Abstract
A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) is essential for the initiation of aerial mycelium formation in Streptomyces griseus. amfR is one of the genes which, when cloned on a low-copy-number plasmid, suppresses the aerial mycelium-negative phenotype of an A-factor-deficient mutant of S. griseus. Disruption of the chromosomal amfR gene resulted in complete abolition of aerial mycelium formation, indicating that amfR is essential for the onset of morphogenesis. Cloning and nucleotide sequencing of the region upstream of amfR predicted an operon consisting of orf5, orf4, and amfR. Consistent with this idea, Northern blotting and S1 mapping analyses suggested that these three genes were cotranscribed mainly by a promoter (PORF5) in front of orf5. Furthermore, PORF5 was active only in the presence of A-factor, indicating that it is A-factor dependent. Gel mobility shift assays showed the presence of a protein (AdpB) able to bind PORF5 in the cell extract from an A-factor-deficient mutant but not from the wild-type strain. AdpB was purified to homogeneity and found to bind specifically to the region from −72 to −44 bp with respect to the transcriptional start point. Runoff transcriptional analysis of PORF5 with purified AdpB and an RNA polymerase complex isolated from vegetative mycelium showed that AdpB repressed the transcription in a concentration-dependent manner. It is thus apparent that AmfR as a switch for aerial mycelium formation and AdpB as a repressor for amfR are members in the A-factor regulatory cascade, leading to morphogenesis.
The gram-positive bacterial genus Streptomyces shows characteristic morphological differentiation, which is a useful target for the genetic analysis of procaryotic multicellular differentiation (5–7). On solid media, Streptomyces spp. form branched, multinucleoid substrate hyphae during vegetative growth. In response to nutritional limitation, older parts of the substrate hyphae produce aerial mycelium. After septa have been formed at regular intervals along the hyphae, long chains of uninucleoid spores are formed.
Our approach to studying morphological differentiation in Streptomyces spp. has been focused on a hormonal substance, A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone), which is essential for the initiation of aerial mycelium formation and streptomycin production in Streptomyces griseus (9, 18). An A-factor-deficient mutant of S. griseus shows a bald (Bld) phenotype, deficient in both aerial mycelium and spore formation, and exogenous supply of A-factor at an extremely low concentration to this strain induces aerial mycelium and spore formation (13, 14). Taking advantage of the Bld phenotype caused by A-factor deficiency, we have cloned genes that cause aerial mycelium and spore formation by using this mutant strain as a host (12). This strategy brought forth three sets of genes, amfR, amfA, and amfB (30); orf1590 (1); and amfC (20). Among these, orf1590 was also shown by Babcock and Kendrick (1, 2) to complement a Bld phenotype of one class of mutants of S. griseus. amfR encodes a protein similar to the response regulators of two-component regulatory systems, and the amfA and amfB products are members of a family of bacterial membrane translocators. Ma and Kendall (21) cloned an amfRAB homologous locus, named ram, from Streptomyces coelicolor A3(2), as a gene cluster that caused accelerated aerial mycelium formation in Streptomyces lividans.
In this study we show that amfR is essential for aerial growth. Identification of a protein (AdpB) able to bind the promoter of the amfR operon only in the absence of A-factor and repressor-like behavior of AdpB observed in runoff transcription experiments strongly suggest that AmfR and AdpB are important members in the A-factor regulatory cascade, leading to aerial mycelium formation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. griseus IFO 13350, obtained from the Institute of Fermentation, Osaka, Japan, was the parental strain of an A-factor-deficient mutant strain, HH1 (16). This mutant strain, obtained by incubation at 37°C, was defective in aerial mycelium formation and streptomycin production because of its A-factor deficiency. Plasmid pIJ486 (carrying thiostrepton resistance) (32) has a copy number of 40 to 100 per genome. Plasmids pIJ922 (11) and pTMA1 (31) (both carrying thiostrepton resistance) have a copy number of 1 to 2 per genome, as judged by agarose gel electrophoresis (30). pKU206 (carrying thiostrepton resistance), an SCP2*-derived plasmid, was obtained from H. Ikeda (17). DNA was manipulated in Escherichia coli JM109 [Δ(lac-pro) thi-1 endA1 gyrA96 hsdR17 relA1 recA1/F′ traD36 proAB lacIq lacZΔM15] (33) by cloning in pUC18 and pUC19 (33). S. griseus strains were grown in Bennett-glucose medium (containing the following [grams per liter]): yeast extract [Difco Laboratory], 1; meat extract [Kyokuto Co.], 1; N. Z. amine [Wako Pure Chemical], 2; and glucose, 10, (pH 7.2), YMPG medium (9), and nutrient agar medium (Difco Laboratory). Growth conditions for E. coli strains were as described by Maniatis et al. (22).
General recombinant DNA techniques.
Restriction endonucleases and other DNA-modifying enzymes were purchased from Takara Shuzo Co., Kyoto, Japan. [α-32P]dCTP at 110 TBq/mmol for nucleotide sequencing by the M13-dideoxynucleotide method with M13mp18 and M13mp19 (33) and for the Takara multiprime DNA labeling system and [γ-32P]ATP at >220 TBq/mmol for 5′-end labeling of probe DNA for primer extension and gel mobility shift assays were purchased from Amersham International. Thiostrepton was a gift from Asahi Chemical Industry, Shizuoka, Japan. DNA manipulations in E. coli, Southern hybridization, and colony hybridization were as described by Maniatis et al. (22), and those in Streptomyces spp. were as described by Hopwood et al. (11).
Cloning and subcloning of the region upstream from amfR.
The previously cloned 9-kb Sau3AI fragment on pSPO1 was inserted at the BamHI site of pIJ487 (30). The SmaI site of pUC19 was first changed into an NcoI site by use of an 8-mer NcoI linker (pUC-Nco), and the 1.6-kb BamHI-NcoI fragment was cloned between the BamHI and NcoI sites of pUC-Nco to generate plasmid pUC-A. The 1.6-kb BamHI-NcoI fragment from pUC-A was used as a 32P-labeled hybridization probe, and an approximately 6.5-kb NcoI fragment giving a positive signal by Southern hybridization against the chromosomal DNA from S. griseus IFO 13350 was detected. The 6.5-kb NcoI fragment was cloned by colony hybridization at the NcoI site of pUC-Nco to generate pUC-F. The nucleotide sequence of the 2-kb region upstream from amfR was determined and combined with the previous sequence data.
The 3.5-kb EcoRI-NcoI fragment containing orf5, orf4, and amfR (the orf5-orf4-amfR operon) (see Fig. 1) was recovered as an EcoRI fragment from pUC-F and cloned at the EcoRI site of pTMA1 to generate pAFL1. For construction of pAFL1Δ, the EcoRI fragment from pUC-F was first cloned onto pUC19. This plasmid was cleaved by PmaCI, and 8-mer BglII linkers were attached to the ends. After digestion with BglII, it was religated to form a circular DNA. The DNA fragment with an internal deletion of the 384-bp PmaCI fragment was recovered as an EcoRI fragment and finally cloned onto pTMA1. For construction of pAFL2, the 3.5-kb EcoRI fragment from pUC-F was partially digested with Eco47III and the 1.9-kb EcoRI-Eco47III fragment was cloned between the EcoRI and SmaI sites of pUC19. The fragment was recovered as an EcoRI-HindIII fragment and inserted into pTMA1. For construction of pSL6, the 1.6-kb BamHI-NcoI fragment was recovered as a BamHI-EcoRI fragment from pUC-A and inserted between the BamHI and EcoRI sites of pTMA1. For construction of pSH11 and pAFL11, the 316-bp EcoRI-Eco47III fragment carrying the promoter region in front of ORF5 was first cloned between the EcoRI and SmaI sites of pUC19, then recovered as an EcoRI-HindIII fragment, and inserted into pIJ487 and pTMA1, respectively.
