Abstract
Expression of the principal chaperones of the heat shock stimulon of Streptomyces albus G are under the negative control of different repressors. The dnaK operon is regulated by hspR, the last gene of the operon (dnaK-grpE-dnaJ-hspR). hsp18, encoding a member of the small heat shock protein family, is regulated by orfY, which is in the opposite orientation upstream of hsp18. The groES-groEL1 operon and the groEL2 gene are regulated differently. They present tandem copies of the CIRCE element found in the 5′ region of many heat shock genes and shown to act in Bacillus subtilis as an operator for a repressor encoded by hrcA (hrc stands for heat regulation at CIRCE). We report the identification in S. albus of a new heat shock operon containing hrcA and dnaJ homologs. Disruption of hrcA increased the transcription of the groES-groEL1 operon and of the groEL2 gene. These features were lost when the mutant was complemented in trans by an intact copy of hrcA. Despite considerable accumulation of the GroE chaperones in the hrcA mutant, there was no effect on formation of the aerial mycelium and sporulation, indicating that neither hrcA nor the level of groE gene expression is directly involved in the regulation of Streptomyces morphological differentiation.
Streptomyces ssp. are filamentous soil bacteria belonging to the high-G+C branch of the gram-positive eubacteria. These bacteria have a complex morphological cycle. On solid media, spores germinate and form a basal mycelium. This is followed by aerial mycelium development, and septation of the mycelium leads to spore formation. Streptomyces spp. are metabolically active during the stationary phase and produce a very large variety of secondary metabolites. We are interested in studying the heat shock response in Streptomyces because heat shock proteins (HSPs) may be involved in physiological and morphological differentiation. Nutritional imbalances activate a developmental program involving the heat shock stress regulon (33). This suggests that the regulatory systems that control HSPs are involved in Streptomyces development. We are also interested in identifying the principal HSPs of Streptomyces, because they could be used to facilitate protein overproduction and secretion (45). Manipulation of the hsp genes may make it possible to develop Streptomyces hosts that produce homologous or heterologous proteins more efficiently.
Scores of differentiation mutants have been obtained in Streptomyces in recent years. They fall into two main classes: “bald” mutants, unable to produce aerial mycelia, and “white” mutants, unable to sporulate (4). Several mutations have been characterized, but none are related to the hsp genes. However, the cascade of events leading to the change from basal mycelium to aerial mycelium formation has not been elucidated.
The heat shock response and its regulation were first studied in detail in Escherichia coli. There are two heat shock regulons, positively regulated at the level of transcription by sigma factors. The largest regulon is controlled by ς32, and the smallest regulon is controlled by ς24 (for recent reviews see references 3, 11, and 48).
However, recent results obtained with other bacteria show a different situation. In Bacillus subtilis, the heat shock response of which has also been studied in detail, three classes of heat shock genes have been described. The number of classes is likely to increase because class III comprises heat shock genes of unknown regulation. The chaperone-encoding genes dnaK and cohort dnaJ and grpE and the chaperonin-encoding genes groEL and groES are ubiquitous. These are class I genes in B. subtilis, and their thermoregulation depends on a repressor and an operator CIRCE (for controlling inverted repeat of chaperone expression) (35, 49). CIRCE elements have been identified more than 50 times in 28 eubacterial species (17). CIRCE has been reported to be associated with the dnaK and/or groEL genes or their cohort, grpE, dnaJ, and groES. In B. subtilis, the gene of the dnaK operon furthest upstream, formerly called orf39, has been shown to encode the repressor of the dnaK and groEL operons. This protein binds to CIRCE-bearing DNA fragments (47), and orf39 has been renamed hrcA (for heat regulation at CIRCE).
It is likely that all microorganisms with CIRCE contain a gene similar to hrcA. Genes similar to hrcA have been identified by chance by sequencing the dnaK operon and during the sequencing of the entire genomes of organisms.
In Streptomyces spp., neither hrcA nor CIRCE is associated with the dnaK operon, the regulation of which depends on another repressor, HspR (2, 13). CIRCE is associated with two groEL genes (groEL1 and groEL2) (25). groEL1 forms an operon with a groES gene, whereas groEL2 maps to another location on the chromosome (7, 34) and is not associated with a groES gene. Transcription of these genes is thermoregulated. Transcription of the groES-groEL1 operon generates two mRNAs: a 500-base groES transcript and a 2,200-base polycistronic groES-groEL1 transcript. The transcript of groEL2 is 1,900 bases long (15). Two copies of CIRCE are present in tandem upstream from groESL1 and groEL2. One copy is located between the transcriptional and translational start sites, and the other overlaps the −35 hexamer of the promoter (7). The presence of CIRCE elements and the detection of a CIRCE DNA-binding protein in gel retardation assays (8) suggests that an hrcA-like gene is present in Streptomyces.
We report here the cloning and characterization of the Streptomyces albus hrcA gene. The effects of disrupting hrcA show that it is the principal regulator of groEL expression and that neither hrcA nor the level of groEL is directly involved in control of the Streptomyces cell cycle.
MATERIALS AND METHODS
Bacterial strains, media, and plasmids.
