently conjugated to the amino group of Lys residue of a substrate protein by E3 Ub ligases.[ 4 ] There are currently four E3 subtypes identified: homologous with E6‐associated protein C‐terminus (HECT)‐, really interesting new gene (RING)‐, U‐box‐, and RING‐ between‐RING (RBR)‐type. RING‐ and U‐box‐type E3s directly facilitate Ub transfer from E2 to the substrate protein. Whereas, HECT‐ and RBR‐type E3s can form a thioester bond between their active site cysteine and Ub before transferring Ub to the substrate protein (Figure 1A).[ 5 , 6 ]
Figure 1.
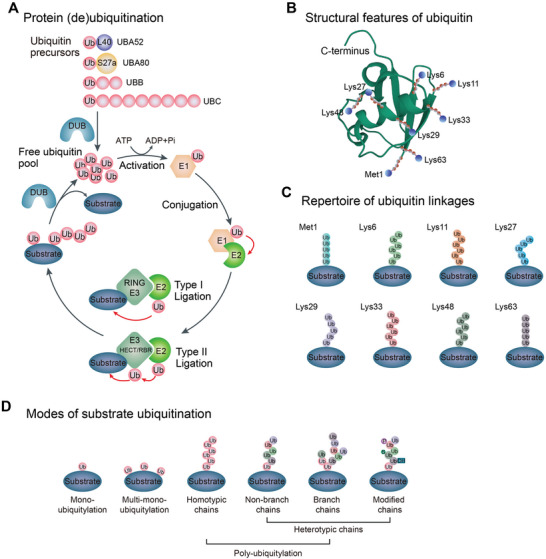
The ubiquitination cascade and the complexity of ubiquitin modifications. A) Schematic showing the key events in ubiquitination and deubiquitination. Ubiquitin (Ub) is produced by cleavage of Ub precursors via deubiquitylating enzymes (DUBs). Ubiquitin precursors include four different genes: UBB and UBC genes code for a polyubiquitin precursor and UBA52 and UBA80 genes code for a single copy of Ub fused to the ribosomal proteins L40 (L) and S27a (S), respectively. Ubiquitin is activated by the E1‐activating enzyme in an ATP‐dependent manner. The E2‐conjugating enzyme catalyzes the conjugation of E1‐bound Ub to itself. Finally, E3 ligase directly (RING E3 ligases) or indirectly (HECT/RBR E3 ligases) transfers E2‐bound Ub to a substrate, forming a thioester linkage between the carboxyl group of glycine in Ub and the ε‐amino group of lysine in the substrate. Ubiquitin molecules are removed from substrates by DUBs and recycled. B) Schematic representation of the structure of Ub. Stick representation showing the seven Lys residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) and Met1 of Ub (Protein Data Bank ID: 1UBQ[ 612 ]), and amino groups that are modified with Ub during the formation of the chain are marked with blue spheres. C) The repertoire of ubiquitin linkages, Met1, and seven Lys residues in Ub can form specific chain linkages with distinct conformations and exert specific (or unknown) functions. D) A substrate can be modified by mono‐, multi‐mono‐ or poly‐ubiquitin. Ubiquitin chains are colored according to linkage type, and each type of chain represents a distinct posttranslational modification. Polyubiquitin includes homotypic chains and heterotypic chains. Heterotypic chains are either branched or modified, such as SUMOylation (S), phosphorylation (P), or hydroxylation (OH), etc.
Ubiquitin function mainly relies on seven Lys residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) and an N‐terminal methionine (Met1) (Figure 1B), which facilitates intricate ubiquitination linkages (Figure 1C).[ 7 ] Ub is attached as a monomer or multiple mono‐ubiquitin adducts to the same or different residues of substrate proteins. To add complexity, poly‐ubiquitination can generate homotypic chains, or heterotypic, or branched chains. What is more, ubiquitin moieties can also be modified by other PTMs (Figure 1D).[ 8 , 9 ] Each linkage reconciles distinctive signaling pathways to shape the fate of substrate proteins.[ 10 , 11 ] The most typical Lys48‐ or Lys63‐linked chains perform either direct proteasomal degradation of substrate proteins through the ubiquitin‐proteasome system (UPS) or non‐degradative roles in administering cellular localization, protein‐protein interactions, and signaling transduction.[ 10 , 12 ] In contrast, the remaining ‘atypical’ linkages are less studied till now. Generally, Met1‐linked linear chains participate in inflammatory processes and apoptosis. Lys6‐linkages are implicated in DNA damage response. Lys11‐linkages have been reported to regulate proteasomal degradation, cell cycle, or membrane trafficking. Lys27‐linkages play a role in protein secretion, DNA damage repair, and mitochondrial damage response. Lys29‐linkages are associated with proteasomal degradation, innate immune response, and AMP‐activated protein kinase (AMPK) signaling regulation. Lys33‐linkages can influence innate immune response and intracellular trafficking.[ 6 , 9 ]
Ubiquitinated substrates are acknowledged by a myriad of proteins involving ubiquitin‐binding domains (UBDs), which interrelate with the surface patches on Ub.[ 13 ] Ubiquitination is reversed by peptidases termed deubiquitylating enzymes or deubiquitinases (DUBs) through hydrolysis of linkages between Ub moieties or between Ub and substrate.[ 14 , 15 ] DUBs trim Ub molecules from substrates to produce a free Ub pool which guarantees Ub recycling. Ubiquitination and deubiquitination regulate many aspects of human cell biology and physiology. Dysregulation in these processes leads to various clinical implications, including cancer and infection diseases.[ 16 , 17 ] This review presents the fundamental understanding of DUBs, and then outlines the mechanisms and Janus‐faced roles of DUBs in determining the occurrence and development of tumor and immune disorders. We also discuss the therapeutic potential and clinical translation of DUB inhibitors based on the cellular mechanisms.
2. The Characteristics of DUBs
2.1. DUB Superfamily and Structure
More than 100 DUBs are identified in humans and classified into 9 superfamilies according to the sequence and domain conservation: ubiquitin‐specific proteases (USPs), ovarian tumor proteases (OTUs), ubiquitin C‐terminal hydrolases (UCHs), Machado‐Joseph domain‐containing proteases (MJDs, also known as Josephins), JAMM/MPN domain‐associated Zn‐depend metalloproteases (JAMMs, also known as MPN+), the motif interacting with ubiquitin (MIU)‐containing novel DUB family (MINDYs), monocyte chemotactic protein‐induced proteins (MCPIPs), permuted papain fold peptidases of dsRNA viruses and eukaryotes (PPPDEs), and zinc finger (ZnF) containing ubiquitin peptidase 1 (ZUP1) (Figure 2 ).
Figure 2.
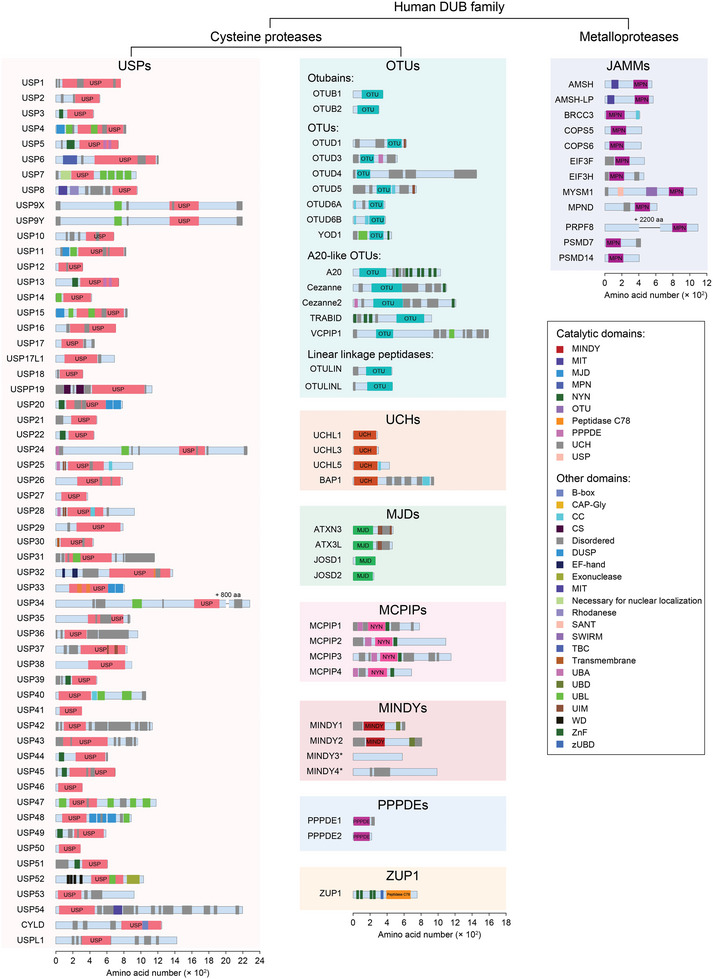
Subfamilies and domain structure of the human DUBs. The human DUBs are classified into six families of cysteine proteases, including ubiquitin‐specific proteases (USPs), ovarian tumor proteases (OTUs), ubiquitin C‐terminal hydrolases (UCHs), Machado–Joseph domain‐containing proteases (MJDs) or Josephins, monocyte chemotactic protein‐inducing proteins (MCPIPs), the motif interacting with ubiquitin (MIU)‐containing novel DUB family (MINDYs), permutated papain fold peptidase of DsRNA viruses and eukaryotes (PPPDEs), zinc finger containing Ubiquitin Peptidase 1 (ZUP1) or zinc finger with UFM1‐specific peptidase domain protein/C6orf113/ZUP1 (ZUFSP); and one metalloprotease subfamily, i.e., JAB1/MPN/MOV34 metalloenzymes (JAMMs, also known as MPN+). CAP‐Gly, cytoskeleton‐associated protein‐glycine‐rich; CC, coiled‐coil; CS, CHORD‐containing proteins and SGT1; MIT, the microtubule interacting and trafficking; MPN, Mpr1/Pad1 N‐terminal; MIT, the microtubule interacting and trafficking; NYN, Nedd4‐BP1, YacP nucleases; SANT, SWI3, ADA2, N‐CoR and TFIIIB DNA‐binding; SWIRM, SWI3P, RSC8P and MOIRA; TBC, Tre2–Bub2–Cdc16; UBA, ubiquitin‐associated; UBD, ubiquitin‐binding domain; UBL, Ubiquitin‐like; UIM, ubiquitin‐interacting motif; ZnF, zinc finger. Numbers with a "+" indicate the number of amino acids is not shown in the sequence. "*" indicates there is no catalytic domain annotated.
DUBs generally contain one or more binding domains for substrate recognition, for example, the ZnF motif, ubiquitin‐like domain (UBL), coiled‐coil (CC) domain, and ubiquitin‐interacting motif (UIM), etc., which cooperates with an indispensable catalytic domain to precisely govern hydrolysis. According to the hydrolysis mechanism of Ub chains, the DUB families identified so far mostly belong to cysteine proteases, except the JAMM family which is considered as zinc‐metalloprotease (Figure 2, Figure 4A).[ 18 , 19 ] The DUBs with cysteine protease activity commonly exhibit a catalytic pocket comprising cysteine, histidine, and an acidic residue. The JAMM family DUBs are characterized as Zinc (Zn)‐dependent metalloproteases, coordinating zinc ions with aspartate, histidine, and serine residues to facilitate the recruitment of water molecules and initiate hydrolysis of isopeptide bonds.[ 20 , 21 , 22 , 23 ]
Figure 4.
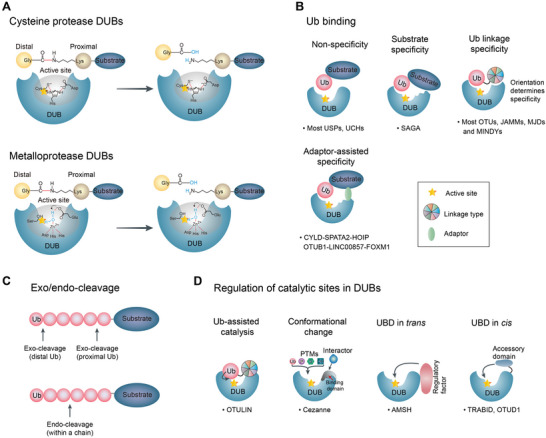
Catalytic DUB mechanisms of cleavage and recognition. A) Cysteine protease DUBs commonly include a catalytic pocket composed of cysteine, histidine, and an acidic residue. JAMM family DUBs are Zinc (Zn)‐dependent metalloproteases. A Zn atom is essential for the active site that is coordinated by conserved catalytic residues and a water molecule. Red represents a fragile isopeptide. B) Principles of substrate recognition and linkage specificity according to a variety of Ub binding and regulation catalytic sites of DUBs. Most members of the USP and UCL subfamilies have nonspecific interactions with the substrate (without a motif). Conversely, SAGA has substrate specificity through motif interactions with the substrate protein. Ub linkage specificity is acquired so that only one linkage type is identified by Ub‐binding sites to catalyze DUB activity. Many members of OTU‐, JAMM‐, Josephin‐, and MINDY families have linkage specificity. In addition, some DUBs recognize targeted proteins with the assistance of adaptors or scaffolds. C) The characteristics of Ub‐binding sites in DUBs determine whether polyubiquitin is cleaved from the distal‐ or the proximal end (exo‐cleavage) or within the en bloc chain (endo‐cleavage). D) Regulation of catalytic sites in DUBs. Ub complements the enzymatic activity by directly participating in the active site, and other post‐translational modifications (SUMOylation, phosphorylation, and hydroxylation) are also key features in regulating DUB activity. Furthermore, regulatory factor in trans (between two molecules) or accessory domain in cis (within a molecule) exerts a boosting effect on the catalytic activity. The star indicates the active site.
The first USP structure study was USP7 in the presence and absence of substrate. USP7 exhibits a conserved three‐domain architecture, consisting of fingers, palm, and thumb. The leaving ubiquitin moiety is specifically coordinated by the fingers, positioning its C terminus within the active site situated between the palm and the thumb. The binding of ubiquitin aldehyde induces a significant conformational change in the active site, resulting in the realignment of catalytic triad residues to facilitate catalysis (Figure 3A).[ 24 ] Based on the analogy with inhibitor complexes of papain‐like enzymes, researchers proposed a model to elucidate the binding mechanism of UCH DUBs to ubiquitinated substrate. The unliganded structure of UCHL3 revealed that its active site cleft is concealed by two distinct segments of the enzyme, implying a regulatory mechanism to restrict non‐specific hydrolysis. Substrate binding leads to a conformational change. Ubiquitin vinyl methyl ester (UbVME, an inhibitor) forms a covalent adduct with the catalytic active site cysteine of UCHL3 (Figure 3B).[ 25 , 26 ]
Figure 3.
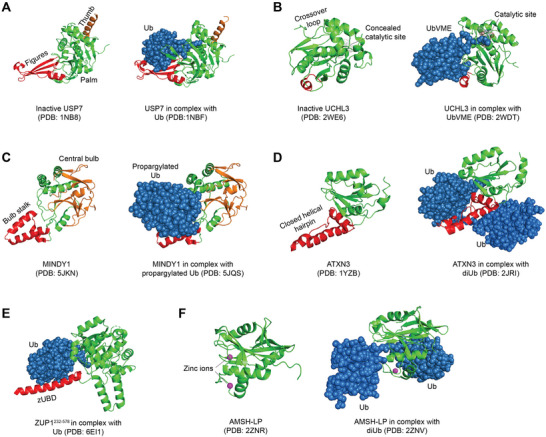
Representative crystal structures of DUBs in cartoon. The crystal structures of USP7 (A), UCHL3 (B), MINDY1 (C), ATXN3 (D), ZUP1231–576 (E), AMSH‐LP (F) in the absence (left panel) or presence (right panel) of Ub substrates/inhibitors were adapted from protein data bank (PDB) using PyMOL software.
The crystal structure study of the N‐terminal OTU deubiquitinase domain of A20 elucidated distinct characteristics compared to other DUBs, but still having a similar essential catalytic core. The analysis of conserved surface regions enables the prediction of ubiquitin‐binding sites for both the proximal and distal ubiquitin molecules. In addition, structural and biochemical analysis revealed a novel architectural arrangement of the catalytic triad that was potentially found in a subset of OTU domains including Cezanne and TRABID.[ 27 ] The crystal structure of MINDY‐1 in complex with propargylated Ub reveals conformational changes that reposition the active site for catalysis. MINDY1 exhibits a preference for cleaving lengthy polyUb chains and operates by selectively trimming the chains from the distal ends (Figure 3C).[ 28 ] The significant discovery of MJD DUBs, particularly ATXN3, demonstrated selective Ub esterase activity implying their specialized role in specifically modifying non‐lysine ubiquitinated substrates (Figure 3D).[ 29 ] ZUP1, the sole human member of the deubiquitinase family, harbors multiple UBDs responsible for its specific action on K63‐linked chains.[ 30 ] MINDYs and ZUP1 family members were newly identified, but their cellular function is poorly understood (Figure 3E).[ 28 ] PPPDE1 is also a newly discovered deubiquitinase enzyme that belongs to the cysteine isopeptidase family, and it plays a regulatory role in ubiquitin activity through its binding affinity for both K48 and K63 linkages.[ 31 ]
The JAMM metalloprotease structures represent the sole current instance in which a deubiquitinating enzyme has been crystallized with a ubiquitin chain bound across its active site. The JAMM motif coordinates two zinc ions, with one of them serving as a catalyst to activate a water molecule for attacking the isopeptide bond. The amino group is subsequently liberated from the charged catalytic intermediate through a mechanism akin to that of cytidine deaminase (Figure 3F).[ 32 ]
Generally, DUB's catalytic activity can be evaluated by a method based on the hydrolysis of the substrate conjugated with ubiquitin 7‐amido‐4‐methylcoumarin (Ub‐AMC) and the determination of fluorogenic substrate AMC in vitro. However, this method is subject to the throughput range of identification, and has not been widely applied to identify DUBs on a large scale. Additionally, whether other types of proteases, such as serine or asparate, execute DUB‐like functions might discover novel DUB subclasses. The development of innovative technology is extremely necessary for the identification of DUB superfamily and the exploration of their diverse functions in the future.[ 33 , 34 ]
2.2. The Ubiquitin Linkage Specificity of DUBs
DUBs are a class of proteases responsible for cleaving peptide or isopeptide bonds between linked ubiquitin molecules or between ubiquitin and a modified protein. The diverse linkage types, length, and topology give rise to the complexity of Ub chain architectures, which requires a broad variety of individual DUB specificity.[ 35 ] DUB–substrate interactions are thought to be weak and transient in nature state, much like most enzyme–substrate interactions. Overall, most members of the USP and UCH subfamilies have nonspecific interactions with the substrate (without a motif). Whereas, substrate recognition by DUBs is also governed by sequences and motifs outside the conserved catalytic domain. For example, Spt‐Ada‐Gcn5‐acetyltransferase (SAGA) deubiquitination module has substrate specificity through motif interactions with the substrate protein. Ubiquitin linkage specificity is acquired so that only one linkage type is identified by Ub‐binding sites to catalyze DUB activity. Many members of OTU, JAMM, Josephin, and MINDY subfamilies have linkage specificity. Most USPs are recruited to substrates for direct deubiquitination regardless of linkage‐type or architecture.[ 36 , 37 ] Exceptionally, a subset of the USPs, including CYLD and USP30, prefers to Lys63‐ and Met1‐linkage.[ 38 , 39 , 40 ] In contrast, a handful of DUBs carry out the function by targeting specific chain types. JAMM subfamily members such as AMSH, AMSH‐LP, BRCC, and POH1 are generally Lys63 specific; and this is the first confirmed DUB family to display linkage specificity.[ 41 , 42 ] In addition, numerous OTU DUBs are also linkage‐specific,[ 43 ] such as the Lys48 linkage specificity of OTUB1,[ 44 ] the Lys11 linkage specificity of Cezanne (OTUD7B),[ 45 ] the Met1 linkage specificity of OTULIN,[ 46 ] and the Lys29 and Lys33 linkages specificity of TRABID.[ 47 ] The ZUP1 family, with ZUP1 itself as its sole human member, is specific for Lys63‐linkage conferred by multiple UBDs and is associated with genome stability.[ 30 , 48 , 49 , 50 ] Whereas, each MINDY family member exhibits the specificity for Lys48‐linkage, strongly suggesting this family plays a vital role in protein turnover.[ 28 ] In addition to direct recognition, some DUBs recognize targeted proteins with the assistance of adaptors or scaffolds. For instance, LINC00857 acts as a protein scaffold by binding to both FOXM1 and OTUB1, thereby enhancing their interaction and inhibiting FOXM1 degradation through the ubiquitin‐proteasome pathway.[ 51 ] The recruitment of CYLD to HOIP is facilitated by the adaptor protein SPATA2, which acts as a bridge between the DUB and HOIP (Figure 4B).[ 52 ]
The length of Ub chains is distinguishable for linkage types and supplies one more variable. Ub chains can be trimmed by DUBs according to endo‐ or exo‐cleavage activity. Endo‐cleavage activity clears the proximal Ub from a substrate and the unattached Ub chains must be further converted into monoubiquitin. In contrast, exo‐cleavage activity directly produces monoubiquitin from the distal Ub, but multiple actions of DUB are required to remove the en bloc polyubiquitin chain (Figure 4C). Where to cleave, endo or exo, depends on the DUB itself, linkage type, and polyubiquitin identification.[ 35 , 53 ] With the advances in biochemistry and chemical biology, several methods have been developed to profile DUB linkage preference, such as mutation of ubiquitin, middle‐down mass spectrometry, development of antibodies, etc.[ 54 ] What is prominent, Ubiquitin Chain Restriction (UbiCRest) was designed to assess the intrinsic linkage‐specificity of DUBs in a short time. Briefly, a panel of artificially generated and specific linkages was incubated with DUB of interesting. Cleavage products were then resolved on SDS‐PAGE gradient gel and visualized by silver stained or western blot analysis.[ 54 ] Based on UbiCRest analysis of cleavage patterns and Ub linkage products with the treatment of DUB‐specific enzymes in vitro or in cellulo, the appearance of Ub chain composition and architecture on the substrates can be partly outlined.[ 43 , 54 ] Although significant structural information has been disclosed. Predicting the linkage or substrate specificity of DUBs remains challenging.
2.3. The Regulation of DUBs’ Enzymatic Activity
Catalytic activity of DUBs is directly governed by a variety of manners, including substrate‐assisted catalysis, allosteric regulation, and PTMs (Figure 4D).[ 17 ] In some instances, substrate binding directly contributes to enzyme activation, enabling accurate control of specificity in deubiquitination. OTULIN catalytic activity is modulated by a substrate‐assisted mechanism.[ 46 ] It is unable to trim the Lys63‐linkage; however, its enzymatic activity is directly activated through preferential binding of Met1‐linked ubiquitin.[ 46 , 55 ] Likewise, Cezanne undergoes dramatic conformational rearrangements upon interacting with the appropriate polyubiquitin which impairs its turnover and defines its linkage specificity.[ 45 ] In addition, allostery plays a vital role in adjusting the activity of DUBs. One example is that USP1 binds to USP1‐associated factor 1 (UAF‐1) and converts its ineffective state into an active state through conformational changes.[ 56 ] Meanwhile, USP7 catalytic activity is controlled by the interaction of internal domains with herpesvirus‐associated USP (HAUSP).[ 57 ] The last two of five HAUSP ubiquitin‐like domains (HUBLs) and a C‐terminal peptide (CTP) enhance USP7 activity 100‐fold by re‐binding to the so‐called switching loop in the USP domain.[ 57 ]
PTMs also play a crucial role in adjusting catalytic DUB activity in addition to the above‐mentioned PTMs regulation of DUBs localization. Crosstalk between PTMs and the Ub system mainly includes phosphorylation, ubiquitination, SUMOylation, and oxidation.[ 35 , 58 ] Phosphorylation can positively or negatively regulate DUB activity. For instance, USP8 is inhibited by 14‐3‐3 proteins in a phosphorylation‐dependent manner, and dephosphorylation in the cell cycle M phase increases USP8 activity.[ 59 ] Mutations associated with binding motifs in 14‐3‐3 also improve USP8 activity, leading to upregulation of the epidermal growth factor (EGF) receptor signaling pathway which causes Cushing's disease.[ 60 ] CYLD activity is also controlled by phosphorylation in positive and negative ways. Although IKKγ[ 61 ] and IKKε[ 62 ] of the IκB kinase (IKK) family modulate inhibitory phosphorylation, IKKβ phosphorylation activates CYLD.[ 63 ] Coincidentally, A20 modification by IKKβ[ 64 ] enhances A20 activity involving Lys63‐linked chains.[ 65 ] However, the detailed mechanisms of CYLD or A20 regulation by phosphorylation remain unclear, and it is worth noting that the phosphorylation sites are not in their respective catalytic domains. Elucidating the protein interactome of DUBs helps to optimize the fine regulation mechanism of phosphorylation modification in DUB activity and offers novel lights for therapeutic targeting via disrupting this interaction. DUBs can also be ubiquitinated in complexes with E3 ligases. For example, USP30 is ubiquitinated by Parkin in its zinc fingers subdomain, which may impair catalytic activity.[ 66 ] In contrast, monoubiquitinylation of the ATXN3 Josephin domain boosts enzymatic activity by initiating a conformational change.[ 67 ] In addition, poly‐SUMOylation enables OTUB2 to deubiquitinate Yes‐associated protein (YAP) and transcriptional coactivator with a PDZ‐binding motif (TAZ).[ 68 ] Moreover, most DUBs are cysteine proteases harboring a reactive cysteine residue that is vulnerable to be oxidated by reactive oxygen species (ROS). Oxidation attenuates the activity of USP‐, OUT‐, and UCH family members and it may be reversible.[ 69 , 70 , 71 ] Interestingly, the inhibitory or activating modulation of DUBs activity depicts the fascinating intricacy of post‐translational adjustment.
Furthermore, the regulatory factor in trans (between two molecules) or accessory domain in cis (within a molecule) exerts a boosting effect on the catalytic activity of individual DUB through allosteric regulation or promoting the interaction with protein substrates. This endows divergent members of multiple DUB families with miscellaneous regulatory mechanisms and expands the broad range of DUBs regulation in various cellular processes.[ 35 ]
Overall, these aspects coordinately modulate the specificity and catalytic activity of DUBs, forming dynamic and diverse interactions between DUBs and protein substrates. Diverse mechanisms of DUB regulation allow for finely tuned functions that guarantees an appropriate response to the external and internal situation.
2.4. The Abundance of DUBs
In theory, the simplest mechanism to regulate the activity and consequent biological function of a given DUB is regulating its intracellular abundance, which can be controlled by transcription, translation, and degradation. The copy number of DUBs lies on several orders of magnitude from the low hundreds to hundreds of thousands estimated by mass spectrometry datasets.[ 72 ] Some high‐copy number DUBs are required for the maintenance of basic cellular functions, which are analogous to housekeeping genes. In mammalian cells, USP14, 26S proteasome non‐ATPase regulatory subunit 14 (PSMD14), and UCHL5 are associated with the 19S particle lid of proteasome to coordinately ensure the essential preprocessing of protein degradation and maintain sufficient levels of free Ub.[ 73 ] USP14 and UCHL5 trim ubiquitin chains from the distal end, while PSMD14 is responsible for removing whole poly‐UB chain from target proteins degraded by 20S proteasome core particle.[ 74 , 75 ] Loss of PSMD14 activity in purified proteasomes prevents protein degradation.[ 53 ] Two inactive‐ or pseudo‐DUBs, USP39 and PRPF8, have a central role in the mature spliceosome assembly and pre‐mRNA splicing as components of the spliceosome complex.[ 76 , 77 ] In addition, some of the other DUBs, such as USP7, USP8, USP36 are essential for cell survival.[ 78 , 79 , 80 ]
Notably, lower abundance DUBs can be activated in a stimulation‐dependent manner and exert a particular regulatory effect. For instance, OTU family member A20 (also called TNFAIP3) is hardly expressed in unstimulated cells, but its expression is induced by tumor necrosis factor (TNF) and substantially upregulated upon TLR4‐mediated nuclear factor κB (NF‐κB) activation.[ 81 , 82 ] A20 also acts as a negative feedback regulator to inhibit NF‐κB activation, thereby avoiding the overproduction of inflammatory factors. Moreover, A20 protein levels are controlled by MALT1, which hydrolyzes A20 between the N‐terminal catalytic OTU domain and the C‐terminal UBD, thus jeopardizing its function.[ 83 ] Similarly, cylindromatosis (CYLD) is cleaved and destabilized by MALT1.[ 84 , 85 , 86 ] These examples highlight that DUB abundance can be regulated by non‐specific and situation‐dependent ways to impact cellular function. Individual DUB performs specific effects under certain stimuli and different DUBs can cooperate or antagonize each other to maintain the homeostasis of cellular functions.
2.5. The Localization of DUBs
DUBs’ localization can be governed in various ways. Above all, PTMs are a convenient way for cells to affect DUB functional localization.[ 35 ] Phosphorylation catalyzed by casein kinase 2 is essential for nuclear localization of OTUB1 and ATXN3 (a member of MJD family), which exerts a critical effect in regulating cellular metabolism during hypoxic stress.[ 87 ] Similarly, USP10 can be phosphorylated by ataxia‐telangiectasia mutated kinase (ATM) and translocate to the nucleus, which interacts with p53 and inhibits p53 nuclear export and degradation; thereby suppressing tumor cell growth.[ 88 , 89 , 90 ] On the contrary, phosphorylation by protein kinase B (PKB/AKT) excludes USP4 from the nucleus, allowing it to reach the plasma membrane and deubiquitinate transforming growth factor‐β (TGF‐β) receptor I (TβRI). This can stabilize the TβR‐I at the plasma membrane and boost TGF‐β signaling, thereby promoting tumorigenesis.[ 91 ]
Moreover, localization is also controlled by altering DUB interactomes. OTUB1 hydroxylation by factor inhibiting HIF (FIH) changes the OTUB1 interactome and substrates.[ 87 ] Ubiquitination of USP4 regulates its interactome and its roles in DNA damage response,[ 92 ] whereas binding of phosphorylated OTULIN prevents its interaction with linear Ub chain assembly complex (LUBAC).[ 93 , 94 ] Numerous DUBs are produced as multiple splice variants which may localize to different compartments and have distinct half‐lives.[ 53 ] USP35 has one form which localizes to the ER and lipid droplets and other forms are targeted to the cytosol. A short form of USP35 is linked to the mitochondria, but this variant lacks an intact catalytic domain.[ 95 , 96 ]
Thus, DUBs localization not only depends on the signal peptide of DUB itself, but also can be affected by PTMs and DUB interactome. This reveals that domains beyond DUBs’ catalytic sites play a pivotal role in regulating their functions.
3. The Roles of DUBs in Cancer
An increasing number of studies show that DUBs play oncogenic or suppressive functions at multidimensional levels in cancer, which are listed in Table 1 and summarized in Figure 5 . The diverse functions of DUBs mainly act on the instability, conformational changes, and signal transduction of substrates. Here, the multiple roles of DUBs in cancer are elucidated as the following aspects: proteasomal degradation, DNA repair, apoptosis, and metastasis.
Table 1.
