Abstract
Lewy body dementia is the second most common neurodegenerative dementia after Alzheimer’s disease. Disease-modifying therapies for this disabling neuropsychiatric condition are critically needed. To identify drugs associated with the risk of developing Lewy body dementia, we performed a population-based case–control study of 148 170 US Medicare participants diagnosed with Lewy body dementia between 1 January 2008 and 31 December 2014 and of 1 253 043 frequency-matched controls. We estimated odds ratios and 95% confidence intervals for the association of Lewy body dementia risk with 1017 prescription drugs overall and separately for the three major racial groups (Black, Hispanic and White Americans). We identified significantly reduced Lewy body dementia risk associated with drugs used to treat cardiovascular diseases (anti-hypertensives: odds ratio = 0.72, 95% confidence interval = 0.70–0.74, P-value = 0; cholesterol-lowering agents: odds ratio = 0.85, 95% confidence interval = 0.83–0.87, P-value = 0; anti-diabetics: odds ratio = 0.83, 95% confidence interval = 0.62–0.72, P-value = 0). Notably, anti-diabetic medications were associated with a larger risk reduction among Black Lewy body dementia patients compared with other racial groups (Black: odds ratio = 0.67, 95% confidence interval = 0.62–0.72, P-value = 0; Hispanic: odds ratio = 0.86, 95% = 0.80–0.92, P-value = 5.16 × 10−5; White: odds ratio = 0.85, 95% confidence interval = 0.82–0.88, P-value = 0). To independently confirm the epidemiological findings, we looked for evidence of genetic overlap between Lewy body dementia and cardiovascular traits using whole-genome sequence data generated for 2591 Lewy body dementia patients and 4027 controls. Bivariate mixed modelling identified shared genetic risk between Lewy body dementia and low-density lipoprotein cholesterol levels, Type 2 diabetes and hypertension. By combining epidemiological and genomic data, we demonstrated that drugs treating cardiovascular diseases are associated with reduced Lewy body dementia risk, and these associations varied across racial groups. Future randomized clinical trials need to confirm our findings, but our data suggest that assiduous management of cardiovascular diseases may be beneficial in this understudied form of dementia.
Keywords: Lewy body dementia, prescription drugs, Medicare, cardiovascular disease, genomic analysis
Using prescription drug association analyses of a Lewy body dementia case–control cohort in the Medicare database, Scholz et al. demonstrate significantly reduced disease risk in participants treated with cardiovascular risk management drugs. These findings were corroborated by genomic evaluations demonstrating genetic correlations between Lewy body dementia and cardiovascular traits.
Graphical Abstract
Graphical Abstract.
Introduction
Lewy body dementia (LBD) is a neurological disease characterized by fluctuating cognition, parkinsonism, visual hallucinations and rapid eye movement sleep behaviour disorder.1 This form of dementia affects ∼1.4 million people in the USA, substantially burdening the healthcare system and raising major public health concerns. Despite increased attention to LBD, current treatments focus on symptomatic management, and there are no therapies that slow disease progression or delay onset. Aside from ageing and a few genetic variants,2 the risk factors for LBD remain elusive. Additionally, little is known about LBD occurrence in Blacks, Hispanics and other racial/ethnic groups because accurate data on these populations have been lacking. Consequently, there is a need to identify modifiable risk factors and medications that reduce LBD risk or slow progression after symptom onset.
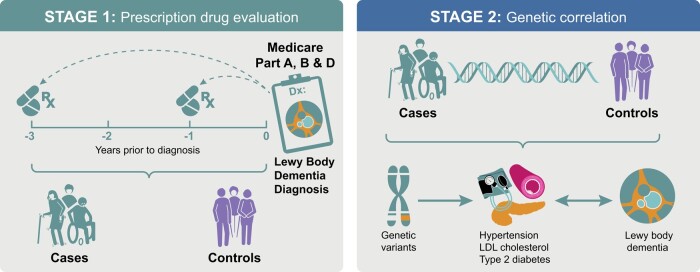
The main objective of the present study was to identify protective drugs that could be repurposed to lower LBD risk and potentially be used as treatments. In the process, we also aimed to shed further light on LBD aetiology, the risk factors driving the disease and possible racial differences. We used a two-stage approach to identify drugs and their corresponding conditions associated with LBD risk (Fig. 1). First, we conducted a large drug screen using a case–control study sampled from the US Medicare claims database. Then, to independently corroborate associations of LBD risk with the conditions treated by the prescription drugs identified in Stage 1, we looked for evidence of genetic overlap between these conditions and LBD. Combining the epidemiological and genomic approaches lessened the impact of possible unmeasured confounders and allowed a mechanistic interpretation of the empirical findings.
Figure 1.
Study design. This graphical representation showcases the two-stage study design. Stage 1 consisted of a prescription drug evaluation using LBD cases and controls sampled from the US Medicare database. In Stage 2, we studied the polygenic overlap between cardiovascular disease traits and LBD using genomic data.
Materials and methods
Study population
We studied LBD cases and frequency-matched controls enrolled in the Medicare claims database between 1 January 2008 and 31 December 2014. Medicare was established in 1965 as a federal health insurance programme for disabled adults and elderly persons living in the USA. We focused on the 2008–14 period because Medicare Part D supplemental plans (covering prescription drugs) became available on 1 January 2006, and Medicare data were not available to us for the time after 2015. To be eligible for the present study, the subjects had to have at least 13 months of Medicare plans Part A/B insurance (covering hospital and medical insurance) and supplementary plan Part D insurance.
Selection of study participants
We identified LBD cases using the International Classification of Diseases-9-CM (ICD-9-CM) code 331.82. Each patient was required to have at least one hospital claim or two outpatient claims 30 days or more apart. Cases were restricted to those with a first LBD claim between 1 January 2008 and 31 December 2014, with ages 66 through 89. We aimed to randomly select 10 control subjects per case (actually matching ratio = 8.5), frequency-matched based on sex, calendar year of case selection and age group at diagnosis/selection (66–69, 70–74, 75–79, 80–84 and 85–89 years).3 Controls were assigned 30 June as the selection date for their selection year. Individuals with claims for the following conditions were ineligible as controls: primary and secondary dementias, mild cognitive impairment, age-related neurodegenerative diseases, psychiatric or organic diseases that may mimic LBD, severe sleep disorders and HIV/AIDS (see Supplementary Table 1 for the exclusion codes). Supplementary Fig. 1 outlines the study participant selection process.
Drug exposure definition and outcome measures
Drug exposure was defined as having a prescription drug claim under the Medicare Prescription Drug Plan (Part D). In each study participant, we examined drug use for up to 1 year and up to 3 years (primary outcome measure) prior to the LBD diagnosis/selection date. We chose these time lags to capture medication use in the disease’s prodromal stages when therapeutic interventions would be most beneficial. As an example of the 3-year lag model, only the available prescription data up to 70 years of age would be analysed in a patient diagnosed with LBD at the age of 73. This approach minimized the possibility of detecting drugs commonly used to treat LBD symptoms (reverse causality). Medications prescribed to fewer than 100 study participants were excluded. Overall, 1321 drugs were tested for associations in the 1-year lag analysis and 1017 in the 3-year lag analysis, with substantial overlap between drugs in the two lag groups (Supplementary Tables 2 and 3).
