Abstract
We have previously reported the isolation and characterization of two filamentous bacteriophages of Vibrio parahaemolyticus, designated Vf12 and Vf33. In this study, to understand the potential of these phages as tools for genetic transmission, we investigated the gene structures of replicative-form (RF) DNAs of their genomes and the distribution of these DNAs on chromosomal and extrachromosomal DNAs. The 7,965-bp nucleotide sequences of Vf12 and Vf33 were determined. An analysis of the overall gene structures revealed that Vf12 and Vf33 had conserved regions and distinctive regions. The gene organization of their conserved regions was similar to that of CTX phage of Vibrio cholerae and coliphage Ff of Escherichia coli, while their distinctive regions were characteristic of Vf12 and Vf33 phage genomes. Southern blot hybridization testing revealed that the filamentous phage genomes integrated into chromosomal DNA of V. parahaemolyticus at the distinctive region of the phage genome and were also distributed on some plasmids of V. parahaemolyticus and total cellular DNAs of one Vibrio damsela and one nonagglutinable Vibrio strain tested. These results strongly suggest the possibilities of genetic interaction among the bacteriophage Vf12 and Vf33 genomes and chromosomal and plasmid-borne DNAs of V. parahaemolyticus strains and of genetic transmission among strains through these filamentous phages.
Vibrio parahaemolyticus is a halophilic marine vibrio which causes gastroenteritis in humans by seafood consumption (4, 11). Although its pathogenic mechanism is not exactly understood, the thermostable direct hemolysin (TDH) and the TDH-related hemolysin have been considered to be its important virulence factors (26, 28). The tdh gene has many variants, and these have been found on plasmid DNAs and chromosomal DNAs (1, 20) and also in other Vibrio strains (10, 36–38). Therefore, it can be assumed that some accessory genetic elements (chromosomal islands, plasmids, bacteriophages, and transposons) can move the genes horizontally as well as vertically through species, clones, chromosomal DNAs, and plasmids. Terai et al. reported the possibility of the genetic transfer of the tdh and tdh-like genes by a transposon-like unit whose transposase activity from insertion sequences has been lost (30).
Nakanishi et al. (18) originally reported on the filamentous phage v6 of V. parahaemolyticus, but they did not carry out a genetic or physiological analysis of this bacteriophage. To find a possible clue to the mystery of V. parahaemolyticus, we previously tried to isolate extrachromosomal elements from 37 strains and revealed that 9 of those strains possessed these elements and that two of the elements were the replicative-form (RF) DNAs of the filamentous phages (29). We designated these two phages Vf12 and Vf33. These filamentous phages are approximately 1,400 nm in length and 7 nm in width and possess one single-stranded circular DNA genome approximately 8.4 kb in size. However, we could not find an association between the possession of the extrachromosomal elements and the Kanagawa phenomenon (beta-hemolytic on Wagatsuma agar medium).
Filamentous bacteriophages have been divided into two classes (16, 17). Class I includes the Escherichia coli phages M13, fd, f1, If1, and IKe, whereas class II includes the phages Pf1 and Pf3, which infect Pseudomonas aeruginosa, and phage Xf, which infects Xanthomonas oryzae. They generally possess a circular, single-stranded DNA genome and can exist in an infected host cell as a double-stranded RF DNA which can be isolated as an extrachromosomal plasmid. Some of them can integrate into chromosomal DNA (12, 35). Infected bacteria continue to produce phage particles for considerable periods without lysis (19). Therefore, it is logical to assume that they might play an important role in horizontal genetic transmission in the same mode as a lambdoid phage carrying the verocytotoxin gene does (5).
Recently, a filamentous phage of Vibrio cholerae, CTX phage, was reported to be a genetic mechanism for the transmission of the cholera toxin gene cluster (ctxAB) (34). This phage integrates into chromosomal DNA via the attRS attachment site or otherwise replicates as a plasmid in strains lacking the attRS site. Since ctxAB is part of the CTX phage structure, this phage can transmit ctxAB horizontally from toxigenic to nontoxigenic V. cholerae strains. Since that study was published, filamentous bacteriophages designated VSK (12), fs1 (7), and fs2 (7) have been isolated from V. cholerae O139. VSK could also integrate into the chromosome, forming a lysogen.
In this study, to assess the possible association between the filamentous phages Vf12 and Vf33 and the mystery of the Kanagawa phenomenon of V. parahaemolyticus, we analyzed the gene structures and the distribution among Vibrio species of Vf12 and Vf33. Although no tdh or trh gene was detected on the two filamentous phage genomes, the phage genome integrated into the chromosomal DNAs of host cells and also into extrachromosomal DNA and other Vibrio species. The results strongly suggested that Vf12 and Vf33 phage genomes could interact with plasmid-borne and chromosomal DNAs of host cells and could play a role in a dynamic mobilization of the pathogenic genes of V. parahaemolyticus by the filamentous phages.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains of the genus Vibrio used in this study are described in Table 1. E. coli K-12 XL1-Blue was used as the host strain for the recombinant plasmid DNA. Luria-Bertani broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 0.1% glucose) was used for the plasmid preparation. Nutrient agar (Nissui Seiyaku, Tokyo, Japan) supplemented with 1.0% NaCl (final concentration of NaCl, 1.5%) was used for the solid culture of V. parahaemolyticus. Luria-Bertani agar plates and 2YT agar (1.6% tryptone, 1.0% yeast extract, 0.5% NaCl, 1.5% agar) plates were used for culture of E. coli strains. Plasmids pUC119 (ampicillin resistant) and pZErO-2.1 (kanamycin resistant; Invitrogen Corporation, San Diego, Calif.) were used as vectors. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml, and kanamycin, 50 μg/ml.
TABLE 1.