The roles of DUBs in cancers.
| DUB | Roles | Target | Function | Consequences |
|---|---|---|---|---|
| USP1 | Promotor | ULK1, EZH2, ID1/2/3, CHK1 | Stabilization | Promote proliferation, metastasis, cancer cell stemness, and regulate DDR[ 117 , 118 , 282 , 283 ] |
| USP2 | Promotor | CCND1, CCNA1, FAS, MDM2, MDM4, TβRI | Stabilization | Promote proliferation and metastasis, evade apoptosis[ 284 , 285 , 286 , 287 , 288 ] |
| USP3 | Promotor | SUZ12 | Stabilization | Boost metastasis[ 289 ] |
| Suppressor | p53 | Stabilization | Inhibit proliferation[ 290 ] | |
| USP4 | Promotor | HDAC2, ARF‐BP1, TβRI, Twist1, β‐catenin, PRL‐3, CYPA, | Stabilization | Promote proliferation and metastasis, decrease sensitivity to chemotherapy[ 91 , 144 , 147 , 291 , 292 , 293 ] |
| USP5 | Promotor | c‐Maf, Slug, β‐catenin | Stabilization | Evade apoptosis, promote metastasis[ 294 , 295 , 296 ] |
| USP6 | Promotor | c‐Jun, Fzd, JAK1 | Stabilization | Promote metastasis and tumor growth[ 297 , 298 , 299 ] |
| USP7 | Promotor | CDC25A, N‐Myc, Ki‐67, HIF‐1α, MDM2, MDM4, PHF8, MDC1, β‐catenin, LSD1, NOTCH1, HDM2 | Stabilization |
Antagonize p53‐involved tumor suppression regulations, Promoting tumor metastasis, decrease drug sensitivity, regulate DDR[ 135 , 300 , 301 , 302 , 303 , 304 , 305 , 306 , 307 , 308 , 309 , 310 , 311 , 312 , 313 , 314 , 315 , 316 , 317 ] |
| Suppressor | PD‐L1 | Stabilization | Anti‐tumor immune response[ 257 ] | |
| USP8 | Promotor | Cx43, AKT, TβRII, PD‐L1 | Stabilization | Promote proliferation and metastasis, inhibit immonotherapy[ 318 , 319 ] |
| USP9X | Promotor | pVHL, CEP131, TRB3, SMAD4, SMURF1, TDRD3, β‐catenin | Stabilization | Promoting proliferation and metastasis, evade apoptosis[ 320 , 321 , 322 , 323 , 324 , 325 , 326 , 327 , 328 , 329 , 330 ] |
| Suppressor | YAP1, TIK, MCL1, XIAP, Ets‐1, IRS‐2, CLASPIN, FBW7, AMOT, ITCH, LATS2, CLASPIN | Stabilization or altered activity | Maintain DNA replication fork stability and inhibit proliferation[ 127 , 321 , 324 , 331 , 332 ] | |
| USP10 | Promotor | SMAD4, Slug, TOP2α, FLT3, G3BP2 | Stabilization or altered activity | Promote tumorigenesis and metastasis[ 333 , 334 , 335 , 336 ] |
| Suppressor | MSH2, p53, SIRT6, p14ARF, AMPKα, PTEN, MSH2 | Stabilization | Suppress tumor progression and increase apoptosis, regulate cellular sensitivity to DNA damage[ 88 , 337 , 338 , 339 ] | |
| USP11 | Promotor | XIAP, cIAP2, RAE1, PPP1CA, NF90, eIF4B, TβRII | Stabilization | Promote proliferation and metastasis, evade apoptosis[ 340 , 341 , 342 , 343 , 344 , 345 , 346 , 347 ] |
| Suppressor | VGLL4, ARID1A, PTEN, Mgl‐1, p21, PML, BRCA2 | Stabilization or altered activity | Inhibit tumorigenesis and metastasis, regulate DDR[ 123 , 124 , 348 , 349 , 350 , 351 , 352 , 353 , 354 ] | |
| USP12 | Promotor | AR, MDK, MDM2 | Stabilization | Maintain malignant traits and promote angiogenesis[ 355 , 356 , 357 ] |
| USP13 | Promotor |
MCL1, c‐Myc, MITF RAP80, ACLY, OGDH |
Stabilization | Promote proliferation and metabolism, chemotherapy resistance[ 358 , 359 , 360 , 361 , 362 ] |
| Suppressor | PTEN, RAP80‐BRCA1 | Stabilization | Anti‐tumor and regulate DDR[ 359 , 363 ] | |
| USP14 | Promotor | PI3K, Dvl, Vimentin, Aurora B, AR | Stabilization | Promote proliferation and metastasis, prevent apoptosis and regulate DDR[ 103 , 105 , 364 , 365 , 366 , 367 ] |
| USP15 | Promotor | TOP2α, MDM2, TβRI, HBx | Stabilization or altered activity | Promote metastasis and inhibit anti‐tumor immunoresponses[ 368 , 369 , 370 , 371 ] |
| Suppressor | Keap1, IRS2, p53 | Stabilization or altered activity | Inhibit tumorigenesis and decrease chemo‐resistance[ 372 , 373 , 374 ] | |
| USP17 | Promotor | CDC25A, Geminin, HAS2, Slug, Snail, Twist, SMAD4, BRD4 | Stabilization | Promote tumorigenesis and metastasis, prevent apoptosis[ 166 , 311 , 375 , 376 , 377 , 378 , 379 ] |
| USP18 | Promotor | ZEB1, RARα, BCL2L1, KRAS | Stabilization | Promote tumorigenesis and metastasis, prevent apoptosis[ 380 , 381 , 382 , 383 ] |
| USP19 | Suppressor | HDAC1/2 | Altered activity | Maintain genomic stability[ 384 ] |
| USP20 | Promotor | β‐catenin | Stabilization | Promote tumorigenesis[ 385 ] |
| Suppressor | TRAF6, Tax, CLASPIN | Stabilization or altered activity | Inhibit tumorigenesis[ 386 , 387 ] | |
| USP21 | Promotor | EZH2, BRCA2, MEK2, PD‐L1 | Stabilization | Promote tumorigenesis, metastasis, immune escape[ 265 , 388 , 389 , 390 , 391 ] |
| Suppressor | Fra‐1, MARK1 | Stabilization | Anti‐tumor[ 392 ] | |
| USP22 | Promotor | CCNB1, CCND1, c‐Myc, FBP1, BMI1, COX‐2, EGFR, H2A, KDM1A, SIRT1, PD‐L1 | Altered activity or stabilization | Promote proliferation, metastasis and cancer stemness, drug resistance, reduce sensitivity to immune therapy, regulateγH2AX‐mediated DDR[ 265 , 359 , 393 , 394 , 395 , 396 , 397 , 398 , 399 , 400 , 401 ] |
| Suppressor | PU.1 | Stabilization | Anti‐differentiation[ 402 ] | |
| USP24 | Promotor | β‐TrCP, MCL‐1 | Stabilization | Promote cancer malignancy[ 403 , 404 ] |
| Suppressor | Bax, E2F4, p300, Securin | Stabilization | Promote apoptosis[ 405 ] | |
| USP26 | Promotor | AR, Snail | Altered activity or Stabilization | Promote proliferation and metastasis[ 406 , 407 ] |
| Suppressor | SMAD7 | Stabilization | Inhibit metastasis[ 406 ] | |
| USP27 | Promotor | Cyclin E | Stabilization | Pro‐tumor effects[ 408 ] |
| USP28 | Promotor | c‐Myc, Fbw7 | Stabilization | Promote tumorigenesis and metastasis[ 409 , 410 , 411 , 412 , 413 ] |
| Suppressor | LIN28A, LSD1, c‐Jun, NICD1, p53 | Stabilization | Arrest cell cycle[ 414 ] | |
| USP29 | Promotor | Claspin, Snail | Stabilization | Promote tumorigenesis and metastasis[ 415 , 416 ] |
| Suppressor | p53 | Stabilization | Inhibit proliferation[ 417 ] | |
| USP30 | Promotor | TOM20, DRP1 | Stabilization | Promote migration, evade apoptosis[ 418 , 419 ] |
| USP33 | Promotor | SP1 | Stabilization | Pro‐tumor effects[ 420 , 421 ] |
| Suppressor | PPM1A, Robo1, β‐arrestin2 | Altered activity or Stabilization | Inhibiting tumor metastasis[ 422 , 423 , 424 ] | |
| USP35 | Suppressor | ABIN‐2 | Stabilization | Inhibit proliferation[ 425 ] |
| USP36 | Promotor | c‐Myc, DHX33, CHD7, H2B | Altered activity or stabilization | Promote tumorigenesis and proliferation[ 426 , 427 , 428 , 429 ] |
| USP37 | Promotor | RARA, Gli1, Snail, 14‐3‐3γ, c‐Myc | Stabilization | Promote proliferation and metastasis[ 430 , 431 , 432 , 433 , 434 ] |
| Suppressor | p27 | Stabilization | Inhibit proliferation[ 435 ] | |
| USP42 | Suppressor | p53 | Stabilization | Inhibit proliferation[ 436 ] |
| USP43 | Promotor | H2B, ZEB1 | Altered activity or Stabilization | Promote proliferation and metastasis[ 156 , 437 ] |
| USP44 | Promotor | H2B, EZH2, Securin | Altered activity or Stabilization | Promote proliferation and metastasis[ 438 , 439 , 440 , 441 ] |
| USP46 | Promotor | CDT2, ENO1 | Stabilization | Promote proliferation and metastasis[ 442 , 443 ] |
| Suppressor | PHLPP | Stabilization | Inhibit proliferation[ 444 ] | |
| USP47 | Promotor | β‐catenin, Snail | Stabilization | Promote proliferation and metastasis[ 445 , 446 , 447 ] |
| USP48 | Promotor | TRAF2, Gli1 | Stabilization | Promote tumorigenesis and metastasis[ 448 ] |
| USP49 | Suppressor | p53, FKBP51 | Stabilization | Inhibit proliferation and regulate DDR[ 449 , 450 ] |
| USP51 | Promotor | ZEB1, FAT4 | Stabilization | Promote proliferation and metastasis[ 451 , 452 ] |
| USP52 | Promotor | ASF1A | Stabilization | Drug resistance[ 453 ] |
| CYLD | Suppressor | Dvl, p53, TRAF2, Bcl‐3, c‐Jun, c‐Fox | Stabilization | Suppress tumorigenesis and metastasis, promote apoptosis, regulate DDR[ 454 , 455 , 456 , 457 , 458 , 459 , 460 ] |
| OTUB1 | Promotor | c‐IAP1, FOXM1, Snail, SLC7A11, PD‐L1, UBC13 | Stabilization | Promote tumorigenesis, metastasis and immune escape, regulate DDR[ 131 , 461 , 462 , 463 , 464 , 465 ] |
| Suppressor | Erα, RPA1, DEPTOR | Stabilization or altered activity | Suppress tumorigenesis[ 466 , 467 , 468 ] | |
| OTUB2 | Promotor | YAP/TAZ, U2AF2 | Stabilization | Promote metastasis[ 68 , 469 ] |
| OTUD1 | Suppressor | YAP, p53, SMAD7 | Stabilization or altered activity | Suppress tumorigenesis cancer stemness and metastasis[ 470 , 471 , 472 ] |
| OTUD2 | Promotor | ITCH | Stabilization | Promote proliferation[ 473 ] |
| OTUD3 | Promotor | GRP78 | Stabilization | Promote tumorigenesis[ 474 ] |
| Suppressor | PTEN, p53 | Stabilization | Inhibit proliferation and induce apoptosis[ 475 , 476 ] | |
| OTUD5 | Suppressor | p53, PDCD5, UBR5 | Stabilization | Promote apoptosis and drug sensitivity[ 477 , 478 , 479 ] |
| OTUD6B | Suppressor | pVHL | Stabilization | Inhibit metastasis[ 480 ] |
| OTUD7A | Suppressor | TRAF6 | Altered activity | Inhibit metastasis[ 481 ] |
| OTUD7B | Promotor | GβL, Aurora A, Cyclin B, EGFR | Stabilization or altered activity | Promote turiogenesis, proliferation, angiogenesis[ 482 , 483 , 484 , 485 ] |
| A20 | Promotor | ERα | Stabilization | Promote proliferation[ 486 ] |
| TRABID | Promotor | EZH2 | Stabilization | Promote proliferation[ 487 ] |
| Suppressor | Twist1 | Altered activity | Inhibit metastasis[ 488 ] | |
| UCHL1 | Promotor | HIF‐1α, TβRI, SMAD2 | Stabilization | Pro‐metastasis[ 169 , 489 ] |
| Suppressor | NOXA, p53 | Stabilization | Inhibit proliferation, induce chemosensitivity[ 141 , 490 ] | |
| UCHL3 | Promotor | TDP1 | Stabilization | Drug resistance[ 491 ] |
| UCHL5 | Promotor | TβRI, PRP19 | Stabilization | Promote tumorigenesis and metastasis[ 107 , 492 ] |
| BAP1 | Suppressor | H2A, γ‐tubulin, Ino80, IP3R3, MCRS1 | Altered activity or Stabilization | Maintain chromosomal stability, inducecell apoptosis[ 493 , 494 , 495 , 496 , 497 ] |
| ATXN3 | Promotor | KLF4 | Stabilization | Promote proliferation[ 498 ] |
| Suppressor | p53 | Stabilization | Induce apoptosis[ 499 ] | |
| ATXN3L | Promotor | KLF5 | Stabilization | Promote proliferation[ 500 ] |
| POH1 | Promotor | E2F1 | Stabilization | Promote proliferation[ 98 ] |
| COPS5 | Promotor | PD‐L1, HK2, Trx, Snail, Survivin, FOXM1, ZEB1 | Stabilization | Reduce sensitivity to immune therapy, promote proliferation and metastasis[ 263 , 501 , 502 , 503 , 504 , 505 ] |
| COPS6 | Promotor | CHIP, CTSL, PD‐L1 | Altered activity | Promote immune escape, promote proliferation and metastasis[ 506 , 507 , 508 ] |
| AMSH | Promotor | Slug | Stabilization | Promote metastasis[ 509 ] |
| BRCC36 | Promotor | NuMA | Altered activity | Pro‐proliferation[ 510 ] |
| MYSM1 | Suppressor | H2A | Altered activity | Anti‐tumor effects[ 511 ] |
Figure 5.
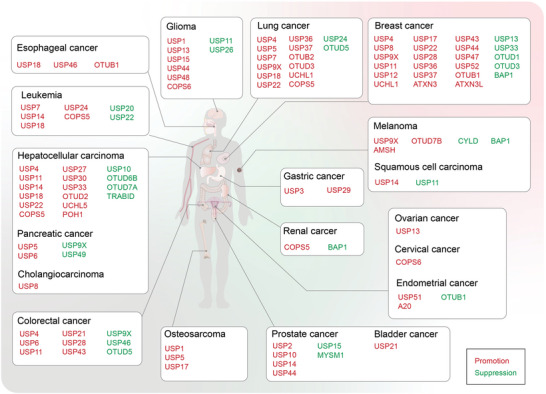
Overview of Janus‐faced roles of DUBs in cancers. DUBs exhibit dual roles in the pathogenesis of diverse cancers, including glioma, lung cancer, breast cancer, melanoma, ovarian cancer, cervical cancer, endometrial cancer, gastric cancer, renal cancer, prostate cancer, bladder cancer, osteosarcoma, colorectal carcinoma hepatocellular carcinoma pancreatic adenocarcinoma cholangiocarcinoma leukemia and esophageal squamous cell carcinoma, affecting multiple organs within the human body. DUBs exhibiting tumor‐promoting function are visually highlighted by red, while those demonstrating tumor‐suppressing function are visually indicated by green.
3.1. DUBs in Tumorigenesis
Proteasomal degradation plays a crucial role in governing the turnover and function of intracellular proteins involving in cancer cell growth and death. Taken the aforementioned proteasome‐related DUBs, USP14, PSMD14, and UCHL5, for instance. The levels of PSMD14 are negatively correlated with the survival of patients with multiple myeloma and its depletion affects the proliferation of multiple myeloma cells.[ 97 ] Elevated nuclear PSMD14 is seen in hepatocellular carcinomas and promotes tumour growth.[ 98 ] Furthermore, PSMD14 has been identified to control the ubiquitination and stability of the oncogene receptor tyrosine kinase ERBB2.[ 99 ] PSMD14 deubiquitinates Ub‐chain at the double‐strand break (DSB) sites to promote the recruitment of RAS associated with diabetes protein 51 (RAD51), which enhances the cellular responses to DSB and undergoes DSB signaling and repair by homologous recombination (HR).[ 100 ] PSMD14 depletion sensitizes and kills cancer cells with a strong reliance on HR.[ 100 ]
USP14 expression is connected with colorectal cancer, intrahepatic bile duct cancer, lung cancer, and ovarian cancer.[ 101 , 102 ] High USP14 expression correlates with poor prognosis in non‐small‐cell lung carcinoma (NSCLC).[ 103 ] Up‐regulation of USP14 in cisplatin‐resistance ovarian cancer contributes to cancer cell proliferation by stabilizing oncoprotein B‐cell lymphoma 6 (BCL6).[ 104 ] Moreover, high USP14 expression guards Aurora‐B (an anti‐apoptotic protein) against degradation and inhibits cell apoptosis induced by chemotherapeutic drugs in leukemic cells.[ 105 ] Similarly, UCHL5 is overexpressed in epithelial ovarian cancer, which is closely associated with advanced tumor progression and poor prognosis.[ 106 ] UCHL5 is also upregulated in hepatocellular carcinoma, and promote cells migration and invasion by deubiquitinating pre‐mRNA‐processing factor 19 (PRP19).[ 107 ] Unlike PSMD14, UCHL5 and USP14 are not fundamental constituents of the proteasome. Interestingly, depletion of either UCHL5 or USP14 alone has no measurable effect on cell viability and structure or proteolytic capacity of the proteasome, but does facilitate cellular protein degradation. In comparison, depletion of both of these DUBs reduces protein degradation, indicating that there are overlapping functions between them.[ 108 ]
3.2. DUBs in Tumor DNA Damage Repair
Dysregulation or loss of DNA repair and DNA damage response pathways (DDR) is one hallmark of cancer due to DSBs; this lethal damaging event must be repaired before cell division occurs.[ 109 , 110 , 111 ] Ubiquitination and deubiquitination closely participate in DNA repair and DDR.[ 112 , 113 ] DUBs perform a multifaceted role in DNA repair during tumorigenesis (Figure 6A and Table 2 ). USP1 regulates Fanconi anemia group D2 protein (FANCD2) ubiquitination which plays a vital role in the Fanconi anemia pathway of DNA crosslink repair.[ 114 , 115 ] USP1 depletion increases voluntary monoubiquitination of FANCD2 and impairs accumulation of the Fanconi anemia core complex at DNA damage sites in a cell cycle‐dependent manner.[ 115 ] USP1 promotes multiple rounds of repair in the S phase by modulating the turnover of FANCD2‐ Fanconi anemia group I protein (FANCI) monoubiquitination at the damaged sites. Similarly, USP1 deubiquitinated the proliferating cell nuclear antigen (PCNA) to govern the error‐prone translesion synthesis repair pathway.[ 116 ] In addition, USP1 activities are involved in regulating a feedback loop to inhibit DDR checkpoint kinase 1(CHK1) activity,[ 117 ] thereby regulating cellular differentiation in osteosarcoma cells via deubiquitination, and impairing the stability of DNA‐binding protein inhibitors.[ 118 ] USP4 can directly regulate DNA end resection. USP4 autodeubiquitylation regulates CtIP recruitment to sites of DNA damage.[ 119 , 120 , 121 ] USP52 can directly interact with CtIP and deubiquitinate it, thereby promoting and facilitate the phosphorylation and activation of CtIP for DNA repair.[ 122 ] USP11 is also a DNA repair‐associated DUB that forms a complex with breast cancer type 2 susceptibility protein (BRCA2), which boosts HR in DSB sites and plays a tumor suppressor role in DDR.[ 123 ] Inhibiting USP11 activity sensitizes cancer cells to olaparid which limits the activity of DDR enzyme poly(ADP‐ribose) polymerase 1 (PARP1).[ 124 ] USP15 deubiquitinates BARD1, and promotes BARD1‐HP1γ interaction, resulting in BRCA1/BARD1 retention at DSBs. Cancer‐associated USP15 mutations increase PARP inhibitor sensitivity in cancer cells.[ 125 ]
Figure 6.
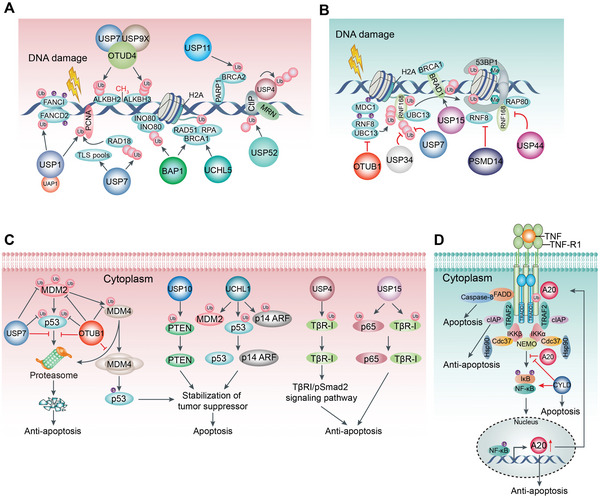
DUBs play multi‐faced roles in tumor‐associated DNA repair and apoptosis. A) Replication‐associated repair is regulated by DUBs. USP1 promotes multiple rounds of repair in the S phase by modulating the turnover of FANCD2‐FANCI monoubiquitination at the damaged sites. Similarly, PCNA is deubiquitinated by USP1 to govern the error‐prone translesion synthesis repair pathway. USP7 indirectly coordinates PCNA deubiquitination. OTUD4 acts as a scaffold to form a complex with USP7 and USP9X which interacts with ALKBH2 and ALKBH3 to suppress their ubiquitination and degradation. ALKBH2 and ALKBH3 preferentially remove methyl groups from single‐stranded and double‐stranded DNA and reverse DNA alkylation. In addition, USP9X upholds the stability of the DNA replication fork and the response of DNA damage checkpoint by modulating the protein claspin in the S phase. BAP1 promotes HR repair by recruiting BRCA1, RAD51, and RPA to DSB sites and reduces ubiquitinated forms of H2A and H2AX. USP11 forms a complex with BRCA2 that boosts HR in DSB sites and limits PARP1 activity. DUBs govern Ub‐dependent signaling, contributing to DNA repair. USP4 autodeubiquitylation regulates CtIP recruitment to sites of DNA damage. USP52 deubiquitinates CtIP, thereby promoting and facilitating the phosphorylation and activation of CtIP for DNA repair. B) Double‐strand breaks (DSBs) promote ATM kinase to phosphorylate histone H2A and MDC1, a mediator of DNA damage checkpoint protein 1. This leads to the recruitment of E3 ligase RNF8 conjugated with E2 enzyme UBC13 that induces the production of Lys63‐linked Ub chains on scaffold proteins (such as RNF168) in response to DNA damage. Several DUBs counteract this ubiquitination in diverse manners including OTUB1, USP34, and USP7, although some are functionally redundant. Additionally, oligomerized 53BP1 is stably recruited by RNF168‐ and RNF8‐regulated chromatin ubiquitination, which is inhibited by PSMD14 and USP44. USP15 deubiquitinates BARD1, and promotes BARD1‐HP1γ interaction, resulting in BRCA1/BARD1 retention at DSBs. C) The role of DUBs in regulating apoptosis. DUBs increase or decrease apoptosis by inhibiting proteasomal degradation of pro‐apoptotic or anti‐apoptotic proteins through deubiquitination. USP7, OTUB1, UCHL1, and USP10 promote apoptosis by enhancing the stability of tumor suppressors such as p53, PTEN, and p14 ARF. Conversely, the oncogene of TβR‐I and p65 are stabilized by USP4 and USP15, which exert an anti‐apoptotic effect. D) Moreover, A20 is part of a negative feedback mechanism inhibiting continuous NF‐κB activation during single TNF stimulation, wherein A20 expression is constitutively activated and prevents cells from apoptosis through stabilizing the Ub network involved in the TNFR1 signaling complex. However, the DUB CYLD antagonizes this process.
Table 2.
The function of DUBs in DNA damage response.
| DUB | Target | Process | Rationale |
|---|---|---|---|
| USP1 | FANCI | Homologous recombination (HR) | Efficient foci formation at sites of DNA damge[ 274 ] |
| Promote homologous recombination[ 512 ] | |||
| PCNA | Translesion synthesis (TLS) | Promote recruitment of TLS polymerases[ 116 ] | |
| USP4 | USP4 itself | HR | USP4 autodeubiquitylation regulates CtIP recruitment to sites of DNA damage[ 119 , 120 , 121 ] |
| USP7 | PCNA | HR | Regulate DNA polymerase η stability[ 513 ] |
| Rad18 | TLS | Maintain Rad18 stability and DNA damage tolerance[ 514 ] | |
| RNF168 | TLS | Stabilize RNF168 and regulate BRCA1 recruitment at DSB sites[ 515 ] | |
| RNF169 | TLS | Support the nuclear function of DSB response protein[ 516 ] | |
| ERCC6 | Nucleotide‐excision repair (NER) | Regulate the stability of ERCC6 for transcription‐coupled NER[ 517 ] | |
| XPC | NER | Regulate NER via deubiquitinating XPC[ 518 ] | |
| USP7S | Mule | Base excision repair (BER) | Prevent Mule self‐ubiquitination and regulate DNA damage and repair[ 519 ] |
| USP9X | MCL1 | HR | Stabilize MCL‐1 and promote homologous recombination[ 329 , 520 ] |
| ALKBH2 | BER | Promote alkylation damage resistance[ 126 ] | |
| USP11 | XPC | NER | Maintain NER capacity via deubiquitinating XPC[ 350 ] |
| H2AX | HR | Maintain the proper status of ubiquitylation γH2AX to repair DSB[ 521 ] | |
| USP15 | BARD1 | BER | USP15 deubiquitinates BARD1, and promotes BARD1‐HP1γ interaction, resulting in BRCA1/BARD1 retention at DSBs[ 125 ] |
| USP24 | DDB2 | NER | Regulate DDB2 stability and homologous recombination repair[ 124 , 522 ] |
| USP34 | RNF168 | HR | Promote ubiquitin signaling at DNA double‐strand breaks[ 523 ] |
| USP45 | ERCC1 | NER | Promote translocation of ERCC1 to foci of DNA damage[ 524 ] |
| USP47 | Polymerase β | BER | Regulate DNA repair and maintain genome integrity[ 525 ] |
| USP52 | CtIP | HR | Remove the ubiquitination of CtIP to facilitate the phosphorylation and activation of CtIP[ 122 ] |
| BAP1 | BRCA1 | HR | Promote DNA repair and cellular recovery from DNA damage[ 526 ] |
| UCHL5 | Ino80 | BER | Regulate transcription and DNA repair[ 527 ] |
| OTUB1 | UBC13 | HR | Suppress DNA damage response[ 131 ] |
| OTUB2 | L3MBTL1 | HR | Fine‐tune DSB speed and regulate DNA repair pathway[ 528 ] |
| OTUD4 | ALKBH2 | BER | Promote alkylation damage resistance via stabilization of AlkB homologues[ 126 ] |
| BRCC36 | BRCA1 | HR | Limit DNA Break Processing and Repair[ 529 ] |
AlkB homolog (ALKBH) 2 and 3 preferentially remove methyl groups from single‐stranded and double‐stranded DNA and reverse DNA alkylation, one of the most common mutagenic events in the cell.[ 126 ] OTUD4 serves as a scaffold to form a complex with USP7 and USP9X which then interacts with ALKBH2 and ALKBH3 to suppress ubiquitination and degradation.[ 126 ] USP9X activity can uphold the stability of the DNA replication fork and DDR checkpoint by modulating the protein claspin in the S phase.[ 127 ] Additionally, USP9X influences radiosensitivity in glioblastoma cells through Myeloid cell leukemia 1 (MCL1)‐dependent or ‐independent mechanisms.[ 128 ] Breast cancer susceptibility gene 1 (BRCA1) associated protein 1 (BAP1) is a nuclear DUB with a UCH domain. It functions as a tumor suppressor on breast cancer growth.[ 129 ] BAP1 promotes HR repair by recruiting BRCA1, RAD51, and replication protein A (RPA) to DSB sites and reducing ubiquitinated forms of H2A and γH2AX.[ 130 ]
The above DUBs all play positive regulatory roles in DNA repair; however, several other DUBs respond to DNA repair with opposing effects (Figure 6B and Table 2). OTUB1 is a Lys48 linkage‐specific DUB; however, OTUB1 overexpression inhibits Lys63 chains by interacting with ubiquitin‐charged E2 enzyme UBC13 and disrupting ring finger protein 168 (RNF168)‐mediated polyubiquitination during DSB repair.[ 131 ] Intriguingly, OTUB1 efficiently limits the DDR mediator/adaptor p53 binding protein 1 (53BP1) foci formation independent of its catalytic activity.[ 131 ] Moreover, PSMD14 cleaves the Lys63 chains to modulate DSB repair signaling, in addition to disassembling the Lys48 chains of proteasomal substrates.[ 41 ] PSMD14 depletion enlarges the formation of the nuclear foci for Ub and 53BP1, indicating that the DSB repair signal propagates without DUB, and PSMD14 estranges the role of RNF8 in Lys63 chains formation and nonhomologous end joining (NHEJ) repair.[ 132 ] In addition, USP44 is an antagonist of RNF168‐regulated Ub conjugates at DSB sites.[ 133 ] The highly abundant USP44 curbs foci formation of ionizing radiation (IR)‐induced Ub conjugates, RNF168, receptor‐associated protein 80 (RAP80), and 53BP1 at DSB sites through antagonizing IR‐ and RNF168‐mediated H2A ubiquitination.[ 133 ]
These examples reveal that DUBs implement multiple regulatory effects on DNA repair and DDR pathways, including recruitment and stability of core regulators, deubiquitination of nucleosome core particle, and protein interactome in a catalytically independent manner at DNA damage sites. However, DUBs perform a multifaceted role in DNA repair during tumorigenesis, further in‐depth research is urgently needed to determine the detailed and fine‐tuned regulatory mechanisms of DUBs.
