Abstract
Papillary renal cell carcinoma (PRCC) is a highly heterogeneous cancer, and PRCC patients with advanced/metastatic subgroup showed obviously shorter survival compared to other kinds of renal cell carcinomas. However, the molecular mechanism and prognostic predictors of PRCC remain unclear and are worth deep studying. The aim of this study is to identify novel molecular classification and construct a reliable prognostic model for PRCC. The expression data were retrieved from TCGA, GEO, GTEx and TARGET databases. CRISPR data was obtained from Depmap database. The key genes were selected by the intersection of CRISPR-Cas9 screening genes, differentially expressed genes, and genes with prognostic capacity in PRCC. The molecular classification was identified based on the key genes. Drug sensitivity, tumor microenvironment, somatic mutation, and survival were compared among the novel classification. A prognostic model utilizing multiple machine learning algorithms based on the key genes was developed and tested by independent external validation set. Our study identified three clusters (C1, C2 and C3) in PRCC based on 41 key genes. C2 had obviously higher expression of the key genes and lower survival than C1 and C3. Significant differences in drug sensitivity, tumor microenvironment, and mutation landscape have been observed among the three clusters. By utilizing 21 combinations of 9 machine learning algorithms, 9 out of 41 genes were chosen to construct a robust prognostic signature, which exhibited good prognostic ability. SERPINH1 was identified as a critical gene for its strong prognostic ability in PRCC by univariate and multiple Cox regression analyses. Quantitative real-time PCR and Western blot demonstrated that SERPINH1 mRNA and protein were highly expressed in PRCC cells compared with normal human renal cells. This study exhibited a new molecular classification and prognostic signature for PRCC, which may provide a potential biomarker and therapy target for PRCC patients.
Keywords: Papillary renal cell carcinoma, CRISPR-Cas9, Depmap, Machine learning, SERPINH1
Highlights
-
•
The 41 key genes were chosen by the intersection of CRISPR-Cas9 screening genes, DEGs, and prognostic genes in PRCC.
-
•
Three clusters with distinct characteristics were identified based on 41 key genes in papillary renal cell carcinoma.
-
•
By using 21 combinations of 9 machine learning algorithms, 9 genes were chosen to construct a robust prognostic signature.
-
•
SERPINH1 may be an important biomarker and a potential therapeutic target for PRCCpatients.
1. Introduction
Renal cell carcinoma (RCC) is the sixth most commonly diagnosed cancer among men and the tenth most common cancer among women. Papillary renal cell carcinoma (PRCC), which makes up 10%–15 % of the RCCs, is the most frequently diagnosed cancer in non-clear cell renal cell carcinoma (ccRCC) with an increasing incidence [1,2]. PRCC is a highly heterogeneous cancer and PRCC patients with advanced/metastatic subgroup showed obviously shorter survival compared to other kinds of RCCs [3]. The WHO classification of tumors of the urinary system in 2016 subclassified PRCCs into two types (type 1 and type 2), however the classification may be challenging for the mixture of two-type areas in well-sampled tumors. Recently, specific phenotypes have been defined in the 2022 WHO classification of tumors of the urinary system instead of type 1 and type 2 [4]. Previous studies showed that PRCC patients with partial nephrectomy had lower 10-year RFS (73 %) compared to ccRCC patients (96.1 %), and the prognosis of PRCCs remained to be poor in advanced or metastatic tumors [[5], [6], [7]].
In recent years, advanced approaches were applied to explore more efficient biomarkers and investigate precision medicine in some cancers. A three-gene risk model was established to predict the prognosis in patients with PRCC [8]. And the immune-related biomarkers were analyzed based on the abundance of different kinds of immune cells in PRCC [9]. However, the effective biomarkers and prognostic predictors of PRCC remain unclear and are worth deep studying. The aim of this study is to identify novel molecular classification and construct a reliable prognostic model for PRCC.
Clustered regularly interspaced short palindromic repeats (CRISPR), which is the defense mechanism in many bacteria and archaea, has been used as an efficient tool for gene editing [10,11]. The researchers can knockout targeted genes and explore therapeutic strategies by CRISPR [12,13]. In order to systematically explore the potential cancer biomarkers, CRISPR-Cas9 was applied to identify the crucial genes for the cancer cells' proliferation or survival. And cancer dependency map was established, bringing new methods for the research on the molecular mechanisms in the development and treatment of cancers [14,15]. Previous studies have proved that the crucial genes selected by CRISPR data in several kinds of cancer had the ability to predict the tumor biomarkers and survival. For example, the CRISPR data derived from the Depmap database were used to determine the key genes associated with hepatocellular carcinoma and ccRCC [16,17]. However, the crucial genes selected by CRISPR data and multiple machine learning in PRCC haven't been reported before. The CRISPR data and multiple machine learning may identify some crucial genes in order to establish molecular subtypes and reliable prognostic model for PRCC.
In this study, the differentially expressed genes (DEGs) and the genes having prognostic predictive capabilities were identified in PRCC based on the open data. And crucial genes in PRCC were found by a large-scale CRISPR-Cas9 screening utilizing the Depmap database. Then, the key genes were got by the intersection of crucial genes, DEGs, and genes related to the survival of PRCC. Subsequently, clustering analysis based on the TCGA cohort was performed to recognize three clusters with the key genes. We compared the tumor microenvironment, expression of immune checkpoint-related genes, mutation frequency, and drug sensitivity among the three clusters. A reliable and robust prognostic signature including 9 genes was then developed using multiple machine learning algorithms based on the key genes. SERPINH1, one of 9 genes in the prognostic signature, was selected because of its strong prognostic predictive ability. The expression of SERPINH1 in PRCC was investigated and pan-cancer analysis was performed. Our study exhibited a new molecular classification and prognostic signature for PRCC, which may offer novel insights into its molecular mechanism and provide a potential therapy target for PRCC.
2. Methods
2.1. Data collection and processing
The RNAseq data and corresponding clinical information were obtained from TCGA database (https://www.cancer.gov/tcga). Normal tissues were retrieved from GTEx database (https://www.gtexportal.org/home/) [18]. The external validation set was obtained from GSE2748 in the GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE2748) [19,20]. And a total number of 28 PRCC patients with complete survival data in the GSE2748 dataset were selected to validate the prognostic model. Supplementary Table S1 presented the clinical data of the 28 PRCC patients. Depmap database (https://depmap.org/portal/) provided information on gene dependency in cancer cell lines. Through the Depmap database, we are able to investigate the gene dependency in multiple types of cell lines by the Chronos score generated by the CRISPR-Cas9 technology. In order to recognize the potential molecular biomarkers of PRCC, the 22Q2 version of the CRISPR file in the Depmap database was downloaded. The genes whose Chronos score was less than −0.6 were defined as crucial genes utilizing the R package “depmap”. The clinical information from TCGA-PRCC cohort was sorted out and organized with the online tool Sangerbox [21].
2.2. Recognition of key genes in PRCC
Based on the PRCC samples from TCGA-PRCC cohort and the normal samples from GTEx database, DEGs were identified by using R package “limma” [22]. Adjusted P < 0.05 and | Log2 (Fold Change) | > 1 were set to be the threshold. To screen the genes related to survival in PRCC, univariate Cox proportional hazards regression and log-rank test were performed to generate the P-value and the hazard ratio of the Kaplan-Meier (KM) curves for each gene. P < 0.05 was considered to be statically significant. In order to obtain the key genes in PRCC, we make the intersection of crucial genes, DEGs, and genes related to the survival of PRCC with the Venn diagrams (https://bioinformatics.psb.ugent.be/webtools/Venn/). KEGG and GO analyses were performed by using the R package “clusterProfiler” [23].