Vibrio strains used for gene distribution analysis and examined by Southern blot hybridization
| Strain (n) | Serotype | Reference or source |
|---|---|---|
| V. parahaemolyticus | ||
| Vp2, Vp25, Vp29 | K6 | 29 |
| Vp12, Vp33 | K12 | 29 |
| Vp1 | K13 | 29 |
| Vp26, Vp34 | K64 | 29 |
| Wp1 (TDH+) | O4:K12 | Clinical isolate |
| Wp28 (TDH−) | Untypeable | Clinical isolate |
| V. fluvialis (1) | Clinical isolate | |
| V. damsela (1) | Clinical isolate | |
| NAG Vibrio (3) | Food | |
| V. cholerae (4) | Food | |
| V. vulnificus (2) | Clinical isolate | |
| V. hollisae (2) | Clinical isolate |
Isolation and cloning of RF DNAs of bacteriophages Vf12 and Vf33.
RF DNAs of Vf12 and Vf33 were isolated from V. parahaemolyticus Vp12 and Vp33, respectively, by the alkaline lysis method of Birnboim and Doly (3). To determine their nucleotide sequences, RF DNAs of Vf12 and Vf33 were digested with the restriction enzymes EcoRI, EcoRV, HincII, HindIII, KpnI, and PstI (see Fig. 1A) and the digested fragments were cloned into the plasmid vector pUC119 or pZErO-2.1. The recombinant clones were used to transform E. coli K-12 XL1-Blue and were selected by resistance to ampicillin (100 μg/ml) or kanamycin (50 μg/ml). All of the restriction enzymes were purchased from TaKaRa Shuzo Co., Ltd. (Kyoto, Japan).
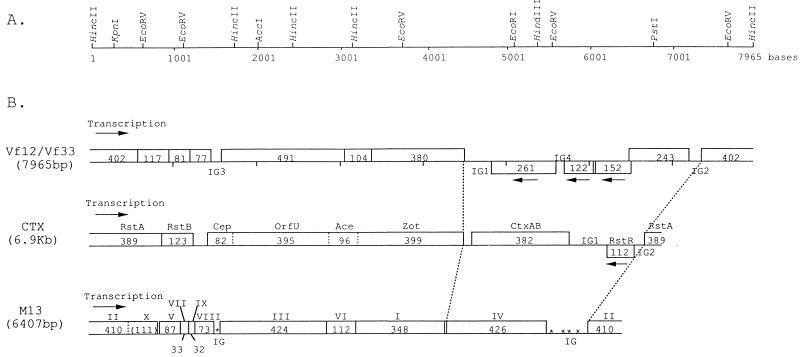
FIG. 1.
Gene structures of bacteriophage Vf12 and Vf33 genomes. (A) Restriction enzyme cleavage map. The circular phage genome is represented in a linear form with the HincII site as point zero and is numbered in the 5′-to-3′ direction of the viral strand. (B) Comparison of the linear ORF maps of filamentous phages Vf12 and Vf33, CTX of V. cholerae, and M13 of E. coli. ORFs are represented as blocks. The numbers in the blocks refer to the number of predicted amino acids, and arrows indicate the transcription directions of genes. The genetic map of CTX phage was designed according to the work of Waldor and Mekalanos (34) and Waldor et al. (35). The genetic map of M13 phage was designed according to the work of Van Wezenbeek et al. (33).
PCR amplification.
RF DNAs (1 to 20 ng) of Vf12 and Vf33 were amplified in a 100-μl reaction mixture containing 200 μM (each) dATP, dTTP, dCTP, and dGTP; 1.0 μM each primer; 2.5 U of Taq DNA polymerase (TaKaRa Shuzo, Shiga, Japan); 10 mM Tris-HCl (pH 8.3); 50 mM KCl; and 1.5 mM MgCl2. By using Program Temp Control System PC-700 (Astec Co., Ltd., Kyoto, Japan), PCR amplifications were initially denatured at 95°C for 3 min and then subjected to 30 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 74°C for 1 min. Oligonucleotide primers used for PCR were purchased from Greiner Japan Co., Ltd. (Kyoto, Japan).
Nucleotide sequencing of the cloned fragments and the PCR products.
Nucleotide sequencing of Vf12 and Vf33 was carried out by using the cloned fragments and PCR products. Initially, the nucleotide sequences of both terminal regions of the cloned fragments were determined by using a fluorescein-labeled M13 universal primer (5′-TGTAAAACGACGGCCAGT-3′) and an M13 reverse primer (5′-CAGGAAACAGCTATGACC-3′) with Dye Primer Cycle Sequencing FS Ready Reaction kits (Perkin-Elmer Japan Co., Ltd., Tokyo, Japan). Next, the nucleotide sequences of the middle regions and each of the connected portions of the cloned fragments were determined by amplifying RF DNAs of Vf12 and Vf33 with synthesized primers. PCR products were sequenced with a TaKaRa Taq Cycle Sequencing core kit (TaKaRa Shuzo Co., Ltd., Kyoto, Japan) and Dye Terminator Cycle Sequencing FS Ready Reaction kits. The nucleotide sequences were analyzed with an ABI 373S DNA sequencer (Perkin-Elmer Japan Co., Ltd., Tokyo, Japan). The MacGenetyx and BLAST Search (1) programs were used for analyzing and searching for homology, and the DNASIS program (Hitachi Software Engineering Co., Ltd., Yokohama, Japan) was used to determine G+C contents.
DNA probes and Southern hybridization.
To determine the distribution of the bacteriophage genomes on chromosomal and extrachromosomal DNAs of V. parahaemolyticus and of other Vibrio strains, Southern hybridization tests were carried out. Total cellular DNAs of Vibrio strains were extracted by the method of Saito and Miura (23). Chromosomal and extrachromosomal DNAs digested or not digested with EcoRV enzyme were electrophoresed on a 1% agarose gel (Agarose ME; Nakarai Chemicals Ltd.), transferred to a nylon membrane (Hybond-N; Amersham Japan, Ltd., Tokyo, Japan) and hybridized with probes under stringent conditions as described by Southern (27). The nine fragments of Vf33 RF DNAs presented in Fig. 3 were purified from an agarose gel and labeled with digoxigenin. The prehybridization, hybridization, and chemiluminescent detection of the nylon membrane blots were done as recommended by the manufacturer (Boehringer GmbH, Mannheim, Germany) with a DIG DNA labeling and detection kit.
