Abstract
The behaviors of both cheZ-deleted and wild-type cells of Escherichia coli were found to be very sensitive to the level of expression of CheZ, a protein known to accelerate the dephosphorylation of the response regulator CheY-phosphate (CheY-P). However, cells induced to run and tumble by the unphosphorylated mutant protein CheY13DK106YW (CheY**) failed to respond to CheZ, even when CheZ was expressed at high levels. Therefore, CheZ neither affects the flagellar motors directly nor sequesters CheY**. In in vitro cross-linking studies, CheY** promoted trimerization of CheZ to the same extent as wild-type CheY but failed to induce the formation of complexes of higher molecular weight observed with CheY-P. Also, CheY** could be cross-linked to FliM, the motor receptor protein, nearly as well as CheY-P. Thus, to CheZ, CheY** looks like CheY, but to FliM, it looks like CheY-P.
Flagellated bacteria, such as Escherichia coli and Salmonella typhimurium, respond to a wide variety of positive or negative environmental stimuli through modulation of the direction of rotation of their flagella. An increase in the concentration of a chemical attractant (e.g., aspartate) or a decrease in the concentration of a chemical repellent (e.g., leucine) promotes counterclockwise rotation, or more persistent smooth swimming. The signal transduction chain includes a kinase, CheA, which phosphorylates a response regulator, CheY. CheY-phosphate (CheY-P), the active form, binds to the motor switch complex and promotes clockwise (CW) rotation, or more frequent tumbling. The turnover of CheY-P is relatively rapid: in vitro, CheY-P autodephosphorylates with a half time of about 14 s (27) and its dephosphorylation is accelerated by CheZ (11); in vivo, physiological measurements indicate that this acceleration should be by about a factor of 10 (26).
Several CheY mutants are active in the absence of phosphorylation. When overexpressed, the mutant protein CheY13DK, for example, produces a tumbly phenotype in a host deleted for cheA (5, 6). However, it does not show enhanced binding to FliM or to CheZ, as does CheY-P (4, 35, 36). Another class of CheY mutants, including CheY106YW and CheY95IV, are hyperactive when expressed at normal levels in an otherwise wild-type background but are inactive in a cheA deletion background (25, 38). In vitro, the latter mutant exhibits enhanced binding to FliM (25), whereas the former shows no change in affinity for FliM (38). The double mutant CheY13DK106YW (CheY**) is active at relatively low concentrations in a host deleted for cheA. In an earlier study, we used this mutant to establish quantitative relationships between intracellular concentrations and statistics of motor switching (24).
It is known from in vitro studies that the phosphatase CheZ does not bind to the switch component FliG, FliM, or FliN (8). However, it is not known whether CheZ has a direct effect on the switch in vivo, because it has not been possible to distinguish this action from inactivation of CheY-P. Early studies of intragenic suppression argued for direct interaction, because the suppression appeared to be allele specific (20, 21), but more recent work has made this less certain (13, 29). In the present study, we used the double mutant CheY** to settle the issue. We studied the effects of CheZ on CheY** and the switch, comparing these results with those obtained with wild-type CheY. CheZ did not affect the behavior of the flagellar motor, either directly or indirectly, e.g., by sequestration of CheY**. As judged in vitro by cross-linking, CheY** induced oligomerization of CheZ to the same extent as CheY (but much less so than CheY-P) and bound to FliM about as well as CheY-P.
MATERIALS AND METHODS
Bacteria and plasmids.
The bacteria and plasmids used are listed in Table 1. Strains HCB900 and HCB915 contain the doubly mutated gene cheY13DK106YW, under control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promotor Ptrc, and a deletion in the adjacent gene, cheZ. The construction of HCB900, a derivative of RP9535 (ΔcheA), has been described by Scharf et al. (24). HCB915, a derivative of RP437 (cheA+), was constructed in parallel. Both strains carry cat (chloramphenicol transacetylase).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genes and/or phenotype | Reference or source | Remarks |
|---|---|---|---|
| Strains | |||
| AW405 | Wild type for chemotaxis | 1 | Wild type for chemotaxis |
| BL21-DE3 | λDE3 (prophage carries T7 gene1) | 32 | Host for FliM expression |
| HCB900a | Δ(cheA)1643 cat+ Ptrc420cheY13DK106YW Δ(cheZ) | 24 | ΔcheA ΔcheZ, with IPTG-inducible cheY13DK106YW |
| HCB901 | HCB900 fliC726 uvrC-279::Tn10 | 24 | Nonmotile derivative of HCB900 |
| HCB915 | cat+ Ptrc420cheY13DK106YW Δ(cheZ) | This work | ΔcheZ, with IPTG-inducible cheY13DK106YW |
| HCB916 | HCB915 fliC726 uvrC-279::Tn10 | This work | Nonmotile derivative of HCB915 |
| HCB917 | HCB915/pBES40/pMS421 | This work | Swarm assay |
| HCB918 | HCB915/pASK75/pMS421 | This work | Swarm assay |