Statistical analysis
Race/ethnicity was derived using the Research Triangle Institute algorithm.4 Conditions and comorbidities were identified using Part B claims and ICD-9-CM codes (restricted to claims at ages ≥65 years, 1 January 2006 and up to the timepoint 1 year before selection). They included dyslipidaemia (ICD-9CM code 272), diabetes (250), stroke (434.91), renal disease (585), hypertension (401), acute myocardial infarction (410) and chronic obstructive pulmonary disease (490, 491, 492, 494 and 496). Obesity was derived from the Chronic Conditions Data Warehouse obesity definition, based on diagnostic and procedural codes (including V85.3 and V85.4). As the frequency of health care visits is likely to affect health outcomes, we accounted for medical surveillance intensity by calculating the average number of physician visits per 6-month interval between 2006 and the selection date, omitting the first and last intervals. We excluded claims from specialists with limited patient contact (radiologists, anaesthesiologists and pathologists).
We used logistic regression models to assess associations [odds ratios (ORs), 95% confidence intervals (CIs)] between the individual-level prescription drugs and LBD risk. Models were adjusted for diagnosis/selection year, age at diagnosis/selection (categorized as 66–69, 70–74, 75–79, 80–84 and 85–89 years), sex, race (White, Black, Hispanic and other/unknown), duration of Medicare Part D coverage divided into quintiles (≤30, 31–42, 43–54, 55–78 and ≥79 months), the average number of physician visits per 6 months period (quintiles: cut points ≤0.75, ≤2.06, ≤5.92 and >5.92 visits), Medicaid eligibility (yes/no), limited income subsidy (yes/no) and conditions/comorbidities (dyslipidaemia, diabetes, stroke, renal disease, hypertension, acute myocardial infarction, obesity and chronic obstructive pulmonary disease).5 To limit the number of false-positive findings, we applied Bonferroni multiple testing corrections with significance thresholds of 3.78 × 10−5 (= 0.05/1321 drugs tested) and 4.92 × 10−5 (= 0.05/1017 drugs) for the 1- and 3-year lag models, respectively.
To assess the combined effects of drugs within the same pharmacologic group, we fitted a two-stage hierarchical logistic regression model separately to the three medication groups for which many individual drugs were significantly associated with lower LBD risk, namely (i) anti-hypertensive drugs, (ii) anti-diabetic drugs and (iii) cholesterol-lowering drugs. Models were also stratified by limited-income subsidy/Medicaid eligibility and/or race/ethnicity. Further details on these models are given in the Supplementary Materials. All analyses were implemented using the SAS/STATS software (version 9.4, SAS Institute Inc., Cary, NC, USA). The study’s use of de-identified Medicare claims was exempt from review by an Institutional Review Board.
Defining shared polygenicity between cardiovascular traits and LBD
For the second stage of the study, we used summary statistics from a genome-wide association study (GWAS) of 2591 well-characterized LBD cases and 4027 controls. These cohorts have been described elsewhere.2 Briefly, LBD patients were diagnosed with pathologically definite (69% of the cohort) or clinically probable disease (31%) according to consensus criteria.1,6 The control participants were selected based on a lack of evidence of cognitive decline in their clinical history and the absence of neurological deficits on neurological examination. Pathologically confirmed control individuals (n = 605) had no evidence of notable neurodegenerative disease on histopathological examination. All study participants were of European ancestry and underwent 150-bp, paired-end whole-genome sequencing using a uniform sequencing, alignment, variant calling and quality control pipeline, as described elsewhere.2
The summary statistics of the GWAS for low-density lipoprotein (LDL) cholesterol (study identifier: ieu-b-110), Type 2 diabetes (ebi-a-GCST006867) and essential (primary) hypertension (ukb-b-14177) were obtained from the MRC IEU Open GWAS Project (https://gwas.mrcieu.ac.uk/).7-9 We applied bivariate causal mixture modelling (as implemented in MiXeR, version 1.3)10 to the summary statistics. This statistical method estimates the number of shared and unique trait-influencing variants between complex traits (i.e. a cardiovascular trait and LBD in the present study) and determines the genetic correlation of the shared variants. The best model with polygenic overlap estimated with MiXeR was compared with models with the least and maximum possible overlap. The best model fit was determined from the lowest point on the negative log-likelihood curve (Fig. 2). A model was considered acceptable when it had a positive minimum and maximum Akaike information criterion compared with the best model, indicating sufficient power in each GWAS dataset to distinguish the estimated polygenic overlap from the constrained models based on minimal and maximum polygenic overlap.10
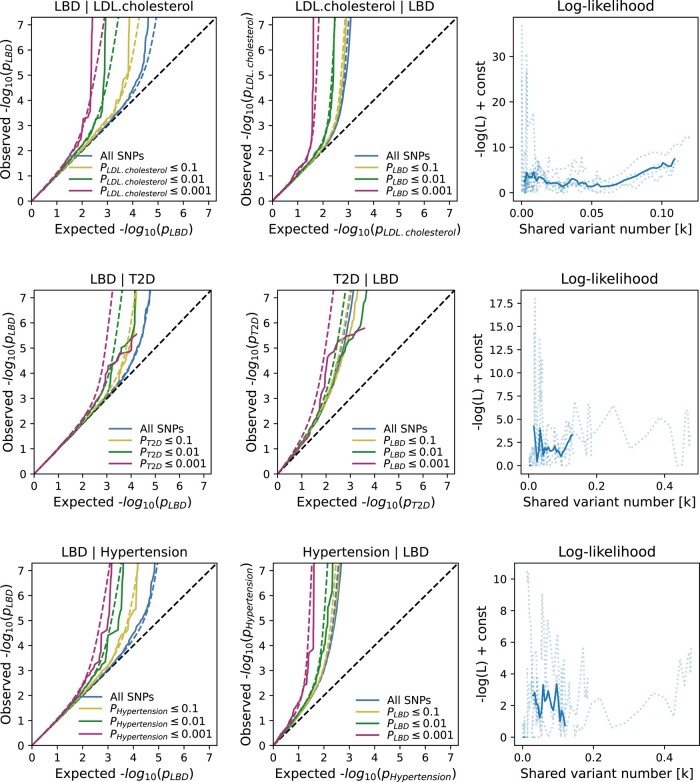
Figure 2.