The S. albus G strain (J1074), deficient in both SalI restriction and modification systems (5), was obtained from the John Innes Culture Collection (Norwich, England). S. albus was routinely cultivated on NE plates (29) or in YEME rich liquid medium (20). When required, thiostrepton and viomycin were added at a concentration of 25 μg/ml, and hygromycin was used at a concentration of 250 μg/ml. The E. coli strain SURE (14) was used as the host for cosmid library preparation, and strain TG1 (12) was used for plasmid construction. E. coli S17.1 (39), carrying an integrated RP4 derivative, was used for intergenic conjugation between E. coli and S. albus. E. coli GM2929 (dam dcm) was used for the extraction of unmethylated DNA. E. coli cells were cultured in Luria broth supplemented with ampicillin (100 μg/ml) when required. pUC19 and M13 vectors (46) were used for cloning and sequencing. pHM11a (28), an E. coli-Streptomyces integrative shuttle vector, was used for complementation of the hrcA mutation. Cosmid vector SuperCos I (9) was obtained from Stratagene.
Preparation of the cosmid library.
Chromosomal DNA from S. albus was partially digested with Sau3AI and ligated into the BamHI site of SuperCos I. The cosmid was packaged by using λ packaging extracts from Stratagene. E. coli SURE was infected with the λ particles, and ampicillin-resistant clones were selected. A total of 1,152 colonies were used to inoculate 150 μl of Luria-Bertani broth containing ampicillin in 12 96-well microtiter plates. After overnight growth, glycerol was added to a final concentration of 20%, and the plates were stored at −70°C.
DNA manipulations.
Degenerate oligonucleotide Dal3 (ACSATCCGSAACGASATGGCSSASCTSGAG, with S being G or C) based on the HrcA consensus region, TIRNEMADLE, was used to probe the cosmid library. The oligonucleotide was radiolabeled with [γ-32P]dATP by using polynucleotide kinase (Pharmacia). The blots were probed overnight by incubation at 57°C with the oligonucleotide. They were then washed with 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) at 57°C and autoradiographed. Four colonies that gave a strong signal were analyzed further. In Southern blots probed with radiolabeled Dal3, three cosmids (H1, H2, and H3) generated similar hybridization patterns. In particular, a 1.2-kb SalI fragment present in the three preparations hybridized with Dal3. This fragment was isolated from agarose gel and inserted into pUC19. The sequence was determined and showed that this fragment contained the 5′ region of a gene similar to hrcA. The fourth cosmid, H4, generated a different hybridization pattern. A 450-bp SalI fragment which hybridized with Dal3 was cloned and sequenced. It contained a stretch of 22 nucleotides identical to the Dal3 probe, accounting for the hybridization signal. However, the rest of the sequence was very different from that of hrcA. The protein encoded by the 450-bp SalI fragment had a sequence similar to that of the NusA protein (SwissProt identification no. P32727) of B. subtilis. There were no significant similarities between the sequences of NusA and HrcA, so we concluded that the signal obtained was due to chance, and cosmid H4 was not analyzed further.
We obtained a complete copy of the hrcA gene by isolating a 3.5-kb SmaI fragment from cosmid H1 and inserting it into pUC19 cut with SmaI, giving pPM3881.
Determination of the DNA sequence.
The EcoRI/HindIII fragment of pPM3881 was inserted into the appropriate sites of M13mp18 or M13mp19. DNA was sequenced by the dideoxy chain method of Sanger et al. with the Sequenase kit (version 2.0; U.S. Biochemical Corp.) and 35S-dATP, the universal M13 primer, and synthetic oligonucleotides (17-mers; Genset).
Construction of an hrcA insertion mutant.
pPM3881 was cut with BclI, and the 1.8-kb BamHI fragment from pIJ39 (41) carrying the tsr gene was inserted into the BclI site of pPM3881 to give pPM3891. pCG3981 was constructed by introducing the oriT-vioR cassette (37) into the single HindIII site of pPM3891. This cassette contains an origin of transfer and makes it possible to select for viomycin resistance. pPM3981 was transferred by conjugation from E. coli to S. albus as previously described (26).
Construction of a dnaJ2 insertion mutant.
pPM3881 was cut with BamHI and AlwNI, and the 980-bp internal fragment of dnaJ2 was isolated, its ends were blunted, and it was ligated into SmaI-digested pUC19, giving pPM3902. The internal dnaJ2 fragment was removed by digestion with KpnI and BamHI and was inserted into KpnI- and BamHI-digested pPM2208 (formerly called pPM935-del [37]) to give pPM3922. pPM3922 was transferred by conjugation from E. coli to S. albus, and thiostrepton-resistant clones were selected.
Secretion of endogenous proteins.
For α-amylase secretion analysis, S. albus wild-type and mutant strains were patched on starch-containing plates and halos of starch hydrolysis were detected by iodine staining. For β-lactamase secretion analysis, the strains were patched on ampicillin-containing plates. They were cultured for various periods of time, and the plates were then spread with ampicillin-sensitive E. coli bacteria. The size of the halos of E. coli growing at the point of contact with the Streptomyces patches indicated the level of ampicillin detoxification by secreted β-lactamase.
Complementation of the hrcA mutation.
A full-length hrcA gene was obtained by PCR with the oligonucleotides OLC45 (5′ CGCATATGCTCAGCGAACGAAGGCTCGAAG 3′) and OLC46 (5′ CGGGATCCTTACGACTCCGCCAGGATCTGTC 3′).
The PCR product was cut with NdeI and BamHI and ligated into pHM11a digested with NdeI and BamHI to give pCG92. pCG92 was transferred by conjugation from E. coli to S. albus. Exconjugants were selected on the basis of hygromycin resistance.
Heat shock protein induction in Streptomyces.
Total proteins, either [35S]methionine-cysteine labeled or unlabeled, were extracted as described previously (16). SDS-polyacrylamide gel electrophoresis (PAGE) (15% polyacrylamide) was performed according to the method of Laemmli (23). The proteins were transferred to Immobilon membranes. GroEL proteins were detected by Western blotting with the ECL Western blotting system (Amersham). Mouse antibodies raised against Mycobacterium leprae GroEL were provided by B. Gicquel (Institut Pasteur).