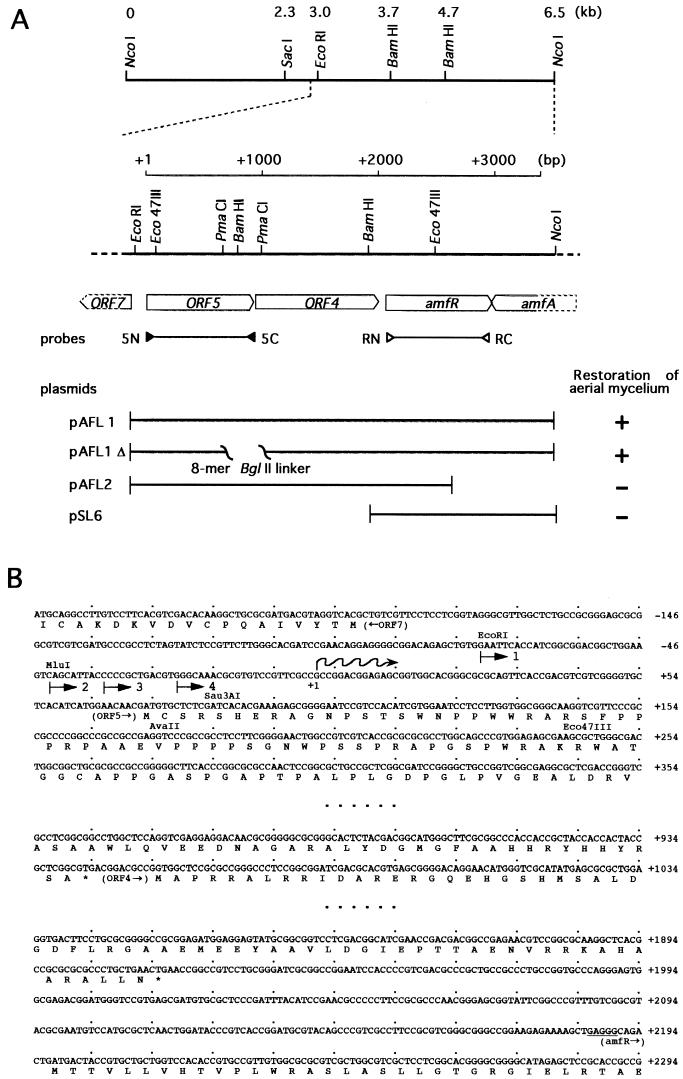
FIG. 1.
Restriction map of the 6.5-kb region containing orf5-orf4-amfR-amfA and nucleotide sequences of part of this region. (A) The directions and extents of the ORFs deduced from the nucleotide sequence are shown just below the restriction map. The positions of a pair of the primers used for probes for Northern hybridization (Fig. 3) are indicated. The fragments on a low-copy-number plasmid and their ability to restore aerial mycelium formation in S. griseus HH1 are shown. (B) The nucleotide sequences of the region including the promoter in front of orf5, the termination codon of orf5 and the initiation codon of orf4, and the termination codon of orf4 and the initiation codon of amfR. Probable ribosome-binding sites for orf5 and amfR are underlined. The transcriptional start point is indicated as +1. Numbers with an arrow indicate the probes used for the gel mobility shift assay (Fig. 4).
The transformants were viewed microscopically as well as macroscopically to determine whether they produced aerial mycelium and spores.
Gene disruption.
The chromosomal amfR gene was disrupted according to the procedure used for amfC disruption (19). The 1.6-kb BamHI-NcoI region carrying the entire amfR gene was first cloned onto pUC19 as a BamHI fragment after the NcoI site had been changed into a BamHI site with an 8-mer BamHI linker. The unique Eco47III site in the middle of the amfR-coding region was changed into a BglII site with an 8-mer BglII linker, and the 1.7-kb BamHI fragment containing the kanamycin resistance (aphII) gene (3) was inserted into the newly created BglII cleavage site to construct a mutagenized amfR gene. The mutagenized amfR sequence with aphII was excised with BamHI as a 3.3-kb fragment and ligated into the BamHI site in pKU206 to construct plasmid pRD1. pKU206 is known to be unstable in S. griseus in the absence of thiostrepton used as a selection marker. pRD1 was introduced by transformation into the wild-type strain, S. griseus IFO 13350. Thiostrepton (20 μg/ml)- and kanamycin (20 μg/ml)-resistant transformants were then cultured at 30°C for 72 h in YMPG liquid medium without thiostrepton and plated onto YMPG agar plates containing 20 μg of kanamycin per ml. Kanamycin-resistant colonies thus obtained were then checked for their sensitivity to thiostrepton, and finally four colonies showing thiostrepton sensitivity and kanamycin resistance were obtained. All four colonies showed a Bld phenotype on YMPG agar plates, and correct disruption of amfR in one of the colonies was confirmed by Southern blot hybridization with the 1.6-kb BamHI-NcoI fragment carrying the intact amfR gene and the 1.7-kb BamHI fragment containing aphII as probes.
Northern blot analysis.
S. griseus IFO 13350 and HH1 were grown at 30°C for 24 to 60 h in YMPG liquid medium. Strain HH1 was grown with or without exogenous A-factor at a final concentration of 50 ng/ml. Total cellular RNA was isolated as described previously (15) and quantified by the absorbance at 260 nm. Northern hybridization was carried out by the formaldehyde method according to Maniatis et al. (22). Fifteen micrograms of RNA was loaded onto each lane, electrophoresed on a 1% agarose gel, and transferred to a Hybond-N+ membrane (Amersham) according to the manufacturer’s recommendation. Hybridization was performed at 65°C for 18 h, and the blot was washed three times with 2× SSC (1× SSC contains 0.15 M sodium citrate and 0.15 M NaCl, pH 8.0) containing 0.1% sodium dodecyl sulfate (SDS) at 65°C for 10 min. The probes were labeled with [α-32P]dCTP by the BcaBEST random primer DNA labelling kit (Takara Shuzo) according to the protocol supplied. The probes were an 890-bp fragment containing the entire orf5 sequence and a 613-bp fragment containing the amfR-coding sequence, both of which were amplified by the standard PCR method with the following oligonucleotide primers (see Fig. 1): 5′-GCCGAATTCGGAACAACGATGTGC (5N) and 5′-CGGCTCGAGCGCCGAGCGGTAGTGG (5C) for orf5 and 5′-GCCGAATTCGACTACCGTGCTGC (RN) and 5′-CGGCTCGAGGATCCAGCCCGCTTCC (RC) for amfR.
S1 mapping.
The 320-bp EcoRI-Eco47III fragment (see Fig. 1) with 32P at the Eco47III end was prepared by standard DNA manipulation and used as a hybridization probe. For clearer resolution of the end of the protected fragment, another 32P-labeled fragment used as a probe was prepared as follows. An 18-mer oligonucleotide, 5′-GATTCCCCGCTCTTTCGT-3′ (nucleotide positions +106 to +89), was labeled with [γ-32P]ATP with T4 polynucleotide kinase. For preparation of a 32P-labeled 130-bp probe, PCR was carried out with this oligonucleotide and a nonlabeled 21-mer oligonucleotide, 5′-GTGGAATTCACCATCGGCGGA-3′ (nucleotide positions −75 to −55), under standard conditions. RNA was prepared from S. griseus IFO 13350 and strain HH1 grown at 30°C for 2 days in YMPG medium. RNA (100 μg) was hybridized with 100,000 Cerenkov cpm of the probe, as described previously (15). Maxam-Gilbert sequencing ladders on a 12% polyacrylamide sequencing gel (23) were generated with the same 32P-labeled fragment.
