Abstract
Escherichia coli growing on glucose in minimal medium controls its metabolite pools in response to environmental conditions. The extent of pool changes was followed through two-dimensional thin-layer chromatography of all 14C-glucose labelled compounds extracted from bacteria. The patterns of metabolites and spot intensities detected by phosphorimaging were found to reproducibly differ depending on culture conditions. Clear trends were apparent in the pool sizes of several of the 70 most abundant metabolites extracted from bacteria growing in glucose-limited chemostats at different growth rates. The pools of glutamate, aspartate, trehalose, and adenosine as well as UDP-sugars and putrescine changed markedly. The data on pools observed by two-dimensional thin-layer chromatography were confirmed for amino acids by independent analysis. Other unidentified metabolites also displayed different spot intensities under various conditions, with four trend patterns depending on growth rate. As RpoS controls a number of metabolic genes in response to nutrient limitation, an rpoS mutant was also analyzed for metabolite pools. The mutant had altered metabolite profiles, but only some of the changes at slow growth rates were ascribable to the known control of metabolic genes by RpoS. These results indicate that total metabolite pool (“metabolome”) analysis offers a means of revealing novel aspects of cellular metabolism and global regulation.
Textbooks describing glucose metabolism in Escherichia coli traditionally concentrate on the pathways for energy generation and the anabolic reactions leading to cellular building blocks. This approach gives the impression that glucose metabolism by E. coli is a biochemical entity with set pathways and clear endpoints. In reality, and as recognized long ago, glucose supplied to E. coli will end up in quite different products depending on environmental conditions. For example, bacteria growing at different pH values produce different metabolic enzymes (16). Another complication is that bacteria supplied with excess glucose convert some of it to acetate even under aerobic conditions, whereas glucose-limited bacteria do not generate this product (30). The fate of glucose is even more complex under anaerobic conditions (40). In general, there is a surprising lack of information on how global aspects of metabolism are affected by factors such as growth rate, stress, or gene-regulatory mutations. To be able to harness and engineer metabolism even in simple organisms (2), information about the environmental effects on metabolism is essential. In this study, we propose a means of studying metabolism, using the concept of the “metabolome”, or total complement of metabolites in a cell, to look at global shifts in cellular function under different growth conditions.
The pools of many individual metabolites as well as intermediates in metabolic pathways have been measured in E. coli in a multitude of earlier studies (see reference 3 for a recent example). Analyses of classes of metabolite-like amino acids (39), cell wall precursors (25), and nucleotides (4) have also been undertaken. However, the environmental factors affecting pool sizes have not been looked at in detail, and certainly not across several classes of metabolite. Given that E. coli can contain up to 1,200 or more low-molecular-weight compounds (20), a more complete view of metabolites and metabolism would be gained by a more inclusive metabolome approach.
Ideally, noninvasive techniques like nuclear magnetic resonance spectroscopy can be used for metabolite analysis, but in practice this approach has been limited to certain culture conditions and classes of metabolites, predominantly phosphorylated compounds (discussed in reference 44). To get an impression of the overall contents of a cell, we preferred to adopt an analysis where all 14C-labelled water-soluble compounds extracted from bacteria are present in a two-dimensional (2-D) space on a thin-layer chromatography (TLC) plate. Although not as instantaneous as noninvasive techniques, cellular labelling is possible under various culture conditions, and the 2-D separation approach permits use of parallel technology to proteome studies, such that quantitation and software developed to match spots in 2-D arrays should also be usable in metabolome analysis (41, 45). The technical advantages and disadvantages of this approach are discussed in light of the results obtained below.
To experimentally test the metabolome strategy, variability of metabolite pools was investigated in bacteria growing in minimal medium with glucose as the sole carbon and energy source. Altered growth rates were achieved with glucose-limited continuous cultures set at different dilution rates. Recent results suggested that endogenous pools of sugars (galactose and maltotriose) involved in gene induction vary considerably at different growth rates in chemostats (13, 32, 33), and so a global analysis under these conditions was also important from the point of view of metabolites that have important gene-regulatory functions.
A particularly important aspect of growth rate regulation is due to the RpoS sigma factor-dependent stress responses of E. coli approaching stationary phase (14, 24). Although the gene-regulatory role of RpoS is well established, the metabolic effects of global gene regulators are far less well defined. The known role of RpoS in regulating trehalose and glycogen synthesis genes (19) already points to a significant influence on cellular metabolism, and expression of RpoS-regulated genes was previously shown to be affected in glucose-limited chemostat cultures growing at slow rates (32). The metabolome analysis described in this study therefore also tested whether cellular metabolism is sensitive to the loss of sigma factor in rpoS mutants. The metabolite shifts observed indicate that the approach used here also opens the way to assessing the function of other global gene regulators whose metabolic effects are largely undefined.
MATERIALS AND METHODS
Materials.
Bacterial cultures were labelled with [U-14C]glucose (303 mCi/mmol), obtained from Amersham Life Science, Sydney, New South Wales, Australia. The TLC plates for separations were Merck silica gel 60 plates with fluorescence indicator (Merck Pty. Ltd., Kilsyth, Victoria, Australia). The solvents and standards for chromatography were all analytical-grade reagents from commercial sources.
Bacterial strains and media.
The E. coli K-12 strain BW2951 [F− araD139 (ΔargF-lac)U169 flbB5301 ptsF25 rbsR deoC1 relA1 rpsL150 Φ(lamB-lacZ)] (31) was used as wild type for labelling experiments. Strain BW2996 contained an additional rpoS::Tn10 mutation but was otherwise identical to BW2951, the rpoS+ strain used and characterized previously (32).
The chemostat and batch media, together with the culture conditions used in this study, were as previously described (9, 11). Minimal medium A (MMA) (28) was the basal salts medium for both chemostat and batch cultures.
Labelling with [14C]glucose and extraction of metabolites.