RNA analysis.
S. albus total RNA was extracted, and Northern blotting was carried out as previously described (38). Highly stringent conditions were used for hybridization, and the blots were washed with 0.5× SSC–0.1% SDS at 65°C. The probes used were S. albus groES, groEL2 (25), and hrcA (this work).
The transcription start site upstream from the hrcA-dnaJ2 operon was determined by primer extension. The hrcA-specific oligonucleotide OLC47 (5′ GCGGTACCCTTGTCGGTCGGGATCGGCCC 3′), radiolabeled with [γ-32P]dATP by using polynucleotide kinase, was used with RNA preparations from heat-shocked and untreated S. albus cultures. Avian myeloblastosis virus (AMV) reverse transcriptase was used according to the manufacturer’s recommendations (Pharmacia). The samples were subjected to electrophoresis in a 5% polyacrylamide–8 M urea gel.
Nucleotide sequence accession number.
The nucleotide sequence of S. albus hrcA has been deposited in GenBank (accession no. AF025656).
RESULTS
Cloning of the hrcA gene of S. albus.
The CIRCE operator sequences are highly conserved among bacterial species, but the sequences of HrcA repressors share few similarities. This makes it difficult to design a nucleotide probe for hrcA, even if the Streptomyces codon bias is used. Streptomyces has a high G+C content, and it tends to favor codons with a G or a C in the third position. However, we mostly used highly degenerate probes. Various oligonucleotides were designed based on the available HrcA sequences. Oligonucleotide Dal3 is based on the peptide TIRN(EDY)MA(DAQV)LE, a consensus sequence derived from the HrcA proteins of B. subtilis (44), Staphylococcus aureus (31), Clostridium acetobutylicum (30), Mycoplasma genitalium (10), and Chlamydia trachomatis (40), and on ygr, a sequence encoding an HrcA-like protein, discovered during the M. leprae sequencing project (cosmid B1937; K. Robison, GenBank). Oligonucleotide Dal3 hybridized strongly to four clones of the S. albus cosmid library. Three of the clones were found to have sequences similar to that of hrcA, whereas the fourth was a false positive.
Nucleotide sequence of S. albus hrcA and dnaJ2.
The nucleotide sequence of hrcA and its downstream and upstream regions (2,521 bp) was determined by using plasmid pPM3881 and subclones in M13 vectors. The sequence was aligned with those of HrcA proteins, and a GTG translation initiation codon was identified, preceded by a GGAGG ribosome-binding site. The hrcA gene encodes a protein of 336 amino acids, with a molecular mass of 36.9 kDa and a sequence 26% identical to that of the B. subtilis HrcA. The deduced peptide sequence contained the sequence TVRNDMAALE rather than the consensus TIRN(EDY)MA(DAQV)LE.
There was also a “dnaJ-like” gene immediately downstream from hrcA. As there was already a dnaJ gene in the dnaK operon of Streptomyces, we called this gene dnaJ2. It encodes a 379-amino-acid polypeptide, with a predicted molecular mass of 40.6 kDa. A sequence with dyad symmetry downstream from dnaJ2 may be a transcriptional terminator. The hrcA gene is preceded by a canonical CIRCE sequence TTGGCACTC 9N GAGTGCCAG 78 bp upstream from the putative GTG translation start site of hrcA.
Transcription of the hrcA-dnaJ2 operon.
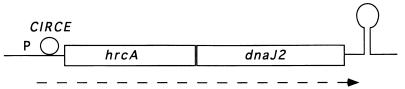
The transcription of hrcA was investigated by Northern blotting with an internal hrcA probe. A 2.4-kb transcript was detected in RNA extracted from heat-shocked cultures (data not shown). An mRNA of this size was predicted from the sequence of an hrcA-dnaJ2 polycistronic transcript, starting near the CIRCE element and ending at the putative transcription terminator. This result also suggests that the transcription of the hrcA-dnaJ2 operon is heat induced, and this was confirmed by a primer extension experiment, as described below. The arrangement of the genes of this new operon is shown in Fig. 1.
FIG. 1.
Schematic representation of the hrcA-dnaJ2 operon. The promoter (P), CIRCE motif (circle), and putative transcription terminator (hairpin) are indicated. Polycistronic mRNA is represented by a dashed arrow.
Determination of the transcription initiation site.
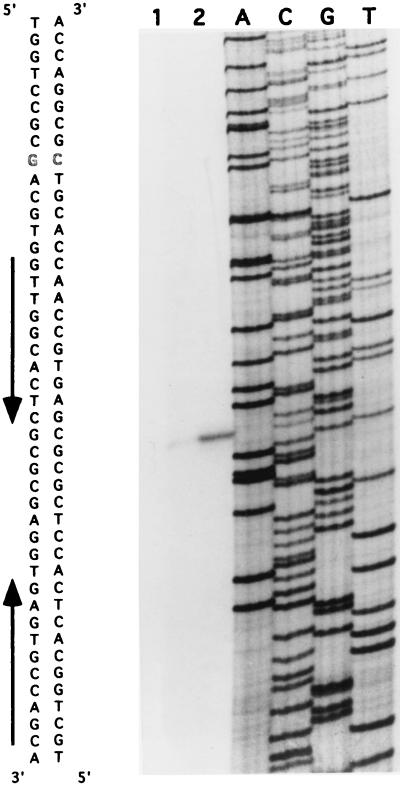
We determined the transcription start site of the hrcA-dnaJ2 operon by primer extension analysis with oligonucleotide OLC47 on RNA isolated from cultures grown at 30°C or heat shocked at 41°C for 20 min (Fig. 2).