Conditional quantile–quantile (Q–Q) plots and shared variant estimates. The conditional Q–Q plots show the relationship between the expected (x-axis) versus the observed (y-axis) −log10 P-values of the single nucleotide polymorphism (SNP) associations in the primary phenotype (e.g. LDL cholesterol) stratified by the P-values in the conditional trait (e.g. LBD). We show four strata: all SNPs, SNPs with two-sided P-values ≤0.1 (orange lines), P-values ≤0.01 (green lines) and P-values ≤0.001 (red lines). The dashed, diagonal line indicates the expected Q–Q plot under the null hypothesis for genome-wide associations. The increasing degree of leftward deflection from the null in the conditional phenotypes indicates polygenic overlap. Using MiXeR modelling of polygenic overlap, all three tested traits [LDL cholesterol, Type 2 diabetes (T2D) and primary hypertension] show genetic overlap when conditioned on LBD. The log-likelihood plots illustrate the goodness-of-model-fit, plotting the negative log-likelihood function against the π12 parameter (number of influencing variants shared between two tested traits).
Results
Demographic characteristics
We included 148 170 LBD cases diagnosed between 1 January 2008 and 31 December 2014 in the Medicare claims database for analysis. Demographic characteristics of the cases and their 1 253 043 matched controls are summarized in Table 1. Approximately half of the study subjects were male [n = 75 442 (50.92%) cases; n = 632 990 (50.52%) controls]. The mean age at the time of the first LBD claim was 79.50 years [standard deviation = 5.91 years, range = 66–89 years], with a similar mean selection age for controls of 79.33 years (standard deviation = 5.97 years, range 66–89 years). The majority of the study participants were non-Hispanic, White individuals [n = 119 856 (80.89%) LBD cases; n = 1 000 875 (79.88%) controls]. The remaining participants were Black [n = 11 542 (7.79%) cases; n = 96 910 (7.73%) controls], Hispanic [n = 11 679 (7.88%) cases; n = 99 539 (7.94%) controls] or had ‘other’ or unknown racial/ethnic backgrounds [n = 5093 (3.44%) cases; n = 55 719 (4.45%) controls]. LBD cases had noticeably higher prevalences of comorbidities compared with controls (Table 1; e.g. 36.82 versus 27.51% had diabetes, 7.51 versus 0.88% had a history of stroke). More Black and Hispanic than White individuals received the limited-income subsidy and were eligible for Medicaid (Supplementary Table 4). The prevalence of diabetes, stroke, hypertension, renal disease and obesity was higher among Black individuals than among White people (Supplementary Table 4).
Table 1.
Demographic characteristics of selected study participants
| Characteristics | LBD cases | Controls | |||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Total | Category | 148 170 | 100% | 1 253 043 | 100% |
| Sex | Male | 75 442 | 50.92 | 632 990 | 50.52 |
| Female | 72 728 | 49.08 | 620 053 | 49.48 | |
| Age at selectiona | 66–69 | 9552 | 6.45 | 85 582 | 6.83 |
| 70–74 | 22 961 | 15.50 | 199 579 | 15.93 | |
| 75–79 | 36 464 | 24.61 | 296 600 | 23.67 | |
| 80–84 | 44 100 | 29.76 | 328 702 | 26.23 | |
| 85–89 | 35 093 | 23.68 | 342 580 | 27.34 | |
| Race/ethnic group | White, non-Hispanic | 119 856 | 80.89 | 1 000 875 | 79.88 |
| Black, non-Hispanic | 11 542 | 7.79 | 96 910 | 7.73 | |
| Hispanic | 11 679 | 7.88 | 99 539 | 7.94 | |
| Other/unknown | 5093 | 3.44 | 55 719 | 4.45 | |
| Selection year | 2008 | 18 187 | 12.27 | 154 411 | 12.32 |
| 2009 | 19 011 | 12.83 | 166 406 | 13.28 | |
| 2010 | 20 244 | 13.66 | 179 449 | 14.32 | |
| 2011 | 23 737 | 16.02 | 209 608 | 16.73 | |
| 2012 | 22 627 | 15.27 | 190 883 | 15.23 | |
| 2013 | 22 062 | 14.89 | 176 526 | 14.09 | |
| 2014 | 22 302 | 15.05 | 175 760 | 14.03 | |
| Socio-economics | Low-income subsidy | 77 843 | 52.54 | 393 320 | 31.39 |
| Medicaidb eligibility | 76 325 | 51.51 | 358 189 | 28.59 | |
| Comorbidities | Dyslipidaemia | 98 838 | 66.71 | 797 227 | 63.62 |
| Diabetes | 54 560 | 36.82 | 344 713 | 27.51 | |
| Stroke | 11 121 | 7.51 | 10 992 | 0.88 | |
| Renal disease | 22 468 | 15.16 | 115 285 | 9.20 | |
| Hypertension | 116 898 | 78.89 | 868 944 | 69.35 | |
| Acute myocardial infarction | 6559 | 4.43 | 37 686 | 3.01 | |
| Obesity | 11 050 | 7.46 | 74 449 | 5.94 | |
| COPD | 35 870 | 24.21 | 205 940 | 16.44 | |
COPD, chronic obstructive pulmonary disease. aAge at selection in LBD cases refers to the age at first LBD diagnosis. bMedicaid is a federal state health insurance programme providing health care coverage to low-income and disabled individuals.
Prescription drug screening
To accommodate the potentially long period from symptom onset to establishing the diagnosis, we focused on prescription data from the 3-year lag analysis, i.e. excluding medications prescribed during the 3 years prior to the LBD diagnosis (or selection date for matched controls). Overall, 1017 drugs were tested in the 3-year model. Medications prescribed to fewer than 100 study participants were excluded from the analysis. As expected, we identified significant associations for the medications used to treat common prodromal features of LBD (Supplementary Table 5). These included anti-depressants and anti-psychotics (e.g. quetiapine OR for the 3-year model = 12.32, 95% CI = 12.27–12.37, P-value = 0), anti-parkinsonian drugs (e.g. carbidopa/levodopa OR = 109.9, 95% CI = 109.86–109.95, P-value = 0), cognition-enhancing drugs (e.g. donepezil OR = 16.0, 95% CI = 15.98–16.04, P-value = 0) and medications used to treat constipation, sleep dysfunction, dysautonomia or neurogenic bladder symptoms. The use of these medications was increased in patients with LBD, confirming that most patients had been symptomatic 3 years before receiving a formal neurological diagnosis of LBD.
In contrast, several cholesterol-lowering, anti-hypertensive and anti-diabetic medications were associated with a significantly reduced risk for LBD after Bonferroni adjustment in the 3-year lag model (Tables 2 and 3; Supplementary Table 6; Fig. 3). We also found evidence for significantly reduced LBD risk with the use of anti-inflammatory agents, glaucoma medications and others. However, we focused the remaining investigations on modifiable cardiovascular risk factors that have been implicated in Alzheimer’s disease, a common form of dementia that has many similarities with LBD.2,11 Similar to the 3-year lag model, the 1-year lag model identified drugs used to treat prodromal LBD symptoms and cardiovascular risk factors as associated with LBD risk with generally the same directions of effects as in the 3-year lag model (Supplementary Table 5). The 1-year lag model showed the overall robustness of the associations in the prodromal phase of LBD and demonstrated that the effect directions were stable over time (Supplementary Fig. 2).