3.3. DUBs in Tumor Apoptosis
Apoptosis‐mediated cell death serves as a pivotal tumor suppressor in cancer progression. DUBs are demonstrated as a main regulator of apoptosis.[ 134 ] USP7 can remove Ub‐chain on Mdm2 conjugated by self‐ubiquitination, thus supporting Mdm2‐dependent p53 degradation.[ 135 , 136 ] USP7 depletion stimulates MDM2 proteasomal degradation and promotes p53‐mediated apoptosis in tumor cells through stabilizing p53.[ 137 , 138 ] In addition, p53 and MDM2 are also regulated by OTUB1, which controls p53 stabilization and activation by disrupting the interaction between p53 and MDM2.[ 139 ] Moreover, OTUB1 can stabilize murine double minute 4 (MDM4) by curbing MDM4 ubiquitination mediated by MDM2. This promotes MDM4 accumulation in the cytoplasm and mitochondria, p53 phosphorylation, and mitochondrial‐mediated apoptosis.[ 140 ] UCHL1 forms a complex with p53/p14 ARF/ MDM2 to deubiquitinates p53, and p14 ARF for p53 signaling and apoptosis triggering in nasopharyngeal carcinoma and breast cancer.[ 141 , 142 ] USP10 is another DUB with a pro‐apoptosis function; it modulates phosphatase and tensin homolog (PTEN) in lung and breast cancer cell lines.[ 143 ] Knockdown of USP10 promotes tumor growth, invasion, and metastasis antagonized by PTEN overexpression (Figure 6C).[ 143 ]
Moreover, USP4 can deubiquitinate and stabilize the TβRI, which plays a positive role in liver tumorigenesis.[ 144 ] Subsequent activation of the TβRI/pSmad2 signaling pathway leads to cell migration and invasion in glioblastoma, breast cancer, liver, and ovarian cancer.[ 145 , 146 , 147 ] In addition, USP15 is up‐regulated in degenerative nucleus pulposus (NP) cells.[ 148 ] Its overexpression reduces AKT phosphorylation and accelerates cell apoptosis by enhancing the ubiquitination of FK506‐binding protein 5 (FKBP5).[ 148 ] Furthermore, NF‐κB p65 (RelA) and TβR‐I are potent targets for USP15 through deubiquitination, which plays multiple roles in mediating tumorigenesis in various carcinomas (Figure 6C).[ 149 ]
On the contrary, several DUBs diminish apoptosis by controlling proteasomal degradation of pivotal anti‐apoptotic protein substrates via deubiquitination. A20 has a potent anti‐inflammatory function as part of a negative feedback mechanism by inhibiting NF‐κB activation.[ 150 ] A20 deficiency increases receptor‐interacting protein kinase 1 (RIPK1) dependent and independent apoptosis in this process. Upon single tumor necrosis factor‐α (TNF‐α) stimulation, A20 expression is then constitutively activated and interacts with the TNF receptor 1 (TNFR1) signaling complex, which prevents apoptosis through stabilizing the Ub network involved in TNFR1 signaling complex.[ 151 ] Intriguingly, this process is independent of A20's E3 ligase‐ and deubiquitylase activities and can be antagonized by another DUB, CYLD (Figure 6D).[ 152 ]
Whereas, some DUBs play a Janus‐faced role in pro‐/anti‐apoptois of cancer, including UCHL5, USP2, USP4, USP7, USP9X, USP10, and USP14.[ 153 , 154 ] The reasons for the opposing roles depend on diverse functions of linkage type for deubiquitination, selective protein substrates associated with oncogenic or tumor suppressive effect, individual interactome, and an ever‐changing microenvironment. It is worth noting that tumor heterogeneity endows extensive differential expression and divergent regulatory roles of DUBs.
3.4. DUBs in Tumor Invasion and Metastasis
The leading cause of cancer‐related mortality is metastasis which involves tumor cell migration, extravasation, circulation, immune escape, intravasation into the adjacent tissues, and cell death.[ 155 ] DUB dysregulation disturbs the level of ubiquitination and function of proteins associated with tumor metastasis which ultimately induces tumor deterioration. For illustration, USP43 is a nuclear‐localized DUB, which is tightly connected with the nucleosome remodeling and deacetylase (NuRD) particle.[ 156 ] The USP43‐NuRD complex synergistically deubiquitinates H2BK120 and represses a host of genes, including epidermal growth factor receptor (EGFR). USP43 thus robustly suppresses breast cancer growth and metastasis.[ 156 ] In breast cancer, USP43 can be phosphorylated on Ser29 by activated AKT and sequestrated in the cytoplasm through interacting with the 14‐3‐3β/ε heterodimer.[ 156 ] Decreased nuclear USP43 leads to EGFR upregulation, which further strengthens the EGFR/PI3K/AKT pathway to accelerate breast cancer progression.[ 156 ] BAP1 deubiquitinates PTEN and stabilizes it, which allows PTEN to negatively mediate AKT kinase activity. BAP1 inhibition causes the reduction of PTEN protein and enhancement of the AKT signaling pathway which accelerates the malignant transformation and metastasis in prostate cancer.[ 157 ] Besides, BAP1 deubiquitinates histone 2A (H2A) ubiquitination on the cystine/glutamate transporter (SLC7A11) promoter and represses SLC7A11expression which causes ferroptosis to control tumor development and metastasis.[ 158 , 159 , 160 ] OTUD1 selectively trims the Lys48‐chains from SMAD7 to prevent it from proteasomal degradation in breast cancer (Figure 7A).[ 161 ] Moreover, OTUD1 eliminates the Lys33‐chains and exposes the PY motif of SMAD7, which can subsequently bind to E3 ligases SMURF2 for TβRI degradation and ubiquitination.[ 162 , 163 ] Consequently, OTUD1 acts dual effects on SMAD7 to mitigate TGF‐β signaling and inhibit metastasis.
Figure 7.
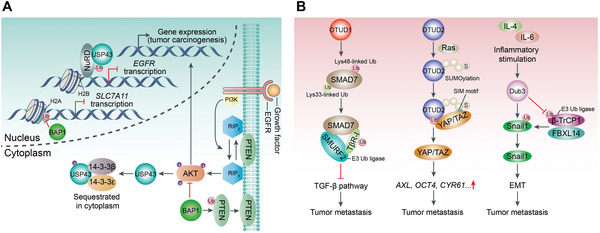
DUBs regulate tumor metastasis. A) USP43 interacts with NuRD to synergistically catalyze H2BK120 deubiquitination and repress EGFR expression which suppresses the activity of the EGFR/PI3K/AKT pathway, as well as the growth and metastasis of cancer. However, USP43 can be phosphorylated by activated AKT, and sequestrated in the cytoplasm to limit USP43 function. Additionally, BAP1 deubiquitinates and stabilizes PTEN (a protein that negatively governs the AKT pathway). Besides, BAP1 deubiquitinates H2A on the SLC7A11 promoter and represses SLC7A11 expression, which causes ferroptosis to control tumor development and metastasis. B) OTUD1 has dual effects on SMAD7 according to the Ub linkage type. OTUD1 selectively cleaves Lys48‐linked Ub chain from SMAD7 to prevent it from proteasomal degradation which promotes tumor metastasis. Moreover, OTUD1 eliminates Lys33‐linked Ub chain from SMAD7, which can be subsequently bound by SMURF2 (E3 Ub‐protein ligases) to degrade TβRI through ubiquitination, thereby inhibiting the TGF‐β pathway and tumor metastasis. SUMOylated OTUB2 interacts with YAP/TAZ by the SUMO‐interacting motif in YAP and TAZ which promotes the accumulation of YAP and TAZ through deubiquitination and the transcriptional activation of AXL, OCT4, CYR61 to increase cell proliferation and metastasis. Dub3 is induced by IL‐4 and IL‐6, and stabilizes Snail1 (a pivotal EMT‐driving transcription factor) through its deubiquitinase activity, and promotes tumor cell migration, invasion, and metastasis.
In contrast, some DUBs promote tumor metastasis (Figure 7B). OTUB2 can be poly‐SUMOylated on lysine 233 and SUMOylated‐OTUB2 can interact with yes‐associated protein (YAP)/transcriptional coactivator with PDZ‐binding motif (TAZ) by the SUMO‐binding motif of YAP and TAZ.[ 68 ] OTUB2 promotes the accumulation of YAP and TAZ through deubiquitination, and their translocation to the nucleus where they interact with the TEA domain family of transcription factors to activate genes that potentiate cell proliferation and metastasis in a Hippo‐independent manner.[ 68 ] Dub3 is an early inducible DUB by cytokines, such as IL‐4 and IL‐6.[ 164 ] In breast cancer, overexpressed Dub3 can stabilize epithelial‐mesenchymal transition (EMT)‐driving transcription factor Snail1[ 165 ] through its deubiquitylase activity to promote tumor cell migration, invasion, and metastasis.[ 166 ] Therefore, Dub3 rapidly responds to inflammatory stimulation and exerts its tumor‐promoting effect by stabilizing Snail1.[ 164 , 166 ] USP7 interacts with N‐methyltransferase enhancer of zeste homolog 2 (EZH2) to eliminate ubiquitination and improve forkhead box protein A1 (FOXA1) stability, which facilitates the progression of prostate cancer.[ 167 , 168 ] Moreover, UCHL1 facilitates TGFβ signaling‐induced breast cancer metastasis by protecting TβRI and SMAD2 from ubiquitination.[ 169 ]
Taken together, the functions of DUBs in cancer can be roughly classified in the following aspects. On the one hand, nuclear‐localized DUBs can directly or indirectly govern the transcription of tumor driver genes through deubiquitylating on histones or forming a complex with cofactors. Additionally, these DUBs also can serve as a scaffold to recruit core regulators for DNA repair. On the other hand, cytosolic‐localized DUBs exert diverse effects by stabilizing classical tumor suppressors or promotors, such as PTEN, TβRI, and p53. This involves the broad crosstalk between EGFR/PI3K/AKT, TGF‐β/SMAD, and NF‐κB signaling. It should be noted that potent inhibitors restraining DUBs activity may exhibit opposite anticancer efficacy in different cancer types.
4. The Roles of DUBs in Immunity
Innate immunity is the first line of host defense against invading viral pathogens. It is mainly activated through the recognition of pathogen‐associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs).[ 170 , 171 ] There are three classes of PRRs: Toll‐like receptors (TLRs), retinoic acid‐inducible gene I (RIG‐I)‐like receptors (RLRs), and DNA sensor cyclic GMP‐AMP (cGAMP) synthase (cGAS).[ 172 ] These receptor‐initiated signaling cascades contribute to the production and secretion of pro‐inflammatory cytokines and type I interferon (IFN) to elicit immune responses.
Given that both excessive increase or decrease in PRRs signaling may cause physiological disorders. PRRs signaling is thus tightly controlled at both spatial and temporal levels in a coordinated manner. For instance, spatially, compartmentalization has been recognized as a major safeguard mechanism that prevents PRRs from activation by self‐DNA/RNA, such as restraining cGAS on the plasma membrane through PI(4,5)P2 binding[ 173 ] or in the nucleus via nucleosomes/BAF association.[ 174 ] In addition, multiple truncated isoforms of MAVS prevent its spontaneous aggregation by interacting with full‐length MAVS and spatially isolating MAVS monomers from each other on the outer mitochondrial membrane.[ 175 ] MAVS could localize on peroxisomes to induce the rapid interferon‐independent expression of defense factors that provide short‐term protection.[ 176 ] Temporally, relative long‐term responses of PRRs signaling are transcriptionally regulated.
PRRs signaling is also fine‐tuned by different PTMs of PRRs and other PRRs signaling components, by which a proper and quick innate‐immune response is triggered under distinct conditions. Ubiquitination widely participates in the innate immune signaling cascades (Figure 8 ). Notably, non‐degradative Met1‐/Lys33‐/Lys63‐linked Ub‐chain governs the critical upstream event of recruiting recognition receptors and activating numerous signal transduction cascades to resist pathogens and maintain immune homeostasis.[ 17 ] Ubiquitination of adaptor TRAF3 with Lys33‐linked Ub‐chain serves to connect innate immune signaling to the cellular trafficking apparatus, which crucially ensured temporal and spatial accuracy in response to innate immune signaling.[ 177 ] Met1‐/Lys63‐linked ubiquitination induces NF‐κB essential modulator (NEMO) compartmentalization to effectively activate NF‐κB signaling.[ 178 ] MAVS activation and aggregation are promoted by K63‐linked ubiquitination upon viral infection.[ 179 ] Therefore, DUBs are rationally involved in the regulation of PRRs signaling as summarized in Table 3 .
Figure 8.
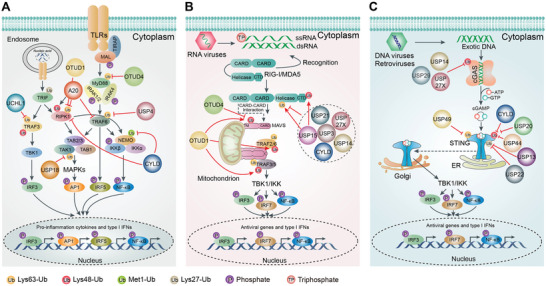
DUBs in innate immune receptor signaling. Innate immunity is activated by signaling cascades associated with pattern recognition receptors (PRRs), including TLRs (A), RLRs (B), and cGAS (C). A) Almost all reported DUBs play a negative regulatory role in TRL signaling, including A20, CYLD, UCHL1, OTUD4, and USP4. These DUBs promote signaling transduction and stabilization of the key mediators in TLR signaling cascades by cleaving the Lys63‐ or Lys48‐linked Ub‐chain from them, such as TRAF6, MyD88, NEMO, TRAF3, and RIP1. This avoids excessive production of pro‐inflammatory cytokines and IFNs. B) Unlike TLR signaling, RLRs can be selectively positively or negatively regulated by DUBs through removal of non‐degradative Lys63‐linked Ub chains or degradative Lys48‐linked Ub chains. CYLD removes Lys63‐linked polyubiquitin chains from RIG‐I, which disrupts the fundamental activation levels and impairs the production of IFN induced by RIG‐I. Additionally, USP3, USP21, USP14, USP27X, and USP15 similarly serve as negative regulators of signaling governed by RIG‐I by removing Lys63‐linked ubiquitination on RIG‐I. However, USP15 also exerts an opposite function through cleaving Lys48‐linked Ub‐chain, which contributes to continuing type I IFN expression. Besides, USP17, USP4, and OTUD4 remove the Lys48‐linked Ub chains from RIG‐I, MDA5, and MAVS, respectively, thereby limiting their proteasomal degradation and facilitating the expression of type I IFN and Antiviral Responses. C) USP14 and USP27X stabilize cGAS by removing the Lys48‐linked Ub‐chain during cGAS‐STING signaling, and promote signaling cascades and type I IFN production. STING can also be stabilized by USP20, CYLD, and USP44 via cleaving Lys48‐linked Ub‐chain. Meanwhile, USP49 removes Lys63‐linked Ub‐chain of STING which leads to decreased interactions between STING and TBK1, and inhibits Antiviral Responses. Moreover, USP13 and USP22 exert a comparable effect on STING and play a negative regulatory role in cGAS‐STING signaling.
Table 3.
The roles of DUBs in immunity.
| DUB | Effect | Target | Linkage | Rationale |
|---|---|---|---|---|
| TLRs signaling | ||||
| USP4 | Inhibition | TRAF6 | Lys63 | Prevent NF‐κB and AP1 signaling[ 187 ] |
| USP10 | Inhibition | TRAF6 | Lys63 | Terminate NF‐κB signaling[ 530 ] |
| Inhibition | NEMO | Met1 | Terminate NF‐κB signaling[ 531 ] | |
| USP18 | Inhibition | TAK1, NEMO | Lys63 | Inhibit TLR/NF‐κB signaling[ 182 ] |
| USP19 | Inhibition | TRIF | Lys27 | Inhibit TLR3/4 signaling[ 532 ] |
| USP25 | Activation | TRAF3/6 | Lys48 | Stabilize TRAF3, promote Antiviral Res..ponses[ 190 , 191 ] |
| OTUB1/2 | Inhibition | TRAF3/6 | Lys63 | Inhibit Antiviral Res..ponses[ 533 ] |
| OTUD1 | Inhibition | TRAF3/6 | Lys48 | Stabilize Smurf1, and promotes the Smurf1‐mediated degradation of MAVS, TRAF3/6[ 534 ] |
| RIPK1 | Lys63 | Inhibit RIPK1‐mediated NF‐κB signaling to suppress colonic inflammation[ 185 ] | ||
| OTUD4 | Inhibition | MyD88 | Lys63 | Inhibit TLR‐mediated NF‐κB signaling[ 186 ] |
| OTUD7B | Inhibition | TRAF3 | Lys48 | Prevent aberrant non‐canonical NF‐κB activation[ 189 ] |
| MYSM1 | Inhibition | TRAF3/6 | Lys63 | Terminate TLR‐induced Antiviral Res..ponses[ 535 ] |
| A20 | Inhibition | TRAF6 | Lys63 | Terminate TLR/NF‐κB activation[ 180 ] |
| NEMO | N/A | Block IKK phosphorylation, or inhibiting NF‐κB activation[ 181 , 536 ] | ||
| UCHL1 | Inhibition | TRAF3 | Lys63 | Inhibit p65 phosphorylation and NF‐κB signaling[ 188 ] |
| CYLD | Inhibition | NEMO | Met1 | Inhibit NF‐κB signaling[ 184 ] |
| RLRs signaling | ||||
| USP3 | Inhibition | RIG‐I | Lys63 | Inhibit type I IFN signaling[ 207 ] |
| USP4 | Activation | RIG‐I | Lys48 | Stabilize RIG‐I, sustain IFN induction[ 214 ] |
| Activation | TRAF6 | Lys48 | Stabilize TRAF6, inhibit EV71 replication[ 537 ] | |
| USP5 | Inhibition | RIG‐I | Lys11/48 | Induce RIG‐I degradation[ 233 ] |
| USP14 | Inhibition | RIG‐I | Lys63 | Inhibit RIG‐I‐triggered type I IFN signaling[ 208 ] |
| USP15 | Inhibition | RIG‐I | Lys63 | Inhibit RIG‐I signaling[ 209 ] |
| Activation | TRIM25 | Lys48 | Stablize TRIM25, then enhance the TRIM25‐ and RIG‐I‐dependent type I IFN production[ 212 ] | |
| USP17 | Activation | RIG‐1 | Lys48 | Stabilize RIG‐I, boost Antiviral Res..ponses[ 213 ] |
| USP21 | Inhibition | RIG‐I | Lys63 | Inhibit RIG‐I‐dependent Antiviral Res..ponses[ 210 ] |
| USP27X | Inhibition | RIG‐I | Lys63 | Inhibit RIG‐I‐dependent Antiviral Res..ponses[ 211 ] |
| CYLD | Inhibition | RIG‐I, TBK1 | Lys63 | Inhibit RIG‐I‐dependent Antiviral Res..ponses[ 206 ] |
| OTUD2 | Inhibition | MAVS | Lys63 | Inhibit IRF3, p65 activation, and IFN‐β production[ 538 ] |
| OTUD3 | Inhibition | MAVS | Lys63 | Inhibit Antiviral Res..ponses[ 539 ] |
| OTUD4 | Activation | MAVS | Lys48 | Stablize MAVS, induce antiviral signaling[ 215 ] |
| cGAS signaling | ||||
| USP1 | Activation | TBK1 | Lys48 | Stablize TBK1, enhance IFN‐β secretion[ 540 ] |
| USP2 | Inhibition | TBK1 | Lys63 | Inhibit TBK1 activation[ 541 ] |
| USP7 | Inhibition | TBK1 | Lys48 | Stabilize TRIM27, inhibit type I IFN signaling[ 542 ] |
| Activation | p65 | Lys48 | Stabilize p65[ 543 ] | |
| USP13 | Inhibition | STING | Lys27/33 | Disrupt the interaction between STING and TBK1, inhibit Antiviral Res..ponses[ 231 ] |
| USP14 | Activation | cGAS | Lys48 | Stabilize cGAS, facilitate antiviral signaling[ 222 ] |
| USP20 | Activation | STING | Lys48 | Stabilize STING, promote Antiviral Res..ponses[ 225 ] |
| USP21 | Inhibition | STING | Lys27/63 | Inhibit type I IFN production[ 232 ] |
| USP22 | Inhibition | STING | Lys27 | Cooperate with USP13, inhibit Antiviral Res..ponses[ 233 ] |
| Activation | IRF3 | Lys48 | Stabilize importin KPNA2 to facilitate IRF3 nuclear translocation[ 544 ] | |
| USP27X | Activation | cGAS | Lys48 | Stabilize cGAS, govern DNA‐mediated signaling[ 223 ] |
| USP29 | Activation | cGAS | Lys48 | Stabilize cGAS, facilitate antiviral signaling[ 545 ] |
| USP38 | Inhibition | TBK1 | Lys33 | Promote TBK1 degradation through NLRP4 signalosome[ 546 ] |
| USP44 | Activation | STING | Lys48 | Stabilize STING, facilitate antiviral signaling[ 227 ] |
| USP49 | Inhibition | STING | Lys63 | Inhibit STING aggregation and TBK1 recruitment[ 230 ] |
| A20 | Inhibition | TBK1 | Lys63 | Cooperate with TAX1BP1 to disrupt TRAF3‐TBK1‐IKKi signaling complex[ 547 ] |
| OTUD1 | Inhibition | IRF3 | Lys6/63 | Inhibit IRF3 nuclear translocation and DNA binding capacity[ 548 , 549 ] |
| CYLD | Activation | STING | Lys48 | Stabilize STING, facilitate antiviral signaling[ 226 ] |
| Th1 and Th17 responses | ||||
| Trabid | Activation | JMJD2D | Lys29 | Promote the production of the Th1 and Th17 subsets of inflammatory T cells[ 237 ] |
| Cezanne | Activation | ZAP70 | Lys48 | Facilitate ZAP activation and inducing TCR signaling[ 239 ] |
| USP4 | Activation | RORγt | Lys48 | Stabilize RORγt, induce rheumatic heart disease[ 236 ] |
| USP10 | Activation | T‐bet | Lys48 | Stabilize T‐bet in Th1 cells, induce asthma[ 240 ] |
| USP17 | Activation | RORγt | Lys48 | Positive regulator of RORγt in Th17 cells[ 550 , 551 , 552 ] |
| USP18 | Activation | TAK1 | Lys63 | Induce differentiation of Th17 cells and cytokines secretion[ 238 ] |
| Treg responses | ||||
| USP7 | Inhibition | FOXP3 | Lys48 | Stabilize FOXP3 in Treg cells, inhibit TNFα‐stimulated NF‐κB activity[ 256 , 543 , 553 ] |
| USP21 | Activation | GATA3 | Lys48 | Stabilize GATA3 in Treg cells, limit inflammatory responses[ 543 , 553 ] |
4.1. DUBs in TLRs Signaling
TLRs are the most extensively researched PRRs (Figure 8A). They are inhibited by several DUBs through deubiquitylating NF‐κB signaling factors such as NEMO, receptor‐interacting serine/threonine‐protein kinase 1 (RIPK1), and tumor necrosis factor (TNF) receptor‐associated factor 6 (TRAF6).[ 150 ] A20 is the best‐identified DUB associated with inflammation. A20 expression is promoted by the NF‐κB pathway and cytokine receptors. In turn, A20 can be part of a negative feedback mechanism inhibiting continued NF‐κB activation, as described above.[ 150 , 152 ] Intriguingly, the role of A20 is not straightforward since it contains an OTU domain and seven ZnF motifs with DUB activity and E3 Ub ligase activity. A20 removes Lys63‐linked polyubiquitin chains from TRAF6, which leads to the termination of TLR‐mediated signaling activity.[ 150 , 180 ] In addition, A20 interacts with ubiquitinated NEMO by its ZnF motifs, which blocks IKK phosphorylation triggered by upstream TGF‐β‐activated kinase 1 (TAK1), and inhibits NF‐κB activation.[ 181 ] USP18 exerts a negative regulatory effect on NF‐κB signaling by employing distinct mechanisms to target TAK1 and NEMO for deubiquitination.[ 182 ] CYLD also negatively modulates TLRs signaling by removing Lys63‐ and Met1‐linked Ub‐chain from RIPK1, TRAF2, and NEMO.[ 61 , 183 , 184 ] OTUD1 physically interacts with RIPK1 and specifically cleaves Lys63‐linked ub‐chain from RIPK1, thereby inhibiting the recruitment of NEMO.[ 185 ] OTUD4 exerts Lys63‐specific DUB activity for myeloid differentiation primary response 88 (MyD88), which is indispensable for all TLRs involved in antiviral immunity, except TLR3.[ 126 ] OTUD4 negatively regulates the TLR‐induced NF‐κB pathway by targeting MyD88. The affinity of OTUD4 to Lys63‐linked chains on MyD88 is increased by an adjacent UIM.[ 186 ]
USP4 is a negative regulator for TLR/IL‐1R signaling and subsequent immune responses by deubiquitinating TRAF6, which is essential for governing downstream TLR signaling.[ 187 ] Additionally, USP4 depletion in growing zebrafish larvae induces pro‐inflammatory cytokines and increases susceptibility to lipopolysaccharide (LPS) challenge.[ 187 ] UCHL1 removes Lys63‐linked Ub‐chain on TRAF3 (another essential regulator in TLR signaling), which decreases the production of pro‐inflammatory cytokines, chemokines, and type I IFN in response to high‐risk human papillomavirus (hrHPV) infection.[ 188 ] TRAF3 is also targeted by OTUD7B through the Lys48‐specific DUB function, which restrains TRAF3 proteolysis and blocks abnormal non‐canonical NF‐κB activity.[ 189 ] Thus, TRAF3 can be deubiquitinated by multiple DUBs and performs distinct effects during viral infection. Intriguingly, USP25 induced by viral infection prevents TRAF3 and TRAF6 from degradation. USP25 knockout mice are more susceptible to viral infection compared with the normal mice.[ 190 , 191 ] USP25 is the only identified DUB that plays a positive regulatory role in the TLRs signaling till now (Table 3).
4.2. DUBs in RLRs Signaling
RLRs can effectively fill in the gaps of most nonimmune cells due to low TLR expression during antiviral immune responses (Figure 8B). RLRs are cytosolic RNA sensors, which include three members: RIG‐I,[ 192 ] melanoma differentiation‐associated protein 5 (MDA5),[ 193 ] and laboratory of genetics and physiology 2 (LGP2).[ 194 ] Lys63‐linked Ub‐chain induced by E3 ligases tripartite motif 25 (TRIM25), RNF135, TRIM4, and TRIM31 promote the stability and oligomerization of RLRs, thereby promoting downstream signaling.[ 179 , 195 , 196 , 197 , 198 , 199 ] However, degradative Lys48‐ and Lys27‐linked Ub‐chain mediated by E3 ligases RNF125, RNF122, CHIP, STUB1, and TRIM40 promote the degradation of RIG‐I and MDA5 to negatively regulate the signaling cascades.[ 200 , 201 , 202 , 203 , 204 ] CYLD is the first DUB confirmed to deubiquitinate RIG‐I through cleavage of its Lys63‐linked Ub‐chain which disrupts the fundamental activation and impairs IFNs production.[ 205 , 206 ] USP3 serves as a negative regulator of RIG‐I signaling via trimming Lys63‐linked Ub on RIG‐I. upon viral infection or ligand stimuli, USP3 interacts with the CARD domain of RIG‐I and cleaves Lys63‐linked Ub on RIG‐I.[ 207 ] However, the detailed mechanism by which USP3 governs RIG‐I in vivo remains unclear. USP14,[ 208 ] USP15,[ 209 ] USP21,[ 210 ] and USP27X,[ 211 ] also negatively regulate RIG‐I signaling through removal of Lys63‐linked Ub on RIG‐I.
Whereas, USP15 exerts a positive regulation of RIG‐I signaling. E3 ligase TRIM25 is in charge of Lys63‐linked poly‐ubiquitination on RIG‐I. USP15 counteracts the LUBAC‐induced degradation of TRIM25, which then contributes to continuing type I IFNs expression.[ 212 ] Therefore, USP15 executes Janus‐faced effects by recognizing diverse substrates or antagonizing the effects of E3 ligase, thereby revealing the rigorous regulation of cellular immune homeostasis. USP17 removes Lys48‐linked Ub‐chain on RIG‐I or MDA5 to facilitate the expression of type I IFNs and Antiviral Responses.[ 213 ] USP4 is another DUB responsible for promoting RIG‐I stability which increases RIG‐I‐induced IFN‐β signaling and reduces VSV replication. Moreover, USP4 protects TRAF6 from degradation and reduces enterovirus 71 (EV71) replication.[ 214 ] DUB screening and biochemical analyses showed that OTUD4 removes Lys48‐linked Ub‐chain of MAVS, thereby limiting MAVS proteasomal degradation and activating downstream signaling in response to viral infection.[ 215 ]
4.3. DUBs in cGAS‐STING Signaling
cGAS is the primary pathway governing the immune response to pathogenic DNA in the cytosol (Figure 8C). Recognition of pathogenic DNA by cGAS activates cGAS to catalyze cGAMP formation.[ 216 , 217 ] cGAMP binds to the endoplasmic reticulum (ER)‐localized adaptor stimulator of IFN genes (STING), which promotes STING dimerization and to perinuclear Golgi.[ 218 , 219 ] TANK binding kinase 1 (TBK1) or IκB kinase (IKK) is then activated by STING, which stimulates phosphorylation and nuclear translocation of the transcription factor IFN regulatory factor 3/7 (IRF3/7), and to a lesser extent nuclear factor‐κB (NF‐κB) for production of pro‐inflammatory cytokines.[ 220 , 221 ]
Lys48‐linked ubiquitination plays a negative role in the cGAS‐STING pathway by promoting proteasomal degradation of cGAS and STING.[ 222 ] USP14 is recruited by E3 ligase TRIM14 to remove the Lys48‐linked Ub‐chain of cGAS and increase its stability. TRIM14 knockout mice are vulnerable to herpes simplex virus‐1 (HSV‐1) infection through damaging type I IFN production.[ 222 ] USP27X directly binds to cGAS and trims Lys48‐linked Ub‐chain during viral infection.[ 211 , 223 ] Cleavage of the Lys48‐linked Ub‐chain in STING can be executed by diverse DUBs. USP20 directly interacts with‐ and cleaves Lys48‐linked Ub‐chain from STING after HSV‐1 infection, thus increasing the stability of STING and inducing cellular Antiviral Res..ponses.[ 224 ] The catalytic activity of USP20 is enhanced by the recruitment of USP18.[ 225 ] Other DUBs, such as CYLD, USP44, are also have positive regulation on STING during DNA virus infection.[ 206 , 226 , 227 , 228 ] USP44‐induced regulation of STING signaling is independent of USP20 and CYLD, suggesting that there is no functional redundancy between the three DUBs.[ 227 ]
Lys27‐, Lys33‐, and Lys63‐linked ubiquitination are implicated in the positive regulation of cGAS‐STING pathway by enhancing dimerization and enzymatic activity of cGAS and STING in response to pathogen DNA.[ 229 ] USP49 removes Lys63‐linked Ub‐chain on STING following HSV‐1 stimulation, which contributes to decreased interactions between STING and TBK1 and inhibition of Antiviral Responses.[ 230 ] In addition, USP13 deconjugates Lys27‐/Lys33‐linked Ub‐chain on STING which also disrupts the interaction between STING and TBK1.[ 231 ] USP13 knockout mice are not susceptible to HSV‐1 infection.[ 231 ] USP21 is recruited to STING by p38‐mediated phosphorylation of USP21 at the late stage of HSV‐1 infection. Then, USP21 negatively regulates the DNA virus‐induced production of type I IFNs by cleaving Lys27‐/Lys63‐linked Ub‐chain on STING.[ 232 ] Furthermore, the Lys27‐linked Ub‐chain on STING is removed by the collaboration of USP22 and USP13 to regulate STING signaling negatively.[ 233 ] Taken together, cGAS‐STING signaling can be elaborately regulated by multiple DUBs to enhance or impair innate Antiviral Res..ponses more accurately (Table 3).
