Abstract
The two-component system (TCS) generally consists of three elements, namely the histidine kinase (HK), response regulator (RR), and histidine phosphotransfer (HP) gene families. This study aimed to assess the expression of TCS genes in P. vulgaris leaf tissue under salt and drought stress and perform a genome-wide analysis of TCS gene family members using bioinformatics methods. This study identified 67 PvTCS genes, including 10 PvHP, 38 PvRR, and 19 PvHK, in the bean genome. PvHK2 had the maximum number of amino acids with 1261, whilst PvHP8 had the lowest number with 87. In addition, their theoretical isoelectric points were between 4.56 (PvHP8) and 9.15 (PvPRR10). The majority of PvTCS genes are unstable. Phylogenetic analysis of TCS genes in A. thaliana, G. max, and bean found that PvTCS genes had close phylogenetic relationships with the genes of other plants. Segmental and tandem duplicate gene pairs were detected among the TCS genes and TCS genes have been subjected to purifying selection pressure in the evolutionary process. Furthermore, the TCS gene family, which has an important role in abiotic stress and hormonal responses in plants, was characterized for the first time in beans, and its expression of TCS genes in bean leaves under salt and drought stress was established using RNAseq and qRT-PCR analyses. The findings of this study will aid future functional and genomic studies by providing essential information about the members of the TCS gene family in beans.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-023-01406-5.
Keywords: Expression profile, Histidine, Melatonin, Phaseolus vulgaris, Two component system (TCS)
Introduction
The bean, Phaseolus vulgaris L., is a new world legume grown as a monoculture crop everywhere from the low-lying tropics to the semi-arid parts of temperate climates (Thung and Rao 1999). Furthermore, it is recognized as the primary food source for millions of people in underdeveloped nations, as well as an essential source of energy, minerals, and total protein globally (Petry et al. 2015; Pereira et al. 2020). One hundred grams of beans contain approximately 20–25 g of protein, 60 g of carbohydrates, and 0.7–1.5 g of fat. It also includes various vitamins, minerals, and phytochemicals (Azarpazhooh and Boye 2012; Terzi et al. 2020).
Plants are subjected to multiple biotic and abiotic burdens throughout their lives as they remain stable in their environment. Drought and soil salinity are two main environmental factors that limit plant growth and development and result in large output losses, especially in arid and semi-arid areas (Raza et al. 2022a, 2023a; Suleymanov et al. 2023). Salinity stress affects plants in three different ways. The first is that the roots cannot obtain enough water from the soil because salt reduces the osmotic potential of the soil. Second, an increase in Na+ and Cl− ions cause the breakdown of cellular organelles, followed by the reduction of protein synthesis and enzyme activity, and finally, decreased nutrition intake and/or transport to shoots (Yavaş and Hussain 2022; Karimi et al. 2023). Since salinity reduces the ability of plants to use water, it slows down the plant's growth rate (Hailu and Mehari 2021). Stomata close and the amount of CO2 between leaf cells reduces under salt stress. Salt stress also decreases the use of NADPH by the Calvin cycle, inhibits the production of chlorophyll and Rubisco, and interferes with the photosynthetic electron transport system (Miao et al. 2023). Drought stress is an important limiting factor in bean cultivation worldwide, especially in marginal and unfavorable environments (Rodiño et al. 2020; Sánchez-Reinoso et al. 2020). More than 60% of bean-producing areas cannot provide the necessary water for growth, and drought-related yield losses of up to 80% have occurred in some regions (Losa et al. 2022). Many metabolic processes are disrupted by drought stress in plants, including photosynthesis, stomatal conductivity, leaf area, reduced biomass, and decreased pod and seed yield (Asfaw et al. 2017; Hussain et al. 2023; Raza et al. 2023b; Yang et al. 2023).
Cytokinin is a key plant hormone that plays a role in virtually all aspects of plant growth and development, such as root and leaf development, plant formation, flowering, and seed germination, as well as responses to environmental stimulants (Liu et al. 2020). A two-component system (TCS) converts the cytokinin signal in plants. This system was initially identified in bacteria, but slime molds, fungi, and plants also use it in other signal transduction pathways (Loomis et al. 1997; Schaller 2000; Stock et al. 2000; Hutchison and Kieber 2002). TCS is typically made up of two gene families: the response regulator (RR) gene family and the histidine kinase (HK) gene family (Stock et al. 2000; Liu et al. 2020). HK proteins can be phosphorylated, transferring a phosphoryl group to a conserved aspartate residue in the RR's response (Rec) domain. The phosphorylated RR protein directly or indirectly regulates downstream gene activity (Stock et al. 2000; Hwang et al. 2012). Plant TCSs are more complex than most bacterial TCSs, and a histidine phosphotransfer (HP) gene family has been discovered to be responsible for an extra phosphorylation step in plants (Urao et al. 1998; Thomason and Kay 2000). The HP protein links the phosphoryl group transfer between HK and RR (Schaller et al. 2008).
Plants have three HK subfamilies: phytochromes, cytokinin receptors, and ethylene receptors. In Arabidopsis, three more HKs (ACKI1, AHK1, and AHK5/CKI2) are not part of any group. The general structure of HKs includes several trans-membrane domains, an input domain located in the N-terminal region, a donor domain with an autophosphorylation site and a conserved histidine residue, and an acceptor domain (Rec) (Hwang et al. 2002). However, because they lack highly conserved residues and motifs, the three ethylene receptors (ERS2, AETR2, and AEIN4) and phytochrome donors are unlikely to display HK action. As a result, they are referred to as distinct HKs (Kiba et al. 2005; Mähönen et al. 2006). Furthermore, the cytokinin receptors AHK2, AHK3, and AHK4 possess a cyclase/HK-associated sensory extracellular domain (CHASE) that cytokinin recognizes. AER1, AETR2, AEIN4, ERS1, and ERS2, are ethylene receptors with ethylene (C2H4) binding domains. Furthermore, phytochrome family members (PHYA, PHYB, PHYC, PHYD, and PHYE) have a chromophore (PHY) binding domain and two PAS folds (Per/Arndt/Sim) (Stock et al. 2000).
The phosphotransfer domain (Hpt) of the HP gene family contains a highly conserved motif (XHQXKGSSXS) required for the transfer of the phosphate group from the Rec domain of HKs to the Rec domain of RRs (Hwang et al. 2002). Another type of HP, known as pseudo-histidine-containing phosphotransfer protein, lacks the conserved histidine residue. This group is not a phosphor-transmitter protein, but it does participate in cytokinin signaling by inhibiting the phosphorus delay from phosphorylated AHP1 to ARR (Mähönen et al. 2006).
The type-A, type-B, and type-C subgroups of the RR family are based on conserved amino acid sequences and domains. Cytokinin response proteins known as type-A RRs have Rec domains in C-terminal extensions. On the other hand, type-B RRs are transcription factors consisting of an N-terminal Rec domain and a C-terminal output domain (Lohrmann et al. 1999). Although type-C RRs have the same domain as type-A RRs, they are not induced by cytokinin (Mizuno 2004). In addition to these RRs, there is a different class of RRs, known as pseudo-RRs (PRRs). PRRs are missing a highly conserved D residue required for phosphorylation (Makino et al. 2000). PRRs contain a specific CCT (Co, Col, and Toc1) motif in their C-terminal extension, vital in regulating circadian rhythms (Más 2008; Tsai et al. 2012).
Melatonin (N-acetyl-5-methoxytryptamine) is found in many different plant specie's leaves, stems, fruits, and seeds (Raza et al. 2022b; Golding and Lee 2023). Seeds and leaves generally have the highest melatonin content than fruit (Xiang et al. 2023). It is an indolic substance that shares structural similarities with other vital substances such as indole-3-acetic acid (IAA), serotonin, and tryptophan (Raza et al. 2022c). Melatonin intermediates exhibit antioxidant properties and have synergistic effects with other antioxidant compounds such as ascorbic acid and glutathione (Ramasamy et al. 2023). Due to its solubility in lipids and water, melatonin functions as a universal hydrophilic and hydrophobic antioxidant (Mir et al. 2022). This property allows it to quickly cross cell membranes and penetrate subcellular spaces (Marcantonini et al. 2022).
The purpose of this study was to analyze the genome-wide TCSs in bean genotypes and determine the effects of melatonin treatment against salt and drought on expressions of TCS genes. This study will contribute significantly to the literature on PvTCS genes and their functions in the context of gene evolution, genomic distribution, and expression patterns under varying environmental conditions. Moreover, these results may help future functional and genomic studies in other plants.
Materials and methods
Detection of PvTCS genes in the bean genome
Protein sequences of HK, HP, and RR genes encoding TCS elements in the bean genome were obtained from the Phytozome v13 database (Goodstein et al. 2012) using Pfam Accession Numbers His kinase A (HisKA) domain (PF00512), histidine kinase-like ATPase (HATPase) domain (PF02518), cyclases/histidine kinases associated sensing extracellular (CHASE) domain (PF03924), histidine phosphotransfer (Hpt) domain (PF01627) and response (Rec) domain (PF00072) in the Pfam database (Mistry et al. 2021). To identify proteins of all possible TCS elements in the bean (Schmutz et al. 2014), the annotated proteins from the P. vulgaris genome were scanned with default parameters, both blastp in the Phytozome v13 database and hidden Markov model (HMM) (http://www.ebi.ac.uk). The presence of HisKA, HATPase, CHASE, HPt, and Rec characteristic domains in the detected sequences was detected using the HMMER scanner. The molecular weight, amino acid numbers, and instability index, which if the index is less than 40, then protein is probably stable. If it is greater then it is probably unstable (Guruprasad et al. 1990) and the theoretical isoelectric point (pI) of the obtained TCS proteins was determined using the “ProtParam tool” (Gasteiger et al. 2005).
Determination of gene structures, chromosomal locations, gene duplications, and conserved motifs of TCS elements in beans
Gene Structure Display Server v2.0 was used to obtain information about the exon–intron structures of genes of TCS elements in beans (Guo et al. 2007). Through a comparison of genome and coding DNA (CDS) sequences, the locations of the bean TCS element genes were established. The Phytozome v13 database was used to establish the chromosomal localizations and sizes of bean TCS genes. In addition, PvTCS genes were marked on the corresponding chromosomes and shaped with the Circos (Krzywinski et al. 2009); TBtools subsequently displayed a syntenic map (Chen et al. 2020). Non-synonymous values (Ka), synonymous values (Ks), and evolutionary strains’ ratios (Ka/Ks) were calculated at the codeml program of the PAML package (Yang 2007) with PAL2NAL v14 online software which is the aligner for PAML (Suyama et al. 2006) using protein and transcript sequence of segmentally and tandemly-duplicated PvTCS gene pairs. In this online software, ‘Remove mismatches’ and ‘selected positions’ were selected as no, and Universal Code (NCBI: transl_table = 1) ‘Removing Gaps’ and ‘Calculate dS and dN’ were selected as yes.
Multiple EM for Motif Elimination (MEME) v5.1.0 tools were used to determine extra-conserved motifs of bean TCS element proteins (Bailey et al. 2006). The limits for the min (2)/max (50) width as well as the max number of motifs (10) were set. In addition, the motif regions were adjusted between 2 and 300. The determined motifs were investigated using the default settings of the InterPro scanner (Quevillon et al. 2005).