FIG. 2.
Determination of the transcription start site of the hrcA-dnaJ2 operon. Primer extension mapping with the AMV reverse transcriptase was carried out with 5′-labeled oligonucleotide OLC47 and RNA from cultures grown at 30°C (lane 1) or subjected to heat shock at 41°C for 20 min (lane 2). The sizes of the fragments were determined by electrophoresis and comparison with sequencing ladders (lanes A, C, G, and T) generated with OLC47 on single-stranded DNA (a subclone from pPM3881 in M13mp18). The relevant DNA sequence is indicated, and the transcription start site is shown by outlined characters. The converging arrows indicate the CIRCE sequence.
A single transcription start site was identified 87 bp upstream from the hrcA translation initiation codon and 6 bp upstream from the CIRCE element. The −35 and −10 hexamers were aligned, and the following similarities were found between the Streptomyces vegetative promoter consensus and the hrcA promoter: TTGACPu17–18ntAgPuPuT for the vegetative promoter and TTGCGC19ntACACT for the hrcA-dnaJ2 promoter, where nt is nucleotides. The space between the −10 and −35 hexamer motifs is exceptionally long, with 19 bp rather than 17 or 18 bp. The −35 hexamer of the hrcA promoter (boldface) is located within a palindromic sequence (underlined), GGTGTGCCCCGGCCTTGCGCTCGGCACACC, which may be involved in the regulation of expression of this gene (see Discussion).
Disruption of the hrcA gene and resulting phenotype.
We disrupted the hrcA gene of S. albus by using a technique involving the conjugative transfer from E. coli to Streptomyces of a plasmid unable to replicate in Streptomyces. Conjugation between E. coli S17-1 containing pPM3891 and S. albus resulted in thiostrepton-resistant exconjugants, which were screened for viomycin sensitivity. The frequency of viomycin-sensitive, thiostrepton-resistant clones (Tsrr Vios) was about 6%. The hrcA gene of these mutants was assumed to be disrupted by a tsr insertion following a double-crossover event at the hrcA locus. The elimination of vector DNA and integration of tsr into the genome was demonstrated by Southern blotting (data not shown).
Growth of the hrcA mutant was not impaired at 30°C on solid rich medium. The formation of the aerial mycelium and sporulation, in particular, were not prevented at 30 or 37°C. However, the mutant grew more slowly at 37°C than the wild type did.
GroEL facilitates the export of some proteins from E. coli (24, 32). Endogenous amylase and β-lactamase secretion was assessed by plate assays, and no obvious effect of GroEL accumulation on the export of these two proteins from the hrcA mutant was observed (results not shown).
Analysis of an hrcA null mutation on the expression of the principal hsp genes.
The levels of GroES and GroEL proteins in the hrcA mutant were assessed by SDS-PAGE (Fig. 3). There were large amounts of a 60- and an 11-kDa protein in the protein extract of the mutant. The overproduced proteins were recognized by anti-GroEL (Fig. 4) and anti-GroES antibodies (data not shown). Anti-GroEL antibodies detected several minor bands for the mutant, which were not detected for the wild-type control. These bands are probably GroEL degradation products. A high level of proteolysis was observed for various proteins in E. coli cells overproducing GroEL or GroEL and GroES (21).
FIG. 3.
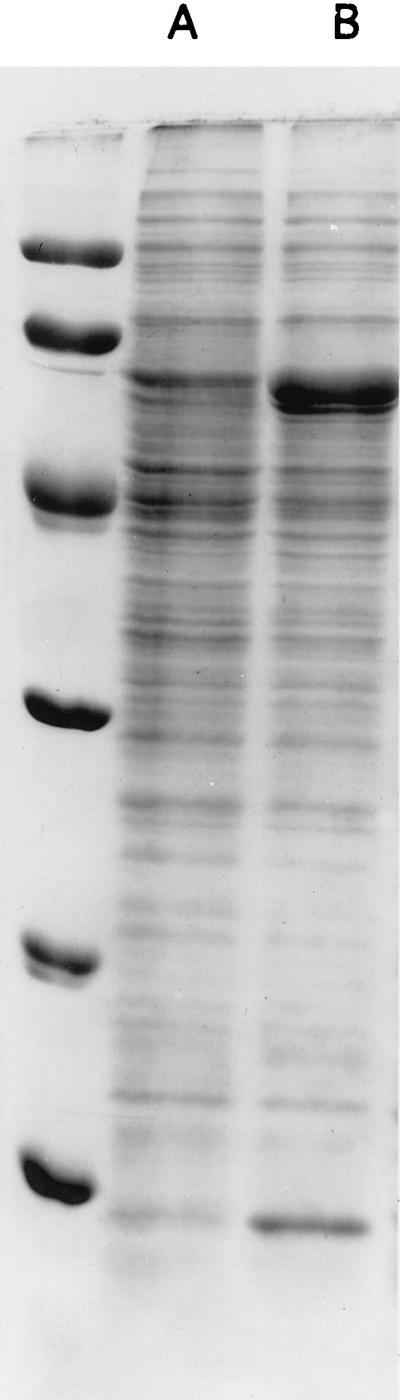
Accumulation of GroEL and GroES in the hrcA mutant. S. albus wild-type (lane A) and hrcA null-mutant (lane B) bacteria were grown at 30°C for 18 h. Protein extracts were prepared and subjected to SDS-PAGE (15% polyacrylamide). The gel was stained with Coomassie blue R250. Molecular mass markers (top to bottom): 94, 67, 43, 30, 20.1, and 14.4 kDa.