Table 2.
Cholesterol-lowering drugs associated with decreased risk for LBD in the 3-year lag model
| Drug name | Exposed cases | Exposed controls | Unexposed cases | Unexposed controls | OR (95% CI) | P-value | Drug class(es) |
|---|---|---|---|---|---|---|---|
| Colesevelam HCL | 447 | 3214 | 123 900 | 908 766 | 0.77 (0.70–0.83) | 2.00E−14 | Bile acid sequestrant |
| Amlodipine/atorvastatin calcium | 692 | 6128 | 123 655 | 905 852 | 0.69 (0.63–0.74) | 1.58E−42 | Calcium channel blocker/HMG-CoA reductase inhibitor |
| Fenofibrate | 360 | 2920 | 123 987 | 909 060 | 0.67 (0.58–0.75) | 2.40E−22 | Fibrate |
| Fenofibrate nanocrystallized | 2726 | 18 577 | 121 621 | 893 403 | 0.74 (0.72–0.77) | 6.46E−86 | Fibrate |
| Fenofibrate, micronized | 414 | 2598 | 123 933 | 909 382 | 0.82 (0.74–0.89) | 2.19E−07 | Fibrate |
| Fenofibric acid (choline) | 156 | 1147 | 124 191 | 910 833 | 0.48 (0.35–0.62) | 8.51E−26 | Fibrate |
| Gemfibrozil | 1816 | 13 254 | 122 531 | 898 726 | 0.80 (0.76–0.83) | 1.17E−36 | Fibrate |
| Atorvastatin calcium | 18 562 | 133 503 | 105 785 | 778 477 | 0.89 (0.88–0.90) | 6.31E−82 | HMG-CoA-reductase inhibitor |
| Ezetimibe/simvastatin | 4466 | 35 904 | 119 881 | 876 076 | 0.79 (0.76–0.81) | 5.13E−107 | Cholesterol absorption inhibitor/HMG-CoA-reductase inhibitor |
| Fluvastatin sodium | 915 | 6408 | 123 432 | 905 572 | 0.91 (0.86–0.95) | 3.89E−05 | HMG-CoA reductase inhibitor |
| Pravastatin sodium | 4897 | 35 149 | 119 450 | 876 831 | 0.90 (0.88–0.92) | 1.02E−21 | HMG-CoA reductase inhibitor |
| Rosuvastatin calcium | 4870 | 37 077 | 119 477 | 874 903 | 0.82 (0.80–0.84) | 1.78E−76 | HMG-CoA reductase inhibitor |
| Simvastatin | 21 867 | 143 640 | 102 480 | 768 340 | 0.97 (0.96–0.99) | 1.17E−05 | HMG-CoA reductase inhibitor |
| Ezetimibe | 4529 | 34 260 | 119 818 | 877 720 | 0.79 (0.77–0.81) | 1.74E−100 | Intestinal cholesterol absorption inhibitor |
| Niacin | 1460 | 10 975 | 122 887 | 901 005 | 0.84 (0.80–0.87) | 1.58E−21 | Niacin |
| Niacin/lovastatin | 220 | 1688 | 124 127 | 910 292 | 0.75 (0.65–0.85) | 7.94E−09 | Niacin/HMG-CoA reductase inhibitor |
Table 3.
Anti-diabetic drugs associated with decreased risk for LBD in the 3-year lag model
| Drug name | Exposed cases | Exposed controls | Unexposed cases | Unexposed controls | OR (95% CI) | P-value | Drug class(es) |
|---|---|---|---|---|---|---|---|
| Acarbose | 174 | 910 | 124 173 | 911 070 | 0.68 (0.55–0.81) | 3.63E−09 | Alpha-glucosidase inhibitor |
| Metformin HCL | 12 503 | 83 176 | 111 844 | 828 804 | 0.83 (0.82–0.85) | 1.48E−110 | Biguanide |
| Sitagliptin phosphate | 1401 | 7704 | 122 946 | 904 276 | 0.82 (0.77–0.86) | 9.77E−20 | Dipeptidyl peptidase-4 inhibitor |
| Sitagliptin phos/metformin HCL | 280 | 1662 | 124 067 | 910 318 | 0.79 (0.70–0.89) | 1.91E−06 | Dipeptidyl peptidase-4 inhibitor/biguanide |
| Exenatide | 320 | 2309 | 124 027 | 909 671 | 0.61 (0.53–0.70) | 8.91E−29 | Glucagon-like peptide-1 receptor agonist |
| Glimepiride | 3260 | 18 889 | 121 087 | 893 091 | 0.88 (0.85–0.91) | 2.04E−19 | Sulphonylurea |
| Glipizide | 5883 | 33 822 | 118 464 | 878 158 | 0.90 (0.88–0.92) | 7.08E−21 | Sulphonylurea |
| Glyburide | 3443 | 19 986 | 120 904 | 891 994 | 0.92 (0.90–0.95) | 2.51E−08 | Sulphonylurea |
| Glipizide/metformin HCL | 104 | 829 | 124 243 | 911 151 | 0.72 (0.57–0.88) | 3.02E−05 | Sulphonylurea/biguanide |
| Glyburide, micro/metformin HCL | 1407 | 9228 | 122 940 | 902 752 | 0.84 (0.80–0.88) | 1.05E−16 | Sulphonylurea/biguanide |
| Glyburide/metformin HCL | 536 | 3946 | 123 811 | 908 034 | 0.70 (0.63–0.77) | 8.91E−23 | Sulphonylurea/biguanide |
| Pioglitazone HCL | 4356 | 27 795 | 119 991 | 884 185 | 0.80 (0.78–0.83) | 5.50E−72 | Thiazolidinedione |
| Rosiglitazone maleate | 2950 | 18 219 | 121 397 | 893 761 | 0.81 (0.78–0.84) | 2.69E−44 | Thiazolidinedione |
| Rosiglitazone HCL/metformin HCL | 291 | 2435 | 124 056 | 909 545 | 0.66 (0.57–0.74) | 4.37E−22 | Thiazolidinedione/biguanide |
| Rosiglitazone/metformin HCL | 339 | 2709 | 124 008 | 909 271 | 0.67 (0.59–0.75) | 9.12E−23 | Thiazolidinedione/biguanide |
| Rosiglitazone/glimepiride | 148 | 1034 | 124 199 | 910 946 | 0.69 (0.57–0.82) | 2.40E−08 | Thiazolidinedione/sulphonylurea |
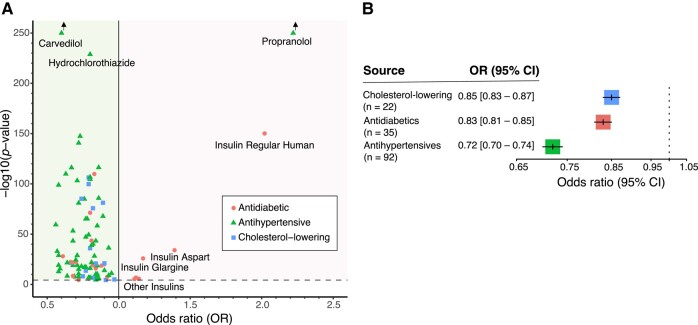
Figure 3.