Phylogenetic analysis and multiple sequence alignment
Phylogenetic analyses were carried out using the Neighbor-joining (NJ) method with a bootstrapping value of 1000 replicates. The protein sequences of the bean TCS elements were aligned with the ClustalW (p-distance) program (online last version/https://www.genome.jp/tools-bin/clustalw) (Thompson et al. 1997) and a phylogenetic tree was drawn by the MEGA v7 tool (Letunic and Bork 2011) and visualized by Interactive tree of life (iTOL) (Letunic and Bork 2011).
Comparative mapping between beans and other species
The orthologs of the TCS genes of A. thaliana (Lamesch et al. 2012), G. max (Schmutz et al. 2010), and P. vulgaris were identified by using the Multiple Collinearity Scan Toolkit (MCScanX) (Wang et al. 2012). Protein sequences of orthologous genes were revealed using Phytozome v13. A Synteny map of TCS genes in these plants was generated using TBtools.
Determination of cis-acting elements of PvTCS genes and subcellular localization of PvTCS proteins
Cis-acting element analysis was made using the PlantCARE database (Lescot et al. 2002) by taking the 5′ upstream region including approximately 2 kb DNA fragments of the PvTCS genes. Subcellular localization was determined using PvTCS protein sequences with WoLF PSORT (last version/https://wolfpsort.hgc.jp/) scanner (organism type: plants) (Horton et al. 2007).
Gene ontology analysis homology modeling, and protein–protein interactions of TCS proteins in bean
The STRING was used to describe protein–protein interactions' physical and functional linkages (Szklarczyk et al. 2021). The confidence score (0.4) and maximum additional interactions (5) were set in the STRING application in the Cytoscape program, and the interactions of PvTCS proteins with each other and with a few other proteins were determined. Furthermore, protein–protein interactions were reshaped with the Cytoscape tool (Shannon et al. 2003). Gene ontology data of PvTCS genes were accessed using the Blast2GO (Conesa et al. 2005) and Omics Box (Götz et al. 2008) tools to retrieve functional information on PvTCS proteins. Three-dimensional (3D) structures, created by comparing the reliability scores of the protein models, and protein homology patterns were determined using the Phyre2 database in the PvTCS protein sequences determined in this study (Kelley et al. 2015).
Identification of Pvu-miRNAs associated with PvTCS genes
All defined miRNA common bean sequences were obtained using PmiREN (Plant miRNA ENcyclopedia, http://www.pmiren.com). miRNA parameters were estimated using default settings in the psRNA Target Server (www.zhaolab.org/psRNATarget) (Zhang 2005). Targeted regions were predicted using PvTCS CDS sequences and Pvu-miRNA sequences. BLASTX online software (with a 1e-10 threshold) was used to compare in silico predicted miRNA targets to common bean-expressed sequence tags in the NCBI.
Tissue-specific in silico gene expression analysis of PvTCS genes
Illumina RNA-seq data to detect the expression of genes of TCS elements in bean leaves were obtained from the NCBI Sequence Read Archive (SRA) (https://www.ncbi.nlm.nih.gov/sra) data library. Accession numbers of SRR957668 (leaf exposed to salt stress), SRR958469 (leaf salt control), SRR8284481 (leaf exposed to drought stress), and SRR8284480 (leaf drought control) determined as a result of detailed examinations were used (Hiz et al. 2014). RPKM (reads per kilobase of exon model per million mapped reads) algorithm was used to normalize gene expression profiles (Mortazavi et al. 2008). RPKM values were transformed to log2 and a heatmap map was obtained with the CIMMiner (https://discover.nci.nih.gov/cimminer/oneMatrix.do).
Plant material, stress, and melatonin applications
Serra and Elkoca-05 common bean cultivars determined in a previous study (Aygören et al. 2022) were used as plant material in this study. PEG 6000 (0 and 20%) and NaCl (0 and 150 mM) were used as drought and salt stress agents, respectively. Cultivation of plants, applications of Melatonin, and stress agents were carried out according to the instructions of Aygören et al. (2022). Melatonin application (0 and 200 mM) was made by spraying the leaves 24 h before applying stresses in the hydroponic system containing 1/10 Hoagland solution. Plants were exposed to salt and drought stress for 9 days and 24 h, respectively. At the end of the period, the leaf tissue of the bean cultivars was taken into liquid nitrogen and stored at − 80 °C. This research was carried out based on a completely randomized design with three replicates. Five plants from each replicate per treatment were selected and bulked. The bulked sample technique was used for molecular analysis in the research.
Isolation of total RNA, cDNA synthesis, and expression analysis
The Trizol® Reagent (Invitrogen Life Technologies, ABD) was used to isolate total RNA. The RNA quality was checked using a spectrophotometer before being photographed on a 1.5% agarose gel. The RevertAid RT Reverse Transcription Kit (Fermentas, ABD) was used to apply cDNA synthesis according to the manufacturer's protocol. Eleven PvTCS genes were preferred for qRT-PCR analysis based on RNAseq data. Table 1 contains information about primers designed for qRT-PCR. The qRT-PCR reactions were carried out on the RotorGene® Q Real-Time PCR System (Corbett Research, Qiagen GmbH, Germany) with the ABT SYBER Green mix (Cat No: Q03-02-01, Ankara, Turkey). The qRT-PCR reaction mixture was 20 μL, of which the cDNA was 200 ng, both forward and reverse primers were 0.4 μL, and the 2X ABT SYBER Green Mix was 10 μL, with the reaction volume being completed to 20 μL with double distilled water. The reaction was performed as follows: 95 °C for 10 min, followed by 40 cycles of 94 °C for 15 s, and 60 °C for 30 s. The β-actin gene from P. vulgaris was used as a housekeeping gene. qRT-PCR data were normalized using the 2−∆∆CT method for relative quantification (Livak and Schmittgen 2001). GraphPad Prism 9 software (http://www.graphpad.com/) was used for statistical analyses based on the two-way ANOVA method and Dunnett's test at 0.05 significant levels. The interaction effect for genotype x treatment was evaluated based on a simple main effect.
Table 1.
The primer sequences used in qRT-PCR
| Gene Name | Forward primer | Reverse primer |
|---|---|---|
| PvPHYA2 | GTGAGTAAGAAGTTGGACGTAGAG | CAGATAGGCGCTGAATATGTAGT |
| PvHK2 | CTTGACTTCCCAAGAGGGTTATT | CTCAGCTCCCTGTCGAATTATG |
| PvHK4 | GTTCCATTTGATCTTCGTTCCATAC | CCCATGACAATATCCGGTACTTTA |
| PvHK6 | CATGCTCTGGCTATCCTTGTATC | CAAATGCTGTACTCTCGGTATATTC |
| PvHK8 | AGTAGAAGATGATTACGGTGAGATG | CTGATCTCGTGGGAAACAGTAG |
| PvHP5 | AGAGTACTGCCTTGTGAAGAATAAA | CACTGAAACTCAGTTCCGTAGG |
| PvHP6 | GAACAAGTGCCTGCTGATTTC | CTCACAGAAAGTCCTGAAGGTAG |
| PvRR12 | CTGAAGAGCCTCCCACTATTTC | TACCCTGTCATTTCTGGCATAC |
| PvRR15 | TGACAAGTGGTGGACAGAAC | GTAGCTCATAGCCTGTCATTCC |
| PvRR18 | AAGTGACGGTGGTGGAAAG | GGCATGGAGTAGTCTGTCATTAT |
| PvRR28 | GGAGGACGACCTGAATTTCTT | GCAACAAGCCTTGGATCATAAC |
| β-actin | TGAGCAAGGAGATTACAGCATTGG | CATACTCTGCCTTCGCAATCCAC |
Results
Detection of TCS genes in bean genome
Current genome-wide analysis revealed the presence of HK, HP, and RR genes encoding TCS elements in the common bean genome. Except for Pv4, the PvTCS genes of the bean were found on Pv1, 2, 3, 5, 6, 7, 8, 9, 10, 11 chromosomes and scaffold_17. The start and stop positions of the PvTCS genes, as well as the molecular weights, amino acid lengths, instability indexes, and theoretical isoelectric points (pI) of the proteins, as calculated by the "ProtParam tool," have been listed in Table S1. PvHK2 had the most amino acids (1261), whereas PvHP8 had the fewest (87). Furthermore, the isoelectric points of PvTCS genes were determined to vary from acid to alkaline. PvRR11 had the lowest value (4.18) while PvPRR10 had the highest (9.15). A great majority of PvTCS genes are unstable.
Determination of gene structures, chromosomal locations, gene duplications, and conserved motifs of TCS elements in beans
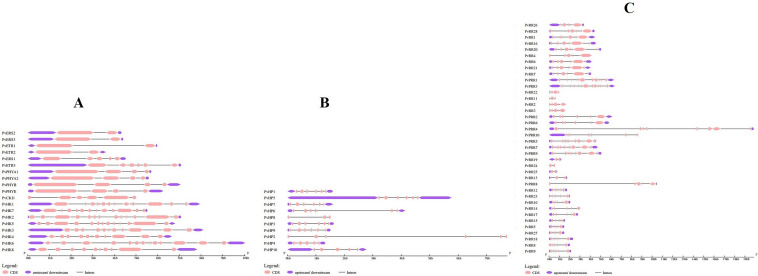
Figure 1 presents chromosomal localizations and sizes of bean TCS gene family members. Figure 2A depicts information regarding the intron–exon numbers of HK gene family members. In PvHK genes, intron numbers vary between 1 and 12 and exon numbers vary between 2 and 13. PvERS2, PvERS3, PvETR1, and PvETR2 genes had the lowest intron number, whereas PvHK2, 5, and 6 genes had the greatest intron number. On the other hand, the PvERS2, PvERS3, PvETR1, and PvETR2 genes had the fewest exons (2), whereas PvHK2, 5, and 6 genes had the most exons (13). Figure 2B shows the intron–exon numbers of the bean HP gene family members as a result of structural analysis using Gene Structure Display Server v2.0. Among PvHP genes, intron numbers vary between 2 and 5 and exon numbers vary between 3 and 6. It was determined that the lowest intron number was in the PvHP8 gene with 2, and the highest intron number was in the PvHP1, 2, 3, 5, 7, and 9 genes. On the other hand, in terms of exon number, the lowest number of exons was detected in the PvHP8 gene with 3, and the highest number of exons with 6 in PvHP1, 2, 3, 5, 7, and 9 genes.
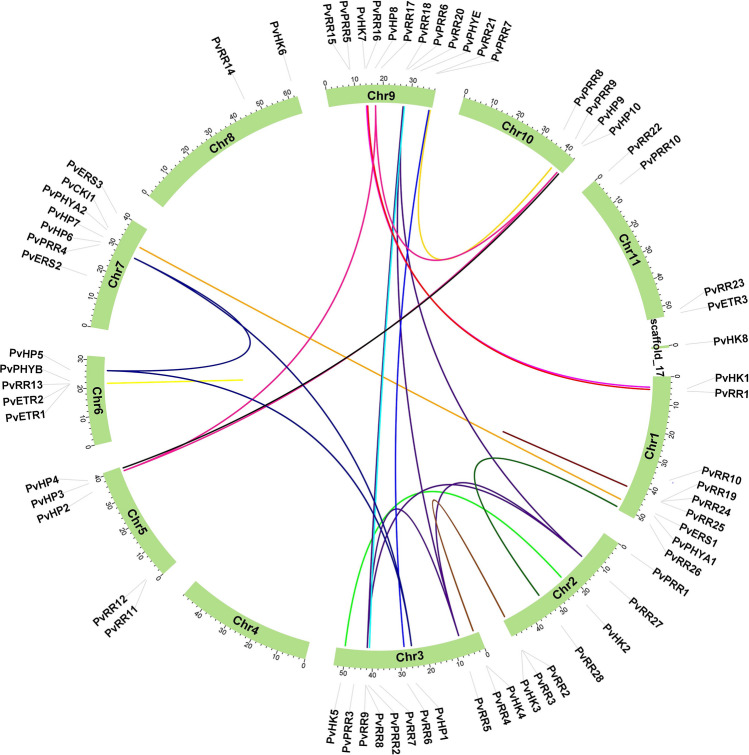
Fig. 1.