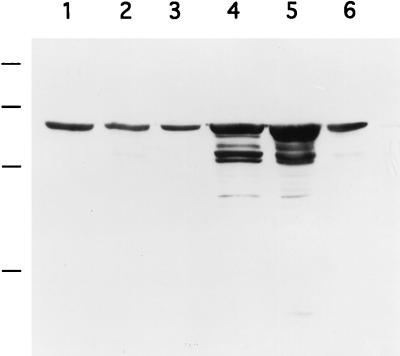
FIG. 4.
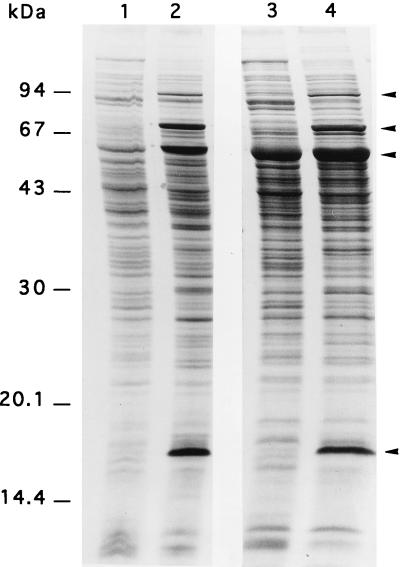
Immunological detection of GroEL. Cultures of the S. albus wild type (lane 1), wild type with vector pHM11a (lane 2), wild type with pCG92 (lane 3), hrcA null mutant (lane 4), hrcA mutant with pHM11a (lane 5), and hrcA mutant complemented by pCG92 (lane 6) were grown at 30°C. Total cell protein extracts were prepared, and equal amounts of protein were applied to each lane. The immunoblot was probed with antibodies raised against M. leprae GroEL. Molecular mass markers (the bars adjacent to lane 1; top to bottom): 94, 67, 43, and 30 kDa.
We scanned the Coomassie blue-stained gel and found that the 60-kDa protein accounted for 19% of total protein in the hrcA mutant and the 11-kDa protein accounted for 5.5%. This provides enough GroES versus GroEL for the formation of the chaperonin machinery. Indeed, in the classical asymmetric GroEL-GroES chaperonin complex (6), 14 subunits of GroEL (56 kDa) interact with 7 subunits of GroES (11 kDa). The predicted protein ratio (wt/wt) between GroEL and GroES in this complex is around 10. The protein ratio is around 5 for a symmetric 14-GroEL–14-GroES complex.
Northern blotting with a groES probe showed that the groES and groES-groEL1 transcripts accumulated in the hrcA mutant at 30°C (Fig. 5). These transcripts did not accumulate in a control in which the mutation was complemented by pCG92, which contains a functional copy of hrcA. The blot was probed with a groEL2-specific probe, and groEL2 transcripts were found to accumulate similarly in the noncomplemented hrcA mutant (data not shown).
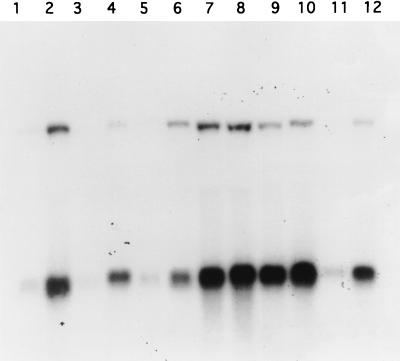
FIG. 5.
Northern blot analysis of the groES and groESL1 transcripts. Total RNA was extracted from cultures of the S. albus wild type (lanes 1 and 2), wild type with vector pHM11a (lanes 3 and 4), wild type with pCG92 (lanes 5 and 6), hrcA null mutant (lanes 7 and 8), hrcA mutant with vector pHM11a (lanes 9 and 10), and hrcA mutant complemented by pCG92 (lanes 11 and 12). The cultures were grown at 30°C (lanes 1, 3, 5, 7, 9, and 11) or shifted to 41°C for 20 min (lanes 2, 4, 6, 8, 10, and 12). RNAs were hybridized with a groES-specific probe which detects both the monocistronic groES and the polycistronic groES-groEL1 products of the groESL1 operon.
The pattern of expression of the principal, CIRCE-independent hsp genes (hsp94, dnaK, and hsp18) was not affected by the hrcA mutation. Labeling experiments with [35S]methionine-cysteine showed that these genes underwent normal thermoinduction (Fig. 6).
FIG. 6.
Effect of the hrcA mutation on the kinetics of HSP synthesis after temperature shift in S. albus. Cultures (20 h after inoculation) of the S. albus wild type (lanes 1 and 2) or hrcA null mutant (lanes 3 and 4) growing at 30°C were divided into 1-ml aliquots, which were cultured at 30°C (lanes 1 and 3) or 41°C (lanes 2 and 4). Five minutes later, [35S]methionine-cysteine was added, and the cultures were labeled by incubation for 20 min. Total protein extracts were prepared and subjected to SDS-PAGE, and the gel was dried and subjected to autoradiography. HSP94, HSP70 (DnaK), HSP56-58 (GroEL), and HSP18 (top to bottom) are indicated by arrows. GroES does not contain methionine or cysteine and therefore is not detected.
Complementation of the hrcA null mutation.