2.3. Identification of three clusters in PRCC
The R package “ConsensusClusterPlus” was used to classify the TCGA-PRCC cohort according to the expression of the key genes [24]. The maximum number of clusters was 6, and 80 % of the total samples were drawn 100 times, clusterAlg = “hc”, innerLinkage = ‘ward.D2’. Heatmap was drawn utilizing the R package “pheatmap”. The KM curve of each cluster was generated with the R package “survival”.
2.4. Drug sensitivity analysis
The R package “oncoPredict” can be utilized for predicting the sensitivity of patients to chemotherapeutic agents [25]. The training set data contained with the R package “oncoPredict” was retrieved from GDSC database (https://www.cancerrxgene.org/). We calculated the response of 198 kinds of drugs in each patient. Lower score represents higher drug sensitivity and better treatment effect.
2.5. Tumor microenvironment analysis among the three clusters
The R package “CIBERSORT” predicted the abundance of different kinds of immune cells in PRCC. In order to compare the infiltration level of the immune cells among the clusters, the R package “ESTIMATE” was applied to calculate the immune score [26]. The expression of immune checkpoint-related genes was compared to investigate the difference of the immune checkpoint blockade (ICB) therapy among the clusters. And Tumor Immune Dysfunction and Exclusion (TIDE) algorithm predicted the ICB response of each sample [27]. The R package “EPIC” was used to compare 8 kinds of cells recruited to the tumor microenvironment among the three clusters [28].
2.6. Mutation analysis among the three clusters
The mutation landscape showed the 15 most frequently mutated genes in PRCC and their mutation frequency in the three clusters. The distribution of different variant types was visualized in the cohort summary plot. The altered genes in the abnormal signaling pathways were summarized. All the analyses above were finished with the R package “maftools” [29].
2.7. Construction of a prognostic signature based on the key genes
First, the samples from the TCGA-PRCC cohort with information on the survival time were reserved. In order to improve the accuracy of the signature, the samples with a survival time of less than 30 days were removed. The selected samples were then divided into a training set and a validation set with a ratio of 7 : 3. The FPKM data retrieved from the TCGA database was normalized to TPM data and changed to log2 (TPM+1) format. GSE2748 was defined as a test set for external validation. Next, we separately applied standardization with z-score normalization to the training set, validation set, and test set. Multiple machine learning algorithms were applied, including RSF, Enet, GBSA, CoxBoost, SuperPC, LASSO, SSVM, plsRcox, and StepCox [30]. The full names of the algorithms were shown in Supplementary Table S2. 21 combinations of 9 machine learning algorithms were utilized to construct the signature based on the key genes. The mean AUC value of each signature was calculated in the validation set and test set to select the signature with the highest predictive capability.
2.8. Single gene analysis
According to the genes in the signature, univariate and multivariate Cox regression analyses were performed. P value < 0.01 was set to be statistically significant. The R package “forestplot” was utilized to draw the forest plot.
TMB data and MSI data were retrieved from previous researches [31,32]. The correlation between the expression of the single gene and MSI or TMB was explored by Spearman's correlation analysis. In order to investigate the relationship between the expression of the single gene and the abundance of different types of immune cells, the immune analysis was performed with the R package “TIMER” [33]. The correlation between the expression of SERPINH1 and other characteristics was visualized by utilizing the “ggstatsplot” R package. P value < 0.05 was set to be statistically significant.
2.9. Pan-cancer analysis
The RNAseq data were from TCGA, GTEx, and TARGET databases. The R package “survival” and log-rank test were used to analyze the prognostic ability of the single gene in multiple kinds of tumors. The GEPIA2 database provided visual representations of variations in gene expression across different stages of tumors in multiple types of tumors [34]. The immunohistochemical staining of SERPINH1 protein in normal renal tubules, PRCC and ccRCC tumor tissues were retrieved from HPA database (https://www.proteinatlas.org/). SERPINH1 antibody (CAB004441) was applied to detect the expression of SERPINH1. The R package “immunedeconv” including 6 algorithms was applied to analyze the correlation between the expression of the single gene and the abundance of the immune cells in 33 types of tumors [35]. The analysis between the expression of the single gene and TMB or MSI of multiple types of tumors was based on the data from previous researches [31,32]. The flow diagram of our research was shown in Fig. 1.
Fig. 1.
The flow chart of our study.
2.10. Culture of cell lines
Cell lines HK-2 (normal human renal cell line), 786-O (human ccRCC cell line), and ACHN (human PRCC cell line) were obtained from ATCC (Manassas, VA, USA). HK-2, 786-O and ACHN cells were cultured in DMEM, RMPI-1640, and MEM medium with 10 % fetal bovine serum (Gibco, Waltham, MA, USA) respectively in a 37 °C, 5 % CO2 incubator. The cells was observed and cultured after passage every 48–72 h.
2.11. Quantitative real-time PCR (qRT-PCR)
TRIzol reagent (Yeasen Biotechnology, Shanghai, China) was used to extract the total RNA of cells. qRT-PCR was conducted on an ABI 7300 real-time PCR instrument (ABI, Carlsbad, CA) using Hieff® qPCR SYBR Green Master Mix (Yeasen Biotechnology) according to the manufacturer's methods. The relative gene expression levels were normalized to those of GAPDH and analyzed by using the 2−ΔΔCT method. SERPINH1: Forward primer 5′-TCAGTGAGCTTCGCTGATGAC -3′, Reverse primer 5′- CATGGCGTTGACTAGCAGGG -3’; GAPDH: Forward primer 5′-GGAGCGAGATCCCTCC AA AAT -3′, Reverse primer 5′- GGCTGTTGTCATACTTCTCATGG -3’.
2.12. Western blot
The cells were lysed using RIPA lysis buffer (Thermo Scientific, Waltham, MA, USA). The supernatant of the lysis was collected and the protein concentration was measured by using the BCA Protein Assay Kit (Thermo Scientific). Proteins from the cells were separated by 10 % SDS-PAGE and transferred to PVDF membranes (Millipore, Burlington, MA, USA). The membranes were incubated overnight at 4 °C with SERPINH1 (1:1000, Cell Signaling Technology, MA, USA) and GAPDH (1:30000, Proteintech, Chicago, USA) antibodies diluted in TBST buffer. Then the membranes were incubated for 1 h with HRP-conjugated anti-rabbit and anti-mouse secondary antibodies (Beijing Zhongshan Jinqiao Biotechnology, Beijing, China) respectively. Immunoreactive proteins were visualized on a Tanon-6300 Multi Image System. The relative expression levels of proteins were quantified with Image J software (v. 1.53, National Institutes of Health, USA) and normalized to those of GAPDH.
2.13. Statistical analysis
Statistical analysis was performed using R, version 4.2.1, and python 3.11.3. Non-parametric test was used to compare the predictive drug sensitivity, immune score, and TIDE score between different clusters and the expression of SERPINH1 in normal and tumor tissues or different clusters. Spearman's correlation analysis was utilized to show the association between the expression of SERPINH1 and the abundance of different kinds of immune cells. The R package “pROC” was performed to calculate the AUC value at 1, 3, and 5 years. Independent samples t-test was applied to detect differences in the results of qRT-PCR and Western blot. P value < 0.05 was considered statistically significant.