Distribution of PvTCS genes in bean chromosomes. *The colored lines indicate segmentally and tandemly duplicated gene pairs. Solid lines indicate segmental duplication, while incomplete lines indicate tandem duplications
Fig. 2.
Intron–exon model of PvHP, PvHK and PvRR gene family members. *Pink boxes represent exons, purple boxes represent upstream and downstream UTRs, and black lines represent introns
While the intron numbers of PvRR genes were between 1 and 10, their exon numbers varied between 2 and 11 (Fig. 2C). It was determined that the least number of introns was in PvRR19, 24, and 25 genes, and the highest number of introns was in PvRR1 and PvRR3 genes. The least number of exons were in PvRR19, 24, and 25 genes with 2, and the highest number of exons were found in PvRR1 and PvRR3 genes with 11.
The rough time of duplication events was calculated using T = Ks/2λ × 10 − 6 Mya (million years ago) in terms of synonymous replacement rates (λ) in beans (Schmutz et al. 2014). The Ka/Ks ratio of PvTCS genes was found to be less than one in the calculated values. All duplicated gene pairs of beans TCS gene family members have undergone negative selection in the evolutionary process. PvRR19–PvRR24 and PvETR1–PvETR2 gene pairs are tandemly duplicated genes. PvHK1–PvHK7, PvHK2–PvHK5, PvHP1–PvHP5, PvHP1–PvHP6, PvHP3–PvHP8, PvHP3–PvHP9, PvHP4–PvHP10, PvHP5–PvHP6, PvHP8–PvHP9, PvPHYA1–PvPHYA2, PvPRR2–PvPRR6, PvPRR7–PvPRR9, PvPRR1–PvPRR16, PvPRR2–PvPRR4, PvPRR26–PvPRR28, PvPRR27–PvPRR8, PvPRR27–PvPRR18, PvPRR27–PvPRR5, PvPRR5–PvPRR8, PvPRR5–PvPRR18, PvPRR6–PvPRR21 and PvPRR8–PvPRR18 gene pairs were found to be segmentally duplicated genes (Table S2).
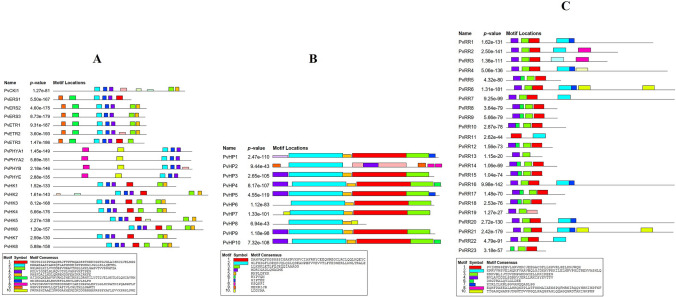
Ten conserved motifs were identified for each of the PvTCS gene family (HK, HP, and RR) members in the bean. Figure 3A–C, respectively, show possible conserved motif schemes for members of PvHK, PvRR, and PvHP gene families. According to the conserved motif analysis results,10 conserved motifs were determined per gene family and the amino acid numbers of the motifs ranged from 6 to 50. In the PvHK gene family members, PvPHYA1, PvPHYA2, and PvPHYE had the least motifs with 5 motifs while PvHK2 and PvCKI1 proteins had the highest motif with 8 motifs. HK-Motif-2, -4, and -7 were also found in all PvHK proteins. The HK-Motif-2 contains the HisKA domain. In the PvHP gene family members, the least motif (3 motifs) was found in the PvHP8 protein, and the highest number of motifs (8 motifs) was in the PvHP2 protein. In addition, HP-Motif-2 was found in the entire PvHP proteins and contained the HPt domain. HP-Motif-1 also contains the HPt domain and is located in all other PvHP proteins except PvHP2 and PvHP8. The best possible matches in the bean TCS genes and the domains in these sequences are given in Table S3. It was determined that the PvRR11 protein had the most minor motifs (2 motifs), while the PvRR6 and PvRR21 proteins had the most motifs (7 motifs). All PvRR proteins contain RR-Motif-1. Moreover, The RR-Motif-1, -3, and -4 motifs include the RESPONSE REGULATORY domain, that is, the RR domain. RR-Motif-3 was found in other PvRR proteins except for the PvRR11 protein, while the RR-Motif-4 was present in other PvRR proteins except for PvRR11 and PvRR23 proteins.
Fig. 3.
Possible conserved motif scheme of HK, RR, and HP gene family members in bean and possible sequence data corresponding to colored box symbols
Phylogenetic analysis and sequence alignment
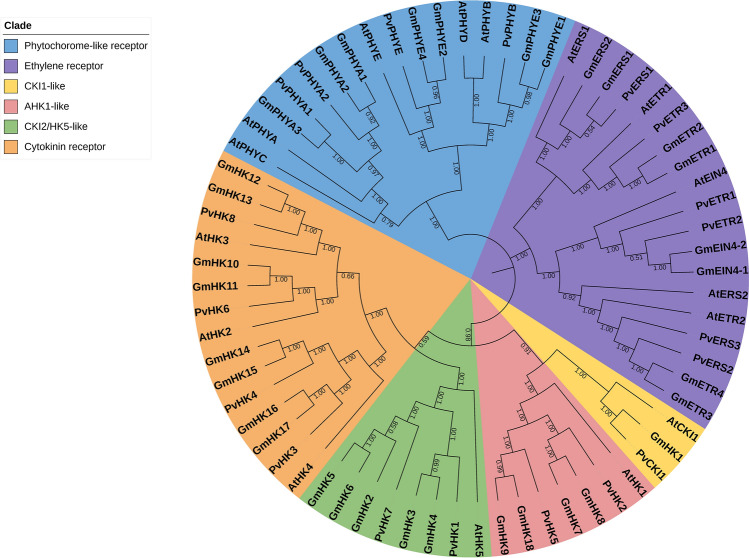
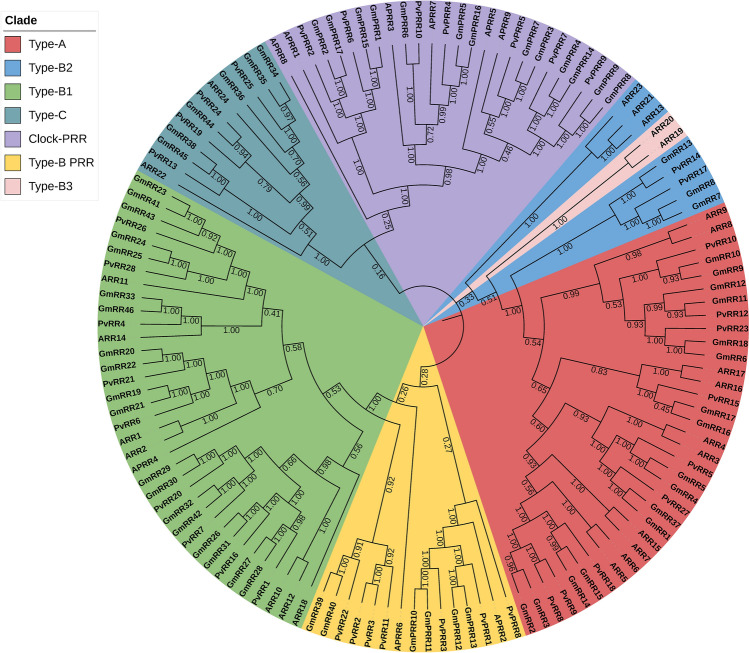
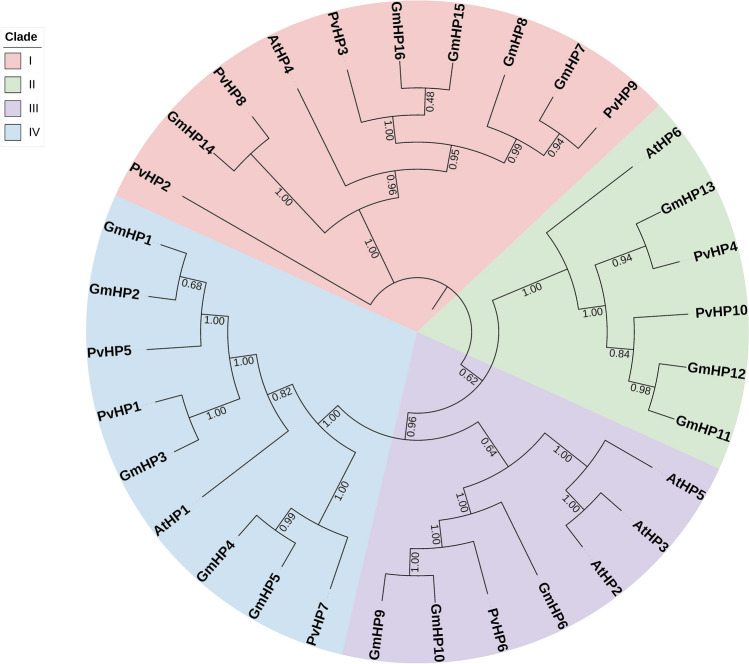
Phylogenetic analyses were performed according to the Neighbor-Joining (NJ) method with 1000 replicated bootstrap values. The protein sequences of the bean, soybean, and Arabidopsis TCS elements were aligned with the ClustalW program (Thompson et al. 1997). Phylogenetic trees were constructed by using the TCS protein sequences of bean, soybean, and Arabidopsis, and the phylogenetic trees of the HK (Fig. 4), RR (Fig. 5), and HP (Fig. 6) gene family members were divided into 6, 7, and 4 groups, respectively.
Fig. 4.
Neighbor-joining phylogenetic tree of P. vulgaris, G. max and A. thaliana HK gene family members. The phylogenetic tree was created using the Mega v7 program and full-length amino acid sequences from soybean, Arabidopsis, and common bean. The tree was divided into 6 classes with a total of 68 proteins, including Phytochorome-like receptor group, Ethylene receptor group, CKI1-like group, AHK1-like group, CKI2/HK5-like group and Cytokinin receptor group. The numbers are bootstrap values based on 1000 replicates
Fig. 5.
Neighbor-joining phylogenetic tree of P. vulgaris, G. max and A. thaliana RR gene family members. The phylogenetic tree was created using the Mega v7 program and full-length amino acid sequences from soybean, Arabidopsis, and common bean. The tree was divided into 7 classes with a total of 135 proteins, including Type-A group, Type-B1 group, Type-B2 group, Type-B3, Type-C group, Clock-PRR group, and Type-B PRR group. The numbers are bootstrap values based on 1000 replicates
Fig. 6.