| HCB919 | HCB916/pBES40/pBES42 | This work | Tethering assay |
| HCB920 | HCB916/pASK75/pBES42 | This work | Tethering assay |
| HCB921 | HCB900/pBES40/pMS421 | This work | Swarm assay |
| HCB922 | HCB901/pBES40/pBES42 | This work | Tethering assay |
| HCB1264 | fliC726 uvrC-279::Tn10 | 24 | Tn10-linked fliC726 derivative of HCB5 |
| RP437 | Wild type for chemotaxis | 19 | Wild type for chemotaxis |
| RP1616 | Δ(cheZ)m67-25 | J. S. Parkinson | cheZ-deleted strain |
| RP3098 | Δ(flhA-flhD) srl::Tn10 | 28 | cheA-, cheY-, and cheZ-deleted strain |
| Plasmids | |||
| pASK75 | bla tetR P/Otet | Biometra | Expression vector with tetA promoter/operator and constitutively expressed tetR |
| pBES38 | bla fliC(St) lacIq | 24 | Source of lacIq and fliC(St) |
| pBES39 | bla tetR P/OtetA cheZ | This work | AHT-inducible cheZ with “strep-tag” |
| pBES40 | bla tetR P/OtetA cheZ | This work | AHT-inducible cheZ |
| pBES42 | lacIqfliC(St) Sm/Sprb oripSC101 | This work | pGBM1 derivative; provides LacI and FliC(St) in HCB919, 920, and 922 |
| pGBM1 | repA(cop) Sm/SproripSC101 | 16 | High-copy-number derivative of pSC101 |
| pHT28 | fliM | 33 | FliM expression |
| pMS421 | lacI Sm/SproripSC101 | Karen McGovern | Provides LacI in HCB917, 918, and 921 |
| pRL22 | bla Ptrp ′cheB cheY cheZ ′flhB oripBR322 | 18 | CheY and CheZ expression |
CheZ was cloned into pASK75 (Biometra) as follows. A 1.6-kb BpmI/BamHI fragment of pRL22, which contains CheZ, was made blunt-ended with Polymerase I Large Fragment (New England Biolabs) and ligated into pASK75, which had been predigested with StuI and XbaI and made blunt-ended, to yield pBES39. This plasmid was then digested with Eco47III and HindIII (to remove a “strep-tag” of the pASK75 vector), made blunt-ended, and religated, resulting in pBES40. This plasmid expresses CheZ from the tetA promoter/operator, which is under control of its repressor, TetR. The tet promoter is inducible by anhydrotetracycline (AHT) (Acros Organics, Fairlawn, N.J.) at concentrations far below those at which AHT acts as an antibiotic.
Plasmids and strains used for expression and isolation of E. coli proteins were pRL22 in RP3098 for CheZ and CheY (18), pXYZ202 in RP3098 for CheY** (24), and pHT28 in BL21-DE3 for FliM (33).
For swarming assays, HCB900 and HCB915 were transformed with both pBES40 (inducible CheZ expression) and pMS421 (constitutive LacI expression), resulting in strains HCB921 and HCB917, respectively.
For tethering studies, a plasmid expressing flagellin FliC(St), which forms sticky filaments, as well as LacI, was constructed to be compatible with pBES40 as follows. The 4.9-kb EcoRI/SalI fragment of pBES38 (24) containing lacIq and fliC(St) was ligated into the EcoRI/SalI sites of pGBM1, a higher-copy-number derivative of pSC101 (16), to yield pBES42. A null fliC mutation was introduced into the genomes of strains HCB900 and HCB915 by transducing Tn10-linked fliC726, giving HCB901 and HCB916. These strains were cotransformed with pBES42 and pBES40 to give HCB922 and HCB919, respectively.
The following were gifts: pGBM1 from Johannes Geiselmann, pHT28 from David Blair, BL21-DE3 from Antje Hofmeister, pMS421 from Karen McGovern, pRBB40 from Bob Bourret, and RP1616 from Sandy Parkinson.
Cell cultures.
Cells were prepared in the same way for tethering assays and quantitative immunoblots, as described by Scharf et al. (24). They were grown in TB (1% tryptone, 0.5% NaCl) for 4 h at 33°C, and then (if required) inducers were added and incubation was continued for 2 h. For example, for tethering assays, IPTG was added to a final concentration of 15 μM, the culture was divided into two parts, and AHT was added to one part at a final concentration of 200 μg/liter.
Behavioral assays.
Swarm assays were performed on soft agar plates (0.3% agar, 1% tryptone, 0.5% NaCl) inoculated with 2.5 μl of a 10-fold-concentrated culture that was grown in TB for 6 h at 30°C. The plates were incubated at temperatures and times indicated in the figure legends and text. Tethering, data acquisition, and analysis were done as described by Scharf et al. (24).
Immunoblots and cross-linking.
Purified CheY13DK was a gift from Phil Matsumura. Purification of CheY, CheY**, and CheZ was described previously (24). FliM was purified as described by Bren et al. (8), but the chromatography step was omitted.
Quantitative immunoblotting of CheY was performed according to Scharf et al. (24). Immunoblots of CheZ were assayed by the same procedure with the following changes. Proteins were separated in a gel with a linear gradient from 10 to 17.5% acrylamide. Nitrocellulose blots were blocked for 1 h and then probed with anti-CheZ monoclonal antibody (MAb) 3D8B5 at a dilution of 1:500 overnight.
The numbers of molecules of CheY or CheZ per cell were determined from the immunoblots and cell numbers (24). Conversion to micromolar concentrations was made from the corresponding dry weights (24), using the cytoplasmic volume per milligram (dry weight) (1.4 μl) given by Stock et al. (31).