3. Results
3.1. Selection of the crucial genes in PRCC
The DEGs were selected based on the tumor samples from the TCGA database and the normal samples from the GTEx database. The volcano plot showed 2365 DEGs in PRCC (Fig. 2A). All 2365 DEGs were shown in Supplementary Table S3. According to the CRISPR data retrieved from the Depmap database, we collected 2170 genes associated with the development of the PRCC. Subsequently, a total of 2347 genes were found to be related to the survival of PRCC patients with univariate Cox regression. 41 genes possessing differential expression and prognostic ability, which were got by interaction of CRISPR screening, DEGs and genes related to the survival of PRCC, were shown by Venn diagram (Fig. 2B). Table 1 showed the name and other information of 41 key genes with differential expression levels.
Fig. 2.
Selection and enrichment analysis of the key genes. (A) The volcano plot showed the DEGs. Red points indicated up-regulated genes and blue points indicated down-regulated genes. (B) The key genes were found in the Venn diagram. The enriched KEGG signaling pathways (C), GO biological processes analysis (D), GO cellular components analysis (E), and GO molecular functions analysis (F) were performed based on the key genes.
Table 1.
The selected 41 key genes.
| Gene Symbol | Ensembl ID | Log2 (Fold Change) | Adjusted P value |
|---|---|---|---|
| KLHL13 | ENSG00000003096 | −1.512876218 | 1.01461E-08 |
| PTPRH | ENSG00000080031 | 1.051454285 | 3.52617E-06 |
| FST | ENSG00000134363 | −1.378058717 | 7.31869E-12 |
| CD93 | ENSG00000125810 | −1.884196804 | 4.11957E-17 |
| CCNA2 | ENSG00000145386 | 1.114144084 | 5.95662E-11 |
| BGN | ENSG00000182492 | −1.124466644 | 0.001089224 |
| CENPW | ENSG00000203760 | 1.124699812 | 1.70404E-13 |
| ROBO4 | ENSG00000154133 | −2.447506181 | 3.28918E-43 |
| CDC45 | ENSG00000093009 | 1.117860484 | 1.18572E-12 |
| TRPM3 | ENSG00000083067 | −1.08735926 | 1.12693E-09 |
| LPL | ENSG00000175445 | −2.675475405 | 1.23627E-25 |
| CRYL1 | ENSG00000165475 | −1.220672831 | 1.03368E-12 |
| RPL38 | ENSG00000172809 | 1.175647986 | 5.1007E-19 |
| TRIP13 | ENSG00000071539 | 1.130164006 | 3.78154E-11 |
| PIMREG | ENSG00000129195 | 1.059680497 | 8.82699E-11 |
| RRM2 | ENSG00000171848 | 1.008074491 | 5.84167E-07 |
| TOP2A | ENSG00000131747 | 1.725432858 | 2.88432E-10 |
| RPS19 | ENSG00000105372 | 1.236225978 | 1.66444E-16 |
| OTOGL | ENSG00000165899 | −1.07298278 | 1.30069E-29 |
| NOTCH3 | ENSG00000074181 | −2.367724235 | 2.79129E-15 |
| TPX2 | ENSG00000088325 | 1.564032755 | 4.15148E-11 |
| BIRC5 | ENSG00000089685 | 1.396591084 | 2.44409E-10 |
| BAMBI | ENSG00000095739 | 1.222115395 | 8.3771E-06 |
| AURKB | ENSG00000178999 | 1.39516125 | 1.99056E-12 |
| SYT7 | ENSG00000011347 | −3.237053542 | 1.88221E-59 |
| TCF7L1 | ENSG00000152284 | −1.802196192 | 1.50265E-23 |
| HOXD10 | ENSG00000128710 | −3.61208281 | 1.02695E-34 |
| PALM3 | ENSG00000187867 | −1.805283261 | 4.93822E-11 |
| PCDH17 | ENSG00000118946 | −1.281681258 | 3.54558E-15 |
| SLC7A1 | ENSG00000139514 | −1.148094619 | 1.0776E-07 |
| GINS2 | ENSG00000131153 | 1.535605246 | 4.41115E-19 |
| DONSON | ENSG00000159147 | 1.028615593 | 1.24042E-12 |
| KCTD15 | ENSG00000153885 | −1.58788661 | 1.39165E-11 |
| MYBL2 | ENSG00000101057 | 1.873844864 | 3.55362E-14 |
| RPL22L1 | ENSG00000163584 | 1.713533521 | 6.06719E-21 |
| TFRC | ENSG00000072274 | −1.148714269 | 5.28267E-07 |
| LHX1 | ENSG00000273706 | −2.630357275 | 1.34491E-25 |
| SERPINH1 | ENSG00000149257 | 1.098599498 | 2.86939E-13 |
| HADH | ENSG00000138796 | −1.54661165 | 2.82557E-31 |
| DTL | ENSG00000143476 | 1.075928307 | 1.38673E-08 |
| KIF20A | ENSG00000112984 | 1.06628196 | 2.66604E-07 |
Then KEGG and GO analyses were performed based on the key genes. KEGG analysis showed that the key genes were mainly associated with pathways in cancers and ribosomes (Fig. 2C). And GO analysis revealed that the key genes were also associated with cell cycle, chromosome, and kinase binding (Fig. 2D–F).
3.2. Identification of three clusters in PRCC based on the key genes and drug sensitivity analysis
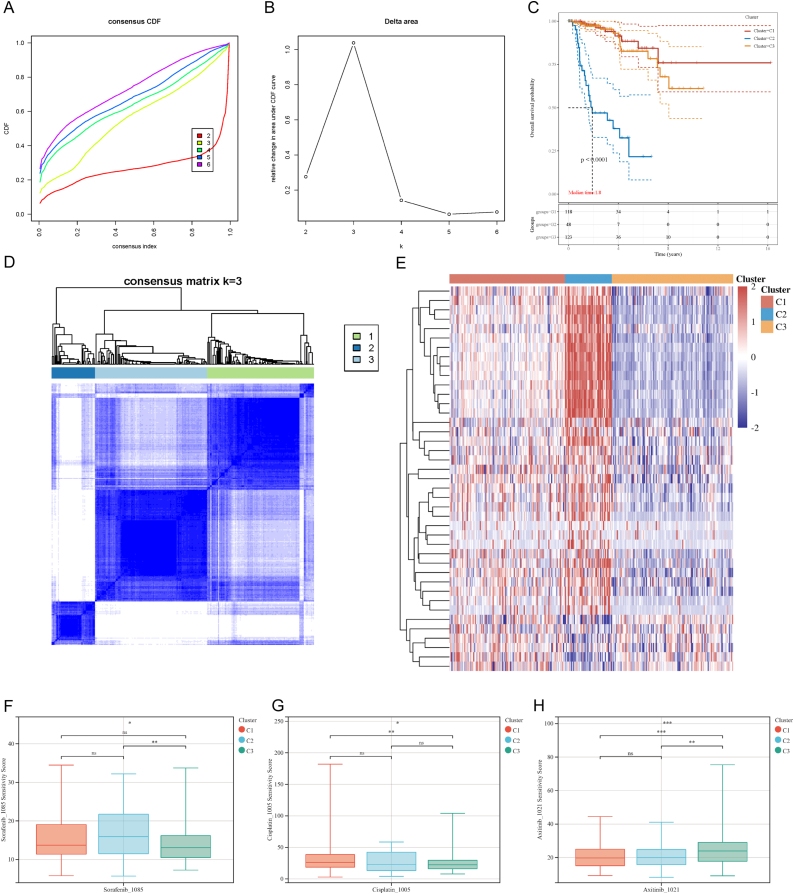
On the basis of expression of key genes in tumor samples, the consensus clustering analysis was conducted. The three clusters (C1, C2, and C3) were identified in the TCGA-PRCC cohort, which showed distinguishing survival and discrepant expression patterns of the key genes (Fig. 3A–D). The heatmap visualized the expression of 41 key genes among the three clusters. And C2 displayed higher expression of the key genes and lower survival than C1 and C3 (Fig. 3E). Table 2 displayed the clinical characteristics of the three clusters and cluster C2 presented the shortest survival and the youngest median age at initial pathologic diagnosis.