Strains BW2951 and BW2996 were cultured in 80-ml chemostats and equilibrated at low (D = 0.1 h−1 culture) or high (D = 0.6 h−1 culture) dilution rate with limiting 0.02% glucose input to the medium. A three-way tap was arranged in the medium feed line to allow a switch from the standard medium reservoir to labelling medium while pumping continued at the same dilution rates. The labelling medium contained 6 ml of MMA and 0.02% total glucose concentration spiked with [U-14C]glucose (300 μCi in the 6-ml labelling mix). The delivery of the labelling medium was followed by flushing of tubing into the vessel with unlabelled medium. The total time of exposure to labelling was 9.5 min for the D = 0.6 h−1 culture and 47 min for the D = 0.1 h−1 culture. Bacteria were then immediately harvested from the chemostat by centrifugation (10 min at 6,000 × g) at 0°C, washed twice with MMA, and resuspended in 1 ml of water.
To label batch cultures, an exponential culture growing in 0.2% glucose in MMA was centrifuged and resuspended in 80 ml to an optical density of 0.3 at 580 nm, to parallel the conditions of the chemostat cultures. This culture at 37°C was incubated with 0.2% glucose mixed with 300 μCi of [14C]glucose, as specified above, for 9.5 min (this was the labelling time for the D = 0.6 h−1 culture). Harvesting and processing of this batch culture were then done as described above for the chemostat cultures.
Extraction of 14C-metabolites.
Concentrated, labelled bacteria were immediately lysed by the addition of 2 volumes of boiling ethanol. Excess ethanol was removed by boiling for 30 min. Debris was removed from extracts by centrifugation in an Eppendorf centrifuge for 10 min. Samples were freeze-dried and resuspended in 70 μl of water. Insoluble material was removed by centrifugation and scintillation counting of the various extracts permitted equivalent loadings to be applied to the TLC analysis plates. Approximately 5 × 105 dpm of labelled material was applied to the plates.
Separation by TLC.
The analysis was carried out on 10- by 10-cm aluminum-backed silica gel plates. The plates were activated at 110°C for 20 min before extracts were spotted and dried by heating 3 min at 80°C. Four pairs of solvent systems were used for 2-D elution of spots applied to the bottom left corner of TLC plates as shown in Fig. 1. Some elutions required two consecutive developments with the same solvent system to achieve a good separation; in these elutions, the plate was dried for 20 min in a ventilated fume hood and then for 3 min at 80°C between different solvent runs. All of the four solvent combinations used were those described by other workers (18, 21, 22, 27, 42): elution of the first dimension twice with pyridine–dioxan–28% (wt/wt) ammonia-water (35:35:15:15, vol/vol) before elution of the second dimension twice with butanol-acetone-acetic acid-water (35:35:7:23, vol/vol) (system A); B-elution twice with propanol–methanol–28% (wt/wt) ammonia-water (60:20:20:20, vol/vol) in the first dimension, followed by elution twice with butanol-acetone-acetic acid-water (35:35:7:23, vol/vol) in the second dimension (system B); development of the first dimension once with butanol–acetone–0.1 M phosphate buffer, pH 5 (40:50:10, vol/vol), and then elution of the second dimension once with butanol–2-propanol–0.5% (wt/vol) boric acid (30:50:20, vol/vol) (system C); and elution of the first dimension twice with chloroform–methanol–28% (wt/wt) ammonia (40:40:20, vol/vol) and then once with butanol–acetic acid–pyridine–37% (wt/wt) formaldehyde (30:30:20:10) (system D).
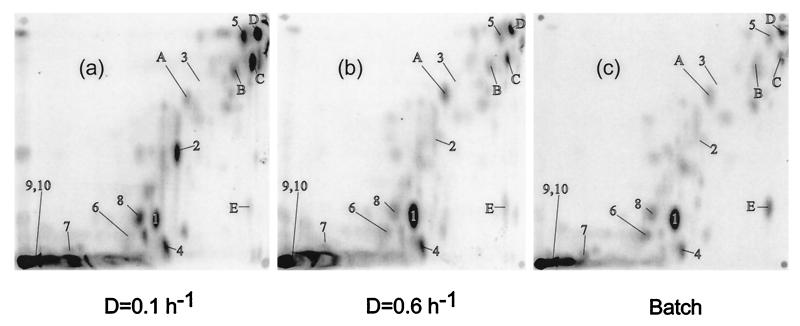
FIG. 1.
The metabolome of E. coli growing on glucose. Extracts were obtained from chemostat-grown bacteria at low (a) or high (b) dilution rate with limiting 0.02% glucose or from an exponentially growing culture with 0.2% glucose (c). The extracts were subjected to TLC in solvent system A, and the phosphorimages were labelled with the positions of spots corresponding to the following: 1, glutamate; 2, trehalose; 3, glucose; 4, UDP-glucose plus UDP-galactose; 5, adenosine; 6, aspartate; 7, lysine; 8, UDP-N-acetylglucosamine; 9, glutathione; 10, putrescine. The lettered spots were unidentified compounds mentioned in the text. The origin of the chromatographs was the bottom left.
Detection of 14C and standards on TLC plates.
Radioactive spots were detected on plates by using a Molecular Dynamics PhosphorImager and recording and processing results with the Molecular Dynamics ImageQuant package. The ImageQuant data were converted to 8-bit TIFF files before import into Canvas version 4 for the preparation of Fig. 1 and 2.