Protein cross-linking experiments were done with EDC (1-ethyl-3-(dimethylaminopropyl)carbodiimide hydrochloride), NHS (N-hydroxysuccinimide) (ICN, Aurora, Ohio), or dimethylsuberimidate dihydrochloride (DMS) (Pierce, Rockford, Ill.), according to the procedures of Blat and Eisenbach (3) and Bren et al. (8) with the following modification: CheZ cross-linked products were assayed on immunoblots as described above. One-fifth of the reaction volume was assayed.
Image enhancement.
Photographs of swarms, immunoblots, and gels were scanned, and contrast was digitally enhanced.
RESULTS AND DISCUSSION
Expression of CheZ from pBES40.
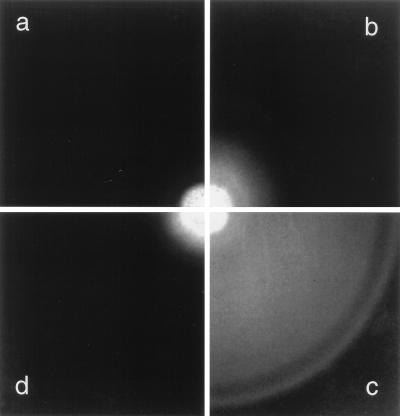
Plasmid pBES40 carries cheZ under tight control of TetR, the repressor of the tetracycline promoter. When this plasmid was introduced into the cheZ deletion strain RP1616 grown in the absence of inducer (AHT), only a slight increase in swarm drift velocities was seen (Fig. 1b). When CheZ was induced, swarm diameters increased substantially and chemotactic rings appeared (Fig. 1c). However, high levels of induction impaired chemotaxis (Fig. 1d). In Fig. 1a, the cells shown were tumbly; in Fig. 1d, they were smooth swimming.
FIG. 1.
Variations of swarm morphology upon expression of CheZ in the cheZ-deletion strain RP1616. Cells were transformed with pBES40 (cheZ inducible with AHT) or its parental plasmid pASK75 (no cheZ) and incubated on soft agar plates containing the following concentrations of AHT: pASK75 at 0 μg/liter (a), pBES40 at 0 μg/liter (b), pBES40 at 2 μg/liter (c), and pBES40 at 20 μg/liter (d). Plates were incubated for 14 h at 30°C. The outer diameter of the swarm in panel c is 46 mm.
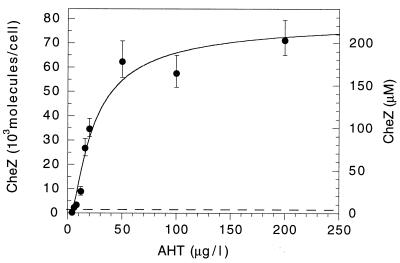
We measured the level of expression of CheZ from pBES40 in another cheZ deletion strain, HCB917, by quantitative immunoblot analysis (Fig. 2). CheZ was not detected (under our standard conditions) at levels of inducer below 4 μg/liter. At higher concentrations, expression increased dramatically, reaching half saturation (about 100 μM CheZ) at about 20 μg/liter. Note that complementation of the cheZ deletion of strain RP1616 carrying pBES40 was achieved at 2 μg of AHT per liter (Fig. 1c). Evidently, relatively little CheZ is required for chemotaxis on swarm plates. We are not able to make a more precise statement, because the level of expression of CheZ in swarm plates (Fig. 1) might not be identical to that observed in liquid media (Fig. 2). However, TetR is constitutively expressed from pBES40, and it seems unlikely that CheZ levels would be significantly higher than those measured for liquid cultures.
FIG. 2.
Quantification of CheZ expressed from pBES40 in HCB917 (a cheZ-deletion strain) as a function of AHT induction. Cells were exposed to AHT for 2 h during exponential growth, harvested, and assayed on immunoblots with anti-CheZ MAb. CheZ levels were calculated as in Table 2. At least two immunoblots were performed for each concentration. Error bars are standard deviations of the means, with the result for each immunoblot weighted equally. The fit is of the form y = exp[C1 − (C2/AHT)], where y is 103 molecules/cell, C1 and C2 are constants, and AHT is in micrograms per liter. The dashed line indicates the level of CheZ measured in strain RP437 (Table 2).
Interference with swarming upon increased expression of CheZ in strain RP1616 has been shown previously by Huang and Stewart (12) and in a recA derivative of RP1616 by Sanna and Simon (23), who used this dependence as a strategy to find cheZ mutants.
Effect of CheZ on swarming in wild-type cells.