Fractionation of cell extracts by DEAE-Toyopearl column chromatography for gel mobility shift assays.
S. griseus HH1 was cultured in 100 ml of YMPG medium at 30°C for 48 h. The mycelium was homogenized with a glass homogenizer and transferred to 1 liter of YMPG medium in a 5-liter flask. After cultivation at 30°C for 48 h, the mycelium was harvested by centrifugation. The mycelium was suspended in 100 ml of buffer A (containing 10 mM Tris-HCl [pH 7.0], 1 mM EDTA, 1 mM dithiothreitol, 10% [vol/vol] glycerol) and disrupted by sonication. The supernatant obtained by centrifugation of the disrupted mycelium at 15,000 × g for 30 min at 4°C was applied to a DEAE-Toyopearl open column (4.6 by 15 cm; Tosoh Corp., Tokyo, Japan) previously equilibrated with buffer A. After the column had been washed with 750 ml of buffer A, proteins were eluted with a linear KCl gradient of 0 to 1 M in a total volume of 200 ml at a flow rate of 2.5 ml/min. The cell extract from strain HH1 grown in the presence of 50 ng of A-factor per ml was similarly fractionated.
Gel mobility shift assay.
DNA-binding determinations by mobility shift assays were done essentially by the method of Chodosh (8). For the standard binding assay, 0.5 to 5 ng of 32P-labeled double-stranded DNA (10,000 to 20,000 cpm) was incubated with 5 to 20 μg of proteins at 30°C for 30 min in binding buffer containing 10 mM Tris-HCl (pH 7.0), 50 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, 10% (vol/vol) glycerol, 1 μg of poly(dI-dC)-poly(dI-dC), and 50 μg of bovine serum albumin per ml in a total volume of 50 μl. After incubation, complexes and free DNA were resolved on nondenaturing polyacrylamide gel containing 6% acrylamide and 0.075% bisacrylamide with running buffer containing 40 mM Tris-HCl (pH 8.0), 20 mM sodium acetate, and 1 mM EDTA. Gels were dried and subjected to autoradiography with a Du Pont Cronex intensifying screen.
Purification of the DNA-binding protein.
S. griseus HH1 was used as a source of the DNA-binding protein. SDS-polyacrylamide gel electrophoresis (PAGE) on slab gels was used to monitor protein purification and to estimate molecular sizes under denaturing conditions. The concentrations of polyacrylamide were 12.5% in separating gels and 4% in stacking gels. The gels were stained with 0.1% Coomassie brilliant blue R-250. During purification, protein concentrations were measured with a Bio-Rad protein assay kit with bovine serum albumin as the standard.
(i) Preparation of cell extract.
S. griseus HH1 was cultured in 100 ml of YMPG medium at 30°C for 48 h. The mycelium was homogenized with a glass homogenizer and transferred to 15 liters of the same medium in a 30-liter jar fermentor. After cultivation at 30°C for 48 h, the mycelium was harvested by centrifugation. Mycelium (wet weight, 300 g) was suspended in 1 liter of buffer A containing 10 mM Tris-HCl (pH 7.0), 1 mM EDTA, 1 mM dithiothreitol, and 10% glycerol, and disrupted by three passages at 700 kg/cm2 through a Manton-Gaulin apparatus (model 15M8TA; Gaurin Corp., Everett, Mass.). The supernatant, obtained by centrifugation of the disrupted mycelium at 10,000 × g for 30 min at 4°C, was used as the cell extract. Proteins (10.2 g) were recovered through this step.
(ii) Ammonium sulfate fractionation.
Solid ammonium sulfate was added to the cell extract to 30% (wt/vol) saturation, and the mixture was gently stirred at 4°C overnight. The precipitate was obtained by centrifugation at 15,000 × g for 30 min, dissolved in 200 ml of buffer A, and dialyzed overnight against buffer A. Proteins (952 mg) were recovered through this step.
(iii) DEAE-Toyopearl column chromatography.
The dialyzed sample was applied to a DEAE-Toyopearl column (3.0 by 40 cm in diameter; Tosoh Corp.) previously equilibrated with buffer A. After the column had been washed with 1 liter of buffer A, proteins were eluted with a linear gradient of KCl from 0 to 1 M in a total volume of 3 liters of buffer A at a flow rate of 2.5 ml/min. Fractions (250 ml) containing DNA-binding activity were pooled and dialyzed overnight against buffer A. Proteins (99 mg) were recovered through this step.
(iv) MonoQ column chromatography.
The dialyzed sample was divided into five portions, each of which was applied to a MonoQ HR 10/10 fast protein liquid chromatography column (Pharmacia) equilibrated with buffer A, because of the small capacity of the column. Proteins were eluted with a linear gradient of 0 to 1 M KCl in buffer A in a total volume of 100 ml at a flow rate of 1 ml/min. One cycle of chromatography gave fractions (30 ml each) containing activity. These fractions, obtained from five cycles of chromatography, were collected and dialyzed overnight against buffer A. Proteins (31 mg) were recovered through this step.
(v) Heparin affinity column chromatography.
The dialyzed sample was divided into three portions, each of which was applied to a HiTrap heparin fast protein liquid chromatography column (Pharmacia) equilibrated with buffer A. Proteins were eluted with a linear gradient of 0 to 1 M KCl in buffer A in a total volume of 100 ml at a flow rate of 0.5 ml/min. One cycle of chromatography gave fractions (5 ml each) containing activity. These fractions, obtained from three cycles of chromatography, were collected and dialyzed overnight against buffer A. Proteins (5 mg) were recovered through this step.
(vi) Nondenaturing PAGE and protein elution.
The above-mentioned sample was concentrated to 2 ml by membrane filtration and separated by nondenaturing PAGE (native PAGE; containing 10 and 2.5% polyacrylamide in the separating and stacking gels, respectively). After identification of the DNA-binding protein by Southwestern blot analysis (see below), a gel piece containing the DNA-binding protein was cut out from each of several gels. The gel pieces were then homogenized with a Teflon homogenizer and suspended in 10 ml of buffer A. The suspension was gently stirred at 4°C for 3 h and centrifuged at 10,000 × g for 20 min to remove gel particles. This elution procedure was repeated three times. The eluates were collected and concentrated by membrane filtration to yield 1 ml of the purified protein solution. The amount of AdpB recovered was 1.2 mg.
Southwestern blotting.
Southwestern blotting assay was done essentially by the method of Miskimins et al. (24). The fraction (4 μg of protein) eluted from the heparin affinity column was divided in two and separated by native PAGE, described above. One lane was stained with Coomassie brilliant blue, and the proteins in the other lane were transferred to a nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany). The membrane was incubated at 30°C for 1 h with 0.1 μg of a 32P-labeled 314-bp EcoRI-Eco47III fragment in 20 ml of the binding buffer used for the gel mobility shift assay. The membrane was washed with the binding buffer containing 0.2 M KCl and subjected to autoradiography with a Du Pont Cronex intensifying screen.
Generation of trimmed fragments for retardation probes.