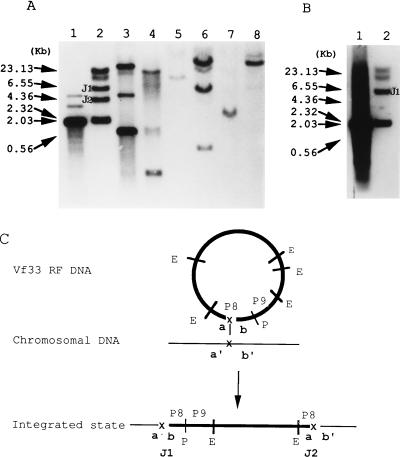
FIG. 3.
Hybridization patterns of the P8 (A) and P9 (B) probes and the mode of integration of the Vf12 and Vf33 genomes (C). (A and B) Vf12 and Vf33 phage genomes are present as the RF and concomitantly integrated into the chromosome. Numbers on the left are the sizes of λ HindIII markers. RF DNAs or total cellular DNAs digested with EcoRV were analyzed by agarose gel electrophoresis, transferred to nylon membranes, and hybridized with the labeled fragments P8 (A) and P9 (B). The chromosomal junction fragments of the Vp33 strain are labeled J1 and J2. (A) Lanes: 1, purified RF DNA of Vf33; 2, total cellular DNA of the Vp33 strain; 3, purified pVp25 DNA; 4, total cellular DNA of the Vp25 strain; 5, total cellular DNA of the Wp1 strain; 6, total cellular DNA of the Wp28 strain; 7, total cellular DNA of V. damsela; 8, total cellular DNA of NAG-Vibrio. (B) Lanes: 1, purified RF DNA of Vf33; 2, total cellular DNA of the Vp33 strain. The fragment labeled J1 has the same size as J1 of panel A, lane 2. (C) The phage genome is represented in a thick circular form, and that of the chromosome is in a thin linear form. P8 and P9 indicate the probes used in panels A and B, respectively. E and P indicate the EcoRV and PstI enzyme sites of the phage genome, respectively. a and b and a′ and b′ indicate the attachment sites of the phage genome and chromosome, respectively. J1 and J2 indicate the junction fragments that are the same as those of panels A and B.
Nucleotide sequence accession numbers.
The nucleotide sequence data for the RF DNAs of Vf12 and Vf33 appear in the DDBJ, EMBJ, and GenBank nucleotide sequence databases with the accession no. AB012574 and AB012573, respectively.
RESULTS
Nucleotide sequencing.
We determined the nucleotide sequences of Vf12 and Vf33 RF DNAs. Both RF DNAs consisted of 7,965 bp.
The entire nucleotide sequences of the genomes of bacteriophages Vf12 and Vf33 were determined by sequencing both the recombinant fragments and PCR products as described in Materials and Methods. All of the nucleotide sequences of the Vf12 and Vf33 genomes were determined in both directions on overlapping DNA fragments. There was only one base difference between the 7,965-nucleotide sequences of the Vf12 and Vf33 genomes. This nucleotide change occurred within the third position of a codon and did not affect the predicted amino acid sequence. We could also find a novel HincII fragment of approximately 703 bp (Fig. 1A). This fragment possessed one AccI site, whereas the other HincII fragment, which was 719 bp in size, did not possess an AccI site. We digested the RF DNA of Vf33 with the HincII enzyme alone or with a combination of the HincII and AccI enzymes and could confirm the existence of the novel HincII fragments.
Gene structure.
Gene structures of Vf12 and Vf33 genomes are presented in Fig. 1B. A computer analysis of the nucleotide sequences of the Vf12 and Vf33 genomes revealed 11 potential open reading frames (ORFs) and four apparently untranslated intergenic regions (IGs) (Fig. 1B). On the basis of their numbers of amino acids, all of the ORFs were designated VPFs (V. parahaemolyticus filamentous phage). Eight VPF ORFs (vpf243, vpf402, vpf117, vpf81, vpf77, vpf491, vpf104, and vpf380) were predicted to be transcribed in one direction, whereas three VPF ORFs (vpf261, vpf122, and vpf152) were predicted to be transcribed in the opposite direction.
The organization and the amino acid numbers of seven VPF ORFs and one IG (vpf402, vpf117, vpf81, vpf77, IG3, vpf491, vpf104, and vpf380) were similar to those of six genes and one IG of CTX phage (rstA, rstB, IG, cep, orfU, ace, and zot) of V. cholerae (34, 35) and seven genes and one IG of M13 phage (genes II/X, V, VIII, III, VI, and I and IG, with IG located between genes VIII and III) of E. coli (33). Therefore, we called the region the conserved region. However, Vf12 and Vf33 phages, like CTX phage, lack a gene corresponding to gene IX of M13 phage, which produces a minor protein at the leading end of the virion.
On the other hand, the region from nucleotide positions 4514 to 7212 included three IGs and four genes (IG1, vpf261, IG4, vpf122, vpf152, vpf243, and IG2). In the corresponding region, M13 phage harbored only gene IV, which encodes an assembly protein, and CTX phage harbored the ctxAB gene, which encodes cholera toxins A and B, IG1, the rstR gene, which encodes the repressor of the expression of the rstA gene, and IG2. Therefore, we called this region of Vf12 and Vf33 the distinctive region.
Amino acid homology search of each VPF.