4.4. DUBs in Inflammatory and Autoimmune Disorders
Chronic inflammatory and autoimmune diseases are generally characterized by a prolonged and persistent pro‐inflammatory state, such as metabolic syndrome, neurodegenerative disease, cardiovascular disease, rheumatoid arthritis (RA), and systemic lupus erythematosus (SLE).[ 234 ] PRR stimulation causes dendritic cells (DCs) to secrete miscellaneous cytokines that regulate the differentiation of CD4+ T cells to different subclasses of T helper (Th) cells, including inducible regulatory T cells (Tregs), T follicular helper cells (Tfh), and Th1, Th2, Th9, and Th17 cells. Th17 cells execute pro‐inflammatory functions via the secretion of pro‐inflammatory cytokines including Interluekin‐17A (IL17A), IL‐17F, and IL‐22.[ 235 ]
USP4 promotes the stability of nuclear receptor retinoid‐elated orphan receptor‐γt (RORγt) in activated Th17 cells, then induces the production of IL‐17A.[ 236 ] Expression of USP4, IL‐17A, and IL‐17F significantly increases in CD4+ T cells from RA patients compared with healthy volunteers.[ 236 ] TRAF‐binding domain (Trabid) is essential for TLR‐regulated expression of IL‐12 and IL‐23 in DCs.[ 237 ] Trabid deubiquitylates and stabilizes lysine demethylase 4D (KDM4D), which promotes histone modification by recruiting the NF‑κB family member REL to the Il12 and Il23 promoters.[ 237 ] Depletion of Trabid in DCs inhibits the production of IL‐12 and IL‐23 and the generation of Th1 and Th17 cells, which neutralizes the stimulation of experimental autoimmune encephalomyelitis (EAE) in mice.[ 237 ] USP18 is also connected with the activity of Th17 cells by promoting the TAK1‐TAK1‐associated binding protein (TAB) interaction; which is fundamental for Th17 cell differentiation and the autoimmune response.[ 238 ]
Moreover, T cell receptor signaling can be governed by Cezanne which deubiquitinates ZAP10 and disrupts the interaction between ZAP10 and negative regulatory phosphatases.[ 239 ] ZAP70 phosphorylation is indispensable for complete activation and downstream phosphorylation of substrates, which enhances T cell signaling. youthful mice with Cezanne knockout have similar naïve‐ and memory‐like T cells compared with normal mice. However, IFN‐γ production in Th1 cells of elderly Cezanne‐deficient mice is reduced.[ 239 ] This demonstrates that the production of pro‐inflammatory cytokines regulated by Cezanne in Th1 cells may be influenced by age stage to varying degrees, but the detailed mechanism remains unclear. T‐bet is critical in the development, differentiation, and function of Th1 cells. USP10 directly binds and inhibits T‐bet ubiquitination. USP10 and IFNG mRNA expression is upregulated in asthmatic patient PBMC, suggesting USP10 may maintain a high level of T‐bet and IFN‐γ to fight against Th2‐dominated inflammation.[ 240 ]
Consequently, various DUBs exert multiple effects on distinct immune cells to cope with complex inflammation and immune responses. This suggests that building genetic models and developing inhibitors for these DUBs is conducive to discovering their therapeutic potential.
5. Virus‐Encoded DUBs in Innate Immunity
Identification and characterization of DUBs encoded by diverse virus families have provided valuable insights into how the viruses deal with elaborate host antiviral mechanisms through perturbing cellular ubiquitin‐dependent processes. Mainly, various signaling molecules of innate immune pathways can be targeted by virus‐encoded DUBs to suppress antiviral immunity and support viral replication.
Most viral DUB activity is mediated by papain‐like proteases (PLP or PLpro). The first identified viral DUB is adenovirus cysteine protease (Avp), which possesses the ability to cleave Lys48‐linked tetraUb chains and ISG15.[ 241 , 242 ] Structural analysis revealed that Avp active site comprises residues His54, Glu71, and Cys122, which closely align with the catalytic triad of papain.[ 241 ] Nairovirus Crimean‐Congo hemorrhagic fever (CCHFV) is a tick‐borne virus and is widely distributed. Individuals infected with CCHF are at risk of multi‐organ failure and hemorrhaging.[ 243 ] The OTU DUB of CCHFV has been identified having deubiquitinating enzyme activity, specifically targeting Lys48 and Lys63 Ub‐chain, as well as ISG15 from cellular proteins.[ 244 ] The structure study revealed that the active site of CCHFV OUT consisted of a catalytic triad, comprising Cys‐His‐Asp residues, arranged in a geometrically competent conformation for catalysis.[ 245 ] The CCHFV OTU was found to suppress IFN‐β response by removing ubiquitin (Ub) from RIG‐I, IRF3.[ 246 ]
Furthermore, the PLpro of severe acute respiratory syndrome coronavirus (SARS‐CoV) possesses deubiquitinating enzyme function and deISGylation activities.[ 247 ] A crystal structure analysis of SARS‐CoV PLpro has confirmed its domain organization to be similar to that of human USP7, comprising “thumb,” “palm,” and “fingers” subdomains that assemble together in a configuration resembling an extended right hand.[ 248 ] The PLpro enzyme exhibits activity towards IRF3 Lys48‐linked Ub‐chain and is implicated in the suppression of cellular innate immune responses.[ 249 , 250 ] In addition, SARS‐CoV PLpro can also removing Lys63‐linked Ub‐chain of TRAF3 and TRAF6, thereby suppresses the TLR7 signaling and the production of IFNs and pro‐inflammatory cytokines.[ 251 ] Seneca Valley Virus (SVV) 3C protease inhibiting the IFN‐β production and promotes virus replication by eliminating ubiquitination of RIG‐I, TBK1, and TRAF3.[ 252 ]
Some virus‐encoded DUBs can negatively act on STING and NF‐κB signaling to manipulate anti‐viral immune responses. Herpes simplex virus (HSV) UL36USP can block type I IFN production by directly interacting with and deubiquitinating Lys48‐linked Ub‐chain of STING.[ 253 ] Human cytomegalovirus (HCMV) pUL48 inhibits PRR‐induced type I IFN signaling through deubiquitinating several key molecules, such as STING, TRAF3, TRAF6, IRAK1, and IRF7.[ 254 ] Epstein‐Barr virus (EBV) large tegument protein, BPLF1, deubiquitinates IκBα, TRAF6, and NEMO, thereby suppressing the NF‐κB signaling activation at multiple levels.[ 255 ]
A growing number of virus‐encoded DUBs have been identified and are recognized to play a pivotal role in antagonizing host immune system as listed in Table 4 . Virus‐encoded DUB‐mediated interaction between virus and host provides a deeper insight into viral pathogenic mechanism. Drugs targeting virus‐encoded DUB has drawn attention towards counteracting virus infection. In addition, harnessing viral DUBs for oncolytic use could be a particular interesting direction for future exploration.
Table 4.
Virus‐encoded DUBs in host innate antiviral immunity.
| Virus family | Virus | Virus‐encoded DUB | Host Target | Linkage | Rationale |
|---|---|---|---|---|---|
| RNA virus | |||||
| Coronaviridae/ alphacoronaviruses | Human coronavirus NL63 | PLP2 | RIG‐I, TBK1, IRF3, STING | Lys48/63 | Negatively regulate antiviral defenses by disrupting the STING‐mediated IFN induction[ 554 ] |
| Mouse hepatitis virus | MHV PLP2 | IRF3, TBK1 | NA | Negatively regulate type I IFN signaling[ 555 , 556 ] | |
| Porcine epidemic diarrhea virus | PEDV PLP2 | RIG‐I, STING | NA | Interfere with the RIG‐I‐ and STING‐mediated signaling[ 557 ] | |
| Coronaviridae/ betacoronaviruses | SARS‐coronavirus (2) | SARS‐CoV PLpro | RIG‐I, TRAF3, STING, TBK1, IRF3, MDA5 | Lys48/63, ISG15 | Negatively regulate IRF3 activation.[ 558 , 559 ] Antagonize ISG15‐dependent activation of MDA5[ 560 ] |
| MERS‐ coronavirus | MERS‐CoV PLpro | IRF3 | Lys48/63, ISG15 | Negatively regulate IRF3 activation[ 561 , 562 ] | |
| Arteriviridae | Porcine reproductive and respiratory syndrome virus | PRRSV PLP2 | RIG‐I | Lys6/11/27/29/33/48/63 | Inhibits RIG‐I‐mediated IFN signaling[ 246 ] |
| IκBα | Inhibit NF‐κB activation[ 563 ] | ||||
| Equine arteritis virus | EAV PLP2 | RIG‐I | NA | Inhibits RIG‐I‐mediated IFN signaling[ 246 ] | |
| Bunyaviridae/ nairoviruses | Crimean‐Congo hemorrhagic fever virus | CCHFV OTU | MDA5, RIG‐I | Lys6/11/48/63, ISG15 | Inactivate RLR‐mediated innate immune signaling[ 246 ] |
| Arteriviridae | Simian hemorrhagic fever virus | SHFV PLP2 | RIG‐I | NA | Inhibits RIG‐I‐mediated IFN signaling[ 246 ] |
| Lactate dehydrogenase‐elevating virus | LDV PLP2 | RIG‐I | NA | Inhibits RIG‐I‐mediated IFN signaling[ 246 ] | |
| Picornaviridae | Dugbe virus | DUGV OTU | NA | Lys48/63 | Negatively regulate type I IFN signaling[ 564 ] |
| Foot‐and‐mouth disease virus | FMDV Lbpro | RIG‐I, TBK1, TRAF3, TRAF6 | Lys48/63 | Negatively regulate type I IFN signaling[ 565 ] | |
| Seneca valley viru | SVV 3Cpro | RIG‐I, TBK1, TRAF3 | NA | block IFN‐β induction[ 252 ] | |
| DNA virus | |||||
| Adenoviridae | Adenovirus | Adenain | Histone H2A | Lys48, ISG15 | Acquire an advantageous property by adenovirus[ 241 ] |
| Herpesviridae/ alphaherpesviruses | Herpes simplex virus | UL36USP | TRAF3 | Lys48/63 | Deubiquitinate TRAF3 and prevent the recruitment of the downstream adaptor TBK1[ 566 ] |
| IκBα | Lys48 | Restrict IκBα degradation and finally abrogate NF‐κB activation[ 567 ] | |||
| STING | Lys48 | Inhibits induction of type I IFN[ 253 ] | |||
| Herpesviridae/ betaherpesviruses | Human cytomegalovirus | pUL36USP | MLKL | Lys63 | Degrade MLKL and inhibit necroptosis[ 568 ] |
| Herpesviridae/ gammaherpesviruses | Epstein‐Barr virus | BPLF1 | PCNA | Lys48/63 | Disrupt the cellular response to DNA damage[ 569 ] |
| SQSTM1/p62 | Regulate selective autophagy, which may promote infection and the production of infectious virus[ 570 ] | ||||
| Cullin | Benefit virus life cycle by inducing a replication‐permissive S‐phase‐like cellular environment[ 571 ] | ||||
| TRAF6 | Deubiquitinate TRAF6 to inhibit NF‐κB signaling[ 572 ] | ||||
| TRAF6, NEMO, IκBα | Deubiquitinate signaling intermediates in the TLR cascade to counteract innate anti‐viral immunity[ 255 ] | ||||
| Kaposi's sarcoma‐associated herpesvirus | KSV ORF64 | RIG‐I | Lys48/63 | Suppresse RIG‐I‐mediated IFN signaling[ 573 ] | |
| Herpesviridae | Hepatitis B virus | HBx | RIG‐I | Lys63 | Evade the induction of IFN and IFN‐induced antiviral effects[ 574 ] |
| RIG‐I, TBK1, CARDIF, TRIF, NEMO, IKKi, IRF3 | Negatively regulate type I IFN production[ 575 ] | ||||
6. DUBs in the Cross‐Talk Between Cancer and Immune Response
The antitumor potential of therapeutically restraining DUB participation in the immune system is being diligently researched given the multiple roles of DUBs in the immune response. USP7 exerts a variety of effects in DNA repair, apoptosis, and tumor metastasis which largely depends on the distinct functions of protein substrates. USP7 stabilizes the expression of Forkhead box protein P3 (FOXP3), thus enhancing the regulatory T cell (Treg) suppressive capacity facilitated by this critical transcription factor.[ 256 ] USP7 depletion prompts the polarization of TAMs (tumor‐associated macrophages) from M2 into M1 by stimulating the P38 MAPK pathway and enhancing the expression of immune checkpoint molecule programmed death ligand 1 (PD‐L1) in the tumor microenvironment.[ 257 ] Accordingly, USP7 inhibition integrated with anti‐PD‐1 immunotherapy could exert a more effective inhibitory effect on tumors. In addition, USP7 and USP21 enhance GATA3‐regulated activity in FOXP3‐expressing cells.[ 258 ] USP21 deficiency in Treg cells reduces FOXP3 expression, which reconciles the expression of Treg signature genes, and diminishes their suppressive activity.[ 259 ] Treg cells limit antitumor immune responses and facilitate tumor survival, therefore, anticancer immunotherapies recommend FOXP3 depletion in Treg cells by targeting USP7 and USP21.
OTUB1 promotes immunosuppression of diverse cancers by targeting PD‐L1 and governs the transcription and post‐transcription of several tumor‐driving factors, including signal transducer and activator of transcription 3 (STAT3), NF‐κB, Hippo, phosphoinositide 3‐kinase (PI3K)/AKT, etc.[ 260 , 261 ] A cohort of cytokines and chemokines are induced by OTUB1, which recruit and modulate the immunosuppressive processes, resulting in cancer immune evasion.[ 261 ] OTUB1 mediates the suppressive function of Treg cells, and the differentiation and maturation of B cells, T cells, and NK cells, which supports the potential role of OTUB1 in the tumor microenvironment.[ 262 ] However, the specific function of OTUB1 and its regulatory network in different tumor microenvironments requires further exploration. Additionally, COP9 signalosome 5 (CSN5), USP8, USP21, and USP22 have been reported to deubiquitinate and stabilized PD‐L1 in cancer cells, thereby suppressing antitumor immunity.[ 263 , 264 , 265 , 266 ]
DUBs‐regulated cross‐talk between tumorigenesis and immune response mainly focuses on the immunosuppression mediated by Treg cells and immune checkpoint. DUBs generally stabilize transcriptional factors to promote the expression of Treg signature genes and tumor‐driving factors. However, the antitumor potential of DUBs in B cells, T cells, and NK cells is relatively limited. Remarkably, as mentioned above, DUBs play a broad regulatory role in various immune signaling cascades, which reveals that there is great potential for exploration in the crosstalk governed by DUBs between cancer and immunity.
7. Therapeutic Applications via Targeting DUBs
Mounting evidence of DUBs in the pathogenesis cancer and immune disorders has brought about the remarkable attractiveness for DUBs as potential therapeutic targets (Table 5 ). There has been a growing rate of progress on successfully screening and identifying potent and specific small molecule inhibitors of DUBs in past years. This highlights that DUBs are certainly druggable and have potential therapeutic targets in diseases ranging from oncology to immunology. The strategy of DUB inhibition can be roughly grouped into two types: conformational stabilization in enzymatic domain and allosteric inhibition in non‐catalytic sites.
Table 5.
Nonexhaustive DUBs inhibitors in anti‐cancer and inflammation.
| DUB | Inhibitor | Characteristics of inhibition | Disease indication |
|---|---|---|---|
| USP1 | ML323 | Highly selective, reversible | Oncology[ 274 ] |
| SJB2‐043 | Highly selective | Oncology[ 576 ] | |
| SJB3‐019A | Highly selective, irreversible | Oncology[ 577 ] | |
| GW7647 | Non‐selective, reversible | Oncology[ 275 ] | |
| Pimozide | Non‐selective, reversible | Oncology[ 275 ] | |
| Rottlerin | Non‐selective, irreversible | Oncology[ 275 ] | |
| USP2 | ML364 | Non‐selective, reversible | Oncology[ 578 ] |
|
LCAHA 6TG |
Non‐selective Non‐selective, irreversible |
Oncology[ 579 ] Oncology[ 580 ] |
|
| USP4 | Vialinin A | Non‐selective | Inflammation and oncology[ 236 , 581 ] |
| USP5 |
WP1130 Vialinin A |
Non‐selective Non‐selective |
|
| USP7 |
FT671 FT827 GNE‐6640 GNE‐6776 Compound 1 Compound 4 P5091 HBX41108 HBX19818 Cpd1 Cpd2 Cpd8 XL188 Ursolic acid |
Highly selective Highly selective Highly selective Highly selective Non‐selective Highly selective Highly selective Non‐selective Non‐selective, irreversible Non‐selective, irreversible Highly selective Highly selective Highly selective Highly selective Non‐selective |
Oncology[ 269 ] Oncology[ 269 ] Oncology[ 583 ] Oncology[ 583 ] Oncology[ 584 ] Oncology[ 585 ] Oncology, immuo‐oncology[ 584 ] Oncology, immuo‐oncology[ 586 , 587 ] Oncology[ 588 ] Oncology[ 589 ] Oncology[ 588 ] Oncology[ 588 ] Oncology[ 590 ] |
| USP8 |
DUBs‐IN‐2 MB7295 |
Highly selective Highly selective |
Oncology[ 591 ] Oncology[ 592 ] |
| USP9X |
WP1130 EOAI3402143 |
Non‐selective, reversible Non‐selective, reversible |
Oncology[ 582 ] Oncology[ 404 ] |
| USP10 | Spautin 1 | Non‐selective | Inflammation[ 593 ] |
| USP11 | MIX | Non‐selective | Oncology[ 268 ] |
| USP13 | Spautin 1 | Non‐selective | Inflammation[ 593 ] |
| USP14 |
IU‐1/analogues b‐AP15 VLX1570 WP1130 Auranofin |
Non‐selective Non‐selective, reversible Non‐selective, reversible Non‐selective Non‐selective |
Oncology[ 599 ] Oncology[ 582 ] Oncology[ 600 ] |
| USP15 | MIX | Non‐selective | Oncology[ 268 ] |
| USP17 | WP1130 | Non‐selective | Oncology[ 582 ] |
| USP20 | GSK2643943A | Highly selective | Oncology[ 601 ] |
| USP24 | EOAI3402143 | Non‐selective, reversible | Oncology[ 404 ] |
| USP25 | AZ1/2/3/4 | Non‐selective, reversible | Oncology[ 602 ] |
| USP28 | AZ1/2/3/4 | Non‐selective, reversible | Oncology[ 602 ] |
| USP30 |
FT385 MF094 MF095 |
Highly selective Highly selective Highly selective |
Oncology[ 603 ] Oncology[ 603 ] Oncology[ 603 ] |
| USP47 | Compound 1 | Non‐selective | Oncology[ 584 ] |
| POH1 |
8‐thioquinoline Capzimin phen |
Non‐selective Highly selective Highly selective |
Oncology[ 604 ] Oncology[ 605 ] Oncology[ 606 ] |
| UCHL1 | LDN‐57444 | Highly selective | Oncology[ 607 ] |
| GK13S | Highly selective | Oncology[ 608 ] | |
| 6RK73 | Highly selective, irreversible | Oncology[ 169 ] | |
| UCHL5 | IU‐1/analogues | Non‐selective | Oncology[ 594 , 595 , 596 ] |
|
b‐AP15 VLX1570 WP1130 Auranofin |
Non‐selective, reversible Non‐selective, reversible Non‐selective Non‐selective |
Oncology[ 599 ] Oncology[ 582 ] Oncology[ 600 ] |
|
| Trabid | NSC112200 | Highly selective | Inflammation and oncology[ 609 , 610 ] |
| CYLD | Subquinocin | Non‐selective | Inflammation[ 611 ] |
| SARS‐CoV PLpro | GRL0617 | Highly selective, reversible | Anti‐infection[ 276 ] |
| DDL‐701 | Highly selective, reversible | Anti‐infection[ 279 ] |
DUB inhibitors maintain the target DUB in an inactive or active conformation by directly interacting with catalytic‐site domains. Antineoplastic agent mitoxantrone (MIX) interacts with USP15 catalytic histidine H862 in a non‐covalent manner, which keeps USP15 catalytic domain in an inactive conformation state.[ 267 , 268 ] Small molecule FT827 forms a covalent bond between its vinyl sulfonamide residue and the catalytic cysteine of USP7, thereby stabilizing USP7 in a catalytically inactivated conformation and disrupting the coupling between Ub and USP7.[ 269 ] CSN5 contains a JAMM domain with a Zn2+ ion that is essential for its catalytic activity. The inhibitor CSN5i‐3 developed by Novartis has a zinc‐binding group for attachment onto the catalytic site, therefore stabilizing CSN5 in an active conformation.[ 270 ] Moreover, VLX1570 is a ring‐expanded version of b‐AP15 that specifically impairs the activity of USP14 and UCHL5 in the 19S regulatory subunit; it is associated with unique conformations of these DUBs.[ 271 ] b‐AP15 exhibits apoptosis‐inducing and antineoplastic activities in various solid tumor models, including multiple myeloma.[ 272 , 273 ]
On the basis of extensive oncogenic functions of USPs family in various cancers, compounds selectively targeting the USPs have been developed to antagonizes their tumorigenic effects. ML323 exerts a selective and reversible effect on the catalytic activity of USP1 in an allosteric manner that misaligns the conformation of Ub‐binding motif.[ 274 ] Unexpectedly, ML323 enhances cisplatin susceptibility in osteosarcoma and NSCLC cells through affecting the DNA damage response.[ 274 ] Moreover, GW7647 and pimozide also restrain USP1 catalytic activity in a non‐selective way. Both of them are allosteric inhibitors outside of enzymatic activity sites of USP1.[ 275 ] In addition to USP1, the catalytic activity of USP7 can also be suppressed by GW7647 and pimozide,[ 275 ] but the detailed functional effects are unclear.
Allosteric inhibitors competitively interact with the Ub‐binding sites of DUBs and hinder substrate recognition. The small molecule GRL0617 was first reported as a competitive non‐covalent inhibitor for a papain‐like protease (PLpro) encoded by SARS‐CoV and MERS‐CoV.[ 276 ] PLpro is a viral DUB that plays a vital role in the regulation of host cellular processes. GRL0617 blocks Ub access to the PLpro catalytic site by occupying the binding pocket for the distal Ub's C‐terminal residues. Additionally, several other inhibitors, such as rac3j, rac3k and rac5c targeting SARS‐CoV PLpro in clinical and preclinical stages have been described elsewhere.[ 277 ] Among them, rac5c also have potent inhibitory effect on SARS2 PLpro, which might be deliberated as a potential therapeutic strategy for COVID‐19.[ 278 ] Another small molecule DDL‐701 is also an effective SARS2 PLpro inhibitor which displayed sustained inhibitory activity.[ 279 ]
Although certain progress has been made in the development of drug targets for DUBs, it is still challenging for researchers to overcome a variety of existing obstacles. First, multiple DUBs are tightly related in similar catalytic pockets, which makes it difficult to develop potent inhibitors that represent selectivity for individual DUBs. Secondly, the interaction between DUB and substrate is not simply one‐to‐one. The same substrate may be targeted by various DUBs, while several DUBs may target multiple substrates. Noteworthily, DUBs exert diametrically opposed roles according to the complex context. Therefore, anti‐cancer efficacies and cytotoxicity for the development of DUB inhibitors should be attentively considered in future clinical trials. Finally, the mechanisms of DUBs regulation are generally complicated, including switch between active and non‐active conformations, allosteric effects, and substrate‐regulated catalysis. This makes it more difficult to carry out the primary screening and biochemical assays.
8. Concluding Remarks
This review summarized the multiple regulatory roles of DUBs in tumorigenesis and immunity. DUBs counteract the biological effects of E3 Ub ligases by removing the Ub chains from downstream substrates. Deubiquitination mediated by DUBs is accomplished alone or in combination with a complex to promote their enzymatic activities. Most DUBs confer some level of specificity regarding their action on diverse substrates or Ub‐linkage types. Indeed, there are still many gaps in knowledge of DUB specificity and mechanisms which limits the development of therapeutic applications in this field.
Various linkage types, lengths, and PTMs contribute to the miscellaneous architecture of polyubiquitin chains. Atypical linkage types of polyubiquitin chains may exert extremely distinctive effects on diverse cellular processes.[ 280 ] However, the topology of polyubiquitin chains and the mechanism of DUB recognition remains unclear. There is a lack of effective tools and methods to reveal the architecture of polyubiquitin chains, which is necessary to precisely identify the cleavage specificity of DUBs on polyubiquitin chains. Furthermore, ubiquitination and deubiquitination usually occur simultaneously (temporally and spatially) due to the close functional interactions and reciprocity between E3 ligases and DUBs. It is worth to elucidate how E3 ligases and DUBs selectively cooperate with key signaling mediators to govern tumor and immune responses. Finally, DUBs are either cysteine proteases or metalloproteases. Therefore, it deserves to investigate whether other cysteine proteases or metalloproteases serve as DUBs to fine‐tune the Ub system and maintain homeostasis. Innovative Ub probes targeting metalloproteases, serine proteases, or aspartate proteases will be helpful to discover novel DUB classes.[ 35 , 281 ]
Taken together, this review provides a glimpse into the Janus‐faced regulatory roles of DUBs in protein turnover and numerous signaling pathways, which is critical for the development of cancer and immunity. The development of innovative approaches will ultimately provide new insights into the mechanics of the ubiquitin system and confer DUBs with broad therapeutic potential.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
J.R., P.Y., and S.L. contributed equally to this work. J.R., P.Y., and S.L. conceived and drafted the manuscript. J.R., P.Y., and S.L. drew the figures. J.R., R.L., X.N., Y.C., and P.Y. discussed the concepts of the manuscript. P.Y., S.L., and Y.W. collected the information and summarized tables. Z.Z., F.Z., and L.Z. provided valuable discussion and revised the manuscript. The authors apologize to those researchers whose related works were not cited in this review.
Acknowledgements
The current work was supported by a special program from the Ministry of Science and Technology of China (2021YFA1101000, 2022YFA1105200), the Chinese National Natural Science Funds (U20A20393, U20A201376, 32125016, 31701234, 91753139, 31925013, 31671457, 31870902, 32070907, 32100699, 32200575, 82273493, 31871405 and 81760041), the Zhejiang Natural Science Fund (LD19C070001), Jiangsu National Science Foundation (19KJA550003), and Fundamental research program of The Shenzhen Science and Technology Innovation Commission (JCYJ20220530144207017).
Biographies
Jiang Ren works as an assistant researcher at the Eighth Affiliated Hospital, Sun Yat‐sen University. His research interests focus on protein posttranslational modifications and liquid‐liquid phase separation.

Peng Yu works as a post‐doctor at Zhongshan Institute for Drug Discovery, Shanghai Institute of Materia Medica, Chinese Academy of Sciences. The main research work is to reveal the structure and function of important membrane protein complexes based on cryo‐EM.

Long Zhang works as an independent PI at Life Sciences Institute, ZheJiang University. His research interest is to elucidate the protein posttranslational modifications and molecular mechanisms underlying cancer metastasis and cancer immunotherapy.

Ren J., Yu P., Liu S., Li R., Niu X., Chen Y., Zhang Z., Zhou F., Zhang L., Deubiquitylating Enzymes in Cancer and Immunity. Adv. Sci. 2023, 10, 2303807. 10.1002/advs.202303807
Contributor Information
Zhenyu Zhang, Email: fcczhangzy1@zzu.edu.cn.
Fangfang Zhou, Email: zhoufangfang@suda.edu.cn.
Long Zhang, Email: l_zhang@zju.edu.cn.