Fig. 3.
Consensus cluster analysis and drug sensitivity analysis. (A, B) CDF and relative change in the area under the CDF curve (CDF Delta area). (C) The KM curves of the clusters. (D) Heatmap described the consensus clustering solution when k = 3. (E) The expression of the key genes in the three clusters. Red color represented high expression, and blue color represented low expression. The predictive drug sensitivity of the three clusters to Sorafenib (F), Cisplatin (G), and Axitinib (H). (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Table 2.
Clinical characteristics among the three clusters in PRCC.
| Characteristics | C1 | C2 | C3 | Total | P value |
|---|---|---|---|---|---|
| n | 118 | 48 | 124 | 290 | |
| Gender | <0.001 | ||||
| FEMALE | 33 (11.38 %) | 22 (7.59 %) | 21 (7.24 %) | 76 (26.21 %) | |
| MALE | 85 (29.31 %) | 26 (8.97 %) | 103 (35.52 %) | 214 (73.79 %) | |
| Age at initial pathologic diagnosis | |||||
| Mean ± SD | 60.08 ± 12.01 | 57.17 ± 13.92 | 64.33 ± 10.59 | 61.42 ± 12.03 | |
| Median [min-max] | 60.00 [28.00,84.00] | 59.00 [28.00,85.00] | 64.00 [40.00,88.00] | 61.00 [28.00,88.00] | |
| Stage | <0.001 | ||||
| Stage I | 82 (30.15 %) | 8 (2.94 %) | 89 (32.72 %) | 179 (65.81 %) | |
| Stage II | 8 (2.94 %) | 4 (1.47 %) | 13 (4.78 %) | 25 (9.19 %) | |
| Stage III | 19 (6.99 %) | 23 (8.46 %) | 10 (3.68 %) | 52 (19.12 %) | |
| Stage IV | 4 (1.47 %) | 9 (3.31 %) | 3 (1.10 %) | 16 (5.88 %) | |
| pathologic_T | <0.001 | ||||
| T1 | 21 (7.24 %) | 8 (2.76 %) | 24 (8.28 %) | 53 (18.28 %) | |
| T1a | 48 (16.55 %) | 4 (1.38 %) | 49 (16.90 %) | 101 (34.83 %) | |
| T1b | 17 (5.86 %) | 4 (1.38 %) | 24 (8.28 %) | 45 (15.52 %) | |
| T2 | 8 (2.76 %) | 5 (1.72 %) | 9 (3.10 %) | 22 (7.59 %) | |
| T2a | 0 (0.0e+0 %) | 2 (0.69 %) | 5 (1.72 %) | 7 (2.41 %) | |
| T2b | 4 (1.38 %) | 0 (0.0e+0 %) | 4 (1.38 %) | 8 (2.76 %) | |
| T3 | 2 (0.69 %) | 5 (1.72 %) | 0 (0.0e+0 %) | 7 (2.41 %) | |
| T3a | 13 (4.48 %) | 17 (5.86 %) | 6 (2.07 %) | 36 (12.41 %) | |
| T3b | 2 (0.69 %) | 1 (0.34 %) | 1 (0.34 %) | 4 (1.38 %) | |
| T3c | 1 (0.34 %) | 0 (0.0e+0 %) | 0 (0.0e+0 %) | 1 (0.34 %) | |
| T4 | 1 (0.34 %) | 1 (0.34 %) | 0 (0.0e+0 %) | 2 (0.69 %) | |
| TX | 1 (0.34 %) | 1 (0.34 %) | 2 (0.69 %) | 4 (1.38 %) | |
| pathologic_N | <0.001 | ||||
| N0 | 60 (20.69 %) | 19 (6.55 %) | 66 (22.76 %) | 145 (50.00 %) | |
| N1 | 6 (2.07 %) | 16 (5.52 %) | 2 (0.69 %) | 24 (8.28 %) | |
| N2 | 1 (0.34 %) | 2 (0.69 %) | 0 (0.0e+0 %) | 3 (1.03 %) | |
| NX | 51 (17.59 %) | 11 (3.79 %) | 56 (19.31 %) | 118 (40.69 %) | |
| pathologic_M | 0.03 | ||||
| M0 | 89 (30.69 %) | 33 (11.38 %) | 83 (28.62 %) | 205 (70.69 %) | |
| M1 | 2 (0.69 %) | 5 (1.72 %) | 3 (1.03 %) | 10 (3.45 %) | |
| MX | 27 (9.31 %) | 10 (3.45 %) | 38 (13.10 %) | 75 (25.86 %) | |
| Status | <0.001 | ||||
| Alive | 110 (38.06 %) | 25 (8.65 %) | 110 (38.06 %) | 245 (84.78 %) | |
| Dead | 8 (2.77 %) | 23 (7.96 %) | 13 (4.50 %) | 44 (15.22 %) | |
Subsequently, the predictive drug sensitivity of the tumor samples was investigated in different clusters. As reported in previous researches, Sorafenib, Cisplatin, and Axitinib, which were generally used in the treatment of RCC, were applied to detect the drug sensitivity of the clusters [[36], [37], [38]]. A higher sensitivity score indicated a higher level of IC50 and lower efficiency. C3 exhibited greater sensitivity to Sorafenib compared to C2 and higher sensitivity to Cisplatin compared to C1 (Fig. 3F and G). Conversely, Axitinib showed a greater effect in C1 and C2 as compared to C3 (Fig. 3H).
3.3. Immune analysis among the clusters
The tumor microenvironment has been proved to play important roles in the development and treatment of all kinds of tumors. Fig. 4A summarized the infiltration of 22 kinds of immune cells in PRCC. ESTIMATE analysis revealed that a higher immune score was observed in C3, indicating more immune cells infiltrating the tumor tissue (Fig. 4B). Then, the immune check-point-related gene expression was investigated among the clusters. C2 showed significantly higher expression of LAG3, PDCD1, and PDCD1LG2 and lower expression of HAVCR2 (Fig. 4C). And C3 exhibited a lower rate of predicted response to ICB therapy as compared to C1 and C2 (Fig. 4D). In addition, the heatmap described the discrepant infiltration level of different kinds of immune cells in the tumor microenvironment among the three clusters (Fig. 4E). The significant decrease in the abundance of endothelial cells, B cells, and CD8+ T cells in C3 may serve as a plausible explanation for the observed lower rate of predicted response to ICB therapy.