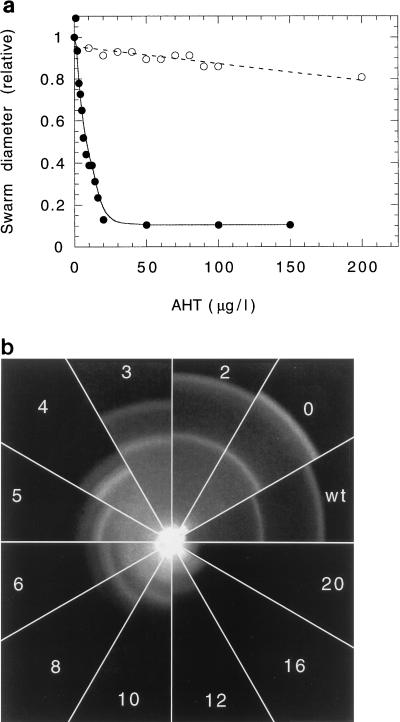
We also examined the effect of cheZ expression from pBES40 in wild-type strain RP437 and found swarm diameters to be remarkably sensitive to AHT (Fig. 3a). This sensitivity was not due to an unspecific antibiotic effect of AHT, which is known to be growth inhibiting at concentrations above 200 μg/liter (Fig. 3a). The swarms obtained in this experiment are shown in Fig. 3b. Chemotactic rings were not seen at concentrations of AHT of 12 μg/liter or higher. These results suggest that the ratio of CheZ to CheY (and hence the rate and/or extent of dephosphorylation of CheY) is important for effective chemotaxis. Huang and Stewart (12) found that swarming in a recA derivative of strain RP437 was impaired upon moderate induction of CheZ from E. coli but less so by induction from S. typhimurium. They concluded that “the chemotaxis system is quite sensitive to the CheY dephosphorylation activity of CheZ.”
FIG. 3.
Variations of swarm size and morphology upon expression of CheZ in wild-type strain RP437. (a) Outer diameters of swarms measured as a function of AHT concentration for cells transformed with pBES40 (closed symbols) or its parental plasmid pASK75 (no cheZ) (open symbols), relative to the diameter observed without AHT. (b) Swarm morphologies for wild-type cells and cells transformed with pBES40. wt, wild-type strain. The numbers are AHT concentrations (in micrograms per liter). Plates were incubated for 9 h at 30°C.
Levels of CheY and CheZ in wild-type cells.
Given this sensitivity to the ratio of CheZ to CheY, we measured the levels of these components in wild-type strains AW405 and RP437 (Table 2). Strain AW405 contained about three times as much of either component as strain RP437, but the CheY/CheZ molar ratios were about the same: 2.3:1. The swarming abilities of both strains were similar (data not shown), suggesting that the absolute values of CheY and CheZ are not as crucial for chemotaxis as their ratios. Kuo and Koshland (14) grew strain RP437 in a minimal medium and found a CheY/CheZ molar ratio of 8:1, with a CheZ concentration of 1 μM (about 900 molecules/cell using their cytoplasmic volume). Thus, their estimate of the number of molecules of CheZ per cell is close to ours, but their estimate of the number of CheY molecules is about 2.5 times higher. Matsumura et al. (17), working with the same strain, reported a 20-fold larger number of molecules of CheZ. The reason for this difference is not known.
TABLE 2.
Concentrations of CheY and CheZ in wild-type and cheZ-deleted strainsa
| Strain | CheY
|
CheZ
|
||
|---|---|---|---|---|
| Molecules/cell | μM | Molecules/cell | μM | |
| AW405 | 6,850 ± 1,300 | 27.2 ± 5.2 | 3,050 ± 580 | 12.1 ± 2.3 |
| RP437 | 2,750 ± 275 | 9.1 ± 0.9 | 1,170 ± 170 | 3.9 ± 0.5 |
| RP1616 | 2,780 ± 4 | 9.2 ± 0.01 | ||
Eight immunoblots (two for RP1616) containing four samples were analyzed for each entry. The values given are means and standard deviations, with the result for each immunoblot weighted equally. Molecules per cell were computed from the molecular mass of each protein (14.3 kDa for CheY and 24 kDa for CheZ) and the measured cell density ([8.7 ± 0.9] × 108/ml for AW405 and [7.6 ± 0.2] × 108/ml for RP437 at an optical density at 610 nm of 0.8). Corresponding concentrations were calculated from the measured dry weight per milliliter of cell culture (0.26 mg/ml for AW405 and 0.27 mg/ml for RP437) and the cytoplasmic volume per milligram (dry weight) (1.4 μl [31]). For RP1616, the cell density and dry weight per milliliter of cell culture were assumed to be the same as for its parental strain, RP437.
Given the recent interest in modeling chemotaxis (7, 10, 15, 30), it might be worth determining the range of concentrations of different chemotaxis proteins needed for effective chemotaxis. However, levels of expression of proteins and cell volumes depend on how cells are grown, so growth conditions would need to be standardized. Using the conversion factor noted earlier (31), we obtained cytoplasmic volumes for strains AW405 and RP437 of 0.42 and 0.50 fl/cell, respectively, in agreement with the value of 0.46 fl/cell found by Brenner and Tomizawa (9) for cells derived from strain C600 grown in Luria broth to a density of 3.7 × 108 cells/ml.
Effect of CheZ on swarming in cells expressing CheY**.