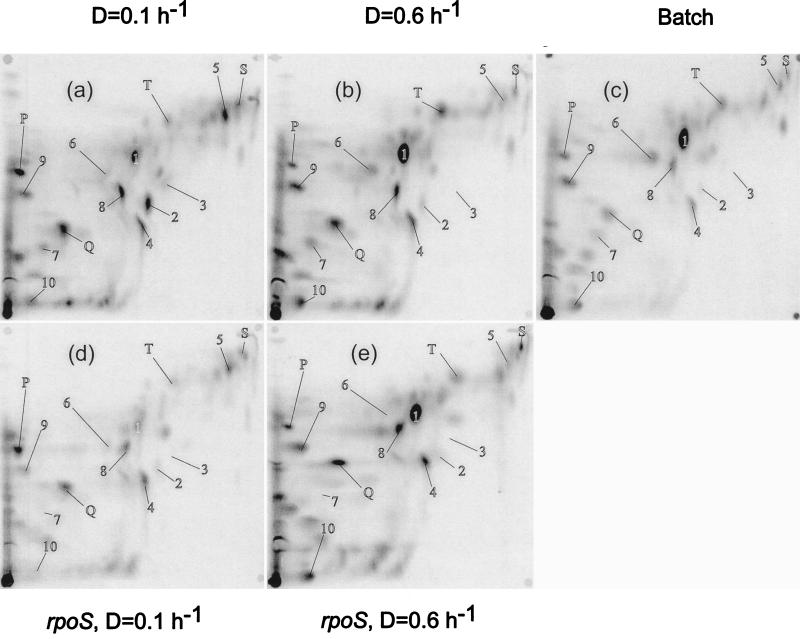
FIG. 2.
The metabolome of E. coli growing on glucose and the effect of an rpoS mutation. The details are as for Fig. 1 except that the extracts were subject to TLC in solvent system B. Panels d and e represent the metabolomes of the rpoS mutant BW2996 growing at low (0.1 h−1) and high (0.6 h−1) dilution rates, respectively.
Chemical detection was used for the metabolites added in spiking experiments for testing comigration with labelled spots. Applied standards were 1 μg for amino acids and polyamines and 4 μg for sugars and other compounds. Amino acids and polyamines were detected by spraying with ninhydrin (300 mg dissolved in 100 ml of ethanol plus 3 ml of acetic acid) and heating for a few min at 120°C. Compounds with vicinal diol groups such as sugars, alditols, and sugar phosphates were visualized with lead tetraacetate (1 g in 100 ml of ethanol); reducing sugars, aldehydes, and ketones were visualized with 4-nitrophenylhydrazine (100 mg in 32% HCl-ethanol [1:9, vol/vol]); nucleosides were visualized under UV light (254 nm).
Amino acid analysis.
Metabolite extracts obtained as described above were derivatized with 9-fluorenylmethyl chloroformate and separated with a GBC AminoMate system (GBC Scientific Equipment, Dandenong, Victoria, Australia). Duplicate analyses on each sample were carried out as a service by the Australian Proteome Analysis Facility, Macquarie University.
RESULTS
The initial metabolome comparisons were of bacteria grown on glucose in minimal medium at three different growth rates. Exponential batch cultures (doubling time of 0.9 h) and glucose-limited chemostats set at D = 0.6 and 0.1 h−1 (doubling times of 1.15 and 7 h) were labelled with [U-14C]glucose and extracted for water-soluble metabolites. The 14C-labelled extracts were applied to silica TLC plates subjected to 2-D elution with different solvent systems in each dimension. The radioactive spots on the plates were visualized by phosphorimaging, as shown in Fig. 1.
In Fig. 1, with TLC plates developed with system A optimized for amino acid separation (22), the resolved and unresolved spots represent all water-soluble carbon-containing compounds of the cell in the 2-D space analyzed. System A resolved not only amino acids but also sugars like glucose and trehalose, as well as metabolites like UDP-sugars. Depending on the sample, 60 to 70 spots could be visualized on the plates in Fig. 1. Comigration is likely to lead to an underestimate of spot numbers present, but the number of spots detected was still low relative to the 1,200 or so possible metabolites in a cell (20). Judging by the amount of label at the origin (more than 45% of the total label on the plate), many metabolites, including most phosphorylated compounds, may have been unresolved in Fig. 1. Also, the low number of spots suggests that only the more abundant metabolites were labelled sufficiently to be detected by the method used. Many metabolic intermediates and signalling molecules, like cyclic AMP, are present at micromolar concentrations in a cell, and most of these are likely to be below the detection limit of the approach in Fig. 1. The detection limit could be estimated from the lack of a distinct galactose spot in samples such as in Fig. 1b, which contains ca. 0.3 mM endogenously produced intracellular galactose in independent enzymatic estimations (10). Hence, only high-abundance metabolites present in millimolar concentration, but including most of the interesting stress metabolites of the cell, are labelled sufficiently to be seen in Fig. 1.
As only 40 to 60% of the extracted label was moved from the origin by the solvent system used for Fig. 1 (and by many other systems tested), different solvent pairings were used to elute compounds not particularly well resolved in Fig. 1. An alternative means for resolving interesting stress metabolites like putrescine and glutathione as well as some nucleotides was with plates eluted in system B (Fig. 2). The total number of individual spots (ca. 70) was not much greater than in Fig. 1, but the metabolites in the unseparated region near the origin in Fig. 1 (system A) were better resolved in system B, and spots labelled 1 to 10 were altogether better separated. In total, we used four different pairs of solvents for resolving metabolites in each sample; 2-D analyses with the other systems (not shown in Fig. 1 and 2) were used to confirm identities and quantities of metabolites separated. Many metabolites stayed at the origin with system C, but it was useful for resolving the high-Rf spots seen with systems A and B. System D, like system B, was useful for sugar phosphates and nucleotides. Many of the spots in system D were less sharp, but the system also permitted good quantitation of glutamate and glutathione. Each of the metabolites analyzed below was resolved and identified (by comigration with standards) in at least two of these four elution systems.