Fig. 4.
Key genes-based clusters were associated with tumor microenvironment and ICB therapy. (A) CIBERSORT analysis showed the abundance of the immune cells in PRCC. Different colors represented different kinds of immune cells. (B) The immune score of the clusters. (C) The expression of the immune checkpoint-related genes exhibited significant differences among the clusters. (D) The predictive ICB therapy response. (E) The abundance of the immune cells in three clusters was shown through the heatmap. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
3.4. Somatic mutation analysis among the clusters
As shown in Fig. 5A, the mutation landscape of TCGA-PRCC cohort presented the top 15 mutated genes. The cohort summary plot showed different kinds of mutant variant types in C1, C2, and C3 (Fig. 5B–D). Missense mutation was the most frequent type of mutation in all the clusters. And the median value of the variants per sample in C2 was lower than the other two clusters. The mutation frequency in nine major oncogenic pathways were then investigated among the three clusters (Fig. 5E–G). RTK-RAS3 pathway exhibited the highest fraction of samples affected. Interestingly, the mutations in the NOTCH pathway were rarely detected in C3 but frequently detected in C1 and C2.
Fig. 5.
Somatic mutation in the clusters. (A) The mutant landscape and the top 15 mutant genes in PRCC. The cohort summary plots showed different variant types in C1 (B), C2 (C), and C3 (D). The mutation frequency was visualized in nine common oncogenic pathways in C1 (E), C2 (F), and C3 (G).
3.5. Development of a prognostic signature based on multiple machine learning algorithms
Considering that the key genes-based clusters were associated with vital characteristics in PRCC, 21 combinations of 9 machine learning algorithms were applied to build a prognostic signature. The AUC values of the prognostic signatures developed by different algorithms were shown in Fig. 6A. According to the average AUC values of the validation set and test set, RSF provided the best result (AUC value = 0.890) and 9 out of 41 genes were chosen to build the signature, including BIRC5, CENPW, KIF20A, MYBL2, RRM2, SERPINH1, TOP2A, TPX2, and TRIP13. And the optimal parameter combinantions of the RSF algorithm was: random_state = 7, n_estimators = 77, min_samples_split = 6, min_samples_leaf = 10, and max_depth = 2. The prognostic model was uploaded to the Github repository (https://github.com/YangYangRes/RSF_PRCC). In order to further validate the signature, the TCGA-PRCC cohort and GSE2748 cohort were divided into high risk group and low risk group based on the median of risk score. The overall survival of the high risk group was markedly lower than that of the low risk group (Fig. 6B and C).
Fig. 6.
A robust prognostic signature was developed based on the key genes. (A) 21 combinations of 9 machine learning algorithms were applied to develop the prognostic signature. The AUC values of each model at 1, 3, and 5 years in the validation set and test set were shown and the prognostic signature based on RSF was selected. The corresponding KM curves for high and low groups in TCGA-PRCC cohort (B) and GSE2748 cohort (C) were visualized. The univariate Cox regression analysis (D) and multiple Cox regression analysis (E) were performed based on the genes included in the prognostic signature.
3.6. Identification of SERPINH1 as a critical gene in PRCC
Univariate and multiple Cox regression analyses were performed using the genes included in the prognostic signature (Fig. 6D and E). All the genes included in the signature were associated with unfavorable overall survival, indicating the correctness of the prognostic signature. SERPINH1 exhibited high hazard ratio value and significant P value (P value < 0.0001) in both univariate and multiple Cox regression analyses. Hence, additional analyses were carried out to investigate the expression of a single gene, namely SERPINH1.
Fig. 7A showed that SERPINH1 expression was higher in the PRCC tumor tissues than in the normal renal tissues. C2 had significantly higher expression of SERPINH1 compared to the other two clusters (Fig. 7B). No relationship was found between the expression of SERPINH1 and TMB score or MSI score (Fig. 7C). Subsequently, the relationship between SERPINH1 expression and the abundance of immune cells were investigated (Fig. 7D–I). The abundance of B cells, CD4+ T cells, CD8+ T cells, neutrophils, and myeloid dendritic cells were significantly positively associated with the expression of SERPINH1, suggesting that SERPINH1 may play a role in the tumor microenvironment in PRCC.
Fig. 7.
The expression of SERPINH1 in PRCC. (A–B) The expression of SERPINH1 in the tumor tissues, normal tissues and three clusters. The relationship between SERPINH1 expression and TMB score or MSI score (C). (D–I) The association between the expression of SERPINH1 and the abundance of immune cells. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
3.7. Pan-cancer analysis for SERPINH1
The prognostic ability of SERPINH1 was evaluated in 33 types of tumors. The full names of the tumors were shown in Supplementary Table S4. The result showed that SERPINH1 was a strong predictor for poor prognosis in 13 types of tumors including GBM, LIHC, PAAD, ACC, CESC, HNSC, ccRCC, PRCC, LGG, LUAD, MESO, SARC, SKCM (P value < 0.01) (Fig. 8A). Subsequently, the correlation between tumor stages and gene expression levels of SERPINH1 was explored among 13 types of tumors, out of which 3 types of tumors exhibited a significant association between the expression of SERPINH1 and tumor stages including PRCC, ACC, and ccRCC (Fig. 8B–D). Immunohistochemistry from HPA database confirmed that compared with normal renal tubules (Fig. 8E), SERPINH1 protein was highly expressed in PRCC and ccRCC tumor tissues (Fig. 8F and G).
Fig. 8.
Pan-cancer analysis for SERPINH1, and SERPINH1 expression in PRCC and ccRCC cells. (A) The prognostic ability of SERPINH1 was evaluated in 33 types of tumors. SERPINH1 expression was associated with the tumor stages in PRCC (B), ACC (C), and ccRCC (D). Immunohistochemistry showed that compared with normal renal tubules (E), SERPINH1 protein was highly expressed in PRCC (F) and ccRCC (G) tumor tissues. (H) Quantitative real-time PCR showed that the expression levels of SERPINH1 mRNA were significantly higher in ACHN PRCC cells and 786-O ccRCC cells than those in normal HK-2 cells. (I–J) Western blot analysis indicated that the relative expression levels of SERPINH1 protein were markedly upregulated in ACHN and 786-O cells compared to normal HK-2 cells. The relative expression levels of proteins were quantified with Image J software and normalized to those of GAPDH. The uncropped figures of SERPINH1 and GAPDH in Western blot were shown in Supplementary Figs. S1 and S2. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
3.8. The high expression of SERPINH1 in PRCC and ccRCC cells
The expression levels of SERPINH1 mRNA in ACHN PRCC cells, 786-O ccRCC cells and normal HK-2 cells were detected by qRT-PCR. As shown in Fig. 8H, the expression levels of SERPINH1 mRNA were significantly higher in ACHN and 786-O cells than those in normal HK-2 cells. Western blot analysis showed that the relative expression levels of SERPINH1 protein were markedly increased in ACHN and 786-O cells compared to normal HK-2 cells (Fig. 8I and J). These results suggest that SERPINH1 mRNA and protein are upregulated in PRCC and ccRCC cells.