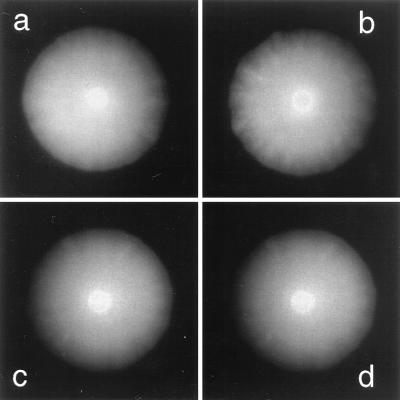
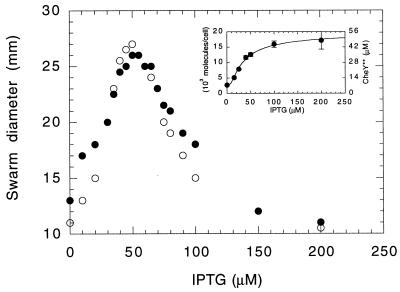
We studied the effect of CheZ on cells expressing CheY**, a protein that induces CW flagellar rotation without phosphorylation (39). Earlier, we used variable expression of this protein to examine control of the direction of flagellar rotation in cells deleted for cheA and cheZ (24). In the present work, we also used cells deleted only for cheZ. Variable expression of CheZ in cells expressing a moderate amount of CheY** had no effect on swarm size, even when cells were grown for a relatively long period of time (Fig. 4). Nor did the swarm size of cells expressing variable amounts of CheY** depend on whether or not the cells expressed CheZ (Fig. 5). In the latter experiment, cells swam smoothly at low concentrations of IPTG and tumbled incessantly at high concentrations. Swarm size was maximal when cells both ran and tumbled (see Wolfe and Berg [37]). Similar results were obtained in a cheA cheZ deletion background with strain HCB900 (data not shown). These results indicate that CheZ has little, if any, direct effect on the behavior of the flagellar motor, nor does it sequester CheY**.
FIG. 4.
Similarities of swarm size and morphology with or without CheZ in strains HCB917 and HCB918, which also express CheY**. Both strains carry pMS421 (which provides LacI); in addition, HCB917 carries pBES40 (inducible cheZ) and HCB918 carries pASK75 (the parental plasmid). CheY was induced with 70 μM IPTG (see Fig. 5). Strains and AHT concentrations are as follows: HCB918, 0 μg/liter (a); HCB917, 0 μg/liter (b); HCB917, 2 μg/liter (c); HCB917, 20 μg/liter (d). Plates were incubated for 40 h at 33°C. Swarm diameters are about 30 mm.
FIG. 5.
Variations in swarm size as a function of the level of expression of CheY**, with (closed symbols, strain HCB917, AHT at 20 μg/liter) or without (open symbols, strain HCB918, AHT at 20 μg/liter) CheZ. Plates were incubated for 40 h at 33°C. (Inset) Level of expression of CheY** in strain HCB917 as measured by immunoblot analysis. Error bars are standard errors, with the result for each sample on an immunoblot weighted equally.
Effect of CheZ on switching behavior of cells expressing CheY**.
The above conclusions were confirmed by measuring the rotational bias of tethered cells expressing small or large amounts of CheZ (Table 3). The strains examined were modified to take advantage of the self-tethering (sticky-filament) property of fliC(St) (24) by introducing a fliC mutation in the chromosome and adding a plasmid, pBES42, carrying both the fliC(St) allele and lacIq. Strain HCB919 carries, in addition, the cheZ plasmid pBES40, and HCB920 carries, in addition, the parental plasmid pASK75. Shifts in CW bias were small: the effect of addition of AHT observed in the absence of CheZ (strain HCB920) was as large as or larger than that observed in the presence of small or large amounts of CheZ (strain HCB919). imilar results were found with the cheA-deleted strain HCB922 (data not shown). We found earlier (see Fig. 3a of reference 24) that the CW bias changes dramatically with CheY** concentration. So again, these results indicate that CheZ has little, if any, direct effect on the behavior of the flagellar motor, nor does it sequester CheY**.
TABLE 3.
CW bias of cells expressing CheY**, with or without CheZa
| Strain | AHT (μg/liter) | CheZ (μM) | CW bias |
|---|---|---|---|
| HCB919 | 0 | <0.2 | 0.61 ± 0.06 |
| 200 | 210 ± 20 | 0.56 ± 0.07 | |
| HCB920 | 0 | 0 | 0.72 ± 0.05 |
| 200 | 0 | 0.50 ± 0.05 |
CheY** was induced with 15 μM IPTG, and the amount of CheY** was determined by immunoblotting with anti-CheY MAb. Concentrations of CheY and CheZ were computed by using the dry weight value of 0.26 mg/ml, determined for the parental strain of HCB919 and HCB920 (HCB901 [24]). For CheY, this gave a concentration of about 9.6 ± 1.4 μM (3,400 ± 500 molecules per cell). Tethering data were collected from two independent sets of experiments. CW bias values are presented as means and standard errors for measurements made on at least 36 cells.
Oligomerization of CheZ in the presence of CheY and CheY**.