To assess the potential function of the product encoded by each VPF of the Vf12 and Vf33 genomes, database searches for proteins similar to the predicted Vf12 and Vf33 polypeptides were carried out. Table 2 shows ratios of amino acid homology with CTX and M13 phages and the function of each protein of CTX and M13 phages.
TABLE 2.
Comparison of ORFs between filamentous bacteriophages Vf12 and Vf33 and CTX or M13
| ORF of Vf12 or Vf33a | CTX phage of V. choleraeb
|
M13 phage of E. colic
|
||||||
|---|---|---|---|---|---|---|---|---|
| ORFd | % (ratio) of homologye
|
Function(s) or nature of homologous proteins | ORF | % (ratio) of homology
|
Functions or nature of homologous proteins | |||
| Identity | Similarity | Identity | Similarity | |||||
| vpf402 | rstA (389) | 39.2 (122/311) | 74.6 (232/311) | Replication, integration? | Gene IIf (410) | Initiates rolling-circle replication; nicks and seals DNA at a specific site | ||
| vpf117 | rstB (123) | 12.9 (11/85) | 47.1 (40/85) | Integration | ||||
| vpf81 | Gene Vf (87) | Single-stranded-DNA-binding protein | ||||||
| vpf77 | cep (82) | 19.5 (15/77) | 63.6 (49/77) | Core DNA-encoded pilin | Gene VIII (73) | 22.1 (17/77) | 62.3 (48/77) | Major coat protein |
| vpf491 | orfUf (395) | Structural protein of virion | Gene IIIf (424) | Minor protein at leading end of virion | ||||
| vpf104 | ace (96) | 26.0 (25/96) | 61.5 (59/96) | Increase short-circuit current or serve as a minor protein | Gene VI (112) | 15.6 (15/96) | 67.7 (65/96) | Minor protein at leading end of virion |
| vpf380 | zot (399) | 23.0 (83/361) | 64.0 (231/361) | Affect intercellular tight junctions or serve as an assembly protein | Gene I (348) | 4.2 (15/361) | 10.2 (37/361) | Assembly protein |
| vpf261 | ||||||||
| vpf122 | rstR (112) | 17.6 (15/85) | 54.1 (52/85) | Regulator, repressor | ||||
| vpf152 | ||||||||
| vpf243 | ||||||||
The number in each VPF ORF designation indicates the number of amino acids of the product of each predicted ORF.
Summarized from the data of Fasano et al. (8), Pearson et al. (22), Trucksis et al. (31), and Waldor et al. (35).
Summarized from the data of Van Wezenbeek et al. (33).
Numbers in parentheses are numbers of amino acids of the product.
Number of identical or similar amino acids in the product/total number of amino acids in the product of the homologous region. The levels of homology were determined with the MaxGenetyx program.
The ORF is a match in size and location, but its product shows no homology to the amino acid sequence of the product of the ORF of Vf12 or Vf33.
The amino acid sequence of vpf402 showed a high similarity (BLAST score = 190) to that of RstA of CTX phage. This product is required for CTX phage replication and possibly integration as well (35). The vpf402 product was also similar to the proteins encoded by orf166 and orf208 of fs1, the filamentous phage of V. cholerae O139 (7), but did not have significant homology with gene II of M13 phage. vpf117 matched closely rstB of CTX phage in size and location, and the predicted proteins also showed 47.1% similarity. A homologous gene is not present in M13 phage. RstB is required for integration of CTX phage (35). By amino acid homology search, it was seen that the vpf81 product was similar to the TraK protein of conjugative plasmid IncP-Beta RP4 of E. coli, which is the single-stranded-DNA-binding protein with a transfer origin (39). vpf81 matched closely in size and location gene V of M13 phage, which encodes the single-stranded-DNA-binding protein (25), but no homology was revealed between the proteins. These VPFs might be required for Vf12 and Vf33 phage DNA replication and integration.
vpf77 corresponded in size and location to cep of CTX phage, which encodes the core-encoded pilin, and gene VIII of M13 phage, which encodes the major coat protein. Like Cep, the NH2 terminus of Vpf77 possesses a hydrophobic signal sequence (22, 34). vpf491, located immediately downstream from vpf77, revealed no significant similarity with any protein. However, the vpf491 product corresponded in size and location to orfU of CTX phage and gene III of M13 phage (31, 34). Vpf491 might be the virion protein necessary for participation in receptor binding, because OrfU of CTX phage and gene III of M13 phage were the minor proteins at the leading end of the virion adsorbing the receptor of the host cell. Perhaps there is great diversity in the structures of these products because the pilus receptors for different filamentous phages vary widely between bacterial species (34). Vpf104 was similar (BLAST score = 55) to Ace (accessory cholera enterotoxin) of CTX phage, Orf93 of bacteriophage Pf3 of P. aeruginosa (15), and the product of gene VI of M13 phage (33). Ace, which is capable of altering cellular ion fluxes, increases the short-circuit current in Ussing chambers and causes fluid secretion in rabbit-ligated ileal loops (31). On the other hand, the ace gene product was similar to the gene VI product of M13 phage, which is the minor protein at the leading end of the virion (33, 34). Vpf380 had significantly more similarity (BLAST score = 103) to Zot (zonula occludens toxin) of V. cholerae (8) than to a family of proteins including the product of gene I of Ff phages of E. coli (2, 33) and the corresponding product of gene I of the filamentous phages Pf3 and Pf1 of P. aeruginosa (9, 15). Zot increases the permeability of the small intestinal mucosa by affecting the structure of the intercellular tight junctions, or zonulae occludens. The zot product was similar to the gene I product, which might be involved in phage assembly and export (34). These genes might be required for phage morphogenesis.