3.9. The relationship between the expression of SERPINH1 and tumor microenvironment, TMB, or MSI
The potential relationship between SERPINH1 and the tumor microenvironment was examined in 33 types of tumors (Fig. 9A). The analysis revealed that SERPINH1 expression was significantly associated with the abundance of CD8+ T cells, CD4+ T cells, macrophages, and endothelial cells in more than half of the tumor types. Notably, the expression of SERPINH1 showed a positive correlation with the abundance of CD8+ T cells and a negative correlation with that of macrophages. Next, the correlation between SERPINH1 and MSI or TMB was analyzed in the tumors (Fig. 9B and C). The expression of SERPINH1 demonstrated a significant association with the TMB score of THYM, ACC, and GBM+LGG, as well as the MSI score of TGCT, and GBM+LGG.
Fig. 9.
The relationship between the expression of SERPINH1 and tumor microenvironment, TMB, or MSI in multiple types of tumors. (A) EPIC analysis revealed the relationship between the expression of SERPINH1 and the abundance of immune cells in 33 types of tumors. The expression of SERPINH1 was related with TMB score (B) and MSI score (C) in multiple types of tumors.
4. Discussion
RCC ranks 13th among the most common types of malignant tumors worldwide and PRCC is the second most prevalent subtype of RCC with an increasing incidence rate [39]. Due to its heterogeneity, PRCC was considered a challenging cancer to treat in clinical practice. Patients with PRCC had limited options in terms of biomarkers and targeted therapies [40]. And lack of clear understanding of the molecular mechanism was thought to be the reason for the unsuccessful treatment of PRCC [41]. Therefore, there is an urgent need to explore the molecular mechanisms and effective biomarkers for PRCC.
The identification of a correlation between gene function and tumor progression can provide crucial information for screening potential biomarkers for targeted therapies in tumors [42]. Oncogenes and tumor suppressor genes play critical roles in both promoting and inhibiting tumor development, respectively, which make them become essential components of the tumor molecular mechanism [43,44]. Therefore, systematically recognizing these genes in malignant tumors, which were defined as crucial genes, may provide new insights into the process of tumor development [45]. Depmap database was established utilizing CRISPR-Cas9 technology to conduct genome-wide gene function inactivation screening across multiple cell lines, which enabled us to use the cancer dependencies of the genes to identify the crucial genes [15].
In the current study, we recognized 41 key genes based on the CRISPR data, differential expression, and prognostic ability in PRCC. Based on the expression of the key genes, three clusters were identified and the results also showed distinct survival, drug sensitivity, clinical characteristics, tumor microenvironment, and mutation landscape among the clusters. Moreover, a reliable prognostic signature was developed utilizing multiple combinations of machine learning algorithms based on the key genes. The signature was tested by an external independent verification set and exhibited good prognostic ability. SERPINH1 was selected from the signature for its strong prognostic ability, and its expressions in PRCC and pan-cancers were also analyzed.
First, three distinct clusters (C1, C2 and C3) were identified in the TCGA-PRCC cohort by consensus cluster analysis based on 41 key genes. C2 had obviously higher expression of the key genes and lower survival than C1 and C3, indicating the key genes as poor prognostic factors for PRCC. Axitinib has been demonstrated to have encouraging efficiency in treating PRCC patients, especially in type 2 PRCC patients [38]. The drug sensitivity analysis suggested that Axitinib may be a potential drug for treating patients in C2.
In this study, we utilized CIBERSORT analysis to evaluate the infiltration of the immune cells in tumor microenvironment. Previous research suggested that PRCC, especially for type 2 PRCC, had the highest expression level of macrophages among renal cancer histological types [46]. Macrophages were critical regulation of tumor with the function of helping stimulate proliferation, metastasis, and angiogenesis, and were possibly related to poor survival [47]. Our results showed that the abundance of macrophages was higher in C3 than in C1 and C2, which had the highest immune score. The abundance of B cells and CD8+ T cells in C1 and C2 was higher than that in C3, which may explain why C1 and C2 had higher predictive ICB therapy response rate. Scholars have also proved the vital role of B cells and CD8+ T cells in immune therapy [33,34].
Previous studies showed that the alteration of the MET gene was observed in 81 % of the type 1 PRCC, and efficient drug targeting the MET/VEGFR2 pathways has been tested [48,49]. Our study revealed the top 15 mutated genes in the TCGA-PRCC cohort and the top 10 mutant genes in three clusters. Based on the mutation landscape, MET was identified as the fourth most frequently mutant gene in the cohort and the second most mutant gene in C1, suggesting its potential as a treatment target for PRCC. Moreover, the mutation frequency of the NOTCH pathway was significantly higher in C1 and C2 when compared with C3. Prior investigations reported that proximal tubules forming with reduced NOTCH signaling have been implicated in the formation of cyst formation, and a few microadenomas contained in the proximal tubules resembling precursors of PRCC [50].
Considering the difficulty in the treatment of advanced PRCC, an accurate prognostic model for PRCC was urgently needed. Accurate prognostic models enable clinicians to make decisions more efficiently [51]. Nomograms are commonly used to predict the prognosis of the patients with tumors with user friendly digital interfaces. They are widely used in multiple types of tumors [52,53]. However, nomograms with good performance may lack clinical practicability [54]. Researchers also developed special models to predict molecular features. For example, m1Ascore was constructed to evaluate the m1A modification pattern of individual patient [55]. Machine learning algorithms have unique advantages in cancer prognosis [56]. But the existing models rarely used machine learning algorithms or the external validation set in PRCC [57,58]. In our study, we developed the prognostic signature utilizing 21 combinations of 9 machine learning algorithms. The external validation set was used to help select the most precise and reliable algorithm. As a result, RSF was selected and applied to develop the signature. Compared to the previous prognostic signature [59,60], this prognostic signature possessed a higher AUC level in the validation set and was more accurate and reliable.
Among the genes included in the prognostic signature, SERPINH1 was selected for its prognostic ability through univariate and multiple Cox regression analysis. SERPINH1 has been found to be involved in the initiation and development of cancer, and regulate cell protease homeostasis as chaperone protein [61]. Previous studies found that SERPINH1 was a predictor for unfavorable prognosis in other types of renal carcinomas [62]. However, the expression and role of SERPINH1 in PRCC still remains unclear. In this research, C2 exhibited the highest expression level of SERPINH1, indicating its contribution to the development of the tumor. And the expression of SERPINH1 presented a significant correlation with the abundance of immune cells. Furthermore, the expression of SERPINH1 was related to the stages, tumor microenvironment, TMB, and MSI across multiple tumors, suggesting its potential as a drug target. And the results of qRT-PCR and Western blot showed that SERPINH1 mRNA and protein were highly expressed in PRCC cells compared with normal human renal cells. Overall, our findings suggested that SERPINH1 may be an important biomarker as well as a potential therapeutic target for PRCC patients.
5. Limitations
Although our study provided valuable insights into PRCC, it also has some limitations that need to be mentioned. A larger number of samples are needed to enhance the accuracy of our prognostic signature. Differences in the patient cohort and experimental cohort may reduce the comparability of the external independent set [63]. And further experimental validation in vitro and in vivo will next be conducted to test the correctness of the molecular classification and prognostic signature.