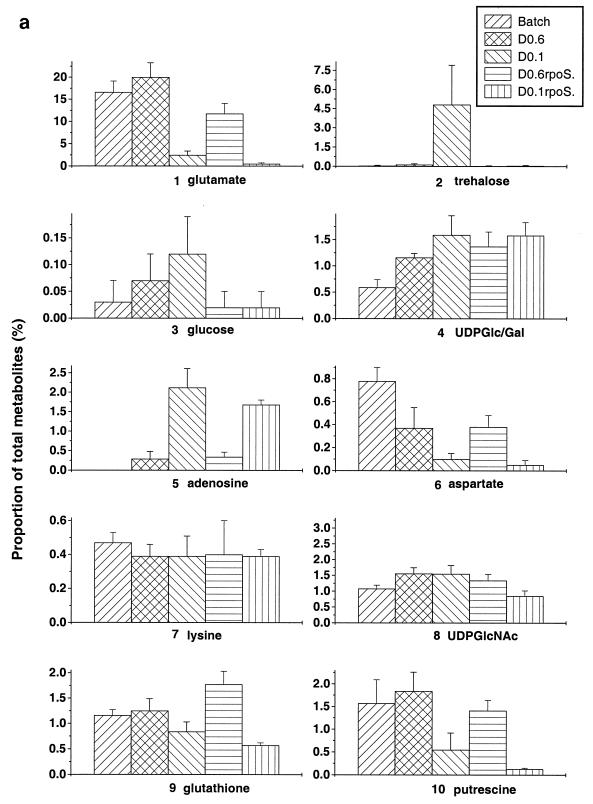
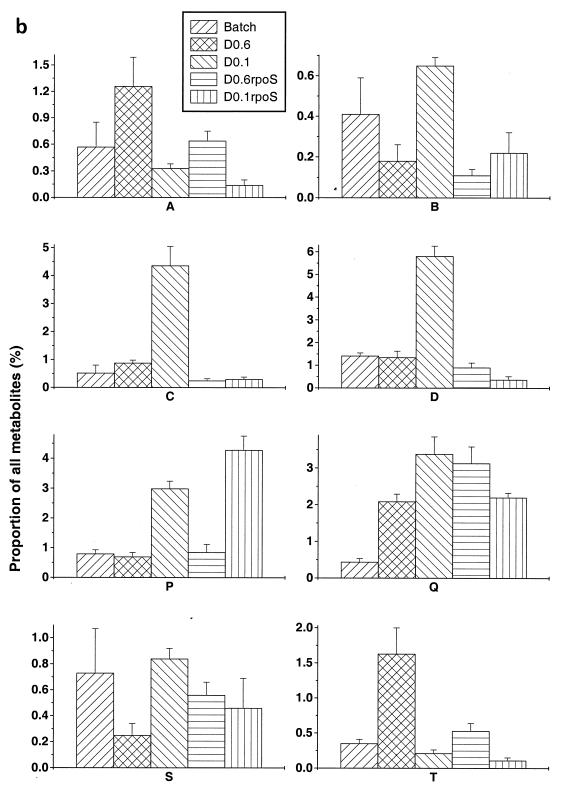
The patterns shown in Fig. 1 and 2 were reproducible in each system, and quantitative data for identified spots (using ImageQuant software) were encouragingly similar for independently grown, independently extracted, and separately analyzed samples. As an indication of the reproducibility, the percentage of each of 10 identified metabolites as a proportion of the applied metabolome was estimated by ImageQuant densitometric analysis and compared in repeat experiments (Fig. 3). This form of quantitation does not provide the absolute concentration of the metabolite in the cell, but the advantage of considering the proportion of the metabolome is that it avoids the problems associated with standardizing the harvesting and extraction conditions for different extracts. The data in Fig. 3, representing the means and standard deviations of four to six estimations for each of the 10 metabolites under each growth condition, suggest that the differences in pool sizes within samples obtained at a particular growth rate were small relative to differences between growth conditions. The identified spots, many of which significantly increase or decrease in intensity with various conditions, are numbered 1 to 10 in Fig. 1 and 2. There were obvious trends in pool changes, increasing or decreasing with growth rate on glucose. The significance of these trends is considered in Discussion.
FIG. 3.
Changes in pool sizes as determined by metabolome analysis. The spots corresponding to the compounds identified (a) and unidentified (b) in Fig. 1 and 2 were quantitated with ImageQuant software in four to six independent determinations.
Differences in spot intensities were found with many compounds besides those identified as metabolites 1 to 10. The lettered spots in Fig. 1 and 2 did not comigrate with known standards but, as shown in Fig. 3b, also gave differences in intensity under different growth conditions. The trends shown by compounds such as spots A, B, S, and T were unlike those exhibited by the identified metabolites, in that these pools were either maximal or minimal at intermediate growth rates. The arrowed spot E is not included in Fig. 3 quantitations, as this is one spot which did significantly vary in concentration between experiments. Although visual comparison of Fig. 1a, b, and c gives the impression that spot E is heavier in the batch sample, this pattern was not maintained in other experiments. This variability in spot E intensities was the exception rather than the rule in these studies.
Given the considerable change in metabolite pools at low growth rate, metabolome analysis was repeated with bacteria containing an rpoS::Tn10 mutation. As shown in Fig. 2d and e, the changes in metabolite pools for some but not all compounds were sensitive to the rpoS mutation. In general, the difference between wild type and mutant was more marked at the lower dilution rate; this was to be expected, as rpoS expression is not significant at the higher dilution rate (13, 32). The trehalose spot disappeared in the rpoS mutant at D = 0.1 h−1, but the glutamate pool size decreased at low growth rate despite the mutation. If anything, glutamate pools were even lower in the mutant. As discussed below, some but by no means all metabolic changes at slow growth rate are identifiably related to rpoS control.