The Vpf122 product has a similarity to the 8.4-kDa Cro protein of lambdoid phage HK022. The Cro protein of the lambdoid phage is the repressor that regulates transcription (21). vpf122 is also similar to rstR of CTX phage in size and transcriptional direction. rstR encodes a CTX phage repressor (35). Search with the Vpf243, Vpf152, and Vpf261 sequences did not reveal significant homologies with any proteins. These genes were distinctive to Vf12 and Vf33 phages and might play a role in the autonomous replication and regulation of the replicons.
Repeat sequences.
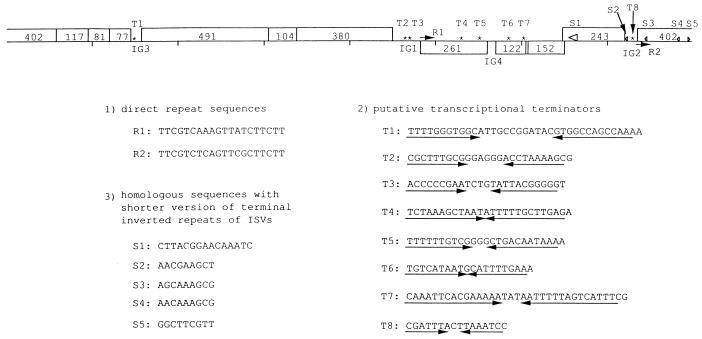
We found three different groups of repeat sequences in the genomes of Vf12 and Vf33 (Fig. 2). They were the sequences of the direct repeats (R1 and R2); the sequences of the inverted repeats (T1 to T8), which were putative transcription terminators; and sequences similar to the shorter 9-bp versions (5′-AACAAATCC-3′ and 5′-GGCTTTGTT-3′) of the 18-bp terminal inverted repeat sequences (5′-GGCTTTGTTGCGTAAATC-3′ and 5′-GATTTACGCAACAAAGCC-3′) of the insertion-like elements (ISVs) which flank the tdh gene (S1 to S5) (30). Many of the repeat sequences were in the region peculiar to Vf12 and Vf33.
FIG. 2.
Repeat sequences of the Vf12 and Vf33 genomes, which consist of (i) direct repeat sequences (R1, R2, and arrows), (ii) inverted repeat sequences (T1 to T8 and asterisks), and (iii) repeated sequences similar to the shorter versions of the 18-bp terminal inverted repeats of the ISVs (S1 to S5). S1 (large open arrowhead) is similar to a part of the inverted repeats (5′-GATTTACGCAACAAAGCC-3′). S2 to S5 indicate the homologous sequences with the shorter versions of the inverted repeats, which are indicated by ◃ (5′-AACAAAGCC-3′) and ▹ (5′-GGCTTTGTT-3′).
Two direct repeat sequences, R1 (5′-TTCGTCAAAGTTATCTTCTT-3′) in the 3′-terminal portion of vpf261 and R2 (5′-TTCGTCTCAGTTCGCTTCTT-3′) in IG2, flanked the distinctive region of the Vf12 and Vf33 genomes. The inverted repeat sequences which form stem-loop structures are assumed to be transcriptional terminators (T1 to T8) (Fig. 2), and many of them are located in the distinctive region and in three IGs: IG1, IG2, and IG3. This localization of terminators suggests that transcription starts upstream from vpf243, vpf402, and vpf491 and terminates downstream from vpf243 (T8 in IG2), vpf77 (T1 in IG3), and vpf380 (T2 and T3 in IG1), respectively. The regulation of transcription in the conserved region seems similar to that of M13 phage. In the distinctive region, putative transcriptional terminators were located in vpf261 (T4 and T5) and vpf122 (T6 and T7). The three genes (vpf261, vpf122, and vpf152) located in the region might be transcribed separately. Furthermore, sequences similar to the shorter versions of the terminal inverted repeat sequences of ISVs were found in vpf243 and vpf402 (S1 to S5) (Fig. 2). We could not find any repeat sequence in IG4.
G+C content.
The G+C content of the complete nucleotide sequences of the Vf12 and Vf33 genomes was 45.7%. The G+C content of the nucleotide sequence of the distinctive region from the 4,780th to the 6,700th position was extremely low, 37%, whereas that of the sequence of the conserved region was 49%.
Southern blot hybridization analysis.
To assess the potential of the filamentous phages Vf12 and Vf33 of being genetic transmitters like CTX phage of V. cholerae, we investigated the integration of the Vf12 and Vf33 genomes into chromosomal DNAs of host cells and their distribution within V. parahaemolyticus and other Vibrio species by Southern hybridization analysis with nine labeled probes (P1 to P9) (see Fig. 4) of Vf33 RF DNA.
FIG. 4.
Distribution of Vf12 and Vf33 genomes in extrachromosomal and chromosomal DNAs of V. parahaemolyticus and total cellular DNAs of other Vibrio species detected by Southern blot hybridization analysis. The cloned fragments used as probes (P1 to P9) are represented by solid lines with two arrowheads. + and − indicate the regions which hybridized and did not hybridize with probes, respectively. (A) Distribution on extrachromosomal DNAs of pVp1, pVp2, pVp25, pVp26, and pVp34. The numbers in parentheses refer to the sizes of the extrachromosomal DNAs. (B) Distribution on total cellular DNAs of V. parahaemolyticus. (C) Distribution on total cellular DNAs of V. damsela and NAG-Vibrio.