References
- 1. Ciechanover A., Biochem. Soc. Trans. 2003, 31, 474. [DOI] [PubMed] [Google Scholar]
- 2. Dubois M. L., Meller A., Samandi S., Brunelle M., Frion J., Brunet M. A., Toupin A., Beaudoin M. C., Jacques J.‐F., Lévesque D., Scott M. S., Lavigne P., Roucou X., Boisvert F.‐M., Nat. Commun. 2020, 11, 1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buetow L., Huang D. T., Nat. Rev. Mol. Cell Biol. 2016, 17, 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zheng N., Shabek N., Annu. Rev. Biochem. 2017, 86, 129. [DOI] [PubMed] [Google Scholar]
- 5. Pickart C. M., Eddins M. J., Biochim. Biophys. Acta. 2004, 1695, 55. [DOI] [PubMed] [Google Scholar]
- 6. Yang Q., Zhao J., Chen D., Wang Y., Mol. Biomed. 2021, 2, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kwon Y. T., Ciechanover A., Trends Biochem. Sci. 2017, 42, 873. [DOI] [PubMed] [Google Scholar]
- 8. Kirschner L. S., Stratakis C. A., Biochem. Biophys. Res. Commun. 2000, 270, 1106. [DOI] [PubMed] [Google Scholar]
- 9. Akutsu M., Dikic I., Bremm A., J. Cell Sci. 2016, 129, 875. [DOI] [PubMed] [Google Scholar]
- 10. Swatek K. N., Komander D., Cell Res. 2016, 26, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ciechanover A., Nat. Rev. Mol. Cell Biol. 2005, 6, 79. [DOI] [PubMed] [Google Scholar]
- 12. Komander D., Rape M., Annu. Rev. Biochem. 2012, 81, 203. [DOI] [PubMed] [Google Scholar]
- 13. Husnjak K., Dikic I., Annu. Rev. Biochem. 2012, 81, 291. [DOI] [PubMed] [Google Scholar]
- 14. Nijman S. M., Luna‐Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R., Cell 2005, 123, 773. [DOI] [PubMed] [Google Scholar]
- 15. Amerik A. Y., Hochstrasser M., Biochim. Biophys. Acta. 2004, 1695, 189. [DOI] [PubMed] [Google Scholar]
- 16. Harrigan J. A., Jacq X., Martin N. M., Jackson S. P., Biochim. Biophys. Acta. 2018, 17, 57. [Google Scholar]
- 17. Zong Z., Zhang Z., Wu L., Zhang L., Zhou F., Adv. Sci. 2021, 8, 2002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chapman H. A., Riese R. J., Shi G. P., Annu. Rev. Physiol. 1997, 59, 63. [DOI] [PubMed] [Google Scholar]
- 19. Komander D., Subcell. Biochem. 2010, 54, 69. [DOI] [PubMed] [Google Scholar]
- 20. Coleman K. E., Huang T. T., Mol. Cell 2018, 70, 10.1016/j.molcel.2018.03.022. [DOI] [PubMed] [Google Scholar]
- 21. Pak C. W., Kosno M., Holehouse A. S., Padrick S. B., Mittal A., Ali R., Yunus A. A., Liu D. R., Pappu R. V., Rosen M. K., Mol. Cell 2016, 63, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Clague M. J., Barsukov I., Coulson J. M., Liu H., Rigden D. J., Urbé S., Physiol. Rev. 2013, 93, 1289. [DOI] [PubMed] [Google Scholar]
- 23. Park H. B., Kim J. W., Baek K. H., Int. J. Mol. Sci. 2020, 21,33375030 [Google Scholar]
- 24. Hu M., Li P., Li M., Li W., Yao T., Wu J. W., Gu W., Cohen R. E., Shi Y., Cell 2002, 111, 1041. [DOI] [PubMed] [Google Scholar]
- 25. Johnston S. C., Larsen C. N., Cook W. J., Wilkinson K. D., Hill C. P., EMBO J. 1997, 16, 3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Artavanis‐Tsakonas K., Weihofen W. A., Antos J. M., Coleman B. I., Comeaux C. A., Duraisingh M. T., Gaudet R., Ploegh H. L., J. Biol. Chem. 2010, 285, 6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Komander D., Barford D., Biochem. J. 2008, 409, 77. [DOI] [PubMed] [Google Scholar]
- 28. Abdul Rehman S. A., Kristariyanto Y. A., Choi S. Y., Nkosi P. J., Weidlich S., Labib K., Hofmann K., Kulathu Y., Mol. Cell 2016, 63, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Seki T., Gong L., Williams A. J., Sakai N., Todi S. V., Paulson H. L., J. Biol. Chem. 2013, 288, 17145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hermanns T., Pichlo C., Woiwode I., Klopffleisch K., Witting K. F., Ovaa H., Baumann U., Hofmann K., Nat. Commun. 2018, 9, 799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xie X., Wang X., Jiang D., Wang J., Fei R., Cong X., Wei L., Wang Y., Chen H., Biochem. Biophys. Res. Commun. 2017, 488, 291. [DOI] [PubMed] [Google Scholar]
- 32. Sato Y., Yoshikawa A., Yamagata A., Mimura H., Yamashita M., Ookata K., Nureki O., Iwai K., Komada M., Fukai S., Nature 2008, 455, 358. [DOI] [PubMed] [Google Scholar]
- 33. Sui X., Wang Y., Du Y.‐X., Liang L.‐J., Zheng Q., Li Y.‐M., Liu L., Chem. Sci. 2020, 11, 12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cho J., Park J., Kim E. E., Song E. J., Int. J. Mol. Sci. 2020, 21,33375030 [Google Scholar]
- 35. Mevissen T. E. T., Komander D., Annu. Rev. Biochem. 2017, 86, 159. [DOI] [PubMed] [Google Scholar]
- 36. Faesen A. C., Luna‐Vargas M. P., Geurink P. P., Clerici M., Merkx R., van Dijk W. J., Hameed D. S., El Oualid F., Ovaa H., Sixma T. K., Chem. Biol. 2011, 18, 1550. [DOI] [PubMed] [Google Scholar]
- 37. Ye Y., Scheel H., Hofmann K., Komander D., Mol. BioSyst. 2009, 5, 1797. [DOI] [PubMed] [Google Scholar]
- 38. Sato Y., Okatsu K., Saeki Y., Yamano K., Matsuda N., Kaiho A., Yamagata A., Goto‐Ito S., Ishikawa M., Hashimoto Y., Tanaka K., Fukai S., Nat. Struct. Mol. Biol. 2017, 24, 911. [DOI] [PubMed] [Google Scholar]
- 39. Gersch M., Gladkova C., Schubert A. F., Michel M. A., Maslen S., Komander D., Nat. Struct. Mol. Biol. 2017, 24, 920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sato Y., Goto E., Shibata Y., Kubota Y., Yamagata A., Goto‐Ito S., Kubota K., Inoue J., Takekawa M., Tokunaga F., Fukai S., Nat. Struct. Mol. Biol. 2015, 22, 222. [DOI] [PubMed] [Google Scholar]
- 41. Cooper E. M., Cutcliffe C., Kristiansen T. Z., Pandey A., Pickart C. M., Cohen R. E., EMBO J. 2009, 28, 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McCullough J., Row P. E., Lorenzo O., Doherty M., Beynon R., Clague M. J., Urbé S., Curr. Biol. 2006, 16, 160. [DOI] [PubMed] [Google Scholar]
- 43. Mevissen T. E., Hospenthal M. K., Geurink P. P., Elliott P. R., Akutsu M., Arnaudo N., Ekkebus R., Kulathu Y., Wauer T., El Oualid F., Freund S. M., Ovaa H., Komander D., Cell 2013, 154, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Juang Y.‐C., Landry M.‐C., Sanches M., Vittal V., Leung C. C. Y., Ceccarelli D. F., Mateo A.‐R. F., Pruneda J. N., Mao D. Y. L., Szilard R. K., Orlicky S., Munro M., Brzovic P. S., Klevit R. E., Sicheri F., Durocher D., Mol. Cell 2012, 45, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mevissen T. E. T., Kulathu Y., Mulder M. P. C., Geurink P. P., Maslen S. L., Gersch M., Elliott P. R., Burke J. E., van Tol B. D. M., Akutsu M., Oualid F. E., Kawasaki M., Freund S. M. V., Ovaa H., Komander D., Nature 2016, 538, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Keusekotten K., Elliott P. R., Glockner L., Fiil B. K., Damgaard R. B., Kulathu Y., Wauer T., Hospenthal M. K., Gyrd‐Hansen M., Krappmann D., Hofmann K., Komander D., Cell 2013, 153, 1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Licchesi J. D. F., Mieszczanek J., Mevissen T. E. T., Rutherford T. J., Akutsu M., Virdee S., Oualid F. E., Chin J. W., Ovaa H., Bienz M., Komander D., Nat. Struct. Mol. Biol. 2012, 19, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kwasna D., Abdul Rehman S. A., Natarajan J., Matthews S., Madden R., De Cesare V., Weidlich S., Virdee S., Ahel I., Gibbs‐Seymour I., Kulathu Y., Mol. Cell 2018, 70, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hewings D. S., Heideker J., Ma T. P., AhYoung A. P., El Oualid F., Amore A., Costakes G. T., Kirchhofer D., Brasher B., Pillow T., Popovych N., Maurer T., Schwerdtfeger C., Forrest W. F., Yu K., Flygare J., Bogyo M., Wertz I. E., Nat. Commun. 2018, 9, 1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Haahr P., Borgermann N., Guo X., Typas D., Achuthankutty D., Hoffmann S., Shearer R., Sixma T. K., Mailand N., Mol. Cell 2018, 70, 165. [DOI] [PubMed] [Google Scholar]
- 51. Zhang W., Qian W., Gu J., Gong M., Zhang W., Zhang S., Zhou C., Jiang Z., Jiang J., Han L., Wang X., Wu Z., Ma Q., Wang Z., Cancer Lett. 2023, 552, 215976. [DOI] [PubMed] [Google Scholar]
- 52. Schlicher L., Brauns‐Schubert P., Schubert F., Maurer U., Cell Death Differ. 2017, 24, 1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Clague M. J., Urbé S., Komander D., Nat. Rev. Mol. Cell Biol. 2019, 20, 338. [DOI] [PubMed] [Google Scholar]
- 54. Hospenthal M. K., Mevissen T. E. T., Komander D., Nat. Protoc. 2015, 10, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rivkin E., Almeida S. M., Ceccarelli D. F., Juang Y. C., MacLean T. A., Srikumar T., Huang H., Dunham W. H., Fukumura R., Xie G., Gondo Y., Raught B., Gingras A. C., Sicheri F., Cordes S. P., Nature 2013, 498, 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Villamil M. A., Chen J., Liang Q., Zhuang Z., Biochemistry 2012, 51, 2829. [DOI] [PubMed] [Google Scholar]
- 57. Faesen A. C., Dirac A. M., Shanmugham A., Ovaa H., Perrakis A., Sixma T. K., Mol. Cell 2011, 44, 147. [DOI] [PubMed] [Google Scholar]
- 58. Kessler B. M., Edelmann M. J., Cell Biochem. Biophys. 2011, 60, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mizuno E., Kitamura N., Komada M., Exp. Cell Res. 2007, 313, 3624. [DOI] [PubMed] [Google Scholar]
- 60. Reincke M., Sbiera S., Hayakawa A., Theodoropoulou M., Osswald A., Beuschlein F., Meitinger T., Mizuno‐Yamasaki E., Kawaguchi K., Saeki Y., Tanaka K., Wieland T., Graf E., Saeger W., Ronchi C. L., Allolio B., Buchfelder M., Strom T. M., Fassnacht M., Komada M., Nat. Genet. 2015, 47, 31. [DOI] [PubMed] [Google Scholar]
- 61. Reiley W., Zhang M., Wu X., Granger E., Sun S. C., Mol. Cell Biol. 2005, 25, 3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hutti J. E., Shen R. R., Abbott D. W., Zhou A. Y., Sprott K. M., Asara J. M., Hahn W. C., Cantley L. C., Mol. Cell 2009, 34, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Thein S., Pham A., Bayer K. U., Tao‐Cheng J. H., Dosemeci A., Biochem. Biophys. Res. Commun. 2014, 450, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hutti J. E., Turk B. E., Asara J. M., Ma A., Cantley L. C., Abbott D. W., Mol. Cell Biol. 2007, 27, 7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wertz I. E., Newton K., Seshasayee D., Kusam S., Lam C., Zhang J., Popovych N., Helgason E., Schoeffler A., Jeet S., Ramamoorthi N., Kategaya L., Newman R. J., Horikawa K., Dugger D., Sandoval W., Mukund S., Zindal A., Martin F., Quan C., Tom J., Fairbrother W. J., Townsend M., Warming S., DeVoss J., Liu J., Dueber E., Caplazi P., Lee W. P., Goodnow C. C., et al., Nature 2015, 528, 370. [DOI] [PubMed] [Google Scholar]
- 66. Bingol B., Tea J. S., Phu L., Reichelt M., Bakalarski C. E., Song Q., Foreman O., Kirkpatrick D. S., Sheng M., Nature 2014, 510, 370. [DOI] [PubMed] [Google Scholar]
- 67. Todi S. V., Winborn B. J., Scaglione K. M., Blount J. R., Travis S. M., Paulson H. L., EMBO J. 2009, 28, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang Z., Du J., Wang S., Shao L., Jin K., Li F., Wei B., Ding W., Fu P., van Dam H., Wang A., Jin J., Ding C., Yang B., Zheng M., Feng X. H., Guan K. L., Zhang L., Mol. Cell 2019, 73, 7. [DOI] [PubMed] [Google Scholar]
- 69. Lee J. G., Baek K., Soetandyo N., Ye Y., Nat. Commun. 2013, 4, 1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kulathu Y., Garcia F. J., Mevissen T. E., Busch M., Arnaudo N., Carroll K. S., Barford D., Komander D., Nat. Commun. 2013, 4, 1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cotto‐Rios X. M., Békés M., Chapman J., Ueberheide B., Huang T. T., Cell Rep. 2012, 2, 1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Clague M. J., Heride C., Urbé S., Trends Cell Biol. 2015, 25, 417. [DOI] [PubMed] [Google Scholar]
- 73. Finley D., Annu. Rev. Biochem. 2009, 78, 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Song Y., Ray A., DAS D. S., Chauhan D., Anderson K. C., Blood 2015, 126, 1811. [Google Scholar]
- 75. Verma R., Aravind L., Oania R., McDonald W. H., Yates J. R., Koonin E. V., Deshaies R. J., Science 2002, 298, 611. [DOI] [PubMed] [Google Scholar]
- 76. Green C. M., Li Z., Smith A. D., Novikova O., Bacot‐Davis V. R., Gao F., Hu S., Banavali N. K., Thiele D. J., Li H., Belfort M., PLoS Biol. 2019, 17, e3000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Plaschka C., Lin P. C., Charenton C., Nagai K., Nature 2018, 559, 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. He Y., Wang S., Tong J., Jiang S., Yang Y., Zhang Z., Xu Y., Zeng Y., Cao B., Moran M. F., Mao X., J. Biol. Chem. 2020, 295, 2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Niendorf S., Oksche A., Kisser A., Löhler J., Prinz M., Schorle H., Feller S., Lewitzky M., Horak I., Knobeloch K. P., Mol Cell Biol 2007, 27, 5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Richardson L. A., Reed B. J., Charette J. M., Freed E. F., Fredrickson E. K., Locke M. N., Baserga S. J., Gardner R. G., Cell Rep. 2012, 2, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hymowitz S. G., Wertz I. E., Nat. Rev. Cancer 2010, 10, 332. [DOI] [PubMed] [Google Scholar]
- 82. Yuk J. M., Kim T. S., Kim S. Y., Lee H. M., Han J., Dufour C. R., Kim J. K., Jin H. S., Yang C. S., Park K. S., Lee C. H., Kim J. M., Kweon G. R., Choi H. S., Vanacker J. M., Moore D. D., Giguère V., Jo E. K., Immunity 2015, 43, 80. [DOI] [PubMed] [Google Scholar]
- 83. Coornaert B., Baens M., Heyninck K., Bekaert T., Haegman M., Staal J., Sun L., Chen Z. J., Marynen P., Beyaert R., Nat. Immunol. 2008, 9, 263. [DOI] [PubMed] [Google Scholar]
- 84. Legarda D., Justus S. J., Ang R. L., Rikhi N., Li W., Moran T. M., Zhang J., Mizoguchi E., Zelic M., Kelliher M. A., Blander J. M., Ting A. T., Cell Rep. 2016, 15, 2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. O'Donnell M. A., Perez‐Jimenez E., Oberst A., Ng A., Massoumi R., Xavier R., Green D. R., Ting A. T., Nat. Cell Biol. 2011, 13, 1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Staal J., Driege Y., Bekaert T., Demeyer A., Muyllaert D., Van Damme P., Gevaert K., Beyaert R., EMBO J. 2011, 30, 1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Scholz C. C., Rodriguez J., Pickel C., Burr S., Fabrizio J. A., Nolan K. A., Spielmann P., Cavadas M. A., Crifo B., Halligan D. N., Nathan J. A., Peet D. J., Wenger R. H., Von Kriegsheim A., Cummins E. P., Taylor C. T., PLoS Biol. 2016, 14, e1002347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yuan J., Luo K., Zhang L., Cheville J. C., Lou Z., Cell 2010, 140, 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Mueller T., Breuer P., Schmitt I., Walter J., Evert B. O., Wüllner U., Hum Mol Genet 2009, 18, 3334. [DOI] [PubMed] [Google Scholar]
- 90. Herhaus L., Perez‐Oliva A. B., Cozza G., Gourlay R., Weidlich S., Campbell D. G., Pinna L. A., Sapkota G. P., Sci. Signal. 2015, 8, ra35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang L., Zhou F., Drabsch Y., Gao R., Snaar‐Jagalska B. E., Mickanin C., Huang H., Sheppard K. A., Porter J. A., Lu C. X., ten Dijke P., Nat. Cell Biol. 2012, 14, 717. [DOI] [PubMed] [Google Scholar]
- 92. Wijnhoven P., Konietzny R., Blackford A. N., Travers J., Kessler B. M., Nishi R., Jackson S. P., Mol. Cell 2015, 60, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Schaeffer V., Akutsu M., Olma M. H., Gomes L. C., Kawasaki M., Dikic I., Mol. Cell 2014, 54, 349. [DOI] [PubMed] [Google Scholar]
- 94. Elliott P. R., Nielsen S. V., Marco‐Casanova P., Fiil B. K., Keusekotten K., Mailand N., Freund S. M., Gyrd‐Hansen M., Komander D., Mol. Cell 2014, 54, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wang Y., Serricchio M., Jauregui M., Shanbhag R., Stoltz T., Di Paolo C. T., Kim P. K., McQuibban G. A., Autophagy 2015, 11, 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Leznicki P., Natarajan J., Bader G., Spevak W., Schlattl A., Abdul Rehman S. A., Pathak D., Weidlich S., Zoephel A., Bordone M. C., Barbosa‐Morais N. L., Boehmelt G., Kulathu Y., J. Cell Sci. 2018, 131, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Song Y., Ray A., Das D. S., Samur M. K., Carrasco R. D., Munshi N. C., Chauhan D., Anderson K. C., Blood 2016, 128, 4469. [Google Scholar]
- 98. Wang B., Ma A., Zhang L., Jin W. L., Qian Y., Xu G., Qiu B., Yang Z., Liu Y., Xia Q., Liu Y., Nat. Commun. 2015, 6, 8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Liu H., Buus R., Clague M. J., Urbé S., PLoS One 2009, 4, e5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kakarougkas A., Ismail A., Katsuki Y., Freire R., Shibata A., Jeggo P. A., Nucleic Acids Res. 2013, 41, 10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wang Y., Wang J., Zhong J., Deng Y., Xi Q., He S., Yang S., Jiang L., Huang M., Tang C., Liu R., Med Oncol 2015, 32, 379. [DOI] [PubMed] [Google Scholar]
- 102. Wu N., Liu C., Bai C., Han Y. P., Cho W. C., Li Q., Int. J. Mol. Sci. 2013, 14, 10749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sharma A., Almasan A., Int. J. Mol. Sci. 2020, 21, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Shen J., Hong L., Chen L., Biochem. Biophys. Res. Commun. 2020, 524, 683. [DOI] [PubMed] [Google Scholar]
- 105. Jung H., Kim B. G., Han W. H., Lee J. H., Cho J. Y., Park W. S., Maurice M. M., Han J. K., Lee M. J., Finley D., Jho E. H., Oncogenesis 2013, 2, e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang L., Chen Y. J., Xu K., Wang Y. Y., Shen X. Z., Tu R. Q., Tumor Biol. 2014, 35, 11427. [DOI] [PubMed] [Google Scholar]
- 107. Fang Y., Fu D., Tang W., Cai Y., Ma D., Wang H., Xue R., Liu T., Huang X., Dong L., Wu H., Shen X., Biochim. Biophys. Acta. 2013, 1833, 559. [DOI] [PubMed] [Google Scholar]
- 108. Koulich E., Li X., DeMartino G. N., Mol. Biol. Cell 2008, 19, 1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Jackson S. P., Bartek J., Nature 2009, 461, 1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. O'Connor M. J., Mol. Cell 2015, 60, 547. [DOI] [PubMed] [Google Scholar]
- 111. Hanahan D., Weinberg R. A., Cell 2000, 100, 57. [DOI] [PubMed] [Google Scholar]
- 112. Jackson S. P., Durocher D., Mol. Cell 2013, 49, 795. [DOI] [PubMed] [Google Scholar]
- 113. Jacq X., Kemp M., Martin N. M., Jackson S. P., Cell Biochem Biophys 2013, 67, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Nijman S. M., Huang T. T., Dirac A. M., Brummelkamp T. R., Kerkhoven R. M., D'Andrea A. D., Bernards R., Mol. Cell 2005, 17, 331. [DOI] [PubMed] [Google Scholar]
- 115. Castella M., Jacquemont C., Thompson E. L., Yeo J. E., Cheung R. S., Huang J. W., Sobeck A., Hendrickson E. A., Taniguchi T., PLoS Genet. 2015, 11, e1005563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Huang T. T., Nijman S. M., Mirchandani K. D., Galardy P. J., Cohn M. A., Haas W., Gygi S. P., Ploegh H. L., Bernards R., D'Andrea A. D., Nat. Cell Biol. 2006, 8, 339. [DOI] [PubMed] [Google Scholar]
- 117. Guervilly J. H., Renaud E., Takata M., Rosselli F., Hum. Mol. Genet. 2011, 20, 2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Williams S. A., Maecker H. L., French D. M., Liu J., Gregg A., Silverstein L. B., Cao T. C., Carano R. A., Dixit V. M., Cell 2011, 146, 918. [DOI] [PubMed] [Google Scholar]
- 119. Takeda S., Nakamura K., Taniguchi Y., Paull T. T., Mol. Cell 2007, 28, 351. [DOI] [PubMed] [Google Scholar]
- 120. Liu H., Zhang H., Wang X., Tian Q., Hu Z., Peng C., Jiang P., Wang T., Guo W., Chen Y., Li X., Zhang P., Pei H., Cell Rep. 2015, 13, 93. [DOI] [PubMed] [Google Scholar]
- 121. Wijnhoven P., Konietzny R., Blackford A. N., Travers J., Kessler B. M., Nishi R., Jackson Stephen P., Mol. Cell 2015, 60, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Gao M., Guo G., Huang J., Kloeber J. A., Zhao F., Deng M., Tu X., Kim W., Zhou Q., Zhang C., Yin P., Luo K., Lou Z., Nat. Commun. 2020, 11, 5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Schoenfeld A. R., Apgar S., Dolios G., Wang R., Aaronson S. A., Mol. Cell Biol. 2004, 24, 7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Wiltshire T. D., Lovejoy C. A., Wang T., Xia F., O'Connor M. J., Cortez D., J. Biol. Chem. 2010, 285, 14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Peng Y., Liao Q., Tan W., Peng C., Hu Z., Chen Y., Li Z., Li J., Zhen B., Zhu W., Li X., Yao Y., Song Q., Liu C., Qi X., He F., Pei H., Nat. Commun. 2019, 10, 1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Zhao Y., Majid M. C., Soll J. M., Brickner J. R., Dango S., Mosammaparast N., EMBO J. 2015, 34, 1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. McGarry E., Gaboriau D., Rainey M. D., Restuccia U., Bachi A., Santocanale C., Cancer Res. 2016, 76, 2384. [DOI] [PubMed] [Google Scholar]
- 128. Wolfsperger F., Hogh‐Binder S. A., Schittenhelm J., Psaras T., Ritter V., Bornes L., Huber S. M., Jendrossek V., Rudner J., Cell Death Dis. 2016, 7, e2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Jensen D. E., Proctor M., Marquis S. T., Gardner H. P., Ha S. I., Chodosh L. A., Ishov A. M., Tommerup N., Vissing H., Sekido Y., Minna J., Borodovsky A., Schultz D. C., Wilkinson K. D., Maul G. G., Barlev N., Berger S. L., Prendergast G. C., F. J. Rauscher, 3rd , Oncogene 1998, 16, 1097. [DOI] [PubMed] [Google Scholar]
- 130. Ismail I. H., Davidson R., Gagné J. P., Xu Z. Z., Poirier G. G., Hendzel M. J., Cancer Res. 2014, 74, 4282. [DOI] [PubMed] [Google Scholar]
- 131. Nakada S., Tai I., Panier S., Al‐Hakim A., Iemura S., Juang Y. C., O'Donnell L., Kumakubo A., Munro M., Sicheri F., Gingras A. C., Natsume T., Suda T., Durocher D., Nature 2010, 466, 941. [DOI] [PubMed] [Google Scholar]
- 132. Butler L. R., Densham R. M., Jia J., Garvin A. J., Stone H. R., Shah V., Weekes D., Festy F., Beesley J., Morris J. R., EMBO J. 2012, 31, 3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Mosbech A., Lukas C., Bekker‐Jensen S., Mailand N., J. Biol. Chem. 2013, 288, 16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Choi H. S., Baek K. H., Cell Mol Life Sci 2022, 79, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Li M., Brooks C. L., Kon N., Gu W., Mol. Cell 2004, 13, 879. [DOI] [PubMed] [Google Scholar]
- 136. Li M., Chen D., Shiloh A., Luo J., Nikolaev A. Y., Qin J., Gu W., Nature 2002, 416, 648. [DOI] [PubMed] [Google Scholar]
- 137. Schauer N. J., Liu X., Magin R. S., Doherty L. M., Chan W. C., Ficarro S. B., Hu W., Roberts R. M., Iacob R. E., Stolte B., Giacomelli A. O., Perera S., McKay K., Boswell S. A., Weisberg E. L., Ray A., Chauhan D., Dhe‐Paganon S., Anderson K. C., Griffin J. D., Li J., Hahn W. C., Sorger P. K., Engen J. R., Stegmaier K., Marto J. A., Buhrlage S. J., Sci. Rep. 2020, 10, 5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Zhou J., Wang J., Chen C., Yuan H., Wen X., Sun H., Med. Chem. 2018, 14, 3. [DOI] [PubMed] [Google Scholar]
- 139. Sparks A., Dayal S., Das J., Robertson P., Menendez S., Saville M. K., Oncogene 2014, 33, 4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Chen Y., Wang Y. G., Li Y., Sun X. X., Dai M. S., OncoTargets Ther. 2017, 8, 11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Li L., Tao Q., Jin H., van Hasselt A., Poon F. F., Wang X., Zeng M. S., Jia W. H., Zeng Y. X., Chan A. T., Cao Y., Clin. Cancer Res. 2010, 16, 2949. [DOI] [PubMed] [Google Scholar]
- 142. Xiang T., Li L., Yin X., Yuan C., Tan C., Su X., Xiong L., Putti T. C., Oberst M., Kelly K., Ren G., Tao Q., PLoS One 2012, 7, e29783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Sun J., Li T., Zhao Y., Huang L., Sun H., Wu H., Jiang X., Mol. Cell Biochem. 2018, 441, 1. [DOI] [PubMed] [Google Scholar]
- 144. Qiu C., Liu Y., Mei Y., Zou M., Zhao Z., Ye M., Wu X., Aging 2018, 10, 2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Li F., Hu Q., He T., Xu J., Yi Y., Xie S., Ding L., Fu M., Guo R., Xiao Z. J., Niu M., Cancers 2020, 12, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Heo M. J., Kim Y. M., Koo J. H., Yang Y. M., An J., Lee S. K., Lee S. J., Kim K. M., Park J. W., Kim S. G., OncoTargets Ther. 2014, 5, 2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Li T., Yan B., Ma Y., Weng J., Yang S., Zhao N., Wang X., Sun X., Cell Death Dis. 2018, 9, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Yu B., Shen B., Ba Z., Liu Z., Yuan J., Zhao W., Wu D., J. Cell Mol. Med 2020, 24, 13813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Zhou L., Jiang H., Du J., Li L., Li R., Lu J., Fu W., Hou J., Exp. Mol. Med. 2018, 50, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Wertz I. E., O'Rourke K. M., Zhou H., Eby M., Aravind L., Seshagiri S., Wu P., Wiesmann C., Baker R., Boone D. L., Ma A., Koonin E. V., Dixit V. M., Nature 2004, 430, 694. [DOI] [PubMed] [Google Scholar]
- 151. Priem D., Devos M., Druwé S., Martens A., Slowicka K., Ting A. T., Pasparakis M., Declercq W., Vandenabeele P., van Loo G., Bertrand M. J. M., Cell Death Dis. 2019, 10, 692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Lork M., Verhelst K., Beyaert R., Cell Death Differ. 2017, 24, 1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Antao A. M., Tyagi A., Kim K. S., Ramakrishna S., Cancers 2020, 12,33375055 [Google Scholar]
- 154. Cheng J., Guo J., North B. J., Wang B., Cui C. P., Li H., Tao K., Zhang L., Wei W., Biochim. Biophys. Acta. Rev. Cancer 2019, 1872, 188312. [DOI] [PubMed] [Google Scholar]
- 155. Steeg P. S., Nat. Med. 2006, 12, 895. [DOI] [PubMed] [Google Scholar]
- 156. He L., Liu X., Yang J., Li W., Liu S., Liu X., Yang Z., Ren J., Wang Y., Shan L., Guan C., Pei F., Lei L., Zhang Y., Yi X., Yang X., Liang J., Liu R., Sun L., Shang Y., Cell Res. 2018, 28, 934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Deng R., Guo Y., Li L., He J., Qiang Z., Zhang H., Chen R., Wang Y., Zhao X., Yu J., Mol. Oncol. 2021, 15, 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Carbone M., Harbour J. W., Brugarolas J., Bononi A., Pagano I., Dey A., Krausz T., Pass H. I., Yang H., Gaudino G., Cancer Discov. 2020, 10, 1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Dixon S. J., Lemberg K. M., Lamprecht M. R., Skouta R., Zaitsev E. M., Gleason C. E., Patel D. N., Bauer A. J., Cantley A. M., Yang W. S., B. Morrison, 3rd , Stockwell B. R., Cell 2012, 149, 1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Zhang Y., Shi J., Liu X., Feng L., Gong Z., Koppula P., Sirohi K., Li X., Wei Y., Lee H., Zhuang L., Chen G., Xiao Z. D., Hung M. C., Chen J., Huang P., Li W., Gan B., Nat. Cell Biol. 2018, 20, 1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L., Mol. Cell 2000, 6, 1365. [DOI] [PubMed] [Google Scholar]
- 162. Zhang L., Huang H., Zhou F., Schimmel J., Pardo C. G., Zhang T., Barakat T. S., Sheppard K. A., Mickanin C., Porter J. A., Vertegaal A. C., van Dam H., Gribnau J., Lu C. X., ten Dijke P., Mol. Cell 2012, 46, 650. [DOI] [PubMed] [Google Scholar]
- 163. Park S. H., Jung E. H., Kim G. Y., Kim B. C., Lim J. H., Woo C. H., Mol. Cells 2015, 38, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Viñas‐Castells R., Beltran M., Valls G., Gómez I., García J. M., Montserrat‐Sentís B., Baulida J., Bonilla F., de Herreros A. G., Díaz V. M., J. Biol. Chem. 2010, 285, 3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Mazzolini R., Gonzàlez N., Garcia‐Garijo A., Millanes‐Romero A., Peiró S., Smith S., García de Herreros A., Canudas S., Nucleic Acids Res. 2018, 46, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Wu Y., Wang Y., Lin Y., Liu Y., Wang Y., Jia J., Singh P., Chi Y. I., Wang C., Dong C., Li W., Tao M., Napier D., Shi Q., Deng J., Evers B. M., Zhou B. P., Nat. Commun. 2017, 8, 14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Park S. H., Fong K. W., Kim J., Wang F., Lu X., Lee Y., Brea L. T., Wadosky K., Guo C., Abdulkadir S. A., Crispino J. D., Fang D., Ntziachristos P., Liu X., Li X., Wan Y., Goodrich D. W., Zhao J. C., Yu J., Sci. Adv. 2021, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Lee J. E., Park C. M., Kim J. H., Genet. Mol. Biol. 2020, 43, e20190338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Liu S., González‐Prieto R., Zhang M., Geurink P. P., Kooij R., Iyengar P. V., van Dinther M., Bos E., Zhang X., Le Dévédec S. E., van de Water B., Koning R. I., Zhu H. J., Mesker W. E., Vertegaal A. C. O., Ovaa H., Zhang L., Martens J. W. M., Ten Dijke P., Clin. Cancer Res. 2020, 26, 1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Kumar H., Kawai T., Akira S., Int. Rev. Immunol. 2011, 30, 16. [DOI] [PubMed] [Google Scholar]