For generation of a deletion series from the 314-bp EcoRI-Eco47III fragment, the original 314-bp fragment was first cloned into pUC19 between the EcoRI and HincII sites and then cleaved with EcoRI. The EcoRI ends were digested with exonuclease III at 37°C for 10 to 60 s. The exonuclease III-treated fragments were blunt ended with mung bean nuclease and Klenow fragment, and an 8-mer EcoRI linker was attached to both ends. After EcoRI digestion, they were religated to form circular DNAs. Plasmids containing fragments with appropriate sizes were screened for by agarose gel electrophoresis, and the terminal ends of deletions were determined by nucleotide sequencing. The EcoRI-Sau3AI fragment was recovered from each of the plasmids, and both ends were 32P labeled with T4 polynucleotide kinase for gel retardation assays.
Run off transcription.
The in vitro transcriptional analysis was done according to the procedure described previously (28). An RNA polymerase complex mainly with ςB promoter specificity, prepared from exponentially growing S. griseus cells (28), was used. The EcoRI-Eco47III and EcoRI-AvaII fragments (see Fig. 7) were purified by agarose gel electrophoresis and used as the templates. A reaction mixture containing the RNA polymerase complex, the DNA template (0.5 pmol), [α-32P]UTP, nucleoside triphosphate mixture, heparin, and various amounts of AdpB was incubated at 30°C for 30 min to allow a single round of transcription. The transcripts were separated by 8 M urea-PAGE (28). The transcription products were detected by autoradiography. The EcoRI-Eco47III and EcoRI-AvaII fragments were expected to yield 244- and 177-bp runoff transcripts, respectively.
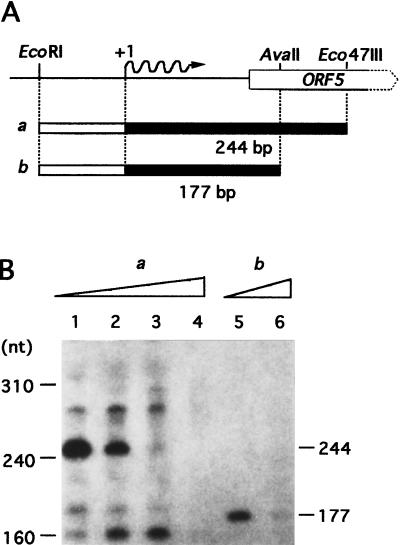
FIG. 7.
In vitro transcription from PORF5. (A) Physical map of the DNA fragments used as the templates for runoff transcription. The EcoRI-Eco47III and the EcoRI-AvaII fragments were 316 and 249 bp, respectively. The expected transcripts, 244 and 177 nucleotides (nt) are depicted as solid bars. (B) The transcripts obtained with the two templates (a in lanes 1 to 4 and b in lanes 5 and 6) in the absence of AdpB (lanes 1 and 5) and the presence of 0.1 μg (lane 2), 1 μg (lane 3), or 10 μg (lanes 4 and 6) of AdpB were analyzed by 8 M urea-PAGE.
Nucleotide sequence accession number.
The newly edited sequence of the 2-kb region upstream from amfR combined with previous sequence data has been registered in the DDBJ, EMBL, and GenBank databases under accession no. AB006206.
RESULTS
Cloning and nucleotide sequencing of a region upstream from amfR.
In order to clone the region upstream from amfR, we used the 1.6-kb NcoI-BamHI fragment containing the whole coding sequence of amfR (Fig. 1) as the 32P-labeled hybridization probe against NcoI-digested chromosomal DNA of S. griseus and detected a signal of 6.5 kb. The 6.5-kb NcoI fragment was cloned into pUC-Nco by the standard DNA manipulation, including colony hybridization. The restriction map of the cloned 6.5-kb fragment is shown in Fig. 1.
We determined the nucleotide sequence of the approximately 2-kb region upstream from amfR. Computer-aided FRAME analysis (4) on this region predicted two complete open reading frames (ORFs) (ORF4, with 319 amino acids, and ORF5, with 289 amino acids) in the same direction as AmfR and a truncated ORF (ORF7) in the opposite direction to that of AmfR (Fig. 1). In front of amfR, orf5, and orf4, possible ribosome-binding sequences, GAGGG, GGAA, and GGA, respectively, are present at appropriate positions. ORF7 is also preceded by a sequence, GGAGGAA. Neither ORF4 nor ORF5 showed any homology with proteins registered in the Swiss-Prot data bank. On the other hand, the truncated ORF7 represents the 7Fe ferredoxin of S. griseus (29), because the 56 amino acids of ORF7 deduced from the nucleotide sequence are identical to the NH2-terminal portion of the ferredoxin, consisting of 105 amino acids.
Disruption of the chromosomal amfR gene.
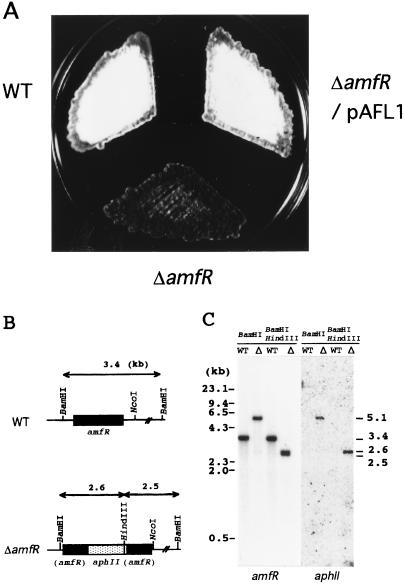
Whether amfR was essentially required for aerial mycelium formation had been unclear, since amfR was cloned as a suppressor for the aerial mycelium-negative phenotype of the A-factor-deficient mutant strain S. griseus HH1 (30). We disrupted the chromosomal amfR gene by means of double crossover, with the kanamycin resistance (aphII) gene as a selection marker to determine its role in morphogenesis. The insertion of aphII by this strategy does not interfere with transcription and translation of orf4 and orf5, because amfR is the third gene in this operon. Southern hybridization with the aphII sequence as a probe against the BamHI-digested and BamHI-plus-HindIII-digested chromosomal DNAs identified four true disruptants. All four disruptants showed a Bld phenotype (Fig. 2). Introduction of pAFL1 carrying amfR and its upstream region on a low-copy-number plasmid restored sporulation to the same level as that of the wild-type strain. It is therefore concluded that amfR plays an essential role in aerial mycelium and spore formation.
FIG. 2.
Phenotypes of the amfR disruptant (A), schematic representation of the disrupted amfR (B), and Southern hybridization analysis of the chromosome of the amfR disruptant (C). (A) Patches were photographed after 5 days of growth at 28°C on YMPG medium. S. griseus IFO13350 (wild type [WT]) formed abundant spores, and the amfR mutant strain (ΔamfR) exhibited a Bld phenotype. The amfR disruptant harboring plasmid pAFL1 carrying the intact amfR gene with its upstream promoter region restored aerial mycelium and spore formation as abundantly as the wild-type strain. (B) Chromosomal amfR is disrupted by aphII. (C) The chromosomal DNAs from strain IFO 13350 (WT) and the amfR disruptant (Δ) were digested with BamHI or BamHI plus HindIII and hybridized with the amfR and aphII probes.
Phenotypes conferred on S. griseus HH1 by amfR together with its upstream region.