At first, to investigate whether Vf12 and Vf33 genomes integrate into the chromosomal DNA of a host cell, total cellular DNA containing Vf33 RF DNA and chromosomal DNA were hybridized with nine labeled probes of Vf33 RF DNA. Undigested chromosomal and Vf33 RF DNAs hybridized with all nine probes (data not shown), suggesting that the Vf33 genome exists also on chromosomal DNA. To elucidate the integration of the Vf33 genome into chromosomal DNA, we searched junction fragments of Vf33 genomic and chromosomal DNA. EcoRV-digested purified Vf33 RF DNA and total cellular DNA hybridized with nine probes, and the hybridizing patterns were compared. When the DNAs were probed with P1 to P7, the hybridizing patterns of total cellular DNAs were identical to those of purified Vf33 RF DNA. However, when the DNAs were probed with P8 and P9, which were located on an EcoRV-digested 2.1-kb fragment of the Vf33 genome, the hybridizing pattern of total cellular DNA revealed additional bands when it was compared to that of Vf33 RF DNA. When hybridized with probe P8, Vf33 RF DNA showed one hybridizing band at 2.1 kb whereas total cellular DNA showed two additional hybridizing bands at 4.4 and 6 kb, which were named J2 and J1, respectively (Fig. 3A, lanes 1 and 2). When hybridized with probe P9, purified Vf33 RF DNA showed one hybridizing band at 2.1 kb whereas total cellular DNA showed one additional hybridizing band at 6 kb, which was identical in size to J1 of the hybridizing band obtained with the P8 probe (Fig. 3B, lanes 1 and 2). These results show that the Vf33 genome integrated into chromosomal DNA of the host cell at the region of P8 and that the region of probe P8 was divided into two terminal portions. Therefore the region of probe P9 is located on one terminal portion (Fig. 3C).
Next, to investigate the distribution on extrachromosomal and chromosomal DNAs of V. parahaemolyticus and other Vibrio species, the extrachromosomal DNAs and total cellular DNAs of all strains (Table 1), digested or not digested with EcoRV, were used for hybridization. Probe P6 was not found on any genetic element tested, except for RF DNAs and chromosomal DNAs of Vf12 and Vf33 (Fig. 4). The other eight probes hybridized in places with some extrachromosomal DNAs (Fig. 4A) and with all of the chromosomal DNAs of V. parahaemolyticus (Fig. 4B). The region of probe P8 hybridized with all chromosomal DNAs of V. parahaemolyticus and also with those of one V. damsela strain and one nonagglutinable-Vibrio strain (NAG-Vibrio strain) (Fig. 3, lanes 4 to 8, and Fig. 4). When probe 8 was hybridized with purified extrachromosomal DNA of pVp25 and chromosomal DNA of the Vp25 strain, there appeared to be more products and the patterns of the purified extrachromosomal and chromosomal DNAs were different (Fig. 3A, lanes 3 and 4). This indicates that sequences similar to those found in Vf12 and Vf33 are integrated into the chromosome of the Vp25 strain. One Vibrio fluvialis strain, two Vibrio hollisae strains, two NAG-vibrio strains, four V. cholerae strains, and two Vibrio vulnificus strains were also tested with these probes, but no bands were detected (data not shown).
DISCUSSION
We determined the nucleotide sequences and analyzed the gene structures of the RF DNAs of Vf12 and Vf33, two filamentous bacteriophages of V. parahaemolyticus. The gene organization and amino acid numbers of the products of VPF ORFs of the region from vpf402 to vpf380 of the Vf12 and Vf33 genomes, designated the conserved region, were similar to those of CTX phage of V. cholerae and those of M13 phage of E. coli. On the other hand, the gene organization and amino acid numbers of the products of VPF ORFs of the region from vpf260 to vpf243, designated the distinctive region, were peculiar to Vf12 and Vf33 phage genomes.
In the conserved region, the amino acid sequences of the products of VPF ORFs were more homologous to those of CTX phage genes than to those of M13 phage genes. The amino acid sequence of the product of vpf380 was homologous to that of the product of zot of CTX phage. Southern hybridization testing indicated that sequences homologous to vpf380 were present in some plasmids (pVp25, pVp26, pVp34, and pVp2) and some chromosomal DNAs of V. parahaemolyticus. It is not yet known whether the vpf380 product is active as a toxin in a manner other than that of an assembly protein like the zot product; however, Zot-like activity has been reported for E. coli (32). The actual functions of the vpf380 product must be further investigated.
In CTX phage, the sole rstA gene could not be subcloned while the fragment containing both the rstA and rstR genes could be. The rstR gene was transcribed divergently from other genes (35). A similar result from subcloning experiments was obtained by subcloning the vpf243 and vpf122 genes of Vf12 and Vf33 phages. That is, the sole vpf243 gene could not be subcloned and the vpf243 gene could be subcloned only when in existence with the vpf122 gene, which is also divergently transcribed like the rstR gene of CTX phage. Therefore, the vpf122 gene product has a function similar to that of the rstR gene product, which is a regulator of CTX phage. The amino acid sequence of the vpf402 gene product, though this gene could be subcloned singly, was homologous to that of the rstA gene product of CTX phage. The vpf243 gene product, which had no homology with any protein in the database, or both the vpf243 and vpf402 products may have a function similar to that of the rstA gene product, which is required for replication of CTX phage.
The region of CTX phage of V. cholerae corresponding to the distinctive region of Vf33 phage possessed both the ctxAB gene cluster, which encodes the cholera toxin, and the divergently transcribed rstR gene described above. The distinctive region of Vf12 and Vf33 phages did not contain the tdh or related genes, whereas it showed some distinctive features. First, the G+C content of this region was extremely low, 37%, compared with those of the remaining region of the Vf33 genome and chromosomal DNA of V. parahaemolyticus. Second, 20-bp-long direct repeat sequences flanked this region (R1 and R2) (Fig. 2). These results suggest the possibility that this region was transmitted from species other than V. parahaemolyticus. Third, this region and adjacent regions had a sequence similar to the shorter 9-bp versions of the 18-bp terminal inverted repeats of the ISVs which flanked the tdh gene (30). Fourth, Southern blot hybridization analysis showed that the P8 fragment was the hot spot for the integration of the Vf33 phage genome into chromosomal DNA of the host cell and that this region was widely spread in V. parahaemolyticus as well as in NAG-Vibrio and V. damsela strains. These findings suggest the ability of the Vf33 phage genome to integrate widely into Vibrio strains which possess the hot-spot portion. By the way, the tdh gene of V. parahaemolyticus has many variants and tdh-like genes were found in other Vibrio species, for example, the NAG-Vibrio V. hollisae. These facts suggest that the filamentous phage Vf33 might play a role in genetic transmission among Vibrio species. Fifth, the region of probe P6 was not detected on any genetic elements. This region was assumed to be required for the development of phages.