- 171. Mortensen E. S., Fenton K. A., Rekvig O. P., Am. J. Pathol. 2008, 172, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. O'Neill L. A., Bowie A. G., Nat. Rev. Immunol. 2007, 7, 353. [DOI] [PubMed] [Google Scholar]
- 173. Barnett K. C., Coronas‐Serna J. M., Zhou W., Ernandes M. J., Cao A., Kranzusch P. J., Kagan J. C., Cell 2019, 176, 1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Guey B., Wischnewski M., Decout A., Makasheva K., Kaynak M., Sakar M. S., Fierz B., Ablasser A., Science 2020, 369, 823. [DOI] [PubMed] [Google Scholar]
- 175. Qi N., Shi Y., Zhang R., Zhu W., Yuan B., Li X., Wang C., Zhang X., Hou F., Nat. Commun. 2017, 8, 15676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Dixit E., Boulant S., Zhang Y., Lee A. S. Y., Odendall C., Shum B., Hacohen N., Chen Z. J., Whelan S. P., Fransen M., Nibert M. L., Superti‐Furga G., Kagan J. C., Cell 2010, 141, 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Miao Y., Wu J., Abraham S. N., Immunity 2016, 45, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Goel S., Oliva R., Jeganathan S., Bader V., Krause L. J., Kriegler S., Stender I. D., Christine C. W., Nakamura K., Hoffmann J. E., Winter R., Tatzelt J., Winklhofer K. F., Life Sci. Alliance 2023, 6, e202201607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Liu B., Zhang M., Chu H., Zhang H., Wu H., Song G., Wang P., Zhao K., Hou J., Wang X., Zhang L., Gao C., Nat. Immunol. 2017, 18, 214. [DOI] [PubMed] [Google Scholar]
- 180. Boone D. L., Turer E. E., Lee E. G., Ahmad R. C., Wheeler M. T., Tsui C., Hurley P., Chien M., Chai S., Hitotsumatsu O., McNally E., Pickart C., Ma A., Nat. Immunol. 2004, 5, 1052. [DOI] [PubMed] [Google Scholar]
- 181. Skaug B., Chen J., Du F., He J., Ma A., Chen Z. J., Mol. Cell 2011, 44, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Yang Z., Xian H., Hu J., Tian S., Qin Y., Wang R. F., Cui J., Sci. Rep. 2015, 5, 12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Zhang J., Stirling B., Temmerman S. T., Ma C. A., Fuss I. J., Derry J. M., Jain A., J. Clin. Invest. 2006, 116, 3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Fulda S., Rajalingam K., Dikic I., EMBO Mol. Med. 2012, 4, 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Wu B., Qiang L., Zhang Y., Fu Y., Zhao M., Lei Z., Lu Z., Wei Y. G., Dai H., Ge Y., Liu M., Zhou X., Peng Z., Li H., Cui C. P., Wang J., Zheng H., Liu C. H., Zhang L., Cell Mol. Immunol. 2022, 19, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Zhao Y., Mudge M. C., Soll J. M., Rodrigues R. B., Byrum A. K., Schwarzkopf E. A., Bradstreet T. R., Gygi S. P., Edelson B. T., Mosammaparast N., Mol. Cell 2018, 69, 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Zhou F., Zhang X., van Dam H., Ten Dijke P., Huang H., Zhang L., J. Biol. Chem. 2012, 287, 11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Karim R., Tummers B., Meyers C., Biryukov J. L., Alam S., Backendorf C., Jha V., Offringa R., van Ommen G. J., Melief C. J., Guardavaccaro D., Boer J. M., van der Burg S. H., PLoS Pathog. 2013, 9, e1003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Hu H., Brittain G. C., Chang J. H., Puebla‐Osorio N., Jin J., Zal A., Xiao Y., Cheng X., Chang M., Fu Y. X., Zal T., Zhu C., Sun S. C., Nature 2013, 494, 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Zhong B., Liu X., Wang X., Liu X., Li H., Darnay B. G., Lin X., Sun S. C., Dong C., Sci Signal 2013, 6, ra35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Lin D., Zhang M., Zhang M. X., Ren Y., Jin J., Zhao Q., Pan Z., Wu M., Shu H. B., Dong C., Zhong B., Proc. Natl. Acad. Sci. USA 2015, 112, 11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T., Nat. Immunol. 2004, 5, 730. [DOI] [PubMed] [Google Scholar]
- 193. Kang D. C., Gopalkrishnan R. V., Wu Q., Jankowsky E., Pyle A. M., Fisher P. B., Proc. Natl. Acad. Sci. USA 2002, 99, 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Saito T., Hirai R., Loo Y. M., Owen D., Johnson C. L., Sinha S. C., Akira S., Fujita T., M. Gale, Jr. , Proc. Natl. Acad. Sci. USA 2007, 104, 582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Zeng W., Sun L., Jiang X., Chen X., Hou F., Adhikari A., Xu M., Chen Z. J., Cell 2010, 141, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Jiang X., Kinch L. N., Brautigam C. A., Chen X., Du F., Grishin N. V., Chen Z. J., Immunity 2012, 36, 959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Kuniyoshi K., Takeuchi O., Pandey S., Satoh T., Iwasaki H., Akira S., Kawai T., Proc. Natl. Acad. Sci. USA 2014, 111, 5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Yan J., Li Q., Mao A. P., Hu M. M., Shu H. B., J. Mol. Cell Biol. 2014, 6, 154. [DOI] [PubMed] [Google Scholar]
- 199. Paz S., Vilasco M., Werden S. J., Arguello M., Joseph‐Pillai D., Zhao T., Nguyen T. L., Sun Q., Meurs E. F., Lin R., Hiscott J., Cell Res. 2011, 21, 895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Narayan K., Waggoner L., Pham S. T., Hendricks G. L., Waggoner S. N., Conlon J., Wang J. P., Fitzgerald K. A., Kang J., J. Virol. 2014, 88, 10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Zhou P., Ding X., Wan X., Liu L., Yuan X., Zhang W., Hui X., Meng G., Xiao H., Li B., Zhong J., Hou F., Deng L., Zhang Y., Nat. Commun. 2018, 9, 1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Pan D., Yu R., Xu H., García de Abajo F. J., Nat. Commun. 2017, 8, 1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Zhao K., Zhang Q., Li X., Zhao D., Liu Y., Shen Q., Yang M., Wang C., Li N., Cao X., J. Immunol. 2016, 196, 1209. [DOI] [PubMed] [Google Scholar]
- 204. Wang W., Jiang M., Liu S., Zhang S., Liu W., Ma Y., Zhang L., Zhang J., Cao X., Proc. Natl. Acad. Sci. USA 2016, 113, 9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Génin P., Cuvelier F., Lambin S., Côrte‐Real Filipe J., Autrusseau E., Laurent C., Laplantine E., Weil R., PLoS Pathog. 2015, 11, e1004877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Friedman C. S., O'Donnell M. A., Legarda‐Addison D., Ng A., Cárdenas W. B., Yount J. S., Moran T. M., Basler C. F., Komuro A., Horvath C. M., Xavier R., Ting A. T., EMBO Rep. 2008, 9, 930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Cui J., Song Y., Li Y., Zhu Q., Tan P., Qin Y., Wang H. Y., Wang R. F., Cell Res. 2014, 24, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Li H., Zhao Z., Ling J., Pan L., Zhao X., Zhu H., Yu J., Xie B., Shen J., Chen W., Eur. J. Immunol. 2019, 49, 42. [DOI] [PubMed] [Google Scholar]
- 209. Zhang H., Wang D., Zhong H., Luo R., Shang M., Liu D., Chen H., Fang L., Xiao S., Sci. Rep. 2015, 5, 11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Fan Y., Mao R., Yu Y., Liu S., Shi Z., Cheng J., Zhang H., An L., Zhao Y., Xu X., Chen Z., Kogiso M., Zhang D., Zhang H., Zhang P., Jung J. U., Li X., Xu G., Yang J., J. Exp. Med. 2014, 211, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Tao X., Chu B., Xin D., Li L., Sun Q., PLoS Pathog. 2020, 16, e1008293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Pauli E. K., Chan Y. K., Davis M. E., Gableske S., Wang M. K., Feister K. F., Gack M. U., Sci. Signal. 2014, 7, ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Chen R., Zhang L., Zhong B., Tan B., Liu Y., Shu H. B., Cell Res. 2010, 20, 802. [DOI] [PubMed] [Google Scholar]
- 214. Wang L., Zhao W., Zhang M., Wang P., Zhao K., Zhao X., Yang S., Gao C., J. Virol. 2013, 87, 4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Liuyu T., Yu K., Ye L., Zhang Z., Zhang M., Ren Y., Cai Z., Zhu Q., Lin D., Zhong B., Cell Res. 2019, 29, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Dempsey A., Bowie A. G., Virology 2015, 479–480, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Sun L., Wu J., Du F., Chen X., Chen Z. J., Science 2013, 339, 786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Ishikawa H., Ma Z., Barber G. N., Nature 2009, 461, 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Ishikawa H., Barber G. N., Nature 2008, 455, 674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y. T., Grishin N. V., Chen Z. J., Science 2015, 347, aaa2630. [DOI] [PubMed] [Google Scholar]
- 221. Dobbs N., Burnaevskiy N., Chen D., Gonugunta V. K., Alto N. M., Yan N., Cell Host Microbe 2015, 18, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Chen M., Meng Q., Qin Y., Liang P., Tan P., He L., Zhou Y., Chen Y., Huang J., Wang R. F., Cui J., Mol. Cell 2016, 64, 105. [DOI] [PubMed] [Google Scholar]
- 223. Guo Y., Jiang F., Kong L., Li B., Yang Y., Zhang L., Liu B., Zheng Y., Gao C., J. Immunol. 2019, 203, 2049. [DOI] [PubMed] [Google Scholar]
- 224. Zhang M. X., Cai Z., Zhang M., Wang X. M., Wang Y., Zhao F., Zhou J., Luo M. H., Zhu Q., Xu Z., Zeng W. B., Zhong B., Lin D., J. Immunol. 2019, 202, 2397. [DOI] [PubMed] [Google Scholar]
- 225. Zhang M., Zhang M. X., Zhang Q., Zhu G. F., Yuan L., Zhang D. E., Zhu Q., Yao J., Shu H. B., Zhong B., Cell Res. 2016, 26, 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Zhang L., Wei N., Cui Y., Hong Z., Liu X., Wang Q., Li S., Liu H., Yu H., Cai Y., Wang Q., Zhu J., Meng W., Chen Z., Wang C., PLoS Pathog. 2018, 14, e1007435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Zhang H. Y., Liao B. W., Xu Z. S., Ran Y., Wang D. P., Yang Y., Luo W. W., Wang Y. Y., PLoS Pathog. 2020, 16, e1008178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Kovalenko A., Chable‐Bessia C., Cantarella G., Israël A., Wallach D., Courtois G., Nature 2003, 424, 801. [DOI] [PubMed] [Google Scholar]
- 229. Tsuchida T., Zou J., Saitoh T., Kumar H., Abe T., Matsuura Y., Kawai T., Akira S., Immunity 2010, 33, 765. [DOI] [PubMed] [Google Scholar]
- 230. Ye L., Zhang Q., Liuyu T., Xu Z., Zhang M. X., Luo M. H., Zeng W. B., Zhu Q., Lin D., Zhong B., PLoS Pathog. 2019, 15, e1007680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Sun H., Zhang Q., Jing Y. Y., Zhang M., Wang H. Y., Cai Z., Liuyu T., Zhang Z. D., Xiong T. C., Wu Y., Zhu Q. Y., Yao J., Shu H. B., Lin D., Zhong B., Nat. Commun. 2017, 8, 15534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Chen Y., Wang L., Jin J., Luan Y., Chen C., Li Y., Chu H., Wang X., Liao G., Yu Y., Teng H., Wang Y., Pan W., Fang L., Liao L., Jiang Z., Ge X., Li B., Wang P., J. Exp. Med. 2017, 214, 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Liu Q., Wu Y., Qin Y., Hu J., Xie W., Qin F. X., Cui J., Sci. Adv. 2018, 4, eaar2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Rioux J. D., Abbas A. K., Nature 2005, 435, 584. [DOI] [PubMed] [Google Scholar]
- 235. Bettelli E., Oukka M., Kuchroo V. K., Nat. Immunol. 2007, 8, 345. [DOI] [PubMed] [Google Scholar]
- 236. Yang J., Xu P., Han L., Guo Z., Wang X., Chen Z., Nie J., Yin S., Piccioni M., Tsun A., Lv L., Ge S., Li B., J. Immunol. 2015, 194, 4094. [DOI] [PubMed] [Google Scholar]
- 237. Jin J., Xie X., Xiao Y., Hu H., Zou Q., Cheng X., Sun S. C., Nat. Immunol. 2016, 17, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Liu X., Li H., Zhong B., Blonska M., Gorjestani S., Yan M., Tian Q., Zhang D. E., Lin X., Dong C., J. Exp. Med. 2013, 210, 1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Hu H., Wang H., Xiao Y., Jin J., Chang J. H., Zou Q., Xie X., Cheng X., Sun S. C., J. Exp. Med. 2016, 213, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Pan L., Chen Z., Wang L., Chen C., Li D., Wan H., Li B., Shi G., Biochem. Biophys. Res. Commun. 2014, 449, 289. [DOI] [PubMed] [Google Scholar]
- 241. Balakirev M. Y., Jaquinod M., Haas A. L., Chroboczek J., J. Virol. 2002, 76, 6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Ding J., McGrath W. J., Sweet R. M., Mangel W. F., EMBO J. 1996, 15, 1778. [PMC free article] [PubMed] [Google Scholar]
- 243. Bente D. A., Forrester N. L., Watts D. M., McAuley A. J., Whitehouse C. A., Bray M., Antiviral Res. 2013, 100, 159. [DOI] [PubMed] [Google Scholar]
- 244. Frias‐Staheli N., Giannakopoulos N. V., Kikkert M., Taylor S. L., Bridgen A., Paragas J., Richt J. A., Rowland R. R., Schmaljohn C. S., Lenschow D. J., Snijder E. J., García‐Sastre A., Virgin H. W. t., Cell Host Microbe 2007, 2, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. James T. W., Frias‐Staheli N., Bacik J. P., Levingston Macleod J. M., Khajehpour M., García‐Sastre A., Mark B. L., Proc. Natl. Acad. Sci. USA 2011, 108, 2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. van Kasteren P. B., Beugeling C., Ninaber D. K., Frias‐Staheli N., van Boheemen S., García‐Sastre A., Snijder E. J., Kikkert M., J. Virol. 2012, 86, 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Barretto N., Jukneliene D., Ratia K., Chen Z., Mesecar A. D., Baker S. C., J. Virol. 2005, 79, 15189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Ratia K., Saikatendu K. S., Santarsiero B. D., Barretto N., Baker S. C., Stevens R. C., Mesecar A. D., Proc. Natl. Acad. Sci. USA 2006, 103, 5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Devaraj S. G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C. J., Tseng C. T., Baker S. C., Li K., J. Biol. Chem. 2007, 282, 32208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Matthews K., Schäfer A., Pham A., Frieman M., Virol. J. 2014, 11, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Li S. W., Wang C. Y., Jou Y. J., Huang S. H., Hsiao L. H., Wan L., Lin Y. J., Kung S. H., Lin C. W., Int. J. Mol. Sci. 2016, 17,28025500 [Google Scholar]
- 252. Xue Q., Liu H., Zhu Z., Yang F., Xue Q., Cai X., Liu X., Zheng H., Antiviral Res. 2018, 160, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Bodda C., Reinert L. S., Fruhwürth S., Richardo T., Sun C., Zhang B. C., Kalamvoki M., Pohlmann A., Mogensen T. H., Bergström P., Agholme L., O'Hare P., Sodeik B., Gyrd‐Hansen M., Zetterberg H., Paludan S. R., J. Exp. Med. 2020, 217, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Kumari P., Saha I., Narayanan A., Narayanan S., Takaoka A., Kumar N. S., Tailor P., Kumar H., Cell Death Dis. 2017, 8, e3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. van Gent M., Braem S. G. E., de Jong A., Delagic N., Peeters J. G. C., Boer I. G. J., Moynagh P. N., Kremmer E., Wiertz E. J., Ovaa H., Griffin B. D., Ressing M. E., PLoS Pathog. 2014, 10, e1003960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. van Loosdregt J., Fleskens V., Fu J., Brenkman A. B., Bekker C. P., Pals C. E., Meerding J., Berkers C. R., Barbi J., Gröne A., Sijts A. J., Maurice M. M., Kalkhoven E., Prakken B. J., Ovaa H., Pan F., Zaiss D. M., Coffer P. J., Immunity 2013, 39, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Dai X., Lu L., Deng S., Meng J., Wan C., Huang J., Sun Y., Hu Y., Wu B., Wu G., Lovell J. F., Jin H., Yang K., Theranostics 2020, 10, 9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Zhang J., Chen C., Hou X., Gao Y., Lin F., Yang J., Gao Z., Pan L., Tao L., Wen C., Yao Z., Tsun A., Shi G., Li B., J. Biol. Chem. 2013, 288, 9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Li Y., Lu Y., Wang S., Han Z., Zhu F., Ni Y., Liang R., Zhang Y., Leng Q., Wei G., Shi G., Zhu R., Li D., Wang H., Zheng S. G., Xu H., Tsun A., Li B., Nat. Commun. 2016, 7, 13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Liao Y., Yang M., Wang K., Wang Y., Zhong B., Jiang N., Cancer Lett. 2022, 526, 248. [DOI] [PubMed] [Google Scholar]
- 261. Herhaus L., Al‐Salihi M., Macartney T., Weidlich S., Sapkota G. P., Nat. Commun. 2013, 4, 2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Zhang W., Qiu W., Front. Mol. Biosci. 2020, 7, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Lim S. O., Li C. W., Xia W., Cha J. H., Chan L. C., Wu Y., Chang S. S., Lin W. C., Hsu J. M., Hsu Y. H., Kim T., Chang W. C., Hsu J. L., Yamaguchi H., Ding Q., Wang Y., Yang Y., Chen C. H., Sahin A. A., Yu D., Hortobagyi G. N., Hung M. C., Cancer Cell 2016, 30, 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Huang X., Zhang Q., Lou Y., Wang J., Zhao X., Wang L., Zhang X., Li S., Zhao Y., Chen Q., Liang T., Bai X., Cancer Immunol. Res. 2019, 7, 1580. [DOI] [PubMed] [Google Scholar]
- 265. Yang S., Yan H., Wu Y., Shan B., Zhou D., Liu X., Mao X., Zhou S., Zhao Q., Xia H., Am. J. Transl. Res. 2021, 13, 12763. [PMC free article] [PubMed] [Google Scholar]
- 266. Xiong W., Gao X., Zhang T., Jiang B., Hu M.‐M., Bu X., Gao Y., Zhang L.‐Z., Xiao B.‐L., He C., Sun Y., Li H., Shi J., Xiao X., Xiang B., Xie C., Chen G., Zhang H., Wei W., Freeman G. J., Shu H.‐B., Wang H., Zhang J., Nat. Commun. 2022, 13, 1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Ward S. J., Gratton H. E., Indrayudha P., Michavila C., Mukhopadhyay R., Maurer S. K., Caulton S. G., Emsley J., Dreveny I., J. Biol. Chem. 2018, 293, 17362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Burkhart R. A., Peng Y., Norris Z. A., Tholey R. M., Talbott V. A., Liang Q., Ai Y., Miller K., Lal S., Cozzitorto J. A., Witkiewicz A. K., Yeo C. J., Gehrmann M., Napper A., Winter J. M., Sawicki J. A., Zhuang Z., Brody J. R., Mol. Cancer Res. 2013, 11, 901. [DOI] [PubMed] [Google Scholar]
- 269. Turnbull A. P., Ioannidis S., Krajewski W. W., Pinto‐Fernandez A., Heride C., Martin A. C. L., Tonkin L. M., Townsend E. C., Buker S. M., Lancia D. R., Caravella J. A., Toms A. V., Charlton T. M., Lahdenranta J., Wilker E., Follows B. C., Evans N. J., Stead L., Alli C., Zarayskiy V. V., Talbot A. C., Buckmelter A. J., Wang M., McKinnon C. L., Saab F., McGouran J. F., Century H., Gersch M., Pittman M. S., Marshall C. G., et al., Nature 2017, 550, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Schlierf A., Altmann E., Quancard J., Jefferson A. B., Assenberg R., Renatus M., Jones M., Hassiepen U., Schaefer M., Kiffe M., Weiss A., Wiesmann C., Sedrani R., Eder J., Martoglio B., Nat. Commun. 2016, 7, 13166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. D'Arcy P., Brnjic S., Olofsson M. H., Fryknäs M., Lindsten K., De Cesare M., Perego P., Sadeghi B., Hassan M., Larsson R., Linder S., Nat. Med. 2011, 17, 1636. [DOI] [PubMed] [Google Scholar]
- 272. Wang X., D'Arcy P., Caulfield T. R., Paulus A., Chitta K., Mohanty C., Gullbo J., Chanan‐Khan A., Linder S., Chem. Biol. Drug Des. 2015, 86, 1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Tian Z., D'Arcy P., Wang X., Ray A., Tai Y. T., Hu Y., Carrasco R. D., Richardson P., Linder S., Chauhan D., Anderson K. C., Blood 2014, 123, 706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Liang Q., Dexheimer T. S., Zhang P., Rosenthal A. S., Villamil M. A., You C., Zhang Q., Chen J., Ott C. A., Sun H., Luci D. K., Yuan B., Simeonov A., Jadhav A., Xiao H., Wang Y., Maloney D. J., Zhuang Z., Nat. Chem. Biol. 2014, 10, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Chen J., Dexheimer T. S., Ai Y., Liang Q., Villamil M. A., Inglese J., Maloney D. J., Jadhav A., Simeonov A., Zhuang Z., Chem. Biol. 2011, 18, 1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliji S., Chaudhuri R., Fu W., Prabhakar B. S., Johnson M. E., Baker S. C., Ghosh A. K., Mesecar A. D., Proc. Natl. Acad. Sci. USA 2008, 105, 16119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Schauer N. J., Magin R. S., Liu X., Doherty L. M., Buhrlage S. J., J. Med Chem 2020, 63, 2731. [DOI] [PubMed] [Google Scholar]
- 278. Klemm T., Ebert G., Calleja D. J., Allison C. C., Richardson L. W., Bernardini J. P., Lu B. G., Kuchel N. W., Grohmann C., Shibata Y., Gan Z. Y., Cooney J. P., Doerflinger M., Au A. E., Blackmore T. R., van der Heden van Noort G. J., Geurink P. P., Ovaa H., Newman J., Riboldi‐Tunnicliffe A., Czabotar P. E., Mitchell J. P., Feltham R., Lechtenberg B. C., Lowes K. N., Dewson G., Pellegrini M., Lessene G., Komander D., EMBO J. 2020, 39, 106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Campagna J. J., Jagodzinska B., Alvarez P. A., Yeun C., Spilman P., Enquist K. M., Cohn W., Fajtová P., O'Donoghue A. J., Arumugaswami V., Li M. M. H., Damoiseaux R., John V., bioRxiv 2022, [Google Scholar]
- 280. Kulathu Y., Komander D., Nat. Rev. Mol. Cell Biol. 2012, 13, 508. [DOI] [PubMed] [Google Scholar]
- 281. Lange S. M., Armstrong L. A., Kulathu Y., Mol. Cell 2022, 82, 15. [DOI] [PubMed] [Google Scholar]
- 282. Raimondi M., Cesselli D., Di Loreto C., Marra F. La, Schneider C., Demarchi F., Autophagy 2019, 15, 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283. Ma L., Lin K., Chang G., Chen Y., Yue C., Guo Q., Zhang S., Jia Z., Huang T. T., Zhou A., Huang S., Cancer Res. 2019, 79, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Zhao Y., Wang X., Wang Q., Deng Y., Li K., Zhang M., Zhang Q., Zhou J., Wang H. Y., Bai P., Ren Y., Zhang N., Li W., Cheng Y., Xiao W., Du H. N., Cheng X., Yin L., Fu X., Lin D., Zhou Q., Zhong B., Cell Rep. 2018, 22, 2442. [DOI] [PubMed] [Google Scholar]
- 285. Kim J., Kim W. J., Liu Z., Loda M., Freeman M. R., Cell Cycle 2012, 11, 1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Shan J., Zhao W., Gu W., Mol. Cell 2009, 36, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Benassi B., Flavin R., Marchionni L., Zanata S., Pan Y., Chowdhury D., Marani M., Strano S., Muti P., Blandino G., Loda M., Cancer Discov. 2012, 2, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Graner E., Tang D., Rossi S., Baron A., Migita T., Weinstein L. J., Lechpammer M., Huesken D., Zimmermann J., Signoretti S., Loda M., Cancer Cell 2004, 5, 253. [DOI] [PubMed] [Google Scholar]
- 289. Wu X., Liu M., Zhu H., Wang J., Dai W., Li J., Zhu D., Tang W., Xiao Y., Lin J., Zhang W., Sun Y., Zhang Y., Chen Y., Li G., Li A., Xiang L., Liu S., Wang J., J Exp Clin Cancer Res 2019, 38, 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Fu S., Shao S., Wang L., Liu H., Hou H., Wang Y., Wang H., Huang X., Lv R., Biochem. Biophys. Res. Commun. 2017, 492, 178. [DOI] [PubMed] [Google Scholar]
- 291. Xing C., Lu X. X., Guo P. D., Shen T., Zhang S., He X. S., Gan W. J., Li X. M., Wang J. R., Zhao Y. Y., Wu H., Li J. M., Cancer Res. 2016, 76, 83. [DOI] [PubMed] [Google Scholar]
- 292. Yun S. I., Kim H. H., Yoon J. H., Park W. S., Hahn M. J., Kim H. C., Chung C. H., Kim K. K., Mol. Oncol. 2015, 9, 1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Zhang X., Berger F. G., Yang J., Lu X., EMBO J. 2011, 30, 2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Xue S., Wu W., Wang Z., Lu G., Sun J., Jin X., Xie L., Wang X., Tan C., Wang Z., Wang W., Ding X., Front Pharmacol. 2020, 11, 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Li X. Y., Wu H. Y., Mao X. F., Jiang L. X., Wang Y. X., Biochem. Biophys. Res. Commun. 2017, 492, 48. [DOI] [PubMed] [Google Scholar]
- 296. Wang S., Juan J., Zhang Z., Du Y., Xu Y., Tong J., Cao B., Moran M. F., Zeng Y., Mao X., Cell Death Dis. 2017, 8, e3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Quick L., Young R., Henrich I. C., Wang X., Asmann Y. W., Oliveira A. M., Chou M. M., Cancer Res. 2016, 76, 5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Madan B., Walker M. P., Young R., Quick L., Orgel K. A., Ryan M., Gupta P., Henrich I. C., Ferrer M., Marine S., Roberts B. S., Arthur W. T., Berndt J. D., Oliveira A. M., Moon R. T., Virshup D. M., Chou M. M., Major M. B., Proc. Natl. Acad. Sci. USA 2016, 113, E2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299. Li L., Yang H., He Y., Li T., Feng J., Chen W., Ao L., Shi X., Lin Y., Liu H., Zheng E., Lin Q., Bu J., Zeng Y., Zheng M., Xu Y., Liao Z., Lin J., Lin D., Mol. Cell Biol. 2018, 38, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Zhang C., Lu J., Zhang Q. W., Zhao W., Guo J. H., Liu S. L., Wu Y. L., Jiang B., Gao F. H., Int. J. Biochem. Cell Biol. 2016, 79, 209. [DOI] [PubMed] [Google Scholar]
- 301. Song M. S., Salmena L., Carracedo A., Egia A., Lo‐Coco F., Teruya‐Feldstein J., Pandolfi P. P., Nature 2008, 455, 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Chen S. T., Okada M., Nakato R., Izumi K., Bando M., Shirahige K., J. Biol. Chem. 2015, 290, 21713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Franqui‐Machin R., Hao M., Bai H., Gu Z., Zhan X., Habelhah H., Jethava Y., Qiu L., Frech I., Tricot G., Zhan F., J. Clin. Invest. 2018, 128, 2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Tavana O., Li D., Dai C., Lopez G., Banerjee D., Kon N., Chen C., Califano A., Yamashiro D. J., Sun H., Gu W., Nat. Med. 2016, 22, 1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Jin Q., Martinez C. A., Arcipowski K. M., Zhu Y., Gutierrez‐Diaz B. T., Wang K. K., Johnson M. R., Volk A. G., Wang F., Wu J., Grove C., Wang H., Sokirniy I., Thomas P. M., Goo Y. A., Abshiru N. A., Hijiya N., Peirs S., Vandamme N., Berx G., Goosens S., Marshall S. A., Rendleman E. J., Takahashi Y. H., Wang L., Rawat R., Bartom E. T., Collings C. K., Van Vlierberghe P., Strikoudis A., et al., Clin. Cancer Res. 2019, 25, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306. Morra F., Luise C., Merolla F., Poser I., Visconti R., Ilardi G., Paladino S., Inuzuka H., Guggino G., Monaco R., Colecchia D., Monaco G., Cerrato A., Chiariello M., Denning K., Claudio P. P., Staibano S., Celetti A., OncoTargets Ther. 2015, 6, 12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Cai J. B., Shi G. M., Dong Z. R., Ke A. W., Ma H. H., Gao Q., Shen Z. Z., Huang X. Y., Chen H., Yu D. D., Liu L. X., Zhang P. F., Zhang C., Hu M. Y., Yang L. X., Shi Y. H., Wang X. Y., Ding Z. B., Qiu S. J., Sun H. C., Zhou J., Shi Y. G., Fan J., Hepatology 2015, 61, 1603. [DOI] [PubMed] [Google Scholar]
- 308. Yi L., Cui Y., Xu Q., Jiang Y., Oncol Rep 2016, 36, 2935. [DOI] [PubMed] [Google Scholar]
- 309. Du Z., Song J., Wang Y., Zhao Y., Guda K., Yang S., Kao H. Y., Xu Y., Willis J., Markowitz S. D., Sedwick D., Ewing R. M., Wang Z., Sci Signal 2010, 3, ra80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Novellasdemunt L., Foglizzo V., Cuadrado L., Antas P., Kucharska A., Encheva V., Snijders A. P., Li V. S. W., Cell Rep. 2017, 21, 612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Hernández‐Pérez S., Cabrera E., Salido E., Lim M., Reid L., Lakhani S. R., Khanna K. K., Saunus J. M., Freire R., Oncogene 2017, 36, 4802. [DOI] [PubMed] [Google Scholar]
- 312. Su D., Ma S., Shan L., Wang Y., Wang Y., Cao C., Liu B., Yang C., Wang L., Tian S., Ding X., Liu X., Yu N., Song N., Liu L., Yang S., Zhang Q., Yang F., Zhang K., Shi L., J. Clin. Invest. 2018, 128, 4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Wang Q., Ma S., Song N., Li X., Liu L., Yang S., Ding X., Shan L., Zhou X., Su D., Wang Y., Zhang Q., Liu X., Yu N., Zhang K., Shang Y., Yao Z., Shi L., J. Clin. Invest. 2016, 126, 2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Meulmeester E., Maurice M. M., Boutell C., Teunisse A. F., Ovaa H., Abraham T. E., Dirks R. W., Jochemsen A. G., Mol. Cell 2005, 18, 565. [DOI] [PubMed] [Google Scholar]