Neighbor-joining phylogenetic tree of P. vulgaris, G. max and A. thaliana HP gene family members. The phylogenetic tree was created using the Mega v7 program and full-length amino acid sequences from soybean, Arabidopsis, and common bean. The tree was divided into 4 classes with 32 proteins, including Group I, Group II, Group III, and Group VI. The numbers are bootstrap values based on 1000 replicates
Comparative mapping between beans and other species
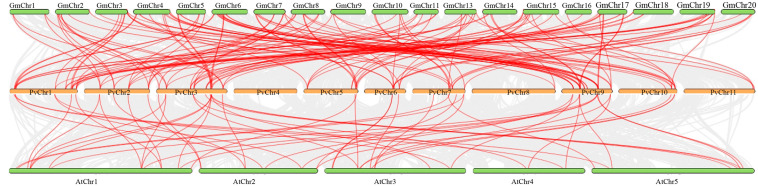
Synteny analysis, one of the elements of comparative genomics, is essential for determining the evolutionary relationship between genomes. Figure 7 show synteny maps for bean with G. max and bean with A. thaliana, respectively. For RR, HK, and HP, 83, 31, and 26 syntenic relationships, respectively, were determined between the bean and G. max. In contrast, this number of synthetic associations between beans and Arabidopsis was determined as 36 for RR genes, 9 for HK genes, and 6 for HP genes.
Fig. 7.
TCS element synteny map for bean/G. max and bean/Arabidopsis. The upper green boxes represent the soybean chromosomes (GmChr), the middle orange boxes the bean chromosomes (PvChr), and the lower green boxes the Arabidopsis chromosomes (AtChr). The red lines show the orthology in the PvTCS and GmTCS genes and PvTCS and AtTCS genes
Intracellular localization and promoter analysis of the genes of the bean TCS elements
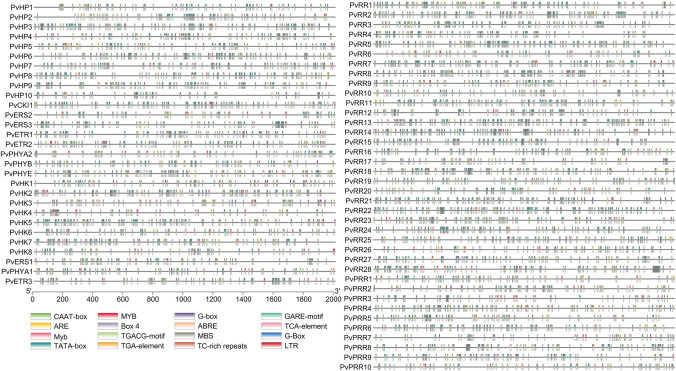
The intracellular localization of each gene of the bean TCS elements is given in Table S4. Moreover, 16 for HK genes, 15 for HP genes, and 14 for RR genes different cis-acting elements were detected in the promoter regions of the bean (Fig. 8).
Fig. 8.
Cis-acting elements in the promoter regions of PvHK, PvHP, and PvRR genes. This analysis was created in TBtools program by selecting cis-acting elements that function under various stress conditions
Homology modeling of TCS proteins and protein–protein interactions in beans
Three-dimensional homology modeling of PvTCS proteins was visualized with the help of the Phyre2 database using TCS protein sequences (Kelley et al. 2015). The best 3D images were generated by analyzing the protein model reliability ratios, and 3D images of HP proteins were given in Table 2, RR proteins in Table 3, and HK proteins in Table 4. The confidence interval was taken as > 90% in the 3D homology modeling of proteins (Incili et al. 2023). Protein–protein interactions (PPI) represent many biological processes such as cell control, intercellular communication, and metabolic development (Braun and Gingras 2012). The STRING database was used to determine functional and physical interactions for ppi, and the data obtained were classified and integrated with the confidence score (Fig. S1).
Table 2.
3D images of bean HP proteins
Table 3.
3D images of bean RR proteins
Table 4.
3D images of bean HK proteins
Tissue-specific in silico gene expression analysis of PvTCS genes
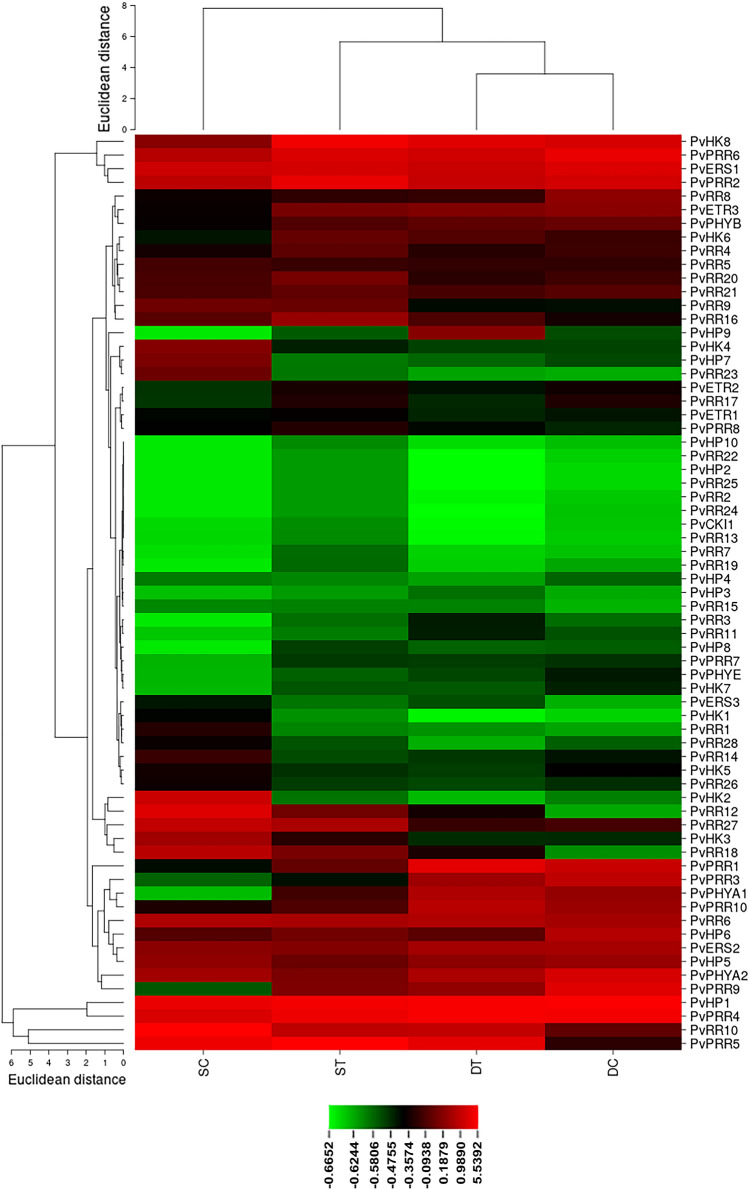
The PvTCS gene family members' expression was analyzed using data from specific tissue libraries (RNAseq). When the expression of the HK genes in the bean leaves subjected to salt stress was analyzed, the expression of the PvHK1, PvERS3, and PvHK2 genes decreased, while the expression of the PvPHYA1, PvHK8, and PvHK6 genes increased as well as the expression of the PvCKI1 gene did not change relative to the control. In response to drought stress, PvHK2 gene expression decreased whereas PvERS3 gene expression increased compared to the control. Other genes exhibited no significant variations (Fig. 9).
Fig. 9.
In silico expression of HK, HP, and RR genes in bean leaves under drought and salt stress treatment. The expression values of each PvTCS gene identified in the study were obtained from RNA-Seq data, including salt-treatment (ST), salt-control (SC), drought-treatment (DT), and drought-control (DC)
The expression levels of PvHP7, PvHP4, and PvHP10 genes were reduced, while the expression of PvHP3 and PvHP8 genes was raised in the digital gene expression of HP genes in salt stress. The expression levels of the other genes did not significantly change. In response to drought stress, the PvHP3, PvHP8, and PvHP9 genes expression increased, while the PvHP4, PvHP6, and PvHP7 genes expression decreased compared to the control. There were no important changes in how other genes were expressed (Fig. 9).
In salt stress, the expression levels of PvPRR1, PvRR3, PvRR12, PvRR14, PvRR15, PvRR18, PvRR23, PvRR26 and PvRR28 genes decreased, the expressions of PvRR7, PvPRR7, and PvPRR9 genes increased, and the expressions of the other genes were increased compared to the control. In drought stress, the expressions of PvPRR1, PvPRR5, PvPRR8, PvRR3, PvRR10, PvRR12, PvRR15, PvRR16, PvRR18, PvRR22, and PvRR24 genes increased while decreasing the expressions of PvPRR9, PvRR16, PvRR17, and PvRR28 genes (Fig. 9).
Expression profiling of TCS genes in bean
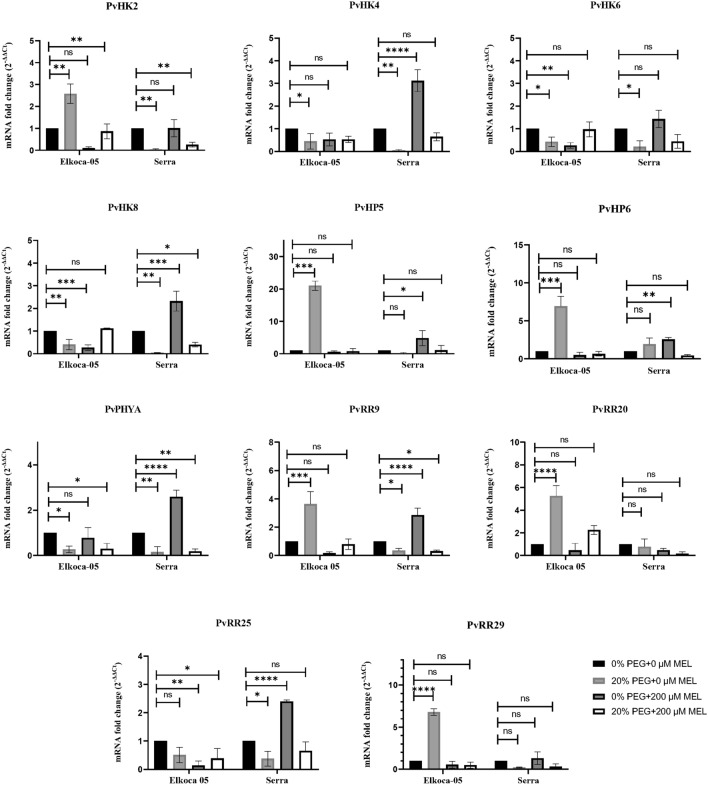
qRT-PCR primers for PvHK2, PvHK4, PvHK6, PvHK8, PvPHYA, PvHP5, PvHP6, PvRR12, PvRR15, PvRR18, and PvRR28 genes were designed, and the expression levels of these genes in Elkoca-05 and Serra varieties treated with salt (0 and 150 mM NaCl), drought (0 and 20% PEG 6000), and melatonin (0 and 200 mM MEL) were determined by quantitative real-time PCR (Fig. 10).
Fig. 10.
Expression of PvTCS genes in Elkoca-05 and Serra cultivars under drought stress. qRT-PCR was used to analyze the expression patterns of eleven PvTCS genes under salt, drought, and melatonin treatments in Elkoca-05 and Serra. Real-time PCR was used to calculate relative transcript levels using β-Actin as an internal standard. This study was conducted using an all-random design with three replicates. The scale of the relative expression levels is shown by the Y-axes. Abiotic stress and melatonin treatments were displayed for each gene of Elkoca-05 and Serra on X-axes. The bars indicate means ± SD (n = 3). ****, ***, **, and * indicate significant at p < 0.0001, p < 0.001, p < 0.01 and p < 0.05 between means, respectively based Dunnet test whereas ns indicates non-significant p > 0.05
Drought stress and melatonin treatment altered the expression of some of the PvTCS genes in both cultivars (Fig. 10). When the effect of drought stress alone was evaluated, in Elkoca-05, a significant increase was observed in the expression of PvHK2, PvHP5, PvHP6, PvRR12, PvRR18, and PvRR28 genes, while it happened a decrease in the expression of PvHK4, PvHK6, PvHK8, and PvPYHA genes. However, no change was observed in the expression of the PvRR15 gene. In the Serra, on the other hand, A significant decrease was observed in the expression of PvHK2, PvHK4, PvHK6, PvHK8, PvPYHA, PvRR15, and PvRR28 genes. No changes were observed in the expression of the other genes.