Associations of cardiovascular drugs with LBD. (A) This plot shows the adjusted logistic regression results of cardiovascular disease treatment drugs associated with LBD risk (n = 64). The ORs are plotted on the x-axis and the −log10 P-values are on the y-axis. The threshold for significance is indicated by a dashed line. The green background colour highlights drugs associated with decreased LBD risk, and the red background colour highlights drugs with increased LBD risk. Among the drugs with increased LBD risk, we point out propranolol, an anti-hypertensive drug that is commonly prescribed for LBD patients due to anti-tremor properties. We also noted insulins as associated with increased risk for disease, which may indicate that metabolic impairment is optimized when prodromal LBD patients seek medical care for cognitive changes. We attributed the increased risk of propranolol and insulins reflecting reverse causality to LBD patients seeking medical care for symptomatic management of neurological changes. (B) Plot showing the effect of the cholesterol-lowering drugs (number of group members g = 22), anti-diabetics (g = 35) and anti-hypertensives (g = 92) on LBD risk based on the 3-year lag model of the US Medicare prescription database.
Hierarchical modelling for drug groups based on the 3-year lag model found a consistent reduction in LBD risk for all classes of anti-hypertensives (e.g. beta-blockers, calcium channel antagonists; Supplementary Table 7), all lipid-lowering drug groups (e.g. fibrates, β-Hydroxy β-methylglutaryl-CoA [HMG-CoA] reductase inhibitors; Supplementary Table 8) and all classes of anti-diabetics (e.g. dipeptidyl peptidase-4 inhibitors, sulphonylureas; Supplementary Table 9). Results were similar for models stratified by limited income subsidy/Medicaid eligibility (yes/no; Supplementary Tables 10–12).
When we stratified the hierarchical analysis of cardiovascular drugs by racial groups (Black, Hispanic and White), they were associated with lower LBD risk across all three groups. However, anti-diabetic drugs had a larger protective effect in Black patients (OR = 0.67, 95% CI = 0.62–0.72, P-value = 0) than in Hispanic (OR = 0.86, 95% CI = 0.80–0.92, P-value = 5.16 × 10−5) and White participants (OR = 0.85, 95% CI = 0.82–0.88, P-value = 0; Supplementary Table 8). To assess the potential effect modification of anti-diabetic drug use by socio-economic factors, we further stratified all hierarchical models by race and limited-income subsidy/Medicaid eligibility (Supplementary Tables 10–12). Among White people, there was no difference in the anti-diabetic estimates, but for Black individuals, the OR was 0.84 (95% CI = 0.72–0.99) for those who did not receive the subsidy and 0.62 (95% CI = 0.57–0.68) for those with limited income subsidy (Supplementary Table 12). Taken together, these findings suggest that racial differences may play a role in the risk for developing LBD, and Black patients may benefit more from stringent glycaemic control.
Genomic evaluations identify shared molecular relationships
We next investigated the shared genetic risk among the cardiovascular traits: hyper-cholesterolaemia, Type 2 diabetes, essential hypertension (as implicated by our epidemiological analyses) and LBD. To do so, we performed bivariate Gaussian mixture modelling of polygenicity using MiXeR, which calculates the genetic overlap independent of the direction of effect, enabling it to capture mixtures of risk effects. The best model fit was ascertained from the lowest point on the negative log-likelihood curve. A model was considered acceptable when it had a positive minimum and maximum Akaike information criterion compared with the best model. We applied this MiXeR methodology to (i) GWAS summary statistics of 2591 well-characterized LBD cases and 4027 controls who had previously undergone whole-genome sequencing2 and (ii) summary statistics of GWASs on LDL cholesterol, Type 2 diabetes and essential (primary) hypertension.7-9
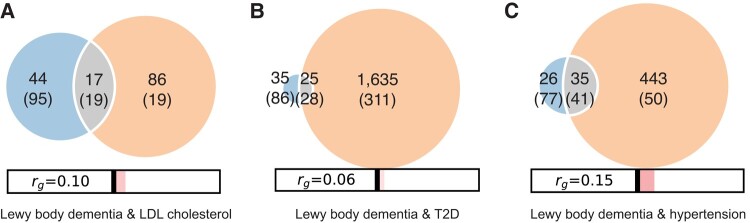
In our data, we found polygenic overlap influencing LBD for the following traits: LDL cholesterol (17 shared variants with a correlation of effect size in the shared polygenic component, ρ12 = 0.49), Type 2 diabetes (25 shared variants, ρ12 = 0.69) and hypertension (34 shared variants, ρ12 = 0.86; Fig. 4). Conditional analyses examining the relationship between LBD and cardiovascular traits were indicative of shared polygenicity (Fig. 2). Model details are presented in Supplementary Table 13.
Figure 4.
Polygenic overlap between cardiovascular traits and LBD. Venn diagrams based on MiXeR modelling from GWASs of cardiovascular traits and LBD (n = 2591 cases and 4027 controls) illustrate the polygenic overlaps between (A) LBD and LDL cholesterol, (B) LBD and T2D and (C) LBD and hypertension. The estimated number of causal variants shared between LBD and each respective cardiovascular trait are shown in the grey intersection. The estimated number of influencing variants that are unique to LBD are shown in blue, while variants uniquely influencing each cardiovascular trait are shown in orange. Standard errors are shown in parentheses. The size of each circle reflects the extent of polygenicity of each trait, with larger circles indicating a bigger number of causal variants. The estimates of genetic correlation, as measured by rg values, are shown at the bottom of each panel. The red bars highlight the significant positive correlations between LBD and each of the three tested cardiovascular traits. T2D, Type 2 diabetes mellitus.
Discussion
In our large, population-based case–control study, drugs used to treat cardiovascular conditions were associated with a lower risk of LBD, a common and fatal neurodegenerative disease. Specifically, we found a significantly reduced risk for developing LBD among individuals treated with cholesterol-lowering, anti-diabetic and anti-hypertensive drugs (Tables 2 and 3; Supplementary Table 6). We focused on drug prescriptions up to 3 years prior to diagnosis to identify medications that might delay the onset or slow the progression of LBD in the prodromal disease stages. We found additional supportive evidence of polygenic overlap with cardiovascular risk factors using a whole-genome sequence dataset obtained from an independent LBD case–control study and summary statistics of cardiovascular traits. Over two-thirds (69%) of the cases in our genetic studies had pathologically confirmed LBD, and the remaining cases were diagnosed with clinically probable LBD (the highest clinical category).2 This is the first study to examine the associations of a broad set of medications with LBD risk and their effects across racial groups based on data from a large population-representative healthcare database.