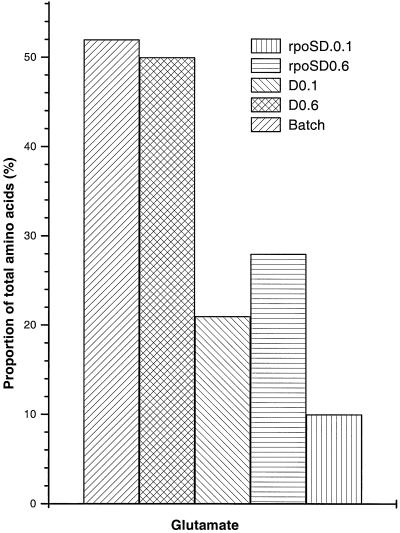
As the growth states tested by metabolome analysis identified significant changes in glutamate pools, chemical amino acid analysis was also carried out to verify the data with extracts from bacteria grown on glucose. As shown in Fig. 4, the changes in the proportion of glutamate found by TLC analysis were mirrored by the quantitation of glutamate by using high- performance liquid chromatography (HPLC) methods. Also as found by metabolome analysis such as that for lysine, there was no great shift in the proportion of other amino acid pools (results not shown). There was one notable exception: the aspartate pool as measured after HPLC gave a large signal, particularly in the D = 0.1 h−1 samples. A high quantity of aspartate was not evident in the metabolome separations by 2-D TLC, and the aspartate spot was not prominent at low dilution rates in either the wild type or the rpoS mutant. Presumably, another unidentified cellular component present in the extract migrates like Asp under the separation conditions used in HPLC.
FIG. 4.
Glutamate pools measured by HPLC analysis. Duplicate samples of the five types of cell extract obtained as for Fig. 2 were analyzed for amino acids by using an AminoMate system. The mean of the glutamate quantities obtained for each type of sample is shown.
DISCUSSION
These results present a new window for viewing cellular function. The metabolome as experimentally described in this study complements strategies such as proteome analyses in a global view of cellular responses. The 2-D TLC approach has the advantage of being able to view the total metabolite pool. Quantitation of individual spots as a proportion of the total pool also avoids problems with variable recoveries and cell volume measurements for comparison of different extracts. 2-D TLC is a simple, inexpensive, and nondestructive procedure from which spots can be recovered for further analysis. System B (Fig. 2) is the best 2-D system for the metabolites identified in this study, but variation of the stationary phase and the solvent systems allows resolution of molecules with different properties. Despite these advantages, the practical compromises involved in the present study warrant further discussion.
The analysis as currently used has the disadvantage that it is neither an instantaneous view of the metabolome nor a complete one, as less than 10% of the estimated number of metabolites are resolved and detected, as shown in Fig. 1 and 2. Of course, many of the 1,200 or so catalogued metabolites in E. coli (20) are present as intermediates in pathways not induced during aerobic growth on glucose, including degradative pathways for other nutrients and anaerobic metabolism. Low-abundance metabolites (which we estimate to be below ca. 0.5 to 1 mM in the cell) are not detected in the current approach. Indeed, in studies using alternative physiological conditions (such as anaerobic metabolism or application of stress states), it will be interesting to see if more of the metabolite pools are detectable or shift significantly.
A limitation of the current method is that metabolites with very fast turnover during the extraction process are not likely to be accurately quantitated. However, previous studies on amino acid pools indicate that the sampling times do not significantly alter the pattern observed with E. coli (39), and we do not see significant variation in the spot patterns in repeat samples extracted at different times, with the exception of spot E to this generalization. In this respect, perhaps it is actually an advantage that only the high-abundance metabolites are detected in Fig. 1 and 2 in that the turnover of these is likely to be slower than that for metabolites like glycolytic intermediates (44). Despite these limitations, it is possible to obtain a reproducible fingerprint of metabolism, particularly of stress-associated metabolites, present under different culture conditions.
Analysis of the same strain growing aerobically on glucose but at different levels of nutrient limitation gave quite different metabolome patterns with the spots analyzed in Fig. 3. The trends seen with individual metabolites range from the expected to the unexpected. Each of the identified metabolites is briefly considered.
Glutamate.
The amino acid glutamate is known to be the most abundant intracellular metabolite and particularly sensitive to osmotic perturbation (6, 39). Glutamate pools increase sharply on osmotic upshock, but the amino acid is slowly replaced by compatible solutes such as trehalose (43). In our studies, the osmolarity of the medium was unchanged during growth and labelling, and so differences in glutamate levels were not osmolarity driven. Glutamate was indeed the most dominant single spot (ca. 20% of all metabolites on the TLC plate, including unresolved spots at the origin) in batch culture or at high dilution rate under glucose limitation. However, the glutamate level dropped dramatically (to less than 3%) at low growth rate. Glutamate was replaced by trehalose as the single most abundant component of the metabolome, consistent with the previous demonstration of trehalose accumulation in slow-growing bacteria (32). Slow-growing bacteria induce RpoS-dependent metabolism such as trehalose synthesis (37). But glutamate pool reduction during slow growth was not RpoS dependent per se, since the rpoS mutant had even less glutamate at low dilution rate. As demonstrated earlier by Senior (36), α-ketoglutarate levels decrease significantly at low dilution rates in glucose-limited chemostats, and so the glutamate pools may reflect a change in tricarboxylic acid cycle regulation at low growth rates.
Trehalose.
As noted above, trehalose accumulation was found solely during very slow growth. As can be seen from the error bars in Fig. 3a, the extent of trehalose accumulation was variable in different cultures at D = 0.1 h−1 but was always much higher than under any other growth condition. Consistent with the RpoS-dependent nature of the biosynthetic otsAB genes (37), trehalose synthesis was abolished in the rpoS mutant.
Glucose.