Our previous study showed that both amfR and amfA on low-copy-number plasmid pIJ922 were required for aerial mycelium formation in S. griseus HH1 as much as for that in the wild-type strain (30). The two genes on pIJ922, with its copy number of 1 to 2, caused abundant aerial mycelium formation, whereas those on high-copy-number plasmid pIJ487, with its copy number of 40 to 100, appeared to inhibit the growth of substrate mycelium, thereby causing less-abundant aerial mycelium formation. The inhibition of growth of substrate mycelium was ascribed as most likely to be caused by overexpression of AmfR (30). Since ORF4 and ORF5, located adjacent to AmfR in the same direction, had been thought to have some functional relationship with AmfR, we wanted to determine whether a region upstream of amfR affected aerial mycelium formation in strain HH1. An unexpected finding, as described below, was that only amfR, when introduced together with a presumptive promoter region in front of orf5, was sufficient to cause aerial mycelium and spore formation in strain HH1.
We performed subcloning experiments as shown at the bottom of Fig. 1A. The 1.6-kb BamHI-NcoI fragment containing amfR (plasmid pSL6) on low-copy-number plasmid pTMA1 was unable to restore aerial mycelium formation in S. griseus HH1. However, the 3.3-kb EcoRI-NcoI fragment containing orf5-orf4-amfR, but not amfA, on pTMA1 (plasmid pAFL1) induced aerial mycelium and spore formation in strain HH1 just as abundantly as in the wild-type strain. Plasmid pAFL1Δ, in which orf5 and orf4 were disrupted by deletion of the indicated 384-bp PmaCI fragment from pAFL1, also restored aerial mycelium and spore formation to the same extent, indicating that neither ORF5 nor ORF4 was required for the restoration. In this plasmid, the insertion of an 8-mer BglII linker changes the reading frame of ORF4 when ORF4 is translated as an ORF5-ORF4 fusion protein from the initiation codon of orf5. In addition, pAFL2, containing orf5 and orf4 but not amfR, failed to restore aerial mycelium formation. These results indicated that amfR together with the region upstream of orf5 was able to restore sporulation in S. griseus HH1 when it was cloned onto the low-copy-number plasmid. The upstream region responsible for aerial mycelium formation in combination with amfR was most likely a presumptive promoter region in front of orf5 (PORF5), because the ORF5- and ORF4-coding sequences caused no detectable effect on the restoration of aerial mycelium formation. Introduction of the EcoRI-Eco47III region in front of orf5 alone either on the multicopy-number (pSH11) or the low-copy-number (pAFL11) plasmid did not restore sporulation in strain HH1 (data not shown).
Our repeated attempts to clone the 3.3-kb EcoRI-NcoI fragment onto the high-copy-number plasmid pIJ487 failed, probably because of a deleterious effect of an excess of AmfR due to the gene dosage effect of amfR, as was observed previously (30). As described below, strong promoter activity of PORF5 was detected.
Northern blot analysis.
The essential involvement of amfR in morphogenesis prompted us to determine the transcription of amfR in response to A-factor by Northern blot analysis. Total RNAs isolated from the A-factor-deficient mutant of S. griseus, strain HH1, cultivated with or without exogenously supplemented A-factor, were hybridized with probe DNA carrying the amfR-coding sequence. The mRNA isolated from strain HH1 grown in the presence of A-factor yielded a hybridization signal of 3.5 kb, whereas no signal was detected in the mRNA isolated from strain HH1 grown in the absence of A-factor (Fig. 3A). These findings suggested that amfR was transcribed in response to A-factor by a promoter located further upstream of its coding region. The gene organization upstream and downstream of amfR (Fig. 1) predicted the presence of a promoter in front of orf5. The presence of a promoter in front of orf5 was also predicted by the above-described subcloning experiments.
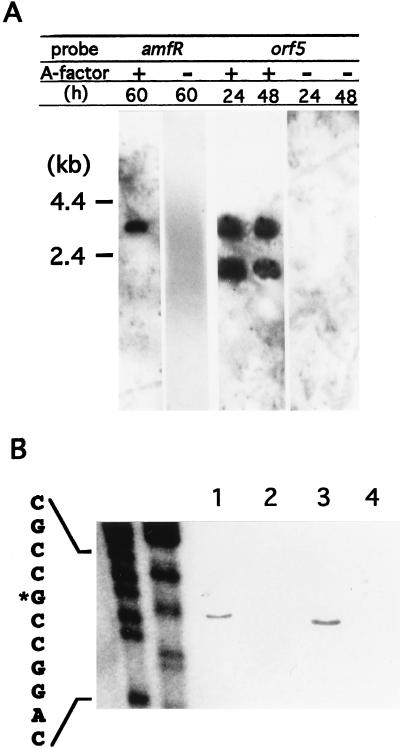
FIG. 3.
Northern blot hybridization for detection of transcripts in the orf5-orf4-amfR region (A) and S1 nuclease mapping for determination of the transcriptional start point in the presence and absence of A-factor (B). S. griseus HH1 (A-factor negative) and the wild-type strain IFO 13350 (A-factor positive) were grown for the indicated periods. Total RNA was isolated from mycelium grown for the indicated period and subjected to Northern hybridization and S1 mapping. (A) When the amfR sequence (see Fig. 1) was used as the 32P probe, a single transcript of 3.5 kb was seen in the mycelium from the wild-type strain but not from strain HH1. When the orf5 sequence (see Fig. 1) was used, two transcripts of 3.5 and 2.2 kb were detected in the mycelium from the wild-type strain but not from strain HH1. (B) The amounts of transcripts directed by PORF5 were examined by S1 nuclease mapping with mRNAs isolated from the wild-type and HH1 strains. The mRNAs were prepared from the wild-type strain grown for 24 h (lane 1) and 48 h (lane 3) and from strain HH1 grown for 24 h (lane 2) and 48 h (lane 4). Protected fragments were analyzed in parallel with sequencing ladders (lanes: left, T plus C; right, A plus G).
We also performed Northern blot hybridization with the orf5-coding region as the 32P probe. In addition to the 3.5-kb signal, a 2.2-kb signal was detected with the mRNA isolated from strain HH1 grown in the presence of A-factor (Fig. 3A). No signal was detected with the mRNA from strain HH1. The 3.5-kb signal supposedly corresponded to the mRNA initiating at PORF5 and covering orf5-orf4-amfR. The 2.2-kb signal presumably represented the mRNA that started at PORF5 and terminated somewhere at the 5′ portion of amfR (Fig. 1A).
The transcriptional organization of orf5-orf4-amfR was also confirmed by promoter-probing experiments with pTMA1 containing the malate dehydrogenase (mdh) gene as the reporter (data not shown). The EcoRI-Eco47III fragment containing the initiation codon of orf5 expressed high MDH activity at 24 h (mid-exponential phase) and 48 h (early stationary phase) after inoculation, whereas the BamHI-Eco47III fragment containing the initiation codon of amfR expressed only very low, but distinct, MDH activity. The MDH activity expressed by the latter was less than 1/10 of that of the former. The weak promoter activity in the BamHI-Eco47III fragment was undetectable in the Northern blot experiment (Fig. 3A). All of these transcriptional analyses of the orf5-orf4-amfR region showed that amfR was transcribed mainly by PORF5.
S1 nuclease mapping.
The above results indicated that the major promoter activity responsible for the transcription of orf5-orf4-amfR was located in front of orf5 (PORF5). The transcriptional start site of PORF5 was determined by S1 nuclease mapping. We used the 320-bp EcoRI-Eco47III fragment including 145 bp of the upstream region from the putative translational start site for ORF5 as the probe by end labeling it at the Eco47III cleavage terminus and hybridized it to mRNAs isolated from wild-type S. griseus IFO 13350 and strain HH1. A single transcriptional start site was detected with the mRNA isolated from the wild-type strain 24 h (mid-exponential phase) and 48 h (early stationary phase) after inoculation but not with that from strain HH1. For clearer resolution of the transcriptional start point, we also used the 130-bp probe (Fig. 3B). The start site was a G that was 72 bp upstream from the putative initiation codon. The presence of the transcript only in the wild-type strain, i.e., in the presence of A-factor, was in agreement with the results of the Northern blot analysis. The identical transcriptional start site was also detected by primer extension analysis with an 18-bp primer (nucleotide positions +106 to +89) (data not shown).