LBD is a spectrum disorder that shares molecular, genetic and clinicopathological features with both Parkinson’s disease and Alzheimer’s disease, the most common form of dementia in the general population.2 Previous studies of Parkinson’s disease and Alzheimer’s disease implicated cardiovascular factors as potentially modifiable contributors to disease risk.12,13 Additional studies on dementia in general (without differentiating the specific forms of the disease) yielded the same results.14,15 The incidence and prevalence of dementia in the population have been decreasing over the last three decades, primarily due to improved cardiovascular risk factor management.16 Similarly, Type 2 diabetes mellitus has been associated with an increased risk for Parkinson’s disease that may be mitigated by anti-diabetic treatment.17-19 Current evidence on how these insights apply to understudied dementia types, such as LBD, is scarce. Our data represent an important step towards filling this knowledge gap and identifying risk factors that are potentially modifiable drivers in the pathogenesis of this complex form of dementia.
We used genomic analyses to confirm the findings arising from the epidemiological part of our study. Applying MiXeR to our recently published GWAS of LBD and summary statistics from cardiovascular traits identified shared genetic risk (Fig. 2). The temporal pattern whereby the cardiovascular risk factors precede LBD onset by decades supports their causal effect. Although more extensive genetic studies are required to confirm these findings, the combination of epidemiological and genetic data implicating cardiovascular factors as determinants for LBD is compelling, especially because of the modifiable nature of these risk factors. Our data suggest that clinical trials testing the efficacy of assiduous cardiovascular risk management in delaying onset and slowing disease progression among LBD patients should be prioritized.
Data on the occurrence of LBD in diverse populations are limited. Notably, our dataset included over 10 000 Black LBD patients, representing 7.74% of participants, which is lower than expected, given that Black people comprise 9% of the US population over 65 years of age.20 Medicare enrolment is relatively complete among racial groups, and the lower percentage could be explained by a lower incidence of LBD among the Black population, as previously suggested in an autopsy study.21 Lower health care utilization may also contribute.22,23 In addition to suggesting that LBD is less common among Black Americans, the associations for cardiovascular medications differed across racial groups in our case–control study. For example, anti-diabetic medications were associated with a 33% lower LBD risk in Black Americans, while they were associated with 15 and 14% lower risk in the White and Hispanic populations, respectively. However, we adjusted models for the number of physician visits and found similar differences when we additionally stratified the analysis by low-income subsidy/Medicaid eligibility within racial groups, indicating that healthcare access alone does not explain the observed racial pattern.
The strengths of our study include the very large number of incident LBD cases; the population-based, US representative design; the inclusion of multiple races/ethnicities; the careful accounting for health care utilization and underlying conditions; the use of hierarchical modelling to elucidate correlations among drugs in the same class; and the use of genomic data to assess the shared molecular drivers between LBD and cardiovascular traits. Furthermore, the genomic analysis helps to alleviate the concern that our findings in the Medicare data are solely due to unmeasured confounding.
Several limitations should be mentioned. First, data derived from electronic health records can be incomplete. Second, the accuracy of the clinical diagnosis across healthcare institutions may vary, and unmeasured confounders are likely present, as the Medicare claims database is designed for administrative rather than research purposes. To mitigate the influence of confounders in this study, we carefully controlled for confounding factors in our analysis by including an extensive list of covariates. However, unmeasured confounding could partly explain differences in associations for anti-diabetic drugs we found for Black participants. As biases (e.g. survival bias, collider bias) can never be entirely dismissed in observational studies, well-designed randomized controlled trials are needed to confirm the findings. Third, another limitation is that we used only 5-year age groups for matching. Fourth, we limited our investigations to the 3-year lag model, examining prescription drug exposures in the prodromal phase of the disease. Longer lag periods should be assessed in future studies to understand the long-term effects of these exposures from the preclinical to the symptomatic stages of the natural history of LBD. Fifth, the sensitivity and specificity of the ICD-9-CM code 311.82 for the diagnosis of LBD is unknown. Sixth, vascular dementia is a common form of dementia that often co-exists with neurodegenerative dementias. It is possible that the protective effect of vascular risk factor management drugs on LBD risk could in part be due to reducing cerebrovascular co-pathologies. Nonetheless, expanding investigations of this understudied disease may be of great public health interest, and these risk factors have not been definitively shown to play a prominent role in this form of dementia. Finally, the power to detect disease associations with less commonly prescribed drugs was limited. To lessen the multiple testing burden by eliminating very rarely prescribed drugs, we excluded medications prescribed to fewer than 100 participants.
Using a hierarchical modelling analysis approach, we found that the LBD risk was lowered not due to a specific cardiovascular risk management drug class (e.g. beta-blockers within the anti-hypertensive drug group) but by all medication classes. This observation supports the veracity of our claims and implies that the observed beneficial effect is not mediated through a secondary effect of a particular treatment subgroup.
Conclusion
Medications treating hypercholesterolaemia, diabetes mellitus and hypertension were associated with lower LBD risk in the US elderly population. Furthermore, based on genomic evaluations, LBD causally shares risk with cardiovascular traits (LDL cholesterol, Type 2 diabetes and hypertension). Our results highlight the utility of combining epidemiological data with genomic information to identify drugs that may modify complex neurodegenerative conditions. Taken together, our findings underline the importance of treating cardiovascular risk factors in LBD patients and suggest that clinical trials or large-scale population studies examining the effects of careful risk factor management on disease onset and progression should be prioritized.
Supplementary Material
Acknowledgements
We thank the patients and families whose help and participation made the genomic work possible. We thank the members of the International Lewy Body Dementia Genomics Consortium (a complete list of site investigators who contributed to the generation of genome sequence data can be found in the Supplementary Materials). We thank the members of the Laboratory of Neurogenetics (NIH) for their collegial support and technical assistance. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). Data used in the preparation of this article were obtained from the AMP PD Knowledge Platform. For up-to-date information on the study, see https://amp-pd.org. AMP PD—a public–private partnership—is managed by the FNIH and funded by Celgene, GSK, the Michael J. Fox Foundation for Parkinson’s Research, the National Institute of Neurological Disorders and Stroke, Pfizer and Verily. Clinical data and biosamples used in the preparation of this article were obtained from the Fox Investigation for New Discovery of Biomarkers (BioFIND), the Harvard Biomarker Study (HBS), the Parkinson’s Progression Markers Initiative (PPMI) and the Parkinson’s Disease Biomarkers Program (PDBP). BioFIND is sponsored by The Michael J. Fox Foundation for Parkinson’s Research (MJFF) with support from the National Institute of Neurological Disorders and Stroke (NINDS). The BioFIND Investigators have not participated in reviewing data analysis or content of the manuscript. For up-to-date information on the study, visit michaeljfox.org/biofind. Harvard NeuroDiscovery Biomarker Study (HBS) is a collaboration of HBS investigators and funded through philanthropy and NIH and non-NIH funding sources. The HBS investigators have not participated in reviewing the data analysis or content of the manuscript. PPMI—a public–private partnership—is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners. The full names of all of the PPMI funding partners can be found at www.ppmi-info.org/fundingpartners. The PPMI investigators have not participated in reviewing the data analysis or content of the manuscript. For up-to-date information on the study, visit www.ppmi-info.org. Parkinson’s Disease Biomarker Program (PDBP) consortium is supported by the NINDS at the National Institutes of Health. A full list of PDBP investigators can be found at https://pdbp.ninds.nih.gov/policy. The PDBP investigators have not participated in reviewing the data analysis or content of the manuscript.