When the expression of genes studied in cultivars of melatonin application alone was evaluated, while a significant decrease was observed in the expression of PvHK6, PvHK8, and PvRR15 genes in Elkoca-05, no change was observed in the expression of other genes. In Serra, there was a significant increase in the expression of PvHK4, PvHK8, PvHP5, PvHP6, PvPYHA, PvRR15, and PvRR28 genes, while no change was detected in the expression of the other genes. When the combined effect of drought and Melatonin treatments was tested, a significant reduction in the expression of PvHK2, PvPYHA, and PvRR15 genes was observed in the Elkoca-05. Still, no change in the expression of other genes was observed. On the other hand, while the expression of PvHK2, PvHK8, PvPYHA, and PvRR28 genes decreased significantly in the Serra, the expression of other genes did not alter.
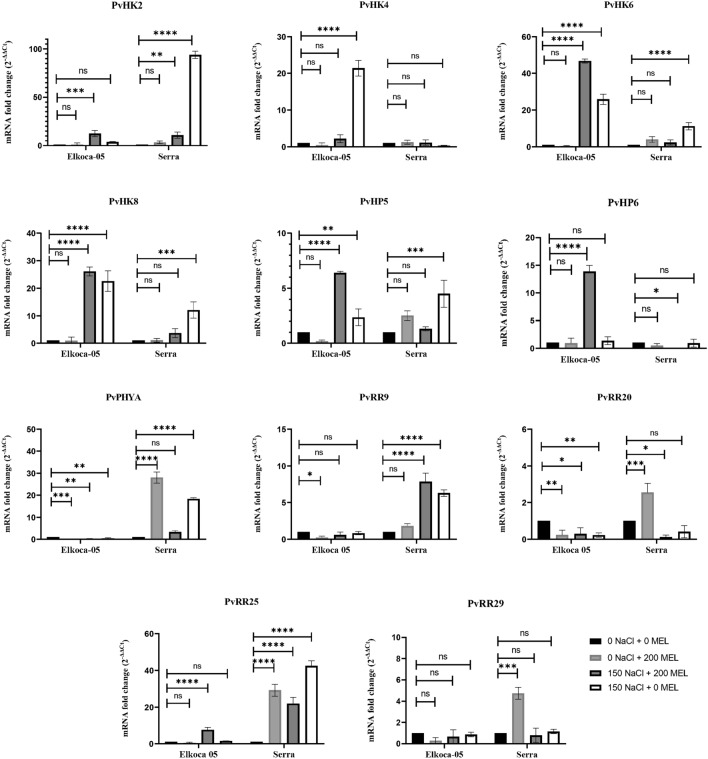
The salt stress and melatonin treatments altered the expression of some PvTCS genes in both cultivars (Fig. 11). When the effects of salt stress alone on the expression of some PvTCS genes in both varieties were evaluated, In Elkoca-05, there was a significant increase in the expression of PvHK4, PvHK6, PvHK8, and PvHP5 genes, and a significant decrease in the expression of PvPYHA, and PvRR12 genes. Moreover, no changes were observed in the expression of other genes. In the Serra, a significant increase in the expression of PvHK2, PvHK6, PvHK8, PvHP5, PvPYHA, PvRR15, and PvRR28 genes was observed while there were no changes in the expression of other genes.
Fig. 11.
Expression of PvTCS genes in Elkoca-05 and Serra cultivars under salt stress. qRT-PCR was used to analyze the expression patterns of eleven PvTCS genes under salt, drought, and melatonin treatments in Elkoca-05 and Serra. Real-time PCR was used to calculate relative transcript levels using β-Actin as an internal standard. This study was conducted using an all-random design with three replicates. The scale of the relative expression levels is shown by the Y-axes. Abiotic stress and melatonin treatments were displayed for each gene of Elkoca-05 and Serra on X-axes. The bars indicate means ± SD (n = 3). ****, ***, **, and * indicate significant at p < 0.0001, p < 0.001, p < 0.01, and p < 0.05 between means, respectively based Dunnet test whereas ns indicates non-significant p > 0.05
When evaluating the expression of some PvTCS genes in cultivars under melatonin treatment alone, there was a change in that it was reduced expression of PvPYHA, PvRR12, and PvRR28 genes in the Elkoca 05. On the other hand, in the Serra, there was increased expression of PvPYHA, PvRR12, PvRR15, and PvRR18 genes.
The combined effect of salt stress and melatonin treatment was evaluated, in the Elkoca-05, PvHK2, PvHK8, PvHP5, PvHP6, and PvRR15 genes showed a significant increase in expression, while there was a significant decrease in the expression of PvPHYA, and PvRR12 genes. On the other hand, in the Serra, a significant increase was observed in the expression of PvHK2, PvRR15, and PvRR28 genes, while a significant decrease was observed in the expression of PvHP6, and PvRR12 genes.
Discussion
The TCS gene family is one of the evolutionary highly conserved signal elements encoded by multiple gene families found in all organisms. Previous research has indicated that this signaling network plays a significant role in various plant processes, including plant growth and development, as well as a variety of stress responses (Lohrmann and Harter 2002). In contrast, even though members of the TCS gene family have been examined in several plant species, no members of this signaling network have been identified in beans. As a result of a genome-wide examination of TCS genes in beans, 67 candidate PvTCS genes, including 10 PvHP genes, 38 PvRR genes, and 19 PvHK genes, were identified in this study. As a result of genome-wide analysis in numerous plants, numerous putative TCS genes were identified, including 55 in Arabidopsis (Hwang et al. 2002), 80 in banana (Dhar et al. 2019), 49 in poplar (Singh and Kumar 2012), 83 in soybean (Mochida et al. 2010), 51 in melon (Liu et al. 2020), 51 in chickpeas (Ahmad et al. 2020), 65 in tomato (He et al. 2016a), and 46 in cucumber (He et al. 2016b).
Segmental and tandem duplications are duplication events that contribute to the development of living things in the evolutionary process. The study for TCS genes determined that both segmental duplication and tandem duplication occurred in the evolutionary process in beans. Similarly, in a study on tomato, it was determined that both duplication types in TCS genes contributed to the evolution process of tomato (He et al. 2016a). In contrast to tomatoes and beans, only segmental duplication was the type of duplication that was effective in the evolutionary process of Arabidopsis, bock choy, and soybean (Schaller et al. 2008; Mochida et al. 2010; Liu et al. 2014). The Ka/Ks ratio determines the selection pressure. During their evolution, duplicated gene pairs may undergo gain-of-function, loss-of-function, and neofunctionalization (Lynch and Conery 2000). Nonsynonymous substitution value (Ka), synonymous substitution value (Ks), and Ka/Ks ratios are often used to help understand the direction of evolution and the selective power in the coding sequence (Li et al. 2009). These algorithms employ distinct models of replacement or mutation based on different assumptions that consider diverse sequence characteristics and permit us to create a variety of evolutionary process predictions (Muse 1996). Ka/Ks > 1 indicates accelerated evolution with positive selection, Ka/Ks = 1 indicates neutral selection, and Ka/Ks < 1 indicates functional restriction due to purifying (negative) selection (İlhan 2018; Kasapoğlu et al. 2020; Kizilkaya et al. 2020).
It has been determined that the Ka/Ks ratio of all TCS duplicate gene pairs in beans is less than 1 and exposed to negative, that is, purifying selection pressure. Moreover, in the duplicate gene pairs identified in melon and tomato, purifying selection pressure occurred as in beans. However, of the duplicate gene pairs detected in tomato, only the SlRR18/SlRR26 duplicate gene pair had a Ka/Ks ratio greater than 1 and was subject to positive selection (He et al. 2016b; Liu et al. 2020).
As a result of the phylogenetic analysis, the phylogenetic tree of the HK gene family members for bean, G. max, and Arabidopsis was divided into 6 groups: Phytochrome-like receptor, Ethylene receptor, CKI1-like, AHK1-like, CKI1/HK5-like, and Cytokinin receptor. As a result of the study, HK gene family members contain HisKA, HATPase, Rec (receiver), CHASE, PAS (Per-ARNT-Sim), PHY (phytochrome), and GAF (cGMP phosphodiesterase/adenylyl cyclase/FhlA) domains in different combinations. All HK gene family members contain the HisKA domain. HK genes consist of three subfamilies cytokinin receptors, ethylene receptors (ETR and ERS), and phytochromes (PHY) (Hwang et al. 2002). Some PvHK genes (PvHK3, -4, -6, and -8) have additional CHASE domains. The CHASE domain is an important region for the recognition and binding of cytokinin, and proteins with this domain have been shown to be involved in cytokinin sensing and signaling (Dhar et al. 2019). There is an additional PAS domain to other domains in the PvPHY genes. The PAS domain is found in many signaling proteins in archaea, bacteria, and eukaryotes and functions as a signal sensor domain (Repik et al. 2000). While there is no CHASE domain in the structure of PETR, PvERS, and PvPHY genes, there is an additional GAF domain. The GAF domain binds molecules such as cAMP and cGMP (Chang and Shockey 1999; Karniol et al. 2005).
The phylogenetic tree of the RR gene family members of beans, G. max, and Arabidopsis is divided into 7 groups as Type-A, Type-B2, Type-B1, Type-C, Clock-PRR, Type-B PRR, and Type-B3. The RR gene family is separated into three subfamilies based on conserved domains. Type A RRs have a conserved D residue and an acceptor domain with a long C-terminal extension. Type B RRs contain a conserved Rec domain and a DNA binding domain (Myb). C-type RRs have the same domain structure as A-type RRs, except for the C-terminal extension. Another class of RRs, known as pseudo-regulators (PRR), contain a CCT motif and a conserved Rec domain at the C-terminus (Ahmad et al. 2020). The CCT motif at the C-terminus of PRRs is critical in regulating circadian rhythms (Más 2008; Tsai et al. 2012). Plant-specific type-B RRs are produced by combining RR and Myb domains (Riaño-Pachón et al. 2008).
The phylogenetic tree of the HP gene family members of the bean, G. max, and Arabidopsis were divided into 4 groups I, II, III, and VI. TCS gene family members also contain a His-containing phosphotransfer (HPt) domain that functions as a signaling module that binds to RRs (Schaller et al. 2008). HP gene family members transfer a phosphate group from the Rec domain of HK genes to the Rec domain of RR genes (Hwang et al. 2002). It contains a highly conserved xHQxKGSSxS motif. However, some HP genes lack the conserved histidine residue and are in a group known as His-containing phosphotransfer proteins. This group cannot function as phosphor transmitter proteins and phosphorylated HP genes regulate Cytokinin signaling by inhibiting phosphorylation in RR genes (Dhar et al. 2019). The phylogenetic analysis performed in this study was based on the classifications made in Arabidopsis (Hwang et al. 2002). The same grouping types are also seen in the phylogenetic analyses performed on Cucumis melo and Cicer arietinum. However, in the study by Mochida et al. (2010), it was concluded that there is no phytochrome-like receptor group in the HK gene family in the G. max phylogenetic tree. There is also no Type C group in the RR gene family. It was determined by new examinations of the development and regeneration of the G. max genome that these two families contain individuals from these groupings (Le et al. 2011).