Detection of an A-factor-responsive DNA-binding protein.
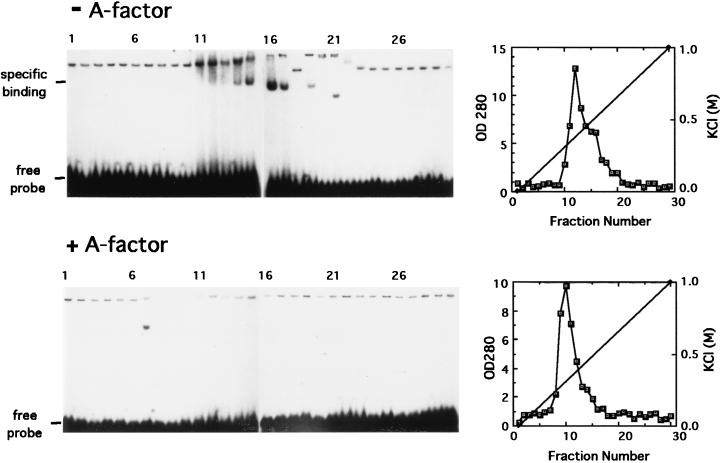
The Northern blot analysis as well as the S1 nuclease mapping experiments indicated that the major transcript covering amfR initiated at PORF5 in positive response to A-factor. Because transcriptional control is often mediated by transcriptional factors that bind operator sites, we examined the presence of an A-factor-dependent DNA-binding protein(s) able to bind the PORF5 sequence. Gel mobility shift assay with the 60-bp EcoRI-MluI fragment (Fig. 1) as the 32P-labeled probe and with cell extracts of strain HH1 cultivated in the presence or absence of exogenously supplied A-factor as a protein source was used for detection of the putative DNA-binding protein. The cell extracts were fractionated by DEAE-Toyopearl column chromatography, and each fraction was subjected to the binding assay. A mobility shift was observed only with the cell extract of strain HH1 cultivated in the absence of exogenous A-factor (Fig. 4), although the gel shift patterns were disturbed due to proteins in various amounts in each fraction. In contrast, no significant shift bands were observed with the cell extract that was prepared from strain HH1 cultivated with exogenous A-factor. We also tested the cell extract similarly prepared from the wild-type strain of S. griseus and found that no significant retardation occurred (data not shown). As described below, the protein causing this mobility shift was found to bind specifically the PORF5 sequence. These results showed that a DNA-binding protein that specifically bound PORF5 is present in strain HH1, an A-factor-deficient mutant, and its production or DNA-binding activity is repressed by exogenously supplemented A-factor.
FIG. 4.
Gel mobility shift assays for detection of a protein able to bind the promoter region of orf5-orf4-amfR. Crude extracts were prepared from the A-factor-deficient mutant strain S. griseus HH1 (upper panel) and strain HH1 grown in the presence of A-factor (lower panel) and fractionated by DEAE column chromatography. Each of the fractions was assayed by gel mobility shift assay with the 62-bp EcoRI-MluI fragment as the probe. OD 280, optical density at 280 nm.
Purification of the A-factor-responsive DNA-binding protein.
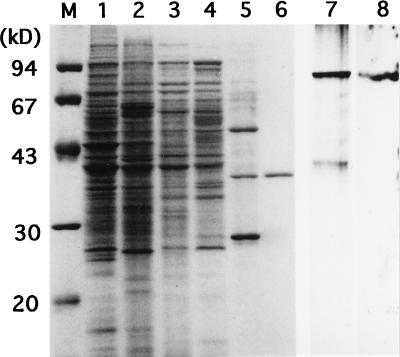
We purified the A-factor-responsive DNA-binding protein (tentatively named AdpB) by monitoring the ability to bind PORF5. Starting with mycelium ([wet] weight, 300 g) of strain HH1 grown at 30°C in 15 liters of YMPG medium, we purified AdpB (1.2 mg) to homogeneity by ammonium sulfate fractionation, three steps of column chromatography, and elution from nondenatured polyacrylamide gel. The affinity column chromatography on heparin was very useful, removing most proteins (Fig. 5, lane 5). The sample containing three major proteins eluted from the heparin column was subjected to native PAGE (lane 7) in order to identify AdpB by Southwestern blotting analysis with the same 32P-labeled probe as in the gel mobility shift assays (lane 8). We eluted the protein giving a positive signal from a gel slice and ran it in SDS-PAGE (lane 6). AdpB purified to homogeneity in this way had an apparent molecular mass of 34 kDa.
FIG. 5.
SDS-PAGE used for monitoring purification of AdpB and Southwestern blotting with the active fraction after heparin affinity column chromatography. Lanes M to 6, SDS-PAGE; lanes 7 and 8, native PAGE. The molecular size markers in lane M were phosphorylase b (94 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), and trypsin inhibitor (20 kDa). Lane 1, the cell lysate from S. griseus HH1 (1 mg of protein); lane 2, after ammonium sulfate fractionation (1 mg of protein); lane 3, after DEAE-Toyopearl chromatography (0.5 mg of protein); lane 4, after Mono Q chromatography (0.5 mg of protein); lane 5, after heparin affinity chromatography (0.2 mg of protein); lane 6, after elution from nondenaturing polyacrylamide gel (0.1 mg of protein); lane 7, nondenaturing gel electrophoresis of the active fraction after heparin affinity chromatography; lane 8, autoradiogram of the same sample as in lane 7 subjected to Southwestern blotting with the 32P-labeled EcoRI-Eco47III fragment.
Our attempt to determine the molecular mass of native AdpB by gel filtration failed, because AdpB appeared to form aggregates readily and eluted in the flowthrough fraction containing miscellaneous proteins with masses of more than 200 kDa. Almost no protein peaks were detected in other fractions examined by spectrophotometry. It is likely that the proteins eluted in the flow-through fraction represent aggregated forms of AdpB. Although the aggregated AdpB protein supposedly had much less activity, its DNA-binding activity, as determined by the gel mobility shift assay and the runoff transcription assay, was detectable as described below.
Gel mobility shift with the purified protein.
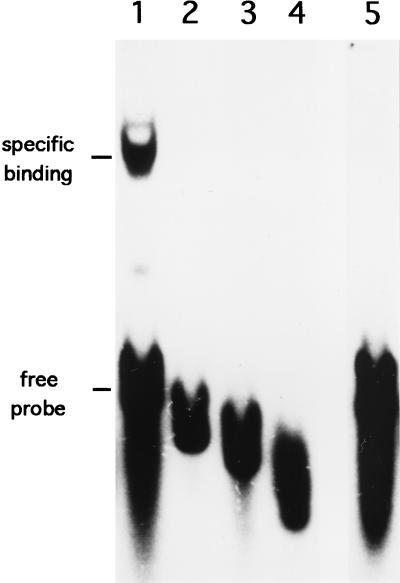
Figure 6 shows the results of a gel shift assay for determination of the DNA-binding activity of the purified AdpB protein. The purified AdpB caused distinct retardation of the 158-bp EcoRI-Sau3AI fragment including the EcoRI-MluI region (lane 1). The binding specificity of AdpB was confirmed by addition of a 100-fold molar excess of cold probe DNA to the reaction mixture, which resulted in the abolishment of the shifted band (lane 5). We also used 32P-labeled probes with various lengths, but none of these were retarded by AdpB (Fig. 5, lanes 2 to 4). The position of the 5′ end of each probe is indicated in Fig. 1B. These results indicate that the sequence essential for the binding of AdpB is located within the 28-bp region between −72 and −44 bp with respect to the transcriptional start point (Fig. 1B).