Appendix I
The American Genome Center
Clifton L. Dalgard, Adelani Adeleye, Anthony R. Soltis, Camille Alba, Coralie Viollet, Dagmar Bacikova, Daniel N. Hupalo, Gauthaman Sukumar, Harvey B. Pollard, Matthew D. Wilkerson and Elisa McGrath Martinez.
Appendix II
International Lewy Body Dementia Genomics Consortium
Sandra E. Black, Ziv Gan-Or, Julia Keith, Mario Masellis, Ekaterina Rogaeva, Alexis Brice, Suzanne Lesage, Georgia Xiromerisiou, Andrea Calvo, Antonio Canosa, Adriano Chio, Giancarlo Logroscino, Gabriele Mora, Reijko Krüger, Patrick May, Daniel Alcolea, Jordi Clarimon, Juan Fortea, Isabel Gonzalez-Aramburu, Jon Infante, Carmen Lage, Alberto Lleó, Pau Pastor, Pascual Sanchez-Juan, Francesca Brett, Dag Aarsland, Safa Al-Sarraj, Johannes Attems, Steve Gentleman, John A. Hardy, Angela K. Hodges, Seth Love, Ian G. McKeith, Christopher M. Morris, Huw R. Morris, Laura Palmer, Stuart Pickering-Brown, Mina Ryten, Alan J. Thomas, Claire Troakes, Marilyn S. Albert, Matthew J. Barrett, Thomas G. Beach, Lynn M. Bekris, David A. Bennett, Bradley F. Boeve, Clifton L. Dalgard, Ted M. Dawson, Dennis W. Dickson, Tanis Ferman, Luigi Ferrucci, Margaret E. Flanagan, Tatiana M. Foroud, Bernardino Ghetti, J. Raphael Gibbs, Alison Goate, David S. Goldstein, Neill R. Graff-Radford, Kejal Kantarci, Horacio Kaufmann, Walter A. Kukull, James B. Leverenz, Qinwen Mao, Eliezer Masliah, Edwin Monuki, Kathy L. Newell, Jose-Alberto Palma, Matthew Perkins, Olga Pletnikova, Alan E. Renton, Susan M. Resnick, Liana S. Rosenthal, Owen A. Ross, Clemens R. Scherzer, Geidy E. Serrano, Vikram G. Shakkottai, Ellen Sidransky, Toshiko Tanaka, Eric Topol, Ali Torkamani, Bryan J. Traynor, Juan C. Troncoso, Randy Woltjer, Zbigniew K. Wszolek and Sonja W. Scholz.
Contributor Information
Sonja W Scholz, Department of Neurology, Johns Hopkins University Medical Center, Baltimore, MD 21287, USA; Neurodegenerative Diseases Research Unit, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD 20892, USA.
Brian E Moroz, Biostatistics Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Sara Saez-Atienzar, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA.
Ruth Chia, Laboratory of Neurogenetics, National Institute on Aging, National Institutes of Health, Bethesda, MD 20814, USA.
Elizabeth K Cahoon, Biostatistics Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Clifton L Dalgard, Department of Anatomy, Physiology and Genetics, Uniformed Services University of the Health Sciences, Bethesda, MD 20814, USA; The American Genome Center, Collaborative Health Initiative Research Program, Uniformed Services University of the Health Sciences, Bethesda, MD 20814, USA.
Daryl Michal Freedman, Biostatistics Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Ruth M Pfeiffer, Biostatistics Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
The American Genome Center:
Clifton L Dalgard, Adelani Adeleye, Anthony R Soltis, Camille Alba, Coralie Viollet, Dagmar Bacikova, Daniel N Hupalo, Gauthaman Sukumar, Harvey B Pollard, Matthew D Wilkerson, and Elisa McGrath Martinez
International Lewy Body Dementia Genomics Consortium:
Sandra E Black, Ziv Gan-Or, Julia Keith, Mario Masellis, Ekaterina Rogaeva, Alexis Brice, Suzanne Lesage, Georgia Xiromerisiou, Andrea Calvo, Antonio Canosa, Adriano Chio, Giancarlo Logroscino, Gabriele Mora, Reijko Krüger, Patrick May, Daniel Alcolea, Jordi Clarimon, Juan Fortea, Isabel Gonzalez-Aramburu, Jon Infante, Carmen Lage, Alberto Lleó, Pau Pastor, Pascual Sanchez-Juan, Francesca Brett, Dag Aarsland, Safa Al-Sarraj, Johannes Attems, Steve Gentleman, John A Hardy, Angela K Hodges, Seth Love, Ian G McKeith, Christopher M Morris, Huw R Morris, Laura Palmer, Stuart Pickering-Brown, Mina Ryten, Alan J Thomas, Claire Troakes, Marilyn S Albert, Matthew J Barrett, Thomas G Beach, Lynn M Bekris, David A Bennett, Bradley F Boeve, Clifton L Dalgard, Ted M Dawson, Dennis W Dickson, Tanis Ferman, Luigi Ferrucci, Margaret E Flanagan, Tatiana M Foroud, Bernardino Ghetti, J Raphael Gibbs, Alison Goate, David S Goldstein, Neill R Graff-Radford, Kejal Kantarci, Horacio Kaufmann, Walter A Kukull, James B Leverenz, Qinwen Mao, Eliezer Masliah, Edwin Monuki, Kathy L Newell, Jose-Alberto Palma, Matthew Perkins, Olga Pletnikova, Alan E Renton, Susan M Resnick, Liana S Rosenthal, Owen A Ross, Clemens R Scherzer, Geidy E Serrano, Vikram G Shakkottai, Ellen Sidransky, Toshiko Tanaka, Eric Topol, Ali Torkamani, Bryan J Traynor, Juan C Troncoso, Randy Woltjer, Zbigniew K Wszolek, and Sonja W Scholz
Supplementary material
Supplementary material is available at Brain Communications online.