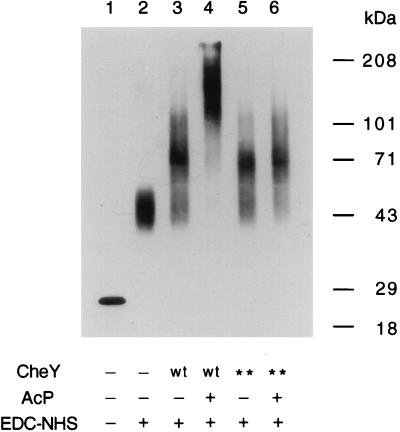
As just seen, CheZ has a great effect on the behavior of cells expressing wild-type CheY, but it has little, if any, effect on cells expressing CheY**, even though CheY** modulates rotational bias. Therefore, the effect of CheZ in wild-type cells must be due to an interaction with CheY itself and/or with an upstream component that controls the phosphorylation of CheY. To test the possibility that CheY** might interact with CheZ, for example, by promoting oligomerization of CheZ (3), we carried out the in vitro cross-linking procedure of Blat and Eisenbach (3) with purified proteins. Cross-linked products were detected on immunoblots of polyacrylamide gels by using anti-CheZ MAbs. Figure 6 shows a comparison of bandsformed in the presence of CheY or CheY** under nonphosphorylating and phosphorylating conditions. In the absence of CheY, CheZ formed a band with an apparent molecular mass of about 46 kDa, which is its dimeric form (Fig. 6, lane 2). In the presence of unphosphorylated CheY, the band shifted to a higher molecular mass, about 72 kDa (Fig. 6, lane 3). This complex did not contain CheY, because it did not appear on immunoblots probed with anti-CheY MAbs (data not shown). Therefore, it must be CheZ in its trimeric form. The addition of acetylphosphate promoted the phosphorylation of CheY, resulting in the formation of products in the 200-kDa range (Fig. 6, lane 4). The presence of CheY was evident in these products when the immunoblots were probed with anti-CheY MAbs (data not shown). These data are very similar to those obtained by Blat and Eisenbach (3), who used radioactively labeled proteins, except that the trimerization of CheZ promoted by CheY (Fig. 6, lane 3) is more pronounced (see also Fig. 5 of reference 2). The addition of CheY** in the absence of acetylphosphate (Fig. 6, lane 5) resulted in the formation of an oligomeric state of CheZ identical to that observed in the presence of unphosphorylated CheY. The result was the same on addition of CheY** in the presence of acetylphosphate (Fig. 6, lane 6); however, we do not know the extent of phosphorylation of CheY** by acetylphosphate at the concentration used (18 mM). The absence of CheY** in these 72 kDa-complexes was confirmed by using anti-CheY MAb as a probe. Thus, in vitro, CheY** has the same effect on CheZ as does unphosphorylated CheY, even though in vivo it also promotes CW flagellar rotation.
FIG. 6.
Immunoblot of cross-linked products of CheZ recorded on film by enhanced chemiluminescence. The molecular mass scale is shown on the right. Symbols on the bottom: +, present; −, absent; wt, wild-type CheY; ∗∗, CheY**; AcP, acetylphosphate (which phosphorylates wild-type CheY); EDC-NHS, cross-linking reagents (see Materials and Methods). Each lane contained 100 ng of CheZ.
Cross-linking of wild-type and mutant CheY to FliM.
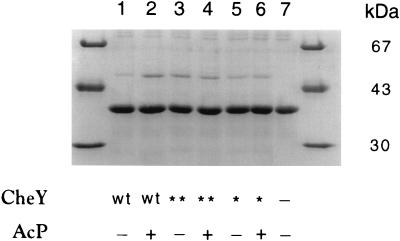
CheY-P is known to bind to the switch component FliM (36). In our earlier work (24), we showed that CheY** promotes switching without being phosphorylated. Here, we demonstrated the affinity of CheY** for FliM by using the in vitro cross-linking assay of Bren et al. (8). Figure 7 shows a polyacrylamide gel after Coomassie staining of the cross-linked products of FliM with wild-type CheY, CheY**, and CheY13DK in the absence or presence of acetylphosphate. The presence of CheY in the bands at 52 kDa was confirmed by immunoblot analysis, and the immunoblot band intensities agreed with those observed with Coomassie staining (data not shown). As expected, increased cross-linking of CheY to FliM occurred when CheY was phosphorylated (Fig. 7, lanes 1 and 2) (see also reference 8). On the other hand, the cross-linked products between CheY** or CheY13DK and FliM were the same under both nonphosphorylating and phosphorylating conditions (Fig. 7, lanes 3 and 4 and lanes 5 and 6). But once again, we do not know the extent of phosphorylation of either mutant protein by acetylphosphate at the concentration used (22 mM). However, the affinity of the double mutant to FliM was about four times higher than that of the single mutant, i.e., about 80% of the affinity of CheY-P to FliM. By this assay, CheY** has nearly the same affinity to FliM as CheY-P.
FIG. 7.
Coomassie-stained polyacrylamide gel of cross-linked products of FliM and CheY. Molecular mass markers appear on the left and right. FliM appears at 38 kDa, and the cross-linked product CheY-FliM appears at 52 kDa. Monomeric CheY (14.3 kDa) is not shown. Symbol not identified in the legend to Fig. 6: ∗, CheY13DK. The cross-linking reagent was DMS. Each lane contained 7 μg of FliM.
Summary.