Due to the bean’s limited genetic and genomic resources, it is crucial to compare it to other plant species. Moreover, the knowledge gained from genes from closely related species is also crucial for establishing the biological functions of their counterparts in beans (Melotto et al. 2011). In the comparative mapping performed between beans and other species, 140 syntenic relationships were detected in the syntenic analysis performed with G. max, while a total of 51 syntenic relationships were found in Arabidopsis. Almost all the TCS genes from G. max and bean were orthologous, indicating that they may have a closer evolutionary link and may have evolved from a common ancestor, consistent with prior findings (Schmutz et al. 2014).
Bean TCS genes were found at 17 locations in the cell, mainly in the nucleus. TCS gene family members are engaged in various biological functions, from stress response to circadian rhythm regulation. TCS genes, distributed almost everywhere in the cell, can affect the total cellular machinery either alone or in combination (Mochida et al. 2010). This control is achieved through cis-regulators in the promoter region. Cis elements activated by various stimuli trigger the expression of many genes. Cis-acting elements trigger other genes related to the maturation or stress (biotic and abiotic) of TCS genes. Many cis-acting elements have been discovered, including the Myb element, which is related to drought and salinity, the MBS element, which is connected to Myb, the gibberellin-sensitive element (GARE-motif), and the abscisic acid-sensitive element (ABRE) (Yamaguchi-Shinozaki and Shinozaki 2005). Type B RR genes containing Myb binding sites regulate the expression of some type A RR genes (Taniguchi et al. 2007). The cis-acting element analysis discovered regulatory elements, including hormone-sensitive and stress-responsive elements. Of the 67 TCS genes, 53 have the ABRE element, 59 have the ARE element, and 18 have the GARE-motif element. In addition, the light-sensitive element Box 4 is found in the 63 TCS gene.
Three-dimensional prediction algorithms of proteins are of great practical value, especially as they enable efficient exploration of sequence space in design applications (Huang et al. 2020). The 3D structure of PvTCS proteins was predicted using homology modeling with a modeling confidence level > 90% (Table 2, 3, and 4). It was determined that all the examined PvHP proteins have alpha helix structures. Most PvRR and PvHK proteins, on the other hand, have several beta-sheet structures in addition to the dominating alpha helix form.
Protein–protein interactions (PPI) are essential mediators in biological processes (Liddington 2004). The three-dimensional arrangement and dynamics of interacting proteins govern most interactions. These interactions are addressed under two sections: permanent and transitory (Nooren and Thornton 2003; Keskin et al. 2008). Some protein–protein interactions are specific to a pair of proteins, whereas others are promiscuous and interact with a wide range of proteins (Bryant et al. 2022). As a result of the PPI analysis of PvHK proteins, it was determined that PvHK proteins interacted mostly with PvHP proteins and cGMP-dependent protein kinase proteins. It was determined that PvHP proteins were mostly in interaction with PvHK and PvRR proteins while PvRR proteins interacted mostly with homeobox proteins, actin-dependent regulatory proteins, and histone-acyl transferase proteins.
MicroRNAs (miRNAs) are a class of small non-coding ribonucleic acids (RNAs) that are present in both animal and plant organisms that negatively regulate gene expression (Hwang et al. 2011; Petrov et al. 2019). The mature miRNAs that are conserved across plants typically consist of 20–22 nucleotides (nt) and originate from stem-loop regions of roughly 70 nt RNA precursors through the action of Dicer-like enzymes, which belong to the RNase III (Jing et al. 2022; Zhao et al. 2023). As a result of microRNA analysis, mir172 and mir390 target HK gene family members, mir164 and mir156 target HP gene family members, and mir166, mir171, and mir395 target RR gene family members (Table S5). The role of miR156, miR164, miR166, miR171, miR172, miR390, and miR395 under stress conditions has been reported in different plant species (Akdoğan et al. 2016; Singroha et al. 2021; Bakhski and Fard 2023; Ghorbanzadeh et al. 2023).
The effects of salt, drought, and melatonin on the expression of TCS genes were examined using in silico and in vitro gene expression analysis. As a result of melatonin application alone, the expression of some genes in the Elkoca-05 cultivar was down-regulated, while these genes were up-regulated in the Serra cultivar. Endogenous melatonin either suppresses or induces the expression of genes encoding important enzymes and transcription factors involved in defense (Weeda et al. 2014). Drought stress caused a decrease in the expression of almost all genes in the Serra cultivar. In the Elkoca-05 cultivar, on the other hand, differences were determined in terms of increase and decrease in the expression of genes. The most significant rise occurred in PvHP5 (about 21-fold), and PvHP6 ( about sevenfold) genes in Elkoca-05 cultivar. Under salt stress, there was a rise in the expression of all genes in the Serra genotype while a decrease in the expression of the PvPHYA (about tenfold), and PvRR20 (about fivefold) genes was observed in the Elkoca-05 cultivar. In the application of melatonin against drought stress, an important decrease in expression of PvHK2 (about fivefold) and PvPYHA (about fivefold) genes was detected in both cultivars. On the other hand, a significant rise in the expression of genes in both cultivars was detected in the application of melatonin against salt stress. It was shown that phytochrome-defective tobacco mutants, especially for phyA and phyAB were more tolerant to salt stress (Yang et al. 2018). Differences in PHYA expression in beans may be due to genotypic differences. In A. thaliana, the ortholog of PvHK2, AHK1 functions in abscisic acid (ABA) signaling and as an osmosensor (Kiba et al. 2004; Pils and Heyl 2009). Accordingly, increased expression of PvHK2 is highly correlated with its potential role in drought stress and osmosis sensing. Kumar and Verslues (2015) found that the mutants in Arabidopsis exhibit specific AHK2 and AHK3-related traits that are also associated with cytokinin signaling for proline buildup and stimulation of NCED3 and P5CS1 gene expression under low water potential circumstances.
The expression of cytokinin receptor genes (PvHK4, PvHK6, and PvHK8) in Serra and Elkoca-05 cultivars was reduced under both stress conditions. In contrast to beans, in rice, drought stress activated HK genes (HK5 and HK3) homologous to Arabidopsis CK receptor genes (AHK2, AHK3, and CRE1) while HK6 homologous to CKI2 was suppressed when exposed to drought stress (Pan et al. 2009). Similarly, soybean HK genes (GmHK10-17) homologous to Arabidopsis CK receptor genes (AHK2, AHK3, and CRE1), such as GmHK12, were induced under dehydration stress while GmHK11 was suppressed (Mochida et al. 2010). Accordingly, it was understood that the genes in the same groups of the phylogenetic tree did not show similar response patterns to abiotic stresses.
Drought stress generally caused a rise in the expression of PvRR genes. PvRR12, PvRR15, and PvRR18 are Type-A RRs and are among the genes whose expression was evaluated in this study while PvRR28 is a Type-B RR. In Arabidopsis, drought enhanced the expression of a subset of Type-A RR genes, ARR5, ARR7, and ARR15, whereas in rice, drought stress inhibited the expression of virtually all genes. Type B RRs do not directly but indirectly participate in cytokinin signaling during the plant life cycle (Ishida et al. 2008). Moreover, type-B RRs regulate the transcription of type-A RRs (Cannon et al. 2004; Garg et al. 2015). It has also been demonstrated that Arabidopsis B-type RRs are involved in drought stress response (Wohlbach et al. 2008). However, comprehensive functional characterization is required to clarify the overall significance of TCS genes in the cell.
Conclusions
As a result of in silico analyses of the bean genome, 67 PvTCS genes including 10 PvHP, 38 PvRR, and 19 PvHK, located on 9 different chromosomes and scaffold_17 of the bean, were determined. Moreover, this gene family member classification was contributed by exon and intron structure, conserved motifs and domains, and phylogeny.
Phylogenetic analysis results show the closest relationships between beans, Arabidopsis, and soybeans. Gene duplication events contributed to the evolution of PvTCS gene family members in the evolutionary process, and it was determined by Ka/Ks analysis that all of these genes were subjected to negative selection in the evolutionary process. The expression changes of TCS genes in bean leaves under salt and drought stress and melatonin treatment of the bean plant were shown by gene expression analysis. Exogenous treatment of melatonin can improve the resistance against a series of stressors drought and salinity which is an answer to the question of how gene expression changes are regulated under stress. This study has revealed that the expression of TCS genes, which are effective in many reactions such as plant growth and development, response to stress, and signal transduction, differ under stress conditions. The results of this study will shed light on functional and genomic studies as well as on metabolic and physiological events such as plant growth and development and signal transduction in salt and drought stress in beans.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors gratefully acknowledge financial support from Atatürk University.
Abbreviations
- CHASE
Cyclases/histidine kinases associated sensing extracellular
- HATPase
Histidine kinase-like ATPase
- HisKA
His kinase A
- HK
Histidine kinase
- HP
Histidine phosphotransfer
- RR
Response regulator
- TCS
Two-component system
Author contributions
Conceptualization, A.G.K, E.I., M.A., and E.Y.; methodology, A.G.K, E.I., M.A., B.I., I.B., and E.Y.; validation, M.S.T., G.A., and A.C.; formal analysis, A.G.K, E.I., M.A., B.I., I.B., and E.Y.; investigation, A.G.K, E.I., M.A., and E.Y.; writing-original draft preparation, A.G.K, E.I., M.A., and E.Y.; writing-review and editing, A.G.K, E.I., M.A., B.I., I.B., M.S.T., G.A., A.C., and E.Y.; visualization, A.G.K, E.I., M.A., B.I., I.B., and E.Y.; supervision, A.G.K, E.I., and M.A.; project administration, A.G.K, E.I., M.A., and E.Y. The published version of the manuscript has been reviewed and approved by all authors.
Funding
This research was funded by Atatürk University Scientific Research Project Coordination Unit with project no: FCD-2021-9813.