6. Conclusions
In summary, our research identified three clusters in PRCC based on 41 key genes which were obtained from the intersection of CRISPR screening, DEGs, and genes related to the survival of PRCC. Significant differences in drug sensitivity, tumor microenvironment, and mutation landscape have been observed among the three clusters. By utilizing 21 combinations of 9 machine learning algorithms, 9 out of 41 genes were chosen to build a robust prognostic signature, including BIRC5, CENPW, KIF20A, MYBL2, RRM2, SERPINH1, TOP2A, TPX2, and TRIP13. The signature was tested by an external independent verification set and exhibited good prognostic ability. SERPINH1 was identified as a critical gene for its strong prognostic ability in PRCC by univariate and multiple Cox regression analyses. And the experiments in vitro at cell level were performed to validate the high expression of SERPINH1 mRNA and protein in PRCC cells. This study exhibited a new molecular classification and prognostic signature for PRCC, which may provide a potential biomarker and therapy target for PRCC patients. However, further experiments in vitro and in vivo are needed to explore the role of SERPINH1 in tumor progression, metastasis, and potential therapeutic targeting for PRCC patients in the forthcoming research.
Funding statement
This work was supported by National Natural Science Foundation of China (Grant No. 82172858), National Undergraduate Training Project for Innovation (Grant No. 202210366025, 202310366023), Key Project of Teaching Reform Research in Anhui Province (Grant No. 2022jyxm718) and Anhui Medical University “Early Involvement of Scientific Research” Training Project (Grant No. 2022-ZQKY-182, 2021-ZQKY-182).
Data availability statement
The data associated with this study could be downloaded at TCGA database (https://www. cancer.gov/tcga), GTEx database (https://www.gtexportal.org/home/), Depmap database (https://depmap.org/portal/), and GDSC database (https://www.cancerrxgene.org/). The code has been deposited at the Github repository (https://github.com/YangYangRes/RSF_PRCC).
CRediT authorship contribution statement
Chang Liu: Writing – original draft, Validation, Software, Methodology, Investigation, Formal analysis. Zhan-Yuan Yuan: Writing – original draft, Software, Resources, Methodology, Investigation, Data curation. Xiao-Xun Zhang: Validation, Software, Resources, Methodology, Investigation, Formal analysis. Jia-Jun Chang: Validation, Software, Methodology, Investigation. Yang Yang: Validation, Software, Methodology, Investigation. Sheng-Jia Sun: Software, Methodology, Investigation, Data curation. Yinan Du: Writing – review & editing, Project administration, Data curation, Conceptualization. He-Qin Zhan: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The results shown here are in part based upon data generated by TCGA database (https://www.cancer.gov/tcga), GTEx database (https://www.gtexportal.org/home/), Depmap database (https://depmap.org/portal/), GDSC database (https://www.cancerrxgene.org/) and HPA database (https://www.proteinatlas.org/).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e23184.
Contributor Information
Yinan Du, Email: duyinannan@126.com.
He-Qin Zhan, Email: heqinzh@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
References
- 1.Capitanio U., Bensalah K., Bex A., et al. Epidemiology of renal cell carcinoma. Eur. Urol. 2019;75:74–84. doi: 10.1016/j.eururo.2018.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perazella M.A., Dreicer R., Rosner M.H. Renal cell carcinoma for the nephrologist. Kidney Int. 2018;94:471–483. doi: 10.1016/j.kint.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 3.Steffens S., Janssen M., Roos F.C., et al. Incidence and long-term prognosis of papillary compared to clear cell renal cell carcinoma--a multicentre study. Eur. J. Cancer. 2012;48:2347–2352. doi: 10.1016/j.ejca.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Lobo J., Ohashi R., Amin M.B., et al. WHO 2022 landscape of papillary and chromophobe renal cell carcinoma. Histopathology. 2022;81:426–438. doi: 10.1111/his.14700. [DOI] [PubMed] [Google Scholar]
- 5.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA. Cancer J. Clin. 2019;69 doi: 10.3322/caac.21551. 2019. [DOI] [PubMed] [Google Scholar]
- 6.Qu Y., Chen H., Gu W., et al. Age-dependent association between sex and renal cell carcinoma mortality: a population-based analysis. Sci. Rep. 2015;5:9160. doi: 10.1038/srep09160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoo S., You D., Jeong I.G., et al. Histologic subtype needs to be considered after partial nephrectomy in patients with pathologic T1a renal cell carcinoma: papillary vs. clear cell renal cell carcinoma. J. Cancer Res. Clin. Oncol. 2017;143:1845–1851. doi: 10.1007/s00432-017-2430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Yan K., Lin J., et al. Three-gene risk model in papillary renal cell carcinoma: a robust likelihood-based survival analysis. Aging (Albany NY) 2020;12:21854–21873. doi: 10.18632/aging.104001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng R., Li J., Zhao H., et al. Identification of potential biomarkers associated with immune infiltration in papillary renal cell carcinoma. J. Clin. Lab. Anal. 2021;35 doi: 10.21203/rs.3.rs-769170/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janik E., Niemcewicz M., Ceremuga M., et al. Various Aspects of a gene editing system-CRISPR- Cas9. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21249604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garneau J.E., Dupuis M.-È., Villion M., et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 12.Ansori A.N., Antonius Y., Susilo R.J., et al. Application of CRISPR-Cas9 genome editing technology in various fields: a review. Narra. J. 2023;3 doi: 10.52225/narra.v3i2.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stefanoudakis D., Kathuria-Prakash N., Sun A.W., et al. The potential Revolution of cancer treatment with CRISPR technology. Cancers. 2023;15 doi: 10.3390/cancers15061813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyers R.M., Bryan J.G., McFarland J.M., et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 2017;49:1779–1784. doi: 10.1101/160861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsherniak A., Vazquez F., Montgomery P.G., et al. Defining a cancer dependency map. Cell. 2017;170 doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Z.-H., Yang D.-L., Wang L., et al. Epigenetic and immune-cell infiltration changes in the tumor microenvironment in hepatocellular carcinoma. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.793343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi B., Ding J., Qi J., et al. Characteristics and prognostic value of potential dependency genes in clear cell renal cell carcinoma based on a large-scale CRISPR-Cas9 and RNAi screening database DepMap. Int. J. Med. Sci. 2021;18:2063–2075. doi: 10.7150/ijms.51703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.GTEx Consortium The Genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X.J., Tan M.-H., Kim H.L., et al. A molecular classification of papillary renal cell carcinoma. Cancer Res. 2005;65:5628–5637. doi: 10.1158/0008-5472.can-05-0533. [DOI] [PubMed] [Google Scholar]
- 21.Shen W., Song Z., Zhong X., et al. Sangerbox: a comprehensive, interaction‐friendly clinical bioinformatics analysis platform. iMeta. 2022;1 doi: 10.1002/imt2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie M.E., Phipson B., Wu D., et al. Limma powers differential expression analyses for RNA- sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu G., Wang L.-G., Han Y., et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeser D., Gruener R.F., Huang R.S. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief. Bioinform. 2021;22 doi: 10.1093/bib/bbab260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newman A.M., Liu C.L., Green M.R., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang P., Gu S., Pan D., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Racle J., de Jonge K., Baumgaertner P., et al. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife. 2017;6 doi: 10.7554/elife.26476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayakonda A., Lin D.-C., Assenov Y., et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pölsterl S. Scikit-survival: a Library for time-to-event analysis built on top of scikit-learn. J. Mach. Learn. Res. 2020;21:8747–8752. https://dl.acm.org/doi/abs/10.5555/3455716.3455928 [Google Scholar]