FIG. 6.
Gel mobility shifts of the regions upstream of orf5 caused by the purified AdpB protein. The 158-bp EcoRI-Sau3AI fragment and a series of fragments trimmed from the EcoRI end (see Fig. 1) were used. Lane 1, the 158-bp fragment (nucleotide positions −71 to 86 in Fig. 1B); lane 2, the 130-bp fragment (positions −43 to 86); lane 3, the 120-bp fragment (positions −34 to 86); lane 4, the 109-bp fragment (positions −22 to 86); lane 5, the 158-bp fragment in the presence of a 100-fold molar excess of the cold probe.
Runoff transcriptional analysis of the A-factor-dependent promoter.
The effect of AdpB on the transcription from PORF5 was determined by runoff transcriptional assay. The RNA polymerase holoenzyme, mainly with ςB promoter specificity partially purified from vegetative mycelium of S. griseus (28), was used. As for templates, we used two different DNA fragments containing the possible AdpB binding sequence and the transcriptional start site, as shown in Fig. 7A. The 316-bp EcoRI-Eco47III fragment (template a) and the 249-bp EcoRI-AvaII fragment (template b) were expected to produce transcripts of 244 and 177 nucleotides, respectively. With template a, a major transcript with the expected size was seen, and the amount of the transcript decreased as the amount of AdpB increased (Fig. 7B). With template b, a transcript with the expected size was seen in the absence of AdpB, and the presence of AdpB in the reaction mixture almost completely abolished it. These results clearly indicate that AdpB represses the in vitro transcription from PORF5, probably by binding to the operator sequence in PORF5.
DISCUSSION
The present study has demonstrated that amfR is essential for morphogenesis in S. griseus and is under the control of A-factor. Disruption of chromosomal amfR resulted in the complete abolishment of aerial mycelium formation, giving a Bld phenotype, which indicates its essential role in the onset of aerial mycelium formation. Nucleotide sequencing and transcriptional analysis of amfR showed that it was mainly transcribed by an A-factor-dependent promoter (PORF5) in front of the orf5-orf4-amfR operon. On the basis of the present study, we can hypothesize the regulation of amfR as follows. In the absence of A-factor, PORF5 does not initiate transcription because of the presence of AdpB that binds the operator site in PORF5 and acts as a repressor. The AdpB binding site is contained in the region from −72 to −44 bp to the transcriptional start point, suggesting that AdpB prevents the RNA-polymerase holoenzyme from recognizing the −35 region and initiating transcription from this promoter. Since the AdpB binding site is an unusual location for an operator site, it could be part of a more complex region; for example, there might be an additional site(s) elsewhere in this region, so that repression could involve DNA looping. The presence of A-factor causes AdpB or its ability to bind PORF5 to disappear, as determined by gel retardation assays, thus leading to transcription starting at PORF5 and reading through into amfR. How the A-factor signal represses the biosynthesis of AdpB or inhibits the repressor-like function of AdpB is unclear. It is apparent that AmfR is a member involved in a decisive step in the A-factor regulatory cascade, leading to the onset of aerial mycelium formation. The details of the mechanism by which AdpB represses PORF5 as a transcription factor will be elucidated when a large amount of AdpB is available by means of recombinant DNA techniques. The mechanism by which the A-factor signal causes DNA-bound AdpB to disappear will also be made clear by examination of the expression of adpB.
AdpB is distinct from ArpA, the A-factor receptor protein which serves as a repressor for both aerial mycelium formation and streptomycin biosynthesis (25). The apparent molecular masses of AdpB and ArpA are 34 and 29 kDa, respectively. The operator site of PORF5 contains no ArpA recognition sequence (26); the consensus sequence recognized and bound by ArpA forms a palindrome 22 bp in length, and one-half of the palindrome is 5′-GG(T/C)CGGT(A/T)(T/C)G(T/G)-3′. In fact, AdpB did not bind the consensus 22-bp oligonucleotide in a gel mobility shift assay (data not shown).
Restoration of aerial mycelium formation by introduction of amfR together with PORF5 on a low-copy-number plasmid into A-factor-deficient mutants can be explained in terms of a gene dosage effect; an increase in the copy number of amfR of even one or two leads to production of AmfR in a larger amount simply due to the gene dosage effect and to titration of AdpB. The failure of pSL6 containing amfR and its own promoter (PAmfR) to cause aerial mycelium formation in strain HH1 may be due to a very weak promoter activity. Since amfR with PORF5 cannot be placed on a high-copy-number plasmid, a moderate amount of AmfR or a phosphorylated form of AmfR appears to be important for healthy growth and normal morphogenesis. The phosphorylated form of AmfR is supposed to be active for the onset of morphogenesis, as was implied through site-directed mutagenesis of an aspartate residue to be phosphorylated (30).
We previously observed that amfR-amfA on the low-copy-number plasmid pTMA1 restored sporulation in strain HH1 (30). In the present study, pSL6 containing only amfR failed to suppress the aerial mycelium-defective phenotype, meaning that an extra copy of amfA compensates for the weak expression of amfR. Although the functions of amfA and amfB, both encoding a membrane translocator, are unclear, these genes may be somehow involved in aerial mycelium formation in combination with amfR. Ma and Kendall (21) also observed that both a response regulator-like protein, RamR, and a membrane translocator-like protein, RamA or RamB, are essential for the acceleration of aerial mycelium formation in S. coelicolor A3 (2).
The operon structure of orf5-orf4-amfR suggests a functional relationship among the products of these genes, although the phenotypes conferred by pAFL1 and pAFL1Δ on strain HH1 are apparently the same. In addition, no changes in the phenotype of the wild-type strain harboring these plasmids are detectable. Nevertheless, it seems possible that ORF5 and ORF4 are involved in the subtle modulation of AmfR in some unknown way under certain growth conditions. Unlike typical two-component regulatory genes, which are closely encoded and cotranscribed, neither ORF5 nor ORF4 has homology with a series of kinases or phosphotransferases.
The fact that amfR encoding a protein belonging to the regulator family of the two-component regulatory systems plays a decisive role in the initiation of aerial mycelium formation is reminiscent of a similarity to spo0A required for the initiation of sporulation in Bacillus subtilis (10). Transcription of spo0A is negatively controlled by a repressor protein, AbrB, which determines the intracellular concentration of Spo0A. Spo0A is the final target of phosphorylation mediated by a so-called phosphorelay, and an increased ratio of the phosphorylated form of Spo0A is supposed to finally determine the onset of sporulation (27). As was implied by our mutational study of a putative phosphorylation site (30), phosphorylation of AmfR is supposed to be essential for its function as a transcriptional regulator. Like Spo0A in B. subtilis (10, 27), AmfR may play a role in widely receiving various signals transmitted through phosphoryl transfers and integrating them into the activation of the downstream-specific genes required for differentiation through its transcriptional control activity, finally leading to the onset of aerial mycelium formation. The precise mechanism can be elucidated by examination of the mode of phosphorylation of AmfR, which is probably mediated by several phosphotransferases encoded by different loci on the chromosome, as in B. subtilis.