Insertion of the tsr resistance gene is likely to have a polar effect on the expression of dnaJ2, the second gene of the operon. We investigated the involvement of hrcA and dnaJ2 in generating the hrcA null phenotype by complementing the mutation with pCG92 in trans. pCG92 contains a copy of hrcA under the control of the constitutive erm promoter from the erythromycin resistance gene of Saccharopolyspora erythraea.
The patterns of groESL1 and groEL2 expression in the hrcA mutant complemented with pCG92 were similar to those of the wild-type strain at 30°C and after heat shock. This shows that HrcA represses the transcription of groESL1 and groEL2. The pHM11a vector alone has no effect on groESL expression. The hrcA gene in pCG92 was constitutively expressed under the control of the erm promoter, so regulation of groESL1 and groEL2 transcription was not directly linked to the level of hrcA transcription. High-level constitutive expression of hrcA did not prevent groES-groEL1 and groEL2 thermoinduction (Fig. 4 and 5). Therefore, regulation probably depends on the modulation of HrcA activity (see Discussion).
Disruption of dnaJ2.
Our attempts to disrupt dnaJ2, the second gene of the operon, were at first unsuccessful. We used a procedure similar to that used to disrupt hrcA but were unable to select the double-crossover event leading to insertion of a cassette into dnaJ2. We then used an internal dnaJ2 fragment to screen for the insertion of the plasmid in a single-recombination event (one crossover), which is more readily selected (37). Clones with disruption of dnaJ2 were selected, and the integration of the plasmid into dnaJ2 was confirmed by Southern blotting. The growth of the dnaJ2 mutant on NE plates was similar to that of the wild type. The pattern of HSPs in SDS-PAGE was also similar to that of the wild-type strain (data not shown). However, in this mutant, of the two deletion-containing copies of dnaJ2 obtained by this procedure, one had a deletion of only 45 bp at its 3′ end. It is not clear whether this deletion is sufficient to prevent the formation of an active gene product.
DISCUSSION
We identified hrcA, which encodes the repressor of the groESL1 and groEL2 genes in S. albus. In an earlier work we reported that groEL1 was regulated transcriptionally and by a thermodependent posttranscriptional regulation (38). Here we show that disruption of the repressor led to groE transcript and protein accumulation at low temperatures, showing that posttranscriptional regulation, by whatever mechanism, did not prevent translation of the accumulated transcripts in the absence of HrcA. Protein synthesis was analyzed by [35S]methionine-cysteine labeling of hrcA mutant cultures grown at 30°C or under heat shock conditions. Temperature had no significant effect on GroEL biosynthesis in the mutant.
The expression of the hrcA gene was thermoinducible. There is a CIRCE sequence at the 5′ end of the hrcA-dnaJ2 transcript, so HrcA probably negatively autoregulates its own synthesis by binding to CIRCE. hrcA transcription may also be subject to positive regulation. The −35 hexamer of the hrcA promoter is located in a palindromic sequence, and the space between the −10 and −35 hexamer motifs is longer than usual: 19 bp rather than 17 or 18 bp. The situation in the mercury resistance gene cluster is similar. In the merT promoter, a 19-bp space between the −10 and −35 hexamers puts these two elements out of alignment with the RNA polymerase regions with which they must interact. Binding of the MerR regulator to a nearby palindromic sequence induces a 33° unwinding, making it possible for the hexamers to align in such a way that they function (18). A similar mechanism has been suggested for the thiostrepton-inducible promoter tipAp in Streptomyces (19). Positive regulation by twisting the promoter may also apply to hrcA.
hrcA was associated with a second copy of dnaJ. The presence of several dnaJ genes is not unusual. There is a second copy of dnaJ, cbpA, in E. coli (42), and there are three dnaJ-like sequences in the small M. genitalium genome (10). The presence of two dnaJ genes suggests that the encoded proteins may have different physiological functions, either alone or in association with DnaK. The dnaJ259 mutation in E. coli, resulting from an H33D change in the HPD tripeptide, prevents interaction with DnaK (43). The HPD motif is present in both DnaJ and DnaJ2, so both may interact with DnaK. DnaJ2 has the canonical organization of all members of the DnaJ family: an amino-terminal J domain, then a glycine-rich domain, zinc fingers composed of repeats of the CXXCXGXG motif, and a low-homology carboxy-terminal region.
The slight inhibition of the growth of the hrcA mutant at 37°C was unexpected because accumulation of GroEL increases E. coli survival at high temperatures (22).
The secretion of heterologous proteins in the hrcA mutant will be analyzed in detail as part of another study.
GroESL is the major modulator of the CIRCE heat shock regulon in B. subtilis (27). The interaction of GroEL and HrcA in this bacterium controls the availability of the repressor form of HrcA. It will be of interest to determine whether one or both of the Streptomyces GroEL proteins interact with HrcA in a similar way. Five groEL genes have been characterized in Bradyrhizobium japonicum. The groESL4 gene is regulated by CIRCE, whereas groESL1 depends on sigma 32 factor for transcription. Knockout mutations in groEL4 resulted in higher levels of groESL4 transcription. This suggests that CIRCE repression depends on the cellular level of GroEL4 (1). HrcA activity may well be regulated by another component in Streptomyces. Indeed, we have shown that overexpression of hrcA, either by thermoinduction or by insertion of the gene in an expression vector, does not lead to repression of CIRCE-regulated genes. In particular, the thermoinduction of hrcA, which is overexpressed at the point at which it stops acting as a repressor, suggests a situation similar to that described for B. subtilis, in which the modulator of HrcA activity is titrated under heat shock conditions (27).