Southern blot hybridization analysis showed that a part of the filamentous phage genome was distributed on extrachromosomal DNAs isolated from V. parahaemolyticus strains (Fig. 3A). It is unclear whether these plasmid DNAs are RF DNAs of phages or defective phages of Vf12 and Vf33 phage genomes, like lambda dv phage (24), or whether other plasmids have acquired some parts of Vf12 and Vf33 genomes. We were not able to detect the plaque-forming activities of the culture supernatants of the strains harboring these extrachromosomal elements (29). Furthermore, we have not investigated the receptor of host cells specific to Vf12 and Vf33 phages. Vf12 and Vf33 phages had lytic activity only on the opaque-type colonies of strains with K-38 capsular antigen, and we could not find any extrachromosomal element in the indicator strain that appeared to encode the receptor pilus-like pili encoded by DNA borne on the F plasmid. With V. cholerae, TCP-pilin formation was affected by many environmental factors and was controlled by the ToxRS and ToxT systems (6). Especially, El Tor-type strains hardly ever formed pili in vitro. However, in vivo, the filamentous phages were easily transferred from the classical type strain to the El Tor-type strain (13). A regulation system equivalent to the ToxRS system of V. cholerae has been reported to exist in V. parahaemolyticus (14). It is interesting how these filamentous phages of V. parahaemolyticus strains behave in vivo. To our knowledge a genetic analysis of filamentous bacteriophages of V. parahaemolyticus has never been reported. Although Vf12 and Vf33 genomes do not possess the tdh gene, the results presented in this report strongly suggest the possibility that these filamentous phages transmit horizontally between species and strains or between chromosomal and extrachromosomal DNAs in V. parahaemolyticus. An analysis of these filamentous phages might give a clue to the solution of mysterious issues of V. parahaemolyticus.
ACKNOWLEDGMENTS
We thank Midori Ogawa and Yoshino Kohi for their technical assistance.
REFERENCES
- 1.Baba K, Shirai H, Terai A, Kumagai K, Takeda Y, Nishibuchi M. Similarity of the tdh gene-bearing plasmids of Vibrio cholerae non-O1 and Vibrio parahaemolyticus. Microb Pathog. 1991;10:61–70. doi: 10.1016/0882-4010(91)90066-j. [DOI] [PubMed] [Google Scholar]
- 2.Beck E, Zink B. Nucleotide sequence and genome organization of filamentous bacteriophages f1 and fd. Gene. 1981;16:35–58. doi: 10.1016/0378-1119(81)90059-7. [DOI] [PubMed] [Google Scholar]
- 3.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blake P A, Weaver R E, Hollis D G. Diseases of humans (other than cholera) caused by vibrios. Annu Rev Microbiol. 1980;34:341–367. doi: 10.1146/annurev.mi.34.100180.002013. [DOI] [PubMed] [Google Scholar]
- 5.Datz M, Janetzki-Mittmann C, Franke S, Gunzer F, Schmidt H, Karch H. Analysis of the enterohemorrhagic Escherichia coli O157 DNA region containing lambdoid phage gene p and Shiga-like toxin structural genes. Appl Environ Microbiol. 1996;62:791–797. doi: 10.1128/aem.62.3.791-797.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dirita V J. Co-ordinate expression of virulence genes by ToxR in Vibrio cholerae. Mol Microbiol. 1992;6:451–458. doi: 10.1111/j.1365-2958.1992.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 7.Ehara M, Shimodori S, Kojima F, Ichinose Y, Hirayama T, Albert M J, Supawat K, Honma Y, Iwanaga M, Amako K. Characterization of filamentous phages of Vibrio cholerae O139 and O1. FEMS Microbiol Lett. 1997;154:293–301. doi: 10.1111/j.1574-6968.1997.tb12659.x. [DOI] [PubMed] [Google Scholar]
- 8.Fasano A, Baudry B, Pumplin D W, Wasserman S S, Tall B D, Ketley J M, Kaper J B. Vibrio cholerae produce a second enterotoxin, which affects intestinal tight junctions. Proc Natl Acad Sci USA. 1991;88:5242–5246. doi: 10.1073/pnas.88.12.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill D F, Short N J, Perham R N, Petersen G B. DNA sequence of the filamentous bacteriophage Pf1. J Mol Biol. 1991;218:349–364. doi: 10.1016/0022-2836(91)90717-k. [DOI] [PubMed] [Google Scholar]
- 10.Honda T, Miwatani T. Multi-toxigenicity of Vibrio cholerae non-O1. In: Ohtomo N, Sack R B, editors. Advances in research on cholera and related diarrheas. Vol. 6. Tokyo, Japan: KTK Scientific Publishers; 1988. pp. 23–32. [Google Scholar]
- 11.Janda J M, Powers C, Bryant R G, Abbott S L. Current perspectives on the epidemiology and pathogenesis of clinically significant Vibrio spp. Clin Microbiol Rev. 1988;1:245–267. doi: 10.1128/cmr.1.3.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kar S, Ghosh R K, Ghosh A N, Ghosh A. Integration of the DNA of a novel filamentous bacteriophage VSK from Vibrio cholerae O139 into the host chromosomal DNA. FEMS Microbiol Lett. 1996;145:17–22. doi: 10.1111/j.1574-6968.1996.tb08550.x. [DOI] [PubMed] [Google Scholar]