ACKNOWLEDGMENTS
This study was supported, in part, by the Nissan Science Foundation, by the “Research for the Future” project of the Japan Society for the Promotion of Science, and by a grant-in-aid (grant no. 09760104) for encouragement of young scientists, Monbu-sho, Japan.
REFERENCES
- 1.Babcock M J, Kendrick K E. Cloning of DNA involved in sporulation of Streptomyces griseus. J Bacteriol. 1988;170:2802–2808. doi: 10.1128/jb.170.6.2802-2808.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babcock M J, Kendrick K E. Transcriptional and translational features of a sporulation gene of Streptomyces griseus. Gene. 1990;95:57–63. doi: 10.1016/0378-1119(90)90413-l. [DOI] [PubMed] [Google Scholar]
- 3.Beck E, Ludwig G, Auerswald E A, Reiss B, Schaller H. Nucleotide sequence and exact localisation of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 4.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 5.Chater K F. Morphological and physiological differentiation in Streptomyces. In: Losick R, Shapiro L, editors. Microbial development. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 89–115. [Google Scholar]
- 6.Chater K F. Sporulation in Streptomyces. In: Smith I, Slepecky R, Setlow P, editors. Regulation of procaryotic development: structural and functional analysis of bacterial sporulation and germination. Washington, D.C: American Society for Microbiology; 1989. pp. 277–299. [Google Scholar]
- 7.Chater K F. Genetics of differentiation in Streptomyces. Annu Rev Microbiol. 1993;47:685–713. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- 8.Chodosh L A. Mobility shift DNA-binding assay using gel electrophoresis. In: Ausubel F M, et al., editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons; 1988. pp. 2.1–2.10. [DOI] [PubMed] [Google Scholar]
- 9.Hara O, Beppu T. Mutants blocked in streptomycin production in Streptomyces griseus—the role of A-factor. J Antibiot. 1982;35:349–358. doi: 10.7164/antibiotics.35.349. [DOI] [PubMed] [Google Scholar]
- 10.Hoch J A. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu Rev Microbiol. 1993;47:441–465. doi: 10.1146/annurev.mi.47.100193.002301. [DOI] [PubMed] [Google Scholar]
- 11.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation in Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 12.Horinouchi S. Streptomyces genes involved in aerial mycelium formation. FEMS Microbiol Lett. 1996;141:1–9. doi: 10.1111/j.1574-6968.2010.02177.x. [DOI] [PubMed] [Google Scholar]
- 13.Horinouchi S, Beppu T. Autoregulatory factors and communication in actinomycetes. Annu Rev Microbiol. 1992;46:377–398. doi: 10.1146/annurev.mi.46.100192.002113. [DOI] [PubMed] [Google Scholar]
- 14.Horinouchi S, Beppu T. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol Microbiol. 1994;12:859–864. doi: 10.1111/j.1365-2958.1994.tb01073.x. [DOI] [PubMed] [Google Scholar]
- 15.Horinouchi S, Furuya K, Nishiyama M, Suzuki H, Beppu T. Nucleotide sequence of the streptothricin acetyltransferase gene from Streptomyces lavendulae and its expression in heterologous hosts. J Bacteriol. 1987;169:1929–1937. doi: 10.1128/jb.169.5.1929-1937.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horinouchi S, Kumada Y, Beppu T. Unstable genetic determinant of A-factor biosynthesis in streptomycin-producing organisms: cloning and characterization. J Bacteriol. 1984;158:481–487. doi: 10.1128/jb.158.2.481-487.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kakinuma S, Takada Y, Ikeda H, Tanaka H, Omura S. Cloning of large DNA fragments, which hybridize with actinorhodin biosynthesis genes, from kalafungin and nanaomycin A methyl ester and identification of genes for kalafungin biosynthesis of the kalafungin producer. J Antibiot. 1989;44:995–1005. doi: 10.7164/antibiotics.44.995. [DOI] [PubMed] [Google Scholar]
- 18.Khokhlov A S, Tovalova I I, Borisova L N, Pliner S A, Schevchenko L A, Kornitskaya E Y, Ivkina N S, Rapoport I A. A-factor responsible for biosynthesis of streptomycin by a mutant strain of Actinomyces streptomycini. Dokl Akad Nauk SSSR. 1967;177:232–235. [PubMed] [Google Scholar]
- 19.Kudo N, Ueda K, Ikeda H, Omura S, Beppu T, Horinouchi S. Plasmid-mediated gene disruption in Streptomyces griseus. Actinomycetologica. 1994;8:17–20. [Google Scholar]
- 20.Kudo N, Kimura M, Beppu T, Horinouchi S. Cloning and characterization of a gene involved in aerial mycelium formation in Streptomyces griseus. J Bacteriol. 1995;177:6401–6410. doi: 10.1128/jb.177.22.6401-6410.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma H, Kendall K. Cloning and analysis of a gene cluster from Streptomyces coelicolor that causes accelerated aerial mycelium formation in Streptomyces lividans. J Bacteriol. 1994;176:3800–3811. doi: 10.1128/jb.176.12.3800-3811.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 23.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 24.Miskimins W K, Roberts M P, McClell A, Ruddle F H. Use of a protein-blotting procedure and a specific DNA probe to identify nuclear proteins that recognize the promoter region of the transferrin receptor gene. Proc Natl Acad Sci USA. 1985;82:6741–6744. doi: 10.1073/pnas.82.20.6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Onaka H, Ando N, Nihira T, Yamada Y, Beppu T, Horinouchi S. Cloning and characterization of the A-factor receptor gene from Streptomyces griseus. J Bacteriol. 1995;177:6083–6092. doi: 10.1128/jb.177.21.6083-6092.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onaka H, Horinouchi S. DNA-binding activity of the A-factor receptor protein and its recognition DNA sequences. Mol Microbiol. 1997;24:991–1000. doi: 10.1046/j.1365-2958.1997.4081772.x. [DOI] [PubMed] [Google Scholar]
- 27.Perego M, Hanstein C, Welsh K M, Djavakhishvili T, Glaser P, Hoch J A. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 28.Shinkawa H, Fujita N, Shiina T, Tanaka K, Takahashi H, Ishihama A, Nimi O. Purification and characterization of RNA polymerase holoenzyme (EςB) from vegetative-phase mycelia of Streptomyces griseus. J Biochem. 1995;118:488–493. doi: 10.1093/oxfordjournals.jbchem.a124934. [DOI] [PubMed] [Google Scholar]
- 29.Trower M K, Marshall J E, Doleman M S, Emptage M H, Sariaslani F S. Primary structure of a 7Fe ferredoxin from Streptomyces griseus. Biochim Biophys Acta. 1990;1037:290–296. doi: 10.1016/0167-4838(90)90027-d. [DOI] [PubMed] [Google Scholar]
- 30.Ueda K, Miyake K, Horinouchi S, Beppu T. A gene cluster involved in aerial mycelium formation in Streptomyces griseus encodes proteins similar to the response regulators of two-component regulatory systems and membrane translocators. J Bacteriol. 1993;175:2006–2016. doi: 10.1128/jb.175.7.2006-2016.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vujaklija D, Ueda K, Hong S-K, Beppu T, Horinouchi S. Identification of an A-factor-dependent promoter in the streptomycin biosynthetic gene cluster of Streptomyces griseus. Mol Gen Genet. 1991;229:119–128. doi: 10.1007/BF00264220. [DOI] [PubMed] [Google Scholar]
- 32.Ward J M, Janssen G R, Kieser T, Bibb M J, Buttner M J, Bibb M J. Construction and characterization of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet. 1986;203:468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]
- 33.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]