Free glucose was not observed in high amounts in the samples, even though the sugar was the extracellular source of the 14C label. This finding was to be expected, since glucose is mostly transported by the phosphoenolpyruvate:sugar phosphotransferase system in batch culture, resulting in phosphorylated intracellular glucose (8). Even at D = 0.6 h−1, where glucose is partly accumulated by the Mgl system without phosphorylation (13), there was no accumulation of glucose. Free intracellular glucose can be phosphorylated by glucokinase (26). If anything, the slight trend was for increased intracellular levels with slower growth. These results do not support the suggestion that free glucose is the source of endogenous inducer in mal gene regulation under glucose limitation (12), as mal induction is highest at D = 0.6 h−1 (13). As with trehalose, glucose pools were somewhat variable but strongly reduced by the rpoS mutation. The metabolic basis for this result is unclear, in that no enzyme dephosphorylating glucose 6-phosphate is known to be under RpoS control. Possibly, higher glucose pools are a result of trehalose recycling (37) when the disaccharide is present at high concentration at low growth rates.
UDP-glucose and UDP-galactose.
The separation techniques used in this study do not resolve UDP-glucose and UDP-galactose with any of the solvent combinations, and so they are considered together. Both of these cellular components are intermediates in cell wall and lipopolysaccharide synthesis, and UDP-glucose has also been proposed to have a regulatory role in stationary-phase gene expression through an unspecified action on RpoS (5). The trend in our results is for the combined pool to increase with decreasing growth rate. The highest concentrations of UDP-sugar were associated with a growth rate (D = 0.1 h−1) known to lead to increased RpoS function (32). But according to the proposal of Böhringer et al. (5), the RpoS level should be negatively controlled by increased UDP-glucose. Three explanations can be offered for the discrepancy. First, other controlling factors override the negative effect of increased UDP-sugar pools in RpoS regulation, which cannot be excluded. Second, an increase in UDP-galactose masks the decrease in UDP-glucose, and so the combined pool that we observe changes in composition; this possibility also cannot be excluded, but since UDP-glucose is the precursor for UDP-galactose, it seems less likely. The third possibility is that the indirect evidence accumulated by Böhringer et al. (5) led to an incorrect conclusion; UDP-glucose levels were not directly measured in that study, based as it was on metabolic-block mutants.
Adenosine.
Pools of free bases are not normally present at high levels in exponentially growing bacteria, but entry into stationary phase results in endogenous and exogenous accumulation of nucleobases in E. coli cultures (35). Figure 3 shows a very strong trend of higher adenosine levels in slow-growing bacteria. The increase in adenosine pools was not abolished in the rpoS mutant and so is independent of this form of stationary-phase control. The accumulation of nucleobases on entry into stationary phase was ascribed to ribosomal degradation (35). This cannot be the entire story because adenosine (and most likely the other bases migrating to the top right corner in Fig. 1) accumulates even in steady-state, slow-growing bacteria which are not degrading ribosomes due to a downshift. Hence the high adenosine pool is typical of slow growth, as is the high concentration of cyclic AMP under these conditions (33).
This study did not address the nature of ATP pools directly and whether the recently postulated growth rate regulation affects levels of nucleoside triphosphates (15). In our TLC, ATP, ADP, and AMP were separated in system B but were too close to overlapping spots for accurate quantitation. It remains to be investigated whether the adenosine/A(N)P pools are coordinately regulated.
Aspartate and lysine.
The spot corresponding to aspartate markedly decreased in intensity with growth rate, being highest in exponentially growing bacteria. In this respect, aspartate pools followed the glutamate pattern, though the D = 0.6 h−1 culture was also affected. In contrast, there was no significant change under any condition with lysine levels. Indeed the constant lysine pattern was true for several other amino acids, both in metabolome analysis and in HPLC assay (results not shown). Therefore, the shifts in glutamate and aspartate pools are more likely to be relevant to general metabolic regulation than protein precursor control.
UDP-N-acetylglucosamine.
The nucleotide sugar UDP-N-acetylglucosamine is an immediate cell envelope precursor. Its concentration, as well as that of the other UDP-sugars examined, increases with decreasing growth rate. It is not intuitively obvious why this should be so, as cell wall synthesis should proceed more slowly in slow-growing bacteria. Whether this has a regulatory significance is also unknown. The rpoS mutation has a partial negative effect on this pool solely at D = 0.1 h−1.
Glutathione.
Levels of glutathione, an important metabolite in maintaining redox balance in cells, are closely linked to oxidative stress responses (7, 17, 34). Our conditions were fully aerobic in each culture investigated, and so major changes in glutathione levels were not expected. The glutathione spot did not show significant changes in intensity except for a small drop in slow-growing bacteria. Previously, an increase in glutathione pools was found in stationary phase (23), but the increase was dependent on the presence of exogenous amino acids and did not occur in glucose minimal media such as the one we used. A possible reason for slightly lower glutathione levels may be the much lower glutamate concentrations during slow growth, as glutamate is required for glutathione synthesis (1).
Putrescine.
Polyamines like putrescine are normally present in high concentration in E. coli, but their pool size is markedly affected by culture conditions (38). As for glutathione, the increase in putrescine concentration in stationary phase in batch culture is influenced by the presence of amino acids like arginine as well as environmental factors such as pH and aeration (38). High osmolarity also reduces putrescine levels (29), a finding confirmed in glucose-limited chemostats run at high osmolarity (result not shown). Putrescine levels were lower at D = 0.1 h−1 under glucose limitation in the rpoS mutant but not as dramatically reduced as trehalose or glucose pools.
Other unidentified metabolites.
Most of the 60 or so other spots in Fig. 1 and 2 are still unidentified. Among these, particular trends were observed with the letter-labelled spots quantitated in Fig. 3, and indeed this is why the lettered spots were chosen for reporting. The first type of trend is the increase of pools with decreasing growth rate. As for trehalose, spots C, D, Q, and P were markedly increased at D = 0.1 h−1. Another example was with spots A and T, in that their pool size was greatest at D = 0.6 h−1, as were galactose and maltotriose pools in earlier studies (10, 31). Yet another pattern was with spots B and S, which had the lowest pool size at this intermediate growth rate.