- 315. Zhang Y., Zhou L., Rouge L., Phillips A. H., Lam C., Liu P., Sandoval W., Helgason E., Murray J. M., Wertz I. E., Corn J. E., Nat. Chem. Biol. 2013, 9, 51. [DOI] [PubMed] [Google Scholar]
- 316. Wu H. T., Kuo Y. C., Hung J. J., Huang C. H., Chen W. Y., Chou T. Y., Chen Y., Chen Y. J., Chen Y. J., Cheng W. C., Teng S. C., Wu K. J., Nat. Commun. 2016, 7, 13644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Biswas K., Philip S., Yadav A., Martin B. K., Burkett S., Singh V., Babbar A., North S. L., Chang S., Sharan S. K., Nat. Commun. 2018, 9, 537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Jing X., Chen Y., Chen Y., Shi G., Lv S., Cheng N., Feng C., Xin Z., Zhang L., Wu J., Cancer Manag Res 2020, 12, 2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Xie F., Zhou X., Li H., Su P., Liu S., Li R., Zou J., Wei X., Pan C., Zhang Z., Zheng M., Liu Z., Meng X., Ovaa H., Ten Dijke P., Zhou F., Zhang L., EMBO J. 2022, 41, 108791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Izrailit J., Jaiswal A., Zheng W., Moran M. F., Reedijk M., Oncogene 2017, 36, 1048. [DOI] [PubMed] [Google Scholar]
- 321. Narayanan N., Wang Z., Li L., Yang Y., Cell Discov. 2017, 3, 16048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Zhu C., Ji X., Zhang H., Zhou Q., Cao X., Tang M., Si Y., Yan H., Li L., Liang T., Feng X. H., Zhao B., J. Biol. Chem. 2018, 293, 1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323. Li L., Liu T., Li Y., Wu C., Luo K., Yin Y., Chen Y., Nowsheen S., Wu J., Lou Z., Yuan J., Oncogene 2018, 37, 2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Xie Y., Avello M., Schirle M., McWhinnie E., Feng Y., Bric‐Furlong E., Wilson C., Nathans R., Zhang J., Kirschner M. W., Huang S. M., Cong F., J. Biol. Chem. 2013, 288, 2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Li X., Song N., Liu L., Liu X., Ding X., Song X., Yang S., Shan L., Zhou X., Su D., Wang Y., Zhang Q., Cao C., Ma S., Yu N., Yang F., Wang Y., Yao Z., Shang Y., Shi L., Nat. Commun. 2017, 8, 14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Engel K., Rudelius M., Slawska J., Jacobs L., Ahangarian Abhari B., Altmann B., Kurutz J., Rathakrishnan A., Fernández‐Sáiz V., Brunner A., Targosz B. S., Loewecke F., Gloeckner C. J., Ueffing M., Fulda S., Pfreundschuh M., Trümper L., Klapper W., Keller U., Jost P. J., Rosenwald A., Peschel C., Bassermann F., EMBO Mol. Med. 2016, 8, 851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Zhao Y., Zhang B., Lei Y., Sun J., Zhang Y., Yang S., Zhang X., Tumor Biol. 2016, 37, 13167. [DOI] [PubMed] [Google Scholar]
- 328. Wu Y., Yu X., Yi X., Wu K., Dwabe S., Atefi M., Elshimali Y., K. T. Kemp, 2nd , Bhat K., Haro J., Sarkissyan M., Vadgama J. V., Cancer Res. 2017, 77, 1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Schwickart M., Huang X., Lill J. R., Liu J., Ferrando R., French D. M., Maecker H., O'Rourke K., Bazan F., Eastham‐Anderson J., Yue P., Dornan D., Huang D. C., Dixit V. M., Nature 2010, 463, 103. [DOI] [PubMed] [Google Scholar]
- 330. Yang B., Zhang S., Wang Z., Yang C., Ouyang W., Zhou F., Zhou Y., Xie C., OncoTargets Ther. 2016, 7, 79515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Pérez‐Mancera P. A., Rust A. G., van der Weyden L., Kristiansen G., Li A., Sarver A. L., Silverstein K. A., Grützmann R., Aust D., Rümmele P., Knösel T., Herd C., Stemple D. L., Kettleborough R., Brosnan J. A., Li A., Morgan R., Knight S., Yu J., Stegeman S., Collier L. S., ten Hoeve J. J., de Ridder J., Klein A. P., Goggins M., Hruban R. H., Chang D. K., Biankin A. V., Grimmond S. M., Wessels L. F., et al., Nature 2012, 486, 266.22699621 [Google Scholar]
- 332. Khan O. M., Carvalho J., Spencer‐Dene B., Mitter R., Frith D., Snijders A. P., Wood S. A., Behrens A., J. Clin. Invest. 2018, 128, 1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333. Ouchida A. T., Kacal M., Zheng A., Ambroise G., Zhang B., Norberg E., Vakifahmetoglu‐Norberg H., Biochem. Biophys. Res. Commun. 2018, 502, 429. [DOI] [PubMed] [Google Scholar]
- 334. Guturi K. K. N., Bohgaki M., Bohgaki T., Srikumar T., Ng D., Kumareswaran R., El Ghamrasni S., Jeon J., Patel P., Eldin M. S., Bristow R., Cheung P., Stewart G. S., Raught B., Hakem A., Hakem R., Nat. Commun. 2016, 7, 12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. Takayama K. I., Suzuki T., Fujimura T., Takahashi S., Inoue S., Mol. Cancer Res. 2018, 16, 846. [DOI] [PubMed] [Google Scholar]
- 336. Weisberg E. L., Schauer N. J., Yang J., Lamberto I., Doherty L., Bhatt S., Nonami A., Meng C., Letai A., Wright R., Tiv H., Gokhale P. C., Ritorto M. S., De Cesare V., Trost M., Christodoulou A., Christie A., Weinstock D. M., Adamia S., Stone R., Chauhan D., Anderson K. C., Seo H. S., Dhe‐Paganon S., Sattler M., Gray N. S., Griffin J. D., Buhrlage S. J., Nat. Chem. Biol. 2017, 13, 1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Ko A., Han S. Y., Choi C. H., Cho H., Lee M. S., Kim S. Y., Song J. S., Hong K. M., Lee H. W., Hewitt S. M., Chung J. Y., Song J., Cell Death Differ. 2018, 25, 1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Lin Z., Yang H., Tan C., Li J., Liu Z., Quan Q., Kong S., Ye J., Gao B., Fang D., Cell Rep. 2013, 5, 1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Zhang M., Hu C., Tong D., Xiang S., Williams K., Bai W., Li G. M., Bepler G., Zhang X., J. Biol. Chem. 2016, 291, 10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340. Stockum A., Snijders A. P., Maertens G. N., PLoS One 2018, 13, e0190513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341. Kapadia B., Nanaji N. M., Bhalla K., Bhandary B., Lapidus R., Beheshti A., Evens A. M., Gartenhaus R. B., Nat. Commun. 2018, 9, 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342. Lee E. W., Seong D., Seo J., Jeong M., Lee H. K., Song J., Cell Death Differ. 2015, 22, 1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343. Zhou Z., Luo A., Shrivastava I., He M., Huang Y., Bahar I., Liu Z., Wan Y., EBioMedicine 2017, 15, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344. Zhang S., Xie C., Li H., Zhang K., Li J., Wang X., Yin Z., Lab Invest 2018, 98, 883. [DOI] [PubMed] [Google Scholar]
- 345. Garcia D. A., Baek C., Estrada M. V., Tysl T., Bennett E. J., Yang J., Chang J. T., Mol. Cancer Res. 2018, 16, 1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346. Zhang C., Xie C., Wang X., Huang Y., Gao S., Lu J., Lu Y., Zhang S., Am. J. Cancer Res. 2020, 10, 1416. [PMC free article] [PubMed] [Google Scholar]
- 347. Sun H., Ou B., Zhao S., Liu X., Song L., Liu X., Wang R., Peng Z., EBioMedicine 2019, 48, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348. Luo Q., Wu X., Nan Y., Chang W., Zhao P., Zhang Y., Su D., Liu Z., Cell Rep. 2020, 30, 98. [DOI] [PubMed] [Google Scholar]
- 349. Park M. K., Yao Y., Xia W., Setijono S. R., Kim J. H., Vila I. K., Chiu H. H., Wu Y., Billalabeitia E. G., Lee M. G., Kalb R. G., Hung M. C., Pandolfi P. P., Song S. J., Song M. S., Nat. Commun. 2019, 10, 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350. Shah P., Qiang L., Yang S., Soltani K., He Y. Y., OncoTargets Ther. 2017, 8, 96522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351. Zhang E., Shen B., Mu X., Qin Y., Zhang F., Liu Y., Xiao J., Zhang P., Wang C., Tan M., Fan Y., Am. J. Cancer Res. 2016, 6, 2901. [PMC free article] [PubMed] [Google Scholar]
- 352. Wu H. C., Lin Y. C., Liu C. H., Chung H. C., Wang Y. T., Lin Y. W., Ma H. I., Tu P. H., Lawler S. E., Chen R. H., Nat. Commun. 2014, 5, 3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353. Deng T., Yan G., Song X., Xie L., Zhou Y., Li J., Hu X., Li Z., Hu J., Zhang Y., Zhang H., Sun Y., Feng P., Wei D., Hu B., Liu J., Tan W., Ye M., Proc. Natl. Acad. Sci. USA 2018, 115, 4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354. Lim K. H., Suresh B., Park J. H., Kim Y. S., Ramakrishna S., Baek K. H., OncoTargets Ther. 2016, 7, 14441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355. McClurg U. L., Chit N., Azizyan M., Edwards J., Nabbi A., Riabowol K. T., Nakjang S., McCracken S. R., Robson C. N., Oncogene 2018, 37, 4679. [DOI] [PubMed] [Google Scholar]
- 356. Burska U. L., Harle V. J., Coffey K., Darby S., Ramsey H., O'Neill D., Logan I. R., Gaughan L., Robson C. N., J. Biol. Chem. 2013, 288, 32641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357. Sheng B., Wei Z., Wu X., Li Y., Liu Z., Cell Death Dis. 2021, 12, 1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358. Zhao X., Fiske B., Kawakami A., Li J., Fisher D. E., Nat. Commun. 2011, 2, 414. [DOI] [PubMed] [Google Scholar]
- 359. Li Y., Luo K., Yin Y., Wu C., Deng M., Li L., Chen Y., Nowsheen S., Lou Z., Yuan J., Nat. Commun. 2017, 8, 15752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360. Han C., Yang L., Choi H. H., Baddour J., Achreja A., Liu Y., Li Y., Li J., Wan G., Huang C., Ji G., Zhang X., Nagrath D., Lu X., Nat. Commun. 2016, 7, 13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361. Fang X., Zhou W., Wu Q., Huang Z., Shi Y., Yang K., Chen C., Xie Q., Mack S. C., Wang X., Carcaboso A. M., Sloan A. E., Ouyang G., McLendon R. E., Bian X. W., Rich J. N., Bao S., J. Exp. Med. 2017, 214, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362. Zhang S., Zhang M., Jing Y., Yin X., Ma P., Zhang Z., Wang X., Di W., Zhuang G., Nat. Commun. 2018, 9, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363. Zhang J., Zhang P., Wei Y., Piao H. L., Wang W., Maddika S., Wang M., Chen D., Sun Y., Hung M. C., Chen J., Ma L., Nat. Cell Biol. 2013, 15, 1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364. Liao Y., Liu N., Hua X., Cai J., Xia X., Wang X., Huang H., Liu J., Cell Death Dis. 2017, 8, e2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365. Zhang B., Li M., Huang P., Guan X. Y., Zhu Y. H., Thorac. Cancer 2017, 8, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366. Song C., Ma R., Yang X., Pang S., Cell Physiol. Biochem. 2017, 42, 965. [DOI] [PubMed] [Google Scholar]
- 367. Zhang Y., Jia J., Jin W., Cao J., Fu T., Ma D., Zhang Y., Pathol. Res. Pract. 2020, 216, 152963. [DOI] [PubMed] [Google Scholar]
- 368. Fielding A. B., Concannon M., Darling S., Rusilowicz‐Jones E. V., Sacco J. J., Prior I. A., Clague M. J., Urbé S., Coulson J. M., Oncogene 2018, 37, 2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369. Su Z. J., Cao J. S., Wu Y. F., Chen W. N., Lin X., Wu Y. L., Lin X., Sci. Rep. 2017, 7, 40246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. Eichhorn P. J., Rodón L., Gonzàlez‐Juncà A., Dirac A., Gili M., Martínez‐Sáez E., Aura C., Barba I., Peg V., Prat A., Cuartas I., Jimenez J., García‐Dorado D., Sahuquillo J., Bernards R., Baselga J., Seoane J., Nat. Med. 2012, 18, 429. [DOI] [PubMed] [Google Scholar]
- 371. Zou Q., Jin J., Hu H., Li H. S., Romano S., Xiao Y., Nakaya M., Zhou X., Cheng X., Yang P., Lozano G., Zhu C., Watowich S. S., Ullrich S. E., Sun S. C., Nat. Immunol. 2014, 15, 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372. Fukushima T., Yoshihara H., Furuta H., Hakuno F., Iemura S. I., Natsume T., Nakatsu Y., Kamata H., Asano T., Komada M., Takahashi S. I., Biochem. Biophys. Res. Commun. 2017, 484, 522. [DOI] [PubMed] [Google Scholar]
- 373. Liu W. T., Huang K. Y., Lu M. C., Huang H. L., Chen C. Y., Cheng Y. L., Yu H. C., Liu S. Q., Lai N. S., Huang H. B., Oncogene 2017, 36, 2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374. Villeneuve N. F., Tian W., Wu T., Sun Z., Lau A., Chapman E., Fang D., Zhang D. D., Mol. Cell 2013, 51, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375. Jin X., Yan Y., Wang D., Ding D., Ma T., Ye Z., Jimenez R., Wang L., Wu H., Huang H., Mol. Cell 2018, 71, 592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376. Song C., Liu W., Li J., Tumor Biol. 2017, 39, 1010428317717138. [DOI] [PubMed] [Google Scholar]
- 377. Lin Y., Wang Y., Shi Q., Yu Q., Liu C., Feng J., Deng J., Evers B. M., Zhou B. P., Wu Y., OncoTargets Ther. 2017, 8, 75127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378. Mehić M., de Sa V. K., Hebestreit S., Heldin C. H., Heldin P., Oncogenesis 2017, 6, e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379. Pereg Y., Liu B. Y., O'Rourke K. M., Sagolla M., Dey A., Komuves L., French D. M., Dixit V. M., Nat. Cell Biol. 2010, 12, 400. [DOI] [PubMed] [Google Scholar]
- 380. Song C., Peng J., Wei Y., Shao J., Chen X., Zhang X., Xu J., Exp. Cell Res. 2021, 409, 112884. [DOI] [PubMed] [Google Scholar]
- 381. Mustachio L. M., Lu Y., Tafe L. J., Memoli V., Rodriguez‐Canales J., Mino B., Villalobos P. A., Wistuba I., Katayama H., Hanash S. M., Roszik J., Kawakami M., Cho K. J., Hancock J. F., Chinyengetere F., Hu S., Liu X., Freemantle S. J., Dmitrovsky E., Mol. Cancer Res. 2017, 15, 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382. Guo Y., Dolinko A. V., Chinyengetere F., Stanton B., Bomberger J. M., Demidenko E., Zhou D. C., Gallagher R., Ma T., Galimberti F., Liu X., Sekula D., Freemantle S., Dmitrovsky E., Cancer Res. 2010, 70, 9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383. Cai J., Liu T., Jiang X., Guo C., Liu A., Xiao X., Exp. Cell Res. 2017, 358, 315. [DOI] [PubMed] [Google Scholar]
- 384. Wu M., Tu H. Q., Chang Y., Tan B., Wang G., Zhou J., Wang L., Mu R., Zhang W. N., OncoTargets Ther. 2017, 8, 2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385. Wu C., Luo K., Zhao F., Yin P., Song Y., Deng M., Huang J., Chen Y., Li L., Lee S., Kim J., Zhou Q., Tu X., Nowsheen S., Luo Q., Gao X., Lou Z., Liu Z., Yuan J., Cell Death Differ. 2018, 25, 1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386. Zhu M., Zhao H., Liao J., Xu X., Nucleic Acids Res. 2014, 42, 13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387. Yasunaga J., Lin F. C., Lu X., Jeang K. T., J. Virol. 2011, 85, 6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388. Li W., Cui K., Prochownik E. V., Li Y., Cell Death Dis. 2018, 9, 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389. Liu J., Kruswick A., Dang H., Tran A. D., Kwon S. M., Wang X. W., Oberdoerffer P., Nat. Commun. 2017, 8, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390. Chen Y., Zhou B., Chen D., OncoTargets Ther. 2017, 10, 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391. Yun S. I., Hong H. K., Yeo S. Y., Kim S. H., Cho Y. B., Kim K. K., Cancers 2020, 12,33375055 [Google Scholar]
- 392. Nguyen H. T., Kugler J. M., Loya A. C., Cohen S. M., OncoTargets Ther. 2017, 8, 64095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393. Qiu G. Z., Mao X. Y., Ma Y., Gao X. C., Wang Z., Jin M. Z., Sun W., Zou Y. X., Lin J., Fu H. L., Jin W. L., Cancer Sci. 2018, 109, 2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394. Zhou A., Lin K., Zhang S., Chen Y., Zhang N., Xue J., Wang Z., Aldape K. D., Xie K., Woodgett J. R., Huang S., Nat. Cell Biol. 2016, 18, 954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395. Wang A., Ning Z., Lu C., Gao W., Liang J., Yan Q., Tan G., Liu J., Front Pharmacol 2017, 8, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396. Xiao H., Tian Y., Yang Y., Hu F., Xie X., Mei J., Ding F., Biochem. Biophys. Res. Commun. 2015, 460, 703. [DOI] [PubMed] [Google Scholar]
- 397. Zhang H., Han B., Lu H., Zhao Y., Chen X., Meng Q., Cao M., Cai L., Hu J., Cancer Lett. 2018, 433, 186. [DOI] [PubMed] [Google Scholar]
- 398. Ling S., Li J., Shan Q., Dai H., Lu D., Wen X., Song P., Xie H., Zhou L., Liu J., Xu X., Zheng S., Mol. Oncol. 2017, 11, 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399. Kim D., Hong A., Park H. I., Shin W. H., Yoo L., Jeon S. J., Chung K. C., J. Cell. Physiol. 2017, 232, 3664. [DOI] [PubMed] [Google Scholar]
- 400. Atanassov B. S., Dent S. Y., EMBO Rep. 2011, 12, 924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401. Gennaro V. J., Stanek T. J., Peck A. R., Sun Y., Wang F., Qie S., Knudsen K. E., Rui H., Butt T., Diehl J. A., McMahon S. B., Proc. Natl. Acad. Sci. USA 2018, 115, E9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402. Melo‐Cardenas J., Xu Y., Wei J., Tan C., Kong S., Gao B., Montauti E., Kirsammer G., Licht J. D., Yu J., Ji P., Crispino J. D., Fang D., Blood 2018, 132, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403. Wang Y. C., Wu Y. S., Hung C. Y., Wang S. A., Young M. J., Hsu T. I., Hung J. J., Nat. Commun. 2018, 9, 3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404. Peterson L. F., Sun H., Liu Y., Potu H., Kandarpa M., Ermann M., Courtney S. M., Young M., Showalter H. D., Sun D., Jakubowiak A., Malek S. N., Talpaz M., Donato N. J., Blood 2015, 125, 3588. [DOI] [PubMed] [Google Scholar]
- 405. Wang S. A., Wang Y. C., Chuang Y. P., Huang Y. H., Su W. C., Chang W. C., Hung J. J., Oncogene 2017, 36, 2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406. Lui S. K. L., Iyengar P. V., Jaynes P., Isa Z., Pang B., Tan T. Z., Eichhorn P. J. A., EMBO Rep. 2017, 18, 797. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 407. Dirac A. M., Bernards R., Mol. Cancer Res. 2010, 8, 844. [DOI] [PubMed] [Google Scholar]
- 408. Dong L., Yu L., Bai C., Liu L., Long H., Shi L., Lin Z., Oncogene 2018, 37, 2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409. Diefenbacher M. E., Popov N., Blake S. M., Schülein‐Völk C., Nye E., Spencer‐Dene B., Jaenicke L. A., Eilers M., Behrens A., J. Clin. Invest. 2014, 124, 3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410. Wu Y., Wang Y., Yang X. H., Kang T., Zhao Y., Wang C., Evers B. M., Zhou B. P., Cell Rep. 2013, 5, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411. Haq S., Das S., Kim D. H., Chandrasekaran A. P., Hong S. H., Kim K. S., Ramakrishna S., Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 599. [DOI] [PubMed] [Google Scholar]
- 412. Schülein‐Völk C., Wolf E., Zhu J., Xu W., Taranets L., Hellmann A., Jänicke L. A., Diefenbacher M. E., Behrens A., Eilers M., Popov N., Cell Rep. 2014, 9, 1099. [DOI] [PubMed] [Google Scholar]
- 413. Popov N., Wanzel M., Madiredjo M., Zhang D., Beijersbergen R., Bernards R., Moll R., Elledge S. J., Eilers M., Nat. Cell Biol. 2007, 9, 765. [DOI] [PubMed] [Google Scholar]
- 414. Fong C. S., Mazo G., Das T., Goodman J., Kim M., O'Rourke B. P., Izquierdo D., Tsou M. F., Elife 2016, 5, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415. Martín Y., Cabrera E., Amoedo H., Hernández‐Pérez S., Domínguez‐Kelly R., Freire R., Oncogene 2015, 34, 1058. [DOI] [PubMed] [Google Scholar]
- 416. Qian W., Li Q., Wu X., Li W., Li Q., Zhang J., Li M., Zhang D., Zhao H., Zou X., Jia H., Zhang L., Yang X. D., Hou Z., Oncogene 2020, 39, 6802. [DOI] [PubMed] [Google Scholar]
- 417. Liu J., Chung H. J., Vogt M., Jin Y., Malide D., He L., Dundr M., Levens D., EMBO J. 2011, 30, 846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418. Gu L., Zhu Y., Lin X., Li Y., Cui K., Prochownik E. V., Li Y., Cancer Res. 2018, 78, 5808. [DOI] [PubMed] [Google Scholar]
- 419. Liang J. R., Martinez A., Lane J. D., Mayor U., Clague M. J., Urbé S., EMBO Rep. 2015, 16, 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420. Buus R., Faronato M., Hammond D. E., Urbé S., Clague M. J., Curr. Biol. 2009, 19, 1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421. Gan Q., Shao J., Cao Y., Lei J., Xie P., Ge J., Hu G., Life Sci. 2020, 261, 118316. [DOI] [PubMed] [Google Scholar]
- 422. Liu H., Zhang Q., Li K., Gong Z., Liu Z., Xu Y., Swaney M. H., Xiao K., Chen Y., OncoTargets Ther. 2016, 7, 81223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423. Yuasa‐Kawada J., Kinoshita‐Kawada M., Rao Y., Wu J. Y., Proc. Natl. Acad. Sci. USA 2009, 106, 14530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424. Lu J., Zhong Y., Chen J., Lin X., Lin Z., Wang N., Lin S., Cell. Physiol. Biochem. 2018, 50, 721. [DOI] [PubMed] [Google Scholar]
- 425. Liu C., Wang L., Chen W., Zhao S., Yin C., Lin Y., Jiang A., Zhang P., OncoTargets Ther. 2015, 6, 27891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426. Mondal T., Juvvuna P. K., Kirkeby A., Mitra S., Kosalai S. T., Traxler L., Hertwig F., Wernig‐Zorc S., Miranda C., Deland L., Volland R., Bartenhagen C., Bartsch D., Bandaru S., Engesser A., Subhash S., Martinsson T., Carén H., Akyürek L. M., Kurian L., Kanduri M., Huarte M., Kogner P., Fischer M., Kanduri C., Cancer Cell 2018, 33, 417. [DOI] [PubMed] [Google Scholar]
- 427. DeVine T., Sears R. C., Dai M. S., Biochem. Biophys. Res. Commun. 2018, 495, 2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428. Sun X. X., He X., Yin L., Komada M., Sears R. C., Dai M. S., Proc. Natl. Acad. Sci. USA 2015, 112, 3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429. Fraile J. M., Campos‐Iglesias D., Rodríguez F., Astudillo A., Vilarrasa‐Blasi R., Verdaguer‐Dot N., Prado M. A., Paulo J. A., Gygi S. P., Martín‐Subero J. I., Freije J. M. P., López‐Otín C., J. Biol. Chem. 2018, 293, 2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430. Qin T., Li B., Feng X., Fan S., Liu L., Liu D., Mao J., Lu Y., Yang J., Yu X., Zhang Q., Zhang J., Song B., Li M., Li L., J Exp Clin Cancer Res 2018, 37, 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431. Cai J., Li M., Wang X., Li L., Li Q., Hou Z., Jia H., Liu S., Front Genet 2019, 10, 1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432. Pan J., Deng Q., Jiang C., Wang X., Niu T., Li H., Chen T., Jin J., Pan W., Cai X., Yang X., Lu M., Xiao J., Wang P., Oncogene 2015, 34, 3957. [DOI] [PubMed] [Google Scholar]
- 433. Yang W. C., Shih H. M., Oncogene 2013, 32, 5167. [DOI] [PubMed] [Google Scholar]
- 434. Kim J. O., Kim S. R., Lim K. H., Kim J. H., Ajjappala B., Lee H. J., Choi J. I., Baek K. H., OncoTargets Ther. 2015, 6, 36551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435. Das C. M., Taylor P., Gireud M., Singh A., Lee D., Fuller G., Ji L., Fangusaro J., Rajaram V., Goldman S., Eberhart C., Gopalakrishnan V., Oncogene 2013, 32, 1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436. Hock A. K., Vigneron A. M., Carter S., Ludwig R. L., Vousden K. H., EMBO J. 2011, 30, 4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437. Ye D. X., Wang S. S., Huang Y., Wang X. J., Chi P., J Cancer 2021, 12, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438. Lan X., Atanassov B. S., Li W., Zhang Y., Florens L., Mohan R. D., Galardy P. J., Washburn M. P., Workman J. L., Dent S. Y. R., Cell Rep. 2016, 17, 2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439. Zou Y., Qiu G., Jiang L., Cai Z., Sun W., Hu H., Lu C., Jin W., Hu G., OncoTargets Ther. 2017, 8, 58231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440. Park J. M., Lee J. E., Park C. M., Kim J. H., Mol. Cells 2019, 42, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441. Liu T., Sun B., Zhao X., Li Y., Zhao X., Liu Y., Yao Z., Gu Q., Dong X., Shao B., Lin X., Liu F., An J., Mol. Cancer Ther. 2015, 14, 2121. [DOI] [PubMed] [Google Scholar]
- 442. Tian M., Zhu R., Ding F., Liu Z., Exp. Cell Res. 2020, 395, 112188. [DOI] [PubMed] [Google Scholar]
- 443. Kiran S., Dar A., Singh S. K., Lee K. Y., Dutta A., Mol. Cell 2018, 72, 823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444. Li X., Stevens P. D., Yang H., Gulhati P., Wang W., Evers B. M., Gao T., Oncogene 2013, 32, 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445. Silvestrini V. C., Thomé C. H., Albuquerque D., de Souza Palma C., Ferreira G. A., Lanfredi G. P., Masson A. P., Delsin L. E. A., Ferreira F. U., de Souza F. C., de Godoy L. M. F., Aquino A., Carrilho E., Panepucci R. A., Covas D. T., Faça V. M., J. Proteomics 2020, 219, 103734. [DOI] [PubMed] [Google Scholar]
- 446. Choi B. J., Park S. A., Lee S. Y., Cha Y. N., Surh Y. J., Sci. Rep. 2017, 7, 15918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447. Shi J., Liu Y., Xu X., Zhang W., Yu T., Jia J., Liu C., Mol Cell Biol 2015, 35, 3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448. Zhou A., Lin K., Zhang S., Ma L., Xue J., Morris S. A., Aldape K. D., Huang S., EMBO Rep. 2017, 18, 1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449. Luo K., Li Y., Yin Y., Li L., Wu C., Chen Y., Nowsheen S., Hu Q., Zhang L., Lou Z., Yuan J., EMBO J. 2017, 36, 1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450. Tu R., Kang W., Yang X., Zhang Q., Xie X., Liu W., Zhang J., Zhang X. D., Wang H., Du R. L., Cell Death Dis. 2018, 9, 553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451. Che X., Jian F., Jia N., Zheng Y., Jiang Y., Feng W., Am. J. Transl. Res. 2019, 11, 2784. [PMC free article] [PubMed] [Google Scholar]
- 452. Zhou Z., Zhang P., Hu X., Kim J., Yao F., Xiao Z., Zeng L., Chang L., Sun Y., Ma L., Am. J. Cancer Res. 2017, 7, 2020. [PMC free article] [PubMed] [Google Scholar]
- 453. Yang S., Liu L., Cao C., Song N., Wang Y., Ma S., Zhang Q., Yu N., Ding X., Yang F., Tian S., Zhang K., Sun T., Yang J., Yao Z., Wu S., Shi L., Nat. Commun. 2018, 9, 1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454. Massoumi R., Chmielarska K., Hennecke K., Pfeifer A., Fässler R., Cell 2006, 125, 665. [DOI] [PubMed] [Google Scholar]
- 455. Brummelkamp T. R., Nijman S. M., Dirac A. M., Bernards R., Nature 2003, 424, 797. [DOI] [PubMed] [Google Scholar]
- 456. Fernández‐Majada V., Welz P. S., Ermolaeva M. A., Schell M., Adam A., Dietlein F., Komander D., Büttner R., Thomas R. K., Schumacher B., Pasparakis M., Nat. Commun. 2016, 7, 12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457. Tauriello D. V., Haegebarth A., Kuper I., Edelmann M. J., Henraat M., Canninga‐van Dijk M. R., Kessler B. M., Clevers H., Maurice M. M., Mol. Cell 2010, 37, 607. [DOI] [PubMed] [Google Scholar]
- 458. Massoumi R., Kuphal S., Hellerbrand C., Haas B., Wild P., Spruss T., Pfeifer A., Fässler R., Bosserhoff A. K., J. Exp. Med. 2009, 206, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459. Miliani de Marval P., Lutfeali S., Jin J. Y., Leshin B., Selim M. A., Zhang J. Y., Cancer Prev. Res. 2011, 4, 851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460. de Jel M. M., Schott M., Lamm S., Neuhuber W., Kuphal S., Bosserhoff A. K., Oncogenesis 2019, 8, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461. Liu T., Jiang L., Tavana O., Gu W., Cancer Res. 2019, 79, 1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462. Zhou H., Liu Y., Zhu R., Ding F., Cao X., Lin D., Liu Z., Oncogene 2018, 37, 3356. [DOI] [PubMed] [Google Scholar]
- 463. Goncharov T., Niessen K., de Almagro M. C., Izrael‐Tomasevic A., Fedorova A. V., Varfolomeev E., Arnott D., Deshayes K., Kirkpatrick D. S., Vucic D., EMBO J. 2013, 32, 1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464. Karunarathna U., Kongsema M., Zona S., Gong C., Cabrera E., Gomes A. R., Man E. P., Khongkow P., Tsang J. W., Khoo U. S., Medema R. H., Freire R., Lam E. W., Oncogene 2016, 35, 1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465. Zhu D., Xu R., Huang X., Tang Z., Tian Y., Zhang J., Zheng X., Cell Death Differ. 2021, 28, 1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466. Stanišić V., Malovannaya A., Qin J., Lonard D. M., O'Malley B. W., J. Biol. Chem. 2009, 284, 16135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467. Zhao L., Wang X., Yu Y., Deng L., Chen L., Peng X., Jiao C., Gao G., Tan X., Pan W., Ge X., Wang P., J. Biol. Chem. 2018, 293, 4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468. Wang G., Li Y., Wang P., Liang H., Cui M., Zhu M., Guo L., Su Q., Sun Y., McNutt M. A., Yin Y., Cell Res. 2015, 25, 1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469. Li J., Cheng D., Zhu M., Yu H., Pan Z., Liu L., Geng Q., Pan H., Yan M., Yao M., Theranostics 2019, 9, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470. Zhang Z., Fan Y., Xie F., Zhou H., Jin K., Shao L., Shi W., Fang P., Yang B., van Dam H., Ten Dijke P., Zheng X., Yan X., Jia J., Zheng M., Jin J., Ding C., Ye S., Zhou F., Zhang L., Nat. Commun. 2017, 8, 2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471. Yao F., Zhou Z., Kim J., Hang Q., Xiao Z., Ton B. N., Chang L., Liu N., Zeng L., Wang W., Wang Y., Zhang P., Hu X., Su X., Liang H., Sun Y., Ma L., Nat. Commun. 2018, 9, 2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472. Piao S., Pei H. Z., Huang B., Baek S. H., Cell Signal. 2017, 33, 22. [DOI] [PubMed] [Google Scholar]