- 31.Thorsson V., Gibbs D.L., Brown S.D., et al. The immune landscape of cancer. Immunity. 2018;48 doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonneville R., Krook M.A., Kautto E.A., et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis. Oncol. 2017;2017 doi: 10.1200/po.17.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li T., Fu J., Zeng Z., et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Z., Kang B., Li C., et al. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sturm G., Finotello F., Petitprez F., et al. Comprehensive evaluation of transcriptome-based cell- type quantification methods for immuno-oncology. Bioinformatics. 2019;35:i436–i445. doi: 10.1093/bioinformatics/btz363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strumberg D. Sorafenib for the treatment of renal cancer, Expert Opin. Pharmacother. 2012;13:407–419. doi: 10.1517/14656566.2012.654776. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J., Ye Z.-W., Tew K.D., et al. Cisplatin chemotherapy and renal function. Adv. Cancer Res. 2021;152:305–327. doi: 10.1016/bs.acr.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Negrier S., Rioux-Leclercq N., Ferlay C., et al. Axitinib in first-line for patients with metastatic papillary renal cell carcinoma: results of the multicentre, open-label, single-arm, phase II AXIPAP trial. Eur. J. Cancer. 2020;129:107–116. doi: 10.1016/j.ejca.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Mendhiratta N., Muraki P., Sisk A.E., et al. Papillary renal cell carcinoma: review. Urol. Oncol. 2021;39:327–337. doi: 10.1016/j.urolonc.2021.04.013. [DOI] [PubMed] [Google Scholar]
- 40.Angori S., Lobo J., Moch H. Papillary renal cell carcinoma: current and controversial issues. Curr. Opin. Urol. 2022;32:344–351. doi: 10.1097/mou.0000000000001000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akhtar M., Al-Bozom I.A., Al Hussain T. Papillary renal cell carcinoma (PRCC): an update. Adv. Anat. Pathol. 2019;26:124–132. doi: 10.1097/pap.0000000000000220. [DOI] [PubMed] [Google Scholar]
- 42.Sheng K.L., Kang L., Pridham K.J., et al. An integrated approach to biomarker discovery reveals gene signatures highly predictive of cancer progression. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-78126-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martín-Lorenzo A., Gonzalez-Herrero I., Rodríguez-Hernández G., et al. Early epigenetic cancer decisions. Biol. Chem. 2014;395:1315–1320. doi: 10.1515/hsz-2014-0185. [DOI] [PubMed] [Google Scholar]
- 44.Bashyam M.D., Animireddy S., Bala P., et al. The Yin and Yang of cancer genes. Gene. 2019;704:121–133. doi: 10.1016/j.gene.2019.04.025. [DOI] [PubMed] [Google Scholar]
- 45.Chen W., Lin Y., Jiang M., et al. Identification of LARS as an essential gene for osteosarcoma proliferation through large-Scale CRISPR-Cas9 screening database and experimental verification. J. Transl. Med. 2022;20:355. doi: 10.1186/s12967-022-03571-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Synnott N.C., Poeta M.L., Costantini M., et al. Characterizing the tumor microenvironment in rare renal cancer histological types. J. Pathol. Clin. Res. 2022;8:88–98. doi: 10.1002/cjp2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeNardo D.G., Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 2019;19:369–382. doi: 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Linehan W.M., Spellman P.T., Ricketts C.J., et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N. Engl. J. Med. 2016;374:135–145. doi: 10.1056/nejmoa1505917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choueiri T.K., Vaishampayan U., Rosenberg J.E., et al. Phase II and biomarker study of the dual MET/VEGFR2 inhibitor foretinib in patients with papillary renal cell carcinoma. J. Clin. Oncol. 2013;31:181–186. doi: 10.1200/jco.2012.43.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Surendran K., Selassie M., Liapis H., et al. Reduced Notch signaling leads to renal cysts and papillary microadenomas. J. Am. Soc. Nephrol. 2010;21:819–832. doi: 10.1681/asn.2009090925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hui D., Maxwell J.P., Pavi C.E. Dealing with prognostic uncertainty: the role of prognostic models and websites for patients with advanced cancer. Curr. Opin. Support. Palliat. Care. 2019;13(4):360–368. doi: 10.1097/spc.0000000000000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J., Mei J., Wang Y., et al. Development of a novel immune-related lncRNA signature as a prognostic classifier for endometrial carcinoma. Int. J. Biosci. 2021;17(2):448–459. doi: 10.7150/ijbs.51207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J., Meng H., Nie S., et al. Identification of a prognostic signature of epithelial ovarian cancer based on tumor immune microenvironment exploration. Genomics. 2020;112(6):4827–4841. doi: 10.1016/j.ygeno.2020.08.027. [DOI] [PubMed] [Google Scholar]
- 54.Balachandran V.P., Gonen M., Smith J.J., et al. Nomograms in oncology: more than meets the eye. Lancet Oncol. 2015;16(4):e173–e180. doi: 10.1016/s1470-2045(14)71116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu J., Chen C., Wang Y., et al. Comprehensive of N1-methyladenosine modifications patterns and immunological characteristics in ovarian cancer. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.746647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Painuli D., Bhardwaj S., Köse U., et al. Recent advancement in cancer diagnosis using machine learning and deep learning techniques: a comprehensive review. Comput. Biol. Med. 2022;146 doi: 10.1016/j.compbiomed.2022.105580. [DOI] [PubMed] [Google Scholar]
- 57.He R., Wang L., Li J., et al. Integrated analysis of a competing endogenous RNA network reveals a prognostic signature in kidney renal papillary cell carcinoma. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.612924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fei H., Chen S., Xu C. Construction autophagy-related prognostic risk signature combined with clinicopathological validation analysis for survival prediction of kidney renal papillary cell carcinoma patients. BMC Cancer. 2021;21:411. doi: 10.1186/s12885-021-08139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yin H., Lin M., Liang S., et al. Ferroptosis-related gene signature predicts prognosis in kidney renal papillary cell carcinoma. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.988867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao Z.Y., Xing C.Q., Zhang T., et al. MicroRNA related prognosis biomarkers from high throughput sequencing data of kidney renal papillary cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2021;25:2235–2244. doi: 10.26355/eurrev_202103_25255. [DOI] [PubMed] [Google Scholar]
- 61.Hirayoshi K., Kudo H., Takechi H., et al. HSP47: a tissue-specific, transformation-sensitive, collagen-binding heat shock protein of chicken embryo fibroblasts. Mol. Cell Biol. 1991;11:4036–4044. doi: 10.1128/mcb.11.8.4036-4044.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qi Y., Zhang Y., Peng Z., et al. SERPINH1 overexpression in clear cell renal cell carcinoma: association with poor clinical outcome and its potential as a novel prognostic marker. J. Cell Mol. Med. 2018;22:1224–1235. doi: 10.1111/jcmm.13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y., Bernau C., Parmigiani G., et al. The impact of different sources of heterogeneity on loss of accuracy from genomic prediction models. Biostatistics. 2020;21:253–268. doi: 10.1093/biostatistics/kxy044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data associated with this study could be downloaded at TCGA database (https://www. cancer.gov/tcga), GTEx database (https://www.gtexportal.org/home/), Depmap database (https://depmap.org/portal/), and GDSC database (https://www.cancerrxgene.org/). The code has been deposited at the Github repository (https://github.com/YangYangRes/RSF_PRCC).