Based on the mobilities in Fig. 1 and 2 and pool trends shown in Fig. 3, it is possible that spots A and T as well as spots B and S represent identical molecules. Without identification and the possibility of testing comigration with standards, this conclusion may be premature. Identification of further spots is obviously necessary for more detailed metabolome comparisons.
In conclusion, each of the trends reported here illustrates that metabolic pools are under strict physiological control. The metabolome approach is evidently a means of discovering these trends. In future, improvements in technology and the study of metabolomes with other stresses should reveal previously unexplored responses of metabolism to environmental regulation.
ACKNOWLEDGMENT
We thank the Australian Research Council for financial support.
REFERENCES
- 1.Apontoweil P, Berends W. Glutathione biosynthesis in Escherichia coli K 12. Properties of the enzymes and regulation. Biochim Biophys Acta. 1975;399:1–9. doi: 10.1016/0304-4165(75)90205-6. [DOI] [PubMed] [Google Scholar]
- 2.Bailey J E. Toward a science of metabolic engineering. Science. 1991;252:1668–1674. doi: 10.1126/science.2047876. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya M, Fuhrman L, Ingram A, Nickerson K W, Conway T. Single-run separation and detection of multiple metabolic intermediates by anion-exchange high-performance liquid chromatography and application to cell pool extracts prepared from Escherichia coli. Anal Biochem. 1995;232:98–106. doi: 10.1006/abio.1995.9954. [DOI] [PubMed] [Google Scholar]
- 4.Bochner B R, Ames B N. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- 5.Böhringer J, Fischer D, Mosler G, Hengge-Aronis R. UDP-glucose is a potential intracellular signal molecule in the control of expression of ςS and ςS-dependent genes in Escherichia coli. J Bacteriol. 1995;177:413–422. doi: 10.1128/jb.177.2.413-422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booth I R, Kleppang K E, Kempsell K E. A genetic locus for the GltII-glutamate transport system in Escherichia coli. J Gen Microbiol. 1989;135:2767–2774. doi: 10.1099/00221287-135-10-2767. [DOI] [PubMed] [Google Scholar]
- 7.Chesney J A, Eaton J W, Mahoney J R. Bacterial glutathione: a sacrificial defense against chlorine compounds. J Bacteriol. 1996;178:2131–2135. doi: 10.1128/jb.178.7.2131-2135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtis S J, Epstein W. Phosphorylation of d-glucose in Escherichia coli mutants defective in glucosephosphotransferase, mannosephosphotransferase, and glucokinase. J Bacteriol. 1975;122:1189–1199. doi: 10.1128/jb.122.3.1189-1199.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Death A, Ferenci T. The importance of the binding-protein-dependent Mgl system to the transport of glucose in Escherichia coli growing on low sugar concentrations. Res Microbiol. 1993;144:529–537. doi: 10.1016/0923-2508(93)90002-j. [DOI] [PubMed] [Google Scholar]
- 10.Death A, Ferenci T. Between feast and famine: endogenous inducer synthesis in the adaptation of Escherichia coli to growth with limiting carbohydrates. J Bacteriol. 1994;176:5101–5107. doi: 10.1128/jb.176.16.5101-5107.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Death A, Notley L, Ferenci T. Derepression of LamB protein facilitates outer membrane permeation of carbohydrates into Escherichia coli under conditions of nutrient stress. J Bacteriol. 1993;175:1475–1483. doi: 10.1128/jb.175.5.1475-1483.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decker K, Peist R, Reidl J, Kossmann M, Brand B, Boos W. Maltose and maltotriose can be formed endogenously in Escherichia coli from glucose and glucose-1-phosphate independently of enzymes of the maltose system. J Bacteriol. 1993;175:5655–5665. doi: 10.1128/jb.175.17.5655-5665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferenci T. Adaptation to life at micromolar nutrient levels: the regulation of Escherichia coli glucose transport by endoinduction and cAMP. FEMS Microbiol Rev. 1996;18:301–317. doi: 10.1111/j.1574-6976.1996.tb00246.x. [DOI] [PubMed] [Google Scholar]
- 14.Foster J W, Spector M P. How Salmonella survive against the odds. Annu Rev Microbiol. 1995;49:145–174. doi: 10.1146/annurev.mi.49.100195.001045. [DOI] [PubMed] [Google Scholar]
- 15.Gaal T, Bartlett M S, Ross W, Turnbough C L, Gourse R L. Transcription regulation by initiating NTP concentration—rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 16.Gale E F, Epps H M R. The effect of pH of the medium during growth on the enzymatic activities of bacteria (Escherichia coli and Micrococcus lysodeikticus) and the biological significance of the changes produced. Biochem J. 1942;36:600–618. doi: 10.1042/bj0360600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grant C M, Dawes I W. Synthesis and role of glutathione in protection against oxidative stress in yeast. Redox Rep. 1996;2:223–229. doi: 10.1080/13510002.1996.11747054. [DOI] [PubMed] [Google Scholar]
- 18.Hamana K, Matsuzaki S, Inoue K. Changes in polyamine levels in various organs of Bombyx mori during its life cycle. J Biochem. 1984;95:1803–1809. doi: 10.1093/oxfordjournals.jbchem.a134793. [DOI] [PubMed] [Google Scholar]
- 19.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 20.Karp P D, Riley M, Paley S M, Pellegrini-Toole A, Krummenacker M. EcoCyc—encyclopedia of Escherichia coli genes and metabolism. Nucleic Acids Res. 1997;25:43–50. doi: 10.1093/nar/26.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klaus R, Ripphahn J. Quantitative dünnschichtchromatographische Analyse von Zuckern, Zuckersäuren und Polyalkoholen. J Chromatogr. 1983;244:99–124. [Google Scholar]