Declarations
Conflicts of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmad B, Azeem F, Ali MA, Nawaz MA, Nadeem H, Abbas A, Batool R, Atif RM, Ijaz U, Nieves-Cordones M, Chung G. Genome-wide identification and expression analysis of two component system genes in Cicer arietinum. Genomics. 2020;112:1371–1383. doi: 10.1016/J.YGENO.2019.08.006. [DOI] [PubMed] [Google Scholar]
- Akdogan G, Tufekci ED, Uranbey S, Unver T. miRNA-based drought regulation in wheat. Funct Integr Genomics. 2016;16:221–233. doi: 10.1007/s10142-015-0452-1. [DOI] [PubMed] [Google Scholar]
- Asfaw A, Ambachew D, Shah T, Blair MW. Trait associations in diversity panels of the two common bean (Phaseolus vulgaris L.) gene pools grown under well-watered and water-stress conditions. Front Plant Sci. 2017;8:733. doi: 10.3389/FPLS.2017.00733/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygören AS, Güneş E, Muslu S, Kasapoglu AG, Yigider E, Aydin M, Büyük I, Ilhan E. Genome-wide analysis and characterization of SABATH gene family in phaseolus vulgaris genotypes subject to melatonin under drought and salinity stresses. Plant Mol Biol Rep. 2022;41:242–259. doi: 10.1007/S11105-022-01363-5/FIGURES/13. [DOI] [Google Scholar]
- Azarpazhooh E, Boye JI. Composition of processed dry beans and pulses. In: Siddiq M, Uebersax MA, editors. Dry beans and pulses production, processing and nutrition. New York: John Wiley & Sons Ltd; 2012. pp. 101–128. [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshi B, Fard EM. The arrangement of MicroRNAs in the regulation of drought stress response in plants: a systematic review. Plant Mol Biol Rep. 2023 doi: 10.1007/s11105-023-01380-y. [DOI] [Google Scholar]
- Braun P, Gingras AC. History of protein–protein interactions: from egg-white to complex networks. Proteomics. 2012;12:1478–1498. doi: 10.1002/PMIC.201100563. [DOI] [PubMed] [Google Scholar]
- Bryant P, Pozzati G, Elofsson A. Improved prediction of protein–protein interactions using AlphaFold2. Nat Commun. 2022;13:1–11. doi: 10.1038/s41467-022-28865-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:1–21. doi: 10.1186/1471-2229-4-10/TABLES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Shockey JA. The ethylene-response pathway: signal perception to gene regulation. Curr Opin Plant Biol. 1999;2:352–358. doi: 10.1016/S1369-5266(99)00004-7. [DOI] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Dhar YV, Lakhwani D, Pandey A, Singh S, Trivedi PK, Asif MH. Genome-wide identification and interactome analysis of members of two-component system in Banana. BMC Genomics. 2019;20:1–15. doi: 10.1186/s12864-019-6050-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Bhattacharjee A, Jain M. Genome-scale transcriptomic insights into molecular aspects of abiotic stress responses in chickpea. Plant Mol Biol Rep. 2015;33:388–400. doi: 10.1007/S11105-014-0753-X/FIGURES/6. [DOI] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook. Berlin: Springer; 2005. [Google Scholar]
- Ghorbanzadeh Z, Hamid R, Jacob F, Mirzaei M, Zeinalabedini M, Abdirad S, Atwell BJ, Haynes PA, Ghaffari MR, Salekdeh GH. MicroRNA profiling of root meristematic zone in contrasting genotypes reveals novel insight into in rice response to water deficiency. J Plant Growth Regul. 2023;42:3814–3834. doi: 10.1007/s00344-022-10842-8. [DOI] [Google Scholar]
- Golding C, Lee H. Study and synthesis of melatonin as a strong antioxidant in plants. Biol Mol Chem. 2023;1:35–44. doi: 10.22034/BMC.2023.417001.1006. [DOI] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/NAR/GKR944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/NAR/GKN176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Chen X, Luo JC. GSDS: a gene structure display server. Yi Chuan = Hereditas / Zhongguo Yi Chuan Xue Hui Bian Ji. 2007;29:1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- Guruprasad K, Reddy BVB, Pandit MW. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng Des Sel. 1990;4:155–161. doi: 10.1093/PROTEIN/4.2.155. [DOI] [PubMed] [Google Scholar]
- Hailu B, Mehari H. Impacts of soil salinity/sodicity on soil-water relations and plant growth in dry land areas: A review. J Nat Sci Res. 2021;12:1–10. doi: 10.7176/jnsr/12-3-01. [DOI] [Google Scholar]
- He Y, Liu X, Ye L, Pan C, Chen L, Zou T, Lu G. Genome-wide identification and expression analysis of two-component system genes in tomato. Int J Mol ScI. 2016;17:1204. doi: 10.3390/IJMS17081204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Liu X, Zou T, Pan C, Qin L, Chen L, Lu G. Genome-Wide identification of two-component system genes in cucurbitaceae crops and expression profiling analyses in Cucumber. Front Plant Sci. 2016;7:899. doi: 10.3389/FPLS.2016.00899/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiz MC, Canher B, Niron H, Turet M. Transcriptome analysis of salt tolerant common bean (Phaseolus vulgaris L.) under saline conditions. PLoS ONE. 2014 doi: 10.1371/journal.pone.0092598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:585–587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.1186/s12864-022-08864-3
- Huang X, Pearce R, Zhang Y. EvoEF2: accurate and fast energy function for computational protein design. Bioinformatics. 2020;36:1135–1142. doi: 10.1093/BIOINFORMATICS/BTZ740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain I, Ayub A, Nayab A, Ashraf MA, Hussain S, Siddiqui MH, Sabir MA, Zulfiqar U, Khan TH. Exogenous application of silicon and zinc attenuates drought tolerance in Eruca sativa L. through increasing chlorophyll pigments, osmoprotectants, and modulating defense mechanisms. J Plant Growth Regul. 2023 doi: 10.1007/s00344-023-11116-7. [DOI] [Google Scholar]
- Hutchison CE, Kieber JJ. Cytokinin signaling in Arabidopsis. Plant Cell. 2002;14:S47–S59. doi: 10.1105/TPC.010444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Chen HC, Sheen J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 2002;129:500–515. doi: 10.1104/PP.005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang EW, Shin SJ, Park SC, Jeong MJ, Kwon HB. Identification of miR172 family members and their putative targets responding to drought stress in Solanum tuberosum. Genes Genomics. 2011;33:105–110. doi: 10.1007/s13258-010-0135-1. [DOI] [Google Scholar]
- Hwang I, Sheen J, Müller B. Cytokinin signaling networks. Annu Rev Plant Biol. 2012;63:353–380. doi: 10.1146/annurev-arplant-042811-105503. [DOI] [PubMed] [Google Scholar]
- İlhan E. Genome-wide analysis of Eucalyptus grandis YABBY transcription factors. Turk Tarim Arast Derg. 2018;5:158–166. [Google Scholar]
- İncili ÇY, Arslan B, Çelik ENY, Ulu F, Horuz E, Baloglu MC, Çağlıyan E, Burcu G, Bayarslan AU, Altunoglu YC. Comparative bioinformatics analysis and abiotic stress responses of expansin proteins in Cucurbitaceae members: watermelon and melon. Protoplasma. 2023;260:509–527. doi: 10.1007/s00709-022-01793-8. [DOI] [PubMed] [Google Scholar]
- Ishida K, Yamashino T, Yokoyama A, Mizuno T. Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 2008;49:47–57. doi: 10.1093/pcp/pcm165. [DOI] [PubMed] [Google Scholar]
- Jing X, Zhang H, Huai X, An Q, Qiao Y. Identification and characterization of miRNAs and PHAS loci related to the early development of the embryo and endosperm in Fragaria × ananassa. BMC Genom. 2022;23:638. doi: 10.1186/s12864-022-08864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R, Merati M, Shayganfar A. Phytochemical characterization of five commercial Vitis vinifera cultivars in response to salinity. Acta Physiol Plant. 2023;45:108. doi: 10.1007/s11738-023-03588-7. [DOI] [Google Scholar]
- Karniol B, Wagner JR, Walker JM, Vierstra RD. Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem J. 2005;392:103–116. doi: 10.1042/BJ20050826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasapoğlu AG, Ilhan E, Kizilkaya D, Hossein-Pour A, Haliloğlu K. Genome-wide analysis of BES1 transcription factor family in sorghum [Sorghum bicolor (L.) Moench] genome. Turk Tarim Arast Derg. 2020;7:85–95. [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin O, Gursoy A, Ma B, Nussinov R. Principles of protein–protein interactions: What are the preferred ways for proteins to interact? Chem Rev. 2008;108:1225–1244. doi: 10.1021/cr040409x. [DOI] [PubMed] [Google Scholar]
- Kiba T, Aoki K, Sakakibara H, Mizuno T. Arabidopsis response regulator, ARR22, ectopic expression of which results in phenotypes similar to the wol cytokinin-receptor mutant. Plant Cell Physiol. 2004;45:1063–1077. doi: 10.1093/PCP/PCH128. [DOI] [PubMed] [Google Scholar]
- Kiba T, Naitou T, Koizumi N, Yamashino T, Sakakibara H, Mizuno T. Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His→Asp phosphorelay circuitry. Plant Cell Physiol. 2005;46:339–355. doi: 10.1093/PCP/PCI033. [DOI] [PubMed] [Google Scholar]
- Kizilkaya D, Kasapoğlu AG, Hosseinpour A, Haliloğlu K, Muslu S, Ilhan E. Genome wide analysis of Sorghum bicolor L. CAMTA transcription factors. Atatürk Üniversitesi Ziraat Fakültesi Dergisi/atatürk Univ. J. Agric. Fac. 2020;51:267–278. [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/GR.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MN, Verslues PE. Stress physiology functions of the Arabidopsis histidine kinase cytokinin receptors. Physiol Plant. 2015;154:369–380. doi: 10.1111/PPL.12290. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, Karthikeyan AS, Lee CH, Nelson WD, Ploetz L, Sing S, Wensel A, Huala E. The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/NAR/GKR1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP. Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res. 2011;18:17–29. doi: 10.1093/DNARES/DSQ032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, de Peer YV, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of LIFE v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:475–478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang Z, Vang S, Yu J, Wong GKS, Wang J. Correlation between Ka/Ks and Ks is related to substitution model and evolutionary lineage. J Mol Evol. 2009;68:414–423. doi: 10.1007/S00239-009-9222-9/TABLES/5. [DOI] [PubMed] [Google Scholar]
- Liddington RC. Structural basis of protein–protein interactions. In: Fu H, editor. Protein-protein interactions: methods and applications. Totowa: Humana Press; 2004. pp. 3–14. [Google Scholar]
- Liu Z, Zhang M, Kong L, Lv Y, Zou M, Lu G, Cao J, Yu X. Genome-wide identification, phylogeny, duplication, and expression analyses of two-component system genes in Chinese cabbage (Brassica rapa ssp. pekinensis) DNA Res. 2014;21:379–396. doi: 10.1093/dnares/dsu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Wang S, Wang X, Yang X, Li Q, Wang C, Chen C, Shi Q, Ren Z, Wang L. Genome-wide characterization of two-component system (TCS) genes in melon (Cucumis melo L.) Plant Physiol Biochem. 2020;151:197–213. doi: 10.1016/J.PLAPHY.2020.03.017. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/METH.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lohrmann J, Harter K. Plant Two-component signaling systems and the role of response regulators. Plant Physiol. 2002;128:363–369. doi: 10.1104/PP.010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann J, Buchholz G, Keitel C, Swere U, Kircher S, Baurle I, Kudla J, Schafer E, Harter K. Differential expression and nuclear localization of response regulator-like proteins from Arabidopsis thaliana. Plant Biol. 1999;1:495–505. doi: 10.1055/S-2007-978544. [DOI] [Google Scholar]
- Loomis WF, Shaulsky G, Wang N. Histidine kinases in signal transduction pathways of eukaryotes. J Cell Sci. 1997;110:1141–1145. doi: 10.1242/JCS.110.10.1141. [DOI] [PubMed] [Google Scholar]