- 13.Lazar S, Waldor M K. ToxR-independent expression of cholera toxin from the replicative form of CTXφ. Infect Immun. 1998;66:394–397. doi: 10.1128/iai.66.1.394-397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Z, Kumagai K, Baba K, Mekalanos J J, Nishibuchi M. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J Bacteriol. 1993;175:3844–3855. doi: 10.1128/jb.175.12.3844-3855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luiten R G, Putterman D G, Schoenmakers J G, Konings R N, Day L A. Nucleotide sequence of the genome of Pf3, an IncP-1 plasmid-specific filamentous bacteriophage of Pseudomonas aeruginosa. J Virol. 1985;56:268–276. doi: 10.1128/jvi.56.1.268-276.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marvin D A, Wiseman R L, Wachtel E J. Filamentous bacterial viruses. XI. Molecular architecture of the class II (pf1, Xf) virion. J Mol Biol. 1974;82:121–138. doi: 10.1016/0022-2836(74)90336-2. [DOI] [PubMed] [Google Scholar]
- 17.Marvin F J. Filamentous bacterial viruses. XII. Molecular architecture of the class I (fd, If1, IKe) virion. J Mol Biol. 1974;88:581–600. doi: 10.1016/0022-2836(74)90409-4. [DOI] [PubMed] [Google Scholar]
- 18.Nakanishi H, Iida Y, Maeshima K, Teramoto T, Hosaka Y, Ozaki M. Isolation and properties of bacteriophages of Vibrio parahaemolyticus. Biken J. 1966;9:149–157. [Google Scholar]
- 19.Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. p. 2327. [Google Scholar]
- 20.Nishibuchi M, Kaper J B. Duplication and variation of the thermostable direct hemolysin (tdh) gene in Vibrio parahaemolyticus. Mol Microbiol. 1990;4:87–99. doi: 10.1111/j.1365-2958.1990.tb02017.x. [DOI] [PubMed] [Google Scholar]
- 21.Oberto J, Weisberg R A. Structure and function of the nun gene and the immunity region of the lambdoid phage HK022. J Mol Biol. 1989;207:675–693. doi: 10.1016/0022-2836(89)90237-4. [DOI] [PubMed] [Google Scholar]
- 22.Pearson G D, Woods A, Chiang S L, Mekalanos J J. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc Natl Acad Sci USA. 1993;90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito H, Miura K. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 2.1–2.125. [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 4.1–4.20. [Google Scholar]
- 26.Shirai H, Ito H, Hirayama T, Nakabayashi Y, Kumagai K, Takeda Y, Nishibuchi M. Molecular epidemiologic evidence for association of thermostable direct hemolysin (TDH) and TDH-related hemolysin of Vibrio parahaemolyticus with gastroenteritis. Infect Immun. 1990;58:3568–3573. doi: 10.1128/iai.58.11.3568-3573.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 28.Takeda Y. Thermostable direct hemolysin of Vibrio parahaemolyticus. Pharmacol Ther. 1983;19:123–146. doi: 10.1016/0163-7258(82)90044-4. [DOI] [PubMed] [Google Scholar]
- 29.Taniguchi H, Sato K, Ogawa M, Udou T, Mizuguchi Y. Isolation and characterization of a filamentous phage, Vf33, specific for Vibrio parahaemolyticus. Microbiol Immunol. 1984;28:327–337. doi: 10.1111/j.1348-0421.1984.tb00684.x. [DOI] [PubMed] [Google Scholar]
- 30.Terai A, Baba K, Shirai H, Yoshida O, Takeda Y, Nishibuchi M. Evidence for insertion sequence-mediated spread of the thermostable direct hemolysin gene among Vibrio species. J Bacteriol. 1991;173:5036–5046. doi: 10.1128/jb.173.16.5036-5046.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trucksis M, Galen J E, Michalski J, Fasano A, Kaper J B. Accessory cholera enterotoxin (Ace), the third toxin of a Vibrio cholerae virulence cassette. Proc Natl Acad Sci USA. 1993;90:5267–5271. doi: 10.1073/pnas.90.11.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uzzau A, Tugizov S, Goldblum G C, Fiore C, Russel R, Pereira L, Fasano A. Abstracts of the 31st US-Japan Cholera & Related Diarrheal Diseases Conference 1995. 1995. Zonula occludens toxin (ZOT): the archetype of a new family of toxins affecting tight junctions (TJ) p. 203. [Google Scholar]
- 33.Van Wezenbeek P M, Hulsebos T J, Schoenmakers J G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980;11:129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]
- 34.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 35.Waldor M K, Rubin E J, Pearson G D, Kimsey H, Mekalanos J J. Regulation, replication, and integration functions of the Vibrio cholerae CTXφ are encoded by region RS2. Mol Microbiol. 1997;24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]
- 36.Yoh M, Honda T, Miwatani T. Production by non-O1 Vibrio cholerae of hemolysin related to thermostable direct hemolysin of Vibrio parahaemolyticus. FEMS Microbiol Lett. 1985;29:197–200. [Google Scholar]
- 37.Yoh M, Honda T, Miwatani T. Purification and partial characterization of a non-O1 Vibrio cholerae hemolysin that cross-reacts with thermostable direct hemolysin of Vibrio parahaemolyticus. Infect Immun. 1986;52:319–322. doi: 10.1128/iai.52.1.319-322.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoh M, Honda T, Miwatani T. Purification and partial characterization of a Vibrio hollisae hemolysin that relates to thermostable direct hemolysin of Vibrio parahaemolyticus. Can J Microbiol. 1986;32:632–636. doi: 10.1139/m86-118. [DOI] [PubMed] [Google Scholar]
- 39.Ziegelin G, Pansegrau W, Strack B, Balzer D, Kroeger M, Kruft U, Lanka E. Nucleotide sequence and organization of genes flanking the transfer origin of promiscuous plasmid RP4. DNA Sequence. 1991;1:303–327. doi: 10.3109/10425179109020786. [DOI] [PubMed] [Google Scholar]