- 22.Kraffczyk F, Helger R, Lang H, Bremer H J. Thin layer chromatographic screening test for amino acid anomalies in urine without desalting using internal standards. Clin Chim Acta. 1971;35:345–351. doi: 10.1016/0009-8981(71)90204-x. [DOI] [PubMed] [Google Scholar]
- 23.Loewen P C. Levels of glutathione in Escherichia coli. Can J Biochem. 1979;57:107–111. doi: 10.1139/o79-013. [DOI] [PubMed] [Google Scholar]
- 24.Loewen P C, Hengge-Aronis R. The role of the sigma factor ςS (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 25.Mengin-Lecreulx D, Flouret B, van Heijenourt J. Pool levels of UDP N-acetylglucosamine and UDP N-acetylglucosamine-enolpyruvate in Escherichia coli and correlation with peptidoglycan synthesis. J Bacteriol. 1983;154:1284–1290. doi: 10.1128/jb.154.3.1284-1290.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer D, Schneiderfresenius C, Horlacher R, Peist R, Boos W. Molecular characterization of glucokinase from Escherichia coli K-12. J Bacteriol. 1997;179:1298–1306. doi: 10.1128/jb.179.4.1298-1306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miksik I, Hodny Z, Deyl Z. Chromatographic separation of glycated nucleotides. J Chromatogr. 1993;612:57–61. doi: 10.1016/0378-4347(93)80367-d. [DOI] [PubMed] [Google Scholar]
- 28.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 29.Munro G F, Hercules K, Morgan J, Sauerbier W. Dependence of the putrescine content of Escherichia coli on the osmotic strength of the medium. J Biol Chem. 1972;247:1272–1280. [PubMed] [Google Scholar]
- 30.Neijssel O M, Tempest D W. The physiology of metabolite over-production. Symp Soc Gen Microbiol. 1979;29:53–82. [Google Scholar]
- 31.Notley L, Ferenci T. Differential expression of mal genes under cAMP and endogenous inducer control in nutrient stressed Escherichia coli. Mol Microbiol. 1995;160:121–129. doi: 10.1111/j.1365-2958.1995.tb02397.x. [DOI] [PubMed] [Google Scholar]
- 32.Notley L, Ferenci T. Induction of RpoS-dependent functions in glucose-limited continuous culture: what level of nutrient limitation induces the stationary phase of Escherichia coli? J Bacteriol. 1996;178:1465–1468. doi: 10.1128/jb.178.5.1465-1468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Notley-McRobb L, Death A, Ferenci T. The relationship between external glucose concentration and cAMP levels inside Escherichia coli—implications for models of phosphotransferase-mediated regulation of adenylate cyclase. Microbiology. 1997;143:1909–1918. doi: 10.1099/00221287-143-6-1909. [DOI] [PubMed] [Google Scholar]
- 34.Penninckx M J, Elskens M T. Metabolism and functions of glutathione in micro-organisms. Adv Microb Physiol. 1993;34:239–301. doi: 10.1016/s0065-2911(08)60031-4. [DOI] [PubMed] [Google Scholar]
- 35.Rinas U, Hellmuth K, Kang R J, Seeger A, Schlieker H. Entry of Escherichia coli into stationary phase is indicated by endogenous and exogenous accumulation of nucleobases. Appl Environ Microbiol. 1995;61:4147–4151. doi: 10.1128/aem.61.12.4147-4151.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Senior P J. Regulation of nitrogen metabolism in Escherichia coli and Klebsiella aerogenes: studies with the continuous-culture technique. J Bacteriol. 1975;123:407–418. doi: 10.1128/jb.123.2.407-418.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strom A R, Kaasen I. Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol Microbiol. 1993;8:205–210. doi: 10.1111/j.1365-2958.1993.tb01564.x. [DOI] [PubMed] [Google Scholar]
- 38.Tabor C W, Tabor H. Polyamines in microorganisms. Microbiol Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tempest D W, Meers J L, Brown C M. Influence of environment on the content and composition of microbial free amino acid pools. J Gen Microbiol. 1970;64:171–185. doi: 10.1099/00221287-64-2-171. [DOI] [PubMed] [Google Scholar]
- 40.Unden G, Becker S, Bongaerts J, Schirawski J, Six S. Oxygen regulated gene expression in facultatively anaerobic bacteria. Antonie Leeuwenhoek. 1994;66:3–23. doi: 10.1007/BF00871629. [DOI] [PubMed] [Google Scholar]
- 41.Vanbogelen R A, Abshire K Z, Moldover B, Olson E R, Neidhardt F C. Escherichia coli proteome analysis using the gene-protein database. Electrophoresis. 1997;18:1243–1251. doi: 10.1002/elps.1150180805. [DOI] [PubMed] [Google Scholar]
- 42.Waldi D. Dünnschicht-Chromatographie einiger Zucker und Zuckeralkohole. J Chromatogr. 1965;18:417–418. doi: 10.1016/s0021-9673(01)80390-3. [DOI] [PubMed] [Google Scholar]
- 43.Welsh D T, Reed R H, Herbert R A. The role of trehalose in the osmoadaptation of Escherichia coli NCIB 9484: interaction of trehalose, K+ and glutamate during osmoadaptation in continuous culture. J Gen Microbiol. 1991;137:745–750. doi: 10.1099/00221287-137-4-745. [DOI] [PubMed] [Google Scholar]
- 44.Weuster-Botz D, de Graaf A A. Reaction engineering methods to study intracellular metabolite concentrations. Adv Biochem Eng Biotechnol. 1996;54:75–108. doi: 10.1007/BFb0102333. [DOI] [PubMed] [Google Scholar]
- 45.Wilkins M R, Pasquali C, Appel R D, Ou K, Golaz O, Sanchez J C, Yan J X, Gooley A A, Hughes G, Humphrey-Smith I, Williams K L, Hochstrasser D F. From proteins to proteomes—large scale protein identification by two-dimensional electrophoresis and amino acid analysis. Bio/Technology. 1996;14:61–65. doi: 10.1038/nbt0196-61. [DOI] [PubMed] [Google Scholar]