Author contributions
R.M.P. and S.W.S.: conceptualization and design. R.M.P., S.W.S. and E.K.C.: supervision. B.E.M. and R.C.: data curation. R.M.P., S.S.-A., R.C., B.E.M., D.M.F. and S.W.S.: formal analysis. S.W.S.: writing—original draft; all authors: writing—review and editing.
Funding
This research was supported by the Intramural Research Program of the US National Institutes of Health (National Cancer Institute, National Institute on Aging and National Institute of Neurological Disorders and Stroke; program #: 1ZIANS003154). B.E.M. was supported by a contract between the National Cancer Institute and Computing & Software Solutions for Science, LLC.
Competing interests
S.W.S. is a member of the Scientific Advisory Council of the Lewy Body Dementia Association and the Multiple System Atrophy Coalition. S.W.S. receives research support from Cerevel Therapeutics. S.W.S. is an editorial board member of JAMA Neurology and the Journal of Parkinson’s Disease. All other authors declare no competing interests.
Data availability
The LBD GWAS summary statistics are publicly available at https://www.ebi.ac.uk/gwas/. The summary statistics of the published genome-wide association studies (GWAS) on LDL cholesterol (study identifier: ieu-b-110), Type 2 diabetes (ebi-a-GCST006867) and essential (primary) hypertension (ukb-b-14177) were obtained from the MRC IEU Open GWAS Project (https://gwas.mrcieu.ac.uk/). To protect the privacy of human subjects, Medicare data are not publicly available; however, data access can be requested by qualified investigators at the Centers for Medicare & Medicaid Services (https://cms.gov). All data needed to evaluate the conclusions in the paper can be found in the paper and the Supplementary Materials.
References
- 1. McKeith IG, Boeve BF, Dickson DW, et al. Diagnosis and management of dementia with Lewy bodies: Fourth consensus report of the DLB Consortium. Neurology. 2017;89(1):88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chia R, Sabir MS, Bandres-Ciga S, et al. Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat Genet. 2021;53(3):294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. De Jonge KE, Jamshed N, Gilden D, Kubisiak J, Bruce SR, Taler G. Effects of home-based primary care on Medicare costs in high-risk elders. J Am Geriatr Soc. 2014;62(10):1825–1831. [DOI] [PubMed] [Google Scholar]
- 4. Eicheldinger C, Bonito A. More accurate racial and ethnic codes for Medicare administrative data. Health Care Financ Rev. 2008;29(3):27–42. [PMC free article] [PubMed] [Google Scholar]
- 5. Pfeiffer RM, Mayer B, Kuncl RW, et al. Identifying potential targets for prevention and treatment of amyotrophic lateral sclerosis based on a screen of Medicare prescription drugs. Amyotroph Lateral Scler Frontotemporal Degener. 2020;21(3–4):235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Emre M, Aarsland D, Brown R, et al. Clinical diagnostic criteria for dementia associated with Parkinson’s disease. Mov Disord. 2007;22(12):1689–1707; quiz 1837. [DOI] [PubMed] [Google Scholar]
- 7. Elsworth B, Lyon M, Alexander T, et al. The MRC IEU OpenGWAS data infrastructure. bioRxiv. 10.1101/2020.08.10.244293 [DOI] [Google Scholar]
- 8. Richardson TG, Sanderson E, Palmer TM, et al. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: A multivariable Mendelian randomisation analysis. PLoS Med. 2020;17(3):e1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xue A, Wu Y, Zhu Z, et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat Commun. 2018;9(1):2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frei O, Holland D, Smeland OB, et al. Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat Commun. 2019;10(1):2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Irwin DJ, Hurtig HI. The contribution of tau, amyloid-beta and alpha-synuclein pathology to dementia in Lewy body disorders. J Alzheimers Dis Parkinsonism. 2018;8(4):444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Norton S, Matthews FE, Barnes DE, Yaffe K, Brayne C. Potential for primary prevention of Alzheimer’s disease: An analysis of population-based data. Lancet Neurol. 2014;13(8):788–794. [DOI] [PubMed] [Google Scholar]
- 13. Kummer BR, Diaz I, Wu X, et al. Associations between cerebrovascular risk factors and Parkinson disease. Ann Neurol. 2019;86(4):572–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hughes D, Judge C, Murphy R, et al. Association of blood pressure lowering with incident dementia or cognitive impairment: A systematic review and meta-analysis. JAMA. 2020;323(19):1934–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Whitmer RA, Sidney S, Selby J, Johnston SC, Yaffe K. Midlife cardiovascular risk factors and risk of dementia in late life. Neurology. 2005;64(2):277–281. [DOI] [PubMed] [Google Scholar]
- 16. Wu YT, Beiser AS, Breteler MMB, et al. The changing prevalence and incidence of dementia over time—Current evidence. Nat Rev Neurol. 2017;13(6):327–339. [DOI] [PubMed] [Google Scholar]
- 17. Brauer R, Wei L, Ma T, et al. Diabetes medications and risk of Parkinson’s disease: A cohort study of patients with diabetes. Brain. 2020;143(10):3067–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chohan H, Senkevich K, Patel RK, et al. Type 2 diabetes as a determinant of Parkinson’s disease risk and progression. Mov Disord. 2021;36(6):1420–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. De Pablo-Fernandez E, Goldacre R, Pakpoor J, Noyce AJ, Warner TT. Association between diabetes and subsequent Parkinson disease: A record-linkage cohort study. Neurology. 2018;91(2):e139–e142. [DOI] [PubMed] [Google Scholar]
- 20. United States Administration on Aging. American Association of Retired Persons . A profile of older Americans. US Dept of Health and Human Services, Administration on Aging; 2017. [Google Scholar]
- 21. Graff-Radford NR, Besser LM, Crook JE, Kukull WA, Dickson DW. Neuropathologic differences by race from the National Alzheimer’s Coordinating Center. Alzheimers Dement. 2016;12(6):669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hou CE, Yaffe K, Perez-Stable EJ, Miller BL. Frequency of dementia etiologies in four ethnic groups. Dement Geriatr Cogn Disord. 2006;22(1):42–47. [DOI] [PubMed] [Google Scholar]
- 23. Kurasz AM, Smith GE, McFarland MG, Armstrong MJ. Ethnoracial differences in Lewy body diseases with cognitive impairment. J Alzheimers Dis. 2020;77(1):165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The LBD GWAS summary statistics are publicly available at https://www.ebi.ac.uk/gwas/. The summary statistics of the published genome-wide association studies (GWAS) on LDL cholesterol (study identifier: ieu-b-110), Type 2 diabetes (ebi-a-GCST006867) and essential (primary) hypertension (ukb-b-14177) were obtained from the MRC IEU Open GWAS Project (https://gwas.mrcieu.ac.uk/). To protect the privacy of human subjects, Medicare data are not publicly available; however, data access can be requested by qualified investigators at the Centers for Medicare & Medicaid Services (https://cms.gov). All data needed to evaluate the conclusions in the paper can be found in the paper and the Supplementary Materials.