This work provides no evidence for a correlation between the Streptomyces cell cycle and groEL regulation. Instead, it complements previous work, in which hspR and orfY, two other regulators of the principal chaperones of Streptomyces, were disrupted or overexpressed (13, 36). In each case, changing the amount of the regulator had no direct effect on the formation of the aerial mycelium or sporulation. Here we show that this is also the case for hrcA. Taken together, these results indicate that the regulatory systems of the main HSPs, GroEL, DnaK, and Hsp18, are not directly involved in Streptomyces development.
ACKNOWLEDGMENTS
We thank P. Servant, V. De Crecy, and W. Schumann for their constant interest in this work. We are grateful to J. Rauzier for his help in sequencing the various hrcA candidates that we obtained. We thank H. Motamedi for supplying pHM11a and B. Gicquel for anti-GroEL and anti-GroES antibodies.
This work was supported by the Pasteur Institute and the CNRS.
REFERENCES
- 1.Babst M, Hennecke H, Fischer H M. Two different mechanisms are involved in the heat-shock regulation of chaperonin gene expression in Bradyrhizobium japonicum. Mol Microbiol. 1996;19:827–839. doi: 10.1046/j.1365-2958.1996.438968.x. [DOI] [PubMed] [Google Scholar]
- 2.Bucca G, Ferina G, Puglia A M, Smith C P. The dnaK operon of Streptomyces coelicolor encodes a novel heat-shock protein which binds to the promoter region of the operon. Mol Microbiol. 1995;17:663–674. doi: 10.1111/j.1365-2958.1995.mmi_17040663.x. [DOI] [PubMed] [Google Scholar]
- 3.Bukau B. Regulation of the Escherichia coli heat-shock response. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 4.Chater K F. Genetics of differentiation in Streptomyces. Annu Rev Microbiol. 1993;47:685–713. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- 5.Chater K F, Wilde L C. Streptomyces albus G mutants defective in the SalGI restriction-modification system. J Gen Microbiol. 1980;116:323–334. doi: 10.1099/00221287-116-2-323. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Roseman A M, Hunter A S, Wood S P, Burston S G, Ranson N A, Clarke A R, Saibil H R. Location of a folding protein and shape changes in GroEL-GroES complexes imaged by cryo-electron microscopy. Nature. 1994;371:261–264. doi: 10.1038/371261a0. [DOI] [PubMed] [Google Scholar]
- 7.Duchêne A-M, Kieser H M, Hopwood D A, Thompson C J, Mazodier P. Characterization of two groEL genes in Streptomyces coelicolor A3(2) Gene. 1994;144:97–101. doi: 10.1016/0378-1119(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 8.Duchêne A-M, Thompson C J, Mazodier P. Transcriptional analysis of groEL genes in Streptomyces coelicolor A3(2) Mol Gen Genet. 1994;245:61–68. doi: 10.1007/BF00279751. [DOI] [PubMed] [Google Scholar]
- 9.Evans G A, Lewis K, Rothenberg B E. High efficiency vectors for cosmid microcloning and genomic analysis. Gene. 1989;79:9–20. doi: 10.1016/0378-1119(89)90088-7. [DOI] [PubMed] [Google Scholar]
- 10.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 11.Georgopoulos C, Liberek K, Zylicz M, Ang D. Properties of the heat shock proteins of Escherichia coli and the autoregulation of the heat shock response. In: Morimoto R I, Tissières A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 209–249. [Google Scholar]
- 12.Gibson T J. Studies on the Epstein-Barr virus genome. Ph.D. thesis. Cambridge, England: Cambridge University; 1984. [Google Scholar]
- 13.Grandvalet C, Servant P, Mazodier P. Disruption of hspR, the repressor gene of the dnaK operon in Streptomyces albus G. Mol Microbiol. 1997;23:77–84. doi: 10.1046/j.1365-2958.1997.1811563.x. [DOI] [PubMed] [Google Scholar]
- 14.Greener A. E. coli SURE clones “unclonable” DNA. Strategies. 1990;3:5–6. [Google Scholar]
- 15.Guglielmi G, Duchêne A-M, Thompson C, Mazodier P. Transcriptional analysis of two different Streptomyces albus groEL-like genes. In: Baltz R H, Hegeman G D, Skatrud P L, editors. Industrial microorganisms: basic and applied molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 17–24. [Google Scholar]
- 16.Guglielmi G, Mazodier P, Thompson C J, Davies J. A survey of the heat shock response in four Streptomyces species reveals two groEL-like genes and three GroEL-like proteins in Streptomyces albus. J Bacteriol. 1991;173:7374–7381. doi: 10.1128/jb.173.22.7374-7381.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hecker M, Schumann W, Volker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 18.Heltzel A, Lee I W, Totis P A, Summers A O. Activator-dependent preinduction binding of sigma-70 RNA polymerase at the metal-regulated mer promoter. Biochemistry. 1990;29:9572–9584. doi: 10.1021/bi00493a011. [DOI] [PubMed] [Google Scholar]
- 19.Holmes D J, Caso J L, Thompson C J. Autogenous transcriptional activation of a thiostrepton-induced gene in Streptomyces lividans. EMBO J. 1993;12:3183–3191. doi: 10.1002/j.1460-2075.1993.tb05987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces. A laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 21.Kandror O, Busconi L, Sherman M, Goldberg A L. Rapid degradation of an abnormal protein in Escherichia coli involves the chaperones GroEL and GroES. J Biol Chem. 1994;269:23575–23582. [PubMed] [Google Scholar]