We have shown that overexpression of CheZ in wild-type cells abolishes chemotaxis (see also reference 12). However, it has no effect on the behavior of cells whose motors are activated for switching by the double mutant CheY**. This is true in both cheA+ and cheA mutant backgrounds. We conclude that CheZ does not affect the motor directly. This is consistent with in vitro studies showing that CheZ does not bind to the switch component FliG, FliM, or FliN (8) and supports the contention that direct interaction between CheZ and the motor has not been established by studies of intragenic suppression (13, 29). We have shown previously, in a cheA mutant background, that rotational bias is a sensitive function of the level of expression of CheY** (24). Therefore, our data also indicate that CheZ does not sequester CheY**. It is possible, a priori, that some CheY** is bound to CheZ. If so, this binding does not interfere with the interaction of CheY** with the switch. We were not able to detect any cross-linking between CheY** and CheZ in vitro. It also is possible that some CheY** is phosphorylated in a cheA+ background, since both CheY13DK and CheY106YW can be phosphorylated by CheA, although only to a limited extent (5, 38). If so, dephosphorylation or sequestration of this form of CheY** by CheZ also has a negligible effect. Our measurements do not address the possibility that CheZ interacts with CheAshort or CheAlong (34), because those interactions would modulate the activity of the kinase, to which our assays are insensitive.
Working in vitro, we found that CheY** can promote trimerization of CheZ to the same extent as wild-type CheY, whereas CheY-P induces the formation of complexes of higher molecular weight containing both CheY and CheZ (3). We also found that CheY** can be cross-linked to the CheY-P receptor, FliM, almost as well as CheY-P itself (36). Cross-linking of CheY13DK to FliM, although detectable, was substantially lower. Therefore, while CheY** interacts with CheZ in a manner similar to CheY, it interacts with FliM in a manner similar to CheY-P. The reasons for this will not be understood in detail until the crystal or nuclear magnetic resonance structures of CheY**, CheY, and CheY-P can be compared.
There are wide discrepancies in the literature on the number of molecules of CheY and CheZ (and their concentrations) present in wild-type cells. Given the large behavioral sensitivity to the CheY/CheZ molar ratio, it is unlikely that modeling of chemotaxis (7, 10, 15, 30) can be very successful until these discrepancies are resolved.
ACKNOWLEDGMENTS
We thank Linda Turner for help with the analysis of tethered cells.
This work was supported by Public Health Service grant AI16478 from the National Institute of Allergy and Infectious Diseases and by the Rowland Institute for Science.
REFERENCES
- 1.Armstrong J B, Adler J, Dahl M M. Nonchemotactic mutants of Escherichia coli. J Bacteriol. 1967;93:390–398. doi: 10.1128/jb.93.1.390-398.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blat Y, Eisenbach M. Mutants with defective phosphatase activity show no phosphorylation-dependent oligomerization of CheZ. J Biol Chem. 1996;271:1232–1236. doi: 10.1074/jbc.271.2.1232. [DOI] [PubMed] [Google Scholar]
- 3.Blat Y, Eisenbach M. Oligomerization of the phosphatase CheZ upon interaction with the phosphorylated form of CheY. J Biol Chem. 1996;271:1226–1231. doi: 10.1074/jbc.271.2.1226. [DOI] [PubMed] [Google Scholar]
- 4.Blat Y, Eisenbach M. Phosphorylation-dependent binding of the chemotaxis signal molecule CheY to its phosphatase, CheZ. Biochemistry. 1994;33:902–906. doi: 10.1021/bi00170a008. [DOI] [PubMed] [Google Scholar]
- 5.Bourret R B, Drake S K, Chervitz S A, Simon M I, Falke J J. Activation of the phophosignaling protein CheY. II. Analysis of activated mutants by 19F NMR and protein engineering. J Biol Chem. 1993;268:13089–13096. [PMC free article] [PubMed] [Google Scholar]
- 6.Bourret R B, Hess J F, Simon M I. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc Natl Acad Sci USA. 1990;87:41–45. doi: 10.1073/pnas.87.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bray D, Bourret R B, Simon M I. Computer simulation of the phosphorylation cascade controlling bacterial chemotaxis. Mol Biol Cell. 1993;4:469–482. doi: 10.1091/mbc.4.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bren A, Welch M, Blat Y, Eisenbach M. Signal termination in bacterial chemotaxis: CheZ mediates dephosphorylation of free rather than switch-bound CheY. Proc Natl Acad Sci USA. 1996;93:10090–10093. doi: 10.1073/pnas.93.19.10090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenner M, Tomizawa J-I. Quantitation of ColE1-encoded replication elements. Proc Natl Acad Sci USA. 1991;88:405–409. doi: 10.1073/pnas.88.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauri D C, Ross J. A model of excitation and adaptation in bacterial chemotaxis. Biophys J. 1995;68:708–722. doi: 10.1016/S0006-3495(95)80232-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hess F, Oosawa K, Kaplan N, Simon M I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 12.Huang C, Stewart R C. CheZ mutants with enhanced ability to dephosphorylate CheY, the response regulator in bacterial chemotaxis. Biochim Biophys Acta. 1993;1202:297–304. doi: 10.1016/0167-4838(93)90019-n. [DOI] [PubMed] [Google Scholar]