- 473. Kim Y., Kim W., Song Y., Kim J. R., Cho K., Moon H., Ro S. W., Seo E., Ryu Y. M., Myung S. J., Jho E. H., Proc. Natl. Acad. Sci. USA 2017, 114, 4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474. Du T., Li H., Fan Y., Yuan L., Guo X., Zhu Q., Yao Y., Li X., Liu C., Yu X., Liu Z., Cui C. P., Han C., Zhang L., Nat. Commun. 2019, 10, 2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475. Yuan L., Lv Y., Li H., Gao H., Song S., Zhang Y., Xing G., Kong X., Wang L., Li Y., Zhou T., Gao D., Xiao Z. X., Yin Y., Wei W., He F., Zhang L., Nat. Cell Biol. 2015, 17, 1169. [DOI] [PubMed] [Google Scholar]
- 476. Pu Q., Lv Y. R., Dong K., Geng W. W., Gao H. D., BMC Cancer 2020, 20, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477. de Vivo A., Sanchez A., Yegres J., Kim J., Emly S., Kee Y., Nucleic Acids Res. 2019, 47, 729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478. Park S. Y., Choi H. K., Choi Y., Kwak S., Choi K. C., Yoon H. G., Cancer Lett. 2015, 357, 419. [DOI] [PubMed] [Google Scholar]
- 479. Luo J., Lu Z., Lu X., Chen L., Cao J., Zhang S., Ling Y., Zhou X., PLoS One 2013, 8, e77682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480. Liu X., Zhang X., Peng Z., Li C., Wang Z., Wang C., Deng Z., Wu B., Cui Y., Wang Z., Cui C. P., Zheng M., Zhang L., Adv. Sci. 2020, 7, 1902040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481. Xu Z., Pei L., Wang L., Zhang F., Hu X., Gui Y., Oncogene 2014, 33, 2836. [DOI] [PubMed] [Google Scholar]
- 482. Spel L., Nieuwenhuis J., Haarsma R., Stickel E., Bleijerveld O. B., Altelaar M., Boelens J. J., Brummelkamp T. R., Nierkens S., Boes M., Cancer Res. 2018, 78, 6621. [DOI] [PubMed] [Google Scholar]
- 483. Bonacci T., Suzuki A., Grant G. D., Stanley N., Cook J. G., Brown N. G., Emanuele M. J., EMBO J. 2018, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484. Pareja F., Ferraro D. A., Rubin C., Cohen‐Dvashi H., Zhang F., Aulmann S., Ben‐Chetrit N., Pines G., Navon R., Crosetto N., Köstler W., Carvalho S., Lavi S., Schmitt F., Dikic I., Yakhini Z., Sinn P., Mills G. B., Yarden Y., Oncogene 2012, 31, 4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485. Wang B., Jie Z., Joo D., Ordureau A., Liu P., Gan W., Guo J., Zhang J., North B. J., Dai X., Cheng X., Bian X., Zhang L., Harper J. W., Sun S. C., Wei W., Nature 2017, 545, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486. Lv Q., Xie L., Cheng Y., Shi Y., Shan W., Ning C., Xie B., Yang B., Luo X., He Q., Zhu Q., Zhang Y., Zhang Z., Wang C., Chen X., Xu C., Cancer Lett. 2019, 442, 137. [DOI] [PubMed] [Google Scholar]
- 487. Zhang P., Wang P. X., Zhao L. P., Zhang X., Ji Y. X., Zhang X. J., Fang C., Lu Y. X., Yang X., Gao M. M., Zhang Y., Tian S., Zhu X. Y., Gong J., Ma X. L., Li F., Wang Z., Huang Z., She Z. G., Li H., Nat. Med. 2018, 24, 84. [DOI] [PubMed] [Google Scholar]
- 488. Zhu Y., Qu C., Hong X., Jia Y., Lin M., Luo Y., Lin F., Xie X., Xie X., Huang J., Wu Q., Qiu X., Piao D., Xing Y., Yu T., Lu Y., Huang Q., Yu C., Jin J., Zhang Z., Cell Death Differ. 2019, 26, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489. Goto Y., Zeng L., Yeom C. J., Zhu Y., Morinibu A., Shinomiya K., Kobayashi M., Hirota K., Itasaka S., Yoshimura M., Tanimoto K., Torii M., Sowa T., Menju T., Sonobe M., Kakeya H., Toi M., Date H., Hammond E. M., Hiraoka M., Harada H., Nat. Commun. 2015, 6, 6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490. Brinkmann K., Zigrino P., Witt A., Schell M., Ackermann L., Broxtermann P., Schüll S., Andree M., Coutelle O., Yazdanpanah B., Seeger J. M., Klubertz D., Drebber U., Hacker U. T., Krönke M., Mauch C., Hoppe T., Kashkar H., Cell Rep. 2013, 3, 881. [DOI] [PubMed] [Google Scholar]
- 491. Liao C., Beveridge R., Hudson J. J. R., Parker J. D., Chiang S. C., Ray S., Ashour M. E., Sudbery I., Dickman M. J., El‐Khamisy S. F., Cell Rep. 2018, 23, 3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492. Wicks S. J., Haros K., Maillard M., Song L., Cohen R. E., Dijke P. T., Chantry A., Oncogene 2005, 24, 8080. [DOI] [PubMed] [Google Scholar]
- 493. Zhang Y., Koppula P., Gan B., Cell Cycle 2019, 18, 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494. Zarrizi R., Menard J. A., Belting M., Massoumi R., Cancer Res. 2014, 74, 6499. [DOI] [PubMed] [Google Scholar]
- 495. Lee H. S., Lee S. A., Hur S. K., Seo J. W., Kwon J., Nat. Commun. 2014, 5, 5128. [DOI] [PubMed] [Google Scholar]
- 496. Bononi A., Giorgi C., Patergnani S., Larson D., Verbruggen K., Tanji M., Pellegrini L., Signorato V., Olivetto F., Pastorino S., Nasu M., Napolitano A., Gaudino G., Morris P., Sakamoto G., Ferris L. K., Danese A., Raimondi A., Tacchetti C., Kuchay S., Pass H. I., Affar E. B., Yang H., Pinton P., Carbone M., Nature 2017, 546, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497. Peng J., Ma J., Li W., Mo R., Zhang P., Gao K., Jin X., Xiao J., Wang C., Fan J., Cancer Lett. 2015, 369, 167. [DOI] [PubMed] [Google Scholar]
- 498. Zou H., Chen H., Zhou Z., Wan Y., Liu Z., Cancer Lett. 2019, 467, 19. [DOI] [PubMed] [Google Scholar]
- 499. Liu H., Li X., Ning G., Zhu S., Ma X., Liu X., Liu C., Huang M., Schmitt I., Wüllner U., Niu Y., Guo C., Wang Q., Tang T. S., PLoS Biol. 2016, 14, e2000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500. Ge F., Chen W., Qin J., Zhou Z., Liu R., Liu L., Tan J., Zou T., Li H., Ren G., Chen C., OncoTargets Ther. 2015, 6, 21369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501. Zhang S., Hong Z., Chai Y., Liu Z., Du Y., Li Q., Liu Q., Biochem. Biophys. Res. Commun. 2017, 488, 101. [DOI] [PubMed] [Google Scholar]
- 502. Huang M., Xiong H., Luo D., Xu B., Liu H., Exp. Cell Res. 2020, 388, 111876. [DOI] [PubMed] [Google Scholar]
- 503. Zhou F., Pan Y., Wei Y., Zhang R., Bai G., Shen Q., Meng S., Le X. F., Andreeff M., Claret F. X., Clin. Cancer Res. 2017, 23, 4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504. Watanabe K., Yokoyama S., Kaneto N., Hori T., Iwakami Y., Kato S., Hayakawa Y., Sakurai H., Fukuoka J., Saiki I., OncoTargets Ther. 2018, 9, 20670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505. Li J., Li Y., Wang B., Ma Y., Chen P., Biochem. Biophys. Res. Commun. 2018, 500, 132. [DOI] [PubMed] [Google Scholar]
- 506. Mao Z., Sang M. M., Chen C., Zhu W. T., Gong Y. S., Pei D. S., Int. J. Biol. Sci. 2019, 15, 1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507. Hou J., Deng Q., Zhou J., Zou J., Zhang Y., Tan P., Zhang W., Cui H., Oncogene 2017, 36, 1134. [DOI] [PubMed] [Google Scholar]
- 508. Su L., Guo W., Lou L., Nie S., Zhang Q., Liu Y., Chang Y., Zhang X., Li Y., Shen H., Mol. Carcinog. 2020, 59, 520. [DOI] [PubMed] [Google Scholar]
- 509. Iwakami Y., Yokoyama S., Watanabe K., Hayakawa Y., Biochem. Biophys. Res. Commun. 2018, 507, 484. [DOI] [PubMed] [Google Scholar]
- 510. Yan K., Li L., Wang X., Hong R., Zhang Y., Yang H., Lin M., Zhang S., He Q., Zheng D., Tang J., Yin Y., Shao G., J. Cell Biol. 2015, 210, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511. Zhu P., Zhou W., Wang J., Puc J., Ohgi K. A., Erdjument‐Bromage H., Tempst P., Glass C. K., Rosenfeld M. G., Mol. Cell 2007, 27, 609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512. Murai J., Yang K., Dejsuphong D., Hirota K., Takeda S., D'Andrea A. D., Mol Cell Biol 2011, 31, 2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513. Qian J., Pentz K., Zhu Q., Wang Q., He J., Srivastava A. K., Wani A. A., Oncogene 2015, 34, 4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514. Zlatanou A., Sabbioneda S., Miller E. S., Greenwalt A., Aggathanggelou A., Maurice M. M., Lehmann A. R., Stankovic T., Reverdy C., Colland F., Vaziri C., Stewart G. S., Oncogene 2016, 35, 965. [DOI] [PubMed] [Google Scholar]
- 515. Zhu Q., Sharma N., He J., Wani G., Wani A. A., Cell Cycle 2015, 14, 1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516. An L., Jiang Y., Ng H. H., Man E. P., Chen J., Khoo U. S., Gong Q., Huen M. S., Proc. Natl. Acad. Sci. USA 2017, 114, E2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517. Schwertman P., Lagarou A., Dekkers D. H., Raams A., van der Hoek A. C., Laffeber C., Hoeijmakers J. H., Demmers J. A., Fousteri M., Vermeulen W., Marteijn J. A., Nat. Genet. 2012, 44, 598. [DOI] [PubMed] [Google Scholar]
- 518. He J., Zhu Q., Wani G., Sharma N., Han C., Qian J., Pentz K., Wang Q. E., Wani A. A., J. Biol. Chem. 2014, 289, 27278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519. Khoronenkova S. V., Dianov G. L., Nucleic Acids Res. 2013, 41, 1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520. Mattoo A. R., Pandita R. K., Chakraborty S., Charaka V., Mujoo K., Hunt C. R., Pandita T. K., Mol. Cell Biol. 2017, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521. Yu M., Liu K., Mao Z., Luo J., Gu W., Zhao W., J. Biol. Chem. 2016, 291, 959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522. Zhang L., Lubin A., Chen H., Sun Z., Gong F., Cell Cycle 2012, 11, 4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523. Sy S. M., Jiang J., Sum O W., Deng Y., Huen M. S., Nucleic Acids Res. 2013, 41, 8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524. Perez‐Oliva A. B., Lachaud C., Szyniarowski P., Muñoz I., Macartney T., Hickson I., Rouse J., Alessi D. R., EMBO J. 2015, 34, 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525. Parsons J. L., Dianova I. I., Khoronenkova S. V., Edelmann M. J., Kessler B. M., Dianov G. L., Mol. Cell 2011, 41, 609. [DOI] [PubMed] [Google Scholar]
- 526. Yu H., Pak H., Hammond‐Martel I., Ghram M., Rodrigue A., Daou S., Barbour H., Corbeil L., Hébert J., Drobetsky E., Masson J. Y., Di Noia J. M., Affar el B., Proc. Natl. Acad. Sci. USA 2014, 111, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 527. Yao T., Song L., Jin J., Cai Y., Takahashi H., Swanson S. K., Washburn M. P., Florens L., Conaway R. C., Cohen R. E., Conaway J. W., Mol. Cell 2008, 31, 909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528. Kato K., Nakajima K., Ui A., Muto‐Terao Y., Ogiwara H., Nakada S., Mol. Cell 2014, 53, 617. [DOI] [PubMed] [Google Scholar]
- 529. Ng H. M., Wei L., Lan L., Huen M. S., J. Biol. Chem. 2016, 291, 16197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530. Wang W., Huang X., Xin H. B., Fu M., Xue A., Wu Z. H., J. Biol. Chem. 2015, 290, 13372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531. Niu J., Shi Y., Xue J., Miao R., Huang S., Wang T., Wu J., Fu M., Wu Z. H., EMBO J. 2013, 32, 3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532. Wu X., Lei C., Xia T., Zhong X., Yang Q., Shu H. B., Nat. Commun. 2019, 10, 4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533. Li S., Zheng H., Mao A. P., Zhong B., Li Y., Liu Y., Gao Y., Ran Y., Tien P., Shu H. B., J. Biol. Chem. 2010, 285, 4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534. Zhang L., Liu J., Qian L., Feng Q., Wang X., Yuan Y., Zuo Y., Cheng Q., Miao Y., Guo T., Zheng X., Zheng H., PLoS Pathog. 2018, 14, e1007067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535. Panda S., Nilsson J. A., Gekara N. O., Immunity 2015, 43, 647. [DOI] [PubMed] [Google Scholar]
- 536. Zilberman‐Rudenko J., Shawver L. M., Wessel A. W., Luo Y., Pelletier M., Tsai W. L., Lee Y., Vonortas S., Cheng L., Ashwell J. D., Orange J. S., Siegel R. M., Hanson E. P., Proc. Natl. Acad. Sci. USA 2016, 113, 1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537. Xu C., Peng Y., Zhang Q., Xu X. P., Kong X. M., Shi W. F., Sci. Rep. 2018, 8, 13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538. Liu C., Huang S., Wang X., Wen M., Zheng J., Wang W., Fu Y., Tian S., Li L., Li Z., Wang X., J. Immunol. 2019, 202, 2957. [DOI] [PubMed] [Google Scholar]
- 539. Zhang Z., Fang X., Wu X., Ling L., Chu F., Li J., Wang S., Zang J., Zhang B., Ye S., Zhang L., Yang B., Lin S., Huang H., Wang A., Zhou F., Mol. Cell 2020, 79, 304. [DOI] [PubMed] [Google Scholar]
- 540. Yu Z., Song H., Jia M., Zhang J., Wang W., Li Q., Zhang L., Zhao W., J. Exp. Med. 2017, 214, 3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541. Zhang L., Zhao X., Zhang M., Zhao W., Gao C., J. Immunol. 2014, 193, 2230. [DOI] [PubMed] [Google Scholar]
- 542. Cai J., Chen H. Y., Peng S. J., Meng J. L., Wang Y., Zhou Y., Qian X. P., Sun X. Y., Pang X. W., Zhang Y., Zhang J., FASEB J. 2018, 32, 5238. [DOI] [PubMed] [Google Scholar]
- 543. Colleran A., Collins P. E., O'Carroll C., Ahmed A., Mao X., McManus B., Kiely P. A., Burstein E., Carmody R. J., Proc. Natl. Acad. Sci. USA 2013, 110, 618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544. Cai Z., Zhang M. X., Tang Z., Zhang Q., Ye J., Xiong T. C., Zhang Z. D., Zhong B., J. Exp. Med. 2020, 217, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545. Zhang Q., Tang Z., An R., Ye L., Zhong B., Cell Res. 2020, 30, 914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546. Lin M., Zhao Z., Yang Z., Meng Q., Tan P., Xie W., Qin Y., Wang R. F., Cui J., Mol. Cell 2016, 64, 267. [DOI] [PubMed] [Google Scholar]
- 547. Parvatiyar K., Barber G. N., Harhaj E. W., J. Biol. Chem. 2010, 285, 14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548. Lu D., Song J., Sun Y., Qi F., Liu L., Jin Y., McNutt M. A., Yin Y., J Autoimmun 2018, 94, 156. [DOI] [PubMed] [Google Scholar]
- 549. Zhang Z., Wang D., Wang P., Zhao Y., You F., J. Immunol. 2020, 204, 1904. [DOI] [PubMed] [Google Scholar]
- 550. Lim K. H., Ramakrishna S., Baek K. H., Cytokine Growth Factor Rev. 2013, 24, 427. [DOI] [PubMed] [Google Scholar]
- 551. Ni Y., Tao L., Chen C., Song H., Li Z., Gao Y., Nie J., Piccioni M., Shi G., Li B., Int. J. Mol. Sci. 2015, 16, 27956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552. Han L., Yang J., Wang X., Wu Q., Yin S., Li Z., Zhang J., Xing Y., Chen Z., Tsun A., Li D., Piccioni M., Zhang Y., Guo Q., Jiang L., Bao L., Lv L., Li B., J. Biol. Chem. 2014, 289, 25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553. Ovaa H., Kessler B. M., Rolén U., Galardy P. J., Ploegh H. L., Masucci M. G., Proc. Natl. Acad. Sci. USA 2004, 101, 2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554. Sun L., Xing Y., Chen X., Zheng Y., Yang Y., Nichols D. B., Clementz M. A., Banach B. S., Li K., Baker S. C., Chen Z., PLoS One 2012, 7, e30802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555. Wang G., Chen G., Zheng D., Cheng G., Tang H., PLoS One 2011, 6, e17192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556. Zheng D., Chen G., Guo B., Cheng G., Tang H., Cell Res. 2008, 18, 1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557. Xing Y., Chen J., Tu J., Zhang B., Chen X., Shi H., Baker S. C., Feng L., Chen Z., J. Gen. Virol. 2013, 94, 1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558. Chen X., Yang X., Zheng Y., Yang Y., Xing Y., Chen Z., Protein Cell 2014, 5, 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559. Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A. R., Tascher G., Geurink P. P., Wilhelm A., van der Heden van Noort G. J., Ovaa H., Müller S., Knobeloch K.‐P., Rajalingam K., Schulman B. A., Cinatl J., Hummer G., Ciesek S., Dikic I., Nature 2020, 587, 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560. Liu G., Lee J. H., Parker Z. M., Acharya D., Chiang J. J., van Gent M., Riedl W., Davis‐Gardner M. E., Wies E., Chiang C., Gack M. U., Nat. Microbiol. 2021, 6, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561. Mielech A. M., Kilianski A., Baez‐Santos Y. M., Mesecar A. D., Baker S. C., Virology 2014, 450–451, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562. Yang X., Chen X., Bian G., Tu J., Xing Y., Wang Y., Chen Z., J. Gen. Virol. 2014, 95, 614. [DOI] [PubMed] [Google Scholar]
- 563. Sun Z., Chen Z., Lawson S. R., Fang Y., J. Virol. 2010, 84, 7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564. Bakshi S., Holzer B., Bridgen A., McMullan G., Quinn D. G., Baron M. D., J. Gen. Virol. 2013, 94, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565. Wang D., Fang L., Li P., Sun L., Fan J., Zhang Q., Luo R., Liu X., Li K., Chen H., Chen Z., Xiao S., J. Virol. 2011, 85, 3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566. Wang S., Wang K., Li J., Zheng C., J. Virol. 2013, 87, 11851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567. Ye R., Su C., Xu H., Zheng C., J. Virol. 2017, 91, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568. Fletcher‐Etherington A., Nobre L., Nightingale K., Antrobus R., Nichols J., Davison A. J., Stanton R. J., Weekes M. P., Proc. Natl. Acad. Sci. USA 2020, 117, 18771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569. Whitehurst C. B., Vaziri C., Shackelford J., Pagano J. S., J. Virol. 2012, 86, 8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570. Ylä‐Anttila P., Gupta S., Masucci M. G., Autophagy 2021, 17, 3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571. Gastaldello S., Hildebrand S., Faridani O., Callegari S., Palmkvist M., Di Guglielmo C., Masucci M. G., Nat. Cell Biol. 2010, 12, 351. [DOI] [PubMed] [Google Scholar]
- 572. Saito S., Murata T., Kanda T., Isomura H., Narita Y., Sugimoto A., Kawashima D., Tsurumi T., J. Virol. 2013, 87, 4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573. Inn K. S., Lee S. H., Rathbun J. Y., Wong L. Y., Toth Z., Machida K., Ou J. H., Jung J. U., J. Virol. 2011, 85, 10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574. Mutz P., Metz P., Lempp F. A., Bender S., Qu B., Schöneweis K., Seitz S., Tu T., Restuccia A., Frankish J., Dächert C., Schusser B., Koschny R., Polychronidis G., Schemmer P., Hoffmann K., Baumert T. F., Binder M., Urban S., Bartenschlager R., Gastroenterology 2018, 154, 1791. [DOI] [PubMed] [Google Scholar]
- 575. Jiang J., Tang H., Protein Cell 2010, 1, 1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576. Mistry H., Hsieh G., Buhrlage S. J., Huang M., Park E., Cuny G. D., Galinsky I., Stone R. M., Gray N. S., D'Andrea A. D., Parmar K., Mol. Cancer Ther. 2013, 12, 2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577. Das D. S., Das A., Ray A., Song Y., Samur M. K., Munshi N. C., Chauhan D., Anderson K. C., Clin. Cancer Res. 2017, 23, 4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578. Davis M. I., Pragani R., Fox J. T., Shen M., Parmar K., Gaudiano E. F., Liu L., Tanega C., McGee L., Hall M. D., McKnight C., Shinn P., Nelson H., Chattopadhyay D., D'Andrea A. D., Auld D. S., DeLucas L. J., Li Z., Boxer M. B., Simeonov A., J. Biol. Chem. 2016, 291, 24628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579. Magiera K., Tomala M., Kubica K., De Cesare V., Trost M., Zieba B. J., Kachamakova‐Trojanowska N., Les M., Dubin G., Holak T. A., Skalniak L., Cell Chem. Biol. 2017, 24, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580. Chuang S. J., Cheng S. C., Tang H. C., Sun C. Y., Chou C. Y., Sci. Rep. 2018, 8, 3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581. Okada K., Ye Y. Q., Taniguchi K., Yoshida A., Akiyama T., Yoshioka Y., Onose J., Koshino H., Takahashi S., Yajima A., Abe N., Yajima S., Bioorg. Med. Chem. Lett. 2013, 23, 4328. [DOI] [PubMed] [Google Scholar]
- 582. Kapuria V., Peterson L. F., Fang D., Bornmann W. G., Talpaz M., Donato N. J., Cancer Res. 2010, 70, 9265. [DOI] [PubMed] [Google Scholar]
- 583. Kategaya L., Di Lello P., Rougé L., Pastor R., Clark K. R., Drummond J., Kleinheinz T., Lin E., Upton J. P., Prakash S., Heideker J., McCleland M., Ritorto M. S., Alessi D. R., Trost M., Bainbridge T. W., Kwok M. C. M., Ma T. P., Stiffler Z., Brasher B., Tang Y., Jaishankar P., Hearn B. R., Renslo A. R., Arkin M. R., Cohen F., Yu K., Peale F., Gnad F., Chang M. T., et al., Nature 2017, 550, 534. [DOI] [PubMed] [Google Scholar]
- 584. Weinstock J., Wu J., Cao P., Kingsbury W. D., McDermott J. L., Kodrasov M. P., McKelvey D. M., Suresh Kumar K. G., Goldenberg S. J., Mattern M. R., Nicholson B., ACS Med. Chem. Lett. 2012, 3, 789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585. Gavory G., O'Dowd C. R., Helm M. D., Flasz J., Arkoudis E., Dossang A., Hughes C., Cassidy E., McClelland K., Odrzywol E., Page N., Barker O., Miel H., Harrison T., Nat. Chem. Biol. 2018, 14, 118. [DOI] [PubMed] [Google Scholar]
- 586. Reverdy C., Conrath S., Lopez R., Planquette C., Atmanene C., Collura V., Harpon J., Battaglia V., Vivat V., Sippl W., Colland F., Chem. Biol. 2012, 19, 467. [DOI] [PubMed] [Google Scholar]
- 587. Colland F., Formstecher E., Jacq X., Reverdy C., Planquette C., Conrath S., Trouplin V., Bianchi J., Aushev V. N., Camonis J., Calabrese A., Borg‐Capra C., Sippl W., Collura V., Boissy G., Rain J. C., Guedat P., Delansorne R., Daviet L., Mol. Cancer Ther. 2009, 8, 2286. [DOI] [PubMed] [Google Scholar]
- 588. Lamberto I., Liu X., Seo H. S., Schauer N. J., Iacob R. E., Hu W., Das D., Mikhailova T., Weisberg E. L., Engen J. R., Anderson K. C., Chauhan D., Dhe‐Paganon S., Buhrlage S. J., Cell Chem. Biol. 2017, 24, 1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589. Di Lello P., Pastor R., Murray J. M., Blake R. A., Cohen F., Crawford T. D., Drobnick J., Drummond J., Kategaya L., Kleinheinz T., Maurer T., Rougé L., Zhao X., Wertz I., Ndubaku C., Tsui V., J. Med. Chem. 2017, 60, 10056. [DOI] [PubMed] [Google Scholar]
- 590. Jing B., Liu M., Yang L., Cai H. Y., Chen J. B., Li Z. X., Kou X., Wu Y. Z., Qin D. J., Zhou L., Jin J., Lei H., Xu H. Z., Wang W. W., Wu Y. L., Acta Pharmacol. Sin. 2018, 39, 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591. Kageyama K., Asari Y., Sugimoto Y., Niioka K., Daimon M., Endocr. J. 2020, 67, 177. [DOI] [PubMed] [Google Scholar]
- 592. Byun S., Lee S. Y., Lee J., Jeong C. H., Farrand L., Lim S., Reddy K., Kim J. Y., Lee M. H., Lee H. J., Bode A. M., Lee K. Won, Dong Z., Clin. Cancer Res. 2013, 19, 3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 593. Liu J., Xia H., Kim M., Xu L., Li Y., Zhang L., Cai Y., Norberg H. V., Zhang T., Furuya T., Jin M., Zhu Z., Wang H., Yu J., Li Y., Hao Y., Choi A., Ke H., Ma D., Yuan J., Cell 2011, 147, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594. Moghadami A. A., Aboutalebi Vand Beilankouhi E., Kalantary‐Charvadeh A., Hamzavi M., Mosayyebi B., Sedghi H., Ghorbani Haghjo A., Nazari Soltan Ahmad S., Cell Stress Chaperones 2020, 25, 909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595. Boselli M., Lee B. H., Robert J., Prado M. A., Min S. W., Cheng C., Silva M. C., Seong C., Elsasser S., Hatle K. M., Gahman T. C., Gygi S. P., Haggarty S. J., Gan L., King R. W., Finley D., J. Biol. Chem. 2017, 292, 19209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596. Lee B. H., Lee M. J., Park S., Oh D. C., Elsasser S., Chen P. C., Gartner C., Dimova N., Hanna J., Gygi S. P., Wilson S. M., King R. W., Finley D., Nature 2010, 467, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597. Oh Y. T., Deng L., Deng J., Sun S. Y., Sci. Rep. 2017, 7, 8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598. Jiang L., Sun Y., Wang J., He Q., Chen X., Lan X., Chen J., Dou Q. P., Shi X., Liu J., J. Exp. Clin. Cancer Res. 2019, 38, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599. Wang X., Mazurkiewicz M., Hillert E. K., Olofsson M. H., Pierrou S., Hillertz P., Gullbo J., Selvaraju K., Paulus A., Akhtar S., Bossler F., Khan A. C., Linder S., D'Arcy P., Sci. Rep. 2016, 6, 26979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600. Liu N., Li X., Huang H., Zhao C., Liao S., Yang C., Liu S., Song W., Lu X., Lan X., Chen X., Yi S., Xu L., Jiang L., Zhao C., Dong X., Zhou P., Li S., Wang S., Shi X., Dou P. Q., Wang X., Liu J., OncoTargets Ther. 2014, 5, 5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601. Seitzberg J. G., Knapp A. E., Lund B. W., Mandrup Bertozzi S., Currier E. A., Ma J. N., Sherbukhin V., Burstein E. S., Olsson R., J. Med. Chem. 2008, 51, 5490. [DOI] [PubMed] [Google Scholar]
- 602. Wrigley J. D., Gavory G., Simpson I., Preston M., Plant H., Bradley J., Goeppert A. U., Rozycka E., Davies G., Walsh J., Valentine A., McClelland K., Odrzywol K. E., Renshaw J., Boros J., Tart J., Leach L., Nowak T., Ward R. A., Harrison T., Andrews D. M., ACS Chem. Biol. 2017, 12, 3113. [DOI] [PubMed] [Google Scholar]
- 603. Rusilowicz‐Jones E. V., Jardine J., Kallinos A., Pinto‐Fernandez A., Guenther F., Giurrandino M., Barone F. G., McCarron K., Burke C. J., Murad A., Martinez A., Marcassa E., Gersch M., Buckmelter A. J., Kayser‐Bricker K. J., Lamoliatte F., Gajbhiye A., Davis S., Scott H. C., Murphy E., England K., Mortiboys H., Komander D., Trost M., Kessler B. M., Ioannidis S., Ahlijanian M. K., Urbé S., Clague M. J., Life Sci. Alliance 2020, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604. Perez C., Li J., Parlati F., Rouffet M., Ma Y., Mackinnon A. L., Chou T. F., Deshaies R. J., Cohen S. M., J. Med. Chem. 2017, 60, 1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605. Li J., Yakushi T., Parlati F., Mackinnon A. L., Perez C., Ma Y., Carter K. P., Colayco S., Magnuson G., Brown B., Nguyen K., Vasile S., Suyama E., Smith L. H., Sergienko E., Pinkerton A. B., Chung T. D. Y., Palmer A. E., Pass I., Hess S., Cohen S. M., Deshaies R. J., Nat. Chem. Biol. 2017, 13, 486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606. Song Y., Li S., Ray A., Das D. S., Qi J., Samur M. K., Tai Y. T., Munshi N., Carrasco R. D., Chauhan D., Anderson K. C., Oncogene 2017, 36, 5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607. Liu Y., Lashuel H. A., Choi S., Xing X., Case A., Ni J., Yeh L. A., Cuny G. D., Stein R. L., P. T. Lansbury, Jr. , Chem. Biol. 2003, 10, 837. [DOI] [PubMed] [Google Scholar]
- 608. Grethe C., Schmidt M., Kipka G. M., O'Dea R., Gallant K., Janning P., Gersch M., Nat. Commun. 2022, 13, 5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609. Pan T., Hua M. H., Lv N., Clin. Exp. Rheumatol. 2022, 40, 1718. [DOI] [PubMed] [Google Scholar]
- 610. Zhang P., Xiao Z., Wang S., Zhang M., Wei Y., Hang Q., Kim J., Yao F., Rodriguez‐Aguayo C., Ton B. N., Lee M., Wang Y., Zhou Z., Zeng L., Hu X., Lawhon S. E., Siverly A. N., Su X., Li J., Xie X., Cheng X., Liu L. C., Chang H. W., Chiang S. F., Lopez‐Berestein G., Sood A. K., Chen J., You M. J., Sun S. C., Liang H., et al., Cell Rep. 2018, 23, 823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611. Yamanaka S., Sato Y., Oikawa D., Goto E., Fukai S., Tokunaga F., Takahashi H., Sawasaki T., Biochem. Biophys. Res. Commun. 2020, 524, 1. [DOI] [PubMed] [Google Scholar]
- 612. Vijay‐Kumar S., Bugg C. E., Cook W. J., J. Mol. Biol. 1987, 194, 531. [DOI] [PubMed] [Google Scholar]