- 22.Kandror O, Goldberg A L. Trigger factor is induced upon cold shock and enhances viability of Escherichia coli at low temperatures. Proc Natl Acad Sci USA. 1997;94:4978–4981. doi: 10.1073/pnas.94.10.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Laminet A A, Ziegelhoffer T, Georgopoulos C, Plückthun A. The Escherichia coli heat shock proteins GroEL and GroES modulate the folding of the β-lactamase precursor. EMBO J. 1990;9:2315–2319. doi: 10.1002/j.1460-2075.1990.tb07403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazodier P, Guglielmi G, Davies J, Thompson C J. Characterization of the groEL-like genes in Streptomyces albus. J Bacteriol. 1991;173:7382–7386. doi: 10.1128/jb.173.22.7382-7386.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazodier P, Petter R, Thompson C. Intergeneric conjugation between Escherichia coli and Streptomyces species. J Bacteriol. 1989;171:3583–3585. doi: 10.1128/jb.171.6.3583-3585.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mogk A, Homuth G, Scholz C, Kim L, Schmid F X, Schumann W. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 1997;16:4579–4590. doi: 10.1093/emboj/16.15.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motamedi H, Shafiee A, Cai S J. Integrative vectors for heterologous gene expression in Streptomyces spp. Gene. 1995;160:25–31. doi: 10.1016/0378-1119(95)00191-8. [DOI] [PubMed] [Google Scholar]
- 29.Murakami T, Anzai H, Imai S, Satoh A, Nagaoka K, Thompson C. The bialaphos biosynthetic genes of Streptomyces hygroscopicus: molecular cloning of the gene cluster. Mol Gen Genet. 1986;205:42–50. [Google Scholar]
- 30.Narberhaus F, Giebeler K, Bahl H. Molecular characterization of the dnaK gene region of Clostridium acetobutylicum, including grpE, dnaJ, and a new heat shock gene. J Bacteriol. 1992;174:3290–3299. doi: 10.1128/jb.174.10.3290-3299.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohta T, Saito K, Kuroda M, Honda K, Hirata H, Hayashi H. Molecular cloning of two new heat shock genes related to the hsp70 genes in Staphylococcus aureus. J Bacteriol. 1994;176:4779–4783. doi: 10.1128/jb.176.15.4779-4783.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips G J, Silhavy T J. Heat-shock proteins DnaK and GroEL facilitate export of LacZ hybrid protein in E. coli. Nature. 1990;344:882–884. doi: 10.1038/344882a0. [DOI] [PubMed] [Google Scholar]
- 33.Puglia A M, Vohradsky J, Thompson C J. Developmental control of the heat shock stress regulon in Streptomyces coelicolor. Mol Microbiol. 1995;17:737–746. doi: 10.1111/j.1365-2958.1995.mmi_17040737.x. [DOI] [PubMed] [Google Scholar]
- 34.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 35.Schulz A, Schumann W. hrcA, the first gene of the Bacillus subtilis dnaK operon encodes a negative regulator of class I heat shock genes. J Bacteriol. 1996;178:1088–1093. doi: 10.1128/jb.178.4.1088-1093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Servant P, Mazodier P. Heat induction of hsp18 gene expression in Streptomyces albus G: transcriptional and posttranscriptional regulation. J Bacteriol. 1996;178:7031–7036. doi: 10.1128/jb.178.24.7031-7036.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Servant P, Thompson C, Mazodier P. Use of new Escherichia coli/Streptomyces conjugative vectors to probe the functions of the two groEL-like genes of Streptomyces albus G by gene disruption. Gene. 1993;134:25–32. doi: 10.1016/0378-1119(93)90170-8. [DOI] [PubMed] [Google Scholar]
- 38.Servant P, Thompson C, Mazodier P. Post-transcriptional regulation of the groEL1 gene of Streptomyces albus. Mol Microbiol. 1994;12:423–432. doi: 10.1111/j.1365-2958.1994.tb01031.x. [DOI] [PubMed] [Google Scholar]
- 39.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 40.Tan M, Wong B, Engel J N. Transcriptional organization and regulation of the dnaK and groE operons of Chlamydia trachomatis. J Bacteriol. 1996;178:6983–6990. doi: 10.1128/jb.178.23.6983-6990.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson C J, Ward J M, Hopwood D A. DNA cloning in Streptomyces: resistance genes from antibiotic producing species. Nature. 1980;286:525–527. doi: 10.1038/286525a0. [DOI] [PubMed] [Google Scholar]
- 42.Ueguchi C, Kakeda M, Yamada H, Mizuno T. An analogue of the DnaJ molecular chaperone in Escherichia coli. Proc Natl Acad Sci USA. 1994;91:1054–1058. doi: 10.1073/pnas.91.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wall D, Zylicz M, Georgopoulos C. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J Biol Chem. 1994;269:5446–5451. [PubMed] [Google Scholar]
- 44.Wetzstein M, Völker U, Dedio J, Löbau S, Zuber U, Schiesswohl M, Herget C, Hecker M, Schumann W. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J Bacteriol. 1992;174:3300–3310. doi: 10.1128/jb.174.10.3300-3310.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wild J, Altman E, Yura T, Gross C A. DnaK and DnaJ heat shock proteins participate in protein export in Escherichia coli. Genes Dev. 1992;6:1165–1172. doi: 10.1101/gad.6.7.1165. [DOI] [PubMed] [Google Scholar]
- 46.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 47.Yuan G, Wong S L. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J Bacteriol. 1995;177:6462–6468. doi: 10.1128/jb.177.22.6462-6468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 49.Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]