- 13.Irikura V M, Kihara M, Yamaguchi S, Sockett H, Macnab R M. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J Bacteriol. 1993;175:802–810. doi: 10.1128/jb.175.3.802-810.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo S C, Koshland D E., Jr Roles of cheY and cheZ gene products in controlling flagellar rotation in bacterial chemotaxis of Escherichia coli. J Bacteriol. 1987;169:1307–1314. doi: 10.1128/jb.169.3.1307-1314.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levin M D, Morton-Firth C J, Abouhamad W N, Bourret R B, Bray D. Origins of individual swimming behavior in bacteria. Biophys J. 1998;74:175–181. doi: 10.1016/S0006-3495(98)77777-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manen D, Pougeon M, Damay P, Geiselmann J. A sensitive reporter gene system using bacterial luciferase based on a series of plasmid cloning vectors compatible with derivatives of pRB322. Gene. 1997;186:197–200. doi: 10.1016/s0378-1119(96)00702-0. [DOI] [PubMed] [Google Scholar]
- 17.Matsumura P, Roman S, Volz K, McNally D. Signalling complexes in bacterial chemotaxis. Symp Soc Gen Microbiol. 1990;46:135–154. [Google Scholar]
- 18.Matsumura P, Rydel J J, Linzmeier R, Vacante D. Overexpression and sequence of the Escherichia coli cheY gene and the biochemical activities of the CheY protein. J Bacteriol. 1984;160:36–41. doi: 10.1128/jb.160.1.36-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parkinson J S. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol. 1978;135:45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson J S, Parker S R. Interaction of the cheC and cheZ gene products is required for chemotactic behavior in Escherichia coli. Proc Natl Acad Sci USA. 1979;76:2390–2394. doi: 10.1073/pnas.76.5.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parkinson J S, Parker S R, Talbert P B, Houts S E. Interactions between chemotaxis genes and flagellar genes in Escherichia coli. J Bacteriol. 1983;155:265–274. doi: 10.1128/jb.155.1.265-274.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanatinia H, Kofoid E C, Morrison T B, Parkinson J S. The smaller of two overlapping cheA gene products is not essential for chemotaxis in Escherichia coli. J Bacteriol. 1995;177:2713–2720. doi: 10.1128/jb.177.10.2713-2720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanna M G, Simon M I. In vivo and in vitro characterization of Escherichia coli protein CheZ gain- and loss-of-function mutants. J Bacteriol. 1996;178:6275–6280. doi: 10.1128/jb.178.21.6275-6280.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scharf B E, Fahrner K A, Turner L, Berg H C. Control of direction of flagellar rotation in bacterial chemotaxis. Proc Natl Acad Sci USA. 1998;95:201–206. doi: 10.1073/pnas.95.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuster M, Abouhamad W N, Silversmith R E, Bourret R B. Chemotactic response regulator mutant CheY95IV exhibits enhanced binding to the flagellar switch and phosphorylation-dependent constitutive signalling. Mol Microbiol. 1998;27:1065–1075. doi: 10.1046/j.1365-2958.1998.00756.x. [DOI] [PubMed] [Google Scholar]
- 26.Segall J E, Ishihara A, Berg H C. Chemotactic signaling in filamentous cells of Escherichia coli. J Bacteriol. 1985;161:51–59. doi: 10.1128/jb.161.1.51-59.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silversmith R E, Appleby J L, Bourret R B. Catalytic mechanism of phosphorylation and dephosphorylation of CheY: kinetic characterization of imidazole phosphates as phosphodonors and the role of acid catalysis. Biochemistry. 1997;36:14965–14974. doi: 10.1021/bi9715573. [DOI] [PubMed] [Google Scholar]
- 28.Smith R A, Parkinson J S. Overlapping genes at the cheA locus of Escherichia coli. Proc Natl Acad Sci USA. 1980;77:5370–5374. doi: 10.1073/pnas.77.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sockett H, Yamaguchi S, Kihara M, Irikura V M, Macnab R M. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J Bacteriol. 1992;174:793–806. doi: 10.1128/jb.174.3.793-806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spiro P A, Parkinson J S, Othmer H G. A model of excitation and adaptation in bacterial chemotaxis. Proc Natl Acad Sci USA. 1997;94:7263–7268. doi: 10.1073/pnas.94.14.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stock J B, Rauch B, Roseman S. Periplasmic space in Salmonella typhimurium and Escherichia coli. J Biol Chem. 1977;252:7850–7861. [PubMed] [Google Scholar]
- 32.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 33.Tang H, Blair D F. Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J Bacteriol. 1995;177:3485–3495. doi: 10.1128/jb.177.12.3485-3495.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, Matsumura P. Characterization of the CheAS/CheZ complex: a specific interaction resulting in enhanced dephosphorylating activity on CheY-phosphate. Mol Microbiol. 1996;19:695–703. doi: 10.1046/j.1365-2958.1996.393934.x. [DOI] [PubMed] [Google Scholar]
- 35.Welch M, Oosawa K, Aizawa S-I, Eisenbach M. Effects of phosphorylation, Mg2+, and conformation of the chemotaxis protein CheY on its binding to the flagellar switch protein FliM. Biochemistry. 1994;33:10470–10476. doi: 10.1021/bi00200a031. [DOI] [PubMed] [Google Scholar]
- 36.Welch M, Oosawa K, Aizawa S-I, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfe A J, Berg H C. Migration of bacteria in semisolid agar. Proc Natl Acad Sci USA. 1989;86:6973–6977. doi: 10.1073/pnas.86.18.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu X, Amsler C D, Volz K, Matsumura P. Tyrosine 106 of CheY plays an important role in chemotaxis signal transduction in Escherichia coli. J Bacteriol. 1996;178:4208–4215. doi: 10.1128/jb.178.14.4208-4215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu, X., and P. Matsumura. Personal communication.