- Losa A, Vorster J, Cominelli E, Sparvoli F, Paolo D, Sala T, Ferrari M, Carbonaro M, Marconi S, Camilli E, Reboul E, Waswa B, Ekesa B, Aragão F, Kunert K. Drought and heat affect common bean minerals and human diet—what we know and where to go. Food Energy Secur. 2022;11:e351. doi: 10.1002/fes3.351. [DOI] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science (1979) 2000;290:1151–1155. doi: 10.1126/SCIENCE.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Mähönen AP, Bishopp A, Higuchi M, Nieminen KM, Kınoshita K, Törmakangas K, Ikeda Y, Oka A, Kakimoto T, Helarıutta Y. Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science (1979) 2006;311:94–98. doi: 10.1126/SCIENCE.1118875/SUPPL_FILE/MAHONEN.SOM.PDF. [DOI] [PubMed] [Google Scholar]
- Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T. Genes encoding pseudo-response regulators: insight into his-to-asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:791–803. doi: 10.1093/PCP/41.6.791. [DOI] [PubMed] [Google Scholar]
- Marcantonini G, Bartolini D, Zatini L, Costa S, Passerini M, Rende M, Luca G, Basta G, Murdolo G, Calafiore R, Galli F. Natural cryoprotective and cytoprotective agents in cryopreservation: A focus on melatonin. Molecules. 2022;27:3254. doi: 10.3390/molecules27103254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P. Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends Cell Biol. 2008;18:273–281. doi: 10.1016/J.TCB.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Melotto M, Monteiro-Vitorello CB, Bruschi AG, Camargo LEA. Comparative bioinformatic analysis of genes expressed in common bean (Phaseolus vulgaris L.) seedlings. Genome. 2011;48:562–570. doi: 10.1139/G05-010. [DOI] [PubMed] [Google Scholar]
- Miao Y, Gao X, Li B, Wang W, Bai L. Low red to far-red light ratio promotes salt tolerance by improving leaf photosynthetic capacity in cucumber. Front Plant Sci. 2023;13:1053780. doi: 10.3389/fpls.2022.1053780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir AR, Alam P, Hayat S. Perspective of melatonin-mediated stress resilience and Cu remediation efficiency of Brassica juncea in Cu-contaminated soils. Front Plant Sci. 2022;13:910714. doi: 10.3389/fpls.2022.910714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/NAR/GKAA913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T. Plant response regulators implicated in signal transduction and circadian rhythm. Curr Opin Plant Biol. 2004;7:499–505. doi: 10.1016/J.PBI.2004.07.015. [DOI] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP. Genome-wide analysis of two-component systems and prediction of stress-responsive two-component system members in soybean. DNA Res. 2010;17:303–324. doi: 10.1093/DNARES/DSQ021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Muse SV. Estimating synonymous and nonsynonymous substitution rates. Mol Biol Evol. 1996;13:105–114. doi: 10.1093/OXFORDJOURNALS.MOLBEV.A025549. [DOI] [PubMed] [Google Scholar]
- Nooren IMA, Thornton JM. Diversity of protein–protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/EMBOJ/CDG359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira WJ, Melo AT, Coelho AS, Rodrigues FA, Mamidi S, Alencar SA, Lanna AC, Valdisser PA, Brondani C, Nascimento-Júnior IR, Borba TC. Genome-wide analysis of the transcriptional response to drought stress in root and leaf of common bean. Genet Mol Biol. 2020;43:e20180259. doi: 10.1590/1678-4685-GMB-2018-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov NM, Stoyanova MI, Gaur RK. Post-transcriptional gene silencing as a tool for controlling viruses in plants. Plant Biotechnol Prog Genom Era. 2019 doi: 10.1007/978-981-13-8499-8_23. [DOI] [Google Scholar]
- Petry N, Boy E, Wirth JP, Hurrell RF. Review: the potential of the common Bean (Phaseolus vulgaris) as a vehicle for iron biofortification. Nutrients. 2015;7:1144–1173. doi: 10.3390/NU7021144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pils B, Heyl A. Unraveling the evolution of cytokinin signaling. Plant Physiol. 2009;151:782–791. doi: 10.1104/PP.109.139188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:116–120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy K, Karuppasami KM, Alagarswamy S, Shanmugam KP, Rathinavelu S, Vellingiri G, Muniyappan U, Kanthan T, Kuppusamy A, Rajendran M, Kathirvel A, Kanagarajan S. Role of melatonin in directing plant physiology. Agron. 2023;13:2405. doi: 10.3390/agronomy13092405. [DOI] [Google Scholar]
- Raza A, Tabassum J, Fakhar AZ, Sharif R, Chen H, Zhang C, Ju L, Fotopoulos V, Siddique KH, Singh RK, Zhuang W. Smart reprograming of plants against salinity stress using modern biotechnological tools. Crit Rev Biotechnol. 2022 doi: 10.1080/07388551.2022.2093695. [DOI] [PubMed] [Google Scholar]
- Raza A, Charagh S, García-Caparrós P, Rahman MA, Ogwugwa VH, Saeed F, Jin W. Melatonin-mediated temperature stress tolerance in plants. GM Crops Food. 2022;13:196–217. doi: 10.1080/21645698.2022.2106111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza A, Salehi H, Rahman MA, Zahid Z, MadadkarHaghjou M, Najafi-Kakavand S, Charagh S, Osman HSİ, Albaqami M, Zhuang Y, Siddique KHM, Zhuang W. Plant hormones and neurotransmitter interactions mediate antioxidant defenses under induced oxidative stress in plants. Front Plant Sci. 2022 doi: 10.3389/fpls.2022.961872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza A, Mubarik MS, Sharif R, Habib M, Jabeen W, Zhang C, Chen H, Chen ZH, Siddique KHM, Zhuang W, Varshney RK. Developing drought-smart, ready-to-grow future crops. Plant Genome. 2023;16:e20279. doi: 10.1002/tpg2.20279. [DOI] [PubMed] [Google Scholar]
- Raza A, Charagh S, Salehi H, Abbas S, Saeed F, Poinern GE, Siddique KHM, Varshney RK. Nano-enabled stress-smart agriculture: Can nanotechnology deliver drought and salinity-smart crops? J Sustain Agric Environ. 2023;2:189–214. doi: 10.1002/sae2.12061. [DOI] [Google Scholar]
- Repik A, Rebbapragada A, Johnson MS, Haznedar JÖ, Zhulin IB, Taylor BL. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol Microbiol. 2000;36:806–816. doi: 10.1046/J.1365-2958.2000.01910.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaño-Pachón DM, Corrêa LGG, Trejos-Espinosa R, Mueller-Roeber B. Green transcription factors: a chlamydomonas overview. Genetics. 2008;179:31–39. doi: 10.1534/GENETICS.107.086090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodiño AP, Riveiro M, De Ron AM. Implications of the symbiotic nitrogen fixation in common bean under seasonal water stress. Agron. 2020;11:70. doi: 10.3390/agronomy11010070. [DOI] [Google Scholar]
- Sánchez-Reinoso AD, Ligarreto-Moreno GA, Restrepo-Díaz H. Evaluation of drought indices to identify tolerant genotypes in common bean bush (Phaseolus vulgaris L.) J Integr Agric. 2020;19:99–107. doi: 10.1016/S2095-3119(19)62620-1. [DOI] [Google Scholar]
- Schaller GE. Histidine kinases and the role of two-component systems in plants. Adv Bot Res. 2000;32:109–148. doi: 10.1016/S0065-2296(00)32023-7. [DOI] [Google Scholar]
- Schaller GE, Kieber JJ, Shiu S-H. Two-component signaling elements and histidyl-aspartyl phosphorelays. Arabidopsis Book. 2008;2008:e0112. doi: 10.1199/tab.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB, Grimwood J, Jenkins J, Shu S, Song Q, Chavarro C, Torres-Torres M. A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet. 2014;46:707–713. doi: 10.1038/ng.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/GR.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida C, Castrucci AML, Lamy-Freund MT. High melatonin solubility in aqueous medium. J Pineal Res. 1994;16:198–201. doi: 10.1111/J.1600-079X.1994.TB00102.X. [DOI] [PubMed] [Google Scholar]
- Singh G, Kumar R. Genome-wide Insilico analysis of plant two component signaling system in woody model plant Populus trichocarpa. Res Plant Biol. 2012;2:13–23. [Google Scholar]
- Singroha G, Sharma P, Sunkur R. Current status of microRNA-mediated regulation of drought stress responses in cereals. Physiol Plant. 2021;172:1808–1821. doi: 10.1111/ppl.13451. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Suleymanov A, Gabbasova I, Komissarov M, Suleymanov R, Garipov T, Tuktarova I, Belan L. Random forest modeling of soil properties in saline semi-arid areas. Agriculture. 2023;13:976. doi: 10.3390/agriculture13050976. [DOI] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/NAR/GKL315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/NAR/GKAA1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A. ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol. 2007;48:263–277. doi: 10.1093/PCP/PCL063. [DOI] [PubMed] [Google Scholar]
- Terzi E, Bilgintürk M, Gündoğan R, Can Karaça A. Fasulye Proteini İzolatının Çeşitli Gıda Ürünlerinin Kalite Özelliklerine Etkisi. Avrupa Bilim Ve Teknoloji Dergisi. 2020 doi: 10.31590/EJOSAT.757599. [DOI] [Google Scholar]
- Thomason P, Kay R. Eukaryotic signal transduction via histidine-aspartate phosphorelay. J Cell Sci. 2000;113:3141–3150. doi: 10.1242/JCS.113.18.3141. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thung M, Rao IM. Integrated management of abiotic stresses. In: Singh SP, editor. Common bean improvement in the twenty-first century. Dordrecht: Springer; 1999. pp. 331–370. [Google Scholar]
- Tsai YC, Weir NR, Hill K, Zhang W, Kim HJ, Shiu SH, Schaller GE, Kieber JJ. Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 2012;158:1666–1684. doi: 10.1104/PP.111.192765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Yamaguchi-Shinozaki K, Shinozaki K. Stress-responsive expression of genes for two-component response regulator-like proteins in Arabidopsis thaliana. FEBS Lett. 1998;427:175–178. doi: 10.1016/S0014-5793(98)00418-9. [DOI] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Tae-ho L, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49–e49. doi: 10.1093/NAR/GKR1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeda S, Zhang N, Zhao X, Ndip G, Guo Y, Buck GA, Fu C, Ren S. Arabidopsis transcriptome analysis reveals key roles of melatonin in plant defense systems. PLoS ONE. 2014;9:e93462. doi: 10.1371/JOURNAL.PONE.0093462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbach DJ, Quirino BF, Sussman MR. Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell. 2008;20:1101–1117. doi: 10.1105/TPC.107.055871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang G, Jia R, Wang F, Wang S, Li Y, Yao Y. Exogenous l-tryptophan treatment increases the concentrations of melatonin and aroma compounds in grape berries and wine. Food Qual Saf. 2023 doi: 10.1093/fqsafe/fyad042. [DOI] [Google Scholar]
- Ya-Jiao PA, Di WA, Ling-Hua ZH, Bin-Ying FU, Zhi-Kang LI. Differential expressions of two-component element genes in rice under drought stress. Acta Agron Sin. 2009;35:1628–1636. doi: 10.1016/S1875-2780(08)60105-4. [DOI] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10:88–94. doi: 10.1016/J.TPLANTS.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/MOLBEV/MSM088. [DOI] [PubMed] [Google Scholar]
- Yang T, Lv R, Li J, Lin H, Xi D. Phytochrome A and B negatively regulate salt stress tolerance of Nicotiana tobacum via ABA–jasmonic acid synergistic cross-talk. Plant Cell Physiol. 2018;59:2381–2393. doi: 10.1093/PCP/PCY164. [DOI] [PubMed] [Google Scholar]
- Yang S, Chu N, Feng N, Zhou B, Zhou H, Deng Z, Shen X, Zheng D. Global responses of autopolyploid sugarcane Badila (Saccharum officinarum L.) to drought stress based on comparative transcriptome and metabolome profiling. Int J Mol Sci. 2023;24:3856. doi: 10.3390/ijms24043856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavaş İ, Hussain S. Recent progress on melatonin-induced salinity tolerance in plants: an overview. Turk JAF Sci Tech. 2022;10:1447–1454. doi: 10.24925/turjaf.v10i8.1447-1454.5125. [DOI] [Google Scholar]
- Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15:336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Li X, Ye C, Huang C, Lv X, Li J. The biogenesis, mechanism and function of the tRNA-derived small RNA (tsRNA): a review compared with microRNA. Am J Cancer Res. 2023;